Background: Exosomes/microvesicles (EMVs) transport protein cargo between cells effecting cell-cell communication.
Results: Alteration of N-link glycans controls recruitment of specific proteins (e.g. EWI-2) into EMVs.
Conclusion: N-Linked glycosylation is a key determinant of glycoprotein sorting into EMVs.
Significance: This is the first demonstration that N-linked glycans mediate protein sorting to EMVs.
Keywords: Carbohydrate Function, Exosome, Galectin, Glycosylation, Intracellular Trafficking, Membrane Trafficking, Tetraspanin, G3BP, IgFS8, Microvesicle
Abstract
Exosomes, also known as microvesicles (EMVs), are nano-sized membranous particles secreted from nearly all mammalian cell types. These nanoparticles play critical roles in many physiological processes including cell-cell signaling, immune activation, and suppression and are associated with disease states such as tumor progression. The biological functions of EMVs are highly dependent on their protein composition, which can dictate pathogenicity. Although some mechanisms have been proposed for the regulation of EMV protein trafficking, little attention has been paid to N-linked glycosylation as a potential sorting signal. Previous work from our laboratory found a conserved glycan signature for EMVs, which differed from that of the parent cell membranes, suggesting a potential role for glycosylation in EMV biogenesis. In this study, we further explore the role of glycosylation in EMV protein trafficking. We identify EMV glycoproteins and demonstrate alteration of their recruitment as a function of their glycosylation status upon pharmacological manipulation. Furthermore, we show that genetic manipulation of the glycosylation levels of a specific EMV glycoprotein, EWI-2, directly impacts its recruitment as a function of N-linked glycan sites. Taken together, our data provide strong evidence that N-linked glycosylation directs glycoprotein sorting into EMVs.
Introduction
Intercellular communication utilizes secreted molecules including nucleotides, lipids, short peptides, and proteins to relay messages between cells. These molecules can be bundled into membrane-bound vesicles or nanoparticles that transmit information (1, 2). Initially identified as a component of reticulocyte maturation, these vesicles are found secreted from nearly all cell types and in physiological fluids (3, 4). Although diverse names have been given to extracellular particles in distinct systems, including microvesicles, exosomes, ectosomes, prostasomes, and tolerasomes, a naming convention remains to be defined (5). The terms exosome (specific to particles derived from multivesicular bodies (6, 7)), and microvesicle (a more general term) are most commonly used and refer to membrane-bound vesicles with 40–100-nm diameters that are released from the cell and carry specific biological messages (herein referred to as EMV)2. Although there is no consensus on EMV protein composition, all EMVs contain tetraspanins such as CD63 and CD81, which are the most commonly identified markers (8). EMVs incorporate cargo both within the vesicle and displayed on the vesicle surface. Typical cargo molecules include membrane proteins and glycoproteins and soluble and membrane-bound cytosolic proteins, mRNA and microRNA (9, 10). EMV content defines their functions, which can vary from presentation of antigens to stimulation of cell differentiation and transmittal of pathogenic information (1, 11). For example, human gliomas shed EMVs containing epidermal growth factor receptor EGFRvIII, a mutant receptor. This receptor is transmitted to nearby non-tumor cells, activating signaling pathways that help support the growing tumor (11). Despite the crucial roles of cargo molecules in the bioactivity of EMVs, the signals controlling protein sorting and recruitment to these particles are not well understood.
Glycosylation is the most common post-translational modification in eukaryotic cells and has been shown to play an important role in protein trafficking to specific membranes, most notably the apical membrane of epithelia (12, 13). Recent work by our laboratory using lectin microarray technology revealed a conserved glycosylation signature for EMVs from a variety of cell types that was distinct from the parent cell membranes. In specific, high mannose, polylactosamine (PolyLacNAc), complex N-linked glycans and α2,6-sialic acids were enriched in EMVs (14, 15). A similar signature was observed by mass spectrometry in EMVs from the ovarian cancer line SKOV3 (16). This suggests that glycans may be a sorting motif for EMVs. Herein we provide strong evidence of a direct role for N-link glycans in the sorting of proteins to EMVs. We demonstrate that recruitment of a specific glycoprotein, EWI-2, is dependent on the number of N-linked glycans attached to the protein core. Overall, our data identify N-linked glycans as a determinant of protein trafficking into EMVs.
EXPERIMENTAL PROCEDURES
General Cell Culture
The Sk-Mel-5 cell line was purchased from the NCI-Frederick, National Institutes of Health. Cells were cultured in growth media (RPMI 1640 (Lonza) supplemented with 10% fetal bovine serum (FBS, v/v, Innovative Research) and 2 mm l-glutamine (Cellgro)). Cells were maintained at 37 °C and 5% CO2.
Cloning of CD63-Venus Construct
The CD63 coding sequence was cloned from pCT-CD63-GFP (System Biosciences) via PCR with following primers: forward (5′-AAAAACTCGAGATGGCGGTGGAAGGAGGAAT-3′)and reverse (5′-AAAAAGCGGCCGCCATCACCTCGTAGCCACTTC-3′).
Venus (17), a YFP variant, was cloned into the EcoRV and HindIII sites of pcDNA3.1B. The CD63-PCR product was then cloned into the XhoI/NotI sites of the Venus pcDNA3.1B to create our CD63-Venus construct (CD63 C terminus linked to Venus). Endo-free DNA was purified using the Endo-free Maxiprep kit (Qiagen).
EWI-2-FLAG Constructs
Cloning
A plasmid containing the EWI-2 coding region was purchased from Origene (SC120151, NM_052868.1). A construct containing EWI-2 with a C-terminal FLAG tag sequence was created via PCR with the following primers: forward (5′-AAAAACTCGAGATGGGCGCCCTCAGGCCCACG-3′) and reverse (5′-AAAAAAAGCTTTCACTTATCGTCGTCATCCTTGTAATCTCCTCCTCCCCGTTTTCGAAGCCTCTTCATGAA-3′).
The EWI-2-FLAG PCR products were digested with XhoI and HindIII restriction enzymes (New England Biolabs) and inserted into plasmid pcDNA3.1B to create the wild type EWI-2 construct (wtEWI-2-FLAG). Site-specific mutagenesis was performed on wtEWI-2-FLAG using the QuikChange Site-Directed Mutagenesis kit from Qiagen with the following primers in succession to create the three glycosylation mutant constructs of EWI-2, N1, N12, and N123: N1 forward (5′-TCCTGCCAGGTGACCGGCTATGAGGGC-3′) and reverse (5′-ATAGCCGGTCACCTGGCAGGAGATGGA-3′); N12 (N1-EWI-2-FLAG was used as template) forward (5′-CTGTGCCAGGTGTCAGGGGCACTTCCC-3′) and reverse (5′-TGCCCCTGACACCTGGCACAGCAGTTC-3′; N123 (N12-EWI-2-FLAG was used as template) forward (5′-CTGTGCCAGATCTCTGTGCGGGGTGGC-3′) and reverse (5′-CCGCACAGAGATCTGGCACAGCAGGGA-3′). Endo-free DNA of all constructs was obtained using the Endo-free Maxiprep kit (Qiagen).
Transfection
For EWI-2 transfection experiments, cells were cultured to ∼60% confluency, the growth medium was replaced with EMV-free media (RPMI 1640 with 10% EMV-free FBS (v/v, FBS was centrifuged overnight at 100,000 × g to remove EMV) and 2 mm l-glutamine), and cells were treated with TransIT 2020 (Mirus)-EWI-2-FLAG plasmid complexes. EMV-free media was used due to issues with cell death under the transfection conditions with serum-free medium and the improved transfection rate observed. Transfected cells were cultured for 48 h to allow plasmid expression before collection of media for isolation of EMV as described below.
Fluorescence Microscopy
Cells were plated on sterile 35 × 10-mm glass-bottomed plates (InVitro Scientific) and grown to ∼75% confluency for imaging. Cells were then washed with HBSS (Cellgro) and fixed in 4% paraformaldehyde (PFA, Electron Microscopy Sciences) at room temperature. After fixation, cells were washed 2× with HBSS before staining. Cells were stained with either fluorescently labeled antibodies (Pacific Blue α-human CD63, 1:100, BioLegend, clone H5C6; FITC α-human CD81, 1:100, BioLegend, clone 5A6; FITC α-FLAG, 1:100, Abcam, polyclonal goat IgG), or biotinylated lectin (biotin-DSA (Datura stramonium), 1:100 dilutions, Vector Laboratories) in HBSS and applied on the cells for 30 min at 37 °C. For biotinylated DSA, strepavidin-Cy5 (1:100 in HBSS, Invitrogen, 15 min, 37 °C) was used to visualize staining. After staining, cells were washed with 2× HBSS and imaged in HBSS to prevent drying.
For imaging with CD63-Venus, cells were grown to ∼60% confluency and transfected with the CD63-Venus construct using TransIT 2020 reagent (Mirus). Transfected cells were cultured for 24 h before staining as previously described.
For chitin inhibition of DSA binding, DSA-biotin was preincubated with chitin hydrolysate (1:4 vol:vol in HBSS, 100-μl total volume, Vector Laboratories) or HBSS only at room temperature for 40 min before use for cell staining as previously described.
For PNGase F and endoglycosidase H treatment, cells were grown to 85% confluency, fixed as previously described, and treated with PNGase F (500 units, 100 μl in reaction buffer (10 μl of 10× reaction buffer in 90 μl of HBSS; New England Biolabs), endoglycosidase H (500 units, 100 μl in reaction buffer; New England Biolabs), or HBSS only at 37 °C for 1 h; slides were then rinsed 1× with 2 ml of HBSS before staining as previously described. For imaging with 1-deoxymannojirimycin (DMJ) treatment, Sk-Mel-5 were grown to ∼60% confluency and treated with DMJ (LC Scientific, 1 mm in growth media) for 48 h before staining as previously described.
For imaging with EWI-2-FLAG, cells were grown to ∼60% confluency and transfected with the EWI-2-FLAG construct using TransIT 2020 reagent (Mirus). Transfected cells were cultured for 24 h before staining as previously described.
For imaging with EWI-2-FLAG under DMJ treatment, cells were grown to ∼60% confluency and treated with DMJ for 24 h. Cells were then transfected in the same media with EWI-2-FLAG construct DNA complex using TransIT 2020 reagent (Mirus). Transfected cells were cultured for another 24 h in DMJ media before staining as previously described.
Fluorescence images were obtained using an inverted microscope (Nikon Eclipse Ti; Photometrics CoolSNAP ES monochrome camera) with a 60× oil immersion lens (NA 1.4) controlled by Nikon NIS Elements AR software. FITC and Venus images were obtained with the dichroic mirror (86012v2, Chroma Technologies) and separate excitation (465–495 nm) and emission (515–555 nm) filters; Cy5 images were obtained with the dichroic mirror (86012v2) and excitation filter (625 to 650 nm) and emission filter (670 nm). Fluorescence images were background-subtracted and thresholded using the NIS Elements AR software.
Colocalization analysis was done using the colocalization module of NIS Elements AR software. The fluorescence images were merged, and labeled cells were selected. The quantitative assessment of colocalized area was performed using Pearson's correlation and Mander's coefficient.
For deconvolution analysis, cells were imaged at multiple z-planes with fluorescence microscopy. Multi-z stack images were processed using the deconvolution module of NIS Elements AR software. A single focal plane is shown.
EMV Isolation
For isolation of untreated EMV, cells were cultured to ∼60% confluency and rinsed 2× with HBSS (Cellgro). Serum-free medium (RPMI 1640 with 2 mm l-glutamine) was then added. Cells were cultured for 48 h before the collection of media. For a typical EMV isolation, media from 4 × 150-mm plates were combined.
EMVs were isolated as previously described (14). In brief, collected media were subjected to differential centrifugation to remove cell debris and large vesicles (300 × g for 15 min, 2000 × g for 20 min, and 10,000 × g for 30 min; all centrifugation steps were done at 4 °C). Clarified supernatant was then subjected to ultracentrifugation at 100,000 × g (Beckman Coulter) at 4 °C for 70 min to obtain an EMV pellet. The pellet was resuspended in PBS, and the suspension was centrifuged at 100,000 × g at 4 °C for 70 min. The pellet was then resuspended in ∼60 μl of PBS, and protein concentration was determined by the microBCA assay (Thermo Scientific).
Membrane Preparation
Cells were scraped off plates in PBS using a cell scraper and pelleted at 300 × g for 15 min. The cell pellet was washed in 1× PBS, resuspended in PBS containing protease inhibitor mixture, and sonicated (5 s, 3 sets, 100% power, 4 °C, Branson sonicator). The cell membranes were then pelleted at 4 °C and 14,000 × g for 1 h. The membrane pellet was washed 1× with PBS, and membranes were resuspended in ∼1 ml of PBS. Protein levels were quantified via Bio-Rad DC assay.
Mass Spectrometry and Data Analysis
Equal amounts of EMV or total cell membrane samples (300 μg) were diluted in HEPES-PPS (10 mm HEPES, pH 7.5, 0.15 m NaCl, 0.1 mm Ca2+, 0.1% PPS Silent Surfactant (Expedeon)) and incubated on ice for 15 min followed by sonication (5s, 100% power, Branson sonicator). Sonicated samples were applied to a DSA-agarose column (DSA-agarose, 0.6 ml, Vector Laboratories) to isolate DSA-binding glycoproteins and complexes from non-DSA-bound proteins. The column was washed with 5 volumes of HEPES and eluted with chitin hydrolysate (1:5 dilution in HEPES, Vector Laboratories). Eluate and unbound fractions were collected for mass spectrometry.
Protein concentrations of the samples were verified using SDS-PAGE and imaged using standard conditions described previously (18). Thirty micrograms of each sample was normalized and suspended in 0.1% Rapigest (Waters Corp.), reduced (10 mm DTT), alkylated (20 mm iodoacetamide), and digested with proteomics grade trypsin (1:25 trypsin:protein) at 37 °C overnight. Digests were separated into 12 fractions using an Agilent OFF gel 3100 fractionator and analyzed using an Agilent 6520 Q-TOF system coupled to a chip cube interface as described previously (18). Data were searched against the SwissProt Mammals subset using Mascot (Matrix Science) and SpectrumMill (Agilent) using standard modifications and parameters (methionine oxidation, carbamidomethylation of cysteine, +2 to +4 charge state; 30 ppm and 0.1 Da for precursor and product ion tolerances, respectively). Result files were uploaded into Scaffold v2.0.3 (Proteome Software) for further analysis, including XTandem! subset database searching, gene ontology annotation, and data visualization (see supplemental Table 1 for details).
Western Blot Analysis
EMV samples and membrane preps with equal amounts of total protein (15 μg) were resolved by SDS-PAGE and transferred to nitrocellulose membranes for Western blot analysis. Membranes were blocked with blocking buffer (10% milk in PBS, pH 7.4, with 0.005% Tween 20 for DSA blot, 10% BSA in PBS, pH 7.4, with 0.005% Tween 20) and probed for CD81 (1:1000, Millipore, clone I.3.3.22, monoclonal mouse IgG), EWI-2 (1:1000, Abcam, polyclonal rabbit IgG), CD63 (1:1000, Millipore, clone RFAC4, monoclonal mouse IgG1), G3BP (1:1000, Abcam, polyclonal rabbit IgG), ADAM10 (1:1000, Abcam, polyclonal rabbit IgG), galectin-3 (1:1000, eBioscience, polyclonal rat IgG(biotinylated)), FLAG (1:1000, Sigma, polyclonal rabbit IgG), or biotin-DSA (1:500, Vector Laboratories). All dilutions were in blocking buffer. Membranes were washed 3× with PBST (PBS, pH 7.4, 0.005% Tween 20) followed by incubation with the appropriate HRP-conjugated secondary antibody (1:2500, goat α-mouse or goat α-rabbit, Bio-Rad) or α-biotin-HRP (1:2500, Cell Signaling Technology) diluted in blocking buffer. All incubations were done at room temperature for 1 h. After washing 3× with PBST, blots were visualized with Supersignal West Femto Chemiluminescent Substrate (Thermo Scientific). For stripping, the blot was incubated with stripping buffer (Thermo Scientific) for 20 min followed by washing 1× with PBS and incubation with blocking buffer (40 min) before probing with a different antibody.
Immunoprecipitation and PNGase F Treatment of Wild Type (WT) EWI-2-FLAG
Wild type EWI-2-FLAG-transfected Sk-Mel-5 membrane preparations (250 μg) were resuspended in lysis buffer (2 mm EDTA, PBS, 1% Brij 97; Sigma) and centrifuged (2500 × g, 5 min, 4 °C) to remove undissolved material. α-FLAG antibody (1:100, Sigma, clone M2, monoclonal mouse IgG) was added to the cleared supernatant. After 1 h at room temperature, protein G beads (50 μl, Pierce Protein G-agarose beads; Thermo Scientific) were added, and the mixture was incubated for 1 h at room temperature. The beads were then spun down at 2500 × g, 1 min, washed 2× with PBS, and resuspended in 30 μl of PBS. The suspension was heated at 98 °C for 5 min to denature proteins and remove them from the beads. The mixture was spun for 2500 × g for 1 min, and the supernatant was collected. The samples (15 μl of supernatant each) were treated with or without PNGase F (2 μl, ∼1000 units, New England Biolabs) following the company's protocol. The samples were analyzed by lectin blot and Western blot analysis as previously described.
DMJ Treatment
Sk-Mel-5 cells cultured to ∼50% confluency were treated with growth media containing the mannosidase inhibitor DMJ (LC Scientific, 1 mm in growth media). Cells were cultured for 24 h after which the medium was removed, and cells were washed 2× with serum-free media (RPMI 1640 with 2 mm l-glutamine). Cells were then cultured in serum-free medium containing 1 mm DMJ for an addition 24 h before EMV isolation as previously described.
Immunoprecipitation of CD81
DMJ-treated or control Sk-Mel-5 membrane preparations (500 μg) were resuspended in lysis buffer (50 mm Tris, 150 mm NaCl, 1% Nonidet P-40; Sigma) and centrifuged (10,000 × g, 10 min, 4 °C) to remove undissolved material. α-CD81 antibody (1:100, Millipore, clone I.3.3.22, monoclonal mouse IgG) was added to the cleared supernatant. After 1 h at room temperature, protein G beads (50 μl, Pierce Protein G-agarose beads; Thermo Scientific) were added, and the mixture was incubated for 1 h at room temperature. The beads were then spun down at 10,000 × g for 1 min, washed 1× with PBS, and heated at 98 °C for 6 min. The samples were analyzed by Western blot analysis as previously described.
EWI-2 Knock-out Experiments
Generation of EWI-2 Knock-out Cell Line
Lentiviral stocks of the Mission shRNA clone against EWI-2 were prepared by the shRNA core service (TRC 1.5 library, TRC# TRCN0000057079; NYU Lagone School of Medicine). Sk-Mel-5 cells were seeded in 96-well plates (Costar, Fisher) with shRNA-containing lentivirus (multiplicity of infection = 3) in growth media. After 24 h growth media were replaced with selection media (growth media containing 1 μg/ml puromycin) for selection. Cells were grown in selection media and propagated a minimum of 5 days before use in experiments and initial analysis by Western blot to confirm knockdown of EWI-2.
EMV Isolation
Cells were grown in selection medium to ∼80% confluency in 150-mm plates. Cells were then washed, and selection medium without FBS was added. After 24 h, medium was collected for EMV isolation as previously described.
RESULTS
EMV Marker Proteins Colocalize with Complex N-Glycans in Discrete Membrane Microdomains
In previous work using the T-cell line Jurkat we demonstrated that glycans enriched in EMVs colocalize with a lipid marker of membrane microdomains associated with EMVs (15). Tetraspanins are a family of conserved transmembrane proteins that is widely observed in endothelial cells and form tetraspanin-enriched microdomains from which EMVs are thought to arise (19–21). The tetraspanins CD63, a glycoprotein, and the non-glycosylated CD81 are the most common markers of EMVs (8). In previous experiments we showed that EMVs isolated from the skin cancer cell line Sk-Mel-5 were enriched in these marker proteins (14). We chose to use Sk-Mel-5 for our EMV experiments throughout this work as high levels of EMVs could be obtained from this cell line, enabling our studies examining the effects of genetic and biochemical manipulation of glycans on EMV protein content.
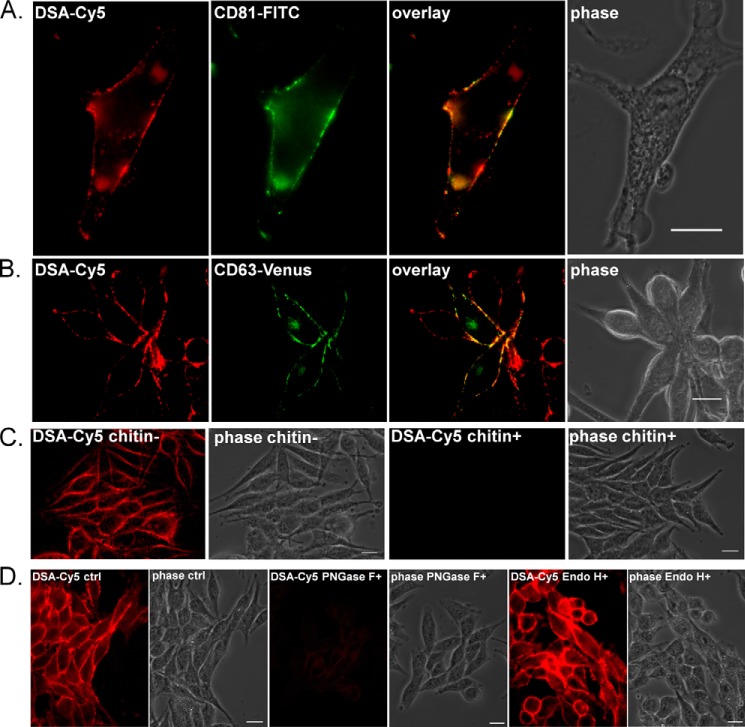
To determine whether EMV-enriched glycans colocalize with protein markers of EMVs in microdomains, we examined the colocalization of the poly/multiantennary LacNAc lectin DSA (22, 23) with the tetraspanins CD63 and CD81 by fluorescence microscopy. DSA was chosen based on our previous work, which showed enrichment of poly/multiantennary LacNAc epitopes in EMVs from Sk-Mel-5 compared with the parent membrane (14). In brief, cells were grown in glass-bottomed dishes and fixed with 4% paraformaldehyde under mild, non-permeabilizing conditions. Cells were then stained with biotinylated DSA followed by staining with Cy5-α-biotin and fluorescently labeled α-CD81 and/or α-CD63 antibodies (Fig. 1, data not shown). In initial experiments we observed diffuse staining with multiple α-CD63 antibodies; therefore, we used a CD63-Venus C-terminal fusion construct for our microscopy studies (Fig. 1B). This construct is similar to others used in the literature for CD63 imaging (24, 25) and colocalizes with CD81 as expected (data not shown). DSA staining was highly colocalized with the EMV-tetraspanin markers, arguing that they stain the same membrane microdomains (Fig. 1). Colocalization between CD81 and DSA-Cy5 showed a very strong correlation (Pearson correlation coefficient (PCC): 0.876; Mander's overlap (MO): 0.941) indicating a highly significant overlap between the two markers (26, 27). Colocalization between Venus-CD63 and DSA-Cy5 was less strong although still highly significant (PCC: 0.662; MO: 0.845), reflecting the internal signal of the expressed Venus-CD63 construct in these non-permeabilized cells. When only cell surface staining was considered, the correlations were in line with those observed for CD81 staining (PCC: 0.875; MO: 0.944). Lectin staining was specific for complex N-linked glycans. As expected, staining was inhibited by chitin hydrolysate (Fig. 1C) (28). In addition, PNGase F, which cleaves all N-glycans, but not endoglycosidase H, which can only cleave hybrid and high mannose N-linked epitopes, abolished the DSA signal (Fig. 1D). These results support the argument that EMVs arise from membrane microdomains with poly/multiantennary LacNAc glycosylation, pointing toward a potential role for complex N-glycans in sorting of proteins to these particles.
FIGURE 1.
EMV markers and complex N-glycans colocalize in discrete cell surface domains. A, fixed Sk-Mel-5 cells (4% PFA) were co-stained with DSA (biotin-DSA, Cy5-α-biotin mAb, red) and CD81 antibody (FITC-α-CD81, green). The cells were not permeabilized and staining was on the cell surface. Cells were imaged with fluorescence microscopy. The two channels were overlapped to show co-localization of CD81 and DSA. PCC was 0.876; MO was 0.941. B, Sk-Mel-5 cells were transfected with CD63-Venus construct (Venus, green), incubated for 24 h at 37 °C, fixed with PFA, and stained with DSA (biotin-DSA, Cy5-α-biotin mAb, red). The cells were imaged in multiple z-planes with fluorescence microscopy and deconvoluted. A single focal plane is shown. The two channels were overlapped to show co-localization of CD63 and DSA. The overlap coefficients were determined for CD63 positive cells only (PCC: 0.662; MO: 0.845). C, fixed Sk-Mel-5 cells (4% PFA) were stained with biotin-DSA preincubated with either chitin hydrolysate (1/5 dilution in HBSS) or HBSS followed by staining with Cy5-α-biotin mAb (red). D, fixed Sk-Mel-5 cells (4% PFA) were treated with PNGase F, endoglycosidase H (Endo H), or HBSS only (ctrl) at 37 °C for 1 h before DSA staining as before (biotin-DSA, Cy5-α-biotin mAb, red). All scale bars are 20 μm.
EWI-2 Is an EMV Glycoprotein Whose Trafficking Is Dependent on Complex N-Glycans
In preliminary experiments, we identified glycoproteins enriched in Sk-Mel-5 EMVs using a DSA pulldown approach coupled with mass spectrometry (supplemental Fig. 1; see supplemental Table 2 for a full list of identified proteins and supplemental Table 3 for a full list of identified peptides). Several glycoproteins were enriched in the DSA-bound fraction from EMV samples including the metallopeptidase ADAM10 and galectin-3-binding protein (G3BP), both of which had been previously identified in EMVs (16, 29), and EWI-2, a CD81 interacting protein (30, 31). We confirmed the enrichment of EWI-2, G3BP, ADAM10, and the EMV markers CD81 and CD63 in Sk-Mel-5 EMVs (Fig. 2). In brief, Sk-Mel-5 EMVs were isolated as previously described by differential centrifugation, and equal amounts by protein of EMV and total cell membrane (TCM) were analyzed by Western blot analysis (14).
FIGURE 2.
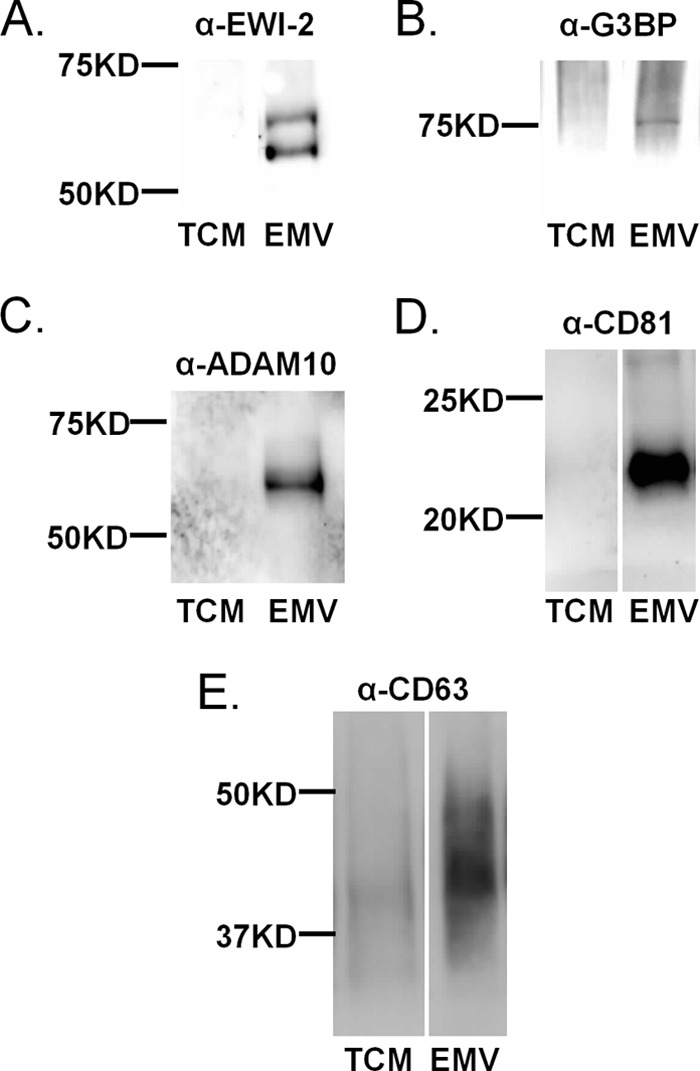
Enrichment of glycoproteins identified by mass spectrometry in EMV. Confirmation of the enrichment of select proteins in Sk-Mel-5 EMVs. Equal amounts of TCM and isolated EMVs (5 μg of protein each) were loaded onto SDS-PAGE gels and analyzed by Western blot analysis with the appropriate antibodies. Equal loading of protein was confirmed by Ponceau staining (data not shown). Samples were analyzed for EMV markers CD81 and CD63 as a control. Western blots are representative of three biological replicates. A, α-EWI-2. B, α-G3BP; C, α-ADAM10. D, α-CD81. E, α-CD63.
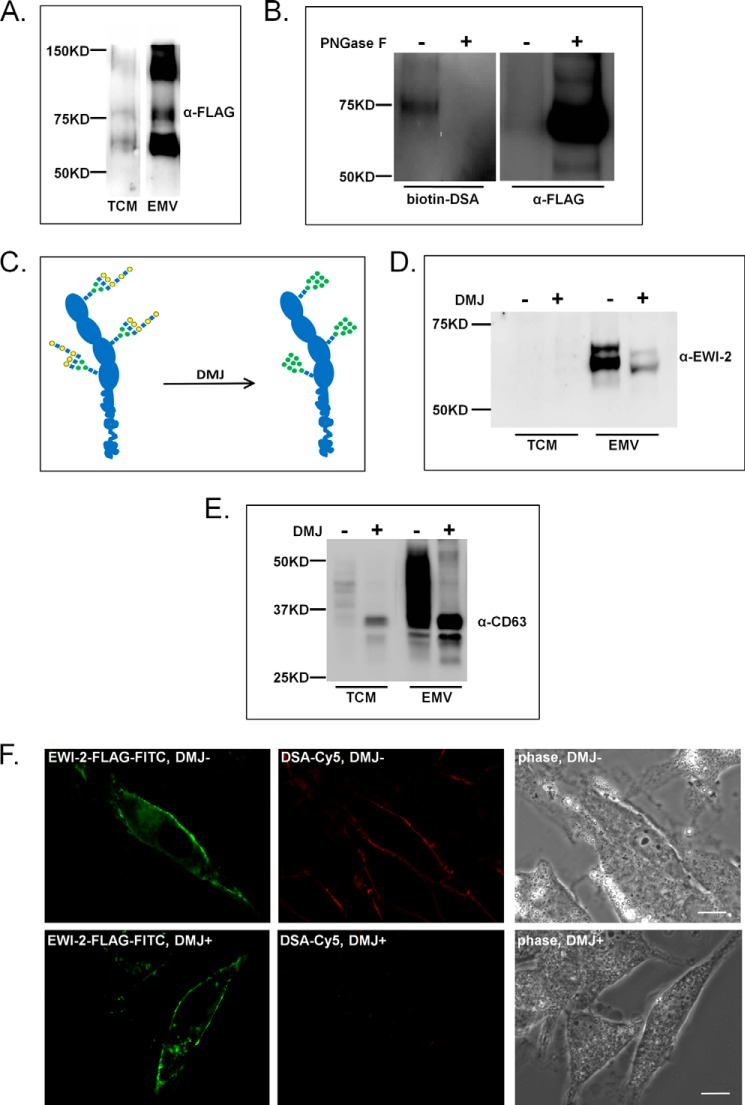
EWI-2, also known as IgSF8, is a member of the immunoglobulin superfamily (IgSF) that plays a role in cell migration (32, 33). EWI-2 has four extracellular Ig-like domains, three of which bear N-glycans, and a single transmembrane domain that directs its known interactions with CD81 (30). We validated the presence of DSA-positive complex N-glycans on EWI-2 (Fig. 3, A and B). Briefly, a C-terminal FLAG-tagged EWI-2 construct was overexpressed in Sk-Mel-5 cells and immunoprecipitated from the total cell membrane fraction with an α-FLAG antibody. This construct has been used in previous studies to examine the glycosylation and cleavage of EWI-2 (32, 34) and showed similar enrichment in Sk-Mel-5 EMVs as the native protein (Fig. 2A and 3A). The EWI-2 immunoprecipitated fraction was divided in two, and one sample was treated with PNGase F, an enzyme that cleaves N-glycans (35), as a control. Equal amounts of the samples were then run on a SDS-PAGE gel and analyzed by lectin blot for DSA staining. The blot was stripped and re-probed for the FLAG tag using a different α-FLAG antibody than that used for the immunoprecipitate. A DSA-positive smear was observed for the FLAG-tagged EWI-2 with a major band at ∼75 kDa. As expected, these bands disappeared in the PNGase F-treated sample, confirming that staining was glycan-dependent. The α-FLAG blot showed a diffuse band for the untreated sample, typical of glycosylated proteins bearing complex glycans. This band condensed to a single, strongly stained band at ∼70 kDa, as expected, for the deglycosylated EWI-2-FLAG construct. Overall our data are in line with previous reports of EWI-2 bearing complex N-glycans in other cell lines (32) and validate the presence of such epitopes on EWI-2 from Sk-Mel-5 cells.
FIGURE 3.
EWI-2 recruitment to EMVs is dependent on complex N-glycans. A, EWI-2 FLAG recruitment to EMVs is similar to native EWI-2. Sk-Mel-5 cells were transfected with a WT EWI-2-FLAG construct. After 48 h, culture medium was collected for EMV isolation, and TCM was isolated from cells. TCM (25 μg) and EMVs (10 μg) were analyzed for FLAG by SDS-PAGE and Western blot analysis. A higher amount of TCM was used to allow for visualization on the blot. Representative blot of three biological replicates is shown. B, EWI-2 glycans contain poly/multiantennary LacNAc structures. Sk-Mel-5 cells were transfected with wt EWI-2-FLAG. TCM was prepared from cells after 48 h. EWI-2-FLAG was immunoprecipitated from TCM samples using a mouse α-FLAG antibody. Immunoprecipitated samples were divided in two and treated either with buffer (PNGase F −) or PNGase F (PNGase F +). Treated and control samples were analyzed by SDS-PAGE and DSA lectin blot analysis. After imaging, the blot was stripped and probed with a rabbit α-FLAG antibody. Western blots shown are representative of two biological replicates. C, schematic representation of the effects of DMJ on EWI-2 glycosylation. Glycan notation follows (57). D, inhibition of complex N-glycan alters EWI-2 trafficking to EMV. Sk-Mel-5 cells were treated with DMJ (1 mm) for 48 h before collection of TCM or EMV (+ DMJ). Untreated Sk-Mel-5 cells were used as a control (− DMJ). TCM and EMV samples (+/− DMJ) were probed for EWI-2. Equal amounts of TCM samples (25 μg of protein) and of EMV (10 μg of protein) were compared by SDS-PAGE and Western blot analysis. Western blots shown are representative of two biological replicates. E, inhibition of complex N-glycan does not alter CD63 levels in EMV. TCM and EMVs from DMJ-treated cells were obtained and analyzed as in D with the exception that blots were probed with an α-CD63 antibody. F, cell surface localization of EWI-2 is not affected by DMJ treatment. Sk-Mel-5 cells were transfected with wt EWI-2-FLAG constructs and treated with either DMJ (1 mm, DMJ +) or buffer (DMJ −) for 48 h. The cells were fixed and co-stained with an α-FLAG antibody (FITC-α-FLAG, green) and DSA (biotin-DSA, Cy5-α-biotin mAb, red). Cells were imaged at multiple z-planes with fluorescence microscopy. Images were then used for deconvolution analysis with NIS Elements. A single focal plane image is shown. The scale bar is 20 μm.
To determine whether the glycosylation state of EWI-2 would alter its recruitment to EMVs, we studied the effects of the mannosidase inhibitor DMJ on protein trafficking to Sk-Mel-5 EMVs (36–38). DMJ is a well known glycan inhibitor that halts N-glycan processing at the high mannose stage, preventing the formation of hybrid and complex N-glycans such as those that contain poly/multiantennary LacNAc (Fig. 3C). In brief, we treated Sk-Mel-5 cells with DMJ (1 mm) for 24 h before changing the medium to serum-free medium containing DMJ. After an additional 24 h, medium was collected, and EMVs were isolated for analysis. Sk-Mel-5 cells treated for 48 h with 1 mm DMJ had normal growth and morphology (Fig. 3F). Furthermore, we observed no differences in the amounts of EMV isolated from DMJ-treated versus untreated cells as quantified by total EMV protein levels, CD63 levels (Fig. 3E), and pellet size. Both EWI-2 and CD63 from DMJ+ EMVs showed a shift in their molecular weights indicating a loss of complex glycosylation (Fig. 3, D and E). Sk-Mel-5 cells treated with DMJ under similar conditions showed the expected loss of DSA binding as observed by fluorescence microscopy (Fig. 3F). A strong reduction in the EWI-2 levels in EMV was observed upon loss of poly/multiantennary LacNAc-containing complex epitopes (∼70% reduction observed; Fig. 3D). This is unlikely to be due to a general effect by DMJ on EWI-2 folding, as EWI-2 FLAG showed no gross relocalization, as would be expected for a protein-folding or cell stress-related trafficking defect upon DMJ treatment (Fig. 3F) arguing for a specific effect of glycosylation on EWI-2 trafficking to EMVs.
EWI-2 Recruitment to EMVs Is Dependent on the Number of N-Linked Glycosylation Sites
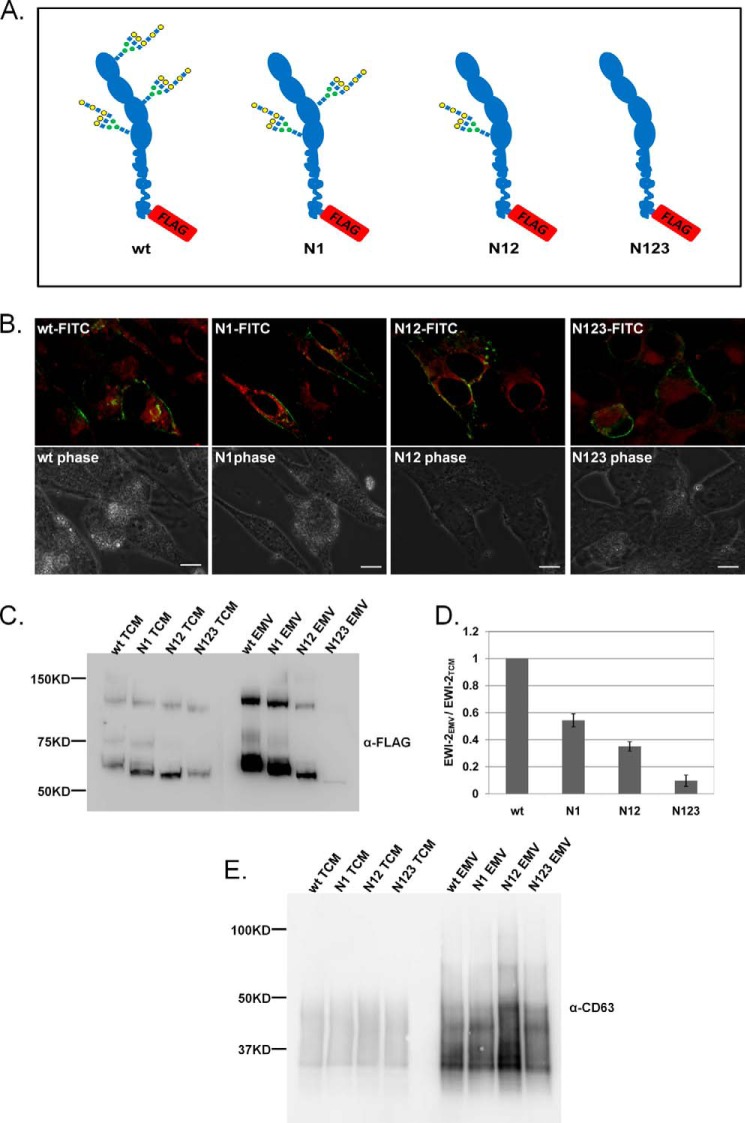
To study the role of N-glycans in trafficking of EWI-2 to EMV in more detail, we created a series of C-terminal FLAG-tagged EWI-2 N-glycan site mutants based on known constructs (32). These constructs replace the asparagines at one (N1), two (N12), or all three (N123) of the N-linked glycosylation sites in the protein with glutamine, which is not glycosylated (Fig. 4A). We overexpressed the FLAG-tagged constructs in Sk-Mel-5 and examined their distribution in cells by fluorescence microscopy to determine whether loss of glycosylation causes misfolding and associated mislocalization of the protein. All constructs showed predominantly cell surface binding and no colocalization with an ER marker (ER tracker, red, Fig. 4B) was observed, arguing that the protein is properly folded and exported to the cell surface.
FIGURE 4.
EWI-2 trafficking to EMV is dependent on the number of N-glycans. A, schematic of WT and mutant (N1, N12, N123) EWI-2 constructs. FLAG tag at the C terminus is shown in red. B, Sk-Mel-5 cells were transfected with wt, N1, N12, or N123 EWI-2 FLAG constructs. The cells were fixed and co-stained with α-FLAG antibody (FITC-α-FLAG, green) and ER-Tracker Red (red). Cells were imaged at varying z-planes with fluorescence microscopy. Images were deconvolved with NIS Elements, and a single focal plane from the deconvoluted images is shown for each. The scale bar is 20 μm. C, Sk-Mel-5 cells were transfected with WT, N1, N12, or N123 EWI-2 FLAG constructs. After 48 h, culture medium was collected for EMV isolation, and TCM was isolated from cells. Equal amounts of TCM samples (25 μg) and of EMV (10 μg) were analyzed for each construct by SDS-PAGE and Western blot analysis (α-FLAG antibody). A representative blot of three biological replicates is shown. D, quantification of all biological replicates of analysis shown in C. Western blots were quantified using GelQuant.NET. For each analysis, EWI-2-FLAG intensity from MV samples was normalized to the corresponding TCM samples to account for differences in expression. All data were then normalized to the WT. The graph shows the average data, and error bars represent the S.E. for the three replicates. E, TCM and EMVs from FLAG construct transfected cells were obtained and analyzed as in C with the exception that blots were probed with an α-CD63 antibody.
We next examined the trafficking of these constructs into EMVs. Sk-Mel-5 cells were transfected with WT, N1, N12, or N123 EWI-2 FLAG constructs, and EMVs and total cell membranes were isolated and analyzed by Western blot using an α-FLAG antibody for detection. No differences in the levels of EMV isolated from the transfected cells were observed. The wild type EWI-2 construct (WT, Fig. 4C) showed strong enrichment in EMV isolated from transfected cells consistent with the recruitment into EMV observed for the native protein (Fig. 2A). N-Glycan mutants of EWI-2 ran at lower molecular weights due to the loss of glycosylation, consistent with published reports (32). A clear glycan-dependent decrease in the levels of EWI-2 recruited to EMV was observed and was directly related to the number of N-glycan sites removed (Fig. 4, C and D). To ensure that this decrease was not due to differences in EWI-2 expression levels between our four constructs, we normalized the expression levels of EWI-2 in EMV to the levels in the TCM for the same construct and compared them to the wild type (WT, N1, N12, N123; Fig. 4D, n = 3 experiments). Removal of even a single N-glycan site from EWI-2 (N1) resulted in an ∼50% loss of EWI-2 trafficking to the EMV. No effect was observed on CD63 trafficking to EMV in the transfected cells (Fig. 4E). These results agree with our previous results and provide strong evidence that N-linked glycans are a determinant of protein trafficking to EMV.
CD81 Trafficking to EMV Is Modulated by Glycosylation in an EWI-2-independent Manner in Sk-Mel-5
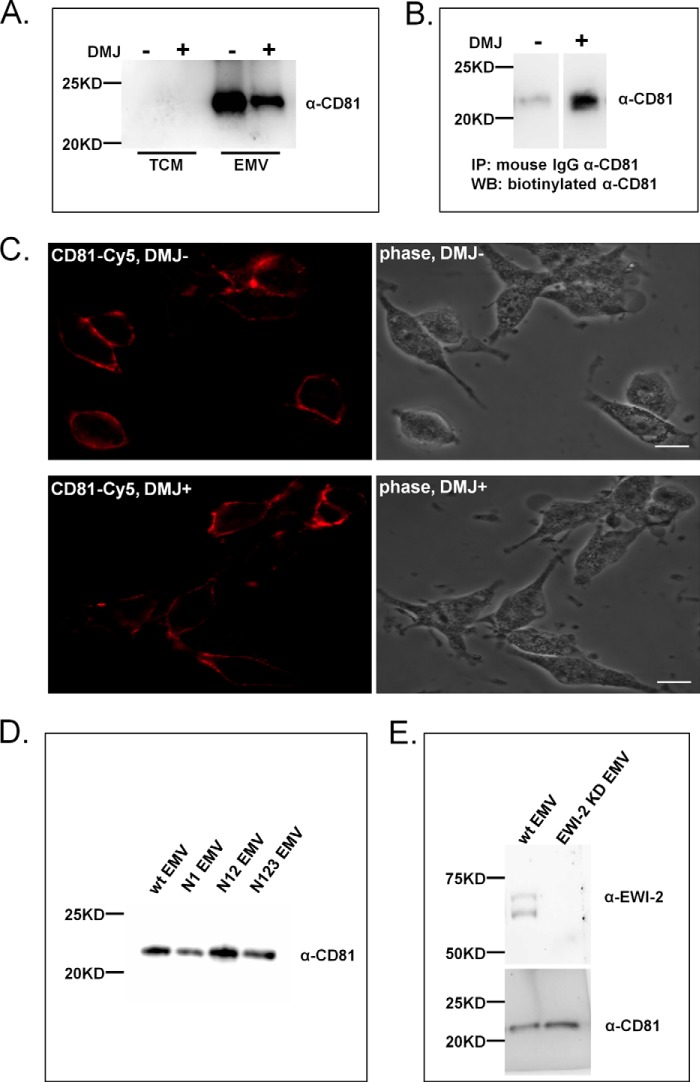
CD81 is commonly used as an EMV marker in tandem with CD63. This use assumes that CD81 levels are related to EMV amounts and not other factors such as glycosylation. In our studies with DMJ we were surprised to observe a strong decrease in the levels of CD81 in EMV upon DMJ treatment (∼64% reduction, Fig. 5A). CD81 is a non-glycosylated tetraspanin, so we tested whether the loss of CD81 was due to a loss of expression of the protein. We immunoprecipitated CD81 from equal amounts of total cell membranes from either control or DMJ-treated cells and looked at protein levels by Western blot analysis. We observed no loss of CD81 in total membranes (Fig. 5B). Analysis of CD81 localization by fluorescence microscopy also showed no loss of CD81 and no gross mislocalization of the protein (Fig. 5C). Taken together this suggested that CD81 might be chaperoned into EMV by a glycosylation-related mechanism. One possibility is that association of CD81 with a glycoprotein might alter its trafficking. Several glycoproteins are known to directly bind CD81 and guide its localization, including EWI-2 (30, 31, 39), EWI-F (40, 41), and CD19 (42–45). The interaction between CD81 and EWI-2 is glycan-independent and is through the transmembrane domain of EWI-2 (32). We tested whether EWI-2 might be responsible for the recruitment of CD81 into EMV in a glycan-dependent manner. We reasoned that overexpression of the EWI-2 glycan site mutants (N1, N12, N123), which alter EWI-2 trafficking to EMV (Fig. 4) but not association with CD81 (32), should have a similar effect on CD81 if CD81 trafficking to EMV were EWI-2-dependent. To the contrary, we found that overexpression of the N1, N12, and N123 mutants had no effect on the levels of CD81 observed in EMV (Fig. 5D), in line with what was observed for CD63 (Fig. 4E). To confirm this finding we generated a stable EWI-2 knock-out cell line using shRNA and examined the levels of CD81 in EMV isolated from these cells. It should be noted that the EWI-2 knock-out cell line produced equivalent levels of EMV as wild type. In concordance with our earlier results, we saw no effect on CD81 recruitment to EMV in the EWI-2 knock-out cell line. Based on this data, EWI-2 is not involved in the observed glycan dependence of CD81 trafficking, a mystery with implications for the use of this tetraspanin as an EMV biomarker.
FIGURE 5.
CD81 recruitment to EMV is glycan-dependent but not EWI-2-dependent. A, inhibition of complex N-glycan alters CD81 trafficking to EMV. Sk-Mel-5 cells were treated with DMJ (1 mm) for 48 h before collection of TCM or EMV (+ DMJ). Untreated Sk-Mel-5 cells were used as a control (− DMJ). TCM and EMV samples (+/− DMJ) were probed for CD81. Equal amounts of TCM samples (25 μg of protein) and of EMV (10 μg of protein) were compared by SDS-PAGE and Western blot analysis. Western blots shown are representative of two biological replicates. B, CD81 was immunoprecipitated from equal amounts (by protein) of DMJ-treated and -untreated TCM samples. Immunoprecipitated samples were run on SDS-PAGE then analyzed for CD81 by Western blot analysis with a different α-CD81 antibody than that used for immunoprecipitation. Western blots shown are representative of two biological replicates. C, cell surface localization of CD81 is not affected by DMJ treatment. Sk-Mel-5 cells were treated with either DMJ (1 mm, DMJ +) or buffer (DMJ −) for 48 h. The cells were fixed and stained with an α-CD81 antibody (α-CD81 biotin, Cy5-α-biotin mAb, red). Cells were imaged with fluorescence microscopy. The scale bar is 20 μm. D, Sk-Mel-5 cells were transfected with WT, N1, N12, and N123 EWI-2-FLAG constructs, and TCM samples were collected. EMV samples were isolated from culture media. Equal amounts of TCM samples (25 μg of protein) and EMV (10 μg of protein) samples were loaded onto SDS-PAGE and analyzed by Western blot analysis for CD81. Western blots shown are representative of three biological replicates. E, a stable EWI-2 knockdown of the Sk-Mel-5 cell line was generated using lentiviral stocks of shRNA. EMV samples were isolated from the EWI-2 knockdown cell line (EWI-2 KD) or from parent Sk-Mel-5 (WT). Equal amounts of EMVs (10 μg) were resolved using SDS-PAGE and analyzed by Western blot analysis for EWI-2 and CD81. Western blots shown are representative of three independent EMV isolations from this cell line.
Glycosylation Alters Recruitment of Galectin-3 and Galectin-3-binding Protein to EMV
Several lectins are known to control glycan-dependent sorting of glycoproteins to specific membranes, including galectins. Galectins are a polyLacNAc-binding lectin family, members of which have been shown to regulate protein trafficking in polarized epithelia (46, 47). Our preliminary mass spectrometry data identified both galectin-3 and the G3BP (also know as Mac-2BP), a known galectin-3 glycoprotein partner, in EMVs from Sk-Mel-5. Galectin-3 was not enriched in EMVs compared with total cell membrane, in contrast to G3BP (Fig. 6, A and B). Altering glycosylation of Sk-Mel-5 cells and the resulting EMVs from complex to high mannose structures by DMJ treatment reduced galectin-3 levels in both the total cell membrane and EMV samples, consistent with the loss of binding epitopes. This provides evidence that in EMVs galectin-3 is associated with the external EMV membrane through glycan binding. Similar results have been observed in other systems for galectins-5 (48) and -9 (49).
FIGURE 6.
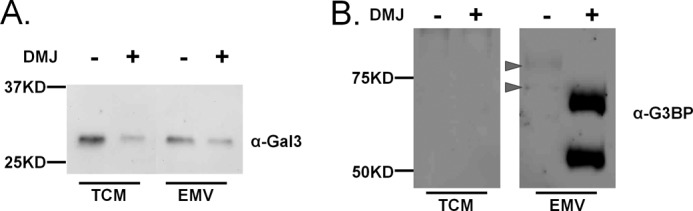
Glycosylation alters galectin-3 and G3BP recruitment to EMV. Sk-Mel-5 cells were treated with DMJ (1 mm) for 48 h before collection of TCM or EMV (+ DMJ). Untreated Sk-Mel-5 cells were used as a control (− DMJ). A, TCM and EMV samples (+/− DMJ) were probed for galectin-3. Equal amounts of TCM and EMV were loaded in each lane (5 μg of protein) and analyzed by SDS-PAGE and Western blot analysis. B, TCM and EMV samples (+/− DMJ) were probed for G3BP. Equal amounts of TCM samples (25 μg) and of EMV (10 μg) were compared by SDS-PAGE and Western blot analysis. The position of glycosylated G3BP is indicated by arrows. Western blots shown are representative of two biological replicates.
In contrast, DMJ treatment greatly enhanced the trafficking of G3BP into EMVs (Fig. 6B). The enhancement in the signal observed was surprising and was not due to increased availability of a cryptic epitope to the antibody. PNGase F cleavage of N-glycans on EMVs from untreated Sk-Mel-5 cells had no effect on the level of antibody binding, although the expected decrease in molecular weight was observed (data not shown). The differential response of G3BP and EWI-2 recruitment into EMVs upon DMJ treatment points to more than one potential mechanism for glycan-dependent protein trafficking to EMVs, in line with what is observed in other protein trafficking pathways, e.g. apical trafficking (12, 13, 50).
DISCUSSION
EMVs are increasingly recognized as a form of long range communication between cells within the body. The nature of this communication depends upon the EMV origin and upon its protein content. The proteins recruited into EMVs are not random; however the determinants underlying protein trafficking to these particles remain to be worked out. Herein, we provide strong evidence that N-linked glycans are one determinant for EMV protein trafficking.
Glycosylation plays a role in the trafficking of discrete subsets of proteins to membrane domains. This has been validated in the classical protein-sorting pathway apical to basolateral trafficking, where multiple glycosylation and other sorting signals exist and are protein-specific determinants (12, 13, 51, 52). Glycosylation has never been shown as a sorting determinant for trafficking to EMVs. In earlier work we demonstrated a conserved glycosylation signature for EMVs that was distinct from the overall parent cell membranes, with EMVs showing enrichment in poly/multiantennary LacNAc, complex N-linked glycans, high mannose, and α2,6-sialic acid epitopes (14, 15). Herein we show that CD63 and CD81, tetraspanins enriched in EMVs that define the membrane domains from which they arise (53, 54), colocalize with high levels of poly/multiantennary LacNAc epitopes (DSA) on melanoma cells as observed by fluorescence microscopy (Fig. 1). The colocalization is highly significant as seen in the Pearson correlation coefficients (PCC = 0.876, DSA/CD81; PCC = 0.875 (membrane only) DSA/CD63). This strongly suggests that EMVs arise from membrane domains that are characterized by the enrichment of these glycans, hinting at a role for glycosylation in protein sorting to these domains.
The mechanism of protein sorting in classical protein trafficking (i.e. apical versus basolateral) is highly dependent on the protein itself as there are multiple potential determinants that can overlap, of which glycosylation is only one (51, 55). To find proteins for which glycosylation is the critical determinant, we identified glycoproteins in EMVs that both bound to a DSA column and were enriched in EMVs (Fig. 2). We identified three glycoproteins, G3BP ADAM-10, and EWI-2. We focused on the trafficking of EWI-2 to EMVs as its glycosylation sites were known and glycan mutants had previously been made (34). We confirmed that EWI-2 contained complex N-glycans and found that inhibition of these glycans by DMJ, which halts glycan processing at the high mannose stage, inhibited recruitment of EWI-2 into EMVs (Fig. 3). It did not, however, alter the gross cell surface localization of EWI-2, arguing that this is not a protein folding issue. Genetic ablation of the glycan attachment sites in EWI-2 also caused a loss of EMV recruitment. Trafficking of EWI-2 to EMVs was proportional to the number of N-glycan sites removed, and again no gross changes in cell surface localization were observed (Fig. 4). Taken together our data provide strong evidence that both the amount and nature of the glycan epitope control EWI-2 recruitment to EMV, arguing that glycosylation is an EMV sorting determinant for this protein.
We expect that multiple signals will exist for protein trafficking to EMVs of which complex N-linked glycosylation is one. A likely mechanism for carbohydrate-mediated glycoprotein trafficking to EMV is through lectin-mediated oligomerization. Lectins are carbohydrate-binding proteins that are often multivalent or oligomeric, enabling them to cross-link glycoproteins. Membrane proteins oligomerized by either self-association domains or through the addition of an exogenous reagent such as a multivalent antibody are selectively sorted to EMVs arguing that oligomerization alone is a sufficient signal to recruit proteins into these structure (53, 56). Several lectins known to be involved in protein trafficking are good candidates for recruiting glycoproteins into EMVs including a family of polyLacNAc binding lectins known as galectins (13). Galectins either contain multiple carbohydrate binding domains or multimerize to create galectin-glycoprotein oligomers that coalesce into “galectin lattices” (46). Multiple galectins, including galectins-3, -4, and -9, have been shown to regulate protein trafficking in polarized epithelia (46, 47). In this work we identified galectin-3 in our EMVs and found that it is associated with them in a glycan-dependent manner (Fig. 6). Previous work also identified galectin-1 in these EMVs. Whether these galectins are responsible for the complex N-glycan-dependent sorting to EMVs observed is the subject of future work but their presence suggests a mechanism for this trafficking event.
One surprising finding in our work was the increased amounts of G3BP (also known as Mac-2BP) found in EMVs when the glycans were shifted to a high mannose cohort by DMJ. This is suggestive that high mannose itself may act as a second potential N-glycan sorting signal. In previous studies we have shown increased high mannose in EMVs (14, 15). It is also consistent with a report that G3BP requires ERGIC-53, a known high mannose binding lectin (58), for secretion (59). ERGIC-53 is recruited to viral particles and required for infectivity of areaviruses, coronaviruses and filoviruses (60). Taken together, this is highly suggestive of a role for ERGIC-53 in recruiting G3BP to EMV, a hypothesis which remains to be tested.
A second surprising finding was the glycan dependence of CD81 recruitment to EMVs (Fig. 5). Treatment of Sk-Mel-5 with DMJ inhibited CD81 levels in EMVs by >60% but had no effect on EMV amounts either by protein or CD63 levels or on CD81 localization as observed by fluorescence microscopy. We initially hypothesized that this change in CD81 could be due to EWI-2-dependent trafficking (33) given the known glycan-independent association between EWI-2 and CD81. Several lines of evidence, most convincingly the lack of change in CD81 levels in EMV from EWI-2 knockdown cell lines, argue that this is not the case. Although we do not understand the glycan dependence of CD81 recruitment, this finding has important implications for the use of this tetraspanin as an independent marker for EMVs.
In conclusion, our work presents the first strong evidence that N-linked glycans are a sorting determinant in EMV cargo protein recruitment and suggests potential mechanisms. Further work is clearly needed to fill in the details of how N-linked glycans control glycoprotein recruitment into EMVs. Given the importance of protein content in the functions of these particles, a deeper understanding is critical to our ability to manipulate, utilize, and understand this important aspect of cellular communication.
Supplementary Material
Acknowledgment
We thank Professor Laurence Cocquerel for help in cloning the EWI-2 mutants.
This work was supported, in whole or in part, by National Institutes of Health Grant 7 DP2 OD004711-02). This work was also supported by Mitzutani Foundation for Glycoscience Grant 130107.

This article contains supplemental Tables 1–3 and Fig. 1.
- EMV
- exosomes, also known as microvesicle
- DMJ
- 1-deoxymannojirimycin
- DSA
- Datura stramonium agglutinin
- PCC
- Pearson correlation coefficient
- MO
- Mander's overlap
- G3BP
- galectin-3-binding protein
- TCM
- total cell membrane
- HBSS
- Hanks' balanced salt solution
- PNGase
- peptide N-glycosidase F
- PFA
- paraformaldehyde.
REFERENCES
- 1. Raposo G., Nijman H. W., Stoorvogel W., Liejendekker R., Harding C. V., Melief C. J., Geuze H. J. (1996) B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 183, 1161–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Johnstone R. M. (2006) Exosomes biological significance: a concise review. Blood Cells. Mol. Dis. 36, 315–321 [DOI] [PubMed] [Google Scholar]
- 3. Simpson R. J., Lim J. W., Moritz R. L., Mathivanan S. (2009) Exosomes: proteomic insights and diagnostic potential. Expert Rev. Proteomics 6, 267–283 [DOI] [PubMed] [Google Scholar]
- 4. Johnstone R. M., Adam M., Hammond J. R., Orr L., Turbide C. (1987) Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 262, 9412–9420 [PubMed] [Google Scholar]
- 5. EL Andaloussi S., Mäger I., Breakefield X. O., Wood M. J. (2013) Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 12, 347–357 [DOI] [PubMed] [Google Scholar]
- 6. Ludwig A. K., Giebel B. (2012) Exosomes: small vesicles participating in intercellular communication. Int. J. Biochem Cell Biol. 44, 11–15 [DOI] [PubMed] [Google Scholar]
- 7. Raposo G., Stoorvogel W. (2013) Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 200, 373–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bobrie A., Colombo M., Raposo G., Théry C. (2011) Exosome secretion: molecular mechanisms and roles in immune responses. Traffic 12, 1659–1668 [DOI] [PubMed] [Google Scholar]
- 9. Mathivanan S., Simpson R. J. (2009) ExoCarta: a compendium of exosomal proteins and RNA. Proteomics 9, 4997–5000 [DOI] [PubMed] [Google Scholar]
- 10. Mathivanan S., Fahner C. J., Reid G. E., Simpson R. J. (2012) ExoCarta 2012: database of exosomal proteins, RNA, and lipids. Nucleic Acids Res. 40, D1241–D1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Al-Nedawi K., Meehan B., Micallef J., Lhotak V., May L., Guha A., Rak J. (2008) Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 10, 619–624 [DOI] [PubMed] [Google Scholar]
- 12. Delacour D., Cramm-Behrens C. I., Drobecq H., Le Bivic A., Naim H. Y., Jacob R. (2006) Requirement for galectin-3 in apical protein sorting. Curr. Biol. 16, 408–414 [DOI] [PubMed] [Google Scholar]
- 13. Vagin O., Kraut J. A., Sachs G. (2009) Role of N-glycosylation in trafficking of apical membrane proteins in epithelia. Am. J. Physiol. Renal Physiol. 296, F459–F469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Batista B. S., Eng W. S., Pilobello K. T., Hendricks-Muñoz K. D., Mahal L. K. (2011) Identification of a conserved glycan signature for microvesicles. J. Proteome Res. 10, 4624–4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krishnamoorthy L., Bess J. W., Jr., Preston A. B., Nagashima K., Mahal L. K. (2009) HIV-1 and microvesicles from T cells share a common glycome, arguing for a common origin. Nat. Chem. Biol. 5, 244–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Escrevente C., Grammel N., Kandzia S., Zeiser J., Tranfield E. M., Conradt H. S., Costa J. (2013) Sialoglycoproteins and N-glycans from secreted exosomes of ovarian carcinoma cells. PLoS ONE 8, e78631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nagai T., Ibata K., Park E. S., Kubota M., Mikoshiba K., Miyawaki A. (2002) A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20, 87–90 [DOI] [PubMed] [Google Scholar]
- 18. Parish L. A., Colquhoun D. R., Ubaida Mohien C., Lyashkov A. E., Graham D. R., Dinglasan R. R. (2011) Ookinete-interacting proteins on the microvillar surface are partitioned into detergent resistant membranes of Anopheles gambiae midguts. J. Proteome Res. 10, 5150–5162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hemler M. E. (2005) Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 6, 801–811 [DOI] [PubMed] [Google Scholar]
- 20. Nazarenko I., Rana S., Baumann A., McAlear J., Hellwig A., Trendelenburg M., Lochnit G., Preissner K. T., Zöller M. (2010) Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. 70, 1668–1678 [DOI] [PubMed] [Google Scholar]
- 21. Perez-Hernandez D., Gutiérrez-Vázquez C., Jorge I., López-Martín S., Ursa A., Sánchez-Madrid F., Vázquez J., Yáñez-Mó M. (2013) The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J. Biol. Chem. 288, 11649–11661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Green E. D., Brodbeck R. M., Baenziger J. U. (1987) Lectin affinity high-performance liquid chromatography: interactions of N-glycanase-released oligosaccharides with leukoagglutinating phytohemagglutinin, concanavalin A, Datura stramonium agglutinin, and Vicia villosa agglutinin. Anal. Biochem. 167, 62–75 [DOI] [PubMed] [Google Scholar]
- 23. Sun Q., Kang X., Zhang Y., Zhou H., Dai Z., Lu W., Zhou X., Liu X., Yang P., Liu Y. (2009) DSA affinity glycoproteome of human liver tissue. Arch. Biochem. Biophys. 484, 24–29 [DOI] [PubMed] [Google Scholar]
- 24. Lee H. K., Finniss S., Cazacu S., Bucris E., Ziv-Av A., Xiang C., Bobbitt K., Rempel S. A., Hasselbach L., Mikkelsen T., Slavin S., Brodie C. (2013) Mesenchymal stem cells deliver synthetic microRNA mimics to glioma cells and glioma stem cells and inhibit their cell migration and self-renewal. Oncotarget 4, 346–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kosaka N., Iguchi H., Hagiwara K., Yoshioka Y., Takeshita F., Ochiya T. (2013) Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J. Biol. Chem. 288, 10849–10859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dunn K. W., Kamocka M. M., McDonald J. H. (2011) A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 300, C723–C742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bolte S., Cordelières F. P. (2006) A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 [DOI] [PubMed] [Google Scholar]
- 28. Crowley J. F., Goldstein I. J., Arnarp J., Lönngren J. (1984) Carbohydrate binding studies on the lectin from Datura stramonium seeds. Arch. Biochem. Biophys. 231, 524–533 [DOI] [PubMed] [Google Scholar]
- 29. Escrevente C., Morais V. A., Keller S., Soares C. M., Altevogt P., Costa J. (2008) Functional role of N-glycosylation from ADAM10 in processing, localization, and activity of the enzyme. Biochim. Biophys. Acta 1780, 905–913 [DOI] [PubMed] [Google Scholar]
- 30. Stipp C. S., Kolesnikova T. V., Hemler M. E. (2001) EWI-2 is a major CD9 and CD81 partner and member of a novel Ig protein subfamily. J. Biol. Chem. 276, 40545–40554 [DOI] [PubMed] [Google Scholar]
- 31. Charrin S., Le Naour F., Labas V., Billard M., Le Caer J.-P., Emile J.-F., Petit M.-A., Boucheix C., Rubinstein E. (2003) EWI-2 is a new component of the tetraspanin web in hepatocytes and lymphoid cells. Biochem. J. 373, 409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Montpellier C., Tews B. A., Poitrimole J., Rocha-Perugini V., D'Arienzo V., Potel J., Zhang X. A., Rubinstein E., Dubuisson J., Cocquerel L. (2011) Interacting regions of CD81 and two of its partners, EWI-2 and EWI-2wint, and their effect on hepatitis C virus infection. J. Biol. Chem. 286, 13954–13965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stipp C. S., Kolesnikova T. V., Hemler M. E. (2003) EWI-2 regulates α3β1 integrin-dependent cell functions on laminin-5. J. Cell Biol. 163, 1167–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rocha-Perugini V., Montpellier C., Delgrange D., Wychowski C., Helle F., Pillez A., Drobecq H., Le Naour F., Charrin S., Levy S., Rubinstein E., Dubuisson J., Cocquerel L. (2008) The CD81 partner EWI-2wint inhibits hepatitis C virus entry. PLoS ONE 3, e1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rakus J. F., Mahal L. K. (2011) New technologies for glycomic analysis: toward a systematic understanding of the glycome. Annu. Rev. Anal. Chem. (Palo Alto Calif) 4, 367–392 [DOI] [PubMed] [Google Scholar]
- 36. Elbein A. (1991) Glycosidase inhibitors: inhibitors of N-linked oligosaccharide processing. FASEB J. 5, 3055–3063 [DOI] [PubMed] [Google Scholar]
- 37. Morelle W., Stechly L., André S., Van Seuningen I., Porchet N., Gabius H. J., Michalski J. C., Huet G. (2009) Glycosylation pattern of brush border-associated glycoproteins in enterocyte-like cells: involvement of complex-type N-glycans in apical trafficking. Biol. Chem. 390, 529–544 [DOI] [PubMed] [Google Scholar]
- 38. Fuhrmann U., Bause E., Legler G., Ploegh H. (1984) Novel mannosidase inhibitor blocking conversion of high mannose to complex oligosaccharides. Nature 307, 755–758 [DOI] [PubMed] [Google Scholar]
- 39. Clark K. L., Zeng Z., Langford A. L., Bowen S. M., Todd S. C. (2001) PGRL is a major CD81-associated protein on lymphocytes and distinguishes a new family of cell surface proteins. J. Immunol. 167, 5115–5121 [DOI] [PubMed] [Google Scholar]
- 40. Stipp C. S., Orlicky D., Hemler M. E. (2001) FPRP, a major, highly stoichiometric, highly specific CD81- and CD9-associated protein. J. Biol. Chem. 276, 4853–4862 [DOI] [PubMed] [Google Scholar]
- 41. Charrin S., Le Naour F., Oualid M., Billard M., Faure G., Hanash S. M., Boucheix C., Rubinstein E. (2001) The major CD9 and CD81 molecular partner. Identification and characterization of the complexes. J. Biol. Chem. 276, 14329–14337 [DOI] [PubMed] [Google Scholar]
- 42. Shoham T., Rajapaksa R., Boucheix C., Rubinstein E., Poe J. C., Tedder T. F., Levy S. (2003) The tetraspanin CD81 regulates the expression of CD19 during B cell development in a postendoplasmic reticulum compartment. J. Immunol. 171, 4062–4072 [DOI] [PubMed] [Google Scholar]
- 43. Horváth G., Serru V., Clay D., Billard M., Boucheix C., Rubinstein E. (1998) CD19 is linked to the integrin-associated tetraspans CD9, CD81, and CD82. J. Biol. Chem. 273, 30537–30543 [DOI] [PubMed] [Google Scholar]
- 44. Bradbury L. E., Kansas G. S., Levy S., Evans R. L., Tedder T. F. (1992) The CD19/CD21 signal transducing complex of human B lymphocytes includes the target of antiproliferative antibody-1 and Leu-13 molecules. J. Immunol. 149, 2841–2850 [PubMed] [Google Scholar]
- 45. Matsumoto A. K., Martin D. R., Carter R. H., Klickstein L. B., Ahearn J. M., Fearon D. T. (1993) Functional dissection of the CD21/CD19/TAPA-1/Leu-13 complex of B lymphocytes. J. Exp. Med. 178, 1407–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Delacour D., Koch A., Jacob R. (2009) The role of galectins in protein trafficking. Traffic 10, 1405–1413 [DOI] [PubMed] [Google Scholar]
- 47. Mishra R., Grzybek M., Niki T., Hirashima M., Simons K. (2010) Galectin-9 trafficking regulates apical-basal polarity in Madin-Darby canine kidney epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 107, 17633–17638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Barrès C., Blanc L., Bette-Bobillo P., André S., Mamoun R., Gabius H. J., Vidal M. (2010) Galectin-5 is bound onto the surface of rat reticulocyte exosomes and modulates vesicle uptake by macrophages. Blood 115, 696–705 [DOI] [PubMed] [Google Scholar]
- 49. Klibi J., Niki T., Riedel A., Pioche-Durieu C., Souquere S., Rubinstein E., Le Moulec S., Moulec S. L., Guigay J., Hirashima M., Guemira F., Adhikary D., Mautner J., Busson P. (2009) Blood diffusion and Th1-suppressive effects of galectin-9-containing exosomes released by Epstein-Barr virus-infected nasopharyngeal carcinoma cells. Blood 113, 1957–1966 [DOI] [PubMed] [Google Scholar]
- 50. Delacour D., Greb C., Koch A., Salomonsson E., Leffler H., Le Bivic A., Jacob R. (2007) Apical sorting by galectin-3-dependent glycoprotein clustering. Traffic 8, 379–388 [DOI] [PubMed] [Google Scholar]
- 51. Mattila P. E., Youker R. T., Mo D., Bruns J. R., Cresawn K. O., Hughey R. P., Ihrke G., Weisz O. A. (2012) Multiple biosynthetic trafficking routes for apically secreted proteins in MDCK cells. Traffic 13, 433–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yamamoto H., Awada C., Hanaki H., Sakane H., Tsujimoto I., Takahashi Y., Takao T., Kikuchi A. (2013) The apical and basolateral secretion of Wnt11 and Wnt3a in polarized epithelial cells is regulated by different mechanisms. J. Cell Sci. 126, 2931–2943 [DOI] [PubMed] [Google Scholar]
- 53. Fang Y., Wu N., Gan X., Yan W., Morrell J. C., Gould S. J. (2007) Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes. PLoS Biol. 5, e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nydegger S., Khurana S., Krementsov D. N., Foti M., Thali M. (2006) Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. J. Cell Biol. 173, 795–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cao X., Surma M. A., Simons K. (2012) Polarized sorting and trafficking in epithelial cells. Cell Res. 22, 793–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shen B., Wu N., Yang J. M., Gould S. J. (2011) Protein targeting to exosomes/microvesicles by plasma membrane anchors. J. Biol. Chem. 286, 14383–14395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Varki A., Cummings R., Esko J. D., Freeze H., Stanley P., Bertozzi C. R., Hart G. W., Etzler M. E. (eds) (2009) Essentials of Glycobiology, 2nd Ed., pp. 1–51, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 58. Neve E. P., Lahtinen U., Pettersson R. F. (2005) Oligomerization and interacellular localization of the glycoprotein receptor ERGIC-53 is independent of disulfide bonds. J. Mol. Biol. 354, 556–568 [DOI] [PubMed] [Google Scholar]
- 59. Chen Y., Hojo S., Matsumoto N., Yamamoto K. (2013) Regulation of Mac-2BP secretion is mediated by its N-glycan binding to ERGIC-53. Glycobiology 23, 904–916 [DOI] [PubMed] [Google Scholar]
- 60. Klaus J. P., Eisenhauer P., Russo J., Mason A. B., Do D., King B., Taatjes D., Cornillez-Ty C., Boyson J. E., Thali M., Zheng C., Liao L., Yates J. R., 3rd, Zhang B., Ballif B. A., Botten J. W. (2013) The intracellular cargo receptor ERGIC-53 is required for the production of infectious arenavirus, coronavirus, and filovirus particles. Cell Host Microbe 14, 522–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.