Background: SAMHD1 is a deoxyribonucleoside triphosphate (dNTP) triphosphohydrolase.
Results: SAMHD1 forms a catalytically active tetramer upon binding of two nucleoside triphosphates with different specificities at two adjacent allosteric sites.
Conclusion: The primary allosteric site selectively binds guanine-containing nucleotides, whereas the secondary site accommodates any dNTP.
Significance: The tetramerization and catalytic activity of SAMHD1 is differentially regulated by different nucleoside triphosphates.
Keywords: Enzyme Mechanism, Nucleic Acid, Protein Crystallization, SAM Domain and HD Domain-containing Protein 1 (SAMHD1), Structural Biology, X-ray Crystallography
Abstract
Sterile α motif and histidine-aspartate domain-containing protein 1 (SAMHD1) plays a critical role in inhibiting HIV infection, curtailing the pool of dNTPs available for reverse transcription of the viral genome. Recent structural data suggested a compelling mechanism for the regulation of SAMHD1 enzymatic activity and revealed dGTP-induced association of two inactive dimers into an active tetrameric enzyme. Here, we present the crystal structures of SAMHD1 catalytic core (residues 113–626) tetramers, complexed with mixtures of nucleotides, including dGTP/dATP, dGTP/dCTP, dGTP/dTTP, and dGTP/dUTP. The combined structural and biochemical data provide insight into dNTP promiscuity at the secondary allosteric site and how enzymatic activity is modulated. In addition, we present biochemical analyses of GTP-induced SAMHD1 full-length tetramerization and the structure of SAMHD1 catalytic core tetramer in complex with GTP/dATP, revealing the structural basis of GTP-mediated SAMHD1 activation. Altogether, the data presented here advance our understanding of SAMHD1 function during cellular homeostasis.
Introduction
SAMHD14 is a potent HIV restriction factor, possessing deoxyribonucleoside triphosphate triphosphohydrolase (dNTPase) activity (1–5). The protein's enzymatic activity, residing within its histidine-aspartate domain (residues 113–626, SAMHD1c), is essential to its role in the innate immune system; current evidence supports a model in which the cellular dNTP pools are maintained at low levels by SAMHD1, thus inhibiting viral reverse transcription in infected immune cells (6–9). Further, recent studies suggest that SAMHD1 has broad antiviral activity, not only against retroviruses but also DNA viruses, due to its catalytic activity (10–12). In addition, nuclease activity has been reported for the histidine-aspartate domain (13), with a phosphomimetic mutant of SAMHD1, T592E, exhibiting a reduction in RNase activity and reduced HIV-1 restriction, in vivo, suggesting an additional dimension to HIV restriction by SAMHD1 (14). Phosphorylation at Thr-592, however, did not alter the dNTPase activity of SAMHD1 (15–18). The essential role of SAMHD1 in innate immunity is further underscored by its association with stroke-related autoimmunity disorders, including Aicardi-Goutières syndrome and systemic lupus erythematosus (19–24).
The dNTPase activity of SAMHD1 is regulated by dGTP, which binds at allosteric sites (2). Several biochemical and structural studies suggest that a tetramer is the active species, with tetramerization induced by dGTP binding, whereas the inactive apo-form interconverts between monomer and dimer (25–27). The active tetramer possesses four allosteric nucleotide binding sites, each capable of accommodating two dGTPs that serve to interlock SAMHD1 chains into the tetrameric state (25, 27). Intriguingly, recent studies have suggested an additional control of SAMHD1 activity via other nucleoside triphosphates (28–30). In particular, the protein can be activated by a mixture of GTP and dNTP. To further explore details of this regulation, we solved the crystal structures of tetrameric SAMHD1c in complex with two sets of different nucleotides. In the first set, the primary allosteric site is only occupied by dGTP, whereas the secondary site is promiscuous for other dNTP (SAMHD1c-dGTP/dNTP). The second set of nucleotide mixtures yielded complexes in which the primary site is occupied by GTP, and the secondary site is occupied by dATP (SAMHD1c-GTP/dATP). Differential binding specificities for nucleosides at the two allosteric sites were verified by biochemical analyses of WT SAMHD1 and several mutants. Together, our results further add to the structural underpinnings for SAMHD1 regulation by nucleoside triphosphates, a critical activity in its cellular functions.
EXPERIMENTAL PROCEDURES
Protein Expression, Purification, and Crystallization
Human SAMHD1 variants (residues 113–626 (SAMHD1c) and 1–626 (SAMHD1fl)) were expressed and purified as described previously (26). Site-directed mutants of SAMHD1c and SAMHDfl were constructed using the QuikChange mutagenesis kit (Agilent). Protein concentrations were determined using molecular extinction coefficients of 62,395 and 76,500 m−1 cm−1 for SAMHD1c and SAMHD1fl, respectively. Purified proteins in 25 mm Tris-HCl buffer, pH 7.5, 150 mm NaCl, 1 mm DTT, 10% glycerol, and 0.02% sodium azide were flash-frozen and stored at −80 °C until used.
Crystallization trials were carried out at room temperature by the microbatch under oil method, using 2-μl drops of protein and 2-μl crystallization solutions, as mentioned below. SAMHD1c H206R/D207N mutant protein (5 mg/ml) was first incubated with 0.5 μm dGTP and subsequently with either 4 mm dATP, dCTP, dTTP, or dUTP for generating co-crystals of SAMHD1c-dGTP/dATP, SAMHD1c-dGTP/dCTP, SAMHD1c-dGTP/dTTP, and SAMHD1c-dGTP/dUTP, respectively. For crystallization of SAMHD1c-GTP/dATP, the protein was incubated with 2 mm GTP and 2 mm dATP. In all cases, well diffracting crystals were obtained with microseeding in 0.1 m SPG (succinic acid, sodium phosphate dibasic, glycine), pH 6.5, 25% PEG 1500, 20% ethylene glycol in the presence of either 0.0012 mm n-hexadecyl-β-d-maltoside, 12 mm n-nonyl-β-d-maltoside, 2% (w/v) CYMAL-1, or 0.12 m NDSB-221 after several days.
Data Collection and Structure Determination
X-ray diffraction data for all crystals were collected at the SER-CAT facility sector 22-BM beam line of the Advance Photon Source at Argonne National Laboratory (Chicago, IL) at a wavelength of 1.00 Å. Diffraction data were processed, integrated, and scaled using d*TREK software (31) and eventually converted to mtz format using the CCP4 package (32).
The structures of SAMHD1c in the presence of dGTP/dATP, dGTP/dCTP, dGTP/dTTP, dGTP/dUTP, or GTP/dATP were determined by molecular replacement, using the previously determined SAMHD1c-dGTP/dGTP complex structure (PDB code 4BZB), as the structural probe in PHASER (33). After generation of the initial model, the chain was rebuilt using the program Coot (34). Iterative refinement was carried out by alternating between manual rebuilding in Coot (34) and refinement in RefMac5 (35) or Phenix (36, 37).
All final models exhibit clear electron density for all residues, except for a short loop (residues 278–283) and the C terminus (residues 600–626), similar to the previous structure of SAMHD1c-dGTP/dGTP (PDB code 4BZB) and SAMHD1c′ (residues 109–626)-dGTP/dATP (PDB code 4MZ7). The final structures are well defined, with the majority of residues located in the favored and allowed regions of the Ramachandran plot and no residues in the disallowed region, as evaluated by MOLPROBITY (38). The extra density in the allosteric and catalytic sites permitted fitting of appropriate GTP or dNTP molecules into each site. A summary of pertinent structural refinement statistics for all structures is provided in Table 1. All structural figures were generated with Chimera (39) or PyMOL (40). The atomic coordinates and diffraction data for structures of SAMHD1c-dGTP/dATP, SAMHD1c-dGTP/dCTP, SAMHD1c-dGTP/dTTP, SAMHD1c-dGTP/dUTP, and SAMHD1c-GTP/dATP have been deposited in the RCSB Protein Data Bank under accession codes 4QFX, 4QFY, 4QFZ, 4QG0, and 4QG1, respectively.
TABLE 1.
Data collection, refinement, and Ramachandran statistics for SAMHD1c tetramers in the presence of dGTP/dATP, dGTP/dCTP, dGTP/dTTP, dGTP/dUTP, and GTP/dATP substrates
Data were obtained from the best diffracting crystal of SAMHD1c H206R/D207N according to crystallization conditions (see “Experimental Procedures” for details) and collected at −180 °C. Crystallographic phases were solved using molecular replacement based on the SAMHD1c-dGTP/dGTP tetramer model (Protein Data Bank code 4BZB). Refinement was carried out using Phenix. Values in parentheses are for the highest resolution shell.
| SAMHD1c-dGTP/dATP | SAMHD1c-dGTP/dCTP | SAMHD1c-dGTP/dTTP | SAMHD1c-dGTP/dUTP | SAMHD1c-GTP/dATP | |
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | P21 | P21 | P21 | P21 | P21 |
| Cell dimensions | |||||
| a, b, c (Å) | 88.56/147.26/98.80 | 87.97/147.02/98.87 | 87.83/146.74/98.94 | 87.86/146.60/98.69 | 87.70/146.72/99.26 |
| α, β, γ (degrees) | 90/114.63/90 | 90/114.31/90 | 90/114.29/90 | 90/114.62/90 | 90/114.76/90 |
| Asymmetric unit content | 4 | 4 | 4 | 4 | 4 |
| Wavelength (Å) | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| Resolution (Å) | 38.34-2.20 (2.28-2.20) | 38.67-2.10 (2.18-2.10) | 33.98-2.30 (2.38-2.30) | 42.91-2.30 (2.38-2.30) | 42.99-2.20 (2.28-2.20) |
| Rmerge | 0.134 (0.500) | 0.108 (0.495) | 0.090 (0.539) | 0.113 (0.487) | 0.074 (0.505) |
| Rmeas | 0.146 (0.544) | 0.117 (0.531) | 0.101 (0.604) | 0.123 (0.531) | 0.089 (0.605) |
| 〈I/σI〉 | 5.7 (2.1) | 7.0 (2.3) | 8.0 (1.7) | 7.2 (2.3) | 6.8 (1.6) |
| Completeness (%) | 99.9 (100.0) | 99.8 (100.0) | 99.9 (99.8) | 99.8 (100.0) | 99.3 (99.7) |
| Redundancy | 6.38 (6.38) | 7.68 (7.68) | 5.09 (5.01) | 6.46 (6.46) | 3.20 (3.18) |
| Mosaicity | 0.65 | 0.43 | 0.78 | 0.51 | 0.78 |
| Refinement | |||||
| Resolution (Å) | 38.34-2.20 (2.26-2.20) | 38.67-2.10 (2.15-2.10) | 33.98-2.30 (2.36-2.30) | 42.91-2.30 (2.36-2.30) | 42.99-2.20 (2.26-2.20) |
| No. of unique reflections | 116,344 | 132,888 | 101,122 | 100,456 | 114,395 |
| No. of test reflectionsa | 2002 | 2009 | 2013 | 2011 | 2000 |
| Rwork/Rfree | 0.228/0.270 (0.257/0.313) | 0.226/0.269 (0.287/0.325) | 0.209/0.250 (0.338/0.390) | 0.197/0.250 (0.279/0.349) | 0.214/0.264 (0.362/0.374) |
| No. of atoms | |||||
| Protein | 15,753 | 15,756 | 15,798 | 15,768 | 15.739 |
| Ligand/ion | 376 | 368 | 336 | 359 | 372 |
| Water | 386 | 394 | 228 | 182 | 191 |
| Average B-factors (Å2) | |||||
| Protein | 42.78 | 38.23 | 68.49 | 64.62 | 66.04 |
| Ligand/ion | 32.00 | 29.63 | 48.69 | 44.41 | 53.20 |
| Water | 40.75 | 26.16 | 46.65 | 26.81 | 58.97 |
| Wilson B (Å2) | 41.64 | 39.06 | 46.35 | 46.88 | 53.00 |
| Root mean square deviations | |||||
| Bond lengths (Å) | 0.005 | 0.005 | 0.005 | 0.007 | 0.006 |
| Bond angles (degrees) | 0.979 | 0.971 | 0.953 | 1.070 | 1.146 |
| MolProbity statisticsb | |||||
| All atom clashscore | 8.99 | 7.14 | 7.50 | 6.83 | 6.20 |
| Rotamer outliers (%) | 2.04 | 2.86 | 2.21 | 2.10 | 1.11 |
| Cβ deviation | 4 | 4 | 4 | 5 | 0 |
| Ramachandranb | |||||
| Favored region (%) | 96.44 | 97.02 | 95.88 | 97.13 | 96.33 |
| Allowed region (%) | 3.29 | 2.98 | 4.02 | 2.77 | 3.40 |
| Outliers (%) | 0.26 | 0.00 | 0.10 | 0.10 | 0.26 |
| PDB accession code | 4QFX | 4QFY | 4QFZ | 4QG0 | 4QG1 |
a Random selection.
b Values were obtained from MOLPROBITY.
Analytical Size Exclusion Column Chromatography and Multiangle Light Scattering (SEC-MALS) of SAMHD1-Ribonucleoside Triphosphate Complexes
Mixtures of SAMHD1fl (0.5 μm) and nucleoside triphosphates, as indicated (NTP, 0–1 mm; dNTP, 0–0.1 mm), were injected into an analytical Superdex200 column at a flow rate of 0.8 ml/min, equilibrated with 20 mm Tris-HCl, pH 7.8, 50 mm NaCl, 5 mm MgCl2, 5% glycerol, and 0.02% sodium azide. The elution profiles were recorded using an in-line fluorometer (Shimadzu) with excitation at 282 nm and emission at 313 nm, as described previously (26). The peak areas of tetramer and dimer/monomer were integrated, and the percentage area of the tetramer was calculated. The molecular masses of SAMHD1fl and SAMHD1fl-nucleoside triphosphate complexes were determined using an analytical Superdex200 column with in-line multiangle light scattering (HELEOS, Wyatt Technology), variable wavelength UV (Agilent 1100 Series, Agilent Technology), and refractive index (Optilab rEX, Wyatt Technology) detector, as described previously (26, 41).
Enzymatic Assay
Deoxyribonucleoside triphosphate triphosphohydrolase (dNTPase) activity of SAMHD1fl WT and mutants (0.1 μm) was measured in a reaction buffer containing 25 mm Tris-HCl, pH 7.8, 50 mm NaCl, 2 mm MgCl2, 5% glycerol, and appropriate concentrations of dNTP and/or GTP (0–2 mm). Reactions were quenched with EDTA (20 mm final concentration) at 2, 4, 6, 8, 16, or 32 min and subjected to HPLC for quantification of deoxyribonucleosides and dNTPs, as described previously (41). Time intervals were chosen to ensure that less than 5% of substrate was converted to product.
RESULTS
The Secondary Allosteric Site of SAMHD1 Is Promiscuous
We previously reported the structure of the dGTP-bound SAMHD1c homotetramer (25). In this structure (referred to as SAMHD1c-dGTP/dGTP), a total of eight dGTP molecules occupy the four allosteric sites, each located at the interface of three subunits of the tetramer. Note that each binding pocket contains two dGTP molecules, one designated as the primary and the other as the secondary nucleotide. In a similar SAMHD1c′ tetramer structure (residues 109–626), co-crystallized in the presence of dGTP and dATP, the authors reported that the secondary allosteric and catalytic sites can be occupied by either dGTP or dATP (27). This prompted us to investigate dNTP binding at the secondary allosteric site, and we crystallized SAMHD1c in the presence of several dNTP combinations: dGTP/dATP, dGTP/dCTP, dGTP/dTTP, and dGTP/dUTP. We used the catalytically inactive SAMHD1c H206R/D207N variant, previously characterized in detail and shown to be essentially identical to the SAMHD1c WT tetramer structure (25).
All structures are, as expected, tetramers and virtually the same, with pairwise heavy atom root mean square deviation values ranging from 0.20 to 0.35 Å (Fig. 1). Similar to the previous structures, SAMHD1c-dGTP/dGTP (PDB entry 4BZB) and SAMHD1c′-dGTP/dATP (PDB entry 4MZ7), all structures comprise a well defined catalytic core, with no density observed for a short loop region (residues 278–283) and the C terminus (residues 600–626).
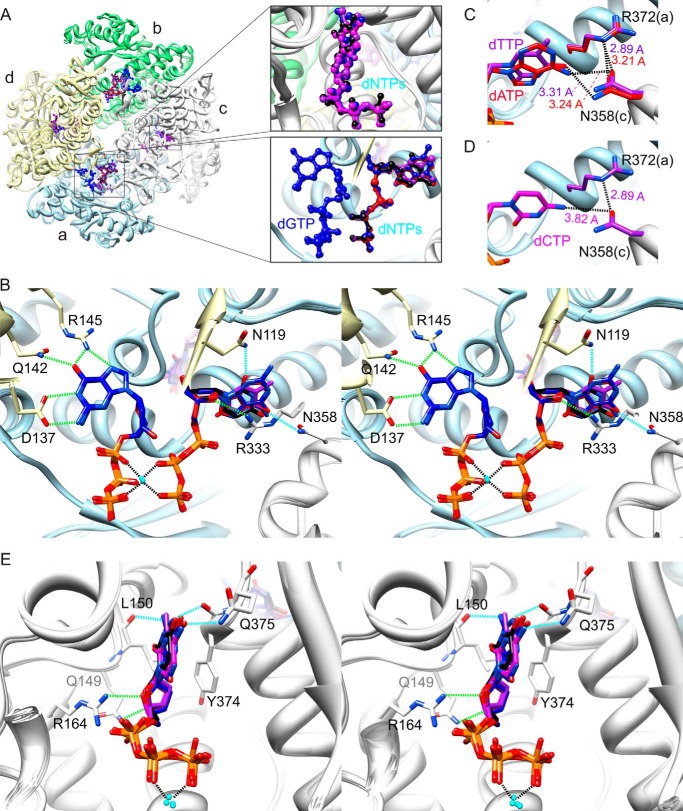
FIGURE 1.
Crystal structures of SAMHD1c-dGTP/dATP, SAMHD1c-dGTP/dCTP, SAMHD1c-dGTP/dTTP, and SAMHD1c-dGTP/dUTP. A, superposition of SAMHD1c structures for different dGTP/dNTPs. The four monomeric subunits (a–d) in each SAMHD1c tetramer are displayed in a ribbon representation and are colored in light blue, light green, light gray, and light yellow, respectively. The tetramer is formed by two dimers, composed of subunits a and d and subunits b and c. The dGTP, dATP, dCTP, dTTP, and dUTP molecules are shown in a ball-and-stick representation and colored in blue, red, magenta, purple, and black, respectively. The different dNTPs occupying the allosteric site between subunits a, c, and d (top right panel) and the catalytic site of subunit c (bottom right panel) are also shown in a magnified view in the inset. B, stereoview of detailed hydrogen bond interactions in the allosteric sites for the superimposed structures of SAMHD1c with different dNTPs. As in A, the overall protein structure is shown in a ribbon representation and colored in light blue, light green, light gray, and light yellow for subunits a, b, c, and d, respectively. The dGTP molecules are shown in a stick representation with the carbon atoms colored in dark blue and other atoms in blue (nitrogen), red (oxygen), and orange (phosphor). Interacting residues are also shown in a stick representation with carbon atoms colored in the corresponding chain color and oxygen and nitrogen atoms colored in red and blue, respectively. dNTP molecules in the secondary allosteric site are also displayed in a stick representation, with the carbon atoms colored in red, magenta, purple, and black for dATP, dCTP, dTTP, and dUTP, respectively, and other atoms in blue (nitrogen), red (oxygen), and orange (phosphor). Conserved and non-conserved hydrogen bonds are represented by dashed green and cyan lines, respectively. C, hydrogen bonds between the base moiety of dTTP and dATP and the protein in the secondary allosteric site. The distances between the O4 of dTTP and the ND2 of Asn-358 and between N6 of dATP and the ND2 of Asn-358 are ∼3.31 and 3.24 Å, respectively. D, model illustrating why dCTP is not found in the second allosteric site. The dCTP molecule was modeled, based on the electron density of dTTP in the SAMHD1c-dGTP/dTTP complex. The distance between N4 of dCTP and OD1 of Asn-358 is ∼3.82 Å, too far for optimal hydrogen bonding. Note that OD1 of Asn-358 forms a hydrogen with to Arg-372, with distances ranging from 2.89 to 3.21 Å in all structures. E, stereoview displaying detailed hydrogen-bond interactions between the different dNTPs and residues in the catalytic site. Coloring is as in B.
Although the overall protein structures are very similar, different dNTP molecules are located in the secondary allosteric and catalytic sites (Fig. 1A and Table 2). In our SAMHD1c-dGTP/dATP structure, the secondary allosteric and catalytic sites are exclusively occupied by dATP and dGTP, respectively. This is distinct from the structure of SAMHD1c′-dGTP/dATP (PDB entry 4MZ7), in which both sites were filled by either dGTP or dATP within the same tetramer (27). In SAMHD1c-dGTP/dCTP structure, on the other hand, the secondary allosteric and catalytic sites are occupied by dGTP and dCTP, respectively. Therefore, it appears that dCTP can readily bind into the catalytic site but less effectively at the secondary allosteric site, different from dATP. In the other two structures, which were crystallized in the presence of dGTP/dTTP or dGTP/dUTP, both dTTP and dUTP nucleotides can occupy the secondary allosteric pocket and the catalytic site.
TABLE 2.
dNTPs in the allosteric and catalytic sites in different SAMHD1c-dNTP/dNTP crystal structures
Atoms of nucleosides and amino acids within hydrogen bonding distance (less than 3.3 Å) are listed.
| dGTP/dATP | dGTP/dCTP | dGTP/dTTP | dGTP/dUTP | |
|---|---|---|---|---|
| Primary allosteric site | dGTP | dGTP | dGTP | dGTP |
| Secondary allosteric site | dATP | dGTP | dTTP | dUTP |
| O3′-N (Asn-119) | O3′-N (Asn-119) | O3′-N (Asn-119) | O3′-N (Asn-119) | |
| O3′-O (Val-156) | O3′-O (Val-156) | O3′-O (Val-156) | O3′-O (Val-156) | |
| O4′-NH1 (Arg-333) | O4′-NH1 (Arg-333) | O4′-NH1 (Arg-333) | O4′-NH1 (Arg-333) | |
| N6-OD1 (Asn-358) | O6-ND2 (Asn-358) | O4-ND2 (Asn-358) | O4-ND2 (Asn-358) | |
| N2-OD2 (Asp-330) | O2-ND2 (Asn-119) | |||
| Catalytic site | dGTP | dCTP | dTTP | dUTP |
| O3′-NE2 (Gln-149) | O3′-NE2 (Gln-149) | O3′-NE2 (Gln-149) | O3′-NE2 (Gln-149) | |
| N2′-O (Leu-150) | ||||
| O4′-NH2 (Arg-164) | O4′-NH2 (Arg-164) | O4′-NH2 (Arg-164) | O4′NH2 (Arg-164) | |
| N1-O (Tyr-374) | ||||
| O6-NE2 (Gln-375) | N4-QE1 (Gln-375) | O4-NE2 (Gln-375) | O4-NE2 (Gln-375) |
The primary allosteric binding pocket in all SAMHD1c-dNTP structures is always occupied by dGTP, exhibiting identical hydrogen bonding interactions between the Watson-Crick and the Hoogsteen (N7) sites of the guanine base and residues Asp-137, Gln-142, and Arg-145, as reported previously (Fig. 1B).
The structural basis for the promiscuity within the secondary allosteric binding pocket is illustrated in Fig. 1B. Because all of the subunits are virtually identical, only the interactions at the secondary allosteric pocket located at the interface between subunits a, c, and d are described, along with those for the dNTPs in the catalytic site of subunit c. Generally, the hydrogen bonding networks between the protein and the nucleotide in the secondary allosteric pocket are very similar to those described earlier for the SAMHD1c-dGTP/dGTP complex (PDB entry 4BZB). In the latter structure, we showed that the terminal guanidinium group (NH1) of Arg-333 (subunit c) hydrogen-bonds with the C4′ hydroxyl group (O4′) of the dGTP sugar moiety. Changing the Arg to Ala abolished dNTPase activity and HIV-1 restriction (25). In all of the current SAMHD1c-dGTP/dNTP structures, the same hydrogen bonding is observed, as well as polar and charge interactions around the triphosphate-Mg2+-triphosphate. In contrast, the interactions with the base moiety are distinct, given the different bases in the individual dNTP molecules. In the SAMHD1c-dGTP/dATP structure, the N6 atom of the adenine base is engaged in hydrogen bonding with the amino group of the Asn-358 side chain (Fig. 1C and Table 2). Similarly, in the SAMHD1c-dGTP/dTTP and SAMHD1c-dGTP/dUTP structures, the O4 atom of the thymine base or the uracil base forms a hydrogen bond with the amino group of the Asn-358 side chain. The absence of dCTP at the secondary allosteric site in SAMHD1c-dGTP/dCTP structure is probably due to the inability of dCTP to form a hydrogen bond with Asn-358 (Fig. 1D) because OD1 of Asn-358 is involved in an interaction with the Nϵ of the Arg-372 side chain (2.9 Å). Indeed, the same orientation of the Asn-358 side chain is seen in all other crystal structures. Comparison of the present structures with the SAMHD1c-dGTP/dGTP structure shows that the second hydrogen bond with the carboxyl group of the Asp-330 side chain is no longer observed in the SAMHD1c-dGTP/dATP, SAMHD1c-dGTP/dTTP, and SAMHD1c-dGTP/dUTP structures. Also, as discussed above, because dGTP occupies the secondary allosteric site in the SAMHD1c-dGTP/dCTP structure, all of the interactions in this site are completely identical to those in the SAMHD1c-dGTP/dGTP structure (PDB entry 4BZB). In addition, a unique hydrogen bond is present in the SAMHD1c-dGTP/dUTP structure between the amino group of the Asn-119 side chain and the O2 atom of the uracil base, whereas the N3 of the adenine ring of dATP or the N2 of the guanine ring of dGTP is not within hydrogen bonding distance. Overall, although different dNTP molecules are bound in the secondary allosteric site, the resulting hydrogen bonding networks exhibit only slight variations, keeping the conformation in the different complexes essentially unchanged.
When different dNTPs are bound in the catalytic site, small but discernable differences can be seen around the binding pockets. Hydrogen bonds between the sugar moiety, namely between the C3′ and C4′ hydroxyl groups and the amino group of the Gln-149 side chain (NE2) and the guanidinium group (NH2) of Arg-164 are conserved in all structures. The amino group of the Gln-375 side chain hydrogen-bonds to all dNTP bases, with the N4 atom of cytosine, the O4 atom of thymine, and the O4 atom of uracil as acceptors, positioning the different dNTP molecules in the active site for productive catalysis (Fig. 1E). Therefore, the Gln-375 side chain can be repositioned to permit SAMHD1c to accommodate all four dNTPs as substrates. In contrast, hydrogen bonding between the backbone carbonyl oxygen of Leu-150 and the base was only observed for guanine bases (Table 2). Similar to the conformational invariance at the secondary allosteric site, for all bound dNTPs in the catalytic site, the triphosphate group interacts with the same basic residues (Arg-164, Arg-206, Lys-312, and Arg-366) as described previously for the SAMHD1c-dGTP/dGTP structure (25). All of the above data suggest that the primary allosteric site is specific for a guanine base, whereas the secondary allosteric site and the catalytic site can interact with a variety of different dNTPs. The absence of dATP in the catalytic site is surprising, because biochemical studies showed that dATP can be hydrolyzed by SAMHD1. A possible explanation may be that the affinity for dATP in the active site is much lower than that for dGTP because fewer hydrogen bond donor/acceptor groups are available in dATP compared with dGTP.
SAMHD1 Tetramerization Is Induced by GTP with dNTP
In addition to dNTPs, a recent report suggested that GTP can also activate SAMHD1, without being hydrolyzed by the enzyme (28). In order to investigate whether GTP can function as an allosteric activator and induce SAMHD1 tetramerization, we determined the quaternary state of SAMHD1fl (molecular mass = 76 kDa) in the presence of GTP and dATP by SEC-MALS (Fig. 2A). Whereas the mixture of GTP and dATP showed SAMHD1fl tetramerization (average molecular mass of the first peak = 320 kDa), GTP or dATP alone did not induce SAMHD1fl tetramerization (see Fig. 2, C and D). For mixtures containing 1 mm GTP and increasing concentrations of different dNTPs, similar results were obtained, suggesting that GTP is indeed an allosteric activator (Fig. 2, B and C; we used the catalytically inactive mutant H210A to prevent hydrolysis of dNTP in the reaction mixture). Among all dNTPs tested, dATP was most potent in inducing tetramerization in combination with GTP, whereas dCTP was least effective, consistent with our crystallographic data, which showed that dCTP does not bind to the secondary allosteric site (Table 2). In summary, dATP, dCTP, dTTP, and dUTP are able to induce SAMHD1 tetramerization in the presence of GTP, albeit with different potencies.
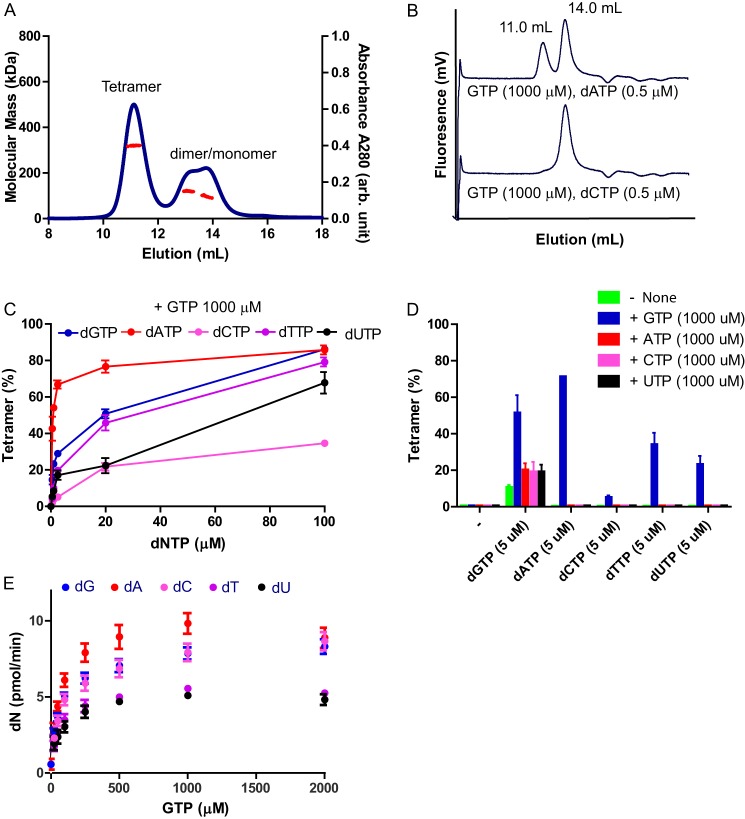
FIGURE 2.
SAMHD1 forms a tetramer with GTP and dNTPs and exhibits enzymatic activity. A, SEC-MALS of SAMHD1fl WT. The protein (25 μm) was preincubated with a mixture of GTP (250 μm) and dATP (250 μm). Molecular masses (red circles) of eluting species are shown across the A280 profile (blue circles). B, analytical gel filtration column chromatography of SAMHD1fl H210A (0.5 μm) with a mixture of GTP and dATP or dCTP. The elution volumes of SAMHD1 tetramer and dimer/monomer, based on SEC MALS experiments, are indicated. The catalytically inactive mutant was used for the assay to prevent hydrolysis of dNTP. C, SAMHD1fl H210A protein was preincubated with a mixture of GTP (at 1000 μm) and each dNTP as indicated (at 0, 0.5, 1.0, 2.5, 20, or 100 μm). The mixtures were separated by size exclusion column chromatography, and the data were analyzed as in B. D, SAMHD1fl H210A protein was preincubated with a mixture of one dNTP (at 5 μm) and/or one NTP (at 1000 μm), as indicated. The mixtures were analyzed as described in B. E, dNTPase activities of WT SAMHD1fl protein was measured using a mixture of dNTPs (each at 5 μm) and increasing concentrations of GTP (0, 25, 50, 100, 250, 500, 1000, and 2000 μm). Each experiment was performed in triplicate. Error bars, S.D. for the individual experiments.
We also tested the ability of other NTPs (ATP, CTP, and UTP) to induce SAMHD1fl tetramerization (Fig. 2D). They enhanced tetramerization in the presence of dGTP by 2-fold, when compared with the reaction mixture containing only dGTP. Here too, GTP was most potent in inducing tetramerization with any dNTP. The SAMHD1fl H206R/D207D mutant exhibited a similar preference with respect to dNTPs and NTPs (data not shown). In addition, in the presence of various dNTPs, GTP increased the dNTPase activity of SAMHD1fl in a dose-dependent manner (Fig. 2E).
Structural Basis of SAMHD1 Activation by GTP
To structurally assess GTP-induced SAMHD1 tetramerization and activation, we co-crystallized SAMHD1c in the presence of GTP and dATP, which, among all of the dNTPs, was the most effective co-stimulator of tetramer formation with GTP (Fig. 2C) and the best substrate (Fig. 2E). The tetrameric SAMHD1c-GTP/dATP structure (Fig. 3, A–C) is very similar to all other dGTP/dNTP complexed SAMHD1c structures (pairwise heavy atom root mean square deviation values ranging between 0.25 and 0.39 Å.
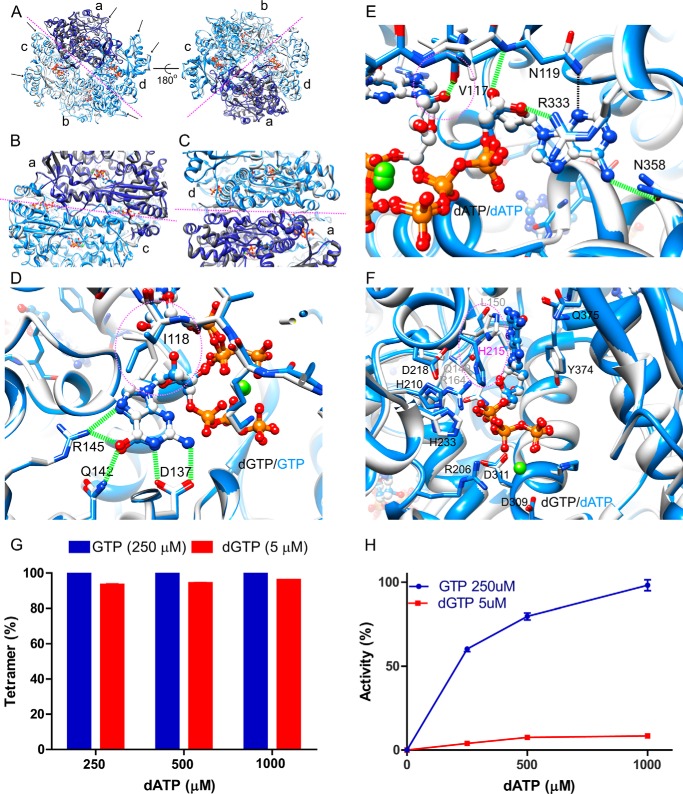
FIGURE 3.
Structural comparison between SAMHD1c-dGTP/dATP and SAMHD1c-GTP/dATP. A–C, superposition of the SAMHD1c-dGTP/dATP (white with subunit a colored dark gray) and SAMHD1c-GTP/dATP structures (blue with subunit a colored dark blue). Overall comparison between the two tetramers is shown in A; the interface between the two dimers is displayed in B; and the interface between the two monomers in a dimer, formed by subunits a and d, is shown in C. The magenta dashed lines indicate the interfaces between the two dimers in the tetramer (A and B) or the two monomers in a dimer C. D and E, superposition of dGTP (carbon atoms in light gray) and GTP (carbon atoms in blue) in the primary allosteric site (D) and in the secondary allosteric site (E) in the structures of SAMHD1c-dGTP/dATP (carbon atoms in light gray) and SAMHD1c-GTP/dATP (carbon atoms in blue). The rotamer change of Ile-118 is illustrated by dashed magenta circles. F, superposition of the catalytic site region in the SAMHD1c-dGTP/dATP and SAMHD1c-GTP/dATP structures. Carbon atoms of dGTP and of dATP are colored white and blue, respectively. Note that the His-215 side chain is in a different orientation in the SAMHD1c-dGTP/dATP (light gray) and SAMHD1-GTP/dATP (blue) structures. G, tetramer formation of the SAMHD1fl H206R/D2077N mutant. The mutant protein was incubated with either a mixture of GTP (250 μm) or dGTP (5 μm) and increasing concentrations of dATP (250–1000 μm). H, dATPase activity of WT SAMHD1fl. Enzymatic activity was measured in the presence of either GTP (250 μm) or dGTP (5 μm) and for increasing concentrations of dATP (250–1000 μm). dATPase activity was normalized relative to that obtained with 250 μm GTP and 1 mm dATP.
Four and eight molecules of GTP and dATP, respectively, are bound in the tetramer, with GTP in the primary allosteric sites. dATP is bound in the secondary allosteric and catalytic sites. The interactions between GTP and the protein are very similar to those observed with dGTP (Figs. 1B and 3D). The guanine base interacts with residues Asp-137, Gln-142, and Arg-145, and five hydrogen bonds to the guanine base are formed. Interestingly, the 2′-hydroxyl group of GTP in the SAMHD1c-GTP/dATP structure is well accommodated in the binding pocket. In the presence of GTP, Ile-118, which is located between the primary and secondary allosteric sites, exhibits a distinct conformation, adopting a different rotamer that permits the additional C2′ hydroxyl group of the GTP ribose to fit into the site (Fig. 3D). In addition, Val-117 also plays an important role, with its backbone carbonyl group accepting a hydrogen bond from the C2′ hydroxyl group of the GTP ribose (Fig. 3E). Note that the dATP molecule found in the secondary allosteric pocket of the SAMHD1c-GTP/dATP structure exhibits the same hydrogen bonding network as seen in the SAMHD1c-dGTP/dATP structure (Fig. 3E).
Comparison of the catalytic site, occupied by dGTP in the SAMHD1c-dGTP/dATP structure and by dATP in the SAMHD1c-GTP/dATP structure, also revealed only minor differences (Fig. 3F). All residues important for catalysis (i.e. Asp-218, His-210, and His-233) exhibit similar conformations, and amino acids that interact with dNTPs, such as Gln-149, Leu-150, Arg-164, Arg-206, Asp-309, Asp-311, Tyr-374, and Gln-375, are also conformationally invariant. Only the His-215 side chain is positioned differently: in the SAMHD1c-dGTP/dATP structure, it flanks the guanine base of dGTP, whereas in the SAMHD1c-GTP/dATP structure, it is flipped and points to the nearest phosphate group of dATP. Interestingly, His-215 exhibits two different side chain conformations in the structures of SAMHD1c′-dGTP/dATP (PDB entry 4MZJ) (27). Whether this side chain flip has consequences for catalysis is not clear at this stage, although SAMHD1 per tetramer is a much more active enzyme in the presence of GTP/dATP, compared with dGTP/dATP (Fig. 3, G and H).
To evaluate whether structurally important residues in the allosteric and catalytic sites are important for enzymatic activity, alanine substitution mutants were generated and interrogated for GTP/dATP-induced tetramerization (Fig. 4A) and catalytic activity (Fig. 4B). Mutations in the primary allosteric site (D137A, Q142A, and R145A) completely abolished tetramerization and catalytic activity. In contrast, single residue changes in the secondary allosteric site (D330A and N358A) did not significantly affect tetramerization or catalysis. Five residues in the catalytic site (Gln-149, His-201, His-215, His-233, and Gln-375) are essential for dNTPase activity but not for nucleoside triphosphate-dependent tetramerization (Fig. 4, A and B).
FIGURE 4.
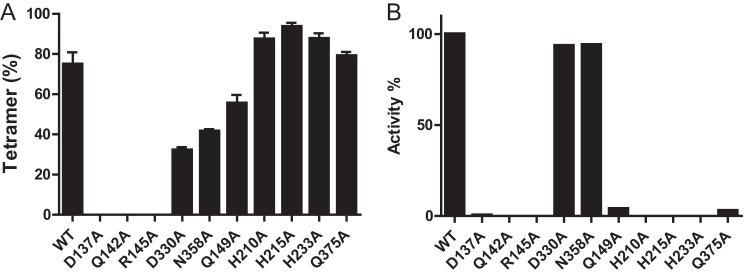
Mutational analysis of allosteric sites and catalytic site residues. A, tetramer formation was determined by size exclusion column chromatography for WT SAMHD1fl and mutants in the presence of 250 μm GTP and 1 mm dATP as described in the legend to Fig. 2, B and C. B, dATPase activities were measured for SAMHD1fl mutants in the presence of 250 μm GTP and 1 mm dATP and normalized with respect to WT. Each experiment was performed in triplicate, and error bars reflect the S.D. in the measurement.
DISCUSSION
SAMHD1 is a dGTP/dNTP- or GTP/dNTP-activated dNTPase that converts dNTPs into deoxynucleosides and triphosphates. It plays an important role in innate immunity, targeting retroviral infection by limiting the cellular dNTP pools, essential for reverse transcription (42–45). Here we elucidated the structural basis for the broad activity toward different dNTP substrates through a series of different enzyme-dNTP/NTP crystal structures (i.e. SAMHD1c-dGTP/dATP, SAMHD1c-dGTP/dCTP, SAMHD1c-dGTP/dTTP, SAMHD1c-dGTP/dUTP, and SAMHD1c-GTP/dATP). These structures reveal that the primary allosteric site is exclusively binding dGTP or GTP, whereas the secondary allosteric and the catalytic sites are promiscuous and can accommodate different dNTPs.
Unique structural features of the two adjacent allosteric nucleotide binding pockets set up distinct ligand binding specificities. Residues Asp-137, Gln-142, and Arg-145 in the primary allosteric site interact with the guanine base via a total of five hydrogen bonds (Figs. 1B and 3D). The adenine base, the other purine, cannot form the same type of hydrogen bonding, because it lacks an NH2 group at the C2 position, and C=O of guanine is replaced with NH2 in the adenine. Any single alanine substitution of these three residues is sufficient to abolish ligand-induced tetramerization and catalytic activity, suggesting a strict conformational requirement for the allosteric site (Fig. 4, A and B). By contrast, in the secondary allosteric site, only one or two hydrogen bonds are formed between the protein and the base of the dNTP (Figs. 1B and 3E), allowing for a variety of dNTPs to be accommodated. Therefore, tetramerization and catalysis are less affected by single alanine mutation in this pocket (Fig. 4, A and B). The ability of SAMHD1 to bind a GTP molecule in the primary allosteric site is facilitated by local plasticity within the first β1-β2 hairpin (residues 113–129), permitting a critical rotameric change of residue Ile-118 that makes room for the additional C2′ hydroxyl of the ribose in GTP (Fig. 3, D and E). In the secondary allosteric site, the phenyl ring of Phe-157 will not permit any C2′ hydroxyl to fit in, thus limiting the sugars to 2-deoxyriboses (25). Despite these unique features around the base and sugar moieties, the triphosphate groups of the two bound nucleoside triphosphates share a Mg2+ ion and engage in multiple charge and hydrogen-bonding interactions with the surrounding basic residues. Other dNTPases, including EF1143 from Enterococcus faecalis and TT1383 from Thermus thermophiles, require two nucleotides as allosteric activators, suggesting similar molecular mechanisms of enzyme activation (46–49).
Because the cellular level of GTP is ∼1000-fold higher than that of dGTP and it is not hydrolyzed by SAMHD1, it has been suggested that GTP is the physiologically relevant, primary activator of SAMHD1 (28–30). NTP levels are relatively constant in the millimolar range throughout the cell cycle, whereas concentrations of dNTPs in cells are tightly regulated and range from 20 nm in resting cells to 15 μm in actively dividing cells (50). Analytical gel filtration analyses suggested that SAMHD1 can form a stable tetramer when the protein is primed with GTP at 1 mm concentration, even for dNTP concentrations below 1 μm (Fig. 2C). Further, dATP is the most potent cofactor for inducing SAMHD1 tetramerization when it is paired with GTP (Fig. 2, B and C), suggesting that these two molecules are likely to be primary activators of SAMHD1 in the cell. Because the concentration of GTP is relatively stable, the quaternary states of SAMHD1 are probably modulated by dATP. Consistent with this notion, SAMHD1-GTP/dATP proved to be by far the most active enzyme when protein-ligand mixtures were compared (Fig. 3, G and H). dATP also functions as an allosteric feedback inhibitor of ribonucleotide reductase, which converts all four ribonucleotide diphosphates to deoxyribonucleotide diphosphates prior to conversion to dNTPs (51). The cellular level of ribonucleotide reductase and its enzymatic activity is tightly regulated during the cell cycle, safeguarding dNTP amounts at appropriate levels for specific cell states (51, 52). A recent study by Franzolin et al. demonstrated that SAMHD1 is involved in the control of dNTP concentrations of cycling cells (53). Therefore, our observation of dATP being the most potent allosteric activator of SAMDH1 suggests that dATP plays a primary role in regulating cellular levels of dNTPs, acting as an activator of SAMHD1 and as an inhibitor of ribonucleotide reductase.
Acknowledgments
We thank Dr. Teresa Brosenitsch for editorial help and Dr. Doowon Lee for technical help with crystallization and data collection. We also thank the staff of the SER-CAT facility sector 22 beam line of the Advance Photon Source at Argonne National Laboratory (Chicago, IL) for help during data collection.
This work was supported, in whole or in part, by National Institutes of Health Grant P50GM82251 (to A. M. G. and J. A.). This work was also supported in part by start-up funds from the University of Pittsburgh School of Medicine (to J. A.).
- SAMHD1
- sterile α motif and histidine-aspartate domain-containing protein 1
- SAMHD1c
- SAMHD1 catalytic core (residues 113–626)
- SAMHD1fl
- SAMHD1 full-length (residues 1–626)
- SAMHD1c′
- SAMHD1 residues 109–626
- SEC-MALS
- analytical size exclusion column chromatography and multiangle light scattering
- PDB
- Protein Data Bank.
REFERENCES
- 1. Berger A., Sommer A. F., Zwarg J., Hamdorf M., Welzel K., Esly N., Panitz S., Reuter A., Ramos I., Jatiani A., Mulder L. C., Fernandez-Sesma A., Rutsch F., Simon V., König R., Flory E. (2011) SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutieres syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 7, e1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Goldstone D. C., Ennis-Adeniran V., Hedden J. J., Groom H. C., Rice G. I., Christodoulou E., Walker P. A., Kelly G., Haire L. F., Yap M. W., de Carvalho L. P., Stoye J. P., Crow Y. J., Taylor I. A., Webb M. (2011) HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382 [DOI] [PubMed] [Google Scholar]
- 3. Hrecka K., Hao C., Gierszewska M., Swanson S. K., Kesik-Brodacka M., Srivastava S., Florens L., Washburn M. P., Skowronski J. (2011) Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Laguette N., Sobhian B., Casartelli N., Ringeard M., Chable-Bessia C., Ségéral E., Yatim A., Emiliani S., Schwartz O., Benkirane M. (2011) SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Powell R. D., Holland P. J., Hollis T., Perrino F. W. (2011) Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J. Biol. Chem. 286, 43596–43600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baldauf H. M., Pan X., Erikson E., Schmidt S., Daddacha W., Burggraf M., Schenkova K., Ambiel I., Wabnitz G., Gramberg T., Panitz S., Flory E., Landau N. R., Sertel S., Rutsch F., Lasitschka F., Kim B., König R., Fackler O. T., Keppler O. T. (2012) SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat. Med. 18, 1682–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Descours B., Cribier A., Chable-Bessia C., Ayinde D., Rice G., Crow Y., Yatim A., Schwartz O., Laguette N., Benkirane M. (2012) SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4(+) T-cells. Retrovirology 9, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lahouassa H., Daddacha W., Hofmann H., Ayinde D., Logue E. C., Dragin L., Bloch N., Maudet C., Bertrand M., Gramberg T., Pancino G., Priet S., Canard B., Laguette N., Benkirane M., Transy C., Landau N. R., Kim B., Margottin-Goguet F. (2012) SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13, 223–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim B., Nguyen L. A., Daddacha W., Hollenbaugh J. A. (2012) Tight interplay among SAMHD1 protein level, cellular dNTP levels, and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. J. Biol. Chem. 287, 21570–21574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. White T. E., Brandariz-Nuñez A., Valle-Casuso J. C., Amie S., Nguyen L., Kim B., Brojatsch J., Diaz-Griffero F. (2013) Contribution of SAM and HD domains to retroviral restriction mediated by human SAMHD1. Virology 436, 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hollenbaugh J. A., Gee P., Baker J., Daly M. B., Amie S. M., Tate J., Kasai N., Kanemura Y., Kim D. H., Ward B. M., Koyanagi Y., Kim B. (2013) Host factor SAMHD1 restricts DNA viruses in non-dividing myeloid cells. PLoS Pathog. 9, e1003481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim E. T., White T. E., Brandariz-Núñez A., Diaz-Griffero F., Weitzman M. D. (2013) SAMHD1 restricts herpes simplex virus 1 in macrophages by limiting DNA replication. J. Virol. 87, 12949–12956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Beloglazova N., Flick R., Tchigvintsev A., Brown G., Popovic A., Nocek B., Yakunin A. F. (2013) Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutieres syndrome and HIV-1 restriction. J. Biol. Chem. 288, 8101–8110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ryoo J., Choi J., Oh C., Kim S., Seo M., Kim S. Y., Seo D., Kim J., White T. E., Brandariz-Nuñez A., Diaz-Griffero F., Yun C. H., Hollenbaugh J. A., Kim B., Baek D., Ahn K. (2014) The ribonuclease activity of SAMHD1 is required for HIV-1 restriction. Nat. Med. 20, 936–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cribier A., Descours B., Valadão A. L., Laguette N., Benkirane M. (2013) Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep. 3, 1036–1043 [DOI] [PubMed] [Google Scholar]
- 16. Welbourn S., Dutta S. M., Semmes O. J., Strebel K. (2013) Restriction of virus infection but not catalytic dNTPase activity is regulated by phosphorylation of SAMHD1. J. Virol. 87, 11516–11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. White T. E., Brandariz-Nuñez A., Valle-Casuso J. C., Amie S., Nguyen L. A., Kim B., Tuzova M., Diaz-Griffero F. (2013) The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe 13, 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. St Gelais C., de Silva S., Hach J. C., White T. E., Diaz-Griffero F., Yount J. S., Wu L. (2014) Identification of cellular proteins interacting with the retroviral restriction factor SAMHD1. J. Virol. 88, 5834–5844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leshinsky-Silver E., Malinger G., Ben-Sira L., Kidron D., Cohen S., Inbar S., Bezaleli T., Levine A., Vinkler C., Lev D., Lerman-Sagie T. (2011) A large homozygous deletion in the SAMHD1 gene causes atypical Aicardi-Goutieres syndrome associated with mtDNA deletions. Eur. J. Hum. Genet. 19, 287–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ramantani G., Häusler M., Niggemann P., Wessling B., Guttmann H., Mull M., Tenbrock K., Lee-Kirsch M. A. (2011) Aicardi-Goutieres syndrome and systemic lupus erythematosus (SLE) in a 12-year-old boy with SAMHD1 mutations. J. Child Neurol. 26, 1425–1428 [DOI] [PubMed] [Google Scholar]
- 21. Ramesh V., Bernardi B., Stafa A., Garone C., Franzoni E., Abinun M., Mitchell P., Mitra D., Friswell M., Nelson J., Shalev S. A., Rice G. I., Gornall H., Szynkiewicz M., Aymard F., Ganesan V., Prendiville J., Livingston J. H., Crow Y. J. (2010) Intracerebral large artery disease in Aicardi-Goutieres syndrome implicates SAMHD1 in vascular homeostasis. Dev. Med. Child Neurol. 52, 725–732 [DOI] [PubMed] [Google Scholar]
- 22. Rice G. I., Bond J., Asipu A., Brunette R. L., Manfield I. W., Carr I. M., Fuller J. C., Jackson R. M., Lamb T., Briggs T. A., Ali M., Gornall H., Couthard L. R., Aeby A., Attard-Montalto S. P., Bertini E., Bodemer C., Brockmann K., Brueton L. A., Corry P. C., Desguerre I., Fazzi E., Cazorla A. G., Gener B., Hamel B. C., Heiberg A., Hunter M., van der Knaap M. S., Kumar R., Lagae L., Landrieu P. G., Lourenco C. M., Marom D., McDermott M. F., van der Merwe W., Orcesi S., Prendiville J. S., Rasmussen M., Shalev S. A., Soler D. M., Shinawi M., Spiegel R., Tan T. Y., Vanderver A., Wakeling E. L., Wassmer E., Whittaker E., Lebon P., Stetson D. B., Bonthron D. T., Crow Y. J. (2009) Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat. Genet. 41, 829–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thiele H., du Moulin M., Barczyk K., George C., Schwindt W., Nürnberg G., Frosch M., Kurlemann G., Roth J., Nürnberg P., Rutsch F. (2010) Cerebral arterial stenoses and stroke: novel features of Aicardi-Goutieres syndrome caused by the Arg164X mutation in SAMHD1 are associated with altered cytokine expression. Hum. Mutat. 31, E1836–E1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xin B., Jones S., Puffenberger E. G., Hinze C., Bright A., Tan H., Zhou A., Wu G., Vargus-Adams J., Agamanolis D., Wang H. (2011) Homozygous mutation in SAMHD1 gene causes cerebral vasculopathy and early onset stroke. Proc. Natl. Acad. Sci. U.S.A. 108, 5372–5377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ji X., Wu Y., Yan J., Mehrens J., Yang H., DeLucia M., Hao C., Gronenborn A. M., Skowronski J., Ahn J., Xiong Y. (2013) Mechanism of allosteric activation of SAMHD1 by dGTP. Nat. Struct. Mol. Biol. 20, 1304–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yan J., Kaur S., DeLucia M., Hao C., Mehrens J., Wang C., Golczak M., Palczewski K., Gronenborn A. M., Ahn J., Skowronski J. (2013) Tetramerization of SAMHD1 is required for biological activity and inhibition of HIV infection. J. Biol. Chem. 288, 10406–10417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhu C., Gao W., Zhao K., Qin X., Zhang Y., Peng X., Zhang L., Dong Y., Zhang W., Li P., Wei W., Gong Y., Yu X. F. (2013) Structural insight into dGTP-dependent activation of tetrameric SAMHD1 deoxynucleoside triphosphate triphosphohydrolase. Nat. Commun. 4, 2722. [DOI] [PubMed] [Google Scholar]
- 28. Amie S. M., Bambara R. A., Kim B. (2013) GTP is the primary activator of the anti-HIV restriction factor SAMHD1. J. Biol. Chem. 288, 25001–25006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hansen E. C., Seamon K. J., Cravens S. L., Stivers J. T. (2014) GTP activator and dNTP substrates of HIV-1 restriction factor SAMHD1 generate a long-lived activated state. Proc. Natl. Acad. Sci. U.S.A. 111, E1843–E1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miazzi C., Ferraro P., Pontarin G., Rampazzo C., Reichard P., Bianchi V. (2014) Allosteric regulation of the human and mouse deoxyribonucleotide triphosphohydrolase sterile α-motif/histidine-aspartate domain-containing protein 1 (SAMHD1). J. Biol. Chem. 289, 18339–18346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pflugrath J. W. (1999) The finer things in x-ray diffraction data collection. Acta Crystallogr. D Biol. Crystallogr. 55, 1718–1725 [DOI] [PubMed] [Google Scholar]
- 32. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 33. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 35. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 36. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 37. Adams P. D., Gopal K., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Pai R. K., Read R. J., Romo T. D., Sacchettini J. C., Sauter N. K., Storoni L. C., Terwilliger T. C. (2004) Recent developments in the PHENIX software for automated crystallographic structure determination. J. Synchrotron Radiat. 11, 53–55 [DOI] [PubMed] [Google Scholar]
- 38. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 40. DeLano W. L. (2012) The PyMOL Molecular Graphics System, version 1.5.0.1, Schrödinger, LLC, New York [Google Scholar]
- 41. DeLucia M., Mehrens J., Wu Y., Ahn J. (2013) HIV-2 and SIVmac accessory virulence factor Vpx down-regulates SAMHD1 enzyme catalysis prior to proteasome-dependent degradation. J. Biol. Chem. 288, 19116–19126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Amie S. M., Noble E., Kim B. (2013) Intracellular nucleotide levels and the control of retroviral infections. Virology 436, 247–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pan X., Baldauf H. M., Keppler O. T., Fackler O. T. (2013) Restrictions to HIV-1 replication in resting CD4+ T lymphocytes. Cell Res. 23, 876–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sze A., Olagnier D., Lin R., van Grevenynghe J., Hiscott J. (2013) SAMHD1 host restriction factor: a link with innate immune sensing of retrovirus infection. J. Mol. Biol. 425, 4981–4994 [DOI] [PubMed] [Google Scholar]
- 45. Wu L. (2013) Cellular and biochemical mechanisms of the retroviral restriction factor SAMHD1. ISRN Biochem. 2013, 728392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vorontsov I. I., Minasov G., Kiryukhina O., Brunzelle J. S., Shuvalova L., Anderson W. F. (2011) Characterization of the deoxynucleotide triphosphate triphosphohydrolase (dNTPase) activity of the EF1143 protein from Enterococcus faecalis and crystal structure of the activator-substrate complex. J. Biol. Chem. 286, 33158–33166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vorontsov I. I., Wu Y., DeLucia M., Minasov G., Mehrens J., Shuvalova L., Anderson W. F., Ahn J. (2014) Mechanisms of allosteric activation and inhibition of the deoxyribonucleoside triphosphate triphosphohydrolase from Enterococcus faecalis. J. Biol. Chem. 289, 2815–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kondo N., Kuramitsu S., Masui R. (2004) Biochemical characterization of TT1383 from Thermus thermophilus identifies a novel dNTP triphosphohydrolase activity stimulated by dATP and dTTP. J. Biochem. 136, 221–231 [DOI] [PubMed] [Google Scholar]
- 49. Kondo N., Nakagawa N., Ebihara A., Chen L., Liu Z. J., Wang B. C., Yokoyama S., Kuramitsu S., Masui R. (2007) Structure of dNTP-inducible dNTP triphosphohydrolase: insight into broad specificity for dNTPs and triphosphohydrolase-type hydrolysis. Acta Crystallogr. D Biol. Crystallogr. 63, 230–239 [DOI] [PubMed] [Google Scholar]
- 50. Kennedy E. M., Gavegnano C., Nguyen L., Slater R., Lucas A., Fromentin E., Schinazi R. F., Kim B. (2010) Ribonucleoside triphosphates as substrate of human immunodeficiency virus type 1 reverse transcriptase in human macrophages. J. Biol. Chem. 285, 39380–39391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hofer A., Crona M., Logan D. T., Sjöberg B. M. (2012) DNA building blocks: keeping control of manufacture. Crit. Rev. Biochem. Mol. Biol. 47, 50–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nordlund P., Reichard P. (2006) Ribonucleotide reductases. Annu. Rev. Biochem. 75, 681–706 [DOI] [PubMed] [Google Scholar]
- 53. Franzolin E., Pontarin G., Rampazzo C., Miazzi C., Ferraro P., Palumbo E., Reichard P., Bianchi V. (2013) The deoxynucleotide triphosphohydrolase SAMHD1 is a major regulator of DNA precursor pools in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 110, 14272–14277 [DOI] [PMC free article] [PubMed] [Google Scholar]