Abstract
Plants contain a sophisticated innate immune network to prevent pathogenic microbes from gaining access to nutrients and from colonizing internal structures. The first layer of inducible response is governed by the plant following the perception of microbe- or modified plant-derived molecules. As the perception of these molecules results in a plant response that can provide efficient resistance toward non-adapted pathogens they can also be described as “defense elicitors.” In compatible plant/microbe interactions, adapted microorganisms have means to avoid or disable this resistance response and promote virulence. However, this requires a detailed spatial and temporal response from the invading pathogens. In agricultural practice, treating plants with isolated defense elicitors in the absence of pathogens can promote plant resistance by uncoupling defense activation from the effects of pathogen virulence determinants. The plant responses to plant, bacterial, oomycete, or fungal-derived elicitors are not, in all cases, universal and need elucidating prior to the application in agriculture. This review provides an overview of currently known elicitors of biological rather than synthetic origin and places their activity into a molecular context.
Keywords: crop protection, elicitors, pathogen effectors, priming, disease resistance
The role of defense elicitors in plant immunity
Plants are under constant threat of microbial pathogen attack. Plant cell walls, cuticles and phytoanticipins are preformed, physical and chemical barriers that limit access of microbes to plant cells (Underwood, 2012; Newman et al., 2013). In addition to these non-inducible defenses, plants recognize and respond to defense elicitors which are signal-inducing compounds perceived by the innate immune system that prime and/or induce defense responses (Henry et al., 2012; Maffei et al., 2012; Newman et al., 2013). Elicitor compounds can be biological in origin, derived from either the plant or the microbe, or can be synthetically generated (Walters et al., 2013). We will focus on elicitors from biological origin (Table 1) rather than synthetic analogs of known signaling or defense molecules such as Bion, acibenzolar-S-methyl (ASM), beta-amino-butyric acid (BABA), and cis-jasmone. Elicitor activity has, for example, been shown for plant-derived cell wall components such as oligogalacturonides (Ferrari et al., 2013), proteinaceous pathogen molecules such as bacterial flagellin (Gomez-Gomez and Boller, 2002), oomycete-derived elicitin INF1 (reviewed in Hein et al., 2009) and non-proteinaceous molecules such as lipopolysaccharides (Erbs and Newman, 2012). However, intact plant- or microbe-derived structures as well as highly-polymerized molecules often tend to result in few recognition responses. In contrast, leakage of metabolites or even minor or partial breakdown of complex host or pathogen molecules leads to the production of eliciting components that are biologically active.
Table 1.
List of plant-, bacterial-, oomycete-, and fungal-derived elicitor compounds, their activity against pathogens and effectiveness in plants.
| Origin | Elicitor compound | Effective toward | Plants effects shown in | References |
|---|---|---|---|---|
| Plant | Oligogalacturonides | Botrytis cinerea, Blumeria graminis | Several | Aziz et al., 2007; Randoux et al., 2010; Galletti et al., 2011 |
| Milsana (giant knotweed) | Botrytis cinerea, Leveillula taurica | Cucumber, tomato | Daayf et al., 1997, 2000; Konstantinidou-Doltsinis et al., 2006 | |
| Burdock fructooligosaccharide | Colletotrichum lagenarium, Botrytis cinerea, TMV | Cucumber, tobacco, tomato | Wang et al., 2009; Guo et al., 2012 | |
| Elicitor peptide 1 (Pep1) | Cochliobolis heterostrophus, Colletotrichum graminicola | Maize | Huffaker et al., 2011 | |
| Carrageenans | Sclerotinia sclerotiorum, TMV | A. thaliana, tobacco | Sangha et al., 2010; Vera et al., 2011 | |
| Fucans | TMV | Tobacco | Vera et al., 2011 | |
| Ulvans | Several | Several | Jaulneau et al., 2011; Vera et al., 2011 | |
| Laminarin | Erwinia carotovora, Plasmopara viticola, Botrytis cinerea, Fusarium solani | Beans, grapevine, tobacco | Craigie, 2011; Vera et al., 2011 | |
| Bacteria | Harpin | Xanthomonas oryzae | Rice | Lee et al., 2001; Li et al., 2012 |
| Lipopeptides | Botrytis cinerea | Tomato | Henry et al., 2012 | |
| Dimethylsulfide | Cochliobolus heterostrophus, Botrytis cinerea | Maize, tobacco | Huang et al., 2012 | |
| Pseudobactin | Botrytis cinerea, Erwinia carotovora | Several | De Vleesschauwer and Höfte, 2009 | |
| Oomycetes | CBEL | Phytophthora parasitica | A. thaliana, tobacco | Mateos et al., 1997; Khatib et al., 2004 |
| Cryptogein | Phytophthora parasitica, Sclerotinia sclerotiorum | Tobacco | Bonnet et al., 1996 | |
| Eicosapentaenoic acid | Phytophthora infestans | Potato | Henriquez et al., 2012 | |
| Pep-13 | Phytophthora spp. | Parsley, potato | Nürnberger et al., 1994; Brunner et al., 2002; Parker, 2003 | |
| INF1 | Phytophthora infestans | Tobacco | Takahashi et al., 2007; Hein et al., 2009; Kawamura et al., 2009 | |
| Fungi | β-glucans | Several | Several | Hahn and Albersheim, 1978; Fu et al., 2011; Falcón-Rodríguez et al., 2012; Henriquez et al., 2012 |
| Chitosan | Several | Several | Kishimoto et al., 2010; Kombrink et al., 2011 | |
| Chitin | Several | Several | El Ghaouth et al., 1994; Copping and Duke, 2007; El Hadrami et al., 2012 | |
| Ergosterol | Botrytis cinerea | Grapevine, tobacco | Laquitaine et al., 2006; Vatsa et al., 2011 | |
| Trichoderma species: xylanases, peptaibol, cerato-platanin family | Pseudomonas syringae, Botrytis cinerea, Colletotrichum graminicola | A. thaliana, cotton, maize | Ron and Avni, 2004; Djonoviç et al., 2007; Viterbo et al., 2007; Yang et al., 2009; de Oliveira et al., 2011 | |
| Cerebrosides | Fusarium spp. | Several | Umemura et al., 2004 | |
| HR-inducing protein | Magnaporthe oryzae | Rice | Chen et al., 2012; Kulye et al., 2012 | |
| PeaT1 | TMV | Tobacco | Zhang et al., 2010, 2011b | |
| PebC1 | Botrytis cinerea | Tomato | Zhang et al., 2010 | |
| PevD1 | TMV | Tobacco | Wang et al., 2012a,b | |
| PemG1 | Pseudomonas syringae, Xanthomonas oryzae | A. thaliana, rice | Qiu et al., 2009; Peng et al., 2011 |
The co-evolution between plants and potential microbial pathogens has been described as a zigzag model by Jones and Dangl (2006) and can also be applied to deducing the biological activity of elicitors (Figure 1). According to the zigzag model, the first inducible responses are a consequence of the perception of chemical elicitors, microbe-associated molecular patterns (MAMPs), pathogen-associated molecular patterns (PAMPs) and/or damage-associated molecular patterns (DAMPs). The latter are also known as danger-associated molecular patterns (Mazzotta and Kemmerling, 2011). MAMPs describe general microbe-derived molecules including those originating from beneficial microbes whereas PAMPs specifically describe molecules from pathogenic microbes such as fungi, oomycetes, and bacteria (Henry et al., 2012; Newman et al., 2013). Thus, PAMPS are a subgroup of MAMPs (Maffei et al., 2012). In contrast, DAMPs are typically plant-derived and are produced after, for example, wounding by insects or herbivores as well as degradation or perturbation of host molecules by microbes (Henry et al., 2012; Newman et al., 2013). All of these molecules, which could universally be described as “patterns that elicit immunity” (PEIs), are often recognized by transmembrane pattern recognition receptors (PRRs) in plant cells (Jones and Dangl, 2006; Maffei et al., 2012; Newman et al., 2013). Upon recognition of MAMP- or DAMP-derived patterns, PTI (PAMP- or pattern-triggered immunity) is activated in the plant and the perceived molecules could be described as immune elicitors. This defense reaction aims to restrict the growth of the intruder and can lead to systemic induced resistance leaving the plant less susceptible to subsequent pathogen attack (Henry et al., 2012).
Figure 1.

Plants recognize chemical elicitors, Microbe-Associated Molecular Patterns (MAMPS) derived from non-pathogenic microbes, Pathogen-Associated Molecular Patterns (PAMPS) derived from pathogens and Damage-Associated Molecular Patterns (DAMPS) that are produced by plants upon insect, herbivore or pathogen attack, via transmembrane Pattern Recognition Receptors (PRRs). The recognition leads to the onset of defense mechanisms referred to as pattern-triggered immunity (PTI). Adapted pathogens secrete effectors that disturb plant defense mechanisms leading to effector-triggered susceptibility (ETS). Plant resistance (R) proteins recognize pathogen effectors and induce effector-triggered immunity (ETI). Treatment of plants with elicitor compounds (chemicals, MAMPs, DAMPs, or PAMPs) in the absence of adapted pathogen leads to priming and/or PTI-based immunity that put plants into an alerted stage of defense that provides some enhanced resistance toward otherwise virulent pathogens. Figure adapted from Henry et al. (2012), and Jones and Dangl (2006).
Systemic induced resistance can be divided into systemic acquired resistance (SAR) or induced systemic resistance (ISR). Systemic acquired resistance is often characterized by localized necrosis, expression of pathogenesis related (PR) protein genes, and involves the salicylic acid (SA) pathway whereas ISR is often triggered by plant growth-promoting rhizobacteria (PGPR) (Walters et al., 2013), is not associated with necrosis and involves the jasmonic acid (JA) and ethylene (ET) pathways (Walters and Heil, 2007; Henry et al., 2012). Typical responses of PTI include cell wall alterations and the production of reactive oxygen species (ROS) which can be directly cytotoxic but also play a role in signaling. Other responses comprise the production of phytoalexins, expression of PR proteins, activation of mitogen activated protein kinase (MAPK) pathways, and defense signaling involving calcium (Ca2+) influx from extracellular spaces and changes in free cytosolic Ca2+ concentrations (Garcion et al., 2007). To counteract the initial plant defense reaction, successful microbes have evolved specialized effectors that perturb recognition of defense elicitors or subsequent plant defense mechanisms to promote effector-triggered susceptibility (ETS). However, if these pathogen effectors are in turn recognized by cognate plant resistance (R) proteins, the second layer of inducible response, effector-triggered immunity (ETI), is initiated that often yields a hypersensitive resistance response (HR) (Jones and Dangl, 2006; Deslandes and Rivas, 2012).
The outcome of plant/microbe interactions can result in symbiosis, disease or disease resistance and is governed by further levels of sophisticated co-evolution. Indeed, it must be recognized that pathogen colonization of plants can generate dynamic pathogenic, mutualistic or parasitic interactions of varying magnitude and specificity. Furthermore, organisms recognized as pathogens in, for example, a crop context, could be benign or even beneficial in another context such as a different host or environment (Newton et al., 2010). It is thus essential for the plant to evaluate the scale of threat and to mount appropriate and proportionate responses. These may range from priming, being ready to respond faster to actual attack, or expression of PTI-based defense mechanisms to yield incompatibility if the microbe/pathogen is unable to suppress these responses. The use of elicitors in agriculture holds the potential to decrease the need for pesticide application by using the plant's own defense system. However, there is a need to understand this process on a molecular level to maximize the efficacy of the treatments.
Inducible defense response in the absence of pathogens
In agricultural practice, elicitor treatments of plants in the absence of virulent pathogens yields a defense response such as priming and/or PTI that is uncoupled from ETS and can provide some protection to subsequent pathogen challenges. Priming is defined as a physiological status of plants leading to faster and stronger activation of defense responses to subsequent biotic and abiotic stresses (reviewed in Conrath et al., 2006; Conrath, 2011; Pastor et al., 2013). Crucially, this is distinct from the level of resistance induction in response to the recognition of true pathogens that are potentially capable of causing disease and where recognition would cause resistance mechanism expression that is more costly to the plant but still proportionate to potential disease cost (Walters and Heil, 2007).
In primed plants, chromatin modifications in the form of methylation and acetylation of histones take place that impact on the interaction of DNA with histones and/or open binding sites for transcriptional co-activators such as WRKY22 and WRKY29 (Eulgem, 2005; Conrath, 2011; Po-Wen et al., 2013). These chromatin modifications in primed plants have been shown to lead to increased expression of transcription factors WRKY6, WRKY29, and WRKY53 after stress exposure (Jaskiewicz et al., 2011). In Arabidopsis thaliana, mRNA and inactive MPK3 and MPK6 accumulate in cells of elicitor-treated plants. Upon exposure to Pseudomonas syringae both MAP kinases are more strongly activated in primed plants than in non-primed plants (Beckers et al., 2009).
When PTI-associated mechanisms are primed by elicitor treatments plants often accumulate ROS and produce a stronger, secondary oxidative burst following pathogen challenge, activate MPKs and stimulate SA-, JA-, and abscisic acid (ABA)-pathways (Beckers et al., 2009; Pastor et al., 2013). Callose deposition, which is potentially also linked to the ABA-pathway, can be enhanced in elicitor-treated plants (Kohler et al., 2002; Flors et al., 2005; Pastor et al., 2013) and elicitor treatment often induces expression of phenylalanine ammonia lyase (PAL) which is required for the production of SA precursors (Chen et al., 2009). In line with SA involvement, pathogenesis-related genes such as PR-1, PR-2, and PR-5 have been implicated with elicitor treatments (Kohler et al., 2002; Conrath et al., 2006). Both priming and the activation of defense mechanisms due to elicitor treatment can lead to a reduction of disease severity when subsequent pathogen attack occurs. Biologically active defense elicitors that either prime plant defenses or initiate PTI responses have been identified from diverse sources. Molecular studies have provided clues to their mechanism and to the processes that govern specificity.
A molecular perspective of elicitor activity in plant immunity
Several studies have shown that elicitor-treated plants show lower infection rates following inoculation with virulent pathogens but responses can vary between plant species (Table 1). In addition to the observed disease reduction, molecular studies are revealing how the elicitor compounds affect gene expression levels in plants and therefore impact on defense responses (Section Plant Genes and Pathways Involved in Elicitor Recognition). Similarly, the diverse mechanisms by which pathogen effectors suppress PTI responses are emerging but, due to the complexity of this research, only selected examples are highlighted in this review.
Plant-derived elicitors
Plant cell walls are composed of cellulose, hemicellulose (cross-linking glycans), pectic polysaccharides, protein, lignin, and a variety of lipids (Wei et al., 2009). Bacteria and fungi can produce cellulases, xylanases, and lignin peroxidases that break down plant cell wall components and common products are β-glucans, xylose, and phenylpropanoid-containing compounds. These break-down products function as plant-derived elicitors and several examples of disease reduction due to the application of plant-derived elicitors exist (Table 1). Well studied plant-derived elicitors include oligogalacturonides (OGs), which are structural components of plant cell walls and are released upon partial degradation of homogalacturonan by microbial polygalacturonases during infection or by plant polygalacturonases induced upon wounding (Ferrari et al., 2013). Plant cell wall-derived OGs are recognized by wall-associated kinase 1 (WAK1) and subsequent signaling is JA-, SA-, and ET-independent (Brutus et al., 2010; Ferrari et al., 2013). A MAP kinase cascade is triggered upon OG perception in A. thaliana, and MPK3 and MPK6 are phosphorylated. However, the importance of these signaling events remains elusive and it has been shown, for example, that lack of MPK3 increases basal susceptibility to Botrytis cinerea but elicitor-induced resistances are not affected (Galletti et al., 2011). In contrast, MPK6 is necessary for OG-induced resistance but does not play a role in basal resistance toward B. cinerea (Galletti et al., 2011).
Bacterial-derived elicitors
In addition to plant-derived elicitors, the application of bacterial-derived elicitors has also been shown to reduce pathogen infection in plants (Table 1). Extracellular polysaccharides (EPS) produced by the bacterial wilt causing pathogen Ralstonia solanacearum have been shown to induce defense responses in tomato (Milling et al., 2011) and lipopolysaccharides (LPS) from Gram-negative bacteria also trigger induced resistance in several other plant species (Dow et al., 2000; Gerber et al., 2004; Desaki et al., 2006; Erbs and Newman, 2012). PGPRs (plant growth-promoting rhizobacteria) can induce resistance in plants by exudating elicitors (De Vleesschauwer and Höfte, 2009 and references therein) and filtrates from cultures of bacteria such as Bacillus subtilis can also elicit crop protection effects (Schönbeck et al., 1980, 1982), though these may be a combination of direct toxicity and/or elicitor recognition events.
The molecular background to bacterial MAMPs, effectors and their plant targets has been reviewed recently (Deslandes and Rivas, 2012) and two well-studied bacterial MAMPs are flagellin and the elongation factor Tu (EF-Tu). Flagellin is recognized in a variety of plant species whereas EF-Tu, one of the most abundant proteins in bacterial cells, and bacterial cold-shock proteins seem to be specifically recognized in Brassicaceae and Solanaceae plants, respectively (Gomez-Gomez and Boller, 2002; Felix and Boller, 2003; Bittel and Robatzek, 2007; Jeworutzki et al., 2010). In both proteins, the N-terminus contains the eliciting site which, for flagellin, can often be described as a 22 amino acid long epitope (flg22), whereas that for EF-Tu is 18 amino acids (Elf18). Flagellin and EF-Tu are recognized by two distinct plant receptors (Gomez-Gomez and Boller, 2002; Kunze et al., 2004). Flagellin is recognized by FLAGELLIN-SENSING 2 (FLS2) whereas EF-Tu is recognized by EF-Tu RECEPTOR (EFR) which has only been found in Brassicaceae (Gomez-Gomez and Boller, 2002; Zipfel et al., 2006). This provides molecular insight into the specificity of elicitors and emphasizes the need to assess candidate defense eliciting compounds in a diverse range of plant species. It is interesting to note that heterologous expression of EFR in Solanaceae plants provides some resistance to bacteria that express EF-Tu (Lacombe et al., 2010) which suggests that downstream signaling cascades could be conserved for different PRRs and in different plant species. Indeed, both FLS2 and EFR are leucine rich repeat receptor like kinases (LRR-RLK) and both interact with BRI1-associated receptor kinase 1 (BAK1) triggering SA-, JA-, and ET-independent signaling (Zipfel et al., 2004, 2006; Chinchilla et al., 2007). Recognition of flg22 and Elf18 leads to an increase in cytosolic Ca2+ and it has been show that early signaling is BAK1-dependent and involves calcium associated plasma membrane anion channel opening (Jeworutzki et al., 2010). Subsequently, a MAP kinase cascade involving MPK3, MPK4, MPK6 and MPK11, and other genes such as Ca2+-dependent proteinase kinases are activated to establish PTI (Zipfel et al., 2004, 2006; Chinchilla et al., 2007; Bethke et al., 2012).
Pathogenic bacteria secrete, amongst others, type III effectors into plant cells to supress PTI and this mechanism has been well studied in the plant pathogen P. syringae (reviewed by Block and Alfano, 2011; Deslandes and Rivas, 2012). These bacterial effectors target a variety of plant genes and metabolites including plasma membrane components like RPM1-interacting protein 4 (RIN4) in A. thaliana (Day et al., 2006). Similarly, host nuclear components are, for example, perturbed by effectors such as PopP2 as well as by transcription-activator like (TAL) type III effectors from Xanthomonas that directly bind to plant DNA and thereby activate gene expression changes that promote virulence and pathogen colonization (Boch et al., 2009; Deslandes and Rivas, 2012; Coll and Valls, 2013). Other examples include the effector HopAl1 that is widely conserved in bacterial plant pathogens and interferes with the MAPK signaling genes MPK3 and MPK6 to supress PTI (Zhang et al., 2007). Furthermore, chloroplast components are also modified by bacterial effectors such as HopI1 that causes remodeling of the chloroplast thylakoid structure and interferes with SA accumulation (Jelenska et al., 2007). As mentioned above, plants have a variety of R genes, the products of which, directly or indirectly, recognize some of these bacterial effectors to elicit ETI (reviewed by Block and Alfano, 2011; Deslandes and Rivas, 2012).
Oomycete-derived elicitors
Oomycetes are taxonomically and structurally distinct from both plants and fungi. Several oomycetes are plant pathogenic and include those from the genus Phytophthora that are responsible for substantial yield losses in crops. Oomycete cell walls consist of cellulose, glycan, and hydroxyproline-rich proteins and several oomycete elicitors have been described (Table 1). For example, necrosis and ethylene-inducing peptide 1 (Nep1)-like proteins (NLP) are recognized in dicots and it has been shown that these proteins trigger a variety of defense responses in A. thaliana (Qutob et al., 2006). Similarly, P. infestans INF1 elicitin causes an HR response in Nicotiana benthamiana (Kamoun et al., 1998) that is dependent on the receptor-like kinase SERK3/BAK1 which, as a central regulator of innate immunity in plants, is required for multiple resistance responses, including those mediated through FLS2 (Heese et al., 2007). Other PTI eliciting molecules from Phytophthora include GP42, a member of the transglutaminase family, and for which the active peptide has been described as Pep-13 (Nürnberger et al., 1994; Brunner et al., 2002), as well as the cellulose binding elicitor lectin (CBEL) that is associated with adhesion to the plant cell (Gaulin et al., 2006; reviewed in Hein et al., 2009).
To suppress PTI during infection, Phytophthora, like other plant pathogens, secretes extracellular and intracellular effectors into plants. Some extracellular effectors encode protease or glucanase inhibitors to prevent, respectively, host protease or glucanase activity in the apoplast (reviewed in Hein et al., 2009; Schornack et al., 2009). Some intracellular effectors contain the canonical RXLR motif and contain an N-terminal signal peptide and a C-terminal effector activity site (Birch et al., 2009).
The modes of action of RXLR effectors in promoting virulence are diverse. For example, it has recently been shown that the P. infestans RXLR effector PexRD2 interacts with the kinase domain of the host MAPKKKε to perturb PTI signaling pathways and to yield ETS responses (King et al., 2014). The RXLR effector PITG_03192, on the other hand, targets two membrane-associated NAC transcription factors that rapidly accumulate following PTI elicitation (McLellan et al., 2013). The effector prevents the release of these NAC transcription factors from the endoplasmic reticulum and subsequent accumulation in the plant nucleus that is typically observed as part of a PTI response. In contrast, the P. infestans RXLR effector Avrblb2 prevents secretion of an immune-associated protease (Bozkurt et al., 2011), whereas two P. sojae RXLRs have been shown to act as silencing suppressors (Qiao et al., 2013). One of the best-characterized intracellular RXLR effectors is Avr3a from P. infestans. Avr3a interacts with and stabilizes the potato E3 ubiquitin ligase CMPG1 and thus perturbs cell death responses triggered by INF1 (Bos et al., 2010) and a range of other pathogen elicitors (Gilroy et al., 2011). Avr3a exists in two forms that both suppress INF1 responses but differ in two amino acids that determine recognition by the potato R gene R3 that subsequently triggers ETI (Armstrong et al., 2005). Finally, several RXLRs from P. infestans act redundantly to suppress flg22-mediated signal transduction and early transcriptional changes (Zheng et al., 2014).
Fungal-derived elicitors
As with plant and oomycete cell walls, break-down products from fungal cell walls, which contain chitin, mannoproteins, and β-glucans, can elicit a range of defense responses as signals of potential colonization (Table 1). Yeast extracts, for example, have widely been used to study defense responses in plants (e.g., Hahn and Albersheim, 1978; Reglinski et al., 1994b, 1995; Suzuki et al., 2005; Khokon et al., 2010). Ergosterol, a fungal cell membrane component, induces defense responses in tobacco, and Cladosporium fulvum host and non-host plant necrosis inducer 1 (CfHNNI1), which shows high homologies to genes encoding bZIP transcription factors, has been shown to induce resistance in tomato and tobacco (Takken et al., 2000; Xu et al., 2012). Similarly, a proteinaceous elicitor called SCLEROTINIA CULTURE FILTRATE ELICITOR1 (SCFE1) has recently been identified from the necrotrophic fungal pathogen Sclerotinia sclerotiorum that induces BAK1-dependent PTI responses upon recognition by the A. thaliana RECEPTOR-LIKE PROTEIN30 (RLP30) (Zhang et al., 2013).
Two of the best studied fungal-derived elicitors are chitin and chitosan, a deacetylated derivative of chitin. Both have been well described as active components that increase resistance to bacterial and fungal pathogens in several plant species including crop plants (El Ghaouth et al., 1994; Copping and Duke, 2007; Kishimoto et al., 2010; El Hadrami et al., 2012; Kombrink et al., 2011). Chitin is detected in plants by a chitin elicitor receptor kinase (CERK1) which is also known as LysMRLK1 (Wan et al., 2008; Kombrink et al., 2011). In A. thaliana, chitin-induced dimerization of AtCERK1 has shown to be necessary for activation of PTI (Liu et al., 2012). In rice, OsCERK1 forms a complex with chitin elicitor binding protein (CEBiP) upon chitin perception and both proteins are critical for chitin-induced signaling (Shimizu et al., 2010). A homolog of OsCEBiP has been identified in barley and HvCEBiP has also been shown to play a role in responses to Magnaporthe oryzae (Tanaka et al., 2010). In contrast to rice, the homolog in A. thaliana, AtCEBiP, binds chitin but does not seem to be required for chitin-induced signaling (Shinya et al., 2012). Chitin-induced PTI is JA-, SA-, and ET-independent but a RING zinc-finger like protein (ATL9) has shown to be induced upon chitin treatment (Berrocal-Lobo et al., 2010).
To suppress these responses, the fungal pathogen C. fulvum has developed two distinct effectors that suppress chitin-induced PTI leading to ETS in planta (de Jonge and Thomma, 2009; de Jonge et al., 2010; Kombrink et al., 2011). The fungal chitin-binding protein Avr4 specifically binds chitin in fungal cell walls and thereby prevents the chitin from degradation by plant chitinases (van den Burg et al., 2006; Wan et al., 2008). Furthermore, the extracellular protein 6 (Ecp6), an effector protein with 3 LysM domains, binds chitin competitively to prevent recognition of chitin by CEBiP (de Jonge and Thomma, 2009; de Jonge et al., 2010). Homologs of Avr4 have been identified in fungi belonging to the class of Dothideomycetes and Ecp6-like genes are widespread within the fungal kingdom (Kombrink et al., 2011). The plant receptor Cf4 is a receptor-like protein (RLP) without kinase activity that recognizes Avr4 (Thomas et al., 1997) and it has recently been shown that SOBIR1, a receptor-like kinase (RLK) from tomato interacts with Cf4 and might be required for Cf4-mediated resistance (Liebrand et al., 2013).
Compared with bacterial and oomycete effectors, the biological function and the targets of fungal effectors remain more elusive (Rafiqi et al., 2012; Liu et al., 2013). This has partly been attributed to the fact that fungal effectors do not seem to have canonical amino acid domains that enable a rapid candidate effector discovery (Rafiqi et al., 2012). Recently, Doehlemann and Hemetsberger (2013) reviewed the current knowledge of effectors from filamentous plant pathogens and compiled a list of known apoplastic effectors and their function. Most fungal effectors are secreted through the fungal endoplasmic reticulum (ER) secretory pathway but the way by which cytoplasmic effector proteins enter the host cells remains unknown (Rafiqi et al., 2012). In the genome of Blumeria graminis, 491 potential effector proteins have been identified but their biological function remains unknown (Pedersen et al., 2012). Similarly, in M. oryzae 15 candidate effector proteins have been identified so far (Liu et al., 2013). An effector protein from Ustilago maydis has been identified as a chorismate mutase, Cmu1, which is required for full virulence. Cmu1 functions by diverting metabolic precursors of the shikimate pathway toward production of aromatic amino acids, and away from the production of SA (Djamei and Kahmann, 2012).
Plant genes and pathways involved in elicitor recognition
Genes up-regulated due to elicitor treatments
In the elicitor research field, the response of plant genes to elicitor treatment is of great interest and several gene expression studies have been conducted. More recently, several microarray studies have been performed in different plant species to gain greater knowledge of the diversity of genes responsive to elicitors (e.g., Medeiros et al., 2009; Kano et al., 2011; Povero et al., 2011; Amelot et al., 2012), albeit knowledge on plant gene expression in response to elicitors has mainly been focused on A. thaliana. As mentioned previously, the presence of the cognate receptors can determine responsiveness to elicitors (Lacombe et al., 2010) and it is thus essential to investigate elicitor effects in diverse crop plants (Nguyen et al., 2010).
As part of this review, we aim to provide an overview of the current knowledge of differentially expressed plant genes following elicitor treatments and identify typically affected plant processes. This will facilitate identification of responses to elicitor application such as plant growth or nutrient metabolisms that are not directly linked to defense but impact on agriculture. For this we have combined over 50 publications to create a list of plant genes that are differentially expressed following the recognition of elicitors (Table S1). Reciprocal BLAST (Basic Local Alignment Search Tool; Altschul et al., 1990) has been used to identify the A. thaliana homologs when the original experiment was performed in a different plant species (>70% identity of nucleotide sequences, E-value < 0.0001). In addition to the publications, PathoPlant®, a database featuring compiled expression data and components of signal transduction pathways related to plant pathogenesis, has been used (Bülow et al., 2004, 2007). This database enables querying differential plant gene expression following diverse pathogen stimuli which, for this study, include Botrytis cinerea, chitin, Erysiphe orontii, Phytophthora infestans, Pseudomonas syringae pv. Maculicola, and Pseudomonas syringae pv. Phaseolicola.
A total of 1592 plant genes that were activated by the recognition of elicitors have been identified (Table S1). Ontological analysis was performed using agriGo (Du et al., 2010). In this analysis, genes of interest are grouped by gene ontology (GO) terms describing biological processes, molecular functions and cellular components (Ashburner et al., 2000). The list of genes of interest is compared to a defined background gene list which, in this study, included the whole genome of A. thaliana as provided by The Arabidopsis Information Resource (TAIR) (Lamesch et al., 2011) to identify GO terms that are significantly over-represented. A total of 762 shared GO terms were identified, comprising 474 biological processes, 206 molecular functions and 82 cellular components (Table S2).
The three most over-represented biological processes were “response to stimulus” (GO:0050896), “multi-organism process” (GO:0051704), and “immune system process” (GO:0002376). The GO term “response to stimulus” contained 36.5% of the elicitor responsive genes in comparison to 10.7% representation in the whole A. thaliana genome. The GO term “multi-organism process” contained 11.1% of the genes in the target gene list, compared to 2.1% of the whole genome, and 6.6% instead of 1% were classed in the GO term “immune system process” (Figure 2).
Figure 2.

Highly significant shared biological processes within Arabidopsis thaliana genes that are induced and overrepresented following the recognition of elicitor compounds (black) in comparison to the whole genome of A. thaliana (gray).
Three of the over-represented molecular functions were “catalytic activity” (GO:0003824), “binding” (GO:0005488), and “molecular transducer activity” (GO:0060089). The GO term “catalytic activity” featured in 48.5% of the elicitor responsive genes in comparison to 25.5% of the whole A. thaliana genome. The GO term “binding” contained 37.8% of the genes in the target gene list in comparison to 29.8% of the whole genome and 3.1% instead of 1.1% were classed in the GO term “molecular transducer activity” (Figure 3).
Figure 3.
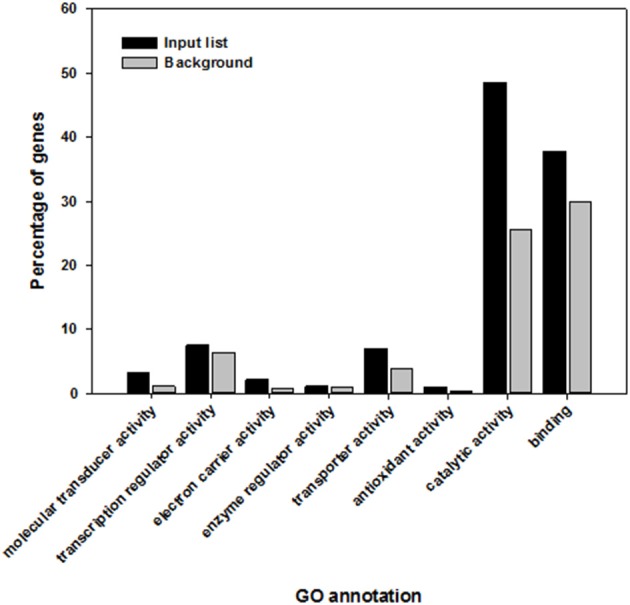
Highly significant shared molecular functions within Arabidopsis thaliana genes that are induced and overrepresented following the recognition of elicitor compounds (black) in comparison to the whole genome of A. thaliana (gray).
The relationships of all over-represented genes in the classification “biological processes” are shown in Figure S1. These include metabolic processes such as amine-, phosphate-, and phytoalexin metabolism; immune system processes and cell death, including regulation of defense response; plant-type hypersensitive response and apoptosis; response to stimuli including JA and SA; systemic acquired resistance and defense responses to fungi and bacteria (Figure S1).
The relationship of all genes over-represented in the GO term “molecular functions” are shown in Figure S2. These functions contain catalytic activity including oxidoreductase, lyase, and kinase activities; and binding activities including ATP and sugar binding (Figure S2). The relationships of all over-represented genes in the classification “cellular components” are shown in Figure S3. All cell parts are involved but the involvements of cell wall and plasma membranes are highly significant (Figure S3).
Targets of pathogen effectors
As noted above, pathogens produce effector molecules to interfere with plant defense responses. An analysis of plant-pathogen protein-protein interactions using A. thaliana and two pathogens, P. syringae and the obligate biotrophic oomycete Hyaloperonospora arabidopsidis revealed 137 A. thaliana proteins that were potentially targeted by pathogen effectors (Mukhtar et al., 2011). A recent review on bacterial effectors listed an additional 22 plant proteins targeted by several bacterial effectors (Deslandes and Rivas, 2012). In the analysis here, genes encoding these 159 proteins were used to search for overlap with the plant genes differentially induced upon elicitor recognition. A total of 23 genes were identified that are both induced by elicitors and targeted by pathogen effectors (Table S3). These comprise receptors such as FLS2 and EFR, genes involved in MAPK cascades like RIPK, MPK3, MPK4, and MPK6, R protein-guarded host proteins such as RIN4 and genes involved in L-phenylalanine biosynthetic process like ADT4 and ADT5 (Table S3). These overlapping genes are grouped into several biological processes, molecular functions and cell parts and their involvements are highlighted with stars in Figures S1–S3. This analysis highlights the complexity of priming and plant immune responses, and the sophisticated interactions with pathogen effectors. It shows that the plant response to elicitor compounds does not only involve genes that are annotated as defense-related but that other metabolic pathways are also involved. We know that elicitor treatments result in positive and negative trade-offs (Walters and Heil, 2007) and therefore expression profiling of some of these genes should result in a better understanding of these responses and how they might be exploited.
“Non-defense” effects of elicitors
As mentioned above, for successful use of elicitors in agriculture it is important to understand their effects not only on plant defense but also on other aspects of plant development and environmental responses. The activation of defense pathways as part of PTI can be very costly to plants but should be less than the potential loss caused by disease if no defense was mounted. However, in the context of crop protection, such costs are unlikely to be acceptable in the absence of known pathogen challenges of a high order. Crop protectants are preferred that enhance the efficacy of PTI assisting a quicker and more effective response when an actual pathogen challenge occurs and therefore is more efficient in its use of resources. This can be achieved through priming if priming is either not costly to the plant or its costs are mitigated by other beneficial means.
Some of the known priming genes are generally regulatory but not necessarily restricted to defense pathways. They regulate signal transduction events, particularly those identified in stress responses or in the GO terms “response to stimulus,” “multi-organism processes,” and “immune system processes,” i.e., the genes disproportionately up-regulated by elicitors highlighted above (see Figures S1, S2). Thus, many non-defense mechanism processes will be affected in their expression by elicitors. These might result in additional costs above those incurred by defense gene expression, but they may also have benefits. An example of a non-defense effect of an elicitor is reduced water use of pepper plants upon treatment with chitosan (Bittelli et al., 2001). Chitosan was also found to affect the net photosynthetic rate of soybean and maize after application (Khan et al., 2002). More general effects on yield, not directly attributable to disease control, were also found from applications of B. subtilis culture filtrates (Dehne et al., 1984; Steiner et al., 1988). Similar effects were recorded for some treatments of yeast cell wall-derived extracts (Reglinski et al., 1994a).
Indeed, on a molecular level, there is evidence of cross talk between the MAPKs involved in PTI and abiotic stress responses. For example, the transcription of MEKK1 is induced by diverse stresses including cold, salt, drought and wounding (Mizoguchi et al., 1998). Conversely, the activation of EDS1/PAD4-dependent signaling during ETI responses can rapidly antagonize ABA signal transduction at the level of Ca2+ signaling (Kim et al., 2011). The overexpression of the gene ACTIVATED DISEASE RESISTANCE1 (ADS1) in A. thaliana, a member of the nucleotide-binding (NB) and leucine-rich repeat (LRR) containing NB-LRR genes, confers both disease resistance (Grant et al., 2003) and drought tolerance, requiring SA, EDS1 and ABA-INSENSITIVE1 (ABI1) (Chini et al., 2004).
Much depends on the basis of determination of costs as we tend to calculate these from an end-user yield perspective. We should also recognize that each of these pathways, whether defense-related or not, is in a complex expression and metabolic network of cross-talk and feedback mechanisms and thus affected by many environmental factors. Those that can be manipulated beneficially and perhaps synergistically fall in the category of nutrition (Walters and Bingham, 2007). Primed plants showed considerably higher fitness than non-primed plants when they were challenged by pathogens without major trade-off effects on growth and seed set (Conrath et al., 2006; van Hulten et al., 2006). Correlation of priming benefits with gene expression profiles may lead to very practical means for developing elicitor-based crop protectants that either off-set any direct costs, increase some aspect of resource use efficiency or specifically enhance other processes beneficial to yield or quality.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful for financial support for this work in part from the Rural & Environment Science & Analytical Services Division of the Scottish Government. Lea Wiesel is supported by the Technology Strategy Board and its Knowledge Transfer Partnership grant KTP008814 with Omex Agriculture Limited.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00655/abstract
References
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Amelot N., de Borne F. D., Clemente H. S., Mazars C., Grima-Pettenati J., Briere C. (2012). Transcriptome analysis of tobacco BY-2 cells elicited by cryptogein reveals new potential actors of calcium-dependent and calcium-independent plant defense pathways. Cell Calcium 51, 117–130. 10.1016/j.ceca.2011.11.010 [DOI] [PubMed] [Google Scholar]
- Armstrong M. R., Whisson S. C., Pritchard L., Bos J. I., Venter E., Avrova A. O., et al. (2005). An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc. Natl. Acad. Sci. U.S.A. 102, 7766–7771. 10.1073/pnas.0500113102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., et al. (2000). Gene Ontology: tool for the unification of biology. Nat. Genet. 25, 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz A., Gauthier A., Bézier A., Poinssot B., Joubert J. M., Pugin A., et al. (2007). Elicitor and resistance-inducing activities of β-1,4 cellodextrins in grapevine, comparison with β-1,3 glucans and α-1,4 oligogalacturonides. J. Exp. Bot. 58, 1463–1472. 10.1093/jxb/erm008 [DOI] [PubMed] [Google Scholar]
- Beckers G. J., Jaskiewicz M., Liu Y., Underwood W. R., He S. Y., Zhang S., et al. (2009). Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 21, 944–953. 10.1105/tpc.108.062158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal-Lobo M., Stone S., Yang X., Antico J., Callis J., Ramonell K. M., et al. (2010). ATL9, a RING zinc finger protein with E3 ubiquitin ligase activity implicated in chitin- and NADPH oxidase-mediated defense responses. PLoS ONE 5:e14426. 10.1371/journal.pone.0014426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke G., Pecher P., Eschen-Lippold L., Tsuda K., Katagiri F., Glazebrook J., et al. (2012). Activation of the Arabidopsis thaliana mitogen-activated protein kinase MPK11 by the flagellin-derived elicitor peptide, flg22. Mol. Plant Microbe Interact. 25, 471–480. 10.1094/MPMI-11-11-0281 [DOI] [PubMed] [Google Scholar]
- Birch P. R., Armstrong M., Bos J., Boevink P., Gilroy E. M., Taylor R. M., et al. (2009). Towards understanding the virulence functions of RXLR effectors of the oomycete plant pathogen Phytophthora infestans. J. Exp. Bot. 60, 1133–1140. 10.1093/jxb/ern353 [DOI] [PubMed] [Google Scholar]
- Bishop G. J., Koncz C. (2002). Brassinosteroids and plant steroid hormone signaling. Plant Cell 14, S97–S110. 10.1105/tpc.001461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittel P., Robatzek S. (2007). Microbe-associated molecular patterns (MAMPs) probe plant immunity. Curr. Opin. Plant Biol. 10, 335–341. 10.1016/j.pbi.2007.04.021 [DOI] [PubMed] [Google Scholar]
- Bittelli M., Flury M., Campbell G. S., Nichols E. J. (2001). Reduction of transpiration through foliar application of chitosan. Agr. Forest Meteorol. 107, 167–175 10.1016/S0168-1923(00)00242-2 [DOI] [Google Scholar]
- Block A., Alfano J. R. (2011). Plant targets for Pseudomonas syringae type III effectors: virulence targets or guarded decoys? Curr. Opin. Plant Biol. 14, 39–46. 10.1016/j.mib.2010.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., Kay S., et al. (2009). Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512. 10.1126/science.1178811 [DOI] [PubMed] [Google Scholar]
- Bonnet P., Bourdon E., Ponchet M., Blein J. P., Ricci P. (1996). Acquired resistance triggered by elicitins in tobacco and other plants. Eur. J. Plant Pathol. 102, 181–192 10.1007/BF01877105 [DOI] [Google Scholar]
- Bos J. I. B., Armstrong M. R., Gilroy E. M., Boevink P. C., Hein I., Taylor R. M., et al. (2010). Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc. Natl. Acad. Sci. U.S.A. 107, 9909–9914. 10.1073/pnas.0914408107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque S., Dutartre A., Hammoudi V., Blanc S., Dahan J., Jeandroz S., et al. (2011). Type-2 histone deacetylases as new regulators of elicitor-induced cell death in plants. New Phytol. 192, 127–139. 10.1111/j.1469-8137.2011.03788.x [DOI] [PubMed] [Google Scholar]
- Bozkurt T. O., Schornack S., Win J., Shindo T., Ilyas M., Oliva R., et al. (2011). Phytophthora infestans effector Avrblb2 prevents secretion of a plant immune protease at the haustorial interface. Proc. Natl. Acad. Sci. U.S.A. 108, 20832–20837. 10.1073/pnas.1112708109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner F., Rosahl S., Lee J., Rudd J. J., Geiler C., Kauppinen S., et al. (2002). Pep-13, a plant defense-inducing pathogen-associated pattern from Phytophthora transglutaminases. EMBO J. 21, 6681–6688. 10.1093/emboj/cdf667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutus A., Sicilia F., Macone A., Cervone F., De Lorenzo G. (2010). A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. U.S.A. 107, 9452–9457. 10.1073/pnas.1000675107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bülow L., Schindler M., Choi C., Hehl R. (2004). PathoPlant®: a database on plant-pathogen interactions. In Silico Biol. 4, 529–536. [PubMed] [Google Scholar]
- Bülow L., Schindler M., Hehl R. (2007). PathoPlant®: a platform for microarray expression data to analyze co-regulated genes involved in plant defense responses. Nucleic Acids Res. 35, D841–D845. 10.1093/nar/gkl835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Zeng H., Qiu D., Guo L., Yang X., Shi H., et al. (2012). Purification and characterization of a novel hypersensitive response-inducing elicitor from Magnaporthe oryzae that triggers defense response in rice. PLoS ONE 7:e37654. 10.1371/journal.pone.0037654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Zheng Z., Huang J., Lai Z., Fan B. (2009). Biosynthesis of salicylic acid in plants. Plant Signal. Behav. 4, 493–496. 10.4161/psb.4.6.8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J. D. G., et al. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500. 10.1038/nature05999 [DOI] [PubMed] [Google Scholar]
- Chini A., Grant J. J., Seki M., Shinozaki K., Loake G. J. (2004). Drought tolerance established by enhanced expression of the CC-NBS-LRR gene, ADR1, requires salicylic acid, EDS1 and ABI1. Plant J. 385, 810–822. 10.1111/j.1365-313X.2004.02086.x [DOI] [PubMed] [Google Scholar]
- Cluzet S., Torregrosa C., Jacquet C., Lafitte C., Fournier J., Mercier L., et al. (2004). Gene expression profiling and protection of Medicago truncatula against a fungal infection in response to an elicitor from green algae Ulva spp. Plant Cell Environ. 27, 917–928 10.1111/j.1365-3040.2004.01197.x [DOI] [Google Scholar]
- Coll N. S., Valls M. (2013). Current knowledge on the Ralstonia solanacearum type III secretion system. Microb. Biotechnol. 6, 614–620. 10.1111/1751-7915.12056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrath U. (2011). Molecular aspects of defence priming. Trends Plant Sci. 16, 524–531. 10.1016/j.tplants.2011.06.004 [DOI] [PubMed] [Google Scholar]
- Conrath U., Beckers G. J., Flors V., Garcia-Agustin P., Jakab G., Mauch F., et al. (2006). Priming: getting ready for battle. Mol. Plant Microbe Interact. 19, 1062–1071. 10.1094/MPMI-19-1062 [DOI] [PubMed] [Google Scholar]
- Copping L. G., Duke S. O. (2007). Natural products that have been used commercially as crop protection agents. Pest Manag. Sci. 63, 524–554. 10.1002/ps.1378 [DOI] [PubMed] [Google Scholar]
- Craigie J. S. (2011). Seaweed extract stimuli in plant science and agriculture. J. Appl. Phycol. 23, 371–393 10.1007/s10811-010-9560-4 [DOI] [Google Scholar]
- Daayf F., Ongena M., Boulanger R., El Hadrami I., Bélanger R. R. (2000). Induction of phenolic compounds in two cultivars of cucumber by treatment of healthy and powdery mildew-infected plants with extracts of Reynoutria sachalinensis. J. Chem. Ecol. 26, 1579–1593 10.1023/A:1005578510954 [DOI] [Google Scholar]
- Daayf F., Schmitt A., Belanger R. R. (1997). Evidence of phytoalexins in cucumber leaves infected with powdery mildew following treatment with leaf extracts of Reynoutria sachalinensis. Plant Physiol. 113, 719–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danna C. H., Millet Y. A., Koller T., Han S. W., Bent A. F., Ronald P. C., et al. (2011). The Arabidopsis flagellin receptor FLS2 mediates the perception of Xanthomonas Ax21 secreted peptides. Proc. Natl. Acad. Sci. U.S.A. 108, 9286–9291. 10.1073/pnas.1106366108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day B., Dahlbeck D., Staskawicz B. J. (2006). NDR1 interaction with RIN4 mediates the differential activation of multiple disease resistance pathways in Arabidopsis. Plant Cell 18, 2782–2791. 10.1105/tpc.106.044693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehne H.-W., Stenzel K., Schönbeck F. (1984). Zur Wirksamkeit induzierter Resistenz unter praktischen Anbaubedingungen III. Reproduktion echter Mehltaupilze auf induziert resistenten Pflanzen. Z. Pflanzenk. Pflanzen. 91, 258–265. [Google Scholar]
- de Jonge R., Peter van Esse H., Kombrink A., Shinya T., Desaki Y., Bours R., et al. (2010). Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 329, 953–955. 10.1126/science.1190859 [DOI] [PubMed] [Google Scholar]
- de Jonge R., Thomma B. P. (2009). Fungal LysM effectors: extinguishers of host immunity? Trends Microbiol. 17, 151–157. 10.1016/j.tim.2009.01.002 [DOI] [PubMed] [Google Scholar]
- de Oliveira A. L., Gallo M., Pazzagli L., Benedetti C. E., Cappugi G., Scala A., et al. (2011). The structure of the elicitor cerato-platanin (CP), the first member of the CP fungal protein family, reveals a double ψβ-barrel fold and carbohydrate binding. J. Biol. Chem. 286, 17560–17568. 10.1074/jbc.M111.223644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desaki Y., Miya A., Venkatesh B., Tsuyumu S., Yamane H., Kaku H., et al. (2006). Bacterial lipopolysaccharides induce defense responses associated with programmed cell death in rice cells. Plant Cell Physiol. 47, 1530–1540. 10.1093/pcp/pcl019 [DOI] [PubMed] [Google Scholar]
- Deslandes L., Rivas S. (2012). Catch me if you can: bacterial effectors and plant targets. Trends Plant Sci. 17, 644–655. 10.1016/j.tplants.2012.06.011 [DOI] [PubMed] [Google Scholar]
- De Vleesschauwer D., Höfte M. (2009). Rhizobacteria-induced systemic resistance. Adv. Bot. Res. 51, 223–281 10.1016/S0065-2296(09)51006-3 [DOI] [Google Scholar]
- Djamei A., Kahmann R. (2012). Ustilago maydis: dissecting the molecular interface between pathogen and plant. PLoS Pathog. 8:e1002955. 10.1371/journal.ppat.1002955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djonoviç S., Vargas W. A., Kolomiets M. V., Horndeski M., Wiest A., Kenerley C. M. (2007). A proteinaceous elicitor Sm1 from the beneficial fungus Trichoderma virens is required for induced systemic resistance in maize. Plant Physiol. 145, 875–889. 10.1104/pp.107.103689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlemann G., Hemetsberger C. (2013). Apoplastic immunity and its suppression by filamentous plant pathogens. New Phytol. 198, 1001–1016. 10.1111/nph.12277 [DOI] [PubMed] [Google Scholar]
- Dow M., Newman M. A., von Roepenack E. (2000). The induction and modulation of plant defense responses by bacterial lipopolysaccharides. Annu. Rev. Phytopathol. 38, 241–261. 10.1146/annurev.phyto.38.1.241 [DOI] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z. (2010). agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38, W64–W70. 10.1093/nar/gkq310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Ghaouth A., Arul J., Grenier J., Benhamou N., Asselin A., Bélanger R. (1994). Effect of chitosan on cucumber plants: suppression of Pythium aphanidermatum and induction of defense reactions. Phytopathology 84, 313–320 10.1094/Phyto-84-313 [DOI] [Google Scholar]
- El Hadrami A., Adam L. R., El Hadrami I., Daayf F. (2012). Chitosan in plant protection. Mar. Drugs 8, 968–987. 10.3390/md8040968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbs G., Newman M. A. (2012). The role of lipopolysaccharide and peptidoglycan, two glycosylated bacterial microbe-associated molecular patterns (MAMPs), in plant innate immunity. Mol. Plant Pathol. 13, 95–104. 10.1111/j.1364-3703.2011.00730.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshraghi L., Anderson J., Aryamanesh N., Shearer B., McComb J., Hardy G. E. S., et al. (2011). Phosphite primed defence responses and enhanced expression of defence genes in Arabidopsis thaliana infected with Phytophthora cinnamomi. Plant Pathol. 60, 1086–1095 10.1111/j.1365-3059.2011.02471.x [DOI] [Google Scholar]
- Eulgem T. (2005). Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci. 10, 71–78. 10.1016/j.tplants.2004.12.006 [DOI] [PubMed] [Google Scholar]
- Falcón-Rodríguez A. B., Wegria G., Cabrera J.-C. (2012). Exploiting plant innate immunity to protect crops against biotic stress: chitosaccharides as natural and suitable candidates for this purpose, in New Perspectives in Plant Protection, ed Bandani A. R. (Rijeka: In Tech Croatia; ), 139–166. [Google Scholar]
- Felix G., Boller T. (2003). Molecular sensing of bacteria in plants. The highly conserved RNA-binding motif RNP-1 of bacterial cold shock proteins is recognized as an elicitor signal in tobacco. J. Biol. Chem. 278, 6201–6208. 10.1074/jbc.M209880200 [DOI] [PubMed] [Google Scholar]
- Ferrari S., Savatin D. V., Sicilia F., Gramegna G., Cervone F., De Lorenzo G. (2013). Oligogalacturonides: plant damage-associated molecular patterns and regulators of growth and development. Front. Plant Sci. 4:49. 10.3389/fpls.2013.00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flors V., Ton J., Jakab G., Mauch-Mani B. (2005). Abscisic acid and callose: team players in defence against pathogens? J. Phytopathol. 153, 377–383 10.1111/j.1439-0434.2005.00987.x [DOI] [Google Scholar]
- Fu Y., Yin H., Wang W., Wang M., Zhang H., Zhao X., et al. (2011). β-1,3-glucan with different degree of polymerization induced different defense responses in tobacco. Carbohyd. Polym. 86, 774–782 10.1016/j.carbpol.2011.05.022 [DOI] [Google Scholar]
- Galletti R., Ferrari S., De Lorenzo G. (2011). Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide- or flagellin-induced resistance against Botrytis cinerea. Plant Physiol. 157, 804–814. 10.1104/pp.111.174003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcion C., Lamotte O., Métraux J.-P. (2007). Mechanisms of defense to pathogens: biochemistry and physiology, in Induced Resistance for Plant Defence, eds Walters D. R., Newton A. C., Lyon G. D. (Oxford: Blackwell Publishing; ), 109–132 10.1002/9780470995983.ch6 [DOI] [Google Scholar]
- Gaulin E., Dramé N., Lafitte C., Torto-Alalibo T., Martinez Y., Ameline-Torregrosa C., et al. (2006). Cellulose binding domains of a Phytophthora cell wall protein are novel pathogen-associated molecular patterns. Plant Cell 18, 1766–1777. 10.1105/tpc.105.038687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber I., Zeidler D., Durner J., Dubery I. (2004). Early perception responses of Nicotiana tabacum cells in response to lipopolysaccharides from Burkholderia cepacia. Planta 218, 647–657. 10.1007/s00425-003-1142-0 [DOI] [PubMed] [Google Scholar]
- Gilroy E. M., Taylor R. M., Hein I., Boevink P., Sadanandom A., Birch P. R. J. (2011). CMPG1-dependent cell death follows perception of diverse pathogen elicitors at the host plasma membrane and is suppressed by Phytophthora infestans RXLR effector AVR3a. New Phytol. 190, 653–666. 10.1111/j.1469-8137.2011.03643.x [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L., Boller T. (2002). Flagellin perception: a paradigm for innate immunity. Trends Plant Sci. 7, 251–256. 10.1016/S1360-1385(02)02261-6 [DOI] [PubMed] [Google Scholar]
- Goupil P., Benouaret R., Charrier O., Ter Halle A., Richard C., Eyheraguibel B., et al. (2012). Grape marc extract acts as elicitor of plant defence responses. Ecotoxicology 21, 1541–1549. 10.1007/s10646-012-0908-1 [DOI] [PubMed] [Google Scholar]
- Grant J. J., Chini A., Basu D., Loake G. J. (2003). Targeted activation tagging of the Arabidopsis NBS-LRR gene, ADR1, conveys resistance to virulent pathogens. Mol. Plant Microbe Interact. 16, 669–680. 10.1094/MPMI.2003.16.8.669 [DOI] [PubMed] [Google Scholar]
- Guo M., Chen K., Zhang P. (2012). Transcriptome profile analysis of resistance induced by burdock fructooligosaccharide in tobacco. J. Plant Physiol. 169, 1511–1519. 10.1016/j.jplph.2012.06.019 [DOI] [PubMed] [Google Scholar]
- Hahn M. G., Albersheim P. (1978). Host-pathogen interactions: XIV. isolation and partial characterization of an elicitor from yeast extract. Plant Physiol. 62, 107–111. 10.1104/pp.62.1.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese A., Hann D. R., Gimenez-Ibanez S., Jones A. M., He K., Li J., et al. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. U.S.A. 104, 12217–12222. 10.1073/pnas.0705306104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein I., Gilroy E. M., Armstrong M. R., Birch P. R. J. (2009). The zig-zag-zig in oomycete?plant interactions. Mol. Plant Pathol. 10, 547–562. 10.1111/j.1364-3703.2009.00547.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriquez M. A., Wolski E. A., Molina O. I., Adam L. R., Andreu A. B., Daayf F. (2012). Effects of glucans and eicosapentaenoic acid on differential regulation of phenylpropanoid and mevalonic pathways during potato response to Phytophthora infestans. Plant Physiol. Biochem. 60, 119–128. 10.1016/j.plaphy.2012.07.027 [DOI] [PubMed] [Google Scholar]
- Henry G., Thonart P., Ongena M. (2012). PAMPs, MAMPs, DAMPs and others: an update on the diversity of plant immunity elicitors. Biotechnol. Agron. Soc. Environ. 16, 257–268. [Google Scholar]
- Huang C.-J., Tsay J.-F., Chang S.-Y., Yang H.-P., Wu W.-S., Chen C.-Y. (2012). Dimethyl disulfide is an induced systemic resistance elicitor produced by Bacillus cereus C1L. Pest Manag. Sci. 68, 1306–1310. 10.1002/ps.3301 [DOI] [PubMed] [Google Scholar]
- Huffaker A., Dafoe N. J., Schmelz E. A. (2011). ZmPep1, an ortholog of Arabidopsis elicitor peptide 1, regulates maize innate immunity and enhances disease resistance. Plant Physiol. 155, 1325–1338. 10.1104/pp.110.166710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi D., Takeda T., Narusaka Y., Totsuka K. (2010). Glutamate fermentation by-product activates plant defence responses and confers resistance against pathogen infection. J. Phytopathol. 158, 668–675 10.1111/j.1439-0434.2010.01678.x [DOI] [Google Scholar]
- Jaskiewicz M., Conrath U., Peterhaensel C. (2011). Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 12, 50–55. 10.1038/embor.2010.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaulneau V., Lafitte C., Corio-Costet M. F., Stadnik M. J., Salamagne S., Briand X., et al. (2011). An Ulva armoricana extract protects plants against three powdery mildew pathogens. Eur. J. Plant Pathol. 131, 393–401 10.1007/s10658-011-9816-0 [DOI] [Google Scholar]
- Jelenska J., Yao N., Vinatzer B. A., Wright C. M., Brodsky J. L., Greenberg J. T. (2007). A J domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. Curr. Biol. 17, 499–508. 10.1016/j.cub.2007.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeworutzki E., Roelfsema M., Anschütz U., Krol E., Elzenga J., Felix G., et al. (2010). Early signaling through the Arabidopsis pattern recognition receptors FLS2 and EFR involves Ca2+-associated opening of plasma membrane anion channels. Plant J. 62, 367–378. 10.1111/j.1365-313X.2010.04155.x [DOI] [PubMed] [Google Scholar]
- Jin W., Wu F., Xiao L., Liang G., Zhen Y., Guo Z., et al. (2012). Microarray-based analysis of tomato miRNA regulated by Botrytis cinerea. J. Plant Growth Regul. 31, 38–46 10.1007/s00344-011-9217-9 [DOI] [Google Scholar]
- Jones J. D., Dangl J. L. (2006). The plant immune system. Nature 444, 323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Kamoun S., van West P., Vleeshouwers V. G. A. A., de Groot K. E., Govers F. (1998). Resistance of Nicotiana benthamiana to Phytophthora infestans is mediated by the recognition of the elicitor protein INF1. Plant Cell 10, 1413–1425. 10.1105/tpc.10.9.1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano A., Hosotani K., Gomi K., Yamasaki-Kokudo Y., Shirakawa C., Fukumoto T., et al. (2011). D-Psicose induces upregulation of defense-related genes and resistance in rice against bacterial blight. J. Plant Physiol. 168, 1852–1857. 10.1016/j.jplph.2011.04.003 [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y., Nishiuchi T., Kodama H., Nakano T., Nishimura K., Shimamura K., et al. (2012). Fungal elicitor-induced retardation and its restoration of root growth in tobacco seedlings. Plant Growth Regul. 66, 59–68 10.1007/s10725-011-9629-3 [DOI] [Google Scholar]
- Kawamura Y., Hase S., Takenaka S., Kanayama Y., Yoshioka H., Kamoun S., et al. (2009). INF1 elicitin activates jasmonic acid- and ethylene-mediated signalling pathways and induces resistance to bacterial wilt disease in tomato. J. Phytopathol. 157, 287–297 10.1111/j.1439-0434.2008.01489.x [DOI] [Google Scholar]
- Khan W. M., Prithiviraj B., Smith D. L. (2002). Effect of foliar application of chitin and chitosan oligosaccharides on photosynthesis of maize and soybean. Photosynthetica 40, 621–624 10.1023/A:1024320606812 [DOI] [Google Scholar]
- Khatib M., Lafitte C., Esquerré-Tugayé M.-T., Bottin A., Rickauer M. (2004). The CBEL elicitor of Phytophthora parasitica var. nicotianae activates defence in Arabidopsis thaliana via three different signalling pathways. New Phytol. 162, 501–510 10.1111/j.1469-8137.2004.01043.x [DOI] [Google Scholar]
- Khokon M., Hossain M. A., Munemasa S., Uraji M., Nakamura Y., Mori I. C., et al. (2010). Yeast elicitor-induced stomatal closure and peroxidase-mediated ROS production in Arabidopsis. Plant Cell Physiol. 51, 1915–1921. 10.1093/pcp/pcq145 [DOI] [PubMed] [Google Scholar]
- Kim T. H., Hauser F., Ha T., Xue S., Böhmer M., Nishimura N., et al. (2011). Chemical genetics reveals negative regulation of abscisic acid signaling by a plant immune response pathway. Curr. Biol. 21, 990–997. 10.1016/j.cub.2011.04.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S. R. F., McLellan H., Boevink P. C., Armstrong M. R., Bukharova T., Sukarta O., et al. (2014). Phytophthora infestans RXLR effector PexRD2 interacts with host MAPKKK? to suppress plant immune signaling. Plant Cell 26, 1345–1359. 10.1105/tpc.113.120055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto K., Kouzai Y., Kaku H., Shibuya N., Minami E., Nishizawa Y. (2010). Perception of the chitin oligosaccharides contributes to disease resistance to blast fungus Magnaporthe oryzae in rice. Plant J. 64, 343–354. 10.1111/j.1365-313X.2010.04328.x [DOI] [PubMed] [Google Scholar]
- Knoth C., Salus M. S., Girke T., Eulgem T. (2009). The synthetic elicitor 3,5-dichloroanthranilic acid induces NPR1-dependent and NPR1-independent mechanisms of disease resistance in Arabidopsis. Plant Physiol. 150, 333–347. 10.1104/pp.108.133678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A., Schwindling S., Conrath U. (2002). Benzothiadiazole-induced priming for potentiated responses to pathogen infection, wounding, and infiltration of water into leaves requires the NPR1/NIM1 gene in Arabidopsis. Plant Physiol. 128, 1046–1056. 10.1104/pp.010744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kombrink A., Sanchez-Vallet A., Thomma B. P. (2011). The role of chitin detection in plant-pathogen interactions. Microb. Infect. 13, 1168–1176. 10.1016/j.micinf.2011.07.010 [DOI] [PubMed] [Google Scholar]
- Konstantinidou-Doltsinis S., Markellou E., Kasselaki A. M., Fanouraki M. N., Koumaki C. M., Schmitt A., et al. (2006). Efficacy of Milsana®, a formulated plant extract from Reynoutria sachalinensis, against powdery mildew of tomato (Leveillula taurica). Biocontrol 51, 375–392 10.1007/s10526-005-5247-1 [DOI] [Google Scholar]
- Koschmann J., Machens F., Becker M., Niemeyer J., Schulze J., Bülow L., et al. (2012). Integration of bioinformatics and synthetic promoters leads to the discovery of novel elicitor-responsive cis-regulatory sequences in Arabidopsis. Plant Physiol. 160, 178–191. 10.1104/pp.112.198259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulye M., Liu H., Zhang Y., Zeng H., Yang X., Qiu D. (2012). Hrip1, a novel protein elicitor from necrotrophic fungus, Alternaria tenuissima, elicits cell death, expression of defence-related genes and systemic acquired resistance in tobacco. Plant Cell Environ. 35, 2104–2120. 10.1111/j.1365-3040.2012.02539.x [DOI] [PubMed] [Google Scholar]
- Kunze G., Zipfel C., Robatzek S., Niehaus K., Boller T., Felix G. (2004). The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16, 3496–3507. 10.1105/tpc.104.026765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe S., Rougon-Cardoso A., Sherwood E., Peeters N., Dahlbeck D., van Esse H. P., et al. (2010). Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat. Biotechnol. 28, 365–369. 10.1038/nbt.1613 [DOI] [PubMed] [Google Scholar]
- Lamesch P., Berardini T. Z., Li D., Swarbreck D., Wilks C., Sasidharan R., et al. (2011). The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210. 10.1093/nar/gkr1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laquitaine L., Gomès E., François J., Marchive C., Pascal S., Hamdi S., et al. (2006). Molecular basis of ergosterol-induced protection of grape against Botrytis cinerea: induction of type I LTP promoter activity, WRKY, and stilbene synthase gene expression. Mol. Plant Microbe Interact. 19, 1103–1112. 10.1094/MPMI-19-1103 [DOI] [PubMed] [Google Scholar]
- Lee J., Klessig D. F., Nürnberger T. (2001). A harpin binding site in tobacco plasma membranes mediates activation of the pathogenesis-related gene HIN1 independent of extracellular calcium but dependent on mitogen-activated protein kinase activity. Plant Cell 13, 1079–1093. 10.1105/tpc.13.5.1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shao M., Zhong W., Yang J., Okada K., Yamane H., et al. (2012). Ectopic expression of Hrf1 enhances bacterial resistance via regulation of diterpene phytoalexins, silicon and reactive oxygen species burst in rice. PLoS ONE 7:e43914. 10.1371/journal.pone.0043914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M., Wan J., Czechowski T., Udvardi M., Stacey G. (2007). Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol. Plant Microbe Interact. 20, 900–911. 10.1094/MPMI-20-8-0900 [DOI] [PubMed] [Google Scholar]
- Liebrand T. W. H., van den Berg G. C. M., Zhang Z., Smit P., Cordewener J. H. G., America A. H. P., et al. (2013). Receptor-like kinase SOBIR1/EVR interacts with receptor-like proteins in plant immunity against fungal infection. Proc. Natl. Acad. Sci. U.S.A. 110, 10010–10015. 10.1073/pnas.1220015110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Liu Z., Song C., Hu Y., Han Z., She J., et al. (2012). Chitin-induced dimerization activates a plant immune receptor. Science 336, 1160–1164. 10.1126/science.1218867 [DOI] [PubMed] [Google Scholar]
- Liu W., Liu J., Ning Y., Ding B., Wang X., Wang Z., et al. (2013). Recent progress in understanding PAMP- and effector-triggered immunity against the rice blast fungus Magnaporthe oryzae. Mol. Plant 6, 605–620. 10.1093/mp/sst015 [DOI] [PubMed] [Google Scholar]
- Livaja M., Palmieri M., von Rad U., Durner J. (2008a). The effect of the bacterial effector protein harpin on transcriptional profile and mitochondrial proteins of Arabidopsis thaliana. J. Proteomics 71, 148–159. 10.1016/j.jprot.2008.04.002 [DOI] [PubMed] [Google Scholar]
- Livaja M., Zeidler D., von Rad U., Durner J. (2008b). Transcriptional responses of Arabidopsis thaliana to the bacteria-derived PAMPs harpin and lipopolysaccharide. Immunobiology 213, 161–171. 10.1016/j.imbio.2007.10.004 [DOI] [PubMed] [Google Scholar]
- Lochman J., Mikes V. (2006). Ergosterol treatment leads to the expression of a specific set of defence-related genes in tobacco. Plant Mol. Biol. 62, 43–51. 10.1007/s11103-006-9002-5 [DOI] [PubMed] [Google Scholar]
- Maffei M. E., Arimura G. I., Mithoefer A. (2012). Natural elicitors, effectors and modulators of plant responses. Nat. Prod. Rep. 29, 1288–1303. 10.1039/c2np20053h [DOI] [PubMed] [Google Scholar]
- Mateos F. V., Rickauer M., Esquerré-Tugayé M.-T. (1997). Cloning and characterization of a cDNA encoding an elicitor of Phytophthora parasitica var. nicotianae that shows cellulose-binding and lectin-like activities. Mol. Plant Microbe Interact. 10, 1045–1053. 10.1094/MPMI.1997.10.9.1045 [DOI] [PubMed] [Google Scholar]
- Mazzotta S., Kemmerling B. (2011). Pattern recognition in plant innate immunity. J. Plant Pathol. 93, 7–17. [Google Scholar]
- McLellan H., Boevink P. C., Armstrong M. R., Pritchard L., Gomez S., Morales J., et al. (2013). An RxLR effector from Phytophthora infestans prevents re-localisation of two plant NAC transcription factors from the endoplasmic reticulum to the nucleus. PLoS Pathog. 9:e1003670. 10.1371/journal.ppat.1003670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros F., Resende M., Medeiros F., Zhang H., Pare P. (2009). Defense gene expression induced by a coffee-leaf extract formulation in tomato. Physiol. Mol. Plant P. 74, 175–183 10.1016/j.pmpp.2009.11.004 [DOI] [Google Scholar]
- Mercier L., Lafitte C., Borderies G., Briand X., Esquerré-Tugayé M.-T., Fournier J. (2001). The algal polysaccharide carrageenans can act as an elicitor of plant defence. New Phytol. 149, 43–51 10.1046/j.1469-8137.2001.00011.x [DOI] [PubMed] [Google Scholar]
- Milling A., Babujee L., Allen C. (2011). Ralstonia solanacearum extracellular polysaccharide is a specific elicitor of defense responses in wilt-resistant tomato plants. PLoS ONE 6:e15853. 10.1371/journal.pone.0015853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T., Ichimura K., Irie K., Morris P., Giraudat J., Matsumoto K., et al. (1998). Identification of a possible MAP kinase cascade in Arabidopsis thaliana based on pairwise yeast two-hybrid analysis and functional complementation tests of yeast mutants. FEBS Lett. 437, 56–60. 10.1016/S0014-5793(98)01197-1 [DOI] [PubMed] [Google Scholar]
- Mukhtar M. S., Carvunis A. R., Dreze M., Epple P., Steinbrenner J., Moore J., et al. (2011). Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333, 596–601. 10.1126/science.1203659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M. A., Sundelin T., Nielsen J. T., Erbs G. (2013). MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front. Plant Sci. 4:139. 10.3389/fpls.2013.00139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton A. C., Fitt B. D. L., Atkins S. D., Walters D. R., Daniell T. J. (2010). Pathogenesis, parasitism and mutualism in the trophic space of microbeplant interactions. Trends Microbiol. 18, 365–373. 10.1016/j.tim.2010.06.002 [DOI] [PubMed] [Google Scholar]
- Nguyen H. P., Chakravarthy S., Velásquez A. C., Mclane H. L., Zeng L. R., Nakayashiki H., et al. (2010). Methods to study PAMP-triggered immunity using tomato and Nicotiana benthamiana. Mol. Plant Microbe Interact. 23, 991–999. 10.1094/MPMI-23-8-0991 [DOI] [PubMed] [Google Scholar]
- Nürnberger T., Nennstiel D., Jabs T., Sacks W. R., Hahlbrock K., Scheel D. (1994). High affinity binding of a fungal oligopeptide elicitor to parsley plasma membranes triggers multiple defense responses. Cell 78, 449–460. 10.1016/0092-8674(94)90423-5 [DOI] [PubMed] [Google Scholar]
- Parker J. E. (2003). Plant recognition of microbial patterns. Trends Plant Sci. 8, 245–247. 10.1016/S1360-1385(03)00105-5 [DOI] [PubMed] [Google Scholar]
- Pastor V., Luna E., Mauch-Mani B., Ton J., Flors V. (2013). Primed plants do not forget. Environ. Exp. Bot. 94, 46–56 10.1016/j.envexpbot.2012.02.013 [DOI] [Google Scholar]
- Pedersen C., van Themaat E. V. L., McGuffin L. J., Abbott J. C., Burgis T. A., Barton G., et al. (2012). Structure and evolution of barley powdery mildew effector candidates. BMC Genomics 13:694. 10.1186/1471-2164-13-694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng D. H., Qiu D. W., Ruan L. F., Zhou C. F., Sun M. (2011). Protein elicitor PemG1 from Magnaporthe grisea induces systemic acquired resistance (SAR) in plants. Mol. Plant Microbe Interact. 24, 1239–1246. 10.1094/MPMI-01-11-0003 [DOI] [PubMed] [Google Scholar]
- Povero G., Loreti E., Pucciariello C., Santaniello A., Di Tommaso D., Di Tommaso G., et al. (2011). Transcript profiling of chitosan-treated Arabidopsis seedlings. J. Plant Res. 124, 619–629. 10.1007/s10265-010-0399-1 [DOI] [PubMed] [Google Scholar]
- Po-Wen C., Singh P., Zimmerli L. (2013). Priming of the Arabidopsis pattern-triggered immunity response upon infection by necrotrophic Pectobacterium carotovorum bacteria. Mol. Plant Pathol. 14, 58–70. 10.1111/j.1364-3703.2012.00827.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y., Liu L., Xiong Q., Flores C., Wong J., Shi J., et al. (2013). Oomycete pathogens encode silencing suppressors. Nat. Genet. 45, 330–333. 10.1038/ng.2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D., Mao J., Yang X., Zeng H. (2009). Expression of an elicitor-encoding gene from Magnaporthe grisea enhances resistance against blast disease in transgenic rice. Plant Cell Rep. 28, 925–933. 10.1007/s00299-009-0698-y [DOI] [PubMed] [Google Scholar]
- Qutob D., Kemmerling B., Brunner F., Küfner I., Engelhardt S., Gust A. A., et al. (2006). Phytotoxicity and innate immune responses induced by Nep1-like proteins. Plant Cell 18, 3721–3744. 10.1105/tpc.106.044180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiqi M., Ellis J. G., Ludowici V. A., Hardham A. R., Dodds P. N. (2012). Challenges and progress towards understanding the role of effectors in plant-fungal interactions. Curr. Opin. Plant Biol. 15, 477–482. 10.1016/j.pbi.2012.05.003 [DOI] [PubMed] [Google Scholar]
- Randoux B., Renard-Merlier D., Mulard G., Rossard S., Duyme F., Sanssené J., et al. (2010). Distinct defenses induced in wheat against powdery mildew by acetylated and nonacetylated oligogalacturonides. Phytopathology 100, 1352–1363. 10.1094/PHYTO-03-10-0086 [DOI] [PubMed] [Google Scholar]
- Reglinski T., Lyon G. D., Newton A. C. (1994a). Assessment of the ability of yeast-derived elicitors to control barley powdery mildew in the field. Z. Pflanzenk. Pflanzen. 101, 1–10. [Google Scholar]
- Reglinski T., Lyon G. D., Newton A. C. (1994b). Induction of resistance mechanisms in barley by yeast-derived elicitors. Ann. Appl. Biol. 124, 509–517 10.1111/j.1744-7348.1994.tb04155.x [DOI] [Google Scholar]
- Reglinski T., Lyon G. D., Newton A. C. (1995). The control of Botrytis cinerea and Rhizoctonia solani on lettuce using elicitors extracted from yeast cell walls. Z. Pflanzenk. Pflanzen. 102, 257–266. [Google Scholar]
- Ron M., Avni A. (2004). The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16, 1604–1615. 10.1105/tpc.022475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangha J. S., Ravichandran S., Prithiviraj K., Critchley A. T., Prithiviraj B. (2010). Sulfated macroalgal polysaccharides lambda-carrageenan and iota-carrageenan differentially alter Arabidopsis thaliana resistance to Sclerotinia sclerotiorum. Physiol. Mol. Plant P. 75, 38–45 10.1016/j.pmpp.2010.08.003 [DOI] [Google Scholar]
- Schönbeck F., Dehne H.-W., Balder H. (1982). Zur Wirksamkeit induzierter Resistenz unter praktischen Anbaubedingungen I. Echter Mehltau an Reben, Gurken und Weizen. Z. Pflanzenk. Pflanzen. 89, 177–184. [Google Scholar]
- Schönbeck F., Dehne H.-W., Beicht W. (1980). Untersuchungen zur Aktivierung unspeziefischer Resistenzmechanismen in Pflanzen. Z. Pflanzenk. Pflanzen. 87, 654–666. [Google Scholar]
- Schornack S., Huitema E., Cano L. M., Bozkurt T. O., Oliva R., Van Damme M., et al. (2009). Ten things to know about oomycete effectors. Mol. Plant Pathol. 10, 795–803. 10.1111/j.1364-3703.2009.00593.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T., Nakano T., Takamizawa D., Desaki Y., Ishii-Minami N., Nishizawa Y., et al. (2010). Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64, 204–214. 10.1111/j.1365-313X.2010.04324.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinya T., Gális I., Narisawa T., Sasaki M., Fukuda H., Matsuoka H., et al. (2007). Comprehensive analysis of glucan elicitor-regulated gene expression in tobacco BY-2 cells reveals a novel MYB transcription factor involved in the regulation of phenylpropanoid metabolism. Plant Cell Physiol. 48, 1404–1413. 10.1093/pcp/pcm115 [DOI] [PubMed] [Google Scholar]
- Shinya T., Motoyama N., Ikeda A., Wada M., Kamiya K., Hayafune M., et al. (2012). Functional characterization of CEBiP and CERK1 homologs in Arabidopsis and rice reveals the presence of different chitin receptor systems in plants. Plant Cell Physiol. 53, 1696–1706. 10.1093/pcp/pcs113 [DOI] [PubMed] [Google Scholar]
- Shoresh M., Yedidia I., Chet I. (2005). Involvement of jasmonic acid/ethylene signaling pathway in the systemic resistance induced in cucumber by Trichoderma asperellum T203. Phytopathology 95, 76–84. 10.1094/PHYTO-95-0076 [DOI] [PubMed] [Google Scholar]
- Steiner U., Oerke E.-C., Schönbeck F. (1988). Zur Wirksamkeit der induzierten Resistenz unter praktischen Anbaubedingungen IV. Befall und Ertrag von Wintergerstensorten mit induzierter Resistenz und nach Fungizidbehandlung. Z. Pflanzenk. Pflanzen. 95, 506–517. 21395787 [Google Scholar]
- Suzuki H., Reddy M. S. S., Naoumkina M., Aziz N., May G. D., Huhman D. V., et al. (2005). Methyl jasmonate and yeast elicitor induce differential transcriptional and metabolic re-programming in cell suspension cultures of the model legume Medicago truncatula. Planta 220, 696–707. 10.1007/s00425-004-1387-2 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Nasir K. H. B., Ito A., Kanzaki H., Matsumura H., Saitoh H., et al. (2007). A high-throughput screen of cell-death-inducing factors in Nicotiana benthamiana identifies a novel MAPKK that mediates INF1-induced cell death signaling and non-host resistance to Pseudomonas cichorii. Plant J. 49, 1030–1040 10.1111/j.1365-313X.2006.03022.x [DOI] [PubMed] [Google Scholar]
- Takken F. L. W., Luderer R., Gabriëls S. H. E. J., Westerink N., Lu R., De Wit P. J. G. M., et al. (2000). A functional cloning strategy, based on a binary PVX-expression vector, to isolate HR-inducing cDNAs of plant pathogens. Plant J. 24, 275–283. 10.1046/j.1365-313x.2000.00866.x [DOI] [PubMed] [Google Scholar]
- Tanaka S., Ichikawa A., Yamada K., Tsuji G., Nishiuchi T., Mori M., et al. (2010). HvCEBiP, a gene homologous to rice chitin receptor CEBiP, contributes to basal resistance of barley to Magnaporthe oryzae. BMC Plant Biol. 10:288. 10.1186/1471-2229-10-288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. M., Jones D. A., Parniske M., Harrison K., Balint-Kurti P. J., Hatzixanthis K., et al. (1997). Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell 9, 2209–2224. 10.1105/tpc.9.12.2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemura K., Tanino S., Nagatsuka T., Koga J., Iwata M., Nagashima K., et al. (2004). Cerebroside elicitor confers resistance to Fusarium disease in various plant species. Phytopathology 94, 813–818. 10.1094/PHYTO.2004.94.8.813 [DOI] [PubMed] [Google Scholar]
- Underwood W. (2012). The plant cell wall: a dynamic barrier against pathogen invasion. Front. Plant Sci. 3:85. 10.3389/fpls.2012.00085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Burg H. A., Harrison S. J., Joosten M. H. A. J., Vervoort J., De Wit P. J. G. M. (2006). Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol. Plant Microbe Interact. 19, 1420–1430. 10.1094/MPMI-19-1420 [DOI] [PubMed] [Google Scholar]
- van Hulten M., Pelser M., van Loon L. C., Pieterse C. M. J., Ton J. (2006). Costs and benefits of priming for defense in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 5602–5607. 10.1073/pnas.0510213103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatsa P., Chiltz A., Luini E., Vandelle E., Pugin A., Roblin G. (2011). Cytosolic calcium rises and related events in ergosterol-treated Nicotiana cells. Plant Physiol. Biochem. 49, 764–773. 10.1016/j.plaphy.2011.04.002 [DOI] [PubMed] [Google Scholar]
- Vera J., Castro J., Gonzalez A., Moenne A. (2011). Seaweed polysaccharides and derived oligosaccharides stimulate defense responses and protection against pathogens in plants. Mar. Drugs 9, 2514–2525. 10.3390/md9122514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viterbo A. D. A., Wiest A. R. I. C., Brotman Y. A. R. I., Chet I. L. A. N., Kenerley C. H. A. R. (2007). The 18mer peptaibols from Trichoderma virens elicit plant defence responses. Mol. Plant Pathol. 8, 737–746. 10.1111/j.1364-3703.2007.00430.x [DOI] [PubMed] [Google Scholar]
- Walters D., Bingham I. (2007). Influence of nutrition on disease development caused by fungal pathogens: implications for plant disease control. Ann. Appl. Biol. 151, 307–324 10.1111/j.1744-7348.2007.00176.x [DOI] [Google Scholar]
- Walters D., Heil M. (2007). Costs and trade-offs associated with induced resistance. Physiol. Mol. Plant P. 71, 3–17 10.1016/j.pmpp.2007.09.008 [DOI] [Google Scholar]
- Walters D. R., Ratsep J., Havis N. D. (2013). Controlling crop diseases using induced resistance: challenges for the future. J. Exp. Bot. 64, 1263–1280. 10.1093/jxb/ert026 [DOI] [PubMed] [Google Scholar]
- Wan J., Zhang X. C., Neece D., Ramonell K. M., Clough S., Kim S., et al. (2008). A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20, 471–481. 10.1105/tpc.107.056754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Wang S., Tan B., Qiu D., Yang X. (2012a). Systemic acquired resistance to Tobacco mosaic virus (TMV) induced by protein elicitor from Verticillium dahliae (PevD1) and its mechanisms in tobacco. J. Agr. Biotechnol. 20, 188–195. 21691787 [Google Scholar]
- Wang B., Yang X., Zeng H., Liu H., Zhou T., Tan B., et al. (2012b). The purification and characterization of a novel hypersensitive-like response-inducing elicitor from Verticillium dahliae that induces resistance responses in tobacco. App. Microbiol. Biotechnol. 93, 191–201. 10.1007/s00253-011-3405-1 [DOI] [PubMed] [Google Scholar]
- Wang F., Feng G., Chen K. (2009). Burdock fructooligosaccharide induces resistance to tobacco mosaic virus in tobacco seedlings. Physiol. Mol. Plant P. 74, 34–40 10.1016/j.pmpp.2009.08.002 [DOI] [Google Scholar]
- Wang S., Durrant W. E., Song J., Spivey N. W., Dong X. (2010). Arabidopsis BRCA2 and RAD51 proteins are specifically involved in defense gene transcription during plant immune responses. Proc. Natl. Acad. Sci. U.S.A. 107, 22716–22721. 10.1073/pnas.1005978107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H., Xu Q., Taylor L. E., 2nd., Baker J. O., Tucker M. P., Ding S. Y. (2009). Natural paradigms of plant cell wall degradation. Curr. Opin. Biotechnol. 20, 330–338. 10.1016/j.copbio.2009.05.008 [DOI] [PubMed] [Google Scholar]
- Xu Y., Chen H., Zhou X., Cai X. (2012). Induction of hypersensitive response and nonhost resistance by a Cladosporium fulvum elicitor CfHNNI1 is dose-dependent and negatively regulated by salicylic acid. J. Integr. Agri. 11, 1665–1674 10.1016/S2095-3119(12)60169-5 [DOI] [Google Scholar]
- Yang D. H., Hettenhausen C., Baldwin I. T., Wu J. (2011). The multifaceted function of BAK1/SERK3: plant immunity to pathogens and responses to insect herbivores. Plant Signal. Behav. 6, 1322–1324. 10.4161/psb.6.9.16438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Zhang H., Li G., Li W., Wang X., Song F. (2009). Ectopic expression of MgSM1, a Cerato-platanin family protein from Magnaporthe grisea, confers broad-spectrum disease resistance in Arabidopsis. Plant Biotechnol. J. 7, 763–777. 10.1111/j.1467-7652.2009.00442.x [DOI] [PubMed] [Google Scholar]
- Zhang J., Shao F., Li Y., Cui H., Chen L., Li H., et al. (2007). A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1, 175–185. 10.1016/j.chom.2007.03.006 [DOI] [PubMed] [Google Scholar]
- Zhang J., Tong Z., Gao Z., Luo C., Qu S., Zhang Z. (2011a). Expression of MhWRKY1 gene induced by the elicitors SA, MeJA, ACC and the apple ring spot pathogen. Sci. Agric. Sin. 44, 990–999. [Google Scholar]
- Zhang W., Yang X., Qiu D., Guo L., Zeng H., Mao J., et al. (2011b). PeaT1-induced systemic acquired resistance in tobacco follows salicylic acid-dependent pathway. Mol. Biol. Rep. 38, 2549–2556. 10.1007/s11033-010-0393-7 [DOI] [PubMed] [Google Scholar]
- Zhang W., Fraiture M., Kolb D., Löffelhardt B., Desaki Y., Boutrot F. F., et al. (2013). Arabidopsis receptor-like protein30 and receptor-like kinase suppressor of BIR1-1/EVERSHED mediate innate immunity to necrotrophic fungi. Plant Cell 25, 4227–4241. 10.1105/tpc.113.117010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yang X., Liu Q., Qiu D., Zhang Y., Zeng H., et al. (2010). Purification of novel protein elicitor from Botrytis cinerea that induces disease resistance and drought tolerance in plants. Microbiol. Res. 165, 142–151. 10.1016/j.micres.2009.03.004 [DOI] [PubMed] [Google Scholar]
- Zheng X., McLellan H., Fraiture M., Liu X., Boevink P. C., Gilroy E. M., et al. (2014). Functionally redundant RXLR effectors from Phytophthora infestans act at different steps to suppress early flg22-triggered immunity. PLoS Pathog. 10:e1004057. 10.1371/journal.ppat.1004057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J. D. G., Boller T., et al. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125, 749–760. 10.1016/j.cell.2006.03.037 [DOI] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S., Navarro L., Oakeley E. J., Jones J. D. G., Felix G., et al. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428, 764–767. 10.1038/nature02485 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


