Background: Germ cells exclusively express PIWI-interacting small RNAs for transposon and gene regulation.
Results: Somatic cells express similar RNAs that do not require known small RNA proteins and that partially complement mRNAs.
Conclusion: These somatic small RNAs represent a novel small RNA population, which potentially regulates mRNA translation.
Significance: Defining novel small RNAs is essential for elucidating the mechanisms that control gene expression.
Keywords: Gene Regulation, RNA, RNA Interference (RNAi), Somatic Cell Genetics, Transposable Element (TE)
Abstract
PIWI-interacting RNAs (piRNAs) are small noncoding RNAs that bind PIWI family proteins exclusively expressed in the germ cells of mammalian gonads. MIWI2-associated piRNAs are essential for silencing transposons during primordial germ cell development, and MIWI-bound piRNAs are required for normal spermatogenesis during adulthood in mice. Although piRNAs have long been regarded as germ cell-specific, increasing lines of evidence suggest that somatic cells also express piRNA-like RNAs (pilRNAs). Here, we report the detection of abundant pilRNAs in somatic cells, which are similar to MIWI-associated piRNAs mainly expressed in pachytene spermatocytes and round spermatids in the testis. Based on small RNA deep sequencing and quantitative PCR analyses, pilRNA expression is dynamic and displays tissue specificity. Although pilRNAs are similar to pachytene piRNAs in both size and genomic origins, they have a distinct ping-pong signature. Furthermore, pilRNA biogenesis appears to utilize a yet to be identified pathway, which is different from all currently known small RNA biogenetic pathways. In addition, pilRNAs appear to preferentially target the 3′-UTRs of mRNAs in a partially complementary manner. Our data suggest that pilRNAs, as an integral component of the small RNA transcriptome in somatic cell lineages, represent a distinct population of small RNAs that may have functions similar to germ cell piRNAs.
Introduction
PIWI-interacting RNAs (piRNAs)2 were discovered by virtue of their ability to bind to PIWI proteins, which are exclusively expressed in germ cells (1–5). Therefore, piRNAs have long been regarded as germ cell-specific small noncoding RNAs (1, 3, 5–8). Two types of piRNAs have been identified based on their genomic origins: repetitive sequence-derived piRNAs and piRNAs derived from nonrepetitive sequences including intragenic, intergenic, or other nonrepetitive regions of the genome (9). piRNAs derived from repetitive sequences are processed through the “ping-pong” mechanism and function to silence transposons by DNA methylation (9, 10). The biogenesis and function of nonrepetitive sequence-derived piRNAs remain largely unknown, although some reports suggest that these piRNAs regulate gene expression at either transcriptional or post-transcriptional levels (1–5, 9, 10).
During male germ cell development, repetitive sequence-derived piRNAs are mainly expressed in primordial germ cells in fetal testes, whereas nonrepetitive sequence-derived piRNAs are predominantly expressed in pachytene spermatocytes and round spermatids in adult testes (9). Therefore, these two types of piRNAs are also called prepachytene and pachytene piRNAs, respectively. Prepachytene piRNAs are produced through the ping-pong mechanism and consist of repetitive sequence-derived primary and secondary transcripts, the production of which involves both MILI (enriched in primary transcripts) and MIWI2 (enriched in secondary transcripts) (6, 9, 11). On the one hand, the ping-pong positive feedback loop inhibits transposon expression through post-transcriptional cleavage of transposon transcripts (11). On the other hand, these transposon-derived piRNAs induce transposon silencing through piRNA-dependent DNA methylation (9). In contrast, pachytene piRNAs associate with MIWI and MILI, creating two subcategories of piRNAs: MILI-associated and MIWI-associated piRNAs (12). These piRNAs do not show ping-pong characteristics such as secondary transcripts that are partially complementary to primary transcripts. Instead, pachytene piRNAs represent largely, if not exclusively, primary transcripts (12).
We and others have reported the existence of piRNA-like RNAs (pilRNAs) in both gonadal and somatic tissues (13–16). However, a detailed characterization of this novel class of small RNAs expressed in somatic cells/tissues has been lacking. Here, we report the identification of hundreds of murine pilRNAs from small RNA libraries of murine testicular Sertoli cells, interstitial cells of Cajal (ICCs) from the murine gastrointestinal tract, and murine small intestine, in addition to millions of murine stomach small RNAs that align to newly defined germ cell piRNA clusters. Our in-depth analyses revealed a high degree of similarities between these somatic pilRNAs and pachytene piRNAs in size, nucleotide composition, clustering, genomic origin, and potential targets. However, unlike pachytene piRNAs, pilRNAs have a distinct ping-pong signature. Finally, biogenesis of pilRNAs is mediated by an unidentified mechanism different from any of the currently known small RNA biogenetic pathways indicative of an entirely novel population of small RNAs.
EXPERIMENTAL PROCEDURES
Knock-out Mouse Lines Used
All animal work was conducted following the animal use protocol approved by the Institutional Animal Care and Use Committee of the University of Nevada, Reno. For FACS-based Sertoli cell purification, control (Amh-Cre; Rosa26-mTmG+/tg), Sertoli cell-specific Dicer conditional knock-out (cKO) (Amh-Cre; Dicerlox/lox; Rosa26-mTmG+/tg), and Sertoli cell-specific Drosha cKO (Amh-Cre; Droshalox/lox; Rosa26-mTmG+/tg) mice were generated as described (16–18). Miwi universal KO mouse line was obtained from the Mutant Mouse Regional Resource Centers. Mov10l1 universal knock-out mice were purchased from the Jackson Laboratory. Miwi2 knock-out mice were generated as described (19).
Tissue Collection and Cell Purification
Tissue samples (brain, colon, heart, kidney, liver, lung, small intestine, spleen, stomach, and testis) were collected from adult wild-type or knock-out C57/BL6 mice. Upon collection, samples were immediately snap frozen in liquid nitrogen followed by storage at −80 °C until RNA isolation. Wild-type Sertoli cells were collected from postnatal day 6 mice as previously reported (16). Amh-cre control and cKO Sertoli cells from 2-day-old mice were isolated by FACS based on GFP expression from the mTmG reporter (20). ICCs were purified from adult mice using FACS as described (21).
qPCR Analyses of Small Noncoding RNAs Expression
Small noncoding RNAs (sncRNAs) were isolated from purified Sertoli cells, ICCs, and small intestine, and sncRNA cDNAs were prepared as previously described (22). qPCR analyses were performed using the 7900HT Fast Real-Time PCR System (Applied Biosystems). A TaqMan-based method was implemented using TaqMan probe (5′-FAM-CTCGGATCCACTAGTC-MGB-3′ (where FAM is 6-carboxyfluorescein and MGB is minor groove binder)) and U6 snRNA as an endogenous control (23). The levels in all somatic tissues were normalized to those in testis and knock-out samples to respective wild-type control samples. Primers for sncRNA qPCR are included in Table 1.
TABLE 1.
Primers used for small RNA qPCR
| sncRNA | ID | Primer |
|---|---|---|
| Endogenous control | U6 | GCAAATTCGTGAAGCGTTCC |
| Known piRNA | piRNA-118029 | GATGGTAGTAGCTGAAGTTCCTTCG |
| Known piRNA | piRNA-126541 | TAGATAGTGAGGACTTGACTCTAGTG |
| Novel piRNA-like | pilRNA-in3 | TGAACTGAATGCAAAGGGGGACAC |
| Novel piRNA-like | pilRNA-in18 | AGCGCCGCTGGTGTAGTGGTATCATG |
| Novel piRNA-like | pilRNA-in87 | ATCTGTGAAATGAGGGAGTTGGACTAGAG |
| NAa | Adapter reverse | CGAATTCTAGAGCTCGAGGCAGG |
a NA, not applicable.
Small RNA Sequencing
Sequencing libraries prepared using sncRNAs from Sertoli cells, ICCs, and small intestine were sequenced on the 454 GS20 platform (454 Life Sciences, Roche, Branford, CT) as previously reported (16). Sequencing data has been deposited to the NCBI Gene Expression Omnibus database with an accession number of GSE48388 (24). 454 Sertoli pilRNA annotations were previously published and can be found with accession number GSE40692 (16).
Two biological replicates of FACS-purified Sertoli cells from control, Dicer cKO, and Drosha cKO testes (n = 2) were sequenced using the Ion Torrent Personal Genome Machine (PGM) (Invitrogen) as described (17, 18). Briefly, sncRNAs were isolated using the mirVana miRNA isolation kit (Invitrogen), and cDNA libraries were generated using the Ion Total RNA-Seq kit v2 (Invitrogen). Emulsion PCR was carried out on the Ion OneTouch (Invitrogen) using the Ion OneTouch 200 template kit v2 (Invitrogen) for cDNA amplification on ion sphere particles. Samples were enriched for positive spheres using the Ion OneTouch enrichment system (Invitrogen). After enrichment, sequencing was performed on the PGM using the Ion PGM 200 sequencing kit (Invitrogen) and Ion 316 chips (Invitrogen). Stomach samples (n = 2) were sequenced in a similar fashion with the exception of 318 chips being used in lieu of 316 chips. Stomach data were submitted to the NCBI Gene Expression Omnibus under accession number GSE53780 (24).
pilRNA Annotation
For 454 sequencing reads, annotation analyses were conducted as described (16). Briefly, alignment to known small RNA libraries (miRBase, the functional RNA database (2010/10/04 16:15:05 GMT+9), the noncoding RNA database (v2.0), the Genomic tRNA database (downloaded January 5, 2011), and Repbase (v14.10)) allowing for up to two mismatches was performed with Sequery v1.0 (16, 26–30). Remaining unmatched sequences were aligned to the mouse genome build mm9 with a perfect match requirement in Sequery v1.0 (31). Sequences that aligned to the genome were categorized as novel endo-siRNA, miRNA, snoRNA, and pilRNA based on length, nucleotide composition, and secondary structures using Sequery v1.0, MIREAP 0.2, and snoREPORT 1.0 (32, 33). Remaining sequences that aligned to the genome but were not categorized were kept as unidentifiable small RNAs.
Sequence reads from Ion Torrent sequencing of Sertoli cell sncRNA libraries were aligned to the mouse genome (build mm9) using the Ion Torrent Server (Suite 2.2) t-map aligner (Invitrogen) (31). Known piRNA-Seq data analysis was performed using Partek Genomic Suite (version 6.6; Partek, St. Louis, MO). Aligned reads were clustered into at least 15 nucleotide groups including overlapping reads. The resulting groups were aligned to mature piRNA sequences from the functional RNA database (2010/10/04 16:15:05 GMT+9), which contains a collection of small RNAs found associated with mouse PIWI proteins (30). Resulting alignments to known germ cell piRNAs were classified as pilRNAs. To account for sequencing depth variation, the abundance of each pilRNA was normalized to the total aligned reads obtained from each sample, multiplied by 1 × 106, and averaged between biological replicates. pilRNAs with ≤1 read in all six libraries were removed from analysis.
Resulting reads from PGM sequencing of wild-type stomach samples were mapped to mm9 using Ion Torrent Server Suite 3.2.1 t-map aligner (Invitrogen) (31). Recently redefined piRNA producing transcript genomic loci were obtained and primary miRNA locations (miRBase release 18) were omitted from these clusters to avoid contaminating miRNA reads (28, 34). In-house python scripts were used to ascertain reads mapped within piRNA clusters and their corresponding nucleotide lengths and composition.
Precursor Transcript RT-PCR
Large RNA from adult wild-type mouse brain and testis was isolated using the mirVana miRNA isolation kit (Invitrogen). RNA was treated with either DNaseI amplification grade (Invitrogen) or RNase A (Sigma-Aldrich). DNaseI treatment was performed according to the manufacturer's recommendations. RNase A treatment was performed at 37 °C for 60 min with 0.5 μg of RNase A in 1X DNaseI reaction buffer, followed by the same reaction conditions used for the DNaseI treatment (without the DNase). Using 1 μg of treated RNA, reverse transcription was performed using the SuperScript III first-strand synthesis system kit with random hexamers (Invitrogen). A reaction volume of 20 μl was used, consisting of 10 μl of 2× GoTaq (Promega), 10 pmol of forward and reverse primer, and 1 μl of cDNA. PCR conditions were as follows: 1 cycle at 95 °C/2min, 28 cycles of amplification (95 °C for 30 s, 58 °C for 30 s, and 72 °C for 45 s), followed by 1 cycle of 72 °C for 5 min. PCR product was run on a 2% agarose gel with the 100-bp DNA ladder H3 (Caisson Labs). Primers and anticipated amplicon sizes for the precursor transcript RT-PCR are included in Table 2.
TABLE 2.
Primers used for precursor transcript RT-PCR
| Cluster | Primer | Primer sequence | Expected amplicon size |
|---|---|---|---|
| bp | |||
| 2-qE1-35981 | Primer 1, forward | CTTGTGTCCTGTGGTTTGGAT | 580 |
| 2-qE1-35981 | Primer 1, reverse | ATGTGAAAATCCATTTGTCTCTGG | |
| 2-qE1-35981 | Primer 2, forward | CCTCTCTTTGGGCTTGCAAGA | 628 |
| 2-qE1-35981 | Primer 2, reverse | AAAAGACAAGTGTTGGGACATCTG | |
| 9-qC-31469 | Primer 1, forward | CATGCTGACATCACTATAAGGA | 721 |
| 9-qC-31469 | Primer 1, reverse | TCCAACATTGTGCACAGATTCT | |
| 9-qC-31469 | Primer 2, forward | AAGGTGGGCTGGCTGAACC | 983 |
| 9-qC-31469 | Primer 2, reverse | GCTCTTGCAGGGTTTTAAGATCA | |
| 17-qA3.3-27363 | Primer 1, forward | CACGTGTCAAGCGGAGACTAG | 793 |
| 17-qA3.3-27363 | Primer 1, reverse | GTCTCTTTGTCTGGAGAGGGG | |
| 17-qA3.3-27363 | Primer 2, forward | TGTTTTCACTATTTTGTGGCTGCG | 696 |
| 17-qA3.3-27363 | Primer 2, reverse | CAGGGAGGAGAGGGCACTT | |
| 17-qA3.3-26735 | Primer 1, forward | TGGTCTAAGCCAGGTGGTTGA | 919 |
| 17-qA3.3-26735 | Primer 1, reverse | AACCCATCACGTATTCCCTAA | |
| 17-qA3.3-26735 | Primer 2, forward | GACACCAGCTGAGACCCTCA | 695 |
| 17-qA3.3-26735 | Primer 2, reverse | GTTCTTCCTATCACCTCGCACT |
pilRNA Post-Transcription Target Analysis
Annotated pilRNA sequences were reverse complement aligned to all known murine 3′-UTRs, 5′-UTRs, and coding sequences (CDSs) using Sequery v1.0 (16). 3′-UTRs, 5′-UTRs, and CDSs were collected from ENSEMBL release 71 (35). Reverse complement analysis was first performed allowing up to two mismatches and then repeated allowing up to five mismatches with no gaps allowed. Gene features aligned to were considered potential post-transcriptional targets of pilRNAs.
Ping-Pong Analysis
Reads were subjected to a pairwise analysis of 5′ ends with a 10- or 11-nt overlap using Sequery v1.0 (16). Reads that reverse complemented at their 5′ ends with a 10-nt overlap were considered ping-pong pilRNAs.
RESULTS
Identification of pilRNAs in Somatic Cells/Tissues
We initially searched for pilRNAs from sequence reads of three small noncoding RNA (sncRNA) libraries prepared using murine Sertoli cells purified from postnatal day 6 testes, ICCs of the murine gastrointestinal tract, and small intestine based on criteria described previously (16). Among 696 nonredundant pilRNAs identified, 117 were from the ICC sncRNA library, 263 were found in Sertoli cells, and 316 were found in the small intestine; ∼40–60% of these pilRNAs have sequences identical to known germ cell piRNAs. The abundance of pilRNAs varied among the three somatic cell sncRNA libraries, with relatively more pilRNAs identified in ICCs than in small intestine or Sertoli cells (Fig. 1A and supplemental Tables S1–S4). The size of germ cell piRNAs ranges from 24 to 32 nt with MILI-, MIWI2-, and MIWI-associated piRNAs being ∼26, 28, and 30 nt long, respectively (6). The lengths of identified pilRNAs ranged from 20 to 32 nt with the majority at 30 nt (Fig. 1B), which is similar to the major size of pachytene/MIWI-associated piRNAs.
FIGURE 1.
Abundance and size of pilRNAs in Sertoli cells, ICCs, small intestine (Sm Int), and stomach. A, abundance of pilRNAs in small noncoding RNA 454 libraries of murine Sertoli cells purified from postnatal day 6 testes, ICCs purified from the gastrointestinal tracts of adult mice, and small intestine from adult mice. B, length distribution of all pilRNAs identified from the three 454 sncRNA libraries of somatic cells/tissue. The dominant size is at 30 nt, which is consistent with that of MIWI-associated/pachytene piRNAs. C, abundance of pilRNAs in small noncoding RNA PGM libraries of murine adult stomach. D, length distribution of all pilRNAs identified from stomach PGM sncRNA libraries. The dominant size is at 30 nt, which is consistent with that of MIWI-associated/pachytene piRNAs. E, scatter plots depicting log2(normalized value + 1) expression between biological replicates from adult mice stomach small RNA. R2 values were at least 0.98, indicating good correlation between biological replicates. Samples had an average depth of 4.4 × 106 aligned reads.
Recent data redefined murine genomic loci of piRNA-producing transcripts using a series of sequencing techniques including small RNA-Seq, sequencing of samples enriched for piRNAs based on 2′-O-methyl-modified 3′ termini, cap analysis of gene expression sequencing, and polyadenylation site sequencing (34). These techniques clarified the termini of transcripts that produced piRNAs and were combined with temporal analyses allowing for the classification of piRNA clusters as prepachytene, hybrid, or pachytene piRNAs (34). Using these redefined clusters and our recent stomach small RNA deep sequencing data, we were able to map ∼1.6 million stomach sncRNA reads to these germ cell piRNA clusters (Fig. 1C and supplemental Table S5). This pilRNA population was the largest classifiable population, taking up 18% of the stomach small RNA sequencing reads. Coinciding with our initial pilRNA results, stomach pilRNAs aligned to the redefined germ cell piRNA clusters were predominantly 30 nt in length, similar to the pachytene/MIWI-associated piRNAs (Fig. 1D). Stomach replicates had a R2 of 0.98, showing good correlation and corroboration of the stomach pilRNA results (Fig. 1E).
Validation of pilRNA Expression in Somatic Tissues
We verified the expression of three novel pilRNAs identified from murine small intestine (piR-in3, piR-in18, and piR-in87) in 10 different tissues using qPCR (Fig. 2). As expected, the two germ cell-expressed piRNAs (piR-118029 and piR-126541) were predominantly detected in the testis. Interestingly, all three small intestine pilRNAs also showed higher expression in the testis than in most of the somatic tissues analyzed. The highest levels of pilRNA-in3 were detected in brain, whereas pilRNA-in18 was most abundant in heart. The top three organs expressing pilRNA-in87 included testis, brain, and heart (Fig. 2).
FIGURE 2.

Validation of pilRNA expression in multiple murine tissues. Levels of three pilRNAs (pilRNA-in3, pilRNA-in18, and pilRNA-in87) identified from the small intestine sncRNA libraries and two germ cell piRNAs (piR-118029 and piR-126541) were determined using qPCR in 10 tissues including nine somatic tissues and the testis. U6 snRNA was used as an endogenous control, and all somatic tissue values are relative to levels of testis expression.
pilRNAs Resemble Pachytene piRNAs
Repeat-associated piRNAs are produced through the ping-pong mechanism involving MILI and MIWI2 in mice (6, 9, 11). These piRNAs include both primary (contain 5′ uracil) and secondary (contain 10th position adenine) piRNA transcripts, and there is a consistent 10-nt complementary overlap between primary and secondary transcripts (11). piRNAs produced through the ping-pong mechanism tend to display an accumulation of secondary piRNAs, which contain an adenine at the 10th nt position (6). We analyzed the pilRNA sequences and found that the 10th nt preference for adenine was absent. In addition, a strong preference for the signature 5′ uracil of primary piRNA transcripts was detected in over 50% of pilRNAs (Fig. 3A). Stomach pilRNAs aligned to redefined germ cell piRNA loci confirmed this data with most reads preferentially containing a 5′ uracil opposed to the 10th nt adenine (Fig. 3B). The nucleotide preferences of novel pilRNAs and pilRNAs that align to known piRNA sequences were investigated separately in ICCs, Sertoli cells, and small intestine (Fig. 4, A–C, respectively). A bias toward a 5′ uracil and a lack of a 10th nt adenine preference was observed in both novel pilRNAs and pilRNAs that align to known piRNA sequences. Together, these data suggest that pilRNAs resemble pachytene piRNAs and are predominantly primary pilRNAs.
FIGURE 3.
Somatic pilRNAs resemble germ cell pachytene piRNAs. A, somatic cell-expressed pilRNAs identified by 454 sequencing display a preference for 5′ uracil but lack enrichment of an adenine at the 10th position. B, PGM sequenced stomach pilRNAs display a preference for 5′ uracil but lack enrichment of an adenine at the 10th position. C, clustering analyses of pilRNAs by aligning pilRNAs to known prepachytene and pachytene piRNA clusters. The number of pilRNAs found within one or both of the two known types of piRNA clusters is indicated. The majority of pilRNAs appear to belong to pachytene piRNA clusters. D, clustering analyses of pilRNAs by aligning pilRNAs to recently redefined prepachytene, hybrid, and pachytene piRNA clusters. The number of pilRNAs found within each category of piRNA clusters is indicated. The majority of stomach pilRNAs belong to pachytene piRNA clusters. Nt, nucleotide; Sm Int, small intestine.
FIGURE 4.

Nucleotide preference of pilRNAs that align to known piRNA and novel pilRNAs. A, ICC pilRNAs show a 5′ uracil preference and no 10th nt preference in both groups. B, Sertoli pilRNAs that align known piRNAs show a 5′ uracil preference for both groups but less so in novel pilRNAs. Both groups show no 10th nt preference. C, small intestine pilRNAs show a 5′ uracil preference and no 10th nt preference in both groups. Nt, nucleotide; Sm Int, small intestine.
pilRNAs Tend to Cluster with Pachytene piRNAs
Clustering is a key characteristic of piRNAs. However, lone piRNAs derived from unique, unclustered genomic locations have been found in large quantities as well (12, 14). To properly classify the pilRNAs identified, we collected genomic coordinates containing murine prepachytene and pachytene piRNA clusters, and pilRNAs were aligned and categorized within these clusters (9, 12). 60 of 117 ICC pilRNAs (∼51%) were found to cluster with pachytene piRNA clusters, whereas none belonged to prepachytene piRNA clusters (Fig. 3C). Although 3 of 263 Sertoli cell pilRNAs were found to belong to prepachytene piRNA clusters, these 3, together with 31 additional Sertoli pilRNAs, also fell into pachytene piRNA clusters (Fig. 3C). Similarly, 117 of 316 small intestine pilRNAs (∼37%) belonged to pachytene piRNA clusters, with only 2 also found in prepachytene piRNA clusters (Fig. 3C). The preferential clustering with pachytene piRNAs again supports the notion that pilRNAs are more like pachytene piRNAs. We confirmed that these results with stomach reads aligned to the redefined germ cell piRNA clusters. Of ∼1.6 million, reads only 7,850 aligned to prepachytene clusters and 27,945 to hybrid clusters, whereas 98% (1.54 million) of the stomach pilRNAs aligned to pachytene clusters (Fig. 3D). Recent data have demonstrated that pachytene piRNA clusters are predominantly intergenic (34). This was found to be true for stomach pilRNAs as well, with an average pilRNA density of 15,800 pilRNA per intergenic cluster in contrast to 422 per nonintergenic cluster (supplemental Table S5).
pilRNA Clusters Are Similar to Pachytene piRNA Clusters
Like pachytene piRNAs, somatic pilRNAs predominantly originate from intergenic genomic loci. Fig. 5A shows an example of two intergenic bidirectional pachytene piRNA clusters (data from NCBI Gene Expression Omnibus accession number GSM1096603) (34). The corresponding expression of stomach pilRNAs is similar to that seen in the pachytene spermatocytes within these clusters. In addition, the stomach pilRNAs that originate from these clusters are largely 30 nt in length and have a preference for 5′ uracil similar to pachytene piRNAs (Fig. 5, B and C). RT-PCR detected the presence of several precursor transcripts in both the stomach and testis, suggesting that these clusters are also transcribed in somatic cells (Fig. 5D).
FIGURE 5.
Examples of stomach pilRNA clusters aligned to germ cell pachytene spermatocyte piRNA clusters. A, genomic view of pachytene spermatocyte and stomach reads aligned to intergenic bidirectional clusters 17-qA3.3–27363 and 17-qA3.3–26735. B, length histogram of stomach pilRNAs originating from clusters 17-qA3.3–27363 and 17-qA3.3–26735. Predominant size of stomach reads aligned to these two clusters is 30 nt, consistent with pachytene piRNAs. C, nucleotide composition of stomach pilRNAs originating from clusters 17-qA3.3–27363 and 17-qA3.3–26735. Stomach pilRNAs have a preference for 5′ uracil indicative of primary processing. D, precursor transcript RT-PCR. Precursor transcripts for germ cell piRNA clusters 2-qE1–35981, 9-qC-31469, 17-qA3.3–27363, and 17-qA3.3–26735 were detected in both stomach (st columns) and testis (tes columns) by two primer sets (Table 2). RNA was either treated with DNase (D columns) or RNase (R columns) as a negative control. All amplicons matched to their anticipated sizes (Table 2).
Although pilRNAs Are Similar to Pachytene piRNAs, pilRNAs Exhibit Ping-Pong Activity
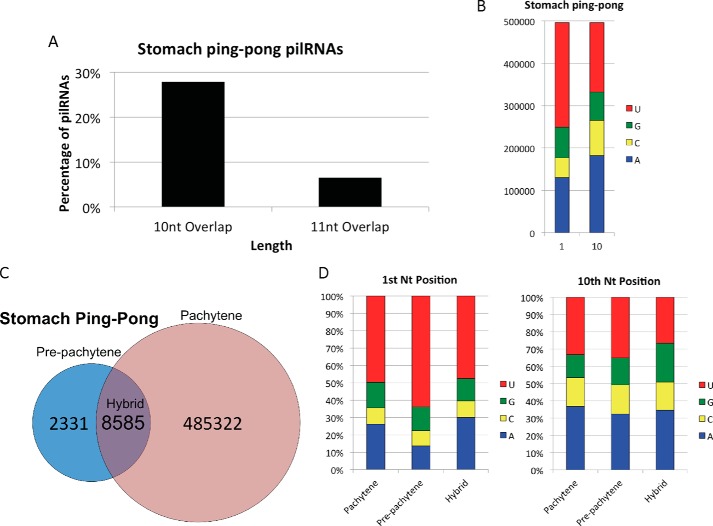
To evaluate whether pilRNAs are in fact products of the ping-pong mechanism, we performed pairwise analyses of the 5′ end of pilRNA pairs. In the small intestine sncRNA library, one single ping-pong match was identified between piR-141849 and pilRNA-in87, which, in addition to having a 10-nt overlap, also had the signature 5′ uracil and 10th nt adenine. The ICC and Sertoli cells did not have any ping-pong signature detected. In contrast, the stomach had a large fraction (28%) of pilRNAs that had the canonical 10-nt overlap at their 5′ ends (Fig. 6A). A similar analysis looking for 11-nt overlaps had markedly reduced alignments (7%), indicating that a 10-nt overlap of 5′ ends is indeed prominent in the stomach pilRNAs. The stomach pilRNAs that generate pairs with a 10-nt overlap also have a preference for a 5′ uracil, as well as the 10th nt adenine indicative of ping-pong activity (Fig. 6B). These ping-pong pilRNAs predominantly align to pachytene clusters (Fig. 6C). In addition, the presence of secondary transcripts with 10th nt adenines do not seem to be dependent on the type of cluster the pilRNA is derived from (Fig. 6D). In fact all ping-pong pilRNAs, regardless of the cluster they originate from, have similar nucleotide profiles.
FIGURE 6.
Although pilRNAs are predominantly like germ cell pachytene piRNAs, pilRNAs still have ping-pong activity. A, results of pairwise alignments of 5′ 10- and 11-nucleotide overlaps in stomach pilRNAs. B, nucleotide composition of ping-pong stomach pilRNAs with 10-nucleotide 5′ overlaps. C, cluster alignments of ping-pong stomach pilRNAs with ping-pong signature. D, nucleotide composition of ping-pong stomach pilRNAs based on cluster classification.
pilRNA Biogenesis Is Independent of Known miRNA or piRNA Biogenetic Pathways
DICER and DROSHA are two RNase III enzymes essential for the biogenesis of the best-studied small RNAs, miRNAs (36, 37). In addition, DICER is also required for the production of endo-siRNAs (37). Because of their necessity in miRNA and/or endo-siRNA production, we examined the effects of DICER and DROSHA depletion on pilRNA expression in murine Sertoli cells. Using Sertoli cells purified from Sertoli cell-specific Dicer (Amh-Cre; Dicerlox/lox; Rosa26-mTmG+/tg) and Drosha (Amh-Cre; Droshalox/lox; Rosa26-mTmG+/tg) conditional knock-out testes, we performed sncRNA deep sequencing (sncRNA-Seq) (17, 38). Sequencing reads aligned to known piRNAs were used for differential pilRNA expression analyses. Although levels of Sertoli cell pilRNAs were slightly altered, almost all pilRNAs were still present at similar levels to control, as shown in the heat map representing the pilRNA transcriptomes of control, Dicer- and Drosha-null Sertoli cells (Fig. 7, A and B, and supplemental Table S6). The persistent expression of pilRNAs in the absence of either DICER or DROSHA suggests that pilRNA production is independent of the two proteins. To see whether pilRNA production is dependent on piRNA biogenetic pathways, we examined the expression of two pilRNAs similar to known piRNAs and three novel pilRNAs in 10 organs of MIWI, MIWI2, and MOV10L1 universal KO mice (Fig. 7C and supplemental Table S7). As expected, the two known piRNAs showed minimal or no expression in the testes of the three KO lines. The three novel pilRNAs were also detected in testes but consistently decreased in the knock-outs. In comparison with the somatic tissues, testis samples had the largest decrease in expression between KOs and controls. This is possibly due to the decrease in germ cell content in KO testis, a phenomenon observed previously in MIWI2 KO lines (19). Despite the fact that there was some differential pilRNA expression between control and KO lines in all somatic tissues, pilRNAs were still detected at variable levels, suggesting that their biogenesis in somatic cells does not rely on the known piRNA biogenetic pathways.
FIGURE 7.
pilRNA production is independent of the known miRNA/endo-siRNA or piRNA biogenetic pathways. A, heat map representing the pilRNA transcriptome in wild-type, Dicer- or Drosha-null murine Sertoli cells based on small RNA sequencing. Expression is based on a Log10 scale. B, scatter plots depicting log2(normalized value) expression between replicates from FACS-purified Sertoli cells of Amh-cre control, Dicer conditional knock-out, and Drosha conditional knock-out mice. R2 values were at least 0.82, indicating relatively good correlation between replicates. Samples had an average depth of ∼450,000 aligned reads. C, heat maps representing qPCR analyses of expression levels of three intestinal pilRNAs (pilRNA-in3, pilRNA-in18, and pilRNA-in87) and two known germ cell piRNAs (piR-118029 and piR-126541) in 10 organs of MIWI, MIWI2, and MOV10L1 global KO mice. U6 snRNA was used as an endogenous control, and values in KO samples were relative to respective values of wild-type samples.
pilRNAs Can Target 3′-UTRs by Partial Annealing
Post-transcriptional regulation is one of the most important functions of small RNAs, e.g. miRNAs and endo-siRNAs (39–41). miRNAs recognize their targets by partial complementary annealing to their target sequences mostly located in 3′-UTRs of mRNAs, whereas endo-siRNAs tend to bind to 3′-UTRs in a perfect complementary manner (40–42). If pilRNAs also function as a post-transcriptional regulator in a manner similar to that of miRNAs and endo-siRNAs, we would expect to see that these pilRNAs could bind mRNAs, especially 3′-UTRs. We therefore performed bioinformatic analyses by aligning ICC, Sertoli, and small intestine pilRNAs in a reverse complementary manner to all known mouse 3′-UTRs, 5′-UTRs, and CDSs (35). We first allowed a 1–2-nt mismatch and found a small number of piRNAs (24 of 263 Sertoli cells and 5 of 316 small intestine pilRNAs) could complementarily anneal to murine mRNAs with the majority (88% Sertoli cells and 80% small intestine) matched to 3′-UTRs (Fig. 8A and supplemental Tables S8 and S9). We then increased the mismatch allowance to 1–5 nt. Interestingly, a drastic increase in alignments was observed in piRNAs of all three tissue-origins, with 12 ICC, 96 Sertoli cells, and 66 small intestine pilRNAs annealed to mRNA sequences (Fig. 8A and supplemental Tables S10–S12). The targeting sites were mainly located in 3′-UTRs, which displayed the highest percentage and were potentially targeted by 67% ICC, 79% Sertoli cells, and 62% small intestine pilRNAs when one to five mismatches were allowed (Fig. 8A). An increase in CDS-targeting pilRNAs was also noted when the mismatch allowance was increased to five, with many of the same pilRNAs annealing to both the CDSs and 3′-UTRs of mRNAs. We examined the number of mRNAs that could potentially be targeted by pilRNAs in a manner of partial complementary annealing, and a preference of potential pilRNA binding sites in 3′-UTRs was observed (Fig. 8B). This was further exemplified by the fact that at least 2.4 times more mRNAs displayed potential 3′-UTR pilRNA binding sites in comparison with those in 5′-UTRs and CDSs combined (Fig. 8B).
FIGURE 8.

Partial complementary alignment of 696 pilRNAs identified in 454 sequencing to all known mRNAs. A, number of pilRNAs potentially targeting 3′-UTRs, 5′-UTRs, and CDSs of all known murine mRNAs. Shown are the results of reverse complementary match of pilRNAs to mRNAs when one to two or one to five mismatches were allowed. B, number of mRNA transcripts potentially targeted within the 3′-UTRs, 5′-UTRs, and CDSs by pilRNAs. Sm Int, small intestine.
DISCUSSION
Given that three PIWI proteins are all expressed exclusively in germ cells in gonads, piRNAs have long been regarded as germ cell-specific (1, 3, 5–8). Several reports have documented the detection of piRNA-like RNAs in somatic cell types (13–16), but systematic characterization of pilRNA structural features, genomic origins, biogenesis, and functions have not been reported. Approximately 6 years ago, we performed sncRNA deep sequencing on two somatic cell types (Sertoli cells purified from the postnatal day 6 testes and interstitial cells of Cajal purified from gastrointestinal tract of adult mice) and somatic murine small intestine tissue using the 454 sequencing platform. We identified a total of 696 pilRNAs from sncRNA libraries of Sertoli cells, ICCs, and small intestine. Our recent Ion Torrent PGM-based sncRNA-Seq analyses on the murine stomach resulted in 1.6 million reads aligned to germ cell piRNA clusters. By analyzing sequence reads from the two types of sncRNA-Seq analyses, we defined the pilRNA transcriptomes in two murine somatic cell types (i.e. Sertoli cell and ICC) and two murine organs (i.e. stomach and small intestine). Interestingly, pilRNAs are very similar to non-repeat-associated intergenic piRNAs expressed abundantly and predominantly in pachytene spermatocytes and round spermatids in adult testes. This notion is supported by the following findings: 1) the major size of pilRNAs is 30 nt, which is closer to that of pachytene piRNAs; 2) pilRNAs largely fall within known pachytene piRNA instead of prepachytene piRNA clusters; and 3) similar to pachytene piRNAs, pilRNAs are mainly derived from intergenic regions (6). In contrast to pachytene piRNAs, which only have ∼2% ping-pong activity, pilRNAs have a definite ping-pong signature with ∼30% of stomach pilRNAs having a 10-nt overlap at their 5′ ends (12). Therefore, we propose that pilRNAs are similar to germ cell pachytene piRNAs but are different in the fact that they have ping-pong activity like prepachytene piRNAs. It remains unknown how pachytene piRNAs are produced and what roles pachytene piRNAs play in spermatogenesis. Elucidation of piRNA biogenesis and functions would help us understand those of pilRNAs, and vice versa.
Although the function of pilRNAs remains elusive, their dynamic, tissue-specific expression patterns strongly suggest that pilRNAs are under strict regulation and thus may play an important role in cellular functions. Given that significantly more hits were observed when one to five mismatches were allowed, pilRNAs may be able to target mRNAs by partial annealing to their binding sites located in 3′-UTRs of mRNAs. In this aspect, pilRNAs or pachytene piRNAs are similar to miRNAs because both pilRNAs/piRNAs and miRNAs can tolerate several mismatches in target recognition. Upon increasing the mismatch allowance from up to two to up to five, an increase in CDS-targeting pilRNAs was also observed. This observation may be the result of CDSs having longer sequences than UTRs, thereby increasing the random chance of a complementary match. Further bioinformatic analyses may reveal the rules that pilRNAs follow to recognize their mRNA targets, and their effects on mRNA fate need to be confirmed experimentally in the future.
Although both DICER and DROSHA are ubiquitously expressed, DROSHA substrates must contain either the stem-loop structure or double-stranded RNA. Because neither pilRNAs nor pachytene piRNAs contain such a structure, it is not surprising to see that inactivation of either Dicer or Drosha, despite changes in pilRNA levels probably caused by secondary effects, does not block pilRNA biogenesis. We know pachytene piRNAs require MIWI and several other proteins including MOV10L1, MITOPLD, and GASZ, which are exclusively expressed in male germ cells (25, 43–45). In addition, our lab has confirmed the lack of MIWI, MIWI2, and MILI expression in several somatic tissues including murine stomach and confirmed the absence of these proteins by Western blot (data not shown). Therefore, it is expected that inactivation of MIWI/MIWI2 or MOV10L1 would not abolish pilRNA production. This was also anticipated, because although pilRNAs are similar to pachytene piRNAs, they do have a key difference in that they have ping-pong mechanism activity. However, given the high degree of similarities between pilRNAs and pachytene piRNAs, similar machinery is likely operating in somatic cells for pilRNA production. Thus, it is critical to identify the proteins responsible for somatic pilRNA production. The elucidation of pilRNA biogenesis will further help define the physiological role of pilRNAs in somatic cell lineages.
In summary, our data confirm that somatic cells indeed express abundant pilRNAs, and these pilRNAs are similar to pachytene piRNAs with the exception of ping-pong activity. pilRNAs are synthesized using a yet to be defined pathway, and pilRNAs may function in a manner similar to miRNAs as post-transcriptional regulators. With more understanding of piRNAs, the biogenesis and function of pilRNAs, the somatic counterparts of germ cell piRNAs, will be elucidated in the near future.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants HD060858, HD071736, and HD074573 (to W. Y.) and COBRE Grant P20-RR18751 (to the University of Nevada Genetic Engineering Center).

This article contains supplemental Tables S1–S12.
- piRNA
- PIWI-interacting RNA
- pilRNA
- piRNA-like small RNA
- ICC
- interstitial cells of Cajal
- cKO
- conditional knock-out
- sncRNA
- small noncoding RNA
- CDS
- coding sequence
- qPCR
- quantitative PCR
- nt
- nucleotide(s)
- PGM
- Personal Genome Machine.
REFERENCES
- 1. Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M. J., Kuramochi-Miyagawa S., Nakano T., Chien M., Russo J. J., Ju J., Sheridan R., Sander C., Zavolan M., Tuschl T. (2006) A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442, 203–207 [DOI] [PubMed] [Google Scholar]
- 2. Girard A., Sachidanandam R., Hannon G. J., Carmell M. A. (2006) A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442, 199–202 [DOI] [PubMed] [Google Scholar]
- 3. Grivna S. T., Beyret E., Wang Z., Lin H. (2006) A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 20, 1709–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lau N. C., Seto A. G., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D. P., Kingston R. E. (2006) Characterization of the piRNA complex from rat testes. Science 313, 363–367 [DOI] [PubMed] [Google Scholar]
- 5. Watanabe T., Takeda A., Tsukiyama T., Mise K., Okuno T., Sasaki H., Minami N., Imai H. (2006) Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 20, 1732–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aravin A. A., Sachidanandam R., Bourc'his D., Schaefer C., Pezic D., Toth K. F., Bestor T., Hannon G. J. (2008) A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 31, 785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deng W., Lin H. (2002) miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell 2, 819–830 [DOI] [PubMed] [Google Scholar]
- 8. Unhavaithaya Y., Hao Y., Beyret E., Yin H., Kuramochi-Miyagawa S., Nakano T., Lin H. (2009) MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J. Biol. Chem. 284, 6507–6519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aravin A. A., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G. J. (2007) Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316, 744–747 [DOI] [PubMed] [Google Scholar]
- 10. Carmell M. A., Girard A., van de Kant H. J., Bourc'his D., Bestor T. H., de Rooij D. G., Hannon G. J. (2007) MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 12, 503–514 [DOI] [PubMed] [Google Scholar]
- 11. Brennecke J., Aravin A. A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G. J. (2007) Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103 [DOI] [PubMed] [Google Scholar]
- 12. Beyret E., Liu N., Lin H. (2012) piRNA biogenesis during adult spermatogenesis in mice is independent of the ping-pong mechanism. Cell Res. 22, 1429–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wei Z., Liu X., Zhang H. (2012) Identification and characterization of piRNA-like small RNAs in the gonad of sea urchin (Strongylocentrotus nudus). Mar. Biotechnol. (NY) 14, 459–467 [DOI] [PubMed] [Google Scholar]
- 14. Yan Z., Hu H. Y., Jiang X., Maierhofer V., Neb E., He L., Hu Y., Hu H., Li N., Chen W., Khaitovich P. (2011) Widespread expression of piRNA-like molecules in somatic tissues. Nucleic Acids Res. 39, 6596–6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ro S., Park C., Song R., Nguyen D., Jin J., Sanders K. M., McCarrey J. R., Yan W. (2007) Cloning and expression profiling of testis-expressed piRNA-like RNAs. RNA 13, 1693–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ortogero N., Hennig G. W., Langille C., Ro S., McCarrey J. R., Yan W. (2013) Computer-assisted annotation of murine Sertoli cell small RNA transcriptome. Biol. Reprod. 88, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu Q., Song R., Ortogero N., Zheng H., Evanoff R., Small C. L., Griswold M. D., Namekawa S. H., Royo H., Turner J. M., Yan W. (2012) The RNase III enzyme DROSHA is essential for microRNA production and spermatogenesis. J. Biol. Chem. 287, 25173–25190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ro S., Ma H. Y., Park C., Ortogero N., Song R., Hennig G. W., Zheng H., Lin Y. M., Moro L., Hsieh J. T., Yan W. (2013) The mitochondrial genome encodes abundant small noncoding RNAs. Cell Res. 23, 759–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bao J., Zhang Y., Schuster A. S., Ortogero N., Nilsson E. E., Skinner M. K., Yan W. (2014) Conditional inactivation of Miwi2 reveals that MIWI2 is only essential for prospermatogonial development in mice. Cell Death Differ. 21, 783–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Muzumdar M. D., Tasic B., Miyamichi K., Li L., Luo L. (2007) A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 [DOI] [PubMed] [Google Scholar]
- 21. Park C., Yan W., Ward S. M., Hwang S. J., Wu Q., Hatton W. J., Park J. K., Sanders K. M., Ro S. (2011) MicroRNAs dynamically remodel gastrointestinal smooth muscle cells. PLoS One 6, e18628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ro S., Yan W. (2010) Small RNA cloning. In RNA Therapeutics: Function, Design, and Delivery (Sioud M., ed) pp. 271–283, Humana Press Inc., Totowa, NJ [Google Scholar]
- 23. Song R., Hennig G. W., Wu Q., Jose C., Zheng H., Yan W. (2011) Male germ cells express abundant endogenous siRNAs. Proc. Natl. Acad. Sci. U.S.A. 108, 13159–13164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Edgar R., Domrachev M., Lash A. E. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Watanabe T., Chuma S., Yamamoto Y., Kuramochi-Miyagawa S., Totoki Y., Toyoda A., Hoki Y., Fujiyama A., Shibata T., Sado T., Noce T., Nakano T., Nakatsuji N., Lin H., Sasaki H. (2011) MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev. Cell 20, 364–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chan P. P., Lowe T. M. (2009) GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 37, D93–D97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jurka J., Kapitonov V. V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J. (2005) Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110, 462–467 [DOI] [PubMed] [Google Scholar]
- 28. Kozomara A., Griffiths-Jones S. (2011) miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39, D152–D157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu C., Bai B., Skogerbø G., Cai L., Deng W., Zhang Y., Bu D., Zhao Y., Chen R. (2005) NONCODE: an integrated knowledge database of non-coding RNAs. Nucleic Acids Res. 33, D112–D115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mituyama T., Yamada K., Hattori E., Okida H., Ono Y., Terai G., Yoshizawa A., Komori T., Asai K. (2009) The Functional RNA Database 3.0: databases to support mining and annotation of functional RNAs. Nucleic Acids Res. 37, D89–D92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Waterston R. H., Lindblad-Toh K., Birney E., Rogers J., Abril J. F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Antonarakis S. E., Attwood J., Baertsch R., Bailey J., Barlow K., Beck S., Berry E., Birren B., Bloom T. (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 [DOI] [PubMed] [Google Scholar]
- 32. Chen X., Li Q., Wang J., Guo X., Jiang X., Ren Z., Weng C., Sun G., Wang X., Liu Y., Ma L., Chen J. Y., Wang J., Zen K., Zhang J., Zhang C. Y. (2009) Identification and characterization of novel amphioxus microRNAs by Solexa sequencing. Genome Biol. 10, R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hertel J., Hofacker I. L., Stadler P. F. (2008) SnoReport: computational identification of snoRNAs with unknown targets. Bioinformatics 24, 158–164 [DOI] [PubMed] [Google Scholar]
- 34. Li X. Z., Roy C. K., Dong X., Bolcun-Filas E., Wang J., Han B. W., Xu J., Moore M. J., Schimenti J. C., Weng Z., Zamore P. D. (2013) An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol. Cell 50, 67–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Flicek P., Amode M. R., Barrell D., Beal K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fairley S., Fitzgerald S., Gil L., Gordon L., Hendrix M., Hourlier T., Johnson N., Kähäri A. K., Keefe D., Keenan S., Kinsella R., Komorowska M., Koscielny G., Kulesha E., Larsson P., Longden I., McLaren W., Muffato M., Overduin B., Pignatelli M., Pritchard B., Riat H. S., Ritchie G. R., Ruffier M., Schuster M., Sobral D., Tang Y. A., Taylor K., Trevanion S., Vandrovcova J., White S., Wilson M., Wilder S. P., Aken B. L., Birney E., Cunningham F., Dunham I., Durbin R., Fernández-Suarez X. M., Harrow J., Herrero J., Hubbard T. J., Parker A., Proctor G., Spudich G., Vogel J., Yates A., Zadissa A., Searle S. M. (2012) Ensembl 2012. Nucleic Acids Res. 40, D84–D90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Rådmark O., Kim S., Kim V. N. (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419 [DOI] [PubMed] [Google Scholar]
- 37. Bartel D. P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 38. Papaioannou M. D., Pitetti J. L., Ro S., Park C., Aubry F., Schaad O., Vejnar C. E., Kühne F., Descombes P., Zdobnov E. M., McManus M. T., Guillou F., Harfe B. D., Yan W., Jégou B., Nef S. (2009) Sertoli cell Dicer is essential for spermatogenesis in mice. Dev. Biol. 326, 250–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee R. C., Feinbaum R. L., Ambros V. (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 [DOI] [PubMed] [Google Scholar]
- 40. Watanabe T., Totoki Y., Toyoda A., Kaneda M., Kuramochi-Miyagawa S., Obata Y., Chiba H., Kohara Y., Kono T., Nakano T., Surani M. A., Sakaki Y., Sasaki H. (2008) Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453, 539–543 [DOI] [PubMed] [Google Scholar]
- 41. Czech B., Malone C. D., Zhou R., Stark A., Schlingeheyde C., Dus M., Perrimon N., Kellis M., Wohlschlegel J. A., Sachidanandam R., Hannon G. J., Brennecke J. (2008) An endogenous small interfering RNA pathway in Drosophila. Nature 453, 798–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lewis B. P., Burge C. B., Bartel D. P. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]
- 43. Ma L., Buchold G. M., Greenbaum M. P., Roy A., Burns K. H., Zhu H., Han D. Y., Harris R. A., Coarfa C., Gunaratne P. H., Yan W., Matzuk M. M. (2009) GASZ is essential for male meiosis and suppression of retrotransposon expression in the male germline. PLoS Genet. 5, e1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Frost R. J., Hamra F. K., Richardson J. A., Qi X., Bassel-Duby R., Olson E. N. (2010) MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc. Natl. Acad. Sci. U.S.A. 107, 11847–11852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zheng K., Xiol J., Reuter M., Eckardt S., Leu N. A., McLaughlin K. J., Stark A., Sachidanandam R., Pillai R. S., Wang P. J. (2010) Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc. Natl. Acad. Sci. U.S.A. 107, 11841–11846 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.