Background: Translesion synthesis DNA polymerases insert nucleotides opposite bulky template lesions not tolerated by replicative DNA polymerases.
Results: Translesion synthesis bypass was studied in the context of the replisome using a purified system.
Conclusion: Only DNA polymerase IV, not DNA polymerase II, was capable of catalyzing replisome-mediated translesion synthesis.
Significance: Lesion bypass can occur readily at stalled replication forks.
Keywords: DNA Damage, DNA Polymerase, DNA Repair, DNA Replication, Genomic Instability
Abstract
A number of different enzymatic pathways have evolved to ensure that DNA replication can proceed past template base damage. These pathways include lesion skipping by the replisome, replication fork regression followed by either correction of the damage and origin-independent replication restart or homologous recombination-mediated restart of replication downstream of the lesion, and bypass of the damage by a translesion synthesis DNA polymerase. We report here that of two translesion synthesis polymerases tested, only DNA polymerase IV, not DNA polymerase II, could engage productively with the Escherichia coli replisome to bypass leading strand template damage, despite the fact that both enzymes are shown to be interacting with the replicase. Inactivation of the 3′ → 5′ proofreading exonuclease of DNA polymerase II did not enable bypass. Bypass by DNA polymerase IV required its ability to interact with the β clamp and act as a translesion polymerase but did not require its “little finger” domain, a secondary region of interaction with the β clamp. Bypass by DNA polymerase IV came at the expense of the inherent leading strand lesion skipping activity of the replisome, indicating that they are competing reactions.
Introduction
The Escherichia coli DNA replication machinery (replisome) is responsible for bidirectional DNA synthesis of the circular chromosome. The replisome is composed of the DNA polymerase (Pol)3 III holoenzyme (HE) that synthesizes the nascent leading and lagging strands, the primase (DnaG) that initiates synthesis of the Okazaki fragments on the lagging strand, and a 5′ → 3′ DNA helicase (DnaB) that unwinds the parental double-stranded DNA template (1).
Lesions blocking replication affect the leading and lagging strand polymerases differently. Blocks on the lagging strand template are thought to be bypassed by a repriming event during Okazaki fragment synthesis downstream of the lesion. This would leave a single-stranded DNA gap containing the lesion that can be repaired postreplicatively (2).
Lesions encountered on the leading strand template cause replisome stalling and lesion bypass may occur via different mechanisms: (a) dissociation (or perhaps enzymatic removal) of the replication machinery at the stalled fork followed by modulation of the fork structure, repair of the lesion, and subsequent replication restart via origin-independent reloading of the replisome (3, 4); (b) lesion skipping, where the replisome only stalls transiently at the lesion and then, via a primase-catalyzed leading strand priming event downstream, resumes coupled leading and lagging strand synthesis, jumping over the lesion (5), leaving both the lesion and a gap in the nascent leading strand behind as substrates for daughter strand gap repair (6); and (c) translesion synthesis (TLS) DNA polymerases could exchange with the blocked replicative polymerase to bypass the lesion, yielding fully replicated DNA that still contains the template damage. Nonmutagenic repair pathways, such as nucleotide excision repair, could then excise and repair the lesion postreplicatively.
E. coli has three TLS polymerases, DNA polymerases (Pols) II, IV (DinB) (7), and V (UmuD′2UmuC) (8), which have different roles in cellular damage repair and lesion bypass capabilities (9). TLS polymerases are error-prone enzymes, generally lacking proofreading 3′ → 5′ exonuclease activity (although Pol II does have one) and having an active site that accommodates bulky adduct-modified bases (10). Pol II and IV are both expressed during normal growth conditions, but neither enzyme appears to play a role in the generation of spontaneous chromosomal mutations (11). All three TLS polymerases are highly induced during the SOS response (9), suggesting their critical participation in a cellular regulatory mechanism to maintain genomic integrity and prevent excessive mutations. Furthermore, all three SOS-induced DNA polymerases have been shown to be involved in lesion-induced or targeted mutagenesis (12).
Unlike Pol V, Pol II and Pol IV are induced early in the SOS response, suggesting a more essential role in TLS bypass. There are 250 molecules of Pol IV per cell (13) and 50–75 molecules of Pol II, compared with the replicative polymerase, Pol III, with 10–20 molecules per cell (14). During the SOS response, Pol II levels increase to 350–1000 molecules per cell, whereas there is a 10-fold increase in Pol IV to 2500 molecules per cell (13), making Pol IV the most abundant polymerase in the cell and suggesting that it could be the default polymerase for bypassing a DNA lesion at stalled replication forks.
Pol IV overexpression results in a mutator phenotype (15–17), an increase in spontaneous −1 frameshifts and spontaneous base substitutions (13, 15), and a decrease in cell viability (16, 18). These observations suggest Pol IV, when present at elevated concentration, can compete with the Pol III HE. Supporting this view, Pol IV has been shown to sequester a moving replisome from the Pol III HE in vitro (19).
Most studies of TLS by Pol IV and II have utilized small oligonucleotide primer templates carrying a lesion on the template strand. These experiments demonstrated that Pol IV could bypass an abasic site tetrahydrofuran (THF) analog lesion, as well as one generated by nitrofurazone (20–22). Pol II was also able to bypass a THF lesion (23–25). On the other hand, Pol IV TLS across a cys-syn cyclopyrimidine dimer (CPD) lesion was inefficient (22).
Using a system that we have described previously, in which replication is reconstituted on templates containing a single lesion in the leading strand template (5, 27), we describe in this report how TLS polymerases interact with the replisome. We find a clear distinction between the bypass ability of Pol II and IV, with only Pol IV being capable of replisome-mediated TLS. Bypass required the ability of Pol IV to bind the β clamp and to accommodate bulky lesions on the template strand. Pol IV-catalyzed TLS came at the expense of leading strand lesion skipping, suggesting that it may be the preferred reaction at stalled replication forks.
EXPERIMENTAL PROCEDURES
DNA Templates
DNA templates containing a site-specific CPD or THF lesion were synthesized using M13-JY13 single-stranded (circular) DNA primed with either the CG212THF or CG224CPD (Table 1) oligonucleotides as described (5, 27). Small oligonucleotide primer templates were prepared by annealing [5′-32P]CG18 to a 1.5-fold excess of CG17THF, CG17CPD, or CG16 (for sequences see Table 1) at 88 °C for 3 min and slow cooling to 24 °C in a water bath. Substrates for PsiI digest tests were prepared in the same way using CG217 annealed to either CG218 or CG219THF. Oligonucleotides were 5′ end-labeled with [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs) and purified through G-50 mini-spin columns (GE Healthcare).
TABLE 1.
Oligonucleotide sequences
| Oligonucleotide | Sequence |
|---|---|
| CG18 | 5′-TTAGACTCCTCAATACGAAGTATG-3′ |
| CG16 | 5′-ACGCTGTCTGCTAACATACTTCGTATTGAGGAGTCTAA-3′ |
| CG17THF | 5′-ACGCTGTCTGXTAACATACTTCGTATTGAGGAGTCTAA-3′ |
| CG17CPD | 5′-ACGCTGTCTG(TT)AACATACTTCGTATTGAGGAGTCTAA-3′ |
| CG212THF | 5′-GGCAAAATCCCTXATAAATCAAAAGAAT-3′, where X = THF |
| CG217 | 5′-ATCTCGGGCTATTCTTTTGATTTATAAGGGATTTTGCCGATTTCGGAACC-3′ |
| CG218 | 5′-GGTTCCGAAATCGGCAAAATCCCTTATAAATCAAAAGAATAGCCCGAGAT-3′ |
| CG219THF | 5′-GGTTCCGAAATCGGCAAAATCCCTXATAAATCAAAAGAATAGCCCGAGAT-3′ |
| CG224CPD | 5′-GGCAAAATCCC(TT)ATAAATCAAAAGAAT-3′, where (TT) = CPD |
Replication Proteins
Replication proteins were purified as described previously (5). A Pol IV overexpression vector (pCG101) was prepared by inserting the dinB ORF into the BamHI and NdeI sites of pET-11a (Novagen). BL21(DE3)(pCG101) cells were grown in rich medium to an A600 of 0.5 and induced with 1 mm isopropyl β-d-thiogalactopyranoside for 2 h. Cells were harvested and lysed, and Pol IV was purified by sequential column chromatography on SP-Sepharose (GE Healthcare), Fast Flow-Phenyl-Sepharose (Pharmacia), and heparin-agarose (Sigma). Pol IV variant proteins Pol IV ΔC5, Pol IV F13V, and Pol IV L1 were gifts of Mike O'Donnell (Rockefeller University), Graham Walker (MIT), and Jérôme Wagner (l'Ecole de Biotechnologie de Strasbourg), respectively. Pol II and Pol II D155A/E157A were the gifts of Myron Goodman (University of Southern California).
TLS on Oligonucleotide Primer Templates
Reaction mixtures (5 μl) containing 50 mm HEPES-KOH (pH 8.0), 30 mm potassium glutamate, 20 nm primer template, 10 mm Mg(OAc)2, 10 mm DTT, 100 μg/ml BSA (NEB), 1 mm ATP, 4% (v/v) glycerol, 250 μm dNTPs, and the indicated concentrations of respective polymerases were incubated at 37 °C for 10 min. Reactions were quenched by the addition of EDTA, formamide, and NaOH to 50 mm, 80%, and 5 mm, respectively, heat-denatured, and analyzed by electrophoresis at 38 W for 40 min through a denaturing, 7 m urea, 20% polyacrylamide gel (19:1, acrylamide:bisacrylamide) using 100 mm Tris borate (pH 8.3), 2 mm EDTA as the electrophoresis buffer. The gel was fixed by soaking in 10% methanol, 7% HOAc, 5% glycerol, dried, exposed to a PhosphorImager screen, and then autoradiographed. The amount of extended primer and unreplicated product were quantified using ImageGauge software (Fuji).
Replication Reactions
Gyrase replication reactions were incubated at 37 °C for 8 min as described previously (28) except that the HEPES-KOH concentration was reduced to 50 mm, and 75 mm potassium glutamate was added to the reaction mixture. Radiolabel was either [α-32P]dATP or [α-32P]dCTP at 4000 cpm/pmol as noted. DNA products were separated by electrophoreses through 0.6% denaturing alkaline agarose gels.
EcoRI replication reactions were as described previously (5) with the following modifications. Reactions were initiated at standard nucleotide concentrations. Elevated nucleotide concentrations and/or Pol IV and Pol II as indicated were added 1 min after EcoRI addition. [α-32P]dCTP was used for standard EcoRI reaction conditions, whereas [α-32P]dATP was used for the pulse-chase reactions. DNA products were digested with PvuI at 37 °C for 12 min after the reactions were quenched.
PsiI Digestion of Duplex Oligonucleotides
Oligonucleotides that matched the sequence of the plasmid surrounding the PsiI restriction digest site were annealed as described above. Reaction mixtures (5 μl) containing 50 mm HEPES-KOH (pH 8.0), 75 mm potassium glutamate, 20 nm duplex oligonucleotide, 10 mm Mg(OAc)2, 10 mm DTT, 100 μg/ml BSA (NEB), 1 mm ATP, 200 μm GTP, CTP, UTP, 40 μm dCTP and dTTP, and 750 μm dATP and dGTP, and the indicated amounts of PsiI were incubated for 12 min at 37 °C. Samples were quenched by the addition of EDTA, formamide, and NaOH were to 50 mm, 80%, and 5 mm, respectively, and the DNA products were processed as described above.
Nucleotide Resolution of Frameshifting and Stalling
To analyze the occurrence of frameshifting, gyrase reactions, quenched as above, were heated at 65 °C for 10 min. Mg(OAc)2 was then added back so that the free concentration of Mg2+ was 10 mm. Samples were then digested with DrdI (NEB) and Acc65I (NEB) in either the presence or absence of PsiI (NEB) for 12 min at 37 °C. Reactions were quenched with EDTA, samples were deproteinized by phenol-CHCl3 extraction, and the DNA was recovered by ethanol precipitation. DNA products were analyzed by electrophoresis at 33 W for 1.8 h through a denaturing 7 m urea, 10% polyacrylamide gel (19:1, acrylamide:bisacrylamide) together with a sequence ladder produced from an oligonucleotide having the same 5′ end as the nascent leading strand at the DrdI site and processed as above. To map the site of leading strand stalling, gyrase reactions were treated and analyzed as above except that they were not digested with Acc65I.
RESULTS
Both Pol II and Pol IV Catalyze TLS on Oligonucleotide Primer Templates
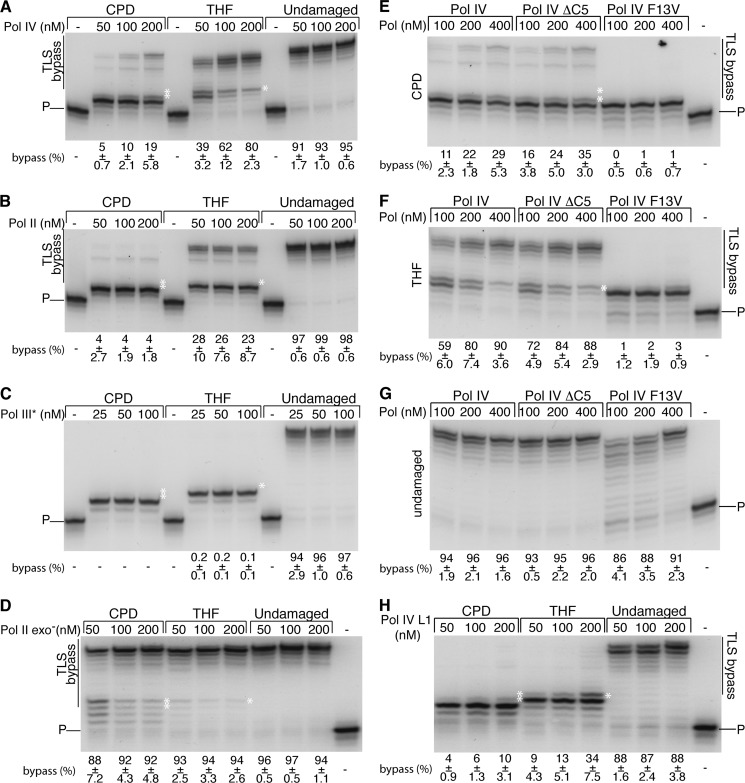
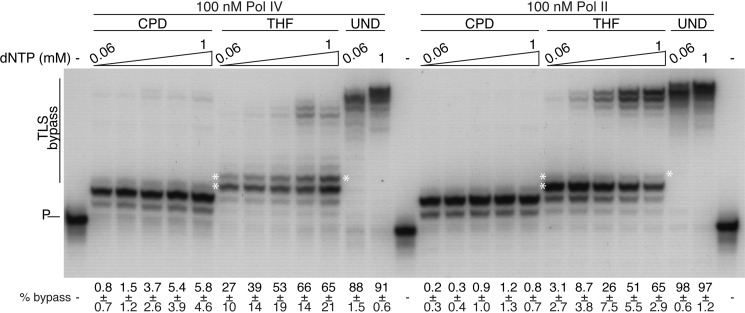
We confirmed the TLS activity of Pol II, Pol IV, and Pol III* (the Pol III HE lacking the β clamp subunit (29)) using oligonucleotide primer templates where the 3′ end of the 5′-[32P]primer was either 3 or 2 nt upstream of the template damage (either a THF or a CPD, respectively) followed by 10 nt of template downstream of the damage. Pol IV and II were both capable of bypassing a THF lesion, whereas Pol IV could also clearly bypass a CPD lesion (Fig. 1, A and B). Increased concentrations improved Pol IV bypass activity (Fig. 1A) but had little effect on Pol II activity (Fig. 1B). The frameshift activity of Pol IV was evident on the THF template, with the completely extended product being shorter than the available template downstream of the lesion (compare the extended products on the CPD and THF templates in Fig. 1A). As expected, Pol III* was unable to bypass either lesion proficiently (Fig. 1C), although a very low amount of bypass was observed with the THF template, as reported previously (22). Elevated dNTP concentrations facilitated both Pol IV and II TLS (Fig. 2). Pol IV TLS of a CPD and a THF was maximally stimulated between 480 and 960 μm and between 240 and 480 μm dNTPs, respectively, whereas Pol II TLS of a THF was maximally stimulated between 480 and 960 μm dNTPs. Pol II bypass of a CPD was minimal.
FIGURE 1.
TLS bypass of THF and CPD lesions by polymerases on oligonucleotide primer templates. The indicated concentrations of DNA polymerases were incubated with the CPD ([5′-32P]CG18:CG17CPD), THF ([5′-32P]CG18:CG17THF), and undamaged ([5′-32P]CG18:CG16) primer templates for 10 min at 37 °C, and the DNA products were processed and analyzed as described under “Experimental Procedures.” The white asterisks correspond to the position of the CPD or THF lesion, accordingly, in the template strand. P denotes the position of the unextended primer. The extent of bypass (mean and standard deviation from three experiments) is given as the percentage of total radioactivity in extended primer species. Representative gels are shown. A, Pol IV bypasses both a THF and CPD lesion. B, Pol II bypasses a THF, but not a CPD, lesion. C, Pol III* does not bypass either a THF or CPD lesion. D, inactivation of the 3′ → 5′ proofreading exonuclease of Pol II stimulates TLS. E, Pol IV ΔC5, but not Pol IV F13V, bypasses a CPD lesion. F, Pol IV ΔC5, but not Pol IV F13V, bypasses a THF lesion. G, Pol IV F13V is active on an undamaged primer template. H, Pol IV L1 (LF domain variant) is active in TLS. Representative gels are shown.
FIGURE 2.
Increasing deoxynucleoside triphosphate concentration stimulates TLS bypass by Pol IV and II on oligonucleotide primer templates. Either Pol IV or Pol II were incubated with the CPD, THF, and undamaged primer templates for 10 min at 37 °C at the indicated concentration of dNTPs (increasing by a factor of 2 from left to right), and the DNA products were processed and analyzed as described under “Experimental Procedures.” The white asterisks correspond to the position of the CPD or THF lesion, accordingly, in the template strand. P denotes the position of the unextended primer. The extent of bypass (mean and standard deviation from three experiments) is given as the percentage of total radioactivity in extended primer species. A representative gel is shown.
Pol IV, but Not Pol II, Catalyzes Replisome-mediated Lesion Bypass
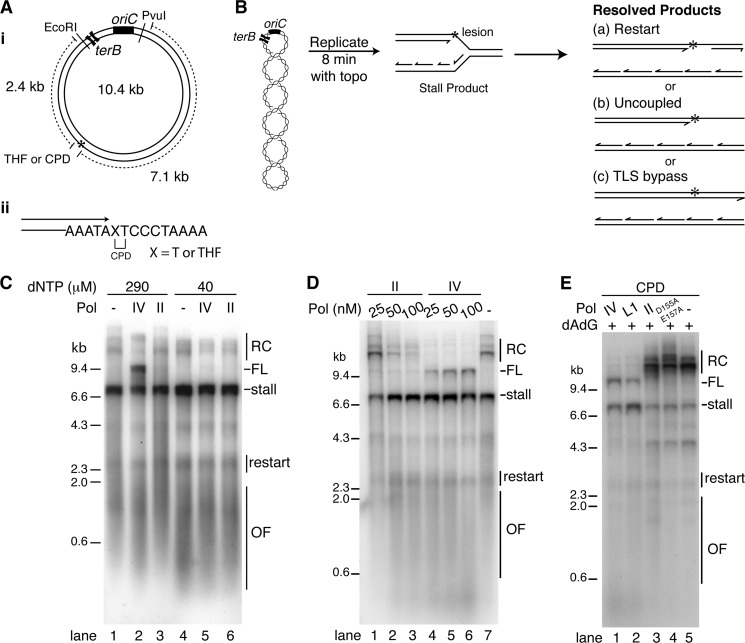
To investigate the interaction of TLS polymerases with the replisome, we utilized a replication system (5) that uses a 10.4-kilobase pair supercoiled DNA template carrying oriC and has either a CPD or THF lesion 7.1 kilobase pairs clockwise from the origin (Fig. 3A, panel i). These two types of lesion were chosen for study because they are the most likely to be encountered by the replisome under normal and UV-stressed conditions. Replication is initiated at oriC in the presence of DnaA, DnaB, DnaC, DnaG, HU, SSB, DNA gyrase, Tus, and the Pol III HE. The fork moving counter-clockwise is blocked by the Tus-TerB complex (30), enabling monitoring of only the encounter of the clockwise-moving fork with the DNA lesion. The DNA products are analyzed by denaturing alkaline agarose gel electrophoresis. The length of the nascent leading strand is diagnostic of the events that happen subsequent to fork stalling (Fig. 3B): replisomes that stall and then skip over the lesion by restarting leading strand synthesis downstream generate a 7.1-kb stall product and restart products in the range of 2.0–2.4 kb (Fig. 3B, panel a). Replisomes that do not restart leading strand synthesis by the time DnaB and the lagging strand polymerase reach the end of the template generate only the leading strand stall product and a full-length lagging strand sister (uncoupled replication (5)) (Fig. 3B, panel b), and if a TLS polymerase successfully engages with the replisome and bypasses the lesion, a full-length, 9.4-kb leading strand will be generated (Fig. 3B, panel c).
FIGURE 3.
Pol IV, but not Pol II, catalyzes replisome-mediated TLS. A, DNA template. Panel i, map showing the positions of the leading strand template lesion relative to the origin of DNA replication, the Ter sites, and the relevant restriction sites used for analysis. Panel ii, DNA sequence about the site of the template lesion. B, replication reaction scheme showing the possible replicated DNA products. C, elevated nucleoside triphosphate concentration is required for Pol IV-catalyzed, replisome-mediated TLS on a THF template. Standard gyrase reactions containing the indicated TLS polymerase (100 nm Pol IV, 20 nm Pol II) and concentrations of dNTPS were incubated, processed, and analyzed as described under “Experimental Procedures.” D, Pol IV, but not Pol II, catalyzes replisome-mediated TLS. Standard gyrase replication reactions containing the THF template and the indicated concentrations of TLS polymerase, standard concentrations of dCTP and TTP, and 750 μm dATP and dGTP were incubated, processed, and analyzed as described under “Experimental Procedures.” E, Pol IV L1 (LF domain variant), but not Pol II D155A/E157A (the 3′ → 5′ exonuclease-defective variant), catalyzes replisome-mediated TLS. Standard gyrase reactions containing the indicated TLS polymerase (100 nm), standard concentrations of dCTP and TTP, and 750 μm dATP and dGTP were incubated, processed, and analyzed as described under “Experimental Procedures.” RC, greater than unit length, rolling circle DNA products that arise from nicks in the DNA template; FL, full-length nascent leading strand spanning the PvuI to EcoRI sites; stall, the nascent leading strand stall product spanning the distance from the PvuI site to the site of the template lesion; restart, nascent leading strands restarted downstream of the damage by leading strand lesion skipping; OF, Okazaki fragments. The extent of bypass in D, lanes 1–7, was (calculated as FL/FL + stall) 0.04 ± 0.01, 0.03 ± 0.01, 0.03 ± 0.01, 0.12 ± 0.04, 0.14 ± 0.03, 0.19 ± 0.04, and 0.03 ± 0.01, respectively (mean and standard deviation from four experiments). The extent of bypass in E, lanes 1 and 2, was 0.24 ± 0.1 and 0.1 ± 0.03, respectively (mean and standard deviation from three experiments). Representative gels are shown.
Under standard reaction conditions (40 μm dNTPs), the addition of either Pol IV or Pol II had no effect on leading strand synthesis (Fig. 3C). Because TLS activity on the oligonucleotide primer templates was more efficient at elevated nucleotide concentrations (Fig. 2), we asked whether elevated dNTP concentrations in the replication reactions would effect TLS. This proved to be the case. Increasing the concentration of dNTPs 7.5-fold supported replisome-mediated TLS by Pol IV, but not Pol II (Fig. 3C). Titrations of Pol IV and II reinforced this conclusion. In the presence of increasing concentrations of Pol IV at standard concentrations of dCTP and TTP and 750 μm dATP and dGTP (see next section), a full-length leading strand product appeared that is indicative of TLS bypass (Fig. 3D, compare lanes 4–6 with lane 7). No such product was observed in the presence of equivalent concentrations of Pol II (Fig. 3D, compare lanes 1–3 with lane 7). Thus, despite the fact that both Pol II and IV have the inherent capacity to bypass a THF lesion (Figs. 1 and 2), only Pol IV is capable of bypass in the presence of the replisome.
It was possible that the 3′ → 5′ proofreading exonuclease of Pol II was preventing bypass by cycling between the removal of the 3′ end of the primer terminus followed by polymerization back to the initial stall position, thereby delaying TLS sufficiently that it no longer was a competitive kinetic pathway with replication restart by leading strand lesion skipping. We therefore tested the activity of Pol II D155A/E157A, a variant defective in the proofreading exonuclease activity (31), in replisome-mediated TLS. As expected, Pol II D155A/E157A was significantly more active (at least 20-fold) in bypass of both a THF and CPD lesion on oligonucleotide primer templates (Fig. 1D); however, this variant still failed to catalyze replisome-mediated TLS (Fig. 3E).
As expected, elevating dNTP concentrations also increased the size of the Okazaki fragments (Fig. 3C, compare lane 1 with lane 4), because the change of the ratio of dNTPs to NTPs affects the efficiency of primer utilization for lagging strand synthesis by the replisome (32). The addition of Pol IV also modulated Okazaki fragment size, shifting the fragments back to smaller products (Fig. 3D, compare lanes 4–6 with lane 7), likely because, as demonstrated previously (19), Pol IV exchanging with the replicative polymerase slows the rate of DNA synthesis and thus the rate at which template is generated (33).
Characterization of Pol IV-catalyzed, Replisome-mediated TLS
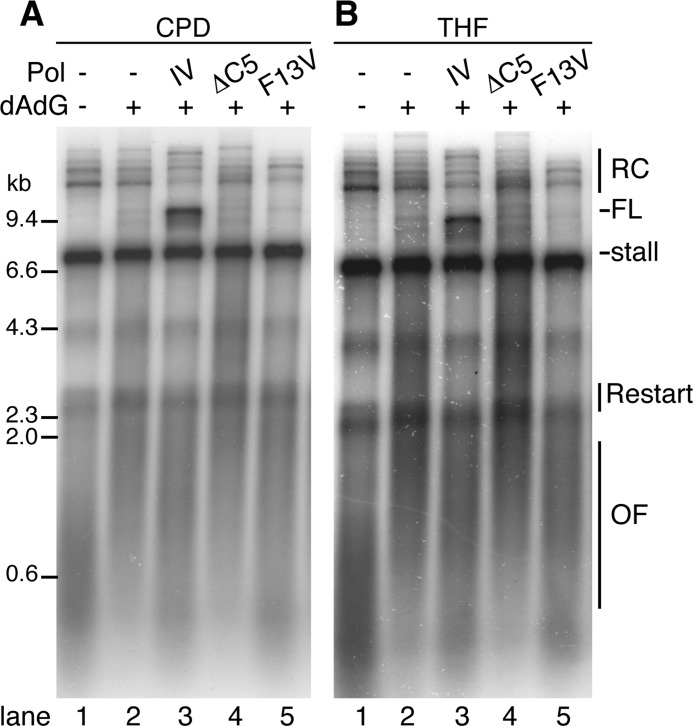
Pol IV TLS does not require that the enzyme associate with the β clamp (Fig. 1). Indeed, a variant enzyme, Pol IV ΔC5, which lacks the C-terminal residues required for interaction with β (19), is just as efficient as the wild type in bypass on oligonucleotide primer templates (Fig. 1, E and F). Replisome-mediated TLS, however, required interaction between Pol IV and β (Fig. 4, A and B). We used another variant, Pol IV F13V, which has a steric site mutation that separates the polymerization activity from the TLS activity of the enzyme (21) (Fig. 1, E–G), to establish that Pol IV, and not the Pol III HE, was the effector of TLS. No full-length leading strand indicative of TLS was observed with this variant enzyme (Fig. 4, A and B), indicating that the interaction between Pol IV and the replisome did not, in some unanticipated manner, endow the Pol III HE with TLS activity. These observations suggest that generation of the full-length leading strand is not a result of Pol IV TLS separable from the action of the replisome—Pol IV ΔC5 is perfectly capable of TLS on the oligonucleotide primer templates (Fig. 1)—but likely results from a polymerase switch that requires Pol IV to interact with β. This conclusion is supported by the fact that whereas Pol IV F13V does not give full-length products, it is still capable of interacting with the Pol III HE in the replisome, as evidenced by its effect on the size of the Okazaki fragments (Fig. 4, A and B).
FIGURE 4.
Replisome-mediated Pol IV-catalyzed TLS bypass requires interaction with β and TLS activity. Standard gyrase replication reactions containing either the CPD (A) or THF template (B), the indicated TLS polymerase at 100 nm, standard concentrations of dCTP and TTP, and 750 μm dATP and dGTP were incubated, processed, and analyzed as described under “Experimental Procedures.” ΔC5, Pol IV ΔC5; F13V, Pol IV F13V. Other abbreviations are as defined in the legend to Fig. 3.
Pol IV contains an extra domain (compared with other bypass polymerases) conserved in all Y family polymerases (34) termed the “little finger” (LF) domain that can engage in a secondary interaction with the β-clamp along its rim (35). We tested the requirement for this domain during replisome-mediated TLS using the Pol IV variant Pol IV L1 (36), where amino acid residues 303VWP305 have been changed to 303SGA305. These mutations disrupt the secondary rim-on interaction with β (36). The Pol IV L1 variant could bypass template damage with ∼50% of the efficiency as the wild type on oligonucleotide primer templates (Fig. 1H), as well as during replisome-mediated TLS (Fig. 3E). Thus, the LF domain of Pol II is not required for, but does stimulate, replisome-mediated TLS.
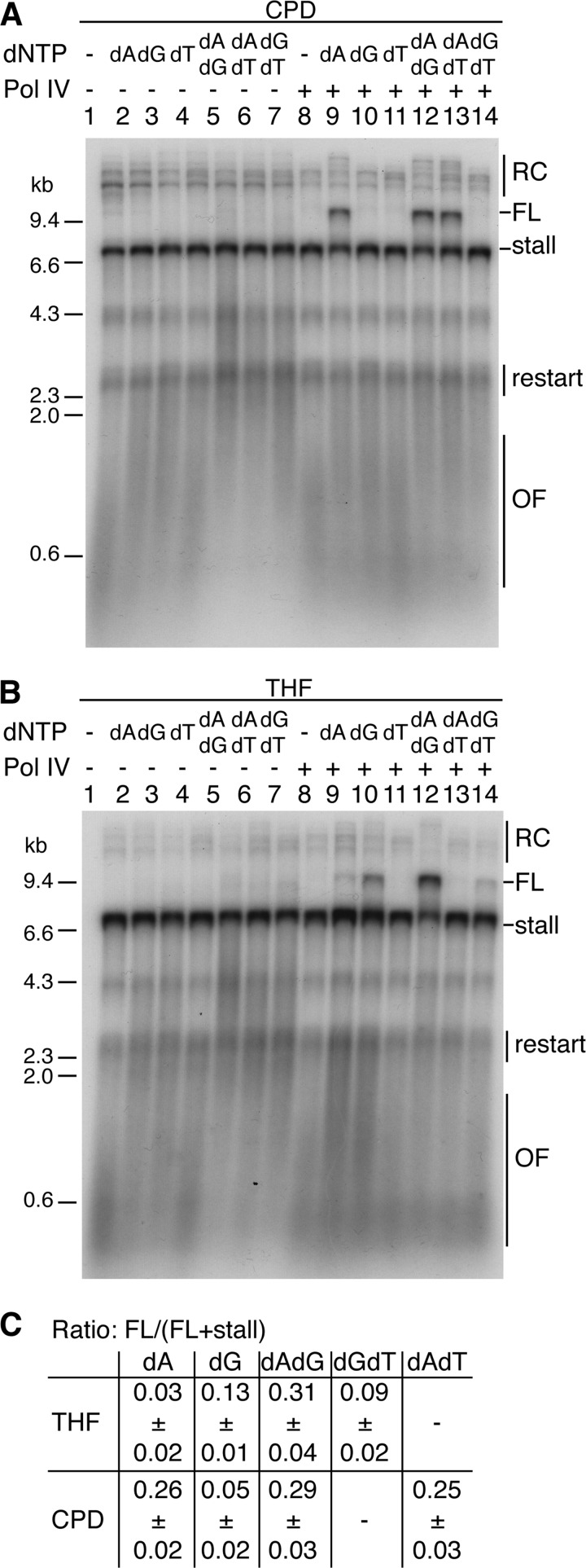
We examined the requirement for elevated nucleotide concentration to determine whether it was of a general nature or specific to the template sequence about the lesion (Fig. 5). Increasing dNTP concentration enhanced replisome-mediated Pol IV TLS, reaching a maximum stimulation at ∼750 μm (data not shown). We therefore tested 750 μm concentrations of single and pairwise combinations of the dNTPs based on the sequence surrounding the lesion (Fig. 3A, panel ii). Note that even under these conditions of elevated nucleotide concentration, neither Pol III (Fig. 5, A and B) nor Pol II (Fig. 3D) manifested TLS.
FIGURE 5.
Nucleotide requirements for Pol IV-catalyzed, replisome-mediated TLS bypass. A and B, standard gyrase replication reactions containing either the CPD template (A) or the THF template (B), and the indicated elevated concentrations (0.75 mm) of dATP, dGTP, or TTP were incubated in either the presence or absence of Pol IV (100 nm), processed, and analyzed as described under “Experimental Procedures.” C, quantification of TLS bypass for reactions containing Pol IV. Given are the means and standard deviations from three experiments. Representative gels are shown. Abbreviations are as defined in the legend to Fig. 3.
The requirements for elevated nucleotide concentrations differed according to the nature of the lesion in the template strand. Bypass of the CPD lesion was effected efficiently in the presence of elevated dATP. Elevated dGTP, not dATP, alone gave significant bypass of the THF lesion, whereas elevating dGTP in addition to dATP gave maximal bypass. These different nucleotide requirements suggested different mechanisms of bypass. Bypass of the CPD seemed likely to be error-free and a product of direct insertion because only elevation of the correct nucleotide was required. Pol IV-catalyzed replisome-mediated TLS bypass of a CPD was quite robust, counter to genetic observations suggesting that this lesion is not a substrate for Pol IV bypass during DNA replication (37). This dissonance could be accounted for if Pol IV bypass of a CPD in the context of the replisome were error-free. On the other hand, bypass of the THF seemed highly likely to be mutagenic. Stimulation of bypass by only the downstream nucleotide, dGTP, implied rearrangement of the primer template, looping out one or both of the dTMP residues on the template strand to generate the frameshift observed in Fig. 1.
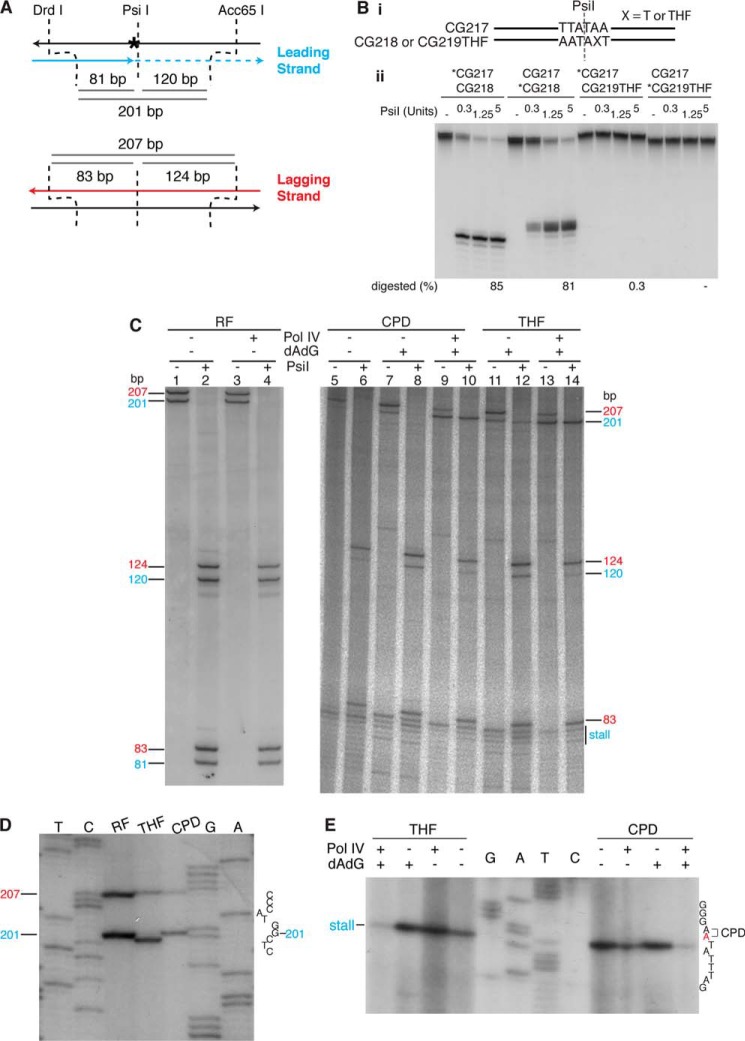
To determine whether TLS was, in fact, contiguous across the lesion and whether frameshifting was occurring on the THF template, we analyzed the DNA products in a small region around the damage as described by Pagès and Fuchs (38) (Fig. 6A). Digestion of the replication products from undamaged DNA templates with DrdI and Acc65I yields a diagnostic 201-nt leading strand and a 207-nt lagging strand that are both susceptible to cleavage by PsiI, located at the site of the lesion (Fig. 6C). Cleavage by PsiI is inhibited by the lesions (Fig. 6B). Thus, persistence of the leading strand fragment after PsiI digestion is indicative of TLS across the lesion. Accordingly, on templates containing either a THF or CPD lesion, a full-length, DrdI-Acc65I leading strand product is generated only in the presence of Pol IV and elevated concentrations of dATP and dGTP (Fig. 6C, CPD and THF). This product is resistant to PsiI cleavage (Fig. 6C, compare lanes 9 and 10 with lanes 13 and 14), indicating that the full-length replicated leading strand product is, in fact, generated by TLS bypass across the lesion. The leading strand band present with the CPD template in the absence of Pol IV at elevated nucleotide concentrations is clearly a background band because it is digested with PsiI (Fig. 6C, lanes 7 and 8), whereas the leading strand band present with the THF template in the absence of Pol IV and presence of elevated nucleotide concentrations is partially resistant to PsiI digestion (Fig. 6C, lanes 11 and 12), indicating the presence of the background band, but, in addition, suggesting some bypass occurred that may represent the known low level of THF TLS by DNA polymerase III (24).
FIGURE 6.
Pol IV-catalyzed, replisome-mediated TLS bypass generates a −1 frameshift on the THF template. A, scheme for analyzing the nascent DNA products. B, a THF lesion inhibits digestion of DNA by the PsiI restriction endonuclease. Panel i, schematic of the two duplex DNA oligonucleotides used as substrates. The DNA sequence is identical to that about the site of the lesion in the template DNA used for replication. Panel ii, different combinations of oligonucleotides were treated with PsiI as indicated, and the DNA products were analyzed by denaturing polyacrylamide gel electrophoresis as described under “Experimental Procedures.” An asterisk on an oligonucleotide name denotes that it was 5′-[32P] end-labeled. C, leading strand replication is contiguous across the site of the template lesion. DNA products generated by replication with either the undamaged, CPD, or THF template either in the presence or absence of Pol IV (100 nm) and the presence or absence of elevated concentrations (0.75 mm) of dATP and dGTP were digested with DrdI and Acc65I either with or without digestion with PsiI as indicated, processed, and analyzed by electrophoresis through a denaturing polyacrylamide gel as described under “Experimental Procedures.” RF, replicative form DNA. D, Pol IV-catalyzed, replisome-mediated TLS bypass generates a −1 frameshift on the THF template. DNA products generated as described for C in the presence of Pol IV (100 nm), and elevated nucleotide concentrations (0.75 mm) were digested with DrdI and Acc65I and analyzed by electrophoresis through a high resolution denaturing 6% polyacrylamide gel (19:1, acrylamide:bisacrylamide) as described under “Experimental Procedures.” T, C, G, and A show a DNA sequencing ladder prepared from undamaged DNA using a primer that has the same 5′ end as the DrdI-digested nascent leading strand DNA. For reasons that are unclear, migration of the digested nascent leading strand from the CPD template was somewhat variable compared with the product from the undamaged template; it could have the same mobility or be a bit slower. We think this could be because of differences in loading volumes necessitated by the differences in the extent of replication with the two templates. E, leading strand replication stalls just 5′ of the template lesion. DNA products generated using either the THF or CPD templates, either in the presence or absence of Pol IV (100 nm) and either in the presence or absence of elevated concentrations (0.75 mm) of dATP and dGTP were prepared and analyzed as in D except they were digested only with DrdI.
A different, higher resolution gel analysis was performed where the DrdI- and Acc65I-digested products were electrophoresed side by side with a DNA sequence ladder. On this gel, it is clear that leading strands made from the THF template were 1 nt shorter than those made from either the CPD or undamaged templates (Fig. 6D), confirming, as suggested above, that bypass at the THF generates a −1 frameshift, most likely by a form of dNTP-stabilized misalignment, whereas bypass of the CPD occurs by direct insertion. Using the same method, we confirmed that the leading strand stalls on both the THF and CPD templates at the nucleotide just 5′ of the template lesion (Fig. 6E).
TLS Likely Occurs by Polymerase Exchange
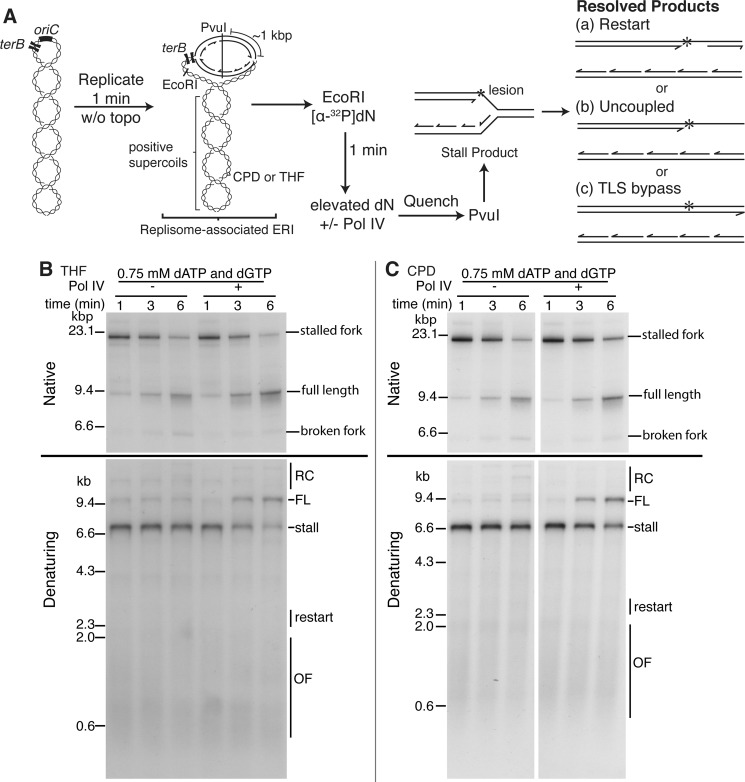
It was possible that the observed generation of full-length leading strand occurred in steps whereby leading and lagging strand synthesis uncoupled, Pol IV bypassed the lesion, and the downstream portion of the leading strand was then completed by free Pol III HE in the reaction mixture. The fact that exonuclease-defective Pol II does not support bypass in the replication system makes this possibility very unlikely—if the uncoupled, partially replicated leading strand sister duplex was present in the reaction, given the concentration of Pol II, bypass would certainly occur and a full-length leading strand would be observed. To examine this possibility in another manner, we used a variation of the replication system that allows for the detection of the uncoupled product that one would expect to be generated as an intermediate. Here (Fig. 7A), the replication system is synchronized by initiation in the absence of DNA gyrase, generating an early replication intermediate paused by the accumulation of positive supercoils. Replication forks are released by rapid digestion with EcoRI. Labeled nucleotide is added for a short period after digestion and then chased with an 100-fold excess of cold nucleotide. Under these conditions, uncoupled products can be detected by native gel electrophoresis (27).
FIGURE 7.
Stalled nascent leading strand is chased into full-length product during Pol IV-catalyzed, replisome-mediated TLS bypass. A, scheme of the replication reaction. B and C, pulse-chase EcoRI replication reactions containing [α-32P]dATP using either the THF (B) or CPD (C) template either in the presence or absence of Pol IV (100 nm) were incubated, processed, and analyzed as described under “Experimental Procedures.” Times are post chase. ERI, early replication intermediate; topo, topoisomerase. Other abbreviations are as defined in the legend to Fig. 3.
In the presence or absence of Pol IV under elevated nucleotide conditions, duplex stalled fork products are efficiently chased to duplex full-length product, as observed by native gel electrophoresis using either the THF or CPD template (Fig. 7, B and C, Native). Denaturing gel electrophoresis conditions confirmed that in the presence of Pol IV in these reactions, the stalled leading strand product was chased directly to full-length, leading strand product (Fig. 7, B and C, Denaturing). Thus, the absence of any detectable uncoupled product argues that the full-length, leading strand product is being generated via TLS bypass likely mediated by a switch between the replicative polymerase and Pol IV via an interaction with the β-clamp. In addition, the kinetics of appearance of full-length duplex DNA product in the presence and absence of Pol IV are identical (Fig. 7, B and C), indicating that Pol IV-catalyzed bypass did not significantly delay progress of the replisome, arguing that once TLS is accomplished, leading strand synthesis is resumed by Pol III. It is important to note that full-length restart products expected in the absence of Pol IV and the full-length TLS product migrate in identical positions on the native gels. Using a replication system similar to the one described here and a template containing a N2-dG leading strand template adduct, Ikeda et al. (39) also concluded that bypass involved a switch from Pol III to Pol IV and back again to Pol III. Single molecule studies on oligonucleotide primer templates (in the absence of a replisome) carrying the same lesion also support the same conclusion (40).
Leading Strand Lesion Skipping by Replication Restart and TLS Are Competing Bypass Mechanisms
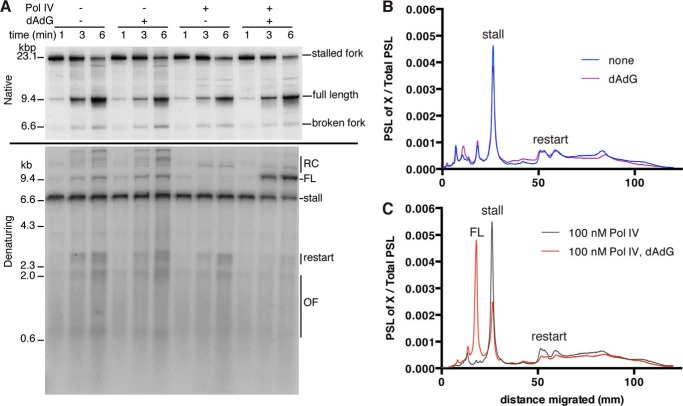
Exchange of Pol IV directly with the stalled leading strand polymerase in the replisome predicts that the fraction of stalled leading strands that are restarted after leading strand repriming should decrease concomitantly. Restart products are not visualized in the protocol used above because they arise after the chase is initiated. Therefore, to visualize both the full-length leading strand generated by TLS and the restart product generated by leading strand lesion skipping, we used the same protocol (Fig. 7A) with [α-32P]dCTP present throughout the course of the reaction subsequent to release of the replication forks by EcoRI digestion (Fig. 8).
FIGURE 8.
Pol IV-catalyzed, replisome-mediated TLS bypass occurs at the expense of leading strand lesion skipping. A, EcoRI replication reactions using the THF template either in the presence or absence of elevated concentrations (0.75 mm) of dATP and dGTP and either in the presence or absence of Pol IV (100 nm), as indicated, were incubated, processed, and analyzed as described under “Experimental Procedures.” Times are post EcoRI addition. B and C, gel lane traces prepared by PhosphorImager analysis comparing the 6-min lanes in A for reactions either in the absence (B) or presence (C) of Pol IV either in the presence or absence of elevated concentrations of dATP and dGTP. PSL, photo stimulated luminescence; RC, greater than unit length, rolling circle DNA products that arise from nicks in the DNA template; FL, full-length nascent leading strand spanning the PvuI to EcoRI sites; stall, the nascent leading strand stall product spanning the distance from the PvuI site to the site of the template lesion; restart, nascent leading strands restarted downstream of the damage by leading strand lesion skipping.
At the initial time point, the stalled fork represents the bulk of the products formed under any condition used and the majority of these products are converted to full-length duplex products after 6 min of incubation (Fig. 8A, Native). Denaturing gel electrophoresis of the products demonstrates that in the absence of elevated nucleotide, either in the presence or absence of Pol IV, as well as in the presence of elevated nucleotide and the absence of Pol IV, significant levels of leading strand restart products appear between 3 and 6 min of incubation post EcoRI cleavage (Fig. 8A, Denaturing). However, in the presence of elevated nucleotide and Pol IV, the generation of restart products is markedly reduced as full-length TLS product accumulates, and the amount of stall product is reduced simultaneously (Fig. 8, B and C). These observations imply a competition between restart and TLS during lesion bypass. Note that in the analysis of this reaction, the DNA products are digested with EcoRI and PvuI; thus, the aberrant rolling circle DNA products are digested to full-length size, accounting for their apparent accumulation in the absence of Pol IV in the presence and absence of elevated nucleotide concentrations; however, unlike in the presence of Pol IV, the stall product is not reduced during the course of the reaction (Fig. 8A, Denaturing).
Ikeda et al. (39) reached the opposite of our conclusion, i.e. that restart and TLS did not compete. We note, however, that in their reactions, oriC replication was ongoing for 5 min before the addition of Pol IV, and uncoupled products are clearly visible in their gels, suggesting that the observed bypass may not have been replisome-associated.
DISCUSSION
Bypass of DNA template damage by translesion synthesis is an essential survival mechanism for all organisms, allowing DNA replication to proceed past the point of damage so that the cell cycle can be completed and cell division accomplished, even at the expense of the misincorporated nucleotides that can be inserted opposite the damage by the generally error-prone TLS DNA polymerases. Some DNA lesions are corrected by either the base excision or nucleotide excision repair pathways either pre- or postreplicatively; however, it seems likely that occasions arise with some frequency that require lesion bypass coincident with DNA replication. We have described here the reconstitution in vitro of TLS polymerase-catalyzed lesion bypass in concert with replisome-catalyzed DNA replication.
Differential Action of Pol II and Pol IV in Concert with the Replisome
Whereas both Pol II and Pol IV were proficient for lesion bypass on small oligonucleotide primer templates containing a lesion, only Pol IV was capable of bypassing either a THF or a CPD lesion during reconstituted replication reactions using oriC templates. Although TLS polymerase-catalyzed lesion bypass on small oligonucleotide primer templates was improved at elevated nucleotide concentrations, no additional cofactors were required. In contrast, the TLS capacity of Pol IV during replisome-mediated lesion bypass was dependent on an interaction with the β-clamp processivity subunit of the Pol III HE and elevated nucleotide concentration specific to the sequence context about the lesion.
However, having TLS activity and being able to interact with β was clearly insufficient for TLS bypass during concerted replication, because Pol II was unable to catalyze bypass in our reactions. Binding of Pol IV (41) and Pol II (42) to β at primer termini mediated by the clamp-loading DnaX complex increases the processivity of both enzymes significantly. Furthermore, both enzymes are able to switch with Pol III.
All E. coli DNA polymerases can interact with the β clamp. This led to the question of whether one DNA polymerase could switch with another during DNA synthesis. Indiani et al. (43) demonstrated, using complementary strand synthesis on a primed M13 single-stranded template as an assay, that Pol IV could switch with a stalled Pol III, but not one that was moving. These same authors demonstrated (19), using a rolling circle DNA replication system, that both Pol II and IV could support slow DNA replication with the DnaB helicase and could remodel a moving Pol III HE replisome by slowing it down.
That such interactions between β and the different polymerases can also occur is supported by studies in vivo. In a dnaN159 strain containing a temperature-sensitive β-clamp, Pol II was shown to block lethal access of Pol IV to the fork (44), and the mutagenic potential of Pol IV has been shown to be limited by the presence of Pol II (45–47). Furthermore, Pol II and IV outcompete Pol V for an interaction with the β-clamp, thereby essentially eliminating UV-induced mutagenesis. Pol V-dependent UV- and methyl methanesulfonate-induced mutagenesis was restored by inactivation of Pol II and IV in the dnaN159 strain (44). It is possible that these phenotypes are because of the inability of the β-clamp mutant to coordinate the action of Pol III with those of Pol II, IV, and V. The impaired clamp-DNA interaction could influence the exposed surfaces of the clamp and thus accessibility for different polymerases (44).
The differential action of Pol II and IV in replisome-mediated TLS cannot be explained completely by the lack of a LF domain in Pol II. Inactivating the interaction between the Pol IV LF domain only reduced TLS by ∼50%, indicating that whereas it stimulated TLS, it was not required for it. It is therefore likely that there are more subtle differences in the manner in which Pol II and IV interact with the β-clamp that are not obvious at the moment and that contribute to a productive (i.e. TLS) interaction.
That the binding between Pol IV and II to a β clamp occupied by Pol III is different is supported by some more recent studies with the dnaN159 mutant strain (48). Under conditions of SOS induction, the increase in Pol IV concentration makes the strain more sensitive to UV light, and increasing Pol IV concentration by an additional 4-fold impedes growth of the strain. On the other hand, increasing Pol II concentration to 8-fold greater than the SOS-induced concentration did not impede growth. The authors confirmed that Pol II switched equally well with either a stalled or moving Pol III in vitro and concluded that Pol II does not interact with a replisome in the same manner as Pol IV. Thus, it seems that whereas interaction of Pol II and IV with a Pol III replisome does indeed slow progression (Ref. 19 and references therein), this effect may not be the best readout of activity, i.e. TLS bypass.
Polymerase Switching as the Underlying Mechanism for TLS Bypass
Replisome-mediated TLS occurs by a switch between the leading strand Pol III stalled at the lesion and Pol IV. At some point after TLS, control of DNA synthesis is returned to Pol III, although the length of time that Pol IV remains engaged on the primer terminus remains unknown. Interestingly, in their single molecule studies of primer extension, Kath et al. (40) demonstrated that Pol IV can remain bound for very long periods—hundreds of seconds, suggesting that the repair patch length could be quite long. Whether these observations will apply to replisome-mediated TLS as well remains to be determined. The original “tool belt” model for polymerase switching on the β clamp invoked the dimeric nature of β and suggested that two polymerases were bound simultaneously, one in each hydrophobic cleft, and the switch might occur by rotation of the β subunit (43). A subsequent structure of β bound to DNA showed that it was tilted by 22° from the helical axis of the duplex DNA, indicating that the two hydrophobic clefts were not equivalent and that binding of a second polymerase to the available cleft of a β already bound to a polymerase on the DNA was unlikely (47). In addition, using an engineered β dimer that contained only one hydrophobic cleft, Heltzel et al. (44) demonstrated that a single cleft on the β dimer was sufficient to observe Pol IV switching with Pol III, in a reaction that required binding of Pol IV to both the cleft and the rim of β. These authors suggested that productive polymerase switching proceeded by Pol IV bound to the rim of β near the hydrophobic cleft displacing the stalled Pol III from that cleft. However, our data indicate that the LF domain is not required for replisome-mediated TLS, so direct competition between the accessible binding site on β is likely to operate as well.
Competing Reactions at Stalled Replication Forks
Stalled forks can be processed in a number of different ways as summarized in the Introduction. Is there a master regulator that acts to channel reactions down certain pathways, or is the decision more of a kinetic one, governed by affinities of various proteins and the time constants of the reactions? We have recently demonstrated that replication fork regression catalyzed by either RecG or RuvAB can occur at stalled forks when the replisome is present on the DNA and have suggested that most regression is actually postreplicative (49, 50). We have shown here that replisome-mediated TLS and leading strand lesion skipping compete directly. This observation suggests that the switch to Pol IV at the stalled fork happens very quickly. Fuchs et al. (51) suggested that the affinity of Pol IV for β loaded on DNA and the high intracellular concentrations of Pol IV were tempting reasons to speculate that Pol IV might constitutively be part of the replisome, possibly assisting the replicative polymerase during synthesis across difficult sequence contexts. Our current data could be construed as being consistent with this proposal. The concentrations of template, Pol III HE, and Pol IV in a newborn E. coli cell growing in rich medium (∼4 fl) with an average of two chromosomes/cell—roughly 0.8 nm, 16 nm (20 copies/cell), and 100 nm (250 copies/cell), respectively—are very similar to the concentrations used in the replication system described herein. Furthermore, the intracellular dNTP concentrations under normal growth conditions have been determined to be ∼900 μm for a similarly sized E. coli cell grown in rich medium (26), more than sufficient for replisome-mediated, Pol IV-catalyzed TLS bypass of DNA lesions.
Acknowledgments
We thank Myron Goodman, Mike O'Donnell, Jérôme Wagner, and Graham Walker for reagents and John Petrini for advice on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM34557 (to K. J. M.).
- Pol
- DNA polymerase
- HE
- holoenzyme
- TLS
- translesion synthesis
- THF
- tetrahydrofuran
- CPD
- cyclopyrimidine dimer
- LF
- little finger
- nt
- nucleotide(s).
REFERENCES
- 1. McHenry C. S. (2011) DNA replicases from a bacterial perspective. Annu. Rev. Biochem. 80, 403–436 [DOI] [PubMed] [Google Scholar]
- 2. Rupp W. D., Wilde C. E., 3rd, Reno D. L., Howard-Flanders P. (1971) Exchanges between DNA strands in ultraviolet-irradiated Escherichia coli. J. Mol. Biol. 61, 25–44 [DOI] [PubMed] [Google Scholar]
- 3. Heller R. C., Marians K. J. (2006) Replisome assembly and the direct restart of stalled replication forks. Nat. Rev. Mol. Cell Biol. 7, 932–943 [DOI] [PubMed] [Google Scholar]
- 4. Heller R. C., Marians K. J. (2005) The disposition of nascent strands at stalled replication forks dictates the pathway of replisome loading during restart. Mol. Cell 17, 733–743 [DOI] [PubMed] [Google Scholar]
- 5. Yeeles J. T., Marians K. J. (2011) The Escherichia coli replisome is inherently DNA damage tolerant. Science 334, 235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rupp W. D. (1996) DNA repair mechanisms. in Escherichia coli and Salmonella Cellular and Molecular Biology (Neidhart F. C., Curtiss R., 3rd, Ingraham J. L., Lin E. C. C., Low K. B., Magazanik B., Reznikoff W. S., Riley M., Schaechter M., Umbarger H. E., eds) pp. 2277–2294, ASM, Washington, D. C. [Google Scholar]
- 7. Wagner J., Gruz P., Kim S. R., Yamada M., Matsui K., Fuchs R. P., Nohmi T. (1999) The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol. Cell 4, 281–286 [DOI] [PubMed] [Google Scholar]
- 8. Tang M., Shen X., Frank E. G., O'Donnell M., Woodgate R., Goodman M. F. (1999) UmuD′(2)C is an error-prone DNA polymerase, Escherichia coli pol V. Proc. Natl. Acad. Sci. U.S.A. 96, 8919–8924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fijalkowska I. J., Schaaper R. M., Jonczyk P. (2012) DNA replication fidelity in Escherichia coli: a multi-DNA polymerase affair. FEMS Microbiol. Rev. 36, 1105–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Friedberg E. C., Wagner R., Radman M. (2002) Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science 296, 1627–1630 [DOI] [PubMed] [Google Scholar]
- 11. Rangarajan S., Woodgate R., Goodman M. F. (1999) A phenotype for enigmatic DNA polymerase II: a pivotal role for pol II in replication restart in UV-irradiated Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 96, 9224–9229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Napolitano R., Janel-Bintz R., Wagner J., Fuchs R. P. (2000) All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J. 19, 6259–6265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim S. R., Matsui K., Yamada M., Gruz P., Nohmi T. (2001) Roles of chromosomal and episomal dinB genes encoding DNA pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol. Genet. Genomics 266, 207–215 [DOI] [PubMed] [Google Scholar]
- 14. McHenry C., Kornberg A. (1977) DNA polymerase III holoenzyme of Escherichia coli: purification and resolution into subunits. J. Biol. Chem. 252, 6478–6484 [PubMed] [Google Scholar]
- 15. Kim S. R., Maenhaut-Michel G., Yamada M., Yamamoto Y., Matsui K., Sofuni T., Nohmi T., Ohmori H. (1997) Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc. Natl. Acad. Sci. U.S.A. 94, 13792–13797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kuban W., Banach-Orlowska M., Bialoskorska M., Lipowska A., Schaaper R. M., Jonczyk P., Fijalkowska I. J. (2005) Mutator phenotype resulting from DNA polymerase IV overproduction in Escherichia coli: preferential mutagenesis on the lagging strand. J. Bacteriol. 187, 6862–6866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wagner J., Nohmi T. (2000) Escherichia coli DNA polymerase IV mutator activity: genetic requirements and mutational specificity. J. Bacteriol. 182, 4587–4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Uchida K., Furukohri A., Shinozaki Y., Mori T., Ogawara D., Kanaya S., Nohmi T., Maki H., Akiyama M. (2008) Overproduction of Escherichia coli DNA polymerase DinB (Pol IV) inhibits replication fork progression and is lethal. Mol. Microbiol. 70, 608–622 [DOI] [PubMed] [Google Scholar]
- 19. Indiani C., Langston L. D., Yurieva O., Goodman M. F., O'Donnell M. (2009) Translesion DNA polymerases remodel the replisome and alter the speed of the replicative helicase. Proc. Natl. Acad. Sci. U.S.A. 106, 6031–6038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kobayashi S., Valentine M. R., Pham P., O'Donnell M., Goodman M. F. (2002) Fidelity of Escherichia coli DNA polymerase IV. Preferential generation of small deletion mutations by dNTP-stabilized misalignment. J. Biol. Chem. 277, 34198–34207 [DOI] [PubMed] [Google Scholar]
- 21. Jarosz D. F., Godoy V. G., Delaney J. C., Essigmann J. M., Walker G. C. (2006) A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature 439, 225–228 [DOI] [PubMed] [Google Scholar]
- 22. Tang M., Pham P., Shen X., Taylor J. S., O'Donnell M., Woodgate R., Goodman M. F. (2000) Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature 404, 1014–1018 [DOI] [PubMed] [Google Scholar]
- 23. Becherel O. J., Fuchs R. P. (2001) Mechanism of DNA polymerase II-mediated frameshift mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 98, 8566–8571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paz-Elizur T., Takeshita M., Goodman M., O'Donnell M., Livneh Z. (1996) Mechanism of translesion DNA synthesis by DNA polymerase II. Comparison to DNA polymerases I and III core. J. Biol. Chem. 271, 24662–24669 [DOI] [PubMed] [Google Scholar]
- 25. Wang F., Yang W. (2009) Structural insight into translesion synthesis by DNA Pol II. Cell 139, 1279–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gon S., Napolitano R., Rocha W., Coulon S., Fuchs R. P. (2011) Increase in dNTP pool size during the DNA damage response plays a key role in spontaneous and induced-mutagenesis in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 108, 19311–19316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yeeles J. T., Marians K. J. (2013) Dynamics of leading-strand lesion skipping by the replisome. Mol. Cell 52, 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hiasa H., Marians K. J. (1994) Primase couples leading- and lagging-strand DNA synthesis from oriC. J. Biol. Chem. 269, 6058–6063 [PubMed] [Google Scholar]
- 29. Wickner W., Schekman R., Geider K., Kornberg A. (1973) A new form of DNA polymerase 3 and a copolymerase replicate a long, single-stranded primer-template. Proc. Natl. Acad. Sci. U.S.A. 70, 1764–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hill T. M., Marians K. J. (1990) Escherichia coli Tus protein acts to arrest the progression of DNA replication forks in vitro. Proc. Natl. Acad. Sci. U.S.A. 87, 2481–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cai H., Yu H., McEntee K., Kunkel T. A., Goodman M. F. (1995) Purification and properties of wild-type and exonuclease-deficient DNA polymerase II from Escherichia coli. J. Biol. Chem. 270, 15327–15335 [DOI] [PubMed] [Google Scholar]
- 32. Zechner E. L., Wu C. A., Marians K. J. (1992) Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. II. Frequency of primer synthesis and efficiency of primer utilization control Okazaki fragment size. J. Biol. Chem. 267, 4045–4053 [PubMed] [Google Scholar]
- 33. Wu C. A., Zechner E. L., Marians K. J. (1992) Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork: I. multiple effectors act to modulate Okazaki fragment size. J. Biol. Chem. 267, 4030–4044 [PubMed] [Google Scholar]
- 34. Ling H., Boudsocq F., Woodgate R., Yang W. (2001) Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell 107, 91–102 [DOI] [PubMed] [Google Scholar]
- 35. Bunting K. A., Roe S. M., Pearl L. H. (2003) Structural basis for recruitment of translesion DNA polymerase Pol IV/DinB to the beta-clamp. EMBO J. 22, 5883–5892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wagner J., Etienne H., Fuchs R. P., Cordonnier A., Burnouf D. (2009) Distinct beta-clamp interactions govern the activities of the Y family Pol IV DNA polymerase. Mol. Microbiol. 74, 1143–1151 [DOI] [PubMed] [Google Scholar]
- 37. Ippoliti P. J., Delateur N. A., Jones K. M., Beuning P. J. (2012) Multiple strategies for translesion synthesis in bacteria. Cells 1, 799–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pagès V., Fuchs R. P. (2003) Uncoupling of leading- and lagging-strand DNA replication during lesion bypass in vivo. Science 300, 1300–1303 [DOI] [PubMed] [Google Scholar]
- 39. Ikeda M., Furukohri A., Philippin G., Loechler E., Akiyama M. T., Katayama T., Fuchs R. P., Maki H. (2014) DNA polymerase IV mediates efficient and quick recovery of replication forks stalled at N2-dG adducts. Nucleic Acids Res. 42, 8461–8472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kath J. E., Jergic S., Heltzel J. M., Jacob D. T., Dixon N. E., Sutton M. D., Walker G. C., Loparo J. J. (2014) Polymerase exchange on single DNA molecules reveals processivity clamp control of translesion synthesis. Proc. Natl. Acad. Sci. U.S.A. 111, 7647–7652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wagner J., Fujii S., Gruz P., Nohmi T., Fuchs R. P. (2000) The beta clamp targets DNA polymerase IV to DNA and strongly increases its processivity. EMBO Rep. 1, 484–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bonner C. A., Stukenberg P. T., Rajagopalan M., Eritja R., O'Donnell M., McEntee K., Echols H., Goodman M. F. (1992) Processive DNA synthesis by DNA polymerase II mediated by DNA polymerase III accessory proteins. J. Biol. Chem. 267, 11431–11438 [PubMed] [Google Scholar]
- 43. Indiani C., McInerney P., Georgescu R., Goodman M. F., O'Donnell M. (2005) A sliding-clamp toolbelt binds high- and low-fidelity DNA polymerases simultaneously. Mol. Cell 19, 805–815 [DOI] [PubMed] [Google Scholar]
- 44. Heltzel J. M., Maul R. W., Scouten Ponticelli S. K., Sutton M. D. (2009) A model for DNA polymerase switching involving a single cleft and the rim of the sliding clamp. Proc. Natl. Acad. Sci. U.S.A. 106, 12664–12669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Banach-Orlowska M., Fijalkowska I. J., Schaaper R. M., Jonczyk P. (2005) DNA polymerase II as a fidelity factor in chromosomal DNA synthesis in Escherichia coli. Mol. Microbiol. 58, 61–70 [DOI] [PubMed] [Google Scholar]
- 46. Gawel D., Pham P. T., Fijalkowska I. J., Jonczyk P., Schaaper R. M. (2008) Role of accessory DNA polymerases in DNA replication in Escherichia coli: analysis of the dnaX36 mutator mutant. J. Bacteriol. 190, 1730–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Georgescu R. E., Kim S.-S., Yurieva O., Kuriyan J., Kong X.-P., O'Donnell M. (2008) Structure of a sliding clamp on DNA. Cell 132, 43–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Heltzel J. M., Maul R. W., Wolff D. W., Sutton M. D. (2012) Escherichia coli DNA polymerase IV (Pol IV), but not Pol II, dynamically switches with a stalled Pol III* replicase. J. Bacteriol. 194, 3589–3600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gupta S., Yeeles J. T., Marians K. J. (2014) Regression of replication forks stalled by leading-strand template damage II: RecA-catalyzed regression is inhibitied by SSB. J. Biol. Chem. 289, 28388–28398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gupta S., Yeeles J. T., Marians K. J. (2014) Regression of replication forks stalled by leading-strand template damage: I. both RecG and RuvAB catalyze regression, but RuvC cleaves the holliday junctions formed by RecG preferentially. J. Biol. Chem. 289, 28376–28387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fuchs R. P., Fujii S., Wagner J. (2004) Properties and functions of Escherichia coli: Pol IV and Pol V. Adv. Protein Chem. 69, 229–264 [DOI] [PubMed] [Google Scholar]