Abstract
Primary metabolism affects all phenotypical traits of filamentous fungi. Particular examples include reacting to extracellular stimuli, producing precursor molecules required for cell division and morphological changes as well as providing monomer building blocks for production of secondary metabolites and extracellular enzymes. In this review, all annotated genes from four Aspergillus species have been examined. In this process, it becomes evident that 80–96% of the genes (depending on the species) are still without verified function. A significant proportion of the genes with verified metabolic functions are assigned to secondary or extracellular metabolism, leaving only 2–4% of the annotated genes within primary metabolism. It is clear that primary metabolism has not received the same attention in the post-genomic area as many other research areas—despite its role at the very centre of cellular function. However, several methods can be employed to use the metabolic networks in tandem with comparative genomics to accelerate functional assignment of genes in primary metabolism. In particular, gaps in metabolic pathways can be used to assign functions to orphan genes. In this review, applications of this from the Aspergillus genes will be examined, and it is proposed that, where feasible, this should be a standard part of functional annotation of fungal genomes.
Keywords: Aspergillus, primary metabolism, functional genomics, metabolic networks
INTRODUCTION
The research in primary metabolism in filamentous fungi has a long and proud history. Perhaps the most known example is the groundbreaking research in the genetics of folic acid biosynthesis in Neurospora crassa published by Beadle and Tatum in the early 1940s [1, 2]. This work eventually led to the formulation of the Nobel prize-awarded ‘one gene–one enzyme’ hypothesis.
Identification and characterization of genes involved in primary metabolism has made significant progress since then, not least in the genus of Aspergillus, employed for the production of primary metabolites since the 1930s [3]. With the introduction of Aspergillus nidulans as a model for eukaryotic cell development by Guido Pontecorvo in the 1950s [4], functional genetics has been pushed forward. For the Aspergillus genus as a whole, it can be argued that the study of primary metabolism is of particular importance for all of the phenotypes and applications of Aspergilli. A thorough and holistic understanding of primary metabolism is a necessary basis for understanding physiology-based regulatory responses impacting other traits. Prime examples are processes related to cellular morphology, the life cycle and cell cycle of the cells, but also other traits associated with growth and development in general, e.g. secondary metabolism, enzyme secretion for degradation of extracellular nutrients.
In this review, I will describe the status quo of the functional genetics and genomics of metabolism in the four best characterized Aspergillus species, namely, Aspergillus oryzae—a traditional food fermenter [5], Aspergillus fumigatus—arguably one of the world’s most lethal fungal pathogens [6, 7], Aspergillus niger—an industrial workhorse for enzyme production and primary metabolism [8] and A. nidulans—the primary model organism of the genus [4, 9]. These four species represent the main areas of diversity in the applications of Aspergilli.
OVERVIEW OF FUNCTIONAL ANNOTATION OF ASPERGILLUS SPECIES DEMONSTRATES SPECIES-SPECIFIC FOCUS AREAS FOR GENE CHARACTERIZATION
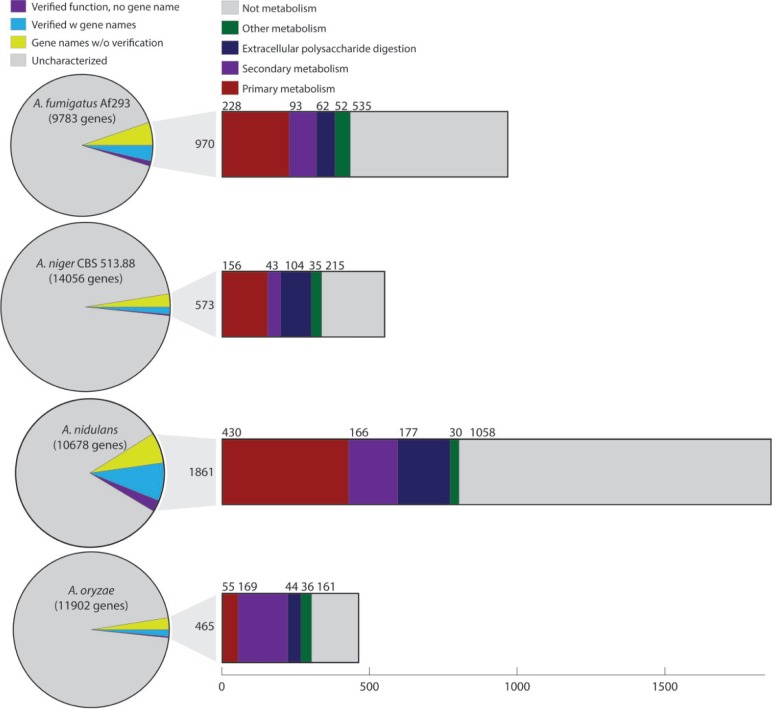
To thoroughly examine the status of the characterization of Aspergillus genes, annotation data were extracted from the world’s currently most comprehensive and extensively curated database of Aspergillus genomics, the AspGD.org [10]. Figure 1 below summarizes the review of these data.
Figure 1:
Overview of characterized genes and their function in A. fumigatus, A. niger, A. nidulans and A. oryzae. Pie charts show total number of genes; the area of the pie is proportional to the number of annotated genes. Bar charts show the distribution of functions related to different types of metabolism and non-metabolic functions. ‘Other metabolism’ denotes genes with functions not relevant in the other categories, primarily proteases. Data are summarized from AspGD.org annotation tables [10].
In general, the percentage of uncharacterized genes in these four Aspergilli is still high. Even for the most characterized model organism, A. nidulans, there are >80% uncharacterized genes, whereas the industrial species A. niger and A. oryzae have >96% uncharacterized genes. These percentages are high and illustrate the necessity for further characterization of the genes of these fungi. For comparison, the genome of the world’s most characterized microbe—Escherichia coli—has been estimated to be 34% uncharacterized, whereas mouse—Mus musculus—is comparable with A. niger and A. oryzae, in that 96% of the genes are still uncharacterized [11].
The phenotype and application of the fungus affects the direction of the research conducted in the organism, and therefore also the types of genes characterized. As Figure 1 clearly shows, the two cell factories A. oryzae and A. niger have a higher percentage of the characterized genes within metabolism. Aspergillus niger has—due to applications within citric acid production—been a model for characterization of much of central metabolism. The genetics and physiology behind the citric acid fermentation has been addressed in several reviews. Reference [12] can be recommended in particular.
When examining the characterized genes, it is significant that 50% of the characterized genes involved in metabolism, are not in primary metabolism. For A. oryzae, this is as high as 80%. The majority of these are within secondary metabolism (the synthesis of bioactive compounds not essential to growth) and extracellular enzymes – both categories extensively reviewed by others previously [13–15] and in other reviews in this issue. These categories have been the object of more focused efforts leading to the verification of the function of a number of genes comparable with those found in the entire diversity of primary metabolism. Considering the diversity of the metabolic pathways in primary metabolism, primary metabolism in Aspergilli has been relatively neglected.
The disparity between the characterization of primary versus the remaining parts of metabolism is to a large extent driven by genomics. The majority of the characterization of genes in Aspergillus primary metabolism has been conducted prior to the publication of the relevant genomes (as can be summarized by metabolic networks for A. nidulans, A. oryzae and A. niger [16–18]). While not large in numbers, it should be emphasized that some highly relevant work on crucial catabolic pathways has been made in recent years (see e.g. refs [19, 20] where ‘missing links’ in sugar catabolism are discovered). In contrast, the number of genes with assigned functions in secondary metabolism and secreted enzymes has exploded with the publication of the genomes [6, 21–23]. While the detailed description of this is out of scope for this review (see other articles in this issue), the common features of secondary metabolism and secreted enzymes for polysaccharide degradations make them highly suited for functional genomics approaches and algorithms in bioinformatics [14, 24–27].
NETWORK APPROACHES TO FUNCTIONAL GENOMICS OF PRIMARY METABOLISM
In many ways, the diversity of fungal primary metabolism (a wealth of catabolic pathways, capability to biosynthesize all important vitamins and cofactors) is what makes the primary metabolism of filamentous fungi fascinating and worthy of further study. This diversity of function is, however, also what makes high-throughput annotation of metabolic genes difficult compared with the more homogenous types of enzymes found in secondary metabolism and secreted carbohydrate-active enzymes. Both groups concern a more homogeneous set of substrates and products. One saving feature that still makes primary metabolism especially well suited for functional genomics is the knowledge of biosynthetic pathways accumulated from other microbes or even plants or animals. Curated and systematic versions of these networks can be accessed in general pathway databases, such as KEGG [28] or the MetaCyc/BioCyc databases [29]. Using the network for annotation can also alleviate the problem that function inferred by sequence homology (as employed in the above databases) can be misleading. Experience shows that manual curation, supplemented with experimental evidence, is still required for absolute accuracy. This can be employed in post-genomic functional genomic set-ups, where underlying metabolic network should provide a scaffold for annotation, e.g. one can assume that all enzymes in an essential pathway should be present in the genome, and use that knowledge to find best gene candidates for all steps in the pathway [30]. Recent publications have shown that is possible to automate this process for fungal species, and generate high-quality gapless metabolic networks [31].
These information-heavy network-based approaches holds a lot of promise for improving the annotation of current genomes in the Aspergilli, and should also be a preferred method for the initial annotation of primary metabolism genes in future genomes from fungal species. Notable examples are studies where the primary metabolism has been reconstructed and used to assign function to orphan genes. One such study is the metabolic reconstruction of A. nidulans, where function was tentatively assigned to 472 orphan genes, increasing the number of genes included in the model to 666 [16]. A similar model for A. oryzae includes tentative and verified annotation of 1314 genes [17]. Finally, a reconstruction of the A. niger metabolic network includes 871 genes [18]. An alternative reconstruction of the metabolic network of A. niger includes a larger (although not fully specified) number of genes [32]. However, based on experience from current ongoing efforts with updating and improving the A. niger iMA871 model [18] by the author and co-workers, these numbers will change significantly when developments in annotation and gene calling are included. More recently published modelling and network reconstruction efforts in other fungi such as N. crassa confirm this trend towards being able to identify more genes [33].
Other notable efforts are efforts using comparative genomics across multiple species to define the most likely candidates for a given gene function. A prime example of this is the work by Flipphi and co-workers [34], where genes for central metabolism were examined in eight different Aspergillus/Neosartorya species. This allowed the assignment of gene candidates for ∼155 enzymatic functions in each of the species.
CONCLUSION
It is argued that research in primary metabolism in these fungi—although highly important—has decreased, in particular compared with efforts within secondary and extracellular metabolism. However, the application of functional genomics to the study of Aspergillus species should accelerate gene identification in primary metabolism and make it possible to continue to provide crucial understanding in this topic. In particular, approaches based on the topology of metabolic networks in tandem with ‘omics’-based analysis of the fungi hold a great promise for continuing this development.
Key points.
Less than 20% of the genes of Aspergillus species are characterized.
Only a fraction of the characterized genes are within primary metabolism.
Relatively few new genes within primary metabolism are being characterized.
Holistic application of metabolic networks is a feasible way of functional genomics for a large number of genes within primary metabolism.
FUNDING
The author gratefully acknowledges funding from the Novo Nordisk Foundation - Programme for Biotechnology-based Synthesis and Production Research.
Biography
Associate Professor Mikael R. Andersen is leader of a research group working within omics-based approaches to designing and engineering cells for industrial biotechnology.
References
- 1.Tatum EL, Beadle GW. Genetic control of biochemical reactions in neurospora: an “aminobenzoicless” mutant. Proc Natl Acad Sci USA. 1942;28:234–43. doi: 10.1073/pnas.28.6.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beadle GW, Tatum EL. Genetic control of biochemical reactions in neurospora. Proc Natl Acad Sci USA. 1941;27:499–506. doi: 10.1073/pnas.27.11.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papagianni M. Advances in citric acid fermentation by Aspergillus niger: biochemical aspects, membrane transport and modeling. Biotechnol Adv. 2007;5:244–63. doi: 10.1016/j.biotechadv.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Pontecorvo G, Roper JA, Hemmons LM, et al. The genetics of Aspergillus nidulans. Adv Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- 5.Machida M, Asai K, Sano M, et al. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–61. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- 6.Nierman WC, Pain A, Anderson MJ, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–6. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 7.Gibbons JG, Beauvais A, Beau R, et al. Global transcriptome changes underlying colony growth in the opportunistic human pathogen Aspergillus fumigatus. Eukaryot Cell. 2012;11:68–78. doi: 10.1128/EC.05102-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andersen MR, Salazar MP, Schaap PJ, et al. Comparative genomics of citric-acid-producing Aspergillus niger ATCC 1015 versus enzyme-producing CBS 513.88. Genome Res. 2011;21:885–97. doi: 10.1101/gr.112169.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galagan JE, Calvo SE, Cuomo C, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–15. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 10.Arnaud MB, Cerqueira GC, Inglis DO, et al. The Aspergillus Genome Database (AspGD): recent developments in comprehensive multispecies curation, comparative genomics and community resources. Nucleic Acids Res. 2012;40:D653–9. doi: 10.1093/nar/gkr875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karp PD, Keseler IM, Shearer A, et al. Multidimensional annotation of the Escherichia coli K-12 genome. Nucleic Acids Res. 2007;35:7577–90. doi: 10.1093/nar/gkm740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karaffa L, Kubicek CP. Aspergillus niger citric acid accumulation: do we understand this well working black box? Appl Microbiol Biotechnol. 2003;61:189–96. doi: 10.1007/s00253-002-1201-7. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez JF, Somoza AD, Keller NP, Wang CCC. Advances in Aspergillus secondary metabolite research in the post-genomic era. Nat Prod Rep. 2012;29:351–71. doi: 10.1039/c2np00084a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coutinho PM, Andersen MR, Kolenova K, et al. Post-genomic insights into the plant polysaccharide degradation potential of Aspergillus nidulans and comparison to Aspergillus niger and Aspergillus oryzae. Fungal Genet Biol. 2009;46:S161–9. doi: 10.1016/j.fgb.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 15.De Souza WR, de Gouvea PF, Savoldi M, et al. Transcriptome analysis of Aspergillus niger grown on sugarcane bagasse. Biotechnol Biofuels. 2011;4:40. doi: 10.1186/1754-6834-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.David H, Ozçelik IS, Hofmann G, Nielsen J. Analysis of Aspergillus nidulans metabolism at the genome-scale. BMC Genomics. 2008;9:163. doi: 10.1186/1471-2164-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vongsangnak W, Olsen P, Hansen K, et al. Improved annotation through genome-scale metabolic modeling of Aspergillus oryzae. BMC Genomics. 2008;9:245. doi: 10.1186/1471-2164-9-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersen MR, Nielsen ML, Nielsen J. Metabolic model integration of the bibliome, genome, metabolome and reactome of Aspergillus niger. Mol Syst Biol. 2008;4:178. doi: 10.1038/msb.2008.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mojzita D, Herold S, Metz B, et al. L-xylo-3-hexulose reductase is the missing link in the oxidoreductive pathway for D-galactose catabolism in filamentous fungi. J Biol Chem. 2012;287:26010–18. doi: 10.1074/jbc.M112.372755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mojzita D, Vuoristo K, Koivistoinen OM, et al. The “true” L-xylulose reductase of filamentous fungi identified in Aspergillus niger. FEBS Lett. 2010;584:3540–4. doi: 10.1016/j.febslet.2010.06.037. [DOI] [PubMed] [Google Scholar]
- 21.Galagan JE, Calvo SE, Cuomo C, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–15. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 22.Machida M, Asai K, Sano M, et al. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–61. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- 23.Pel HJ, de Winde JH, Archer DB, et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol. 2007;25:221–31. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- 24.Blin K, Medema MH, Kazempour D, et al. antiSMASH 2.0–a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41:W204–12. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cantarel BL, Coutinho PM, Rancurel C, et al. The carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–8. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersen MR, Giese M, de Vries RP, Nielsen J. Mapping the polysaccharide degradation potential of Aspergillus niger. BMC Genomics. 2012;13:313. doi: 10.1186/1471-2164-13-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersen MR, Nielsen JB, Klitgaard A, et al. Accurate prediction of secondary metabolite gene clusters in filamentous fungi. Proc Natl Acad Sci USA. 2013;110:E99–107. doi: 10.1073/pnas.1205532110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stelzer M, Sun J, Kamphans T, et al. An extended bioreaction database that significantly improves reconstruction and analysis of genome-scale metabolic networks. Integr Biol (Camb) 2011;3:1071–86. doi: 10.1039/c1ib00008j. [DOI] [PubMed] [Google Scholar]
- 29.Caspi R, Altman T, Billington R, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2014;42:D459–71. doi: 10.1093/nar/gkt1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thiele I, Palsson BØ. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc. 2010;5:93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pitkänen E, Jouhten P, Hou J, et al. Comparative genome-scale reconstruction of gapless metabolic networks for present and ancestral species. PLoS Comput Biol. 2014;10:e1003465. doi: 10.1371/journal.pcbi.1003465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun J, Lu X, Rinas U, Zeng AP. Metabolic peculiarities of Aspergillus niger disclosed by comparative metabolic genomics. Genome Biol. 2007;8:R182. doi: 10.1186/gb-2007-8-9-r182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dreyfuss JM, Zucker JD, Hood HM, et al. Reconstruction and validation of a genome-scale metabolic model for the filamentous fungus Neurospora crassa using FARM. PLoS Comput Biol. 2013;9:e1003126. doi: 10.1371/journal.pcbi.1003126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flipphi M, Sun J, Robellet X, et al. Biodiversity and evolution of primary carbon metabolism in Aspergillus nidulans and other Aspergillus spp. Fungal Genet Biol. 2009;46:S19–44. doi: 10.1016/j.fgb.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 35.Colabardini AC, Ries LNA, Brown NA, et al. Functional characterization of a xylose transporter in Aspergillus nidulans. Biotechnol Biofuels. 2014;7:46. doi: 10.1186/1754-6834-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]