Background: Cholesterol-loaded macrophages are key mediators of atherosclerosis and demonstrate increased SMPDL3A expression and secretion.
Results: SMPDL3A expression is stimulated by cholesterol, liver X receptor ligands, and cAMP and hydrolyzes nucleotide phosphates.
Conclusion: SMPDL3A is a nucleotide phosphodiesterase secreted from cholesterol-loaded macrophages.
Significance: SMPDL3A possesses a novel enzymatic activity with potential relevance to atherosclerosis.
Keywords: Atherosclerosis, Cholesterol, Macrophage, Metalloenzyme, Phosphodiesterases
Abstract
Cholesterol-loaded foam cell macrophages are prominent in atherosclerotic lesions and play complex roles in both inflammatory signaling and lipid metabolism, which are underpinned by large scale reprogramming of gene expression. We performed a microarray study of primary human macrophages that showed that transcription of the sphingomyelin phosphodiesterase acid-like 3A (SMPDL3A) gene is up-regulated after cholesterol loading. SMPDL3A protein expression in and secretion from primary macrophages are stimulated by cholesterol loading, liver X receptor ligands, and cyclic AMP, and N-glycosylated SMPDL3A protein is detectable in circulating blood. We demonstrate for the first time that SMPDL3A is a functional phosphodiesterase with an acidic pH optimum. We provide evidence that SMPDL3A is not an acid sphingomyelinase but unexpectedly is active against nucleotide diphosphate and triphosphate substrates at acidic and neutral pH. SMPDL3A is a major source of nucleotide phosphodiesterase activity secreted by liver X receptor-stimulated human macrophages. Extracellular nucleotides such as ATP may activate pro-inflammatory responses in immune cells. Increased expression and secretion of SMPDL3A by cholesterol-loaded macrophage foam cells in lesions may decrease local concentrations of pro-inflammatory nucleotides and potentially represent a novel anti-inflammatory axis linking lipid metabolism with purinergic signaling in atherosclerosis.
Introduction
The intracellular accumulation of lipoprotein-derived lipids, particularly cholesterol, transforms macrophages into foam cells, and the arterial retention of these cells is a hallmark of early atherogenesis. Foam cells develop a phenotype distinct from their monocyte precursors, which includes secretion of an array of pro-inflammatory cytokines (attracting further immune cells to the lesion) and deposition of extracellular matrix proteins, promoting additional retention of lipoproteins (1). The cellular responses of the macrophage to cholesterol loading represent a complex overlapping pattern of lipid metabolism, storage and transport, inflammatory signaling and responses, extracellular matrix interactions, and apoptotic pathways (2). Inhibition and reversal of macrophage foam cell formation are considered key candidates for therapeutic targeting, and thus identifying the complex perturbations in gene expression that underpin the transformation of macrophage to foam cell is essential.
To further understand the changes in gene expression occurring after foam cell formation, we conducted an oligonucleotide microarray study using cholesterol-loaded primary human monocyte-derived macrophages (HMDMs)2 as a foam cell model. We observed that expression of the gene SMPDL3A (sphingomyelin phosphodiesterase, acid-like 3A) is strongly up-regulated by cholesterol loading. SMPDL3A is named for its sequence homology to the well characterized acid sphingomyelinase (aSMase, encoded by the SMPD1 gene), with which it shares 31% amino acid identity. Acid SMase hydrolyzes sphingomyelin to produce phosphorylcholine and ceramide, a key signaling molecule in several apoptotic and stress-related pathways (3). aSMase is capable of hydrolyzing sphingomyelin in oxidized LDL particles, transforming them into a more aggregation-prone form that is more readily retained by arterial proteoglycans (4, 5). The activity of aSMase appears pro-atherogenic in vivo, where it promotes the retention of lipoprotein in the subendothelium and contributes to the growth of atherosclerotic plaques in the aortae of mice (6).
SMPDL3A was originally identified as a gene up-regulated in bladder tumor versus healthy urothelium tissue (7), and more recently it has been shown to be regulated by liver X receptor (LXR) ligands in human macrophage cell lines (8, 9). Beyond these observations, little is known about the biochemistry or function of SMPDL3A. Based on the strong up-regulation of SMPDL3A expression in cholesterol-loaded macrophages and the established involvement of its homolog aSMase in the progression of atherosclerosis, we explored the biology of this poorly characterized gene further. In this study, we report that cellular expression and secretion of SMPDL3A are not only increased by cholesterol and synthetic LXR ligands in primary human macrophages but also by cyclic AMP. We present the first experimental evidence that SMPDL3A is a functional metallophosphoesterase and, despite having no detectable activity toward sphingomyelin, possesses a nucleotide phosphodiesterase activity, particularly strongly against nucleotide triphosphates. Indeed, we show that SMPDL3A is the major nucleotide phosphodiesterase secreted by human THP-1 macrophages after LXR stimulation. This unexpected activity, together with its up-regulation in cholesterol-loaded macrophages, indicates the regulation and enzymology of SMPDL3A are distinct from aSMase and may play a novel role in the pathobiology of atherosclerosis.
EXPERIMENTAL PROCEDURES
Materials
All p-nitrophenol-conjugated substrates, nucleotide phosphates (ATP, ADP, AMP, cAMP, GTP, GDP, cGMP, UTP, UDP, and CTP), β-methyl cyclodextrin, cholesterol, 22-hydroxycholesterol, T-0901317, phorbol 12-myristate 13-acetate (PMA), dibutyryl-cAMP (Bt2cAMP) and forskolin were obtained from Sigma. White cell buffy coat concentrates and human serum were supplied by the New South Wales Red Cross Blood Transfusion Service, Sydney, Australia. LDL, acetylated LDL (AcLDL), and lipoprotein-deficient serum (LPDS) were prepared as described (10). Soluble cholesterol/β-methyl cyclodextrin (cholesterol-CD) complexes were prepared as described previously (11). Rabbit polyclonal anti-human SMPDL3A (ab68533) and anti-human CD39 (ab108248) antibodies was obtained from Abcam. Rabbit polyclonal anti-mouse SMPDL3A (ARP52244) and anti-human SMPDL3B (ARP33875) antibodies were purchased from Aviva Systems Biology. AlexaFluor 488- and 555-conjugated F(ab′)2 antibodies were obtained from Cell Signaling Technologies. [3H]Choline methyl-labeled sphingomyelin was obtained from PerkinElmer Life Sciences. Unlabeled bovine brain sphingomyelin was purchased from Avanti Polar Lipids.
Cell Culture
Human monocytes were isolated from white cell concentrates of healthy donors as described previously (12). After differentiation for 3 days in the presence of 25 ng/ml M-CSF, the cells were washed and enriched with cholesterol by incubation in RPMI 1640 medium including 10% (v/v) lipoprotein-deficient serum (LPDS) and either acetylated LDL (50 μg protein/ml) for 2 days or cholesterol-CD (20 μg cholesterol/ml) for 24 h. In some experiments, cells were treated with T-0901317 (1 μm) or 22-hydroxycholesterol (10 μm) for 24 h.
CHO-Trex cells (Invitrogen) were cultured in Ham's F-12 complete medium with 10% (v/v) fetal bovine serum and HEK-Trex cells (Invitrogen) in DMEM complete medium with 10% (v/v) fetal bovine serum.
THP-1 cells were maintained in RPMI 1640 medium supplemented with 10% (v/v) fetal bovine serum. THP-1 monocytes were differentiated into macrophages by addition of PMA at a final concentration of 50 ng/ml for 72 h prior to experimental setup.
Microarray Analysis
Total RNA was isolated using RNeasy reagent (Qiagen), amplified using the MessageAmp aRNA kit (Ambion), and indirectly labeled using Alexa 555 dye (Invitrogen). Reference aRNA was pooled from the test aRNA samples and labeled with the Alexa 647 dye (Invitrogen). 5 μg of Alexa 555-labeled aRNA was mixed with 5 μg of Alexa 647-labeled aRNA. The aRNA mix was hybridized using a 50% formamide slide stack against Compugen human oligonucleotide microarrays (Institute for Molecular Bioscience, University of Queensland). Samples included HMDM from two separate donors, and hybridizations were performed in technical replicates. Hybridization signals were collected using a G2565BA microarray scanner (Agilent). Intensity and background data for each spot were calculated using Imagene (BioDiscovery Inc., El Segundo, CA). Microarray data were analyzed using the GeneSpring software (Agilent).
Quantitative Real Time PCR Measurements
Reverse transcription was performed according to the manufacturer's protocol for SuperScript III First Strand cDNA synthesis kit (Invitrogen). Quantitative reverse transcriptase-PCR (QRT-PCR) was performed using SensiMix SYBR (Bioline) on a Corbett Rotorgene 3000 machine and analyzed using Rotor-Gene 6 Version 6.0 (Build 27). Primer pairs (supplemental Table 1) used for the amplification reaction of various genes from cDNAs were designed using NCBI PrimerBlast. Changes in gene expression levels were determined by normalizing mRNA levels of the gene of interest to the mRNA level of the housekeeping gene (β-actin or HPRT) and quantified using the ΔCt method. Melting curve analysis was performed to confirm production of a single product in each reaction.
Generation of a Monoclonal Antibody to Human SMPDL3A
Mice were immunized with a purified synthetic peptide corresponding to amino acids 316–327 of human SMPDL3A (sequence FQYDPRDYKLLD), and a monoclonal hybridoma cell line (6E3G4A1) producing IgG1 antibodies was established from these animals (EZBioLab, IN).
HPLC Analysis of Cellular Cholesterol
Cells were lysed in ice-cold 0.2 n sodium hydroxide, and free cholesterol and cholesteryl ester were determined by reverse phase HPLC after extraction into methanol/hexane as described previously (10). Cholesterol concentration was expressed relative to cellular protein, as determined by BCA assay (Pierce).
Western Blotting
Protein samples were prepared by boiling in SDS-PAGE sample buffer containing 1% SDS, 100 mm DTT, and 60 mm Tris·HCl, pH 6.8. Samples were separated on 4–12% gradient bis-Tris gels (Invitrogen) and electroblotted onto nitrocellulose membranes using iBlot transfer apparatus (Invitrogen). Membranes were blocked by incubation in phosphate-buffered saline (PBS) containing 4% (w/v) skim milk and 0.1% (v/v) Tween 20 for 1 h at room temperature. SMPDL3A Western blots were probed with either undiluted 6E3G4A1 hybridoma supernatant or commercial primary antibody diluted 1:1000 in blocking buffer overnight, followed by either anti-mouse or anti-rabbit IgG-horseradish peroxidase-conjugated secondary antibody for 1 h (Jackson ImmunoResearch). Blots were visualized using enhanced chemiluminescence reagents (GE Healthcare) and either LAS-4000 mini (Fuji) or Gel Doc XR+ (Bio-Rad) CCD imaging systems. Quantitation of immunoreactive bands was performed using Image Lab Version 4.1 software (Bio-Rad). To measure specifically the SMPDL3A protein levels in cell lysates, the ∼52-kDa reactive band was chosen for quantitation.
For Western blotting of secreted proteins in conditioned media, cells were washed with PBS to remove traces of serum proteins, incubated with fresh serum-free medium containing experimental treatment for 24 h, before the medium was harvested and spun down to remove any cellular debris, and loaded neat onto gels with SDS-PAGE sample buffer. Where required, secreted proteins were either concentrated by acetone precipitation (1 volume of conditioned media to 4 volumes of cold acetone at −20 °C for 1 h, centrifugation, air-drying, and resuspension in SDS-PAGE sample buffer) or by a Microcon 10-kDa centrifugal filter device (Millipore). Human and mouse plasma samples were analyzed after dilution into SDS-PAGE sample buffer and loading the equivalent of 200 nl of plasma per well.
Construction of SMPDL3A Expression Plasmid
A plasmid bearing the full-length sequence-verified human SMPDL3A open reading frame (IMAGE clone 100009862; Open Biosystems) was used as a template for all cloning experiments. A tetracycline-inducible C-terminal His6-tagged SMPDL3A expression construct was produced by amplifying the SMPDL3A ORF using the HindIII-containing primers 5′-GAGAAAGCTTACCATGGCGCTGGTGCG-3′ and 5′-TCTCAAGCTTCTAGTGATGGTGATGGTGATGGTAATTGTGCTTTAT-3′. The resulting PCR product was digested and ligated into the HindIII site of pcDNA 4/TO (Invitrogen) to create pcDNA 4/TO-SMPDL3A6His. The correct sequence and orientation of this construct were confirmed by DNA sequencing.
Cellular Expression of SMPDL3A
The pcDNA 4/TO-SMPDL3A-His6 construct was transfected into CHO-Trex cells (Invitrogen) using FuGENE 6 reagent (Roche Applied Science). Transfected cells were maintained under blasticidin and Zeocin (Invitrogen) selection, and a stable clonal cell line was established (CHO-SMPDL3A). Expression of SMPDL3A was induced by addition of tetracycline (1 μm) and overnight (∼16 h) incubation. A stable tetracycline-inducible HEK cell line was created in a similar manner by transfecting the pcDNA 4/TO-SMPDL3A-His6 plasmid into HEK-Trex cells (Invitrogen).
Purification of Secreted Recombinant SMPDL3A from CHO-SMPDL3A Cells
CHO-SMPDL3A cells were plated onto 25-cm2 cell culture dishes and grown to confluency in F-12 media with 10% FCS. Cells were washed three times with PBS, and the media were replaced with serum-free F-12 containing 1 μm tetracycline to induce SMPDL3A expression. After 24 h, the conditioned media were harvested, filtered through a 0.22-μm filter, and adjusted to pH 8.0 (with 20 mm Tris) and 500 mm final NaCl concentration. Media were loaded onto a HisTrap nickel-agarose column (GE Healthcare) connected to an NGC chromatography system (Bio-Rad). The column was washed with 10 column volumes of wash buffer (20 mm Tris, pH 8.0, 500 mm NaCl), followed by a gradient of 0–500 mm imidazole over 20 column volumes. Eluted fractions were analyzed by Western blotting (6E3G4A1 anti-SMPDL3A monoclonal) and enzyme assay, using ATP and p-nitrophenol thymidine monophosphate substrates.
Knockdown of SMPDL3A Expression
Anti-human SMPDL3A (catalog number 1225141) and universal negative control scrambled siRNA (Stealth siRNA) were purchased from Invitrogen. 50 pmol of anti-SMPDL3A or scrambled siRNA negative control were transfected into 1 × 106 PMA-differentiated THP-1 human macrophages using Lipofectamine RNAiMAX liposomes (Invitrogen) according to the manufacturer's recommendations. Cells were incubated with siRNA for 24 h, before replacement with serum-free RPMI 1640 media containing experimental treatments. Efficiency of SMPDL3A siRNA knockdown was assessed using QRT-PCR and anti-SMPDL3A Western blotting.
Removal of N-Linked Glycans
N-Linked glycans were enzymatically removed using PNGaseF (New England Biolabs) according to the manufacturer's instructions. For denaturing conditions, glycoprotein samples were heated to 100 °C for 10 min and then incubated with PNGaseF (500 units) for 1 h at 37 °C. For native conditions, glycoprotein samples were treated directly with 500 units of PNGaseF for 16 h at 37 °C. In each case, control samples were subjected to identical treatment but without addition of PNGaseF enzymes. Relative molecular masses of control and PNGaseF-digested glycoprotein samples were measured by SDS-PAGE and Western blotting.
Confocal Microscopy
Cells were fixed using cold 4% (v/v) paraformaldehyde in PBS for 15 min, followed by a 30-min incubation in permeabilization buffer (0.5% (w/v) saponin in PBS) supplemented with 10% (v/v) whole serum of the same species as the secondary antibody. Cells were probed for 1 h at room temperature with primary antibody, washed, and then incubated with F(ab′)2 fragment antibodies (Cell Signaling Technology) conjugated to either AlexaFluor 488 or AlexaFluor 555 for 1 h at room temperature. Finally, cells were washed, mounted with Mowiol 4–88, and imaged using an Olympus FV1000 confocal laser scanning microscope and a ×60 oil immersion lens. False coloring, merging, and analysis of images were performed using ImageJ 1.46 software (National Institutes of Health).
Phosphodiesterase Activity Assays
Phosphodiesterase activities were determined using a panel of p-nitrophenol-conjugated synthetic substrates. Substrate cleavage was monitored by measuring the increase in absorbance at 405 nm. Absorbances were converted to molar values using an extinction coefficient for p-nitrophenol of 18,000 m−1 cm−1. Substrates tested were as follows: p-nitrophenol phosphate (p-NPP), bis-(p-nitrophenyl) phosphate (bis-p-NPP), p-nitrophenylphosphorylcholine (p-NPPC), and p-nitrophenyl thymidine 5′-monophosphate (p-NP-TMP). Activity was assayed with 125 mm Tris·HCl (for pH 7.0 and above) or 125 mm sodium acetate (below pH 7.0) final concentration buffers and 20 mm p-nitrophenyl-conjugated substrate. Where indicated, reactions were supplemented with metal ion (5 mm). All reactions occurred at 37 °C.
Sphingomyelinase Assays
Sphingomyelinase activity was determined in cell lysates and conditioned media using 3H-radiolabeled sphingomyelin as described previously (13). Briefly, choline [3H]methyl-labeled sphingomyelin (final concentration, 0.1 μCi/ml) and unlabeled bovine brain sphingomyelin (final concentration 500 μm) in ethanol were dried under nitrogen and reconstituted in 0.6% Triton X-100 and reaction buffer (100 mm sodium acetate, 5 mm Zn2+, pH 5.5, for acidic reaction conditions, or 100 mm Tris, 5 mm Mg2+, pH 7.5, for neutral conditions) with sonication. For assays of cell lysates, 10 μl of cell lysates, adjusted to each contain 50 μg of total protein, were mixed with 90 μl of sphingomyelin reaction buffer. For assays of conditioned media, 50 μl of media were mixed with 50 μl of sphingomyelin reaction buffer. Reactions were allowed to proceed for 2 h at 37 °C before quenching and extraction of liberated [3H]choline by addition of 375 μl of a 1:2 v/v chloroform/methanol mixture. Samples were vortexed, and an additional 125 μl of chloroform was added, vortexed again, and 125 μl of water was added. Samples were centrifuged at 14,000 rpm for 1 min, and the upper aqueous layer was collected and analyzed by 3H scintillation counting.
For serum supplementation studies, pooled human serum from healthy donors was first heat-inactivated (HI) by incubation at 56 °C for 30 min. Choline [3H]methyl-labeled sphingomyelin (0.5 μCi/ml final concentration) was equilibrated into serum lipoproteins by mixing with HI serum at room temperature for 1 h. 50 μl of equilibrated HI serum/[3H]sphingomyelin was mixed with 50 μl of tetracycline-induced CHO SMPDL3A conditioned media and incubated for 2 h at 37 °C before extraction and scintillation counting of liberated [3H]choline as described above. A negative control supplemented with water instead of conditioned media was included to demonstrate heat inactivation of endogenous serum sphingomyelinase activity.
Sphingomyelinase activity was additionally assayed in conditioned media using an Amplex Red enzymatically coupled fluorescence assay (Invitrogen). Background levels of choline, which would result in spuriously high baseline readings, were eliminated from samples by buffer exchange (10-kDa Centricon centrifugal concentrator; Millipore). To begin the reaction, 100 μl of buffer-exchanged medium was mixed with 100 μl of sphingomyelinase assay buffer, which contained either 100 mm Tris, pH 7.4, or 100 mm sodium acetate, pH 5.5, and 5 mm Mg2+ or 5 mm Zn2+. Final reagent concentrations, common to all conditions, were 50 μm Amplex Red, 200 milliunits of horseradish peroxidase, 20 milliunits of choline oxidase, 800 milliunits of alkaline phosphatase, 250 μm sphingomyelin, and 0.1% Triton X-100. Assays were incubated at 37 °C for the indicated time. Amplex Red fluorescence was measured using a BMG FLUOstar plate reader (excitation 544 nm/emission 590 nm).
Mass Spectrometric Measurement of Cellular Lipid Composition
HEK-SMPDL3A cells were grown to near confluence in 75-cm2 flasks and treated with or without 1 μm tetracycline for 24 h (n = 6 for each condition). Cells were harvested by scraping into PBS, pelleted, resuspended in Tris-buffered saline, pH 7.0, supplemented with 100 μm butylated hydroxytoluene to prevent lipid oxidation, and lysed using an ultrasonic probe. Protein concentrations of crude cell lysates were determined by BCA assay, and expression of SMPDL3A after tetracycline induction was confirmed by Western blotting with 6E3G4A1 antibody. Total lipids were extracted using the method of Bligh and Dyer (14). Extracted cellular lipids were analyzed by liquid chromatography-tandem electrospray mass spectrometry as described previously (15). Lipid concentrations were normalized and expressed relative to extracted protein concentration.
Nucleotide Phosphodiesterase Activity Measurement
The activity toward nucleotide phosphate substrates of concentrated cell-conditioned media from recombinant SMPDL3A-expressing CHO cells was determined using a Malachite Green inorganic phosphate release assay (Biomol Green, Enzo). Excess free phosphate in the media, which could interfere with the Malachite Green assay, was removed by buffer exchange using a 10-kDa cutoff Centricon device (Millipore). Reaction buffer included Tris·HCl (125 mm, pH 7.5), Mg2+ (5 mm), and nucleotide phosphates (200 μm) as indicated, at a final volume of 100 μl. Reactions were incubated at 37 °C for 1 h, terminated by adding 100 μl of Biomol Green reagent, and incubated for 15 min at room temperature before measuring absorbance at 620 nm. Absorbance values were converted to molarity by generation of a sodium phosphate standard curve.
HPLC Measurement of Adenosine Phosphates
Concentrations of individual adenosine phosphate species arising from the hydrolysis of ATP by SMPDL3A were determined by reversed phase HPLC. Nucleotide phosphates were separated on a 25-cm × 4.6-mm C18 column (Phenomenex) using isocratic elution with a 0.1 m KH2PO4, pH 6, mobile phase at 1 ml/min flow rate, with UV detection at 254 nm on a PerkinElmer Life Sciences Series 200 HPLC system. Pure ATP, ADP, and AMP standards were run at the start of each experiment to determine elution times and response factors.
Human and Mouse Ethics
Ethics approval for human blood collection was obtained from the Sydney Local Health District and the relevant collection site (Royal Prince Alfred Hospital), and written informed consent was obtained from all donors. C57/Bl6 mouse serum was collected with the approval of the University of New South Wales Animal Care and Ethics Committee.
RESULTS
SMPDL3A Expression and Secretion Is Up-regulated by Cholesterol Loading and LXR Agonists in Primary Human Macrophages
To identify genes differentially regulated in human macrophages by cholesterol accumulation, oligonucleotide microarray analysis was performed on HMDMs obtained from two separate donors ± cholesterol loading by incubation with acetylated LDL (AcLDL) for 48 h (supplemental Table 2). SMPDL3A was among the most highly up-regulated genes that passed all quality filters, with expression increased by 4.35- and 3.35-fold in the two individual donors, respectively. To validate the microarray data, HMDMs from five different donors were incubated ± AcLDL as above, and SMPDL3A mRNA expression was determined by quantitative RT-PCR. Expression of SMPDL3A mRNA in cholesterol-loaded cells was significantly increased in all five donors relative to nonloaded controls (mean 3.97-fold increase; Fig. 1A).
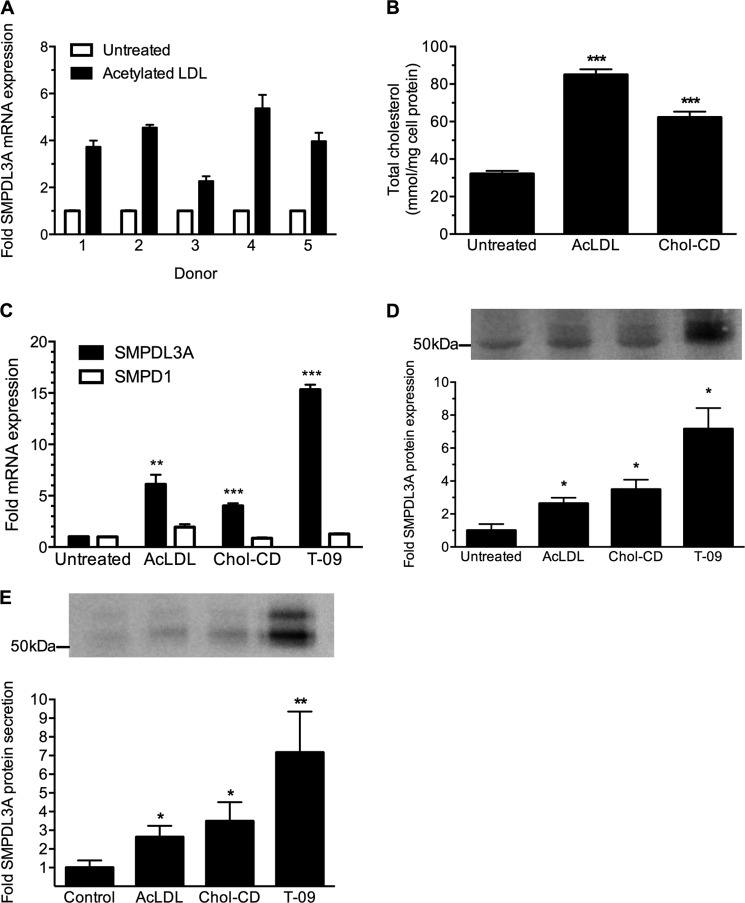
FIGURE 1.
SMPDL3A expression in cholesterol-loaded human monocyte-derived macrophages. A, SMPDL3A mRNA expression. HMDM from five individual healthy donors were incubated for 48 h in RPMI 1640 medium containing 10% (v/v) LPDS ± AcLDL (50 μg/ml), then mRNA expression measured by QRT-PCR. SMPDL3A mRNA was normalized against the housekeeping gene (β-actin) and expression in cholesterol-loaded HMDM expressed relative to nonloaded cells. Data are means ± S.D. of four technical replicates. B, free cholesterol and cholesteryl ester mass were measured using reverse phase HPLC and normalized for cell protein. Data are means ± S.D. for three determinations. C, SMPDL3A mRNA expression and comparison with SMPD1. HMDM were treated with AcLDL(50 μg/ml; 48 h), cholesterol/β-methyl cyclodextrin (Chol-CD) (20 μg/ml; 24 h), or T-090317 (T-09, 1 μm; 24 h) and then mRNA expression measured by QT-RT-PCR. SMPDL3A mRNA was normalized against the housekeeping gene (β-actin), and expression was expressed relative to untreated cells. Data are means ± S.D. of four determinations. D, SMPDL3A protein expression. HMDM cells treated as in C. Western blot analysis of SMPDL3A in cell lysates (15 μg of protein/lane) using 6E3G4A1 monoclonal antibody. Data are means ± S.D. of three determinations. Panel inset shows a representative blot. E, SMPDL3A protein secretion. HMDM¼ cells washed with serum-free media and then treated as above but using serum-free media. Conditioned media were concentrated and analyzed by anti-SMPDL3A Western blot. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (unpaired two-tailed t-tests).
To further demonstrate this increase in SMPDL3A mRNA expression was due to cholesterol accumulation, HMDMs were also separately loaded using soluble cholesterol-β-methyl cyclodextrin complexes, which transfer cholesterol directly to cells independently of receptor-mediated uptake. Effective cholesterol loading of HMDMs under the above conditions was confirmed using HPLC analysis of total cellular cholesterol (Fig. 1B). As for AcLDL loading, SMPDL3A mRNA levels were significantly increased in HMDM by cholesterol-CD loading (Fig. 1C), demonstrating SMPDL3A mRNA expression is sensitive to increased cellular cholesterol levels, regardless of the mechanism of sterol uptake.
To measure levels of SMPDL3A protein, a custom monoclonal antibody directed against a polypeptide sequence unique to human SMPDL3A was created (6E3G4A1; supplemental Fig. 1). Using this antibody, expression of SMPDL3A protein in HMDM cell lysates was measured by Western blotting. This identified two closely spaced bands of ∼52–54 kDa (Fig. 1D, inset), consistent with the predicted molecular mass of SMPDL3A (51.26 kDa). Levels of intracellular SMPDL3A protein were significantly higher in cells loaded with cholesterol using AcLDL or cholesterol-CD, reflecting the transcriptional response (Fig. 1D).
The up-regulation of SMPDL3A mRNA and protein expression in primary human macrophages by cholesterol loading led us to examine the sequence of the putative SMPDL3A promoter region for sterol or lipid-related regulatory motifs. Bioinformatic analysis of the genomic nucleic acid sequence upstream of the SMPDL3A ORF using MatInspector software (16) identified a potential LXR element 160 bp upstream from the transcription start site (sequence AGGTCAGGCGAACTGA). LXR elements were recognized by LXR-retinoid X receptor heterodimeric transcription complexes after activation by binding to oxysterol ligands and are important for the regulation of several key cholesterol-responsive genes, such as ABCA1 and ABCG1 (17). Treatment of HMDMs with the synthetic LXR ligand T-0901317 strongly stimulated SMPDL3A mRNA (Fig. 1C) and protein (Fig. 1D) expression, confirming that SMPDL3A expression is LXR-responsive. SMPDL3A mRNA was also up-regulated by treatment with a second LXR ligand, 22-hydroxycholesterol (data not shown). These results are consistent with other studies published while this work was underway, which demonstrated that SMPDL3A mRNA expression is increased by LXR ligands in the immortalized human leukemic THP-1 macrophage cell line and in undifferentiated human peripheral blood mononuclear cells (8, 9).
To determine whether SMPDL3A was secreted from HMDMs, cells were incubated for 24 h in serum-free medium, and the media were then harvested, concentrated by acetone precipitation, and analyzed by Western blotting. Two closely spaced bands (∼52–55 kDa) of SMPDL3A were detected (Fig. 1E), similar to those observed in HMDM cellular lysates (Fig. 1B). As for cellular mRNA and protein expression, secretion of SMPDL3A was significantly increased by cholesterol loading with either AcLDL or cholesterol-CD and treatment with LXR agonist.
SMPDL3A Expression and Secretion Is Regulated by Intracellular cAMP
MatInspector promoter region analysis also revealed the presence of putative cyclic AMP-responsive elements, suggesting that SMPDL3A expression may also be regulated by cellular cAMP (potential cAMP-response element-binding protein site at −281 bp upstream of transcription start site TGACATCA). To investigate this, HMDMs were treated for 24 h with Bt2cAMP (a membrane-permeable analog of cAMP) and forskolin (an activator of adenylyl cyclase, which increases cAMP production) before measurement of SMPDL3A protein expression. Treatment with Bt2cAMP, forskolin, or forskolin combined with Bt2cAMP led to significant up-regulation in the expression of SMPDL3A mRNA (Fig. 2A), intracellular protein levels (Fig. 2B), and secretion (Fig. 3C), indicating expression of SMPDL3A is indeed sensitive to cAMP levels and that the cyclic AMP-responsive elements in the promoter region of SMPDL3A may play a functional role in primary human macrophages.
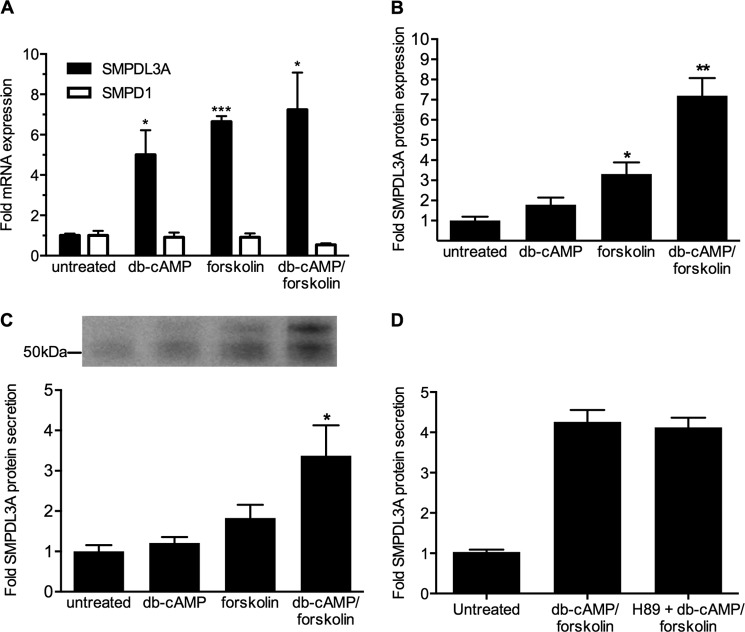
FIGURE 2.
SMPDL3A mRNA expression, protein expression, and secretion is regulated by cellular cAMP. A, SMPDL3A mRNA expression and comparison with SMPD1. HMDM were incubated for 24 h in RPMI 1640 medium containing 10% FCS plus dibutyryl-cAMP (db-cAMP, 1 mm) and/or forskolin (100 μm) as indicated. mRNA was measured by QRT-PCR and normalized to β-actin as described above. Data are means ± S.D. of four determinations. B, SMPDL3A protein expression. HMDM cells treated as in A. Western blot analysis of SMPDL3A in cell lysates (15 μg of protein/lane). Data are means ± S.D. of three determinations. C, SMPDL3A protein secretion. HMDM cells washed with serum-free media and then treated as above but using serum-free media. Conditioned media were concentrated and analyzed by anti-SMPDL3A Western blot. D, effect of PKA inhibition on SMPDL3A secretion. THP-1 macrophages were pretreated with 10 μm H89 for 1 h prior to stimulation with 1 mm Bt2cAMP, 100 μm forskolin for 24 h. Conditioned media were concentrated and analyzed by anti-SMPDL3A Western blot. Data are means ± S.D. of three determinations. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (unpaired two-tailed t tests).
FIGURE 3.
Amino acid sequence comparison of SMPDL3A with acid sphingomyelinase and SMPDL3B. The amino acid sequences of human SMPDL3A and close human homologs, SMPDL3B and aSMase (encoded by the SMPD1 gene), were aligned using ClustalW2. Asterisks indicate exact sequence matches, and colons indicate conservative substitutions. Known or predicted signaling regions are shaded in light gray and saposin domains in dark gray. Phosphodiesterase consensus motifs are indicated using open boxes. Conserved residues known or predicted to be involved in metal binding are indicated by M. Known or predicted N-linked glycosylation sites are indicated by N. The predicted glycosylphosphatidylinositol anchor attachment site at Ala-431 in SMPDL3B is indicated by a light gray shaded box. Uniprot sequence accession numbers are SMPDL3A:Q92484, SMPDL3B:Q92485, and aSMase:P17405.
Many downstream effects of cAMP stimulation are mediated via protein kinase A (PKA) signaling. To determine whether PKA was involved in the cAMP-dependent stimulation of SMPDL3A secretion, THP-1 macrophages were pretreated with H89, a potent PKA inhibitor, prior to stimulation with Bt2cAMP + forskolin. However, H89 had no effect on the secretion of SMPDL3A (Fig. 2D). H89 pretreatment was itself effective, as it significantly reduced the secretion of apolipoprotein E, which is known to be PKA-dependent (data not shown) (18). This result indicates the up-regulation of SMPDL3A secretion by raised cAMP levels is mediated by a PKA-independent pathway.
Comparative Sequence and Expression Analysis of SMPDL3A and Its Homologs
SMPDL3A is named for its amino acid sequence similarity to acid sphingomyelinase (aSMase), encoded by the SMPD1 gene in humans. Alignment of the translated amino acid sequences of SMPDL3A and aSMase using BLAST reveals 31% amino acid identity and 48% overall amino acid similarity. The amino acid sequence of SMPDL3A is also highly similar (41% amino acid identity and 60% similarity) to that of human SMPDL3B (sphingomyelin phosphodiesterase-like 3B), another poorly characterized potential phosphodiesterase family member. SMPDL3A contains a highly conserved example of the classical phosphoesterase G(D/G)NH signature motif (Fig. 3, open boxes) (19), with only a single amino acid (serine 111) deviating from a perfect match to the consensus sequence. ClustalW2 multiple alignment of the amino acid sequences of SMPDL3A, SMPDL3B, and aSMase revealed the key residues of the GD/GNH phosphoesterase motif in SMPDL3A to be perfectly aligned with those of aSMase and SMPDL3B. All five key metal-binding residues predicted from a homology model of aSMase (20) were also found to be conserved and perfectly aligned within the sequences of SMPDL3A, SMPDL3B and aSMase (Fig. 3, indicated by boldface M).
Acid sphingomyelinases (including human aSMase) contain the consensus motif NX3CX3N (where X is any amino acid; indicated in boldface in Fig. 3), which is predicted to be directly involved in the recognition of the phosphorylcholine headgroup of their sphingomyelin substrates (20). In human aSMase, the central cysteine (Cys-385) of the motif is also involved in a disulfide bridge with Cys-431 (21). SMPDL3A and SMPDL3B both deviate from this consensus sequence, containing a tyrosine substitution of the central cysteine residue, suggesting this region may differ from aSMase both in terms of three-dimensional structure and substrate recognition.
aSMase contains a sphingolipid activator protein (saposin) domain in its N-terminal region (Fig. 3, dark gray shading) (22), followed by a proline-rich linker. Saposins disrupt membrane structure, facilitating access to individual phospholipid molecules (23). In aSMase, this domain also plays a role in stabilizing the structure of the enzyme (24). Notably, both SMPDL3A and SMPDL3B lack any sequence homology to either the saposin domain of aSMase or to the individual saposin A–D polypeptides. aSMase also contains an N-terminal 46-amino acid signal peptide with a distinctive leucine-alanine repeat motif, which directs the translated aSMase polypeptide into the endoplasmic reticulum lumen and is required for its secretion (Fig. 3, light gray shading) (25). Signal sequence analysis using the SignalP 4.1 algorithm (26) also predicts the presence of N-terminal signaling peptides in both SMPDL3A and SMPDL3B, consistent with the secretion of SMPDL3A into the media of human macrophages (Figs. 1E and 2C). However, there is limited homology between these two predicted signaling peptides or with the signal region of aSMase, including a lack of the signature leucine-alanine repeat motif. The C-terminal domains of SMPDL3A and SMPDL3B also display limited similarity to each other. Analysis of this region with the PredGPI algorithm (27) predicts a high confidence glycosylphosphatidylinositol anchor attachment point at Ala-431 in the sequence of SMPDL3B (Fig. 3, shaded box), which has recently been experimentally confirmed (28). However, no such modification is predicted or has yet been described experimentally for SMPDL3A.
The aSMase is an N-glycoprotein bearing five discrete asparagine-linked glycans (Fig. 3, indicated by bold N), with glycosylation of Asn-395 and Asn-520 being critical for normal secretion and function (29). SMPDL3A contains seven potential N-linked glycosylation motifs (defined by the pattern NX(S/T), where X is any amino acid except proline). Notably, two of these motifs (Asn-222 and Asn-356) are perfectly aligned with the functionally important Asn-395 and Asn-520 N-linked sites of aSMase. In summary, comparison of the sequences of SMPDL3A and SMPDL3B with their homolog aSMase reveals that both proteins are likely functional members of the metallophosphoesterase enzyme family, although with a potentially altered substrate preference to aSMase and with distinct cellular trafficking and localization.
Because of the similarity of SMPDL3A to aSMase, particularly in their metallophosphodiesterase catalytic domains, we examined the possibility that expression of these two proteins is co-regulated in response to cholesterol loading, LXR ligand, or cAMP treatments. In contrast to the marked up-regulation of SMPDL3A mRNA in HMDMs after cholesterol loading or stimulation with LXR ligands, aSMase/SMPD1 mRNA was more moderately up-regulated by acetylated LDL loading and entirely unaffected by either cholesterol/β-methyl cyclodextrin or T-0901317 treatment (Fig. 1C). Expression of SMPDL3B mRNA was extremely low in untreated HMDMs and was not increased further after cholesterol loading (data not shown). No change in SMPDL3B protein levels in HMDMs or THP-1 macrophages after treatment with T-0901317 was detected (supplemental Fig. 1, A and B). Expression of aSMase/SMPD1 mRNA was similarly unaffected by treatment with Bt2cAMP and forskolin (Fig. 2A), indicating insensitivity to elevated cellular cAMP levels. Together, these results show that despite overall sequence similarity, expression of SMPDL3A and its homologs is regulated by distinct stimuli.
SMPDL3A Is an N-Linked Glycoprotein and Is Present in Human and Mouse Blood
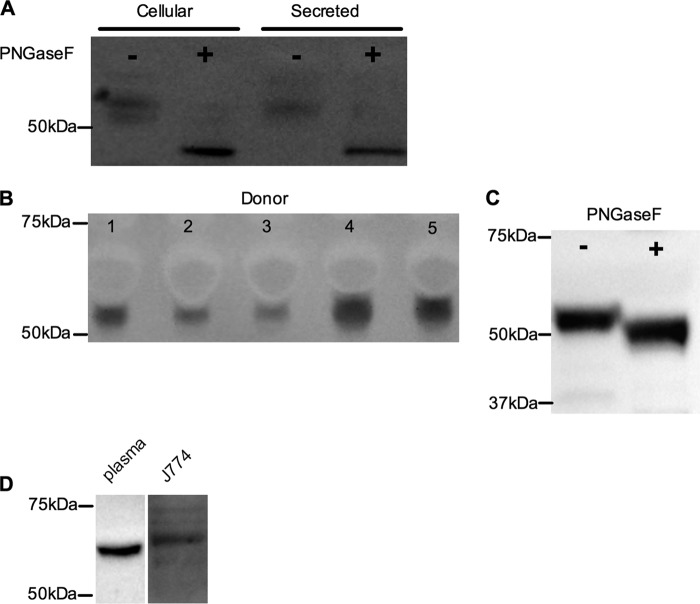
As our sequence analysis of SMPDL3A predicts multiple sites of N-linked glycosylation, we considered that one or more bands of SMPDL3A identified by Western blot might represent glycosylated forms of the protein. To test this, whole cell lysates and conditioned media from HMDMs stimulated with 1 μm T-0901317 were digested with PNGaseF, an enzyme that removes N-linked glycans. For both cellular and secreted proteins, this treatment resulted in the disappearance of the original bands and the appearance of a single SMPDL3A band with a lower apparent molecular mass (∼45 kDa), corresponding to the removal of ∼8 kDa of N-linked glycans (Fig. 4A).
FIGURE 4.
SMPDL3A is secreted from HMDMs, N-glycosylated, and present in human serum. A, HMDM cell lysates and conditioned media from T-0901317-treated cells were incubated ± PNGaseF, separated by SDS-PAGE, and analyzed by Western blotting for human SMPDL3A as described under “Experimental Procedures.” B, SMPDL3A in fasted plasma from five healthy volunteers. Equal volumes of plasma (equivalent to 200 nl of neat plasma) were separated by SDS-PAGE and detected by anti-SMPDL3A Western blot. Area of negative staining above SMPDL3A band is due to human serum albumin. C, human plasma is N-glycosylated. 200 nl of neat plasma from a healthy donor was incubated ± PNGaseF and analyzed by anti-SMPDL3A Western blotting. D, SMPDL3A in mouse plasma (equivalent to 200 nl of neat plasma) and conditioned media from J774 macrophages, analyzed using anti-mouse SMPDL3A Western blotting.
As SMPDL3A was found to be secreted from primary macrophages, we hypothesized that it may also circulate in vivo. To investigate this, freshly isolated sera from five healthy, fasted donors were probed using Western blotting with anti-SMPDL3A antibody, revealing a band of similar size to SMPDL3A secreted by HMDMs (Fig. 4B). Interestingly, there was marked variation between donors in the amount of circulating SMPDL3A protein. Digestion of total human serum with PNGaseF reduced the apparent molecular mass of SMPDL3A on a Western blot (Fig. 4C), demonstrating that circulating human SMPDL3A is also N-glycosylated. However, the decrease in apparent molecular mass after PNGaseF digestion of serum SMPDL3A was less than that for SMPDL3A freshly secreted by HMDMs (5 versus 8 kDa), suggesting that either glycan trimming occurs in the circulation or that circulating SMPDL3A originates from a different tissue source to macrophages, which differs in its degree and/or type of N-glycosylation of SMPDL3A.
SMPDL3A is also present in mouse plasma (wild-type C57/black), detected with a commercial antibody specific to murine SMPDL3A (Fig. 4D). In contrast to circulating human SMPDL3A, the apparent molecular mass of mouse plasma SMPDL3A was ∼62 kDa. This mass was consistent with that observed in conditioned media from the mouse J774 macrophage cell line (Fig. 4D). These results further reinforce that the mature mass of secreted SMPDL3A is tissue- and/or species-dependent.
Recombinant Expression and Purification of Human SMPDL3A
To study the biochemical function of SMPDL3A, stable CHO (CHO-SMPDL3A) and HEK (HEK-SMPDL3A) cell lines were created, expressing full-length human SMPDL3A with a C-terminal His6 tag, under the control of a tetracycline (tet)-inducible promoter. Western blot analysis of CHO-SMPDL3A (Fig. 5A) and HEK-SMPDL3A (supplemental Fig. 1C) cell lysates after overnight tet induction revealed a major band of ∼52 kDa apparent molecular mass, consistent with the size of SMPDL3A endogenously expressed in HMDM. A minor band of ∼65 kDa was also observed in tet-induced CHO- and HEK-SMPDL3A lysates. Negligible leaky expression of human SMPDL3A was observed in CHO or HEK-SMPDL3A lysates in the absence of tet induction. Expression of human SMPDL3A had no detectable effect on cell growth or viability in the CHO or HEK cell line (data not shown). In both tet-induced CHO- and HEK-SMPDL3A cells, no additional (nonspecific) bands were observed using the 6E3G4A1 monoclonal antibody, demonstrating the high specificity of this reagent for human SMPDL3A (Fig. 5A and supplemental Fig. 1C).
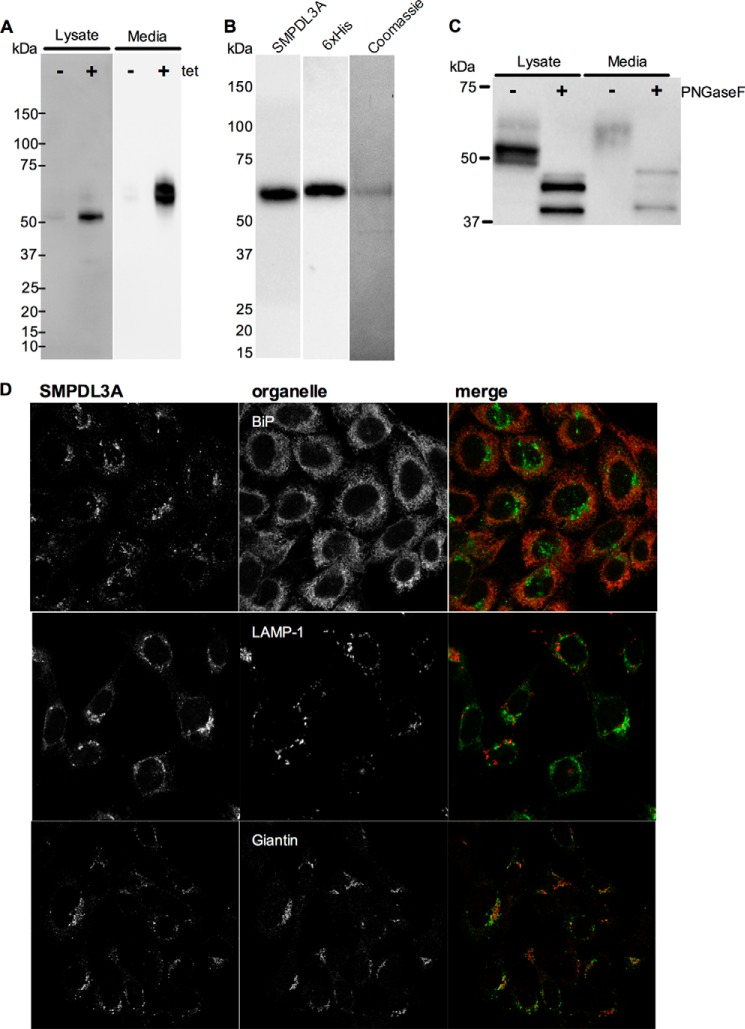
FIGURE 5.
Expression, secretion, purification, glycosylation, and localization of recombinant human SMPDL3A in CHO cells. A, expression of human SMPDL3A in CHO-SMPDL3A cells was induced by addition of ± tetracycline (1 μm, 16 h). Levels of SMPDL3A in cell lysates (10 μg of protein/lane) and conditioned media (20 μl protein/lane) were measured by SDS-PAGE and Western blotting. B, purification of His6-tagged SMPDL3A secreted from tetracycline-induced CHO-SMPDL3A cells with Ni2+ affinity chromatography. Purified material was analyzed using anti-SMPDL3A and anti-His6 Western blotting. Purity analyzed by Coomassie-stained SDS-PAGE. C, N-glycosylation of human SMPDL3A in tetracycline-induced CHO-SMPDL3A cells was analyzed by digestion of whole cell lysates and conditioned media with PNGaseF. Changes in molecular mass of SMPDL3A after PNGaseF digestion were measured by SDS-PAGE and visualized by Western blotting. D, CHO-SMPDL3A cells after induction with 1 μm tetracycline overnight were fixed with 3.7% paraformaldehyde in PBS, stained with a polyclonal antibody directed against human SMPDL3A (Abcam), and visualized with confocal microscopy. Subcellular localization of human SMPDL3A in tetracycline-induced CHO-SMPDL3A cells was determined by co-immunofluorescent staining with antibodies against human SMPDL3A (green) and organelle markers (red) BiP (endoplasmic reticulum), LAMP-1 (lysosome), and giantin (Golgi).
As in primary HMDMs, SMPDL3A was secreted into the media of tet-induced CHO- and HEK-SMPDL3A cells (Fig. 5A and supplemental Fig. 1C). Recombinant secreted SMPDL3A has an apparent molecular mass of ∼65 kDa, corresponding to the size detected in mouse plasma and conditioned murine J774 macrophage media (Fig. 4F). This species also corresponds to the minor intensity 65-kDa band detected in CHO- and HEK-SMPDL3A lysates, possibly representing SMPDL3A transiting through the secretory pathway. SMPDL3A secreted from CHO- and HEK-SMPDL3A cells is 10–15 kDa larger than both the intracellular and secreted forms in HMDM cells and that found in human plasma, further indicating there may be tissue- and/or species-specific differences in the post-translational modification of SMPDL3A.
Recombinant His6-tagged SMPDL3A was purified from the media of tetracycline-induced CHO-SMPDL3A cells using nickel affinity chromatography, yielding a single detectable species on Coomassie Blue SDS-polyacrylamide gel of ∼65 kDa, corresponding to bands detected by Western blotting with either anti-SMPDL3A 6E3G43A1 or His6 antibodies (Fig. 5B).
As in HMDMs, both the intracellular and secreted SMPDL3A from CHO-SMPDL3A are N-glycosylated, based on their decreased apparent molecular masses after PNGaseF digestion (Fig. 5C). The 10–15-kDa difference in apparent molecular mass between SMPDL3A secreted from HMDMs and CHO-SMPDL3A cells was largely lost after PNGaseF treatment, indicating the higher mass of recombinant versus endogenous human SMPDL3A is largely due to N-linked glycans. Both the ∼52- and ∼65-kDa intracellular forms detected in CHO-SMPDL3A cells are glycosylated, as both bands are eliminated after PNGaseF treatment. A small difference in molecular mass between intracellular and secreted SMPDL3A remained after PNGaseF digestion, suggesting other still unknown protein processing or post-translational modifications may be present.
Intracellular Localization of Human SMPDL3A in CHO Cells
Immunofluorescent microscopy of human SMPDL3A expressed in CHO-SMPDL3A cells showed a predominantly perinuclear pattern, with additional small punctate structures visible throughout the cell (Fig. 5D). Co-staining with antibodies against organellar markers revealed the majority of human SMPDL3A to be co-localized with the Golgi marker protein, Giantin. There was little evidence for co-localization of SMPDL3A with the lysosome marker, LAMP-1, in contrast with the typical lysosomal residence of mature intracellular aSMase (30, 31) nor was co-localization with the endoplasmic reticulum marker BiP observed. No indication of SMPDL3A in the plasma membrane was observed, unlike the plasma membrane lipid raft localization in kidney podocytes of homolog SMPDL3B (32). The identity of the multiple small punctate SMPDL3A-positive structures may represent secretory vesicles but requires further investigation to confirm this.
Characterization of the Enzymatic Activity of Recombinant Human SMPDL3A Using Classical Phosphodiesterase Substrates
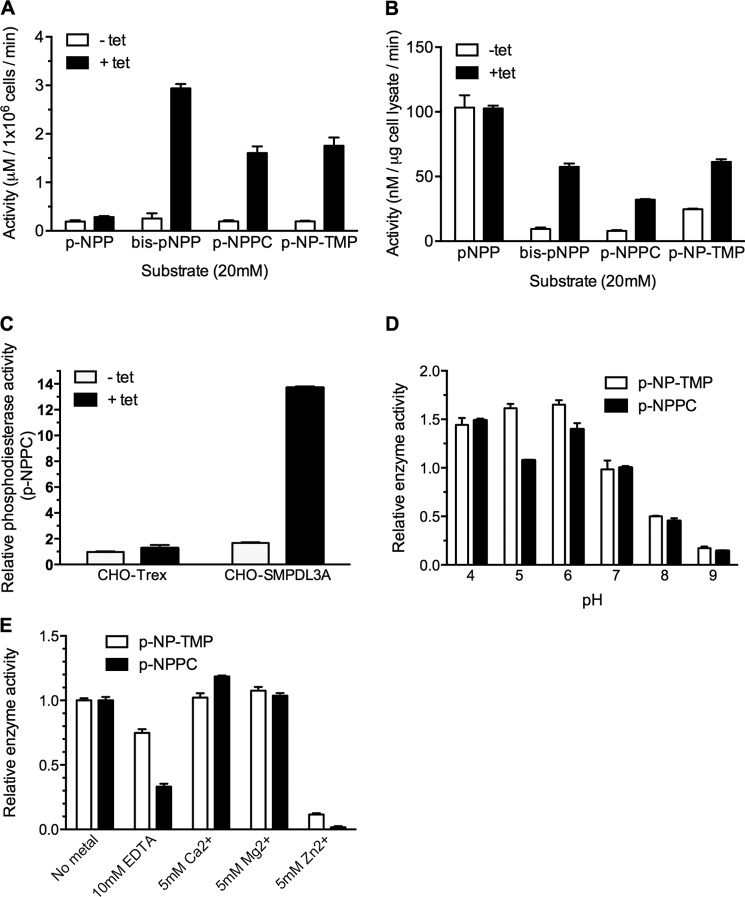
The enzymatic activity of recombinant cellular and secreted human SMPDL3A from tet-induced CHO-SMPDL3A cells was measured using a panel of p-nitrophenol-conjugated synthetic substrates. Hydrolysis of these substrates at the phosphodiester bond releases the p-nitrophenol group, with a corresponding increase in absorbance at 405 nm. For each substrate, cell lysates or conditioned media from noninduced CHO-SMPDL3A cells were used as negative controls. Significantly increased activity was detected in conditioned media from tet-induced cells against the generic phosphodiesterase substrate bis-p-NPP, the phosphorylcholine analog p-NPPC, and the nucleotide analog p-NP-TMP (Fig. 6A), demonstrating secreted SMPDL3A possesses both phosphodiesterase and nucleotide phosphoesterase activities. Negligible activity was detected against the classical alkaline phosphatase substrate (p-NPP), indicating secreted SMPDL3A does not possess phosphatase/monophosphoesterase activity. As expected, high background levels of phosphatase/monophosphoesterase activity against p-NPP were detected in whole cell lysates. Otherwise, data from cell lysates provide the same pattern of SMPDL3A activity for phosphodiester-containing substrates as for conditioned media (Fig. 6B). To rule out possible off-target effects of tetracycline induction, nontransfected parental CHO-Trex cells were treated overnight with 1 μm tetracycline. No significant difference in p-NPPC hydrolyzing activity was observed between CHO-Trex cells with or without tetracycline (Fig. 6C), demonstrating that increased enzymatic activities in CHO-SMPDL3A cells are due to increased SMPDL3A expression, rather than off-target effects of tetracycline induction.
FIGURE 6.
Activity of recombinant human SMPDL3A against synthetic p-nitrophenyl phosphodiester substrates. A, activity of conditioned serum-free media harvested from 1 × 106 control or tetracycline-induced CHO-SMPDL3A cells against p-NPP, bis-p-NPP, p-NPPC, and p-NP-TMP. In each case, the final substrate concentration was 20 mm, in 125 mm Tris, pH 7.0, 5 mm Mg2+. B, activity of cell lysates from control or tetracycline-induced CHO-SMPDL3A cells against substrates listed in A. C, parental nontransfected CHO-Trex or transfected CHO-SMPDL3A cells were treated with or without 1 μm tetracycline, and relative phosphodiesterase activity against 20 mm p-NPPC substrate was measured in whole cell lysates (125 mm Tris, pH 7.0, 5 mm Mg2+, 1 h at 37 °C). Activity values were normalized to noninduced CHO-Trex cell lysates. Effect of pH (D) or EDTA/metal ions (E) on relative activity against 20 mm p-NP-TMP and p-NPPC for purified recombinant SMPDL3A is shown. Activity values were normalized to p-NP-TMP at pH 7 or no metal supplementation, respectively. For all experiments involving cell lysates or conditioned media, tetracycline induction was 1 μm final concentration for 16 h. Data are means ± S.D. of three determinations.
The pH dependence on the activity of secreted recombinant SMPDL3A with both p-NPPC and p-NP-TMP substrates was measured over the range 4.0 to 9.0 (Fig. 6D). Secreted SMPDL3A exhibited optimal activity against both substrates in the range pH 4.0–6.0, with activity being significantly reduced at pH above 7.
The effect of divalent metal ions on secreted SMPDL3A activity was examined, using both p-NPPC and p-NP-TMP as substrates (Fig. 6E). Preincubation of SMPDL3A with 10 mm EDTA for 30 min before the start of the assay inhibited activity relative to unsupplemented samples but did not completely abolish activity. Supplementation of the reaction with either 5 mm Ca2+ or Mg2+ had a moderate stimulatory effect, whereas 5 mm Zn2+ virtually abolished all activity. This result contrasts with the enzymology of aSMase, which utilizes Zn2+ ions as a metal co-factor (13, 33).
Lack of Sphingomyelinase Activity for Recombinant Human SMPDL3A
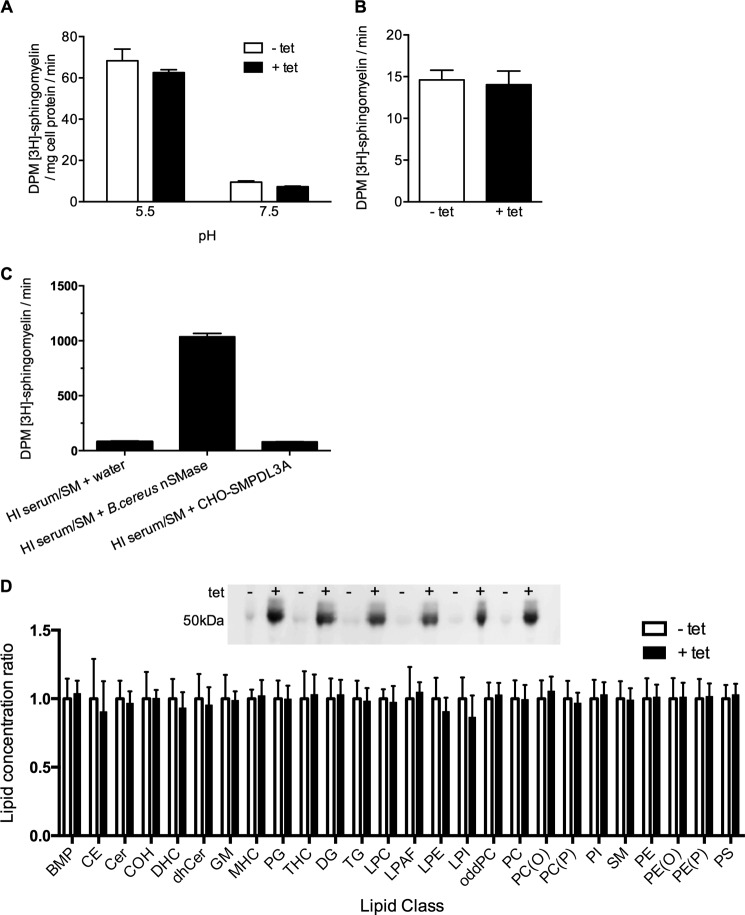
As shown in Fig. 6, cellular and secreted recombinant human SMPDL3A hydrolyzes p-NPPC, a synthetic analog of the phosphorylcholine headgroup of lipids such as sphingomyelin and phosphatidylcholine. To determine whether this translated to activity against authentic sphingomyelin, assays were performed on cell lysates from tetracycline-induced CHO-SMPDL3A cells, using 3H-labeled sphingomyelin/Triton X-100 micelles as a substrate, at both acidic and neutral pH. As expected, both acid and neutral sphingomyelinase activities were detected in both tet-induced and control CHO-SMPDL3A lysates, with acid sphingomyelinase activity being the predominant type (Fig. 7A). However, tet-induced overexpression of SMPDL3A did not result in an increase of either acid or neutral sphingomyelinase activities compared with uninduced controls, indicating that overexpression of SMPDL3A does not detectably contribute to sphingomyelin hydrolysis at either pH.
FIGURE 7.
Sphingomyelinase activity assays of recombinant human SMPDL3A. A, sphingomyelinase activity assay of lysates of CHO-SMPDL3A cells treated ± tetracycline (1 μm, 16 h), using [3H]choline methyl-labeled sphingomyelin/Triton X-100 micelles as substrate under acidic (pH 5.5; 100 mm sodium acetate, 5 mm Zn2+) or neutral (pH 7.5; 100 mm Tris, 5 mm Mg2+) conditions. B, sphingomyelinase activity assay of conditioned serum-free media from CHO-SMPDL3A cells treated ± tetracycline at pH 7.5 under conditions described in A. C, sphingomyelinase activity assay of HI pooled human serum containing 1 μCi if [3H]sphingomyelin (SM). HI serum/sphingomyelin mixtures were supplemented (50% v/v) either with water, 1 unit of B. cereus neutral sphingomyelinase, or conditioned serum-free media from tetracycline-induced (1 μm, 16 h) CHO-SMPDL3A cells. D, lipidomic MS/MS analysis of SMPDL3A-overexpressing human cells. Inset, HEK-SMPDL3A cells were grown to near-confluency in 75-cm2 flasks and treated with or without 1 μm tetracycline for 24 h (n = 6 flasks for each treatment), and whole cell lysates were prepared. Expression of SMPDL3A was examined by Western blotting with 6E3G4A1 anti-SMPDL3A antibodies. Total cellular lipids were extracted from lysates, and lipidomic profiles were generated by tandem LC-MS/MS. Lipid concentrations for each class are normalized to that of the noninduced control cells. No significant differences in the concentrations of any examined lipid class exist between SMPDL3A-induced and -uninduced cells (unpaired two-tailed t test). Lipid class abbreviations are as follows: BMP, bis(monoacylglycerol)phosphate; CE, cholesterol esters; Cer, ceramide; COH, free cholesterol; DHC, dihexosylceramide; dhCer, dihydroceramide; GM, gangliosides; MHC, monohexosylceramide; PG, phosphatidylglycerol; THC, trihexosylceramide; DG, diacylglycerol; TG, triacylglycerol; LPC, lysophosphatidylcholine; LPAF, lyso-platelet-activating factor; LPE, lysophosphatidylethanolamine; LPI, lysophosphatidylinositol; oddPC, odd chain phosphatidylcholine; PC, phosphatidylcholine; PC(O), alkyphosphatidylcholine; PC(P), phosphatidylcholine plasmalogen; PI, phosphatidylinositol; SM, sphingomyelin; PE, phosphatidylethanolamine; PE(O), alkylphosphatidylethanolamine; PE(P), phosphatidylethanolamine plasmalogen; PS, phosphatidylserine.
The activity of SMPDL3A secreted from CHO-SMPDL3A cells was also tested using the [3H]sphingomyelin/Triton X-100 micelle assay at neutral pH (7.4). As for cell lysates, no difference in sphingomyelin hydrolysis was detected in conditioned media collected from tet-induced CHO-SMPDL3A cells versus noninduced controls (Fig. 7B). As SMPDL3A is present in the mammalian circulation (Fig. 4, B–D), we considered the possibility that secreted recombinant SMPDL3A lacked an essential co-factor found only in serum. To test this possibility, conditioned medium from tet-induced CHO-SMPDL3A cells was supplemented with pooled human serum containing lipoprotein-equilibrated [3H]sphingomyelin, which had been HI (56 °C for 30 min) to eliminate any endogenous serum sphingomyelinase activity. However, no increase in [3H]sphingomyelin hydrolysis was detected in conditioned media from tet-induced CHO-SMPDL3A cells supplemented with HI serum versus HI serum alone (Fig. 7C). A positive control reaction supplemented with recombinant Bacillus cereus neutral sphingomyelinase showed abundant cleavage of the [3H]sphingomyelin substrate, indicating that HI human serum did not contain an inhibitor of the hydrolysis reaction.
We also used an alternative method of assaying sphingomyelinase activity, using a commercially available enzymatically coupled assay, based on Amplex Red fluorescence detection of phosphorylcholine released from the hydrolysis of unlabeled sphingomyelin/Triton X-100 micelles. Again, no difference in enzyme activity could be detected between tet-induced and control CHO-SMPDL3A cell lysates or conditioned media samples using this assay at either pH 5.5 or 7.4 (data not shown). Using the Amplex Red kit, we also investigated whether SMPDL3A could utilize phosphatidylcholine (prepared as Triton X-100 micelles at the same concentration as for sphingomyelin). As for sphingomyelin, no difference was detected between tet-induced and control CHO-SMPDL3A-conditioned media with phosphatidylcholine substrate (data not shown).
Artificial overexpression of aSMase in human cells results in increased levels of intracellular ceramide, a product of sphingomyelin hydrolysis, which are detectable using mass spectrometry (34). As it was possible that our in vitro sphingomyelinase assays lacked an essential co-factor or condition only present within an intact cellular environment, we examined whether SMPDL3A overexpression in human cells was associated with increased ceramide and/or reduced sphingolipid levels. A lipidomic profile of 285 individual cellular ceramides, sphingolipid, phospholipid, and sterol species was measured using tandem mass spectrometry in tet-induced HEK-SMPDL3A cells and noninduced controls (n = 6 each for induced and control cells, Fig. 7D, inset). No significant differences were detected in the levels of any assayed lipid species between SMPDL3A-overexpressing HEK cells and controls (Fig. 7D). The unperturbed lipid concentrations in SMPDL3A-overexpressing HEK cells indicates that SMPDL3A likely has no activity against any major natural phospholipid or sphingolipid substrate in this environment. In conclusion, we have been unable to demonstrate that either intracellular or secreted recombinant human SMPDL3A exhibits any enzymatic activity against sphingomyelin or any other phosphorylcholine-containing lipid substrate.
Recombinant Human SMPDL3A Is a Nucleotide Phosphodiesterase
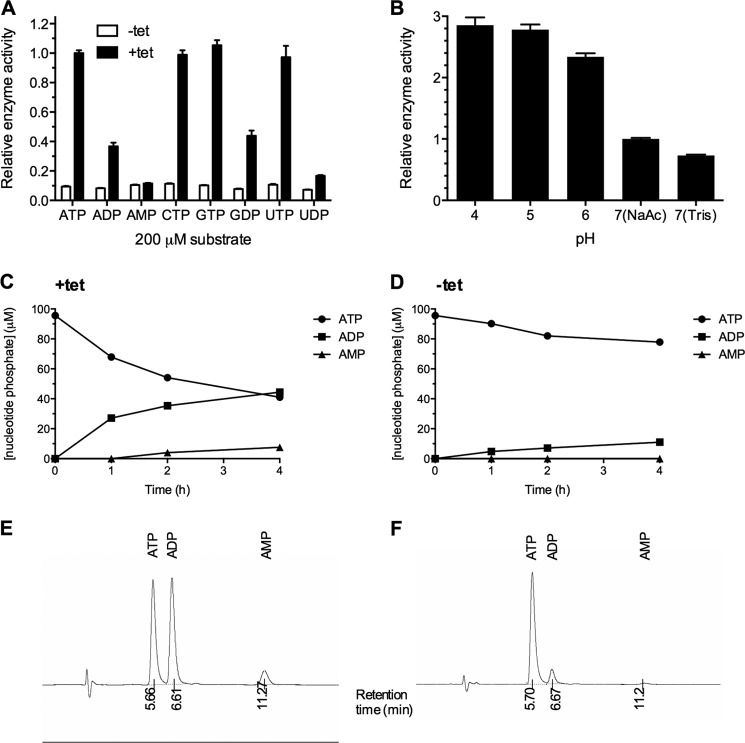
As recombinant human SMPDL3A is capable of utilizing the nucleotide analog p-NP-TMP as a substrate, we also screened the activity of SMPDL3A against a panel of cyclic, mono-, di-, and tri-phosphate nucleotides. Using the malachite green phosphate detection assay, we determined that conditioned media from tet-induced CHO-SMPDL3A cells exhibited significantly increased activity against nucleotide triphosphates (ATP, CTP, GTP, and UTP) relative to media from uninduced control cells (Fig. 8A). All nucleotide triphosphates were hydrolyzed with approximately equal affinity. Significant activity against nucleotide diphosphates (ADP, GDP, and UDP) was also detected in tet-induced versus uninduced CHO-SMPDL3A media, although the level of this activity was substantially lower than that against triphosphate substrates. No significant activity was observed against the nucleotide monophosphate AMP, which is consistent with the lack of activity against the monophosphoester substrate p-nitrophenyl phosphate (Fig. 8A). Despite hydrolysis of the phosphodiester bond in p-NP-TMP, SMPDL3A exhibited negligible phosphodiesterase activity against authentic cyclic nucleotide substrates 3′–5′-cAMP or 3′–5′-cGMP (data not shown). As for synthetic phosphodiester substrates, SMPDL3A exhibited an acidic pH optimum when hydrolyzing ATP as a substrate (pH optimum 4–5). Moderate inhibition of activity was observed at pH 7 using Tris·HCl as a buffer versus sodium acetate (Fig. 8B).
FIGURE 8.
A, nucleotide phosphodiesterase activity of recombinant SMPDL3A against nucleotide phosphate substrates. The activity of concentrated and buffer-exchanged conditioned media from CHO-SMPDL3A cells treated with or without 1 μm tetracycline for 16 h was determined against a panel of nucleotide phosphates. Activity was measured by colorimetric detection at 620 nm of released inorganic phosphate. Concentrated conditioned medium was mixed with reaction buffer in a 1:1 v/v ratio. Final reaction conditions were 100 mm Tris, pH 7.0, 5 mm Mg2+, and 200 μm nucleotide phosphate. Data are means ± S.D. of three determinations. B, pH dependence of purified recombinant SMPDL3A on ATP hydrolytic activity (100 mm sodium acetate, pH 4–7, 100 mm Tris, pH 7). Reverse phase HPLC analysis of reaction products after incubation of ATP with conditioned media from tetracycline-induced (C) and noninduced control (D) CHO-SMPDL3A cells. Reaction mixture was 100 μm ATP, 100 mm sodium acetate, pH 7. Detection of nucleotide products was performed by measuring absorbance at 254 nm. Representative chromatograms showing retention times for nucleotide species for induced and noninduced samples are shown in E and F, respectively.
To determine the position of nucleotide phosphate cleavage by SMPDL3A, reversed-phase HPLC was used to separate and identify the products of ATP hydrolysis by SMPDL3A secreted from tet-induced CHO-SMPDL3A cells at pH 7.0. ADP was the major product of SMPDL3A activity against ATP, accounting for 100% of reaction product at after 1 h, and >85% of total product after 4 h. This indicates that SMPDL3A preferentially hydrolyzes the γ-phosphate bond of ATP, acting as a nucleotide phosphodiesterase. Minor levels of AMP are detected after 4 h of incubation, most likely arising from the further release of a single phosphate from ADP. This is consistent with the ability of SMPDL3A to hydrolyze ADP, albeit at a reduced rate compared with ATP (Fig. 8A). No other nucleotide species, particularly adenosine, were detected on the 254 nm absorbance UV HPLC trace after 4 h of incubation, eliminating the α-phosphate bond of ATP as a significant site of hydrolysis by SMPDL3A. In conclusion, when presented with ATP as a substrate, SMPDL3A acts as a nucleotide phosphodiesterase, predominantly cleaving the γ-phosphate bond.
SMPDL3A Is the Predominant LXR-regulated Secreted Nucleotide Phosphodiesterase in Human Macrophages
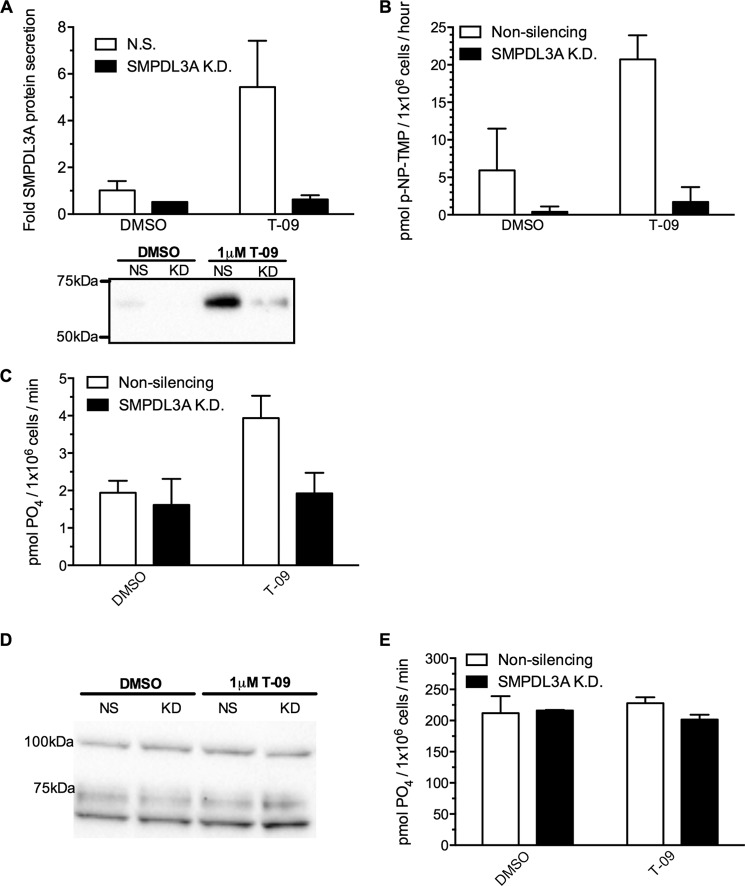
In addition to human SMPDL3A expressed in CHO cells, we also assessed the nucleotide phosphodiesterase activities of SMPDL3A secreted from LXR-stimulated human THP-1 macrophages, using p-NP-TMP and ATP as substrates. The specific contribution of SMPDL3A to enzymatic activity was determined by knocking down SMPDL3A expression with siRNA and comparing activity with scrambled nonsilencing siRNA-treated control cells. As for primary HMDM macrophages, stimulation of THP-1 macrophages with the LXR ligand T-0901317 significantly increased secretion of SMPDL3A above basal levels (Fig. 9A), consistent with previous studies (8, 9). Of note, the molecular mass of secreted SMPDL3A from THP-1 cells is greater than that from HMDMs and is comparable with the recombinant enzyme produced in CHO-SMPDL3A cells (∼65 kDa; Fig. 9A, inset panel). siRNA knockdown of SMPDL3A expression was highly effective, with levels of secreted SMPDL3A falling below those of scrambled nonsilencing siRNA/DMSO-treated control cells, even after LXR stimulation (Fig. 9A).
FIGURE 9.
Secreted SMPDL3A levels and p-NP-TMP/ATP hydrolytic activity from LXR-agonist treated human macrophages. A, PMA differentiated THP-1 macrophages grown in RPMI 1640 medium, 10% FBS were transfected with 50 pmol of SMPDL3A siRNA or scrambled nonsilencing control for 24 h, followed by replacement with serum-free RPMI 1640 medium containing 1 μm T-0901317 or DMSO. Media were harvested after 24 h, concentrated, and analyzed by anti-SMPDL3A Western blot. B, nucleotide phosphoesterase activity from conditioned serum-free media prepared as in A was measured using 20 mm p-NP-TMP as a substrate (reaction buffer 100 mm sodium acetate pH 6.0). Reaction was allowed to proceed overnight at 37 °C, and hydrolysis of the p-NP-TMP substrate was determined by measuring A405 nm. C, phosphate release from ATP in conditioned media as in A measured by Malachite Green assay. Reaction buffer 200 μm ATP, 100 mm Tris·HCl, pH 7.0. D, Western blot analysis of CD39 protein levels in whole cell lysates of THP-1 cells treated as in A. F, cell surface-associated ATP hydrolytic activity of THP-1 cells treated as in A, measured by Malachite Green phosphate detection assay. Reaction conditions were 200 μm ATP, 150 mm NaCl, 2 mm CaCl, 5 mm KCl, 20 mm HEPES, pH 7.4.
After LXR stimulation, secreted nucleotide phosphodiesterase activities against p-NP-TMP and ATP substrates were increased 3.4- and 2-fold versus controls, respectively (Fig. 9, B and C). These increases in secreted nucleotide phosphodiesterase activity appears to be entirely attributable to SMPDL3A, as knockdown of SMPDL3A abolished the effect of LXR stimulation (Fig. 9, B and C). SMPDL3A also appears to be solely responsible for secreted nucleotide phosphodiesterase activity from macrophages under basal nonstimulated conditions, as all activity was abolished in DMSO-treated cells after SMPDL3A knockdown (Fig. 9B).
Macrophages express ectonucleotide triphosphate diphosphohydrolase 1 (ENTPD1, also known as CD39) on their surface (35). CD39 is anchored to the plasma membrane and degrades nucleotide tri- and di-phosphates in the extracellular media (36). To compare the relative activities of cell surface CD39 and secreted SMPDL3A, we also measured cellular CD39 protein expression levels and cell surface-associated hydrolysis of phosphate from ATP from LXR-stimulated THP-1 cells in the same experiment as in Fig. 9, A–C. In contrast to the secretion of SMPDL3A, cellular levels of CD39 protein were unaffected by LXR stimulation with 1 μm T-0901317 (Fig. 9D). siRNA knockdown of SMPDL3A also had no effect on CD39 protein levels. As expected, THP-1 cells exhibited high levels of cell surface ATP hydrolysis (mean activity 214.4 pmol of PO4/1 × 106 cells/min) (Fig. 9E), and this activity was unaffected by knockdown of SMPDL3A. Cell surface-associated ATP hydrolytic activity was also insensitive to LXR stimulation, in contrast to the doubling of SMPDL3A-specific activity (Fig. 9C). In absolute terms, secreted SMPDL3A activity after LXR stimulation is ∼100-fold lower than that occurring at the cell surface at neutral pH.
DISCUSSION
Our microarray study of cholesterol-loaded primary human macrophages led to the identification of SMPDL3A as a novel cholesterol-regulated gene, and we present the first experimental evidence of an enzymatic activity for this protein. Our results confirm bioinformatic predictions that SMPDL3A encodes a functional phosphodiesterase, as demonstrated by its ability to hydrolyze the phosphodiester-containing substrates bis-p-NPP and p-NPPC. Despite significant amino acid sequence homology with aSMase, we were unable to demonstrate that recombinant human SMPDL3A could utilize sphingomyelin as a substrate under standard in vitro assay conditions or affect intracellular sphingolipid, ceramide, or other lipid levels when overexpressed in a human cell line. Unexpectedly, SMPDL3A exhibits a nucleotide phosphodiesterase activity, hydrolyzing the synthetic nucleotide p-NP-TMP and releasing phosphate from natural nucleotide di- and tri-phosphates at an acidic pH optimum. SMPDL3A more efficiently utilizes nucleotide triphosphates versus diphosphates, hydrolyzing them at the γ-phosphate bond. Lack of activity against p-nitrophenol phosphate and the nucleotide monophosphate AMP indicates SMPDL3A does not possess a monophosphoesterase or 5′-adenine monophosphatase activity, such as CD73/5′-nucleotidase (37).
We have for the first time shown that levels of SMPDL3A mRNA, intracellular protein, and secretion are all up-regulated in response to raised cholesterol levels in primary human macrophages. This effect appears to be independent of the means used to deliver cholesterol to the cell, being observed after both scavenger receptor-mediated uptake of acetylated LDL or direct delivery to the plasma membrane using cholesterol-CD complexes. We hypothesize that in the atherosclerotic lesion, densely populated with cholesterol-enriched macrophage foam cells, SMPDL3A expression will also be significantly raised. In support of this hypothesis, microarray analysis of human carotid atherosclerotic plaque tissue by other groups has indicated that SMPDL3A forms part of a gene expression signature that is indicative of inflamed foam cell-rich plaque tissue (38), and SMPDL3A is the fifth most up-regulated mRNA in symptomatic (unstable) plaque tissue obtained from carotid endarterectomy patients (39). Furthermore, our re-analysis of publicly available microarray expression data using the NCBI GEO database and GEO2R tool (40) (accession number GSE21419) reveals that Smpdl3a mRNA is significantly up-regulated in laser-capture microdissected atherosclerotic plaques from apoE−/− mice, versus the adjacent healthy adventitial tissue (41, 42). In this study, neither Smpd1/aSMase nor Smpdl3b mRNA were up-regulated in plaques versus healthy arterial tissue, providing in vivo support of our observation that transcriptional regulation of SMPDL3A is highly distinct from that of its closest homologs. Together, these results indicate that SMPDL3A expression is indeed raised in the atherosclerotic lesions in vivo and may be a specific feature of macrophage foam cells, rather than a generalized tissue response to increased circulating cholesterol levels or development of the lesion. As our results show that secretion of SMPDL3A from HMDMs is also significantly increased after cholesterol loading, this suggests that foam cell-derived SMPDL3A may diffuse into other areas of the plaque and surrounding tissue, creating an SMPDL3A-enriched microenvironment.
We present the first evidence that expression and secretion of SMPDL3A is not only stimulated by LXR agonists but also by raised intracellular cAMP levels. This stimulation was unaffected by pharmacological inhibition of PKA, indicating the involvement of alternative cAMP signaling systems, such as exchange protein directly activated by cAMP (Epac) (43). Further studies could confirm this involvement, by use of Epac mutants and Epac-specific agonists such as 8-(4-chlorophenylthio)-2′-O-Me-cAMP. The contribution of the putative cyclic AMP-responsive element sequences in the SMPDL3A promoter remains to be confirmed by mutational analysis and promoter binding studies.
The physiological significance of cAMP in the regulation of SMPDL3A expression is unknown at present. Levels of CD39 in murine macrophages (44) and human endothelial cells (45) are both increased by raised cAMP concentrations, and this has been hypothesized to play a role in enhancing vascular anti-thrombotic properties. As the nucleotide phosphate substrate specificity of CD39 overlaps with SMPDL3A, there exists the potential for complementary cAMP-driven anti-thrombotic roles at the cell surface and in the extracellular milieu for CD39 and SMPDL3A, respectively.
This study has demonstrated that SMPDL3A is modified by N-linked glycosylation and that the degree of glycosylation varies by cell type, tissue, and species. SMPDL3A contains seven potential N-linked motifs, and differences in the degree of glycosylation may represent variable occupancy of these sites. N-Linked glycosylation of two discrete asparagine residues of aSMase are required for secretion and correct folding of the enzyme, and enzymatic deglycosylation of aSMase results in complete loss of activity (29). The position of both of these asparagine residues are conserved in SMPDL3A, suggesting that glycosylation may play an important role in controlling the behaviors of SMPDL3A as well. Determining the sites and structures of the N-linked glycans of SMPDL3A, as well as their relationship to the cellular origin, secretion, and activity of the enzyme, should be a focus of future studies.
We have shown that SMPDL3A hydrolyzes synthetic phosphodiester and nucleotide di- and tri-phosphate (but not phosphomonoester or nucleotide monophosphate) substrates at acidic and neutral pH, with negligible activity at alkaline pH. SMPDL3A activity is not completely inhibited by preincubation with EDTA (25 and 66% inhibition for p-NP-TMP and p-NPPC substrates, respectively), indicating it is a metalloenzyme, potentially with bound metal ions being relatively inaccessible to direct chelation. SMPDL3A activity is moderately stimulated by Mg2+ and Ca2+ ions but strongly inhibited by Zn2+.
Recombinant human SMPDL3A in a CHO expression system is detected predominantly in the Golgi, with no evidence of lysosomal localization. This set of properties contrasts with those of its homolog aSMase/SMPD1, which is stimulated by Zn2+ and is found intracellularly in the endosomal/lysosomal system (13, 33). Notably, we were unable to demonstrate that recombinant human SMPDL3A was capable of hydrolyzing sphingomyelin, the eponymous substrate of aSMase. Key features in the amino acid sequence of SMPDL3A may account for these divergent properties, including lack of a saposin lipid activator domain, nonconservation of a putative aSMase phosphorylcholine binding domain, and a predicted signaling domain with minimal similarity to that of aSMase. Regulation of the expression of SMPDL3A and aSMase in primary macrophages is also distinct. In contrast to SMPDL3A, we found that expression of aSMase/SMPD1 mRNA is not up-regulated in HMDMs in response to raised cholesterol or cAMP levels nor by treatment with LXR agonists, which is consistent with earlier reports (9). Taken together, these differences in enzymological properties, intracellular localization, and gene regulation indicate that SMPDL3A likely has a distinct physiological role to its homolog aSMase.
It is important to note that our results do not absolutely preclude sphingomyelin or other sphingolipids/phospholipids from being substrates of SMPDL3A. Our assay conditions may not be optimal for this activity, may lack an essential cofactor, or present the sphingomyelin substrate in an inappropriate form. For example, the secreted form of aSMase will not hydrolyze sphingomyelin at neutral pH in the commonly used sphingomyelin/Triton X-100 micelle assay, but it exhibits robust activity when the sphingomyelin substrate is presented as a component of oxidized LDL (4). Similarly, human intestinal alkaline sphingomyelinase activity is dependent on the presence of specific bile salts and is inhibited by Triton X-100/sphingomyelin micelles (46). We are continuing to investigate alternative conditions under which SMPDL3A may hydrolyze sphingomyelin or other lipids.
The ability of SMPDL3A to hydrolyze p-NP-TMP and nucleotide di- and triphosphates at neutral pH is analogous to certain members of the nucleotide phosphodiesterase/pyrophosphatase (47) and ecto-nucleoside triphosphate diphosphohydrolase (48) families. These enzymes are capable of hydrolyzing phosphoanhydride bonds of ATP and ADP to produce AMP, which may subsequently degrade to adenosine (a potent anti-inflammatory signal) by the action of 5′-nucleotidase/CD73 (49, 50). Because these enzymes are either secreted or membrane-anchored with their catalytic domains oriented toward the extracellular space, they play a major role in regulating the levels of nucleotide phosphates in the circulation. Extracellular nucleotides such at ATP and UTP, together with their cellular P2Y/P2X receptors, have become recognized as key modulators of vascular dynamics, platelet aggregation, inflammation, and infection (51). Although levels of cell surface/CD39 ATPase activity are comparatively high in macrophages, several lines of evidence point toward a unique context for SMPDL3A activity. SMPDL3A is secreted and is present in the circulation in soluble form, and although certain CD39 isoforms are shed in a soluble form, CD39 is predominantly cell surface-associated. Expression and secreted activity of SMPDL3A are stimulated by LXR agonists, whereas expression of CD39 protein and cell-surface associated ATP hydrolytic activity in THP-1 macrophages are LXR-insensitive. The substrate preference of secreted SMPDL3A is ATP > ADP, whereas soluble isoforms of CD39, such as CD39-L4, demonstrate a strong preference for nucleotide diphosphates over triphosphates (52). Finally, the optimum pH of SMPDL3A is acidic, whereas for CD39 is 8.0, with activity decreasing by ∼40% at pH 7.0 (53). There is evidence that acidified microenvironments exist within lipid-rich areas of atherosclerotic plaques (54), creating a specific context that would favor both the cholesterol-driven expression and acid-optimal activity of SMPDL3A.
Stimulation of P2 family receptors such as P2X7 by raised extracellular ATP or UTP levels activates multiple pro-inflammatory responses in immune and endothelial cells, including increased release of IL-1β (55), IL-18, and TNFα (56) and chemotactic stimuli such as MCP-1. In the context of atherosclerosis, high fat-fed apoE−/− mice, which were also deficient in the ADP receptors P2Y1 (57) or P2Y12 (58), showed significantly reduced lesion accumulation, as did apoE−/− mice treated with P2Y6-selective antagonists (59). In human patients, both ATP levels and expression of the P2X7 receptor are raised in luminal thrombi and inflammatory infiltrates of carotid artery plaques (60). Together, these results strongly suggest a deleterious role for purinergic signaling in the progression of the atherosclerotic lesion. Given that SMPDL3A is secreted from cholesterol-loaded macrophages, is present in the circulation, and is capable of hydrolyzing ATP, UTP, and other nucleotide phosphates, it is conceivable that SMPDL3A also plays role in purinergic signaling pathways by reducing circulating levels of pro-inflammatory nucleotide phosphates. We have shown that SMPDL3A is largely responsible for increases in p-NP-TMP and ATP hydrolysis observed in the media of LXR-stimulated human macrophages. This suggests SMPDL3A may also be a significant source of secreted nucleotide phosphodiesterase activity in the cholesterol-rich and macrophage-dense environment of the atherosclerotic plaque. This activity, by decreasing the local concentration of nucleotide tri- and di-phosphates (particularly ATP or UTP), may dampen the activation of pro-inflammatory purinergic signaling receptors. If correct, the responsiveness of SMPDL3A in macrophages to synthetic LXR ligands might make it an attractive candidate for therapeutic modulation, and it should be a topic for future investigation.
While our manuscript was in preparation, a study of a novel acid sphingomyelinase-like protein from the bacterium Ralstonia solanacearum, RsASML, was published (61). Consistent with our results for human SMPDL3A, R. solanacearum RsASML exhibited no detectable sphingomyelinase activity but was capable of hydrolyzing both ATP and ADP to AMP (but not AMP to adenosine). Together with our study, the work of Airola et al. (61) strongly supports the idea that SMPDL3A (and possibly the closely related homolog SMPDL3B) possesses an entirely distinct enzymatic activity to acid sphingomyelinase, which is regulated independently, and with potentially unique physiological roles.
Supplementary Material
Acknowledgments
We thank Raani Kumaran, Diana Nawara, Sharon Ngoh, and Jeannette Hallab for technical assistance and Jacquelyn Weir and Ricardo Tan for assistance with lipid mass spectrometry data analysis.
This work was supported in part by a National Heart Foundation postdoctoral fellowship (to M. T.), National Health and Medical Research Council of Australia Program (to W. J, and L. K.), and Principal Research fellowship grants.

This article contains supplemental Fig. S1 and Tables S1 and S2.
- HMDM
- human monocyte-derived macrophage
- AcLDL
- acetylated LDL
- aSMase
- acid sphingomyelinase
- bis-p-NPP
- bis-(p-nitrophenyl) phosphate
- Bt2cAMP
- dibutyryl-cyclic AMP
- LPDS
- lipoprotein-deficient serum
- LXR
- liver X receptor
- PMA
- phorbol 12-myristate 13-acetate
- p-NPP
- p-nitrophenol phosphate
- p-NPPC
- p-nitrophenylphosphorylcholine
- p-NP-TMP
- p-nitrophenyl thymidine 5′-monophosphate
- bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- tet
- tetracycline
- HI
- heat-inactivated
- PNGaseF
- peptide:N-glycosidase F
- QRT
- quantitative RT
- aRNA
- antisense RNA
REFERENCES
- 1. Tabas I. (2010) Macrophage death and defective inflammation resolution in atherosclerosis. Nat. Rev. Immunol. 10, 36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shashkin P., Dragulev B., Ley K. (2005) Macrophage differentiation to foam cells. Curr. Pharm. Des. 11, 3061–3072 [DOI] [PubMed] [Google Scholar]
- 3. Zeidan Y. H., Hannun Y. A. (2010) The acid sphingomyelinase/ceramide pathway: biomedical significance and mechanisms of regulation. Curr. Mol. Med. 10, 454–466 [DOI] [PubMed] [Google Scholar]
- 4. Schissel S. L., Jiang X., Tweedie-Hardman J., Jeong T., Camejo E. H., Najib J., Rapp J. H., Williams K. J., Tabas I. (1998) Secretory sphingomyelinase, a product of the acid sphingomyelinase gene, can hydrolyze atherogenic lipoproteins at neutral pH. Implications for atherosclerotic lesion development. J. Biol. Chem. 273, 2738–2746 [DOI] [PubMed] [Google Scholar]
- 5. Oörni K., Posio P., Ala-Korpela M., Jauhiainen M., Kovanen P. T. (2005) Sphingomyelinase induces aggregation and fusion of small very low-density lipoprotein and intermediate-density lipoprotein particles and increases their retention to human arterial proteoglycans. Arterioscler. Thromb. Vasc. Biol. 25, 1678–1683 [DOI] [PubMed] [Google Scholar]
- 6. Devlin C. M., Leventhal A. R., Kuriakose G., Schuchman E. H., Williams K. J., Tabas I. (2008) Acid sphingomyelinase promotes lipoprotein retention within early atheromata and accelerates lesion progression. Arterioscler. Thromb. Vasc. Biol. 28, 1723–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wright K. O., Messing E. M., Reeder J. E. (2002) Increased expression of the acid sphingomyelinase-like protein ASML3a in bladder tumors. J. Urol. 168, 2645–2649 [DOI] [PubMed] [Google Scholar]
- 8. Pehkonen P., Welter-Stahl L., Diwo J., Ryynänen J., Wienecke-Baldacchino A., Heikkinen S., Treuter E., Steffensen K. R., Carlberg C. (2012) Genome-wide landscape of liver X receptor chromatin binding and gene regulation in human macrophages. BMC Genomics 13, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Noto P. B., Bukhtiyarov Y., Shi M., McKeever B. M., McGeehan G. M., Lala D. S. (2012) Regulation of sphingomyelin phosphodiesterase acid-like 3A gene (SMPDL3A) by liver X receptors. Mol. Pharmacol. 82, 719–727 [DOI] [PubMed] [Google Scholar]
- 10. Kritharides L., Jessup W., Mander E. L., Dean R. T. (1995) Apolipoprotein A-I-mediated efflux of sterols from oxidized LDL-loaded macrophages. Arterioscler. Thromb. Vasc. Biol. 15, 276–289 [DOI] [PubMed] [Google Scholar]
- 11. Kockx M., Dinnes D. L., Huang K.-Y., Sharpe L. J., Jessup W., Brown A. J., Kritharides L. (2012) Cholesterol accumulation inhibits ER to Golgi transport and protein secretion: studies of apolipoprotein E and VSVGt. Biochem. J. 447, 51–60 [DOI] [PubMed] [Google Scholar]
- 12. Kockx M., Guo D. L., Traini M., Gaus K., Kay J., Wimmer-Kleikamp S., Rentero C., Burnett J. R., Le Goff W., Van Eck M., Stow J. L., Jessup W., Kritharides L. (2009) Cyclosporin A decreases apolipoprotein E secretion from human macrophages via a protein phosphatase 2B-dependent and ATP-binding cassette transporter A1 (ABCA1)-independent pathway. J. Biol. Chem. 284, 24144–24154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schissel S. L., Schuchman E. H., Williams K. J., Tabas I. (1996) Zn2+-stimulated sphingomyelinase is secreted by many cell types and is a product of the acid sphingomyelinase gene. J. Biol. Chem. 271, 18431–18436 [DOI] [PubMed] [Google Scholar]
- 14. Bligh E. G., Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 15. Meikle P. J., Wong G., Tsorotes D., Barlow C. K., Weir J. M., Christopher M. J., MacIntosh G. L., Goudey B., Stern L., Kowalczyk A., Haviv I., White A. J., Dart A. M., Duffy S. J., Jennings G. L., Kingwell B. A. (2011) Plasma lipidomic analysis of stable and unstable coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 31, 2723–2732 [DOI] [PubMed] [Google Scholar]
- 16. Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. (2005) MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21, 2933–2942 [DOI] [PubMed] [Google Scholar]
- 17. Im S.-S., Osborne T. F. (2011) Liver X receptors in atherosclerosis and inflammation. Circ. Res. 108, 996–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kockx M., Guo D. L., Huby T., Lesnik P., Kay J., Sabaretnam T., Jary E., Hill M., Gaus K., Chapman J., Stow J. L., Jessup W., Kritharides L. (2007) Secretion of apolipoprotein E from macrophages occurs via a protein kinase A and calcium-dependent pathway along the microtubule network. Circ. Res. 101, 607–616 [DOI] [PubMed] [Google Scholar]
- 19. Koonin E. V. (1994) Conserved sequence pattern in a wide variety of phosphoesterases. Protein Sci. 3, 356–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seto M., Whitlow M., McCarrick M. A., Srinivasan S., Zhu Y., Pagila R., Mintzer R., Light D., Johns A., Meurer-Ogden J. A. (2004) A model of the acid sphingomyelinase phosphoesterase domain based on its remote structural homolog purple acid phosphatase. Protein Sci. 13, 3172–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lansmann S., Schuette C. G., Bartelsen O., Hoernschemeyer J., Linke T., Weisgerber J., Sandhoff K. (2003) Human acid sphingomyelinase. Eur. J. Biochem. 270, 1076–1088 [DOI] [PubMed] [Google Scholar]
- 22. Ponting C. P. (1994) Acid sphingomyelinase possesses a domain homologous to its activator proteins: saposins B and D. Protein Sci. 3, 359–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vaccaro A. M., Salvioli R., Tatti M., Ciaffoni F. (1999) Saposins and their interaction with lipids. Neurochem Res 24, 307–314 [DOI] [PubMed] [Google Scholar]
- 24. Kölzer M., Ferlinz K., Bartelsen O., Hoops S. L., Lang F., Sandhoff K. (2004) Functional characterization of the postulated intramolecular sphingolipid activator protein domain of human acid sphingomyelinase. Biol. Chem. 385, 1193–1195 [DOI] [PubMed] [Google Scholar]
- 25. Ferlinz K., Hurwitz R., Vielhaber G., Suzuki K., Sandhoff K. (1994) Occurrence of two molecular forms of human acid sphingomyelinase. Biochem. J. 301, 855–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 27. Pierleoni A., Martelli P. L., Casadio R. (2008) PredGPI: a GPI-anchor predictor. BMC Bioinformatics 9, 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Masuishi Y., Nomura A., Okayama A., Kimura Y., Arakawa N., Hirano H. (2013) Mass spectrometric identification of glycosylphosphatidylinositol-anchored peptides. J. Proteome Res. 12, 4617–4626 [DOI] [PubMed] [Google Scholar]
- 29. Ferlinz K., Hurwitz R., Moczall H., Lansmann S., Schuchman E. H., Sandhoff K. (1997) Functional characterization of the N-glycosylation sites of human acid sphingomyelinase by site-directed mutagenesis. Eur. J. Biochem. 243, 511–517 [DOI] [PubMed] [Google Scholar]
- 30. Jones I., He X., Katouzian F., Darroch P. I., Schuchman E. H. (2008) Characterization of common SMPD1 mutations causing types A and B Niemann-Pick disease and generation of mutation-specific mouse models. Mol. Genet. Metab. 95, 152–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee C. Y., Tamura T., Rabah N., Lee D.-Y., Ruel I., Hafiane A., Iatan I., Nyholt D., Laporte F., Lazure C., Wada I., Krimbou L., Genest J. (2007) Carboxyl-terminal disulfide bond of acid sphingomyelinase is critical for its secretion and enzymatic function. Biochemistry 46, 14969–14978 [DOI] [PubMed] [Google Scholar]
- 32. Fornoni A., Sageshima J., Wei C., Merscher-Gomez S., Aguillon-Prada R., Jauregui A. N., Li J., Mattiazzi A., Ciancio G., Chen L., Zilleruelo G., Abitbol C., Chandar J., Seeherunvong W., Ricordi C., Ikehata M., Rastaldi M. P., Reiser J., Burke G. W. (2011) Rituximab targets podocytes in recurrent focal segmental glomerulosclerosis. Sci. Transl. Med. 3, 85ra46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schissel S. L., Keesler G. A., Schuchman E. H., Williams K. J., Tabas I. (1998) The cellular trafficking and zinc dependence of secretory and lysosomal sphingomyelinase, two products of the acid sphingomyelinase gene. J. Biol. Chem. 273, 18250–18259 [DOI] [PubMed] [Google Scholar]
- 34. Jenkins R. W., Canals D., Idkowiak-Baldys J., Simbari F., Roddy P., Perry D. M., Kitatani K., Luberto C., Hannun Y. A. (2010) Regulated secretion of acid sphingomyelinase: implications for selectivity of ceramide formation. J. Biol. Chem. 285, 35706–35718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kansas G. S., Wood G. S., Tedder T. F. (1991) Expression, distribution, and biochemistry of human CD39. Role in activation-associated homotypic adhesion of lymphocytes. J. Immunol. 146, 2235–2244 [PubMed] [Google Scholar]
- 36. Knowles A. F. (2011) The GDA1_CD39 superfamily: NTPDases with diverse functions. Purinergic Signal. 7, 21–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zimmermann H. (1992) 5′-Nucleotidase: molecular structure and functional aspects. Biochem. J. 285, 345–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Puig O., Yuan J., Stepaniants S., Zieba R., Zycband E., Morris M., Coulter S., Yu X., Menke J., Woods J., Chen F., Ramey D. R., He X., O'Neill E. A., Hailman E., Johns D. G., Hubbard B. K., Yee Lum P., Wright S. D., Desouza M. M., Plump A., Reiser V. (2011) A gene expression signature that classifies human atherosclerotic plaque by relative inflammation status. Circ. Cardiovasc. Genet. 4, 595–604 [DOI] [PubMed] [Google Scholar]
- 39. Perisic L., Hedin E., Razuvaev A., Lengquist M., Osterholm C., Folkersen L., Gillgren P., Paulsson-Berne G., Ponten F., Odeberg J., Hedin U. (2013) Profiling of atherosclerotic lesions by gene and tissue microarrays reveals PCSK6 as a novel protease in unstable carotid atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 33, 2432–2443 [DOI] [PubMed] [Google Scholar]
- 40. Barrett T., Wilhite S. E., Ledoux P., Evangelista C., Kim I. F., Tomashevsky M., Marshall K. A., Phillippy K. H., Sherman P. M., Holko M., Yefanov A., Lee H., Zhang N., Robertson C. L., Serova N., Davis S., Soboleva A. (2013) NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 41, D991–D995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moos M. P., John N., Gräbner R., Nossmann S., Günther B., Vollandt R., Funk C. D., Kaiser B., Habenicht A. J. (2005) The lamina adventitia is the major site of immune cell accumulation in standard chow-fed apolipoprotein E-deficient mice. Arterioscler. Thromb. Vasc. Biol. 25, 2386–2391 [DOI] [PubMed] [Google Scholar]
- 42. Beer M., Doepping S., Hildner M., Weber G., Grabner R., Hu D., Mohanta S. K., Srikakulapu P., Weih F., Habenicht A. J. (2011) Laser-capture microdissection of hyperlipidemic/ApoE−/− mouse aorta atherosclerosis. Methods Mol. Biol. 755, 417–428 [DOI] [PubMed] [Google Scholar]
- 43. Schmidt M., Dekker F. J., Maarsingh H. (2013) Exchange protein directly activated by cAMP (epac): a multidomain cAMP mediator in the regulation of diverse biological functions. Pharmacol. Rev. 65, 670–709 [DOI] [PubMed] [Google Scholar]
- 44. Liao H., Hyman M. C., Baek A. E., Fukase K., Pinsky D. J. (2010) cAMP/CREB-mediated transcriptional regulation of ectonucleoside triphosphate diphosphohydrolase 1 (CD39) expression. J. Biol. Chem. 285, 14791–14805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Baek A. E., Kanthi Y., Sutton N. R., Liao H., Pinsky D. J. (2013) Regulation of ecto-apyrase CD39 (ENTPD1) expression by phosphodiesterase III (PDE3). FASEB J. 27, 4419–4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Duan R.-D., Bergman T., Xu N., Wu J., Cheng Y., Duan J., Nelander S., Palmberg C., Nilsson A. (2003) Identification of human intestinal alkaline sphingomyelinase as a novel ecto-enzyme related to the nucleotide phosphodiesterase family. J. Biol. Chem. 278, 38528–38536 [DOI] [PubMed] [Google Scholar]
- 47. Stefan C., Jansen S., Bollen M. (2005) NPP-type ectophosphodiesterases: unity in diversity. Trends Biochem. Sci. 30, 542–550 [DOI] [PubMed] [Google Scholar]
- 48. Robson S. C., Sévigny J., Zimmermann H. (2006) The E-NTPDase family of ectonucleotidases: structure function relationships and pathophysiological significance. Purinergic Signal. 2, 409–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Colgan S. P., Eltzschig H. K., Eckle T., Thompson L. F. (2006) Physiological roles for ecto-5′-nucleotidase (CD73). Purinergic Signal. 2, 351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yegutkin G. G., Wieringa B., Robson S. C., Jalkanen S. (2012) Metabolism of circulating ADP in the bloodstream is mediated via integrated actions of soluble adenylate kinase-1 and NTPDase1/CD39 activities. FASEB J. 26, 3875–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Erlinge D., Burnstock G. (2007) P2 receptors in cardiovascular regulation and disease. Purinergic Signal. 4, 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mulero J. J., Yeung G., Nelken S. T., Ford J. E. (1999) CD39-L4 is a secreted human apyrase, specific for the hydrolysis of nucleoside diphosphates. J. Biol. Chem. 274, 20064–20067 [DOI] [PubMed] [Google Scholar]
- 53. Leal D. B., Streher C. A., Neu T. N., Bittencourt F. P., Leal C. A., da Silva J. E., Morsch V. M., Schetinger M. R. (2005) Characterization of NTPDase (NTPDase1; ecto-apyrase; ecto-diphosphohydrolase; CD39; EC 3.6.1.5) activity in human lymphocytes. Biochim. Biophys. Acta 1721, 9–15 [DOI] [PubMed] [Google Scholar]
- 54. Naghavi M., John R., Naguib S., Siadaty M. S., Grasu R., Kurian K. C., van Winkle W. B., Soller B., Litovsky S., Madjid M., Willerson J. T., Casscells W. (2002) pH heterogeneity of human and rabbit atherosclerotic plaques; a new insight into detection of vulnerable plaque. Atherosclerosis 164, 27–35 [DOI] [PubMed] [Google Scholar]
- 55. Ferrari D., Chiozzi P., Falzoni S., Dal Susino M., Melchiorri L., Baricordi O. R., Di Virgilio F. (1997) Extracellular ATP triggers IL-1β release by activating the purinergic P2Z receptor of human macrophages. J. Immunol. 159, 1451–1458 [PubMed] [Google Scholar]
- 56. Hide I., Tanaka M., Inoue A., Nakajima K., Kohsaka S., Inoue K., Nakata Y. (2000) Extracellular ATP triggers tumor necrosis factor-alpha release from rat microglia. J. Neurochem. 75, 965–972 [DOI] [PubMed] [Google Scholar]
- 57. Hechler B., Freund M., Ravanat C., Magnenat S., Cazenave J.-P., Gachet C. (2008) Reduced atherosclerotic lesions in P2Y1/apolipoprotein E double-knockout mice: the contribution of nonhematopoietic-derived P2Y1 receptors. Circulation 118, 754–763 [DOI] [PubMed] [Google Scholar]
- 58. Li D., Wang Y., Zhang L., Luo X., Li J., Chen X., Niu H., Wang K., Sun Y., Wang X., Yan Y., Chai W., Gartner T. K., Liu J. (2012) Roles of purinergic receptor P2Y, G protein-coupled 12 in the development of atherosclerosis in apolipoprotein E-deficient mice. Arterioscler. Thromb. Vasc. Biol. 32, e81–e9 [DOI] [PubMed] [Google Scholar]
- 59. Guns P.-J., Hendrickx J., Van Assche T., Fransen P., Bult H. (2010) P2Y receptors and atherosclerosis in apolipoprotein E-deficient mice. Br. J. Pharmacol. 159, 326–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Piscopiello M., Sessa M., Anzalone N., Castellano R., Maisano F., Ferrero E., Chiesa R., Alfieri O., Comi G., Ferrero M. E., Foglieni C. (2013) P2X7 receptor is expressed in human vessels and might play a role in atherosclerosis. Int. J. Cardiol. 168, 2863–2866 [DOI] [PubMed] [Google Scholar]
- 61. Airola M. V., Tumolo J. M., Snider J., Hannun Y. A. Identification and biochemical characterization of an acid sphingomyelinase-like protein from the bacterial plant pathogen Ralstonia solanacearum that hydrolyzes ATP to AMP but not sphingomyelin to ceramide. PLoS ONE 9, e105830. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.