Background: The bacterial type VI secretion apparatus is a phage tail-like structure that secretes toxins/effectors.
Results: Four components of the type VI secretion system have coevolved and interact.
Conclusion: Distinct classes of type VI secretion systems emerge from the phylogenetic data.
Significance: Molecular understanding of type VI secretion might make it amenable to targeting by antimicrobials.
Keywords: ATPases Associated with Diverse Cellular Activities (AAA), Bacteriophage, Protein Evolution, Protein Secretion, Pseudomonas aeruginosa (P. aeruginosa), ClpV, Type VI Secretion System (T6SS)
Abstract
The type VI secretion system (T6SS) is a bacterial nanomachine for the transport of effector molecules into prokaryotic and eukaryotic cells. It involves the assembly of a tubular structure composed of TssB and TssC that is similar to the tail sheath of bacteriophages. The sheath contracts to provide the energy needed for effector delivery. The AAA+ ATPase ClpV disassembles the contracted sheath, which resets the systems for reassembly of an extended sheath that is ready to fire again. This mechanism is crucial for T6SS function. In Vibrio cholerae, ClpV binds the N terminus of TssC within a hydrophobic groove. In this study, we resolved the crystal structure of the N-terminal domain of Pseudomonas aeruginosa ClpV1 and observed structural alterations in the hydrophobic groove. The modification in the ClpV1 groove is matched by a change in the N terminus of TssC, suggesting the existence of distinct T6SS classes. An accessory T6SS component, TagJ/HsiE, exists predominantly in one of the classes. Using bacterial two-hybrid approaches, we showed that the P. aeruginosa homolog HsiE1 interacts strongly with ClpV1. We then resolved the crystal structure of HsiE1 in complex with the N terminus of HsiB1, a TssB homolog and component of the contractile sheath. Phylogenetic analysis confirmed that these differences distinguish T6SS classes that resulted from a functional co-evolution between TssB, TssC, TagJ/HsiE, and ClpV. The interaction of TagJ/HsiE with the sheath as well as with ClpV suggests an alternative mode of disassembly in which HsiE recruits the ATPase to the sheath.
Introduction
Gram-negative bacteria have evolved various strategies to compete in hostile environments. Among them, secretion systems have attracted a lot of attention because of their clinical importance but also because of their complex architecture and regulation. The conserved bacterial type VI secretion system (T6SS)5 (1, 2) was initially associated with bacteria-host interaction (3–6) but subsequently found to inject toxins into bacterial targets (7–12). These toxins have various activities, the best characterized of which are amidases (13), phospholipases (12), and nucleases (14). However, the list of T6SS toxins and their cognate immunity is expanding (15, 16), whereas their role in bacterial competition during host colonization is beginning to be established (14, 17).
A striking feature of the T6SS is its homology to the tail of the T4 phage. The gp19 tail tube is the conduit through which the phage DNA is injected, whereas the gp5-gp27 heterotrimeric complex is the puncturing device that penetrates the host cell membrane (18). The force needed to push the tube and puncturing device is provided by the contraction of the gp18 sheath (19). In the T6SS, TssB and TssC oligomerize into a sheath-like structure, similar to gp18 (20–24), whereas VgrG and Hcp share features with the puncturing device, gp5-gp27, and the tail tube, gp19, respectively (25).
A fundamental distinction between phage and T6SS is the dynamic nature of injection. The bacteriophage sheath contraction is a singular event, which empties the phage head of its DNA. In this case, return to the original state is unnecessary, and no energy is required to reset the system. In contrast, in T6SS, a series of assembly and contraction events of the TssB/TssC sheath are observed (20, 26, 27), which result in several bursts of toxin injection. ClpV, an AAA+ (ATPase associated with various cellular activities) protein, provides energy for sheath disassembly (21, 27, 28).
AAA+ proteins are hexameric ring-shaped complexes involved in a variety of functions, including protein quality control (29). In the T6SS, ClpV is proposed to disassemble the contracted sheath. In support of this model, it was shown that a Vibrio cholerae clpV mutant has a reduced T6SS-dependent killing activity toward Escherichia coli, suggesting that in this mutant sheath, contraction happens only once, and no subsequent toxin burst occurs (30). Disassembly of the V. cholerae VipA/VipB sheath (TssB/TssC homologs) (20, 21, 27) is dependent on a direct interaction between ClpV and the N-terminal helix of VipB (TssC homolog), which docks into a hydrophobic groove in the N-terminal domain of the ATPase (28).
Pseudomonas aeruginosa is an opportunistic pathogen, which has three T6SSs designated H1- to H3-T6SS (31, 32). Besides 13 conserved core genes, the P. aeruginosa H1-T6SS contains accessory genes, among them tagJ/hsiE1, which is not found in the H2- and H3-T6SSs. Although HsiE1 (also called TagJ) was found to interact with the essential sheath component HsiB1 (TssB/VipA homolog) (33), its exact role in T6SS remains unclear. Here we used a combination of structural and in vivo protein-protein interaction approaches to characterize molecular aspects of the H1-T6SS of P. aeruginosa. Structural characterization of the N domain of ClpV1 and analysis of its interaction with HsiC1 (TssC/VipB homolog) suggest that the H1-T6SS functions differently from the V. cholerae system. We solved the crystal structure of HsiE1 in complex with an N-terminal fragment of HsiB1 and observed that in addition to binding to HsiB1, HsiE1 is capable of interacting with ClpV1. We thus found evidence for distinct T6SS classes, which is confirmed through phylogenetic analysis of the four T6SS components, ClpV, HsiE/TagJ, TssB, and TssC.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Culture Conditions
Strains were cultivated in Luria-Bertani (LB) or Terrific broth at 37 °C. The antibiotics were added at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml. All bacterial strains and plasmids used are listed in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Relevant characteristics/Description | Source/Reference | |
|---|---|---|
| Strain (E. coli) | ||
| One-shotTOP10 | F − mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(araleu) | Invitrogen |
| DHM1 | cya-854 recA1 gyrA96 (NaI) thi1 hsdR17 spoT1 rfbD1 glnV44(AS) | Karimova (41) |
| B834(DE3) | F − ompT hsdSB (rB − mB − ) gal dcm met (DE3) | Laboratory collection |
| Plasmid | ||
| pET28 | Expression vector used for expression of N-terminal 6-histidine tagged proteins | Novagen |
| pET28-hsiE1 | pET28 expressing HsiE1with an N-terminal histidine tag | This study |
| pET28-hsiE1B1 | pET28 expressing HsiE1with an N-terminal histidine tag in tandem with untagged HsiB1 | This study |
| pET28-ClpV1-N | pET28 expressing ClpV1-N with an N-terminal histidine tag | This study |
| pKT25 | BTH vector for fusion of target proteins to B. pertussis cya gene T25 fragment; Plac::cya1–675p15ori, KmR | Karimova et al. (41) |
| pUT18C | BTH vector for fusion of target proteins to B. pertussis cya gene T18 fragment; Plac::cya675–1197pUCori, ApR | Karimova et al. (41) |
| pKT25-zip | Fusion of zip encoding leucine zipper from GCN4 to cya gene T25 fragment in pKT25, KmR | Karimova et al. (41) |
| pUT18C-zip | Fusion of zip encoding leucine zipper from GCN4 to cya gene T18 fragment in pUT18C, ApR | Karimova et al. (41) |
| pKT25-hsiE1 | Fusion of hsiE1 to cya gene T25 fragment in pKT25, KmR | Lossi et al. (33) |
| pUT18C-hsiE1 | Fusion of hsiE1 to cya gene T18 fragment in pUT18C, ApR | Lossi et al. (33) |
| pKT25-hsiB1 | Fusion of hsiB1 to cya gene T25 fragment in pKT25, KmR | Lossi et al. (33) |
| pUT18C-hsiB1 | Fusion of hsiB1 to cya gene T18 fragment in pUT18C, ApR | Lossi et al. (33) |
| pKT25-hsiC1 | Fusion of hsiC1 to cya gene T25 fragment in pKT25, KmR | Lossi et al. (33) |
| pUT18C-hsiC1 | Fusion of hsiC1 to cya gene T18 fragment in pUT18C, ApR | Lossi et al. (33) |
| pKT25-ClpV1 | Fusion of ClpV1 to cya gene T25 fragment in pKT25, KmR | This study |
| pUT18C-ClpV1 | Fusion of ClpV1 to cya gene T18 fragment in pUT18C, ApR | This study |
| pKT25-NterClpV1 | Fusion of NterClpV1 to cya gene T25 fragment in pKT25, KmR | This study |
| pUT18C-NterClpV1 | Fusion of NterClpV1 to cya gene T18 fragment in pUT18C, ApR | This study |
| pKT25-ClpV2 | Fusion of ClpV2 to cya gene T25 fragment in pKT25, KmR | This study |
| pUT18C-ClpV2 | Fusion of ClpV2 to cya gene T18 fragment in pUT18C, ApR | This study |
| pKT25-NterClpV2 | Fusion of NterClpV2 to cya gene T25 fragment in pKT25, KmR | This study |
| pUT18C-NterClpV2 | Fusion of NterClpV2 to cya gene T18 fragment in pUT18C, ApR | This study |
| pKT25-hsiE1Δ1–10 | Fusion of hsiE1Δ1–10 to cya gene T25 fragment in pKT25, KmR | This study |
| pUT18C-hsiE1Δ1–10 | Fusion of hsiE1Δ1–10 to cya gene T18 fragment in pUT18C, ApR | This study |
| pKT25-hsiC1Δ1–33 | Fusion of hsiC1Δ1–33 to cya gene T25 fragment in pKT25, KmR | This study |
| pUT18C-hsiC1Δ1–33 | Fusion of hsiC1Δ1–33 to cya gene T18 fragment in pUT18C, ApR | This study |
| pKT25-hsiC2Δ1–30 | Fusion of hsiC2Δ1–30 to cya gene T25 fragment in pKT25, KmR | This study |
| pUT18C-hsiC2Δ1–30 | Fusion of hsiC2Δ1–30 to cya gene T18 fragment in pUT18C, ApR | This study |
Expression and Protein Purification
The hsiE1 (PA0086) and hsiB1 (PA0083) genes and the sequence corresponding to the first 159 residues of ClpV1 (PA0090) were amplified from P. aeruginosa PAO1 genomic DNA and cloned into pET28. pET28-E1 encodes HsiE1 with an N-terminal His tag cleavable with thrombin. pET28-E1B1 contained hsiE1 preceded by sequence coding for a cleavable N-terminal His tag in frame with hsiB1. pET28-ClpV1-N encodes N-terminally His-tagged ClpV1-N. In all cases, transformed E. coli B834(DE3) cells were grown at 37 °C to an A600 of about 0.6 in Terrific broth. Expression of proteins was induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside, and cells were grown overnight at 18 °C before centrifugation (4,000 × g, 15 min at 4 °C). Cell pellets were resuspended in buffer A (50 mm Tris-HCl, 500 mm NaCl, 20 mm imidazole (pH 8.0)) and lysed by French press after the addition of an anti-protease mixture (Sigma). Cell debris was eliminated by centrifugation (40,000 × g, 40 min).
Proteins were purified by IMAC chromatography using nickel-Sepharose resin (GE Healthcare) equilibrated in buffer A. Proteins were eluted with buffer A containing 500 mm instead of 20 mm imidazole and were further purified by size exclusion chromatography using a HiLoad Superdex 75 column equilibrated in 50 mm Tris-HCl and 250 mm NaCl (pH 8). All chromatographic steps were performed on an ÄKTAprime Plus system (GE Healthcare) at 4 °C. Protein purity was checked by SDS-PAGE. Proteins were concentrated to at least 10 mg/ml using centrifugal concentrators (Millipore) and stored at −80 °C.
Crystallization and Structure Determination
HsiE1 (at 15 mg/ml) crystallized in 30% 2-methyl 2,4-pentanediol, 0.1 m sodium acetate (pH 4.6), and 20 mm CaCl2. For co-crystallization, HsiB1 peptide (MGSTTSSQKFIARNRAPRVQ; Eurogenetec) was added in a 5× molar excess to HsiE1 concentrated to 22 mg/ml. Crystals grew in 50 mm MES (pH 6.5), 100 mm ammonium acetate, 10% glycerol, and 28% PEG 8000. Crystals of HsiE1-HsiB1 fragment grew in 100 mm Tris (pH 8.0), 24% PEG 6000, and 0.2 m CaCl2. ClpV crystals grew in 0.2 m sodium malonate and 20% PEG 3350. Crystals that were not already cryoprotected by the crystallization solution were transferred into buffer containing 25% glycerol.
Data were collected on a MicroMax-007 HF rotating anode (Rigaku) and at Diamond Light Source beamlines I03, I04, and I04-1 and reduced in XDS (34) and iMosflm (35). Structures were solved in Phaser (36). VPA1052 (37) was used as a search model to solve HsiE1, which was in turn used to solve the structure of the HsiE1-HsiB1 complexes. ClpV-N (28) was used as a search model for the ClpV1-N domain. Structures were refined in Refmac5 (38) and phenix.refine (39) and rebuilt in Coot (40) until convergence. Crystallographic statistics are summarized in Table 2. All models and structure factors were deposited to the Protein Data Bank with codes 4UQW (ClpV N domain), 4UQX (HsiE1), 4UQY (HsiE1 + HsiB1 peptide), and 4UQZ (HsiE1 + HsiB1 fragment).
TABLE 2.
Data collection and refinement statistics
| HsiE1 | HsiE1 + HsiB1 peptide | HsiE1 + HsiB1 fragment | ClpV N | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P 21 | P 21 21 21 | P 21 21 21 | P 41 |
| Cell dimensions | ||||
| a, b, c (Å) | 56.8, 45.9, 59.5 | 45.2, 64.0, 94.5 | 52.1, 67.3, 94.0 | 90.5, 90.5, 54.3 |
| α, β, γ (degrees) | 90, 110.0, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 55.9–1.2 (1.22–1.20) | 53.0–1.6 (1.63–1.60) | 54.7–1.6 (1.63–1.60) | 90.5–1.5 (1.53–1.50) |
| Rmerge | 0.055 (0.527) | 0.061 (0.59) | 0.084 (0.442) | 0.08 (0.71) |
| I/σ(I) | 8.2 (1.9) | 10.7 (2.2) | 11.4 (2.7) | 8.8 (1.9) |
| Completeness (%) | 98.1 (98.2) | 99.8 (98.5) | 98.1 (96.0) | 99.9 (99.9) |
| Redundancy | 2.7 (2.7) | 3.2 (3.2) | 5.0 (4.1) | 4.6 (4.7) |
| Refinement | ||||
| No. of reflections | 88,118 (4,483) | 36,309 (1,772) | 43,478 (2,088) | 70,465 (3,476) |
| Rwork / Rfree | 0.151 / 0.158 | 0.177 / 0.205 | 0.188 / 0.221 | 0.159 / 0.181 |
| No. of atoms | ||||
| Protein | 2237 | 2135 | 2191 | 2564 |
| Ligand/ion | 20 | - | 4 | 9 |
| Water | 324 | 197 | 314 | 358 |
| B-Factors | ||||
| Protein | 14.2 | 29.0 | 25.1 | 26.6 |
| Ligand/ion | 23.9 | - | 37.6 | 25.7 |
| Water | 28.9 | 37.5 | 35.7 | 40.2 |
| r.m.s. deviations | ||||
| Bond lengths (Å) | 0.008 | 0.006 | 0.012 | 0.016 |
| Bond angles (degrees) | 1.29 | 1.03 | 1.41 | 1.65 |
| PDB code | 4UQX | 4UQY | 4UQZ | 4UQWx |
a Values in parentheses are for the highest resolution shell.
Bacterial Two-hybrid Assay
The genes of interest were amplified from P. aeruginosa PAO1 genomic DNA, adding appropriate restriction sites. The resulting PCR products were ligated into either bacterial two-hybrid (BTH) plasmid pKT25 or pUT18C, leading to in-frame fusions of the protein of interest with the T25 or T18 subunit of the Bordetella pertussis adenylate cyclase, respectively (41). Recombinant pKT25 and pUT18C plasmids were co-transformed into the reporter E. coli DHM1 strain. Four independent colonies for each co-transformation were inoculated into LB medium supplemented with ampicillin and kanamycin. After overnight growth at 37 °C, 10 μl of each culture were spotted onto MacConkey agar plates with 1% maltose and LB agar plates supplemented with 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-gal), both in the presence of ampicillin, kanamycin, and 1 mm isopropyl 1-thio-β-d-galactopyranoside, and incubated for at least 48 h at 30 °C. The pKT25 and pUT18C derivatives encoding the leucine zipper from GCN4, which readily dimerizes, were used as a positive control in all experiments. The experiments were done at least in duplicate, and a representative result is shown.
For quantification of BTH interactions, β-galactosidase activity from co-transformants picked from X-gal LB agar plates was measured as described previously (42). The β-galactosidase activity is calculated in Miller units.
Bioinformatics Analysis
For analysis of the groove residues, protein sequences were retrieved by BlastP searches using each of ClpV, ClpV1, ClpV2, and ClpV3 as queries. After pruning of duplicates, a total of 1,593 sequences were aligned with MAFFT (43). For the phylogenetic analysis, sequences from 68 T6SSs were retrieved from the Kyoto Encyclopedia of Genes and Genomes. Strains and accession codes are shown in Table 3. Sequences were aligned with MAFFT. For ClpV and TssC, the e-ins-i option (multiple conserved domains and long gaps) was used. TssB and TagJ/HsiE were aligned with default parameters. In all four cases, the Blosum62 scoring matrix was used. Maximum likelihood phylogenies were calculated with phyML (44) with the LG substitution model, no invariable sites, nearest neighbor interchange tree improvement, topology and branch optimization, and aBayes branch support calculation. Trees were visualized with TreeDyn (45). The weblogos were created at WebLogo 3 Web site.
TABLE 3.
List of strains used in the phylogenetic analysis
| Organism | Code | HsiE1/TagJ | HsiB/TssB | HsiC/TssC | ClpV |
|---|---|---|---|---|---|
| Acidovorax citrulli AAC00–1 | aav | Aave_1478 | Aave_1476 | Aave_1477 | Aave_1482 |
| Achromobacter xylosoxidans A8 | axy | AXYL_06394 | AXYL_06397 | AXYL_06396 | AXYL_06389 |
| None | AXYL_05693 | AXYL_05692 | AXYL_05687 | ||
| Aeromonas hydrophila ATCC 7966 | aha | None | AHA_1832 | AHA_1833 | AHA_1841 |
| Agrobacterium tumefaciens C58 | atu | Atu4339 | Atu4342 | Atu4341 | Atu4344 |
| Azoarcus sp. BH72 | azo | azo3899 | azo3895 | azo3896 | azo3903 |
| Bordetella bronchiseptica RB50 | bbr | BB0803 | BB0800 | BB0801 | BB0810 |
| Bradyrhizobium japonicum USDA 6 | bju | None | BJ6T_33630 | BJ6T_33620 | BJ6T_33570 |
| Burkholderia cenocepacia J2315 | bcj | None | BCAL0341 | BCAL0342 | BCAL0347 |
| Burkholderia thailandensis E264 | bte | BTH_II0138 | BTH_II0121 | BTH_II0122 | BTH_II0140 |
| None | BTH_I2964 | BTH_I2963 | BTH_I2958 | ||
| None | BTH_II1901 | BTH_II1900 | BTH_II1895 | ||
| None | BTH_II0870 | BTH_II0869 | BTH_II0864 | ||
| None | BTH_II0258 | BTH_II0259 | BTH_II0264 | ||
| Cronobacter sakazakii ES15 | csk | ES15_3835 | ES15_3846 | ES15_3845 | ES15_3830 |
| ES15_2806 | ES15_2819 | ES15_2818 | ES15_2825 | ||
| Cupriavidus taiwanensis LMG 19424 | cti | None | RALTA_A0608 | RALTA_A0609 | RALTA_A0607 |
| None | RALTA_B1013 | RALTA_B1014 | RALTA_B1019 | ||
| Dechloromonas aromatica RCB | dar | None | Daro_2177 | Daro_2176 | Daro_2171 |
| Delftia acidovorans SPH-1 | del | Daci_3856 | Daci_3850 | Daci_3851 | Daci_3864 |
| Escherichia coli O6:K2:H1 CFT073 | ecc | None | c3385 | c3386 | c3392 |
| Escherichia coli O157:H7 Sakai (EHEC) | ecs | None | ECs0233 | ECs0231 | ECs0223 |
| Edwardsiella piscicida C07-087 | etc | None | ETAC_09445 | ETAC_09440 | ETAC_09415 |
| Francisella tularensis TIGB03 | ftg | None | FTU_1718 | FTU_1717 | FTU_1770 |
| Klebsiella pneumoniae 342 | kpe | None | KPK_3069 | KPK_3068 | KPK_3063 |
| Leptothrix cholodnii SP-6 | lch | None | Lcho_4091 | Lcho_4090 | Lcho_4084 |
| Mesorhizobium loti MAFF303099 | mlo | None | mlr2337 | mlr2338 | mll2335 |
| Methylomicrobium alcaliphilum | mah | MEALZ_1934 | MEALZ_1931 | MEALZ_1932 | MEALZ_1938 |
| Myxococcus xanthus DK 1622 | mxa | None | MXAN_4807 | MXAN_4808 | MXAN_4813 |
| Paracoccus aminophilus JCM 7686 | pami | None | JCM7686_pAMI6p160 | JCM7686_pAMI6p159 | JCM7686_pAMI6p154 |
| Paracoccus denitrificans PD1222 | pde | None | Pden_2443 | Pden_2444 | Pden_2440 |
| Pantoea ananatis LMG 20103 | pam | PANA_2358 | PANA_2366 | PANA_2365 | PANA_2354 |
| None | PANA_4151 | PANA_4150 | PANA_4145 | ||
| Pectobacterium atrosepticum SCRI1043 | eca | None | ECA3445 | ECA3444 | ECA3436 |
| Pelobacter carbinolicus DSM 2380 | pca | None | Pcar_2814 | Pcar_2815 | Pcar_2820 |
| Photorhabdus luminescens TTO1 | plu | None | plu2301 | plu2300 | plu2287 |
| None | plu0372 | plu0371 | plu0363 | ||
| Pseudomonas putida F1 | ppf | None | Pput_2622 | Pput_2623 | Pput_2627 |
| Pseudomonas syringae B728a | psb | None | Psyr_4953 | Psyr_4954 | Psyr_4958 |
| Pseudomonas aeruginosa PA01 | pae | PA0086 | PA0083 | PA0084 | PA0090 |
| None | PA1657 | PA1658 | PA1662 | ||
| None | PA2365 | PA2366 | PA2371 | ||
| Pseudomonas fluorescens SBW25 | pfs | None | PFLU6019 | PFLU6020 | PFLU6025 |
| Rahnella aquatilis HX2 | raa | None | Q7S_25121 | Q7S_25126 | Q7S_25191 |
| None | Q7S_23336 | Q7S_23341 | Q7S_23366 | ||
| Ralstonia eutropha JMP134 | reu | None | Reut_A1733 | Reut_A1732 | Reut_A1727 |
| None | Reut_B5266 | Reut_B5265 | Reut_B5260 | ||
| Ralstonia solanacearum GMI1000 | rso | None | RS01965 | RS01964 | RS01959 |
| Rhizobium leguminosarum 3841 | rle | pRL120471 | pRL120474 | pRL120473 | pRL120476 |
| Rhodobacter sphaeroides 2.4.1 | rsp | None | RSP_3477 | RSP_3478 | RSP_3474 |
| Salmonella typhimurium LT2 | stm | STM0270 | STM0273 | STM0274 | STM0272 |
| Serratia marcescens WW4 | smw | SMWW4_v1c30250 | SMWW4_v1c30140 | SMWW4_v1c30150 | SMWW4_v1c30290 |
| None | SMWW4_v1c31600 | SMWW4_v1c31590 | SMWW4_v1c31540 | ||
| Solibacter usitatus Ellin6076 | sus | Acid_0234 | Acid_0231 | Acid_0232 | Acid_0239 |
| Taylorella equigenitalis MCE9 | teq | TEQUI_0721 | TEQUI_0718 | TEQUI_0719 | TEQUI_0708 |
| Variovorax paradoxus S110 | vap | Vapar_0196 | Vapar_0193 | Vapar_0194 | Vapar_0200 |
| Vapar_0525 | Vapar_0550 | Vapar_0549 | Vapar_0530 | ||
| Vibrio cholerae O1 biovar El Tor N16961 | vch | None | VCA0107 | VCA0108 | VCA0116 |
| Vibrio parahaemolyticus RIMD 2210633 | vpa | VPA1032 | VPA1035 | VPA1034 | VPA1028 |
| None | VP1402 | VP1403 | VP1392 | ||
| Xanthomonas oryzae KACC 10331 | xoo | None | XOO3481 | XOO3480 | XOO3475 |
| XOO3049 | XOO3052 | XOO3051 | XOO3045 | ||
| Xenorhabdus bovienii SS-2004 | xbo | None | XBJ1_2103 | XBJ1_2102 | XBJ1_2096 |
| None | XBJ1_0263 | XBJ1_0264 | XBJ1_0272 | ||
| Yersinia pestis KIM10 | ypk | y1560 | y1545 | y1546 | y1538 |
| None | y2706 | y2705 | y2699 | ||
| None | y3369 | y3368 | y3362 | ||
| None | y3675 | y3674 | y3669 |
RESULTS
Distinct Structural Features of the N-terminal Domain of P. aeruginosa ClpV1
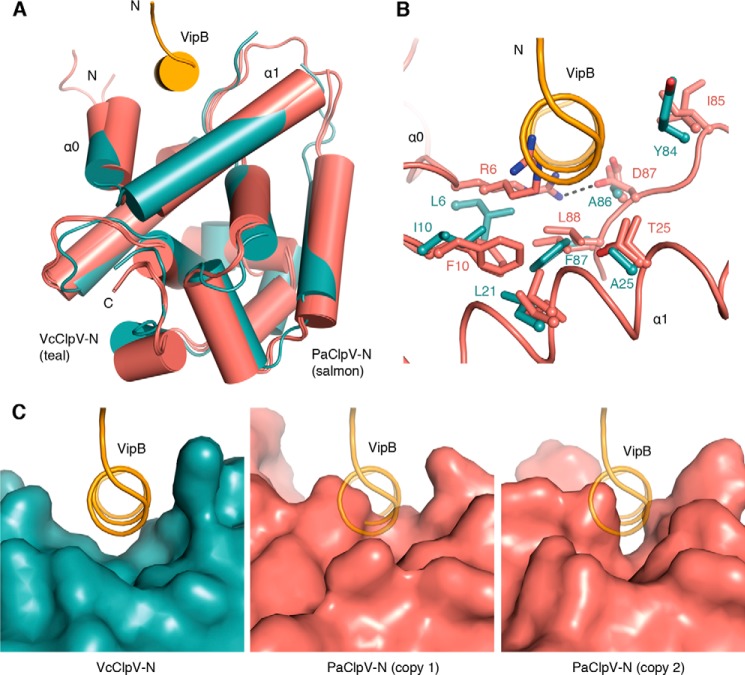
The structure of the N-terminal domain of V. cholerae ClpV (ClpV-N) has previously been solved in complex with a peptide corresponding to the N-terminal helix of the TssC homolog VipB (Table 4) (28). A hydrophobic groove in ClpV-N provides the binding site for VipB. To investigate whether the N-terminal domain of P. aeruginosa ClpV1 (ClpV1-N) can interact with the TssC homolog HsiC1 in a similar manner, we solved the structure of ClpV1-N from crystals diffracting to 1.5 Å, using the Vibrio structure (Protein Data Bank code 3ZRJ) as a model for molecular replacement (Table 2). The asymmetric unit contains two copies of ClpV1-N in nearly identical conformations (r.m.s. deviation for all Cα atoms of 0.5 Å). The overall structure of ClpV1-N is similar to that of the Vibrio ClpV-N, with matching secondary structure elements and an r.m.s. deviation of 1.6 Å for 121 equivalent Cα atoms (Fig. 1A). Both structures possess an N-terminal helix α0 that distinguishes ClpV-type ATPases from other Hsp100 ATPases (e.g. ClpA) (46). A hydrophobic cleft between this helix and helix α1 forms the binding site for VipB in V. cholerae (Fig. 1A).
TABLE 4.
Names of T6SS components used in this study
Given are protein names in the three T6SSs in P. aeruginosa and in V. cholerae. Tss, type six secretion.
| Generic name | P. aeruginosa | V. cholerae | Function |
|---|---|---|---|
| TssB | HsiB1,2,3 | VipA | Sheath component |
| TssC | HsiC1,2,3 | VipB | Sheath component |
| TagJ | HsiE1 | Unknown | |
| ClpV | ClpV1,2,3 | ClpV | Sheath disassembly |
FIGURE 1.
Structural differences between P. aeruginosa ClpV1 and V. cholerae ClpV. A, superposition of two copies of ClpV1-N (salmon) onto V. cholerae ClpV-N (teal) (Protein Data Bank code 3ZRJ) shows overall structural conservation. The peptide mimicking the N terminus of VipB that binds to a groove formed by helices α0 and α1 of ClpV-N is shown as an orange cylinder. The N and C termini of ClpV-N are indicated. B, close-up view of the superposed grooves between helices α0 and α1 in ClpV1-N (salmon) and V. cholerae ClpV-N (teal). The view is rotated clockwise by 20° with respect to A. Residues facilitating binding of VipB in ClpV-N and their counterparts in both copies of ClpV1-N are shown as sticks. In either ClpV1-N copy, two bulky charged residues, Arg6 and Asp87, obstruct the groove. C, surface representations of ClpV-N (left) and the two copies of ClpV1-N (middle and right) show that the binding groove is blocked in P. aeruginosa. The view is as in B.
The residues lining the VipB binding groove in ClpV-N are partially conserved in ClpV1-N. For example, Ile10 and Phe87 lie at the bottom of the groove. The corresponding residues in ClpV1-N are Phe10 and Leu88 (Fig. 1B). However, other residues are not conserved. Tyr84, which packs against Glu22 of VipB in V. cholerae, is Ile85 in P. aeruginosa, a residue that could not participate in the same interaction. The two aliphatic residues Leu6 and Ala86, which sit at the top of the groove in Vibrio ClpV-N, are replaced by large residues of opposite charge in ClpV1-N (i.e. Arg6 and Asp87) (Fig. 1B). In one of the two ClpV1-N molecules in the asymmetric unit, these residues form a salt bridge across the groove, whereas in the other, they point into the groove. Both conformations not only diminish the hydrophobic character of the groove but also are incompatible with binding of an α-helix, as observed in V. cholerae (Fig. 1, B and C). Consistent with the altered conformation of the binding groove, we were unable to obtain a complex of ClpV1-N bound to a peptide corresponding to the N terminus of HsiC1.
Phylogenetic Analysis Identifies Distinct T6SS Classes Based on the ClpV Structure
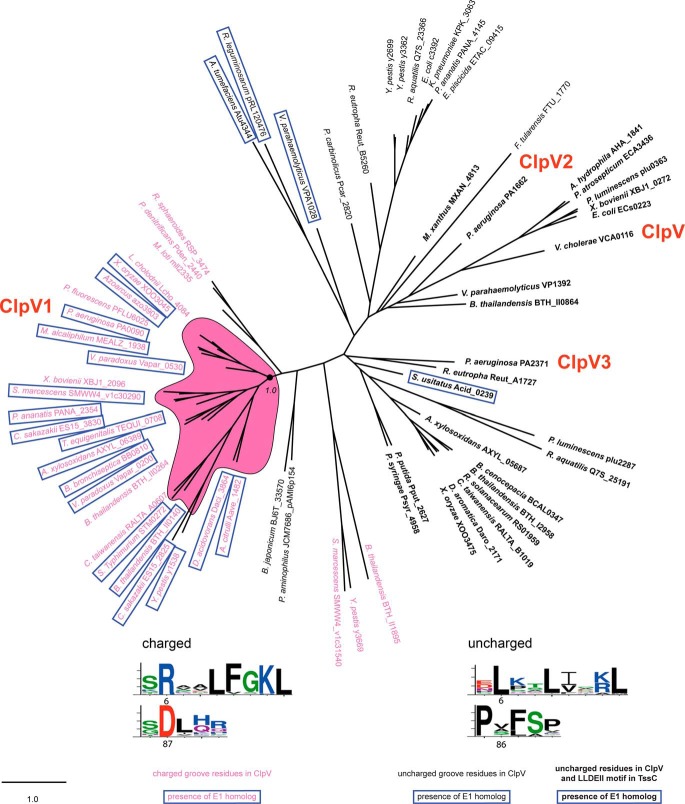
Alignment of ClpV sequences shows a division based on sequence conservation within the binding groove in the N-terminal domain (Fig. 2, weblogos). There are homologs with an arginine and an aspartate (rarely glutamate) as in P. aeruginosa ClpV1 (Arg6 and Asp87, respectively) and homologs with two aliphatic residues as in V. cholerae ClpV (Leu6 and Ala86, respectively). Phylogenetic analysis of ClpV sequences from 68 T6SSs covering a wide range of bacterial species shows evolutionary separation based on the groove residues. ClpV homologs with charged residues group with P. aeruginosa ClpV1 on the left side of the phylogenetic tree (Fig. 2, shown in pink). ClpV homologs with aliphatic residues group with V. cholerae ClpV on the right side of the tree (Fig. 2, shown in black). It is noticeable that the two other P. aeruginosa ClpVs, ClpV2 and ClpV3, which are associated with the H2- and H3-T6SSs, respectively, lack the charged residues found in the hydrophobic groove of ClpV1 and group with V. cholerae ClpV (Fig. 2, shown in black).
FIGURE 2.
Division of T6SSs into distinct phylogenetic classes. Maximum likelihood phylogenetic tree generated from 68 aligned ClpV sequences belonging to the indicated bacterial species. ClpV sequences separate into two classes according to two residues in the hydrophobic groove between helices α0 and α1. Homologs where these residues are uncharged like in V. cholerae are shown in black, whereas homologs with charged residues are shown in pink. Blue boxes indicate the presence of an HsiE1 homolog on the same secretion cluster. Boldface type denotes the presence of the LLDEII motif in the TssC homolog on the same cluster. The branch that is highly enriched in T6SSs containing ClpV homologs with charged residues and HsiE1 homologs is shown against a pink background. The branch support value is shown. The three ClpV homologs from P. aeruginosa and V. cholerae ClpV are indicated in red. The weblogos at the bottom show the sequence conservation of residues in the hydrophobic groove of ClpV, depending on the presence (left) or absence (right) of the pair of charged residues. On the left, the two charged residues are numbered according to the P. aeruginosa ClpV1 sequence as in our structure. On the right, the uncharged residues in the same positions are numbered according to the V. cholerae ClpV sequence, as in the published structure (28). The scale bar shows amino acid changes per site. At the bottom is a brief key to the labeling used.
TssC Has Coevolved with ClpV
In V. cholerae, the peptide corresponding to the N-terminal α-helix of VipB that binds to ClpV contains a short conserved motif (19LLDEIM24) (28). By analyzing the N termini of the P. aeruginosa VipB homologs, HsiC1, HsiC2, and HsiC3, we found this motif to be conserved in C2 (18ILDSII23) and C3 (20LLDEII25) but not C1 (21EFASLL26) (Fig. 3A). Mapping the presence of the LLDEII motif in TssC proteins onto the phylogenetic tree of ClpV sequences suggests that TssC homologs have coevolved with ClpV ATPases (Fig. 2, boldface type). TssC homologs with the LLDEII motif are associated only with ClpV homologs having aliphatic residues in their hydrophobic groove.
FIGURE 3.
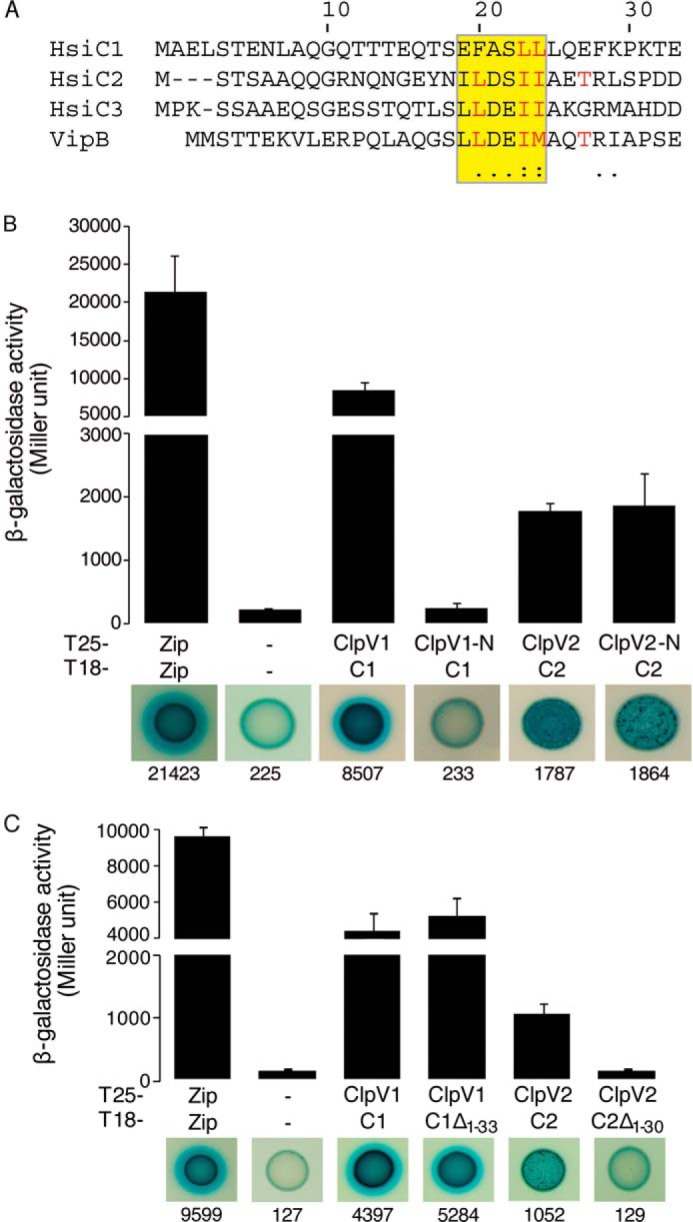
Role of the N-terminal α-helices of HsiC proteins in the interaction with their cognate ClpV. A, sequence alignment of the N terminus (residues 1–33) of the P. aeruginosa proteins HsiC1, HsiC2, and HsiC3 and of VipB, the HsiC homolog of V. cholerae. The conserved binding motif is highlighted. Residues implicated in binding in the ClpV-VipB complex are shown in red. Degrees of conservation are indicated by asterisks, colons, and dots below the alignment. B, BTH assays analyzing the interaction between the N-terminal domain of ClpV1 and full-length HsiC1 and between the N-terminal domain of ClpV2 and full-length HsiC2. A graphical representation of the β-galactosidase activity from co-transformants is shown (top), and images of corresponding spots on X-gal LB agar plates are displayed (bottom). Zip, leucine zipper domain of the yeast transcription factor GCN4. T6SS proteins are HsiC1 (C1), HsiC2 (C2), N-terminal domain of ClpV1 (ClpV1-N), and N-terminal domain of ClpV2 (ClpV2-N). C, ability of truncated forms of HsiC1 and HsiC2 to interact with their cognate ClpV as determined by BTH assays. The combinations of T25/T18 fusion proteins are indicated, and abbreviations are used as described above. Additionally, C1Δ1–33 represents HsiC1 truncated for the first 33 residues (including those forming the first two predicted helices), and C1Δ1–30 indicates HsiC2 truncated for the first 30 residues. Experiments were carried out in duplicate. Error bars, S.E.
To investigate these differences and the interaction between the ClpV and TssC partners, we used the BTH assay. Although full-length ClpV1 interacts with HsiC1, the isolated N-terminal domain does not (Fig. 3B), in contrast to what was observed in V. cholerae (28). The two other P. aeruginosa ClpV proteins display binding grooves that are closely related to the one observed in Vibrio ClpV-N (Phyre homology models; data not shown). In contrast to ClpV1 and HsiC1, the N-terminal domain of ClpV2 interacts with HsiC2 to the same degree as full-length ClpV2 (Fig. 3B), suggesting that the interaction mode between ClpV2 and HsiC2 is similar to the Vibrio ClpV-VipB interaction.
We then engineered an N-terminal truncation of HsiC1 to remove the first 33 residues (HsiC1Δ1–33). HsiC1Δ1–33 interacts with ClpV1 (Fig. 3C), which suggests that the N terminus of HsiC1 is not essential for a stable interaction, further highlighting the difference with V. cholerae. In contrast, the truncation of the corresponding N-terminal residues of HsiC2 (HsiC2Δ1–30) has a detrimental effect (10-fold reduction) on binding to ClpV2 (Fig. 3C). These data provide evidence for at least two distinct classes of TssC homologs that specifically interact with cognate ClpVs, which is consistent with the phylogenetic analysis. One mode of interaction, exemplified by V. cholerae, involves a primary interaction between the ClpV N domain and the N-terminal helix of TssC (VipB/HsiC) within a hydrophobic groove. The other mode involves additional features beyond the N domain of ClpV or additional protein partners, which may modulate the TssC-ClpV interaction.
HsiE1/TagJ Is a Direct T6SS Partner for ClpV1
T6SS clusters vary in genetic organization and are composed of core and accessory genes. The hsiE1/tagJ gene is found in the P. aeruginosa H1-T6SS cluster but is lacking in the H2- and H3-T6SSs or in the V. cholerae T6SS (Table 4). We performed BTH experiments showing that HsiE1 interacts with full-length ClpV1 (Fig. 4). The interaction between ClpV1 and HsiE1 distinguishes P. aeruginosa from V. cholerae where no HsiE/TagJ homolog exists. Analysis of the ClpV phylogenetic tree further showed that when the T6SS involves a TssC protein containing the LLDEII motif, such as in V. cholerae, there is no associated HsiE1 homolog (Fig. 2, boldface type and not boxed). There is only one exception in Solibacter usitatus. This suggests that these two features are mutually exclusive and that HsiE1 plays a role in the interaction between ClpV and the TssC/TssB sheath.
FIGURE 4.
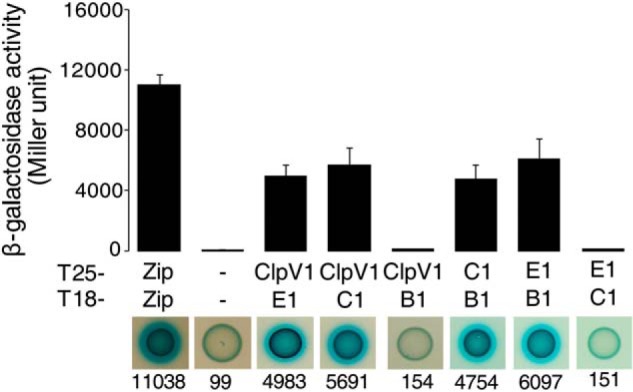
BTH assays showing that ClpV1 interacts with HsiE1 and HsiC1. In the top panel, graphical representations of β-galactosidase activity from co-transformants of E. coli DHM1 cells expressing the indicated proteins fused to the adenylate cyclase T25 or T18 subunit are shown. Images of the corresponding bacterial spots on X-gal LB agar plates are shown in the bottom panel with the corresponding average activity in Miller units indicated below each image. The combinations of T25/T18 fusion proteins are indicated and abbreviated as described in the legend to Fig. 3. B1, HsiB1; E1, HsiE1. Experiments were carried out in triplicate. Error bars, S.E.
Crystal Structure of HsiE1 in Complex with the N Terminus of HsiB1
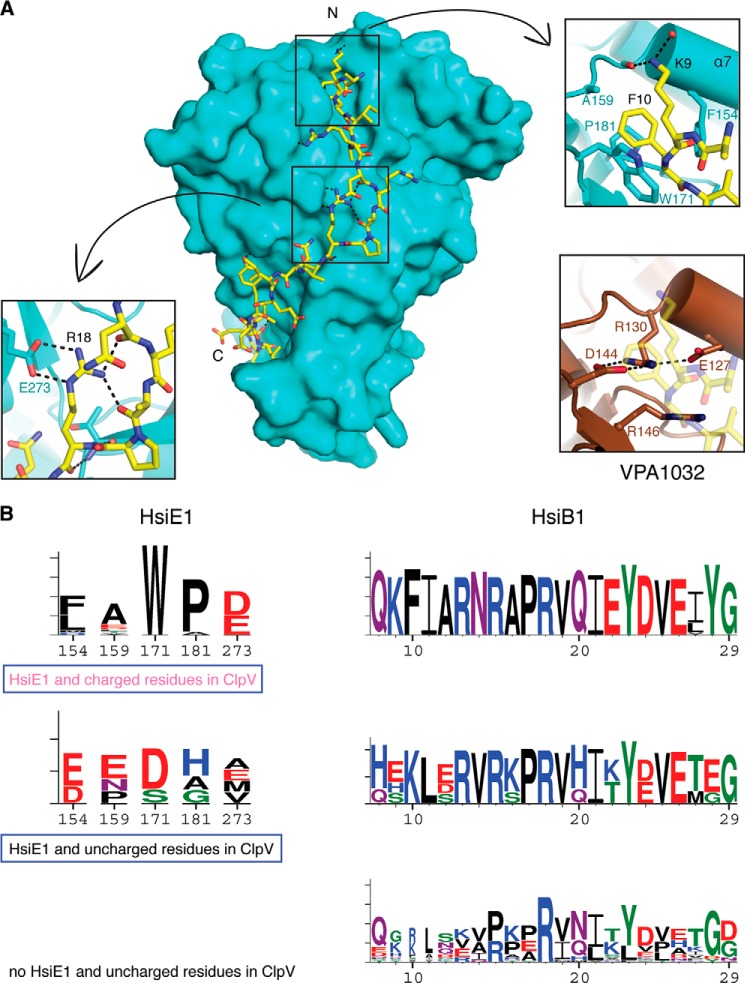
We previously showed that P. aeruginosa HsiE1 interacts with the TssB sheath component HsiB1 (Fig. 4) (33). Here, we solved the HsiE1 structure from crystals diffracting to 1.2 Å (Table 2). Vibrio parahemeolyticus VPA1032, a protein of unknown function (37) with 26% sequence identity to HsiE1 (Protein Data Bank code 1ZBP), served as a model for molecular replacement. Both VPA1032 and HsiE1 belong to the “ImpE family” (47). Their structures are similar, as indicated by the r.m.s. deviation of 1.5 Å over 199 equivalent Cα atoms and the conserved N-terminal tetratricopeptide repeat domain and C-terminal “ImpE fold” (Fig. 5A).
FIGURE 5.
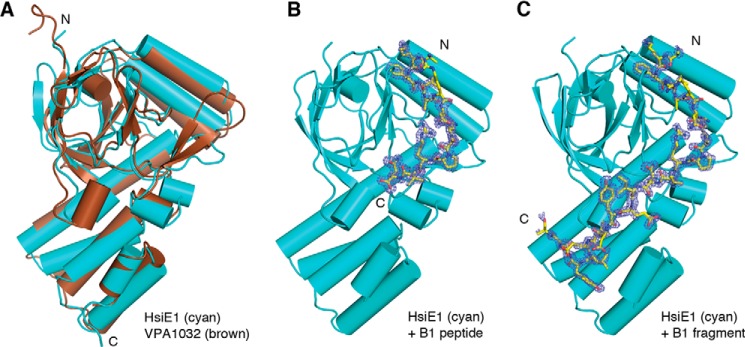
Structure of HsiE1. A, P. aeruginosa HsiE1 (cyan) is structurally similar to VPA1032, the HsiE1 homolog from V. parahemeolyticus (brown). The N and C termini of both proteins are indicated. B, a peptide corresponding to the N terminus of HsiB1 (yellow sticks) binds to HsiE1 (cyan). An Fo − Fc omit map contoured at 3σ is shown as blue mesh around the HsiB1 peptide. The N and C termini of the HsiB1 peptide are indicated. C, an N-terminal fragment of HsiB1 (yellow sticks) binds to HsiE1 (cyan). The panel is designed like B. All three panels display HsiE1 in the same orientation.
We crystallized HsiE1 in complex with a peptide corresponding to the first 20 residues of HsiB1 (1MGSTTSSQKFIARNRAPRVQ20) or an HsiB1 degradation product, which appeared during purification of the E1-B1 complex. Both the E1-B1 peptide and E1-B1 fragment structures were solved by molecular replacement (Table 2 and Fig. 5, B and C). In the E1-B1 complex structures, there is clear electron density for residues 8–20 of the extraneously added B1 peptide (Fig. 5B) and for residues 8–30 of the co-purified B1 fragment (Fig. 5C), respectively. Both structures are nearly identical to the structure of HsiE1 alone, with an r.m.s. deviation of 0.4 Å in either case, and to each other with an r.m.s. deviation of 0.3 Å. The B1 fragment (residues 8–30) wraps around E1 in an extended conformation (Fig. 6A, main panel), following a prominent surface groove that originates in a deep hydrophobic pocket between the 12-stranded β-barrel of the “ImpE fold” and the two α-helices just preceding it (Fig. 6A, left inset). The B1 residue Phe10 nestles in that pocket, which is lined with residues Phe154, Trp171, and Pro181.
FIGURE 6.
Interaction between HsiE1 and HsiB1. A, the HsiB1 fragment binds in a groove on the surface of HsiE1 (main panel). The N and C termini of the HsiB1 fragment are indicated. Two regions of interaction are boxed and magnified (arrows). The top right inset shows the pocket on HsiE1 into which HsiB1 Tyr12 binds and the interaction between HsiB1 Lys9 and helix α7 in HsiE1. The bottom left inset shows the salt bridges between HsiB1 Arg18 and HsiE1 Glu273. The bottom right inset shows the region in VPA1032 corresponding to the HsiB1 binding pocket in HsiE1. HsiB1 is shown in pale yellow. B, weblogos derived from sequence alignments of HsiE1 (left) and HsiB1 (right) homologs. For HsiE1, residues are shown that interact with HsiB1 in our structure. For HsiB1, the residues that are visible in our structure are shown. Residue numbers are indicated and match those in A. The top panels show the sequence conservation in T6SSs containing a ClpV homolog with charged residues and an HsiE1 homolog. The middle panels show the sequence conservation in T6SSs containing a ClpV homolog with uncharged residues and an HsiE1 homolog. This matches the bottom right inset in A. The bottom panel shows the sequence conservation of TssB from T6SSs containing a ClpV homolog with uncharged residues but no HsiE1 homolog. All three sets of panels carry a label that is also found in the phylogenetic tree in Fig. 2.
The amino group of Lys9 of HsiB1 interacts with the negative dipole of helix α7 and forms hydrogen bonds with two carbonyl oxygens at the end of that helix (residues 155 and 157). Another key residue for the interaction is Arg18 of B1, which forms two salt bridges with the carboxyl group of Glu273 in HsiE1 (Fig. 6A, top right inset). In addition, most of the E1-B1 binding interface is hydrophobic in nature, involving Ala12, Gln20, Tyr23, Val25, and Leu27 from B1.
The residues of B1 between 8 and 29 that are involved in specific E1 interactions are highly conserved (Fig. 6B, top). HsiE1 homologs also show conservation of residues that line the hydrophobic pocket with residues 171 and 181 always tryptophan and proline, respectively, and residue 154 always a large hydrophobic residue (e.g. Phe, Leu, or Trp). Residue 273, which forms two salt bridges with the conserved Arg18 in HsiB1, is always glutamic or aspartic acid.
The N Terminus of TssB Homologs Coevolved with HsiE1 Homologs
Because we showed that P. aeruginosa HsiE1 interacts both with ClpV1 and HsiB1, it is tempting to generalize that once HsiE1 binds onto the sheath via an interaction with TssB, it is an ideal docking point for ClpV1 to be recruited to the sheath. There are features in the N termini of TssB proteins that support this idea. The conservation of the N termini of TssB homologs associated with HsiE1 has been described above (Fig. 6B, top). The N termini of TssB homologs not associated with HsiE1 do not show this striking conservation (Fig. 6B, bottom), and there is very little similarity between the two sets. Features in the N termini of TssB homologs, therefore, also appear to differentiate between the two T6SS classes.
DISCUSSION
Repeated injection of T6SS-dependent toxins is essential for effective killing of target cells, and rebuilding of the secretion sheath is necessary in this process. Here, we present molecular details of T6SS components involved in the sheath dynamics and propose the existence of distinct T6SS classes based on their interaction and co-evolution. It has been previously shown that the formation of the contractile sheath of the T6SS requires the co-polymerization of TssB/HsiB1/VipA and TssC/HsiC1/VipB components (Table 4) (20, 21, 23, 24). For sheath disassembly, the AAA+ ATPase ClpV is required (21, 30). In V. cholerae, the ClpV N-domain directly interacts with the VipB component (TssC/HsiC1 homolog) of the sheath via a conserved hydrophobic groove and the N-terminal helix of VipB (28). We identified two key residues that have diverged from their equivalents in V. cholerae in a subset of T6SSs. These residues are of complementary charge and alter the shape and accessibility of the ClpV1-N groove, as shown in the structure of the N-terminal domain of ClpV1 from P. aeruginosa that we solved. The presence of these residues is incompatible with the mode of binding observed for ClpV-VipB. Consequently, we found that the N domain of the Pseudomonas ATPase ClpV1 is not sufficient for interaction with HsiC1, nor is the presence of the N-terminal helix of HsiC1 required for it. This is markedly different from what was observed in V. cholerae, where VipB binds to ClpV via its N-terminal helix containing a LLDEII motif (28). The differences in the interaction between ClpV and the sheath component TssC in the H1-T6SS of P. aeruginosa versus V. cholerae lead us to hypothesize that sheath disassembly differs mechanistically in these systems.
To provide further evidence for this hypothesis, we carried out phylogenetic analysis of T6SSs and mapped our structural and interaction data on the resulting tree. Our phylogenetic tree of ClpV divides into two main branches based on the presence of residues of opposite charge on top of the hydrophobic binding groove of the N domain. We found that TssC sequences divide similarly according to the presence of the N-terminal LLDEII motif, which is not observed in secretion systems where the ClpV homolog contains charged residues in the hydrophobic groove. On these features we could superimpose the observation that the accessory protein TagJ/HsiE co-occurs only with the Pseudomonas ClpV1 type and thus co-evolved with TssC homologs that lack the LLDEII motif.
The importance of the co-evolution between TagJ/HsiE and the two classes of ClpV and TssC is reinforced by two observations. First, we were able to show that HsiE1 directly interacts with ClpV1, which suggests that the function of these two proteins is coordinated. Second, we have shown previously that HsiE1 directly interact with the other sheath component HsiB1/TssB, suggesting that ClpV1 and HsiE1 function is associated with sheath disassembly. Here, we have solved the structure of HsiE1 in complex with residues 8–30 of the P. aeruginosa HsiB1. The N terminus of HsiB1 wraps around HsiE1 in an extended conformation, and all HsiB1 residues visible in the crystal structure are in close proximity to HsiE1. Residues 8–29 are highly conserved in HsiB1 homologs (TssB) from secretion systems containing HsiE1. It is thus clear from our HsiB1-HsiE1 crystal structure that in the context of the intact sheath, the N terminus of HsiB1 would need to be accessible for interaction with HsiE1. In contrast, the interaction between HsiB1/TssB and the other sheath component HsiC1/TssC is mostly mediated by a conserved hydrophobic motif in a helix near the C terminus of TssB (24, 48).
Our phylogenetic analysis does not show a strict division into just two classes of T6SSs. Instead, there are mixed clades and isolated branches. For example, we identified four T6SS clusters containing HsiE1 and a ClpV homolog without charged residues in the groove (Fig. 2, black and boxed), among them V. parahemeolyticus. They are notable for two reasons. First, the N termini of their TssB homologs do not display the strict amino acid conservation (Fig. 6B, middle) seen in T6SSs with a P. aeruginosa-type ATPase and a HsiE1 homolog (Fig. 6B, top). For example, neither Lys9 nor Phe10, two key residues for the interaction with HsiE1 that we identified in the crystal structure of the HsiB1-HsiE1 complex, are present in those TssB homologs. Second, the cognate HsiE1 homologs lack key residues involved in the TssB-HsiE1 interaction (Fig. 6B, middle). From the sequence, we predict the absence of a hydrophobic pocket to accommodate a TssB peptide. The structure of VPA1032, the HsiE1 homolog from V. parahemeolyticus, bears this out (Fig. 6A, bottom right inset). This observation suggests that VPA1032 may not interact with its cognate TssB homolog at all, and this HsiE1 homolog may be an evolutionary vestige whose function has been lost.
All four T6SS components studied here are involved in the dynamics of sheath assembly and disassembly. The close evolutionary link among these four proteins suggests a functional connection. At the same time, the evolutionary data suggest that interactions with sheath components and sheath disassembly differ in different T6SS classes. It is possible that the selective pressure imposed by the environment, target cells, and/or competitors has led to the evolution of subtle mechanical differences that dramatically altered the efficacy and speed of the T6SS machine.
In V. cholerae, a simple system exists. ClpV directly recognizes the sheath component VipB and drives ATP-dependent disassembly after sheath contraction and toxin ejection (Fig. 7A). It is likely that the conformational change associated with contraction (e.g. exposing the N terminus of VipB) is the signal for ClpV binding (28). In P. aeruginosa, the system is more complex. Free ClpV1 can still interact with HsiC1, but HsiE1 is more likely to recruit ClpV1 to the sheath by recognizing the N terminus of HsiB1 (Fig. 7B). It is not clear whether HsiE1 chaperones ClpV1 to the sheath or whether it binds first and provides increased avidity for ClpV1 binding. In either case, proximity of ClpV1 to the sheath would allow the ATPase to recognize and bind HsiC1 and engage in its ATP-dependent disassembly activity.
FIGURE 7.
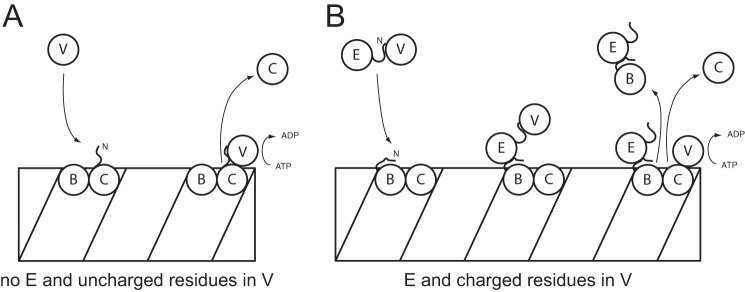
Speculative model for the role of HsiE/TagJ and ClpV in disassembly of the TssB-TssC sheath. A, in V. cholerae, the N terminus of ClpV recognizes the N-terminal helix of the TssC protein VipB and proceeds to disassemble the VipA-VipB sheath. There is no HsiE1 homolog, and the binding groove in the N domain of ClpV contains uncharged residues. B, in P. aeruginosa, HsiE1 recruits the ATPase ClpV1 to the sheath by recognition of the N terminus of the TssB protein HsiB1. Once at the sheath, ClpV can interact with the TssC protein HsiC1 and starts sheath disassembly. This system is distinguished by the presence of HsiE1 and charged residues in the binding groove of the N domain of ClpV. In both panels, B denotes the TssB homolog (VipA or HsiB1), C denotes the TssC homolog (VipB or HsiC1), and V denotes the ClpV homolog (ClpV or ClpV1). In B, E denotes HsiE1.
Because ClpV hexamers bind all along the sheath but in substoichiometric numbers in V. cholerae (21), it is interesting to speculate that disassembly of the sheath leads to the accumulation of oligomeric fragments that would either need to be broken down further for reassembly or cleared from the cell. In P. aeruginosa, HsiE1-mediated disassembly depends on accessible N termini of HsiB1. It has been suggested in previous studies that the N termini of V. cholerae VipA (HsiB) are only accessible at tubule ends (22). The overall similarity of the sheath in P. aeruginosa tempts us to speculate on a two-step mechanism, where the initial explosive disassembly mediated by direct interaction between ClpV1 and HsiC1 leads to the production of sheath fragments. These fragments may offer an increased accessibility of HsiB1 N termini and could then be broken down into their components by sequential extraction of HsiC1 upon targeting of ClpV1 to HsiB1 mediated by HsiE1. This model provides ideas to further elucidate the mechanism of sheath disassembly and how it is regulated within the two different classes of T6SSs identified in this study. The T6SS is used by bacteria to fight against each other in a strategic and efficient manner. We can learn from this molecular process and attempt to reroute it to our advantage by designing T6SS-based antimicrobial strategies against bacterial pathogens.
This work was supported by Medical Research Council (MRC) Program Grant MR/K001930/1.
The atomic coordinates and structure factors (codes 4UQW, 4UQX, 4UQY, and 4UQZ) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- T6SS
- type VI secretion system
- r.m.s.
- root mean square
- BTH
- bacterial two-hybrid.
REFERENCES
- 1. Coulthurst S. J. (2013) The Type VI secretion system: a widespread and versatile cell targeting system. Res. Microbiol. 164, 640–654 [DOI] [PubMed] [Google Scholar]
- 2. Ho B. T., Dong T. G., Mekalanos J. J. (2014) A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15, 9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ma A. T., McAuley S., Pukatzki S., Mekalanos J. J. (2009) Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe 5, 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ma A. T., Mekalanos J. J. (2010) In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc. Natl. Acad. Sci. U.S.A. 107, 4365–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pukatzki S., McAuley S. B., Miyata S. T. (2009) The type VI secretion system: translocation of effectors and effector-domains. Curr. Opin. Microbiol. 12, 11–17 [DOI] [PubMed] [Google Scholar]
- 6. Suarez G., Sierra J. C., Erova T. E., Sha J., Horneman A. J., Chopra A. K. (2010) A type VI secretion system effector protein, VgrG1, from Aeromonas hydrophila that induces host cell toxicity by ADP ribosylation of actin. J. Bacteriol. 192, 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brooks T. M., Unterweger D., Bachmann V., Kostiuk B., Pukatzki S. (2013) Lytic activity of the Vibrio cholerae type VI secretion toxin VgrG-3 is inhibited by the antitoxin TsaB. J. Biol. Chem. 288, 7618–7625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dong T. G., Ho B. T., Yoder-Himes D. R., Mekalanos J. J. (2013) Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 110, 2623–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hood R. D., Singh P., Hsu F., Güvener T., Carl M. A., Trinidad R. R., Silverman J. M., Ohlson B. B., Hicks K. G., Plemel R. L., Li M., Schwarz S., Wang W. Y., Merz A. J., Goodlett D. R., Mougous J. D. (2010) A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Murdoch S. L., Trunk K., English G., Fritsch M. J., Pourkarimi E., Coulthurst S. J. (2011) The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. J. Bacteriol. 193, 6057–6069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Russell A. B., Hood R. D., Bui N. K., LeRoux M., Vollmer W., Mougous J. D. (2011) Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Russell A. B., LeRoux M., Hathazi K., Agnello D. M., Ishikawa T., Wiggins P. A., Wai S. N., Mougous J. D. (2013) Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 496, 508–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Whitney J. C., Chou S., Russell A. B., Biboy J., Gardiner T. E., Ferrin M. A., Brittnacher M., Vollmer W., Mougous J. D. (2013) Identification, structure, and function of a novel type VI secretion peptidoglycan glycoside hydrolase effector-immunity pair. J. Biol. Chem. 288, 26616–26624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ma L. S., Hachani A., Lin J. S., Filloux A., Lai E. M. (2014) Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe 16, 94–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hachani A., Allsopp L. P., Oduko Y., Filloux A. (2014) The VgrG proteins are “a la carte” delivery systems for bacterial type VI effectors. J. Biol. Chem. 289, 17872–17884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Whitney J. C., Beck C. M., Goo Y. A., Russell A. B., Harding B. N., De Leon J. A., Cunningham D. A., Tran B. Q., Low D. A., Goodlett D. R., Hayes C. S., Mougous J. D. (2014) Genetically distinct pathways guide effector export through the type VI secretion system. Mol. Microbiol. 92, 529–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fu Y., Waldor M. K., Mekalanos J. J. (2013) Tn-Seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host Microbe 14, 652–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kanamaru S., Leiman P. G., Kostyuchenko V. A., Chipman P. R., Mesyanzhinov V. V., Arisaka F., Rossmann M. G. (2002) Structure of the cell-puncturing device of bacteriophage T4. Nature 415, 553–557 [DOI] [PubMed] [Google Scholar]
- 19. Aksyuk A. A., Leiman P. G., Kurochkina L. P., Shneider M. M., Kostyuchenko V. A., Mesyanzhinov V. V., Rossmann M. G. (2009) The tail sheath structure of bacteriophage T4: a molecular machine for infecting bacteria. EMBO J. 28, 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Basler M., Pilhofer M., Henderson G. P., Jensen G. J., Mekalanos J. J. (2012) Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483, 182–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bönemann G., Pietrosiuk A., Diemand A., Zentgraf H., Mogk A. (2009) Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 28, 315–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kube S., Kapitein N., Zimniak T., Herzog F., Mogk A., Wendler P. (2014) Structure of the VipA/B type VI secretion complex suggests a contraction-state-specific recycling mechanism. Cell Rep. 8, 20–30 [DOI] [PubMed] [Google Scholar]
- 23. Lossi N. S., Manoli E., Förster A., Dajani R., Pape T., Freemont P., Filloux A. (2013) The HsiB1C1 (TssB-TssC) complex of the Pseudomonas aeruginosa type VI secretion system forms a bacteriophage tail sheathlike structure. J. Biol. Chem. 288, 7536–7548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang X. Y., Brunet Y. R., Logger L., Douzi B., Cambillau C., Journet L., Cascales E. (2013) Dissection of the TssB-TssC interface during type VI secretion sheath complex formation. PLoS One 8, e81074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Leiman P. G., Basler M., Ramagopal U. A., Bonanno J. B., Sauder J. M., Pukatzki S., Burley S. K., Almo S. C., Mekalanos J. J. (2009) Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U.S.A. 106, 4154–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Basler M., Ho B. T., Mekalanos J. J. (2013) Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152, 884–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kapitein N., Bönemann G., Pietrosiuk A., Seyffer F., Hausser I., Locker J. K., Mogk A. (2013) ClpV recycles VipA/VipB tubules and prevents non-productive tubule formation to ensure efficient type VI protein secretion. Mol. Microbiol. 87, 1013–1028 [DOI] [PubMed] [Google Scholar]
- 28. Pietrosiuk A., Lenherr E. D., Falk S., Bönemann G., Kopp J., Zentgraf H., Sinning I., Mogk A. (2011) Molecular basis for the unique role of the AAA+ chaperone ClpV in type VI protein secretion. J. Biol. Chem. 286, 30010–30021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hanson P. I., Whiteheart S. W. (2005) AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6, 519–529 [DOI] [PubMed] [Google Scholar]
- 30. Zheng J., Ho B., Mekalanos J. J. (2011) Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS One 6, e23876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Filloux A., Hachani A., Bleves S. (2008) The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 154, 1570–1583 [DOI] [PubMed] [Google Scholar]
- 32. Mougous J. D., Cuff M. E., Raunser S., Shen A., Zhou M., Gifford C. A., Goodman A. L., Joachimiak G., Ordoñez C. L., Lory S., Walz T., Joachimiak A., Mekalanos J. J. (2006) A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lossi N. S., Manoli E., Simpson P., Jones C., Hui K., Dajani R., Coulthurst S. J., Freemont P., Filloux A. (2012) The archetype Pseudomonas aeruginosa proteins TssB and TagJ form a novel sub-complex in the bacterial Type VI secretion system. Mol. Microbiol. Biol. Crystallogr. 86, 437–456 [DOI] [PubMed] [Google Scholar]
- 34. Kabsch W. (2010) Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McCoy A. J. (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Forouhar F., Kuzin A., Seetharaman J., Lee I., Zhou W., Abashidze M., Chen Y., Yong W., Janjua H., Fang Y., Wang D., Cunningham K., Xiao R., Acton T. B., Pichersky E., Klessig D. F., Porter C. W., Montelione G. T., Tong L. (2007) Functional insights from structural genomics. J. Struct. Funct. Genomics 8, 37–44 [DOI] [PubMed] [Google Scholar]
- 38. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Karimova G., Pidoux J., Ullmann A., Ladant D. (1998) A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Miller J. H. (1992) A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria, pp. 71–80, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 43. Kuraku S., Zmasek C. M., Nishimura O., Katoh K. (2013) aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 41, W22–W28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guindon S., Delsuc F., Dufayard J. F., Gascuel O. (2009) Estimating maximum likelihood phylogenies with PhyML. Methods Mol. Biol. 537, 113–137 [DOI] [PubMed] [Google Scholar]
- 45. Chevenet F., Brun C., Bañuls A. L., Jacq B., Christen R. (2006) TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7, 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schlieker C., Zentgraf H., Dersch P., Mogk A. (2005) ClpV, a unique Hsp100/Clp member of pathogenic proteobacteria. Biol. Chem. 386, 1115–1127 [DOI] [PubMed] [Google Scholar]
- 47. Bladergroen M. R., Badelt K., Spaink H. P. (2003) Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol. Plant Microbe Interact. 16, 53–64 [DOI] [PubMed] [Google Scholar]
- 48. Bröms J. E., Lavander M., Sjöstedt A. (2009) A conserved α-helix essential for a type VI secretion-like system of Francisella tularensis. J. Bacteriol. 191, 2431–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]