Abstract
It is expected that serum protein biomarkers in Duchenne muscular dystrophy (DMD) will reflect disease pathogenesis, progression and aid future therapy developments. Here, we describe use of quantitative in vivo stable isotope labeling in mammals to accurately compare serum proteomes of wild-type and dystrophin-deficient mdx mice. Biomarkers identified in serum from two independent dystrophin-deficient mouse models (mdx-Δ52 and mdx-23) were concordant with those identified in sera samples of DMD patients. Of the 355 mouse sera proteins, 23 were significantly elevated and 4 significantly lower in mdx relative to wild-type mice (P-value < 0.001). Elevated proteins were mostly of muscle origin: including myofibrillar proteins (titin, myosin light chain 1/3, myomesin 3 and filamin-C), glycolytic enzymes (aldolase, phosphoglycerate mutase 2, beta enolase and glycogen phosphorylase), transport proteins (fatty acid-binding protein, myoglobin and somatic cytochrome-C) and others (creatine kinase M, malate dehydrogenase cytosolic, fibrinogen and parvalbumin). Decreased proteins, mostly of extracellular origin, included adiponectin, lumican, plasminogen and leukemia inhibitory factor receptor. Analysis of sera from 1 week to 7 months old mdx mice revealed age-dependent changes in the level of these biomarkers with most biomarkers acutely elevated at 3 weeks of age. Serum analysis of DMD patients, with ages ranging from 4 to 15 years old, confirmed elevation of 20 of the murine biomarkers in DMD, with similar age-related changes. This study provides a panel of biomarkers that reflect muscle activity and pathogenesis and should prove valuable tool to complement natural history studies and to monitor treatment efficacy in future clinical trials.
INTRODUCTION
Duchenne muscular dystrophy (DMD) is the most common and severe form of childhood muscular dystrophy affecting 1 in 3500 boys worldwide (1). Currently, there is no cure for the disease, except corticosteroid treatment that has been shown in several clinical trials to improve muscle strength and respiratory function (2–5), and prolong ambulation for 2 or more years (6–9). These beneficial effects are offset, in part, by side effects, such as weight gain, growth stunting and bone health issues (5).
Fortunately, promising therapeutic strategies for DMD are increasingly under development, with many now in clinical trials (10–13). Perhaps one of the most interesting ones is those aiming to restore the missing dystrophin protein (10,14–16). These include exon skipping strategies with antisense oligonucleotide (17–19), a stop codon read through strategy using the drug Ataluren, also known as PTC124 (11), and viral gene therapy using the adeno-associated virus as a micro dystrophin gene carrier (15,20). VPB15, a novel steroid dissociative drug, has been recently shown to be also a promising anti-inflammatory agent since it improves muscle physiology without side effects (21). As these new-generation therapies move into human clinical trials in DMD, it becomes important to develop minimally invasive outcome measures that can give an early readout of potential drug efficacy.
Currently, most clinical trials for DMD rely on physical assessments such as the 6 min walk test, which is the walking distance a patient can cover in 6 min (22), the North Star Ambulatory assessment (23,24), as well as quantitative muscle strength tests (25). Even though useful, these physical tests are limited to ambulatory patients, often challenging to implement, and could be confounded with patient's willingness, especially in young children. Defining biochemical markers that correlate with the clinical phenotype and the above outcome measures will aid assessment of treatment efficacy in DMD clinical trials.
Serological levels of creatine kinase (CK) have long been used to aid diagnosis of DMD and other muscular dystrophies (26) and remain an important laboratory test for diagnosis. Unfortunately, because CK serum levels can be easily influenced by muscle exertion or trauma, and because of its high abundance in serum of DMD patients, it might not prove sensitive to early treatment response.
Recently, with advances in ‘omics’ technologies (e.g. genomics, proteomics and metabolomics), valuable sets of biomarkers are being discovered in muscle tissue and serum of both DMD patients and animal models. While major biomarkers and molecular mechanism have been discovered by proteomics and genomics studies of dystrophic muscle tissue in the past few years (27), effort toward large-scale biomarkers discovery in body fluids only started emerging in the last 3 years. Indeed, recent studies have shown a clear correlation between specific serum miRNAs and DMD severity in both human patients and animal models (28–30). Other molecular markers that correlated with disease progression include matrix metalloproteinase-9 (MMP9), which was found highly elevated in DMD patients relative to controls and increased with disease severity (31). And more recently, the same group identified fibronectin as potential biomarker for muscular dystrophy (32). Although these preliminary studies were conducted with targeted markers, they clearly support the hypothesis that serum circulating biomarkers can act as indicators of DMD disease progression and potentially as biomarkers to monitor treatment efficacy.
In this study, we carried out comprehensive serum proteome profiling using a combination of stable isotope labeling in mammals (SILAM) strategy (33,34) and high precision mass spectrometry to identify serum circulating protein biomarkers in dystrophin-deficient mdx mice. These were tested across the age range of mdx mice to correlate their association with muscle disease progression. Several of the biomarkers identified in the dystrophin-deficient mouse model also exhibited higher levels in human serum samples collected from DMD patients compared with age-matched healthy controls. Development of a DMD biomarker panel, as opposed to a single biomarker, may prove robust and non-invasive readout tool for muscle tissue, its current disease state and potentially its response to therapy.
RESULTS
Serum protein biomarker discovery using two independent dystrophin-deficient mouse models (mdx-52 and mdx-23)
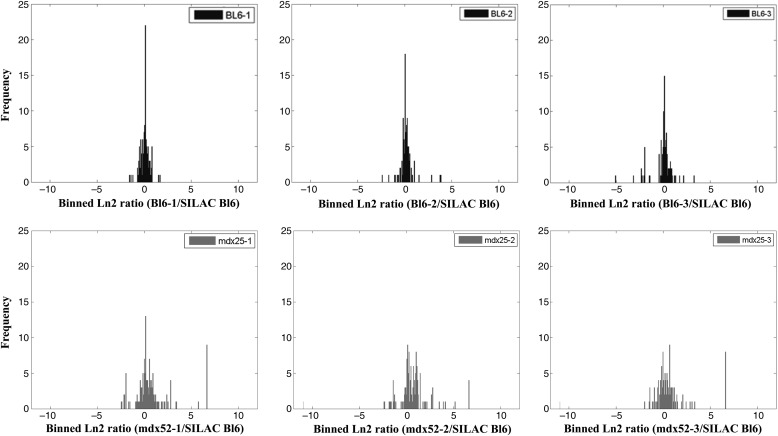
Biomarker discovery was first performed on serum samples from 3-week-old mdx-52 mice (n = 3) and age-matched wild-type controls BL6 (n = 3). Each mdx sample was spiked with an equal amount of 13C6-Lys BL6 labeled serum. Three-week-old mdx mice were chosen because the initial onset of muscle pathogenesis in these mice usually occurs at 3 weeks of age (35). Supplementary Material, Figure S1 shows mass spectra of an elevated protein (e.g. CK M), unchanged protein (e.g. albumin) and decreased protein (e.g. leukemia inhibitory factor receptor) in serum of mdx-52 mouse relative to its wild-type BL6 mouse. This initial experiment led to the identification and quantification of 214 proteins in all samples combined. Keratins and multiple isoform immunoglobulins were removed from the analysis, leaving 192 proteins. These are listed in Supplementary Material, Table S1 with average ratios to internal SILAC standard, spectral count and standard deviation in the mdx-52 mouse versus BL6 groups. Ratio distributions of all quantified proteins in serum of the two mouse groups relative to the internal SILAC-labeled serum proteins are plotted in Figure 1 as log2 of the ratio. While the distribution of protein log2 ratios was around zero (ratio of 1:1) in the serum of all three BL6 to SILAC BL6 pairs, it was wider in the mdx-52 versus SILAC BL6 pairs. This clearly indicates strong alterations in the levels of some proteins in the serum of mdx-52 mice relative to its wild-type counterpart BL6 mice.
Figure 1.
Distribution of protein ratios identified in proteome profiling of sera samples of mdx-52 mice and wild-type BL6 mice spiked at 1:1 ratio with serum aliquots from SILAC-labeled BL6 mouse. Each serum aliquot containing 50 µg of total proteins from 3 weeks old mdx-52 mice (n = 3) or 3 weeks old BL6 mice (n = 3) was spiked with 50 µg serum aliquots from 13C6-Lys labeled BL10 serum and processed for proteome profiling and quantification as described in Materials and Methods. Top panel shows the overall log ratio distribution of all identified unlabeled and labeled peptide pairs in 1:1 serum mixtures of unlabeled BL10 mice relative to 13C6-Lys labeled BL6 internal standard and the bottom panel show similar ratios distribution in 1:1 serum mixture of unlabeled mdx-52 mice relative to 13C6-Lys labeled BL6 internal standard. All distributions showed a normal Gaussian shape with values centered around 1 in BL6 versus SILAC BL6 pairs and wilder distribution in mdx-52 versus SILAC BL6 pairs analyzed. An average of 180–200 proteins were identified and quantified in each analysis.
Similarly, biomarker discovery was performed on serum of the spontaneous allele mdx-23 mice (n = 3) and age-matched wild-type controls BL10 (n = 3). In this second discovery set, 15N labeled wild-type mouse sera were used as a spike-in standard instead of 13C6-Lys labeled serum. The 15N labeling strategy allowed identification and quantitation of 355 proteins from six serum samples combined. Omitting again keratins and the multiple interchangeable immunoglobulin isoforms, 305 proteins are retained for analysis, and these are listed in Supplementary Material, Table S2 with ratios to internal SILAC standard, spectral count and standard deviation in the mdx-23 mice and BL10 mice groups. In general, 15N based proteome profiling led to the identification and quantification of more proteins, 181 more proteins than 13C6-Lys based proteome profiling strategy. This is mainly due to the fact that only Lys containing peptides are quantifiable in 13C6-Lys proteome profiling experiment, while every peptide is quantifiable in 15N based proteome profiling experiments.
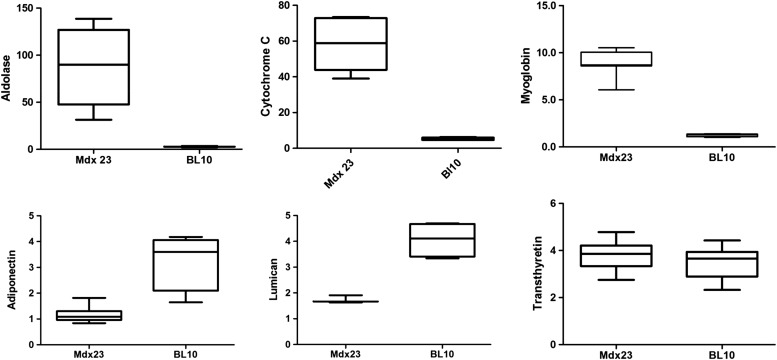
Examples of protein biomarkers whose levels were increased, unchanged or decreased in mdx-23 mice versus wild-type BL10 are shown in Figure 2. Fructose-bisphosphate aldolase A (ALDOA), somatic cytochrome C (CYC) and myoglobin (MYG) were significantly increased in mdx-23 sera relative to wild-type BL10 mice. Adiponectin (ADIPO) and LUM were significantly decreased in serum of mdx-23 mice, while transthyretin remained unchanged between the two groups. Similar analyses of all 305 identified proteins were performed at the peptide level. Proteins that were concordant between the mdx-23 and mdx-52 studies are listed in Table 1 with average fold change relative to wild-type mice, an average peptide spectral count used to quantify each protein and P-values. Elevated proteins in the serum of the dystrophin-deficient mice included myofibril proteins such as titin (TITIN), myosin light chain 1/3 (MYL1), myomesin-3 (MYOM3), myosin-4 (MYH4), myosin-1, filamin-C (FLNC) and actin alpha skeletal (ACTS); glycolytic enzymes such as ALDOA, glyceraldehyde-3-phosphate dehydrogenase (GAPDH or GP3), pyruvate kinase isozymes M1/M2 (KPYM), glycogen phosphorylase (PYGM), l-lactate dehydrogenase A (LDHA), triosephosphate isomerase (TPIS), beta enolase (ENOB), phosphoglycerate mutase 2 (PGAM2); transport proteins such as MYG, CYC and fatty acid-binding protein heart type (FABPH). Other highly elevated proteins in the serum of mdx mice included malate dehydrogenase (MDHC), CK M, thrombospondin-4 (TSP4), serum amyloid A-2 protein, fibrinogen gamma chain (FIBG), parvalbumin (PRVA) and superoxide dismutase. Several IgG kappa and gamma chains were also found significantly increased in the serum of mdx-23 relative to BL10. These were not included in Table 1 but reported in Supplementary Material, Tables S1 and S2.
Figure 2.
Box plots showing levels of six representative proteins in sera samples of mdx-23 mice compared with its healthy counterpart wild-type BL10 mice. Serum samples from 3 weeks old mdx-23 mice (n = 3) and age-matched wild-type BL10 mice (n = 3) were each spiked with fixed amount of 15N-labeled serum samples from wild-type mice as internal standard. Sample mixtures were processed for protein identification and quantification as described in Materials and Methods. Tope panel shows an example of three proteins, aldoalse, cystochrom-c somatic and myoglobin whose level was significantly (P-value < 0.001) higher in serum of mdx-23 mice group compared with the wild-type BL10 mice group. Bottom panels show an example of two proteins, adiponectin and lumican whose level was significantly decreased in serum of the mdx-23 mice group compared with wild-type Bl10 mice (P-valued < 0.001) and transtherytin protein that remained unchanged between the two groups.
Table 1.
List of candidate protein biomarkers whose levels were found significantly altered in the serum of dystrophin-deficient mdx-23 mice (n = 3) versus wild-type BL10 mice (n = 3) and in mdx-52 (n = 3) versus wild-type BL6 (n = 3)
| Accession N° | Locus | Protein name | mdx-23 relative to BL10 (fold change) | mdx-52 relative to BL6 (fold change) | PSC average | P-value |
|---|---|---|---|---|---|---|
| Myofibrillar proteins | ||||||
| A2ASS6 | TITIN | Titin | Unique in mdx-23 | Unique in mdx-52 | 25 | 0.0000 |
| P05977 | MYL1 | Myosin light chain 1/3 | Unique in mdx-23 | Unique in mdx-52 | 4 | 0.0000 |
| Q5SX39 | MYH4 | Myosin-4 | Unique in mdx-23 | Unique in mdx-52 | 3 | 0.0000 |
| A2ABU4 | MYOM3 | Myomesin-3 | Unique in mdx-23 | Unique in mdx-52 | 3 | 0.0000 |
| Q8VHX6 | FLNC | Filamin-C | Unique in mdx-23 | Unique in mdx-52 | 7 | 0.0000 |
| P68134 | ACTS | Actin, alpha skeletal muscle | +1.5-fold | +2.93-fold | 16 | 0.0002 |
| Glycolytic enzymes | ||||||
| P05064 | ALDOA | Fructose-bisphosphate aldolase A | +23-fold | +21-fold | 5 | 0.0479 |
| P16858 | GAPDH or GP3 | Glyceraldehyde-3-phosphate dehydrogenase | Unique in mdx-23 | +4.43-fold | 5 | 0.0000 |
| P52480 | KPYM | Pyruvate kinase isozymes M1/M2 | Unique in mdx-23 | Unique in mdx-52 | 12 | 0.0000 |
| P06151 | LDHA | L-lactate dehydrogenase A | +3.27-fold | +3.2-fold | 3 | 0.0000 |
| P17751 | TPIS | Triosephosphate isomerase | Unique in mdx-23 | +3.1-fold | 3 | 0.0000 |
| Q9WUB3 | PYGM | Glycogen phosphorylase | Unique in mdx-23 | Unique in mdx-52 | 16 | 0.0000 |
| P21550 | ENOB | Beta-enolase | Unique in mdx-23 | nd | 7 | 0.0000 |
| O70250 | PGAM2 | Phosphoglycerate mutase 2 | Unique in mdx-23 | Unique in mdx-52 | 5 | 0.0000 |
| Transport proteins | ||||||
| P04247 | MYG | Myoglobin | +11-fold | +2.9-fold | 7 | 0.0000 |
| P11404 | FABPH | Fatty acid-binding protein, heart type | Unique in mdx-23 | nd | 3 | 0.0000 |
| P62897 | CYC | Cytochrome c, somatic | +55-fold | +20-fold | 5 | 0.0109 |
| Other muscle-specific proteins | ||||||
| P07310 | CK | Creatine kinase M-type | Unique in mdx-23 | Unique in mdx-52 | 8 | 0.0000 |
| P14152 | MDHC | Malate dehydrogenase, cytoplasmic | +57-fold | +1.46-fold | 7 | 0.0001 |
| Q9Z1T2 | TSP4 | Thrombospondin-4 | +7-fold | +1.53-fold | 3 | 0.0000 |
| P32848 | PRVA | Parvalbumin alpha | +13.5-fold | +15-fold | 6 | 0.0000 |
| P05367 | SAA2 | Serum amyloid A-2 protein | +4.2-fold | nd | 12 | 0.0000 |
| Q8VCM7 | FIBG | Fibrinogen gamma chain | +4.16-fold | +3.15-fold | 1 | 0.0000 |
| P20918 | PLMN | Plasminogen | −1.5-fold | −0.2-fold | 28 | 0.0000 |
| Q60994 | ADIPO | Adiponectin | −1.5-fold | Unchanged | 9 | 0.0000 |
| P51885 | LUM | Lumican | −1.5-fold | −0.2-fold | 4 | 0.0006 |
| P42703 | LIFR | Leukemia inhibitory factor receptor | −3.3-fold | −3-fold | 25 | 0.0000 |
PSC, peptide spectral count.
On the other hand, only a few proteins were found moderately decreased in serum of mdx-23 mice relative to wild-type BL10 mice. These included mostly proteins involved in extracellular matrix remodeling and cell proliferation, such as plasminogen (PLMN), ADIPO, lumican (LUM) and leukemia inhibitory factor receptor (LIFR). Perhaps, the most significant decrease was observed for LIFR which was concordant in both mdx-23 and mdx-52 model and decreased by 3-folds in both mouse models relative to their wild-type control.
Age-dependent changes in the level of serum biomarkers in the mdx-23 mouse model
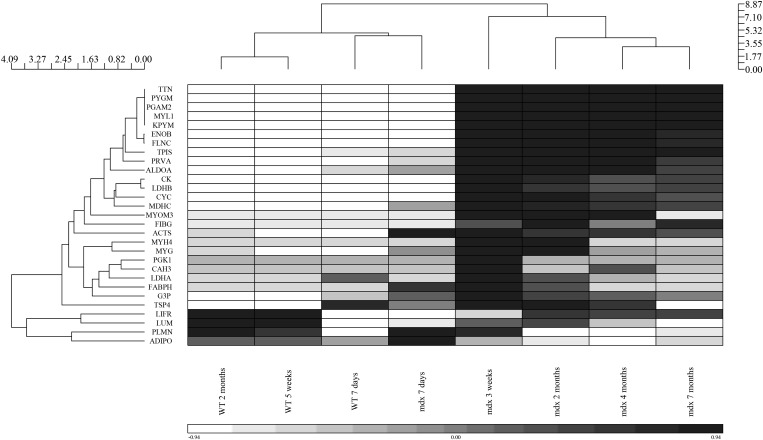
The mdx-23 mouse model shows age-related pathology, with no overt pathology before the age of 3 weeks followed by an acute onset of muscle necrosis at 3 weeks of age, with subsequent successful regeneration and relatively mild dystrophic pathology in most muscle groups in older mice (35). To determine whether the identified candidate biomarkers above are associated with age-dependent muscle pathology in the dystrophin-deficient mdx-23 mouse, we performed quantitative proteome profiling using the 15N SILAM strategy on serum samples of mdx-23 mice at 7 days, 3 weeks, 2 months, 4 months and 7 months of age, focusing on the levels of the biomarkers identified in the discovery set above listed in Table 1. Data were visualized by unsupervised hierarchical clustering of serum biomarkers in mdx-23 and wild-type BL10 mice as a function of age (Fig. 3). A dendrogram of age showed co-clustering of newborn wild-type and mdx mice, co-clustering of 3 weeks and 2 months old mdx mice and co-clustering of 4 months and 7 months old mdx apart from the wild-type mice groups. This clustering by age corresponded well to the established age-related histological patterns. Most of the biomarkers identified above were undetectable in the sera of new born mdx-23 mice then acutely rose in mdx-23 at 3 weeks of age correlating with the active necrotic phase (35). In addition to age-dependent clustering of mdx mice, the heat map revealed three major clusters of biomarkers. A first cluster consisted of biomarkers that acutely rose in sera of the 3 weeks old mdx-23 mouse (necrotic stage) and remained increased throughout the age studied, up to 7 months of age. These included seven glycolytic enzymes (e.g. PYGM, PGAM2, TPIS, KPYM, LDHB, ALDOA and ENOB), three myofibrillar proteins (e.g. TITIN, MYL1 and FLNC) and four other muscle-derived proteins (e.g. CK, MDHC, PRVA and CYC). The second cluster consisted of biomarkers that also acutely rose in blood of 3 weeks old mdx-23 then gradually decreased with age. These included three glycolytic enzymes (e.g. LDHA, PGK1 and GPADH), three myofibriallar proteins (MYOM3, ACTS and MYH4) and three muscle-derived proteins (CAH3, FABPH, MYG and TSP4). Finally, the third cluster consisted of biomarkers that decreased with age in mdx-23 and these were mostly of extracellular origin (e.g. LIFR, LUM, ADIPO, FIBG and PLMN). Even though the majority of these biomarkers rose in blood of mdx mice at 3 weeks of age few of them were, however, found moderately elevated in serum of 7 days old mdx mice, prior to the onset of muscle necrosis (e.g. ACTS, MYH4, ALDOA, GAPDH, MDHC, TSP4 and MYG) and could be classified as pre-necrotic biomarkers.
Figure 3.
Hierarchical clustering of selected candidate protein biomarkers in serum samples of mdx-23 mice across different age group. Serum samples from mdx-23 mice at 7 days, 3 weeks, 2 months, 4 months and 7 months of age (n = 3 per group) were spiked with fixed amount of 15N-labeled serum from the wild-type mouse as internal standard. Similarly serum samples from wild-type Bl10 mice at 7 days, 5 weeks and 2 months of age were spiked with 15N- labeled serum and used as controls. Sample mixtures were processed for proteomes profiling as described in Materials and Methods. Ratios of candidate biomarkers were measured in serum samples across different age groups and then uploaded into Partek software for clustering analysis. SILAC ratio values were natural log transformed, and the color scale is based on how many standard deviations each value deviates from the mean (white for values below the mean, and black for values above the mean). Clustering by columns shows that a select group of biomarkers is able to discriminate wild-type BL10 from mdx-23 mice, and age within each group. Clustering by rows shows consolidation of biomarker ‘classes’ described in the text.
Biomarkers discovery in human serum samples from DMD and healthy controls subjects
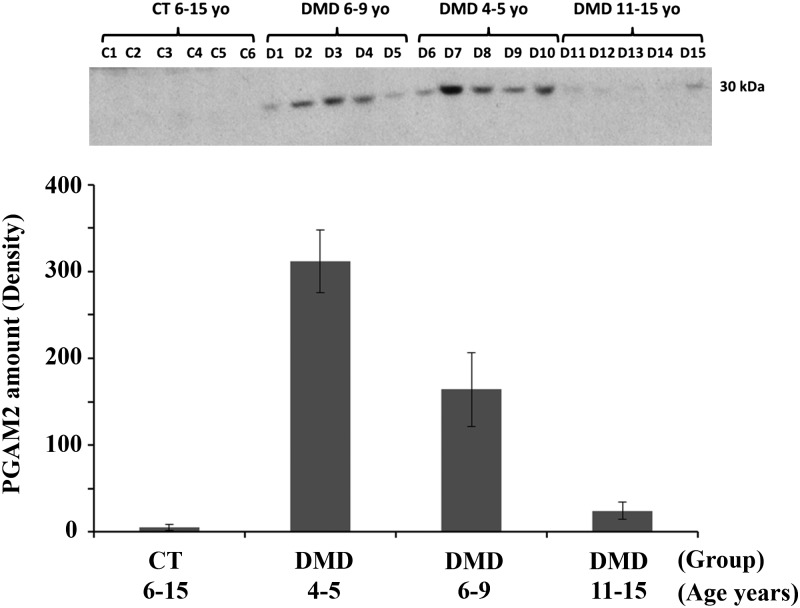
The protein biomarker panel discovered in serum of dystrophin-deficient mouse models was examined in serum samples of DMD (n = 15) and healthy volunteers (n = 6), using a combination of label-free proteome profiling and western blot analysis. Immunoblot analysis was complicated by the serum biomarker typically being a truncated fragment of the full-length protein. Thus, availability of commercial antibodies for the specific fragment was challenging. Of the five antibodies tested against five target candidate biomarkers (FLNC, TITIN, MYOM3, PGAM2 and PGYM), only the antibody against PGAM2 provided clear results. PGAM2 was detected as full-length protein around 30 kDa in all serum samples of the DMD group but at low or undetectable levels in the control group (Fig. 4A). However, unlike in the mdx mouse model, the serum level of PGAM2 rapidly decreased with age in the DMD patient sera. It was significantly higher in younger DMD patients, between the age of 4 and 8 years old, compared with older DMD patients between the age of 11 and 15 years old. PGAM2 was almost undetectable in serum samples of DMD patients 11–15 years of age (Fig. 4B).
Figure 4.
Western blot analysis showing levels of PGAM2 in sera samples of DMD patients and healthy controls. Serum aliquots containing 30 µg total proteins from DMD patients with different age groups (4 to 5, 6–9 and 11–15 years of age, n = 5 per group) and from healthy controls (n = 6) with age ranging from 6 to 15 years old were processed for western blot analysis against PGAM2 as described in Materials and Methods. Top panel shows the actual western blot results of PGAM2 which was detected around its expected molecular mass of 30 kDa and it was present only in serum samples of DMD patients (D1 to D15) and undetectable in serum of healthy controls (CT1 to CT6) . The bottom panel shows the density histogram plot of the PGAM2 band across different age groups. The data clearly show the rapid age-dependent decrease in the serum levels of PGAM2 in DMD patients.
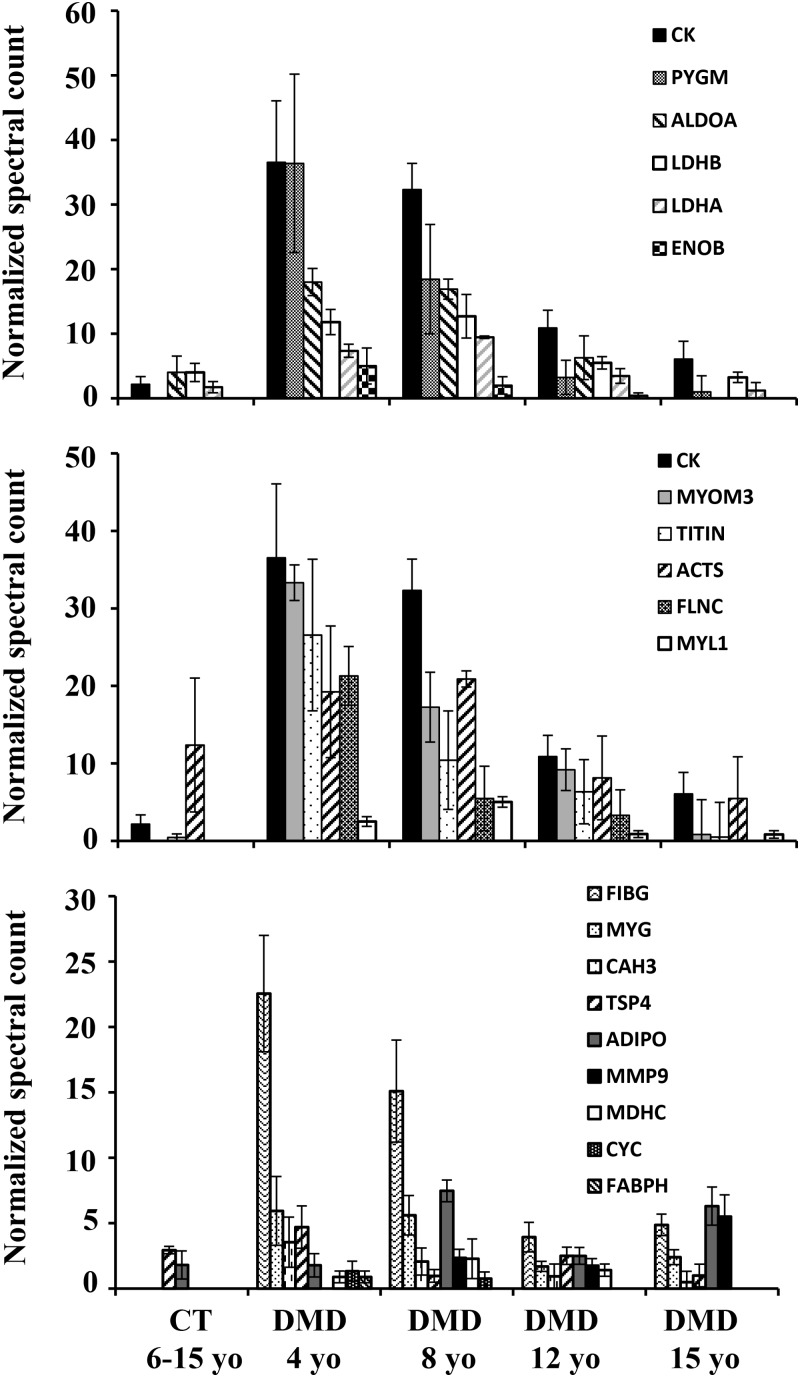
To identify additional biomarkers in DMD patients, label-free proteome profiling was performed on serum samples collected from DMD patients (n = 15) (4–15 years old), and healthy volunteers (n = 3) (6–15 years old). Importantly, of the 23 elevated biomarkers identified in the mdx-23 mouse model, 20 were also found significantly increased, based on spectral count, in sera of DMD patients compared with healthy volunteers (Fig. 5). As in the dystrophin-deficient mouse model, the serum biomarkers identified in DMD patient sera included glycolytic enzymes such as ALDOA, LDHA, LDHB, ENOB and PYGM (Fig. 5A); myofibril proteins such as MYOM3, TITIN, ACTS, FLNC and MYL1 (Fig. 5B) and other muscle-derived protein such as MYG, CAH3, ADIPO, MDHC, CYC, FIBG, TSP4, FABPH and MMP9 (Fig. 5C). All biomarkers, except for ADIPO and MMP9, were highest in younger DMD patients (4–8 years old), then decreased as a function of age in a manner similar to CK while ADIPO and MMP9 increased with age in serum of DMD patients (Fig. 5C).
Figure 5.
Serum levels of candidate biomarkers discovered by label-free proteome profiling in serum samples of DMD patients and healthy control across different age groups. Serum aliquots containing 100 µg total proteins from 12 DMD patients at 4, 8, 12 and 15 years of age (n = 3 per age group) and from 3 healthy controls at 6, 8 and 12 years of age were processed for label-free proteome profiling as described in Materials and Methods. (A) Levels of five glycolytic enzymes in DMD groups and control group. PYGM, glycogen phosphorylase; ALDOA, fructose-bisphosphate aldolase A; LDHB, lactate dehydrogenase B; LDHA, lactate dehydrogenase A; ENOB, beta enolase. (B) Levels of five myofibrillar proteins, in DMD groups and control group. MYOM3, myomesin-3; TITIN, titin; ACTS, alpha skeletal muscle actin; FLNC, filamin-C; MYL1, myosin light chain 1/3 in DMD groups and control group. (C) Levels of nine other muscle-specific proteins in DMD groups and control group. MYG, myoglonin; CAH3, carbonic anhydrase III; ADIPO, adiponectin; TSP4, thrombospondin-4; MDHC, cytosolic malate dehydrogenase; CYC, somatic cytochrome C; MMP9, matrix metalloproteinase-9; FIBG, fibrinogen gamma chain; FABPH, fatty acid-binding protein heart type. A CK level was added along with glycolytic enzyme in (A) and myofibrillar proteins in (B) for trends comparison. It was omitted in the other muscle-specific protein group in (C) due to the large dynamic range between the levels of CK and this class of proteins. Error bars represent standard error obtained from triplicate biological sample.
DISCUSSION
Differential proteome profiling of sera samples of dystrophin-deficient mouse models and DMD patients were conducted using both the SILAM mouse strategy and label-free proteome profiling strategy, respectively. This enabled identification of 20 serum biomarkers that were concordant in sera samples of two independent dystrophin-deficient mouse models (e.g. mdx-23 and mdx-52) and DMD patients. Furthermore, analysis of serum samples across different age groups revealed correlation between the serum levels of these biomarkers and disease stage in both dystrophin-deficient mouse models and in DMD patients.
Most, if not all, the biomarkers identified in the serum of the mdx mouse models were muscle derived. These biomarkers were undetectable in serum of pre-symptomatic young mdx mice (e.g. 7 days of age) and then acutely increased in 3-week-old mdx mouse serum, correlating with the onset of muscle necrosis (35), thereby confirming their validity as a muscle disease state biomarker panel. Interestingly, some proteins including ACTS, ALDOA, GAPDH, MDHC, TSP4, MYG and FABPH were found to be elevated in the serum of mdx mice at 7 days of age, even before the onset muscle necrosis. Presence of these proteins in newborn mdx sera is intriguing. At this time, we could only speculate that this is due to disturbance of an integral mechanism, perhaps via compromised control of permeability of the sarcolemma or exosomal shedding, rather than leakage from catastrophically damaged necrotic muscle fibers. Nevertheless, according to our experience, careful handling is required when collecting blood from newborn mice especially for biomarkers studies. We compared decapitation and heart puncture collection methods using healthy wild-type newborn mice and found that decapitation resulted in contamination with several muscle-derived proteins such as CK and several other glycolytic enzymes. In our hands, the heart puncture method was preferred, giving minimal or no contamination, and was therefore used to collect all mouse sera samples in this study.
These pre-necrotic biomarkers further increased in the blood of mdx mice at 3 weeks of age (necrotic phase). In general, the majority of biomarkers, especially glycolytic enzymes, myofibrillar proteins and other muscle-derived enzymes, acutely increased in serum of mdx mice during the necrotic phase. In DMD patients, where there is no marked separation between pre-necrotic and necrotic phases, this panel of biomarkers was detected in both young and old DMD patients (4–15 years old in this study) with the highest levels seen in 4-year-old DMD patients. In a future study, it would be interesting to examine the level of these biomarkers in newborn DMD patients to determine their value in neonatal screening.
Overall in this study, we identified concordant biomarkers associated with dystrophinopathy between two independent mdx mouse models and DMD patients, further confirming the validity and specificity of this biomarker panel. These mouse/human shared biomarkers can be classified into three categories as described below.
The first category of biomarkers consisted of six muscle-derived proteins (CK, CAH3, MYG, MDHC, FABPH and CYC) and three proteins of extracellular origin (ADIPO, FIBG and TPS4). CK, MYG and CAH3 are well-known DMD markers and were identified long ago to be elevated in sera of DMD patients (26,36,37), while MDHC, TSP4, FABH, CYC, FIBG and ADIPO are relatively novel. As a proof of principle, CK was undetectable in the pre-necrotic phase mdx mice and then acutely increased at 3 weeks of age in agreement with previous studies (38). Its level, however, decreased moderately with age and was still elevated in 7-month-old mdx mice while in DMD patients it decreased sharply with age. Similarly, the level of CAH3 was also historically suggested as a serum biomarker for muscular dystrophy (37) but was not revisited. The CAH3 serum level correlated well with that of CK and also decreased with age in DMD patients.
FABPH, heart type fatty acid-binding protein, has been previously used as a biomarker to monitor the cardiac status in DMD and BMD patients (39) and its level correlated well with that of CK. Even though this protein is named heart type FABPH, it may actually be of skeletal muscle orgin since it was found highly elevated in younger mdx mice and DMD patients whose cardiac muscle should be still healthy. Similarly, CYC was found elevated in 3 weeks old mdx mice and DMD boys and it release into blood circulation could be the result of increased apoptosis due to inflammation and necrosis.
FIBG was also reported to be elevated in serum of DMD patients relative to healthy controls (40) and in mdx mice relative to wild-type mice (41). Here, we further confirm its increased level in serum of both mdx-23 mice and DMD patients relative to their healthy control counterparts. Even though its level decreased with age in DMD patients, it remained relatively elevated in serum of older DMD patients compared with age-matched healthy controls where it was undetectable by mass spectrometry. Fibrinogen is a hexamer composed of two sets of three different subunits (e.g. α, β and γ). It is normally synthesized by liver and is involved in coagulation cascade during blood clot. But it has been also shown to play a major role in inflammation (42). Furthermore, alteration of the inflammatory function of FIBG subunit via missense mutation rescued muscle pathology in the mdx mouse model (43) making it an attractive biomarker and therapeutic target for future investigations.
As for the MDHC, there is only one single publication in 1975 reporting its elevated activity in serum of DMD patients that also decreased with age. At that time, it was not possible to distinguish between mitochondrial isoform (MDHM) and cytosolic isoform (MDHC), on the basis of enzymatic activity. However, the sequence homology between these two isoforms is only 18%, permitting accurate identification by mass spectrometry herein, to reveal MDHC and not MDHM, as the isoform elevated in the serum of dystrophin-deficient mice. Indeed, MDHC was identified with 6 unique tryptic peptides with no overlapping sequence homology with MDHM tryptic peptides. MDHC is highly expressed in skeletal and cardiac muscle (44,45). MDHC is involved in aerobic energy production during muscle contraction by converting malate to oxaloacetate and transporting the resulting NADH equivalent across the mitochondrial membrane (44) and could be a good marker for dystrophinopathies.
TSP4 is an extracellular glycoprotein that is involved in extracellular matrix remodeling. To the best of our knowledge, this is the first time that TSP4 levels have been shown to be elevated in the serum of dystrophin-deficient mouse models, and its increase may be associated with muscle degeneration and regeneration. Indeed, an earlier study by our group showed that expression levels of TSP4 mRNA are significantly increased in the skeletal muscle of DMD and limb-girdle muscular dystrophy patients, compared with normal controls (46). The increased expression of TPS4 in dystrophic muscle is most likely associated with its role in fibrosis and extracellular remodeling, as has been shown in cardiac muscle of the TSP4 knockout mouse model (47). However, the levels of TSP4 were only moderately elevated in serum of younger DMD patients compared with age-matched healthy controls and decreased rapidly with age. Further analyses are needed to confirm its utility as a potential biomarker in DMD patients.
All biomarkers in this first category decreased with age in serum of DMD patients except for ADIPO and MMP9, which actually increased with age. This increase was not related to aging since their levels did not increase in healthy controls with 6, 8 and 15 years of age. ADIPO is a hormone that is exclusively secreted by adipose tissue (48). Thus, its increase in the sera of older DMD patients may reflect the progressive replacement of muscle by fat that is often seen in older DMD patients (49). MMP9, on the other hand, is an extracellular protease and has been shown to be involved in extracellular matrix remodeling, inflammation and fibrosis in a number of diseases, including DMD (50,51). In this study, increased levels of MMP9 in blood circulation of DMD patients became more apparent in older patients. This finding is an agreement with previous study where it has been shown that serum levels of MMP9 increased with age in DMD patients and correlated very well with disease severity, making it an attractive candidate biomarker to monitor disease progression and perhaps response to therapies (31). The fact that MMP9 was not detected or quantified in serum of the mdx mouse model could relate to the less inflammatory status of muscle tissue seen in the mdx mouse relative to DMD patients. Nevertheless, MMP9 was found increased in mdx mouse muscle and it is believed to be involved in muscle pathogenesis (50).
Other biomarkers previously identified in sera samples of mdx mouse and that could belong to this first class of biomarkers included an N-terminal peptide fragment of coagulation factor XIIIa which was reported to be elevated in serum of the mdx mouse relative to the wild-type mouse (52) and the LIFR which was reported to be decreased in serum of mdx mouse relative to the wild-type mouse (41). While the decreased level of LIFR was confirmed in both mdx-23 and mdx 52 mouse models in this study, we were not able to confirm coagulation factor XIIIa perhaps due to the fact that SDS–PAGE was used and low molecular mass fragments were overlooked.
The second category of biomarkers found elevated in serum of mdx and DMD patients consisted of glycolytic enzymes and included PYGM, ALDOA, LDHA, LDHB, ENOB and PGAM2. These enzymes are known to be highly abundant in skeletal muscle and their release into blood seems to correlate with that of CK. The enzyme activities of LDHs, ALDOA, ENOB and PGAM2 were previously reported to be significantly elevated in sera samples of DMD patients but never revisited for further evaluation (26,53,54). Furthermore, the level of PGAM2 was found to be elevated in the serum of DMD patients as early as the fetal stage, thus making it a candidate biomarker for neonatal screening (53). In this study, we identified an additional glycolytic enzyme biomarker, PYGM, whose level was as high as CK in younger DMD boys and then decreased with age. PYGM is one of the major glycolytic enzymes in skeletal muscle that breaks down glycogen to monomeric glucose molecules and controls glycogen metabolism (55) adding it to the panel of glycolytic enzyme biomarkers for DMD.
While the level of these glycolytic enzymes decreased with age in DMD patients, the levels remained elevated in aging mdx mouse, up to 7 months of age. The difference seen between mdx mouse models and DMD patients is perhaps attributable to the maintenance of muscle mass in mdx mice over the age range relative to human patients. In humans, the decrease correlates with a drastic loss of muscle mass, and to some extent reduced exercise because of severely impaired motor function. In the murine model, the maintained high level of these biomarkers is perhaps due to the successful and subsiding degeneration/regeneration cycles (56,57).
The third category of biomarkers consisted of relatively novel myofibrillar proteins and included MYL1, TITIN, MYOM3, FLNC and ACTS. These proteins were undetectable in serum of newborn mdx-23 mice, but rose acutely in the serum of 3-week-old mdx-23 mice. Except for MYOM3, these myofibrillar biomarkers remained elevated in serum of older mdx mice studied herein. Detection of these myofibrillar proteins in serum of 7 months old mdx mice was intriguing and suggests continuous disease activity in old mdx mouse model. However, further serum analysis of older mdx mice as well as age-matched wild-type healthy mice is needed to confirm this hypothesis and to rule out aging effect.
These biomarkers were also confirmed by mass spectrometry analysis as being elevated in serum of DMD patients compared with age-matched healthy subjects. However, their serum levels decreased rapidly with age in DMD patients. The high molecular mass myofibril proteins such as TITIN (3816 kDa molecular mass), FLNC (291 kDa molecular mass) and MYOM3 (161 kDa molecular mass) were all detected as fragments in the serum of dystrophin-deficient mice and DMD patients, with molecular masses ranging between 198 and 260 kDa for TITIN, 110 and 160 kDa for FLNC and around 96 kDa for MYOM3. Their detection in blood circulation seems likely to indicate severe skeletal muscle damage. Both TITIN and MYOM3 are components of the sarcomeres and interact with myosins and MYL1 in striated muscle (58), while FLNC is an actin cross linker protein and is located in the sarcolemma as well as in the sarcomeres (59). The release of these myofibrillar protein fragments into blood circulation is likely a result of proteolytic activity during muscle inflammation and necrosis. This hypothesis is further supported by the fact that their release into the bloodstream was not detected in the mdx mice until 3 weeks of age. In DMD patients however, TITIN and MYOM3 were already present in the serum of DMD patients at 4 years old, probably also being a reflection of the florid inflammation and necrosis at this age (60,61). This is further supported by an earlier study performed on skeletal muscle tissue that revealed dramatic degradation of the titin protein in DMD patients after 5 years of age (62) and is in agreement with a more recent study where it has been shown that specific N-terminal and C-terminal fragments of TITIN were detected in urine samples of both the mdx mouse model and DMD patients (63). In this recent study, it was suggested that TITIN could have been cleaved at its N-terminal and C-terminal extremities by calpains and matrix metalloproteinases during muscle necrosis. However, these low molecular mass TITIN fragments were not detected in DMD sera in this study, probably due to their rapid renal clearance leaving the large middle TITIN fragment in blood circulation. ACTS and MYL1, on the other hand, were detected by mass spectrometry in gel bands corresponding to expected molecular masses of 40 and 20 kDa, respectively, thus indicating their release as intact proteins. This is in agreement with our previous in vitro study showing that cultured dystrophin-deficient myotubes release intact MYL1 via vesicular process into the cultured secretome (64). However, further investigations are needed to verify if MYL1 is also released via vesicles in vivo, by performing proteome profiling on exososomal fractions prepared from serum of dystrophin-deficient mouse models and DMD patients.
To detect these biomarkers in serum of mdx mouse model and DMD patients required further fractionation of serum samples by SDS–PAGE to separate highly abundant proteins such as albumin, transferrin and IgGs from low abundant proteins. Nevertheless, it is possible that other low abundant and valuable biomarkers were still overlooked due the large dynamic range and complex nature of the serum proteome. Thus, new methodologies are needed to perform in-depth serum proteome profiling and discover additional novel biomarkers in the future.
Conclusion
This study identified a valuable panel of serological biomarkers associated with dystrophin deficiency and age-related muscle pathology in two independent dystrophin-deficient mouse models: the naturally occurring mdx-23 and genetically engineered mdx-52. Many of these biomarkers were confirmed in sera of DMD patients, strongly supporting their validity as markers for dystrophinopathies. In our cross-sectional DMD study, all identified biomarkers declined with age along with CK; however, we argue that the behavior of a panel of proteins of different cellular functions better reflects the range of underlying pathological processes. According to this interpretation, we would expect individual biomarkers from this panel will respond differently to different therapies. So, instead of monitoring a single biomarker at a time, we propose, as the ideal, a protocol to monitor a panel of biomarkers simultaneously. In the future, we will pursue this hypothesis, making use of this DMD biomarker panel to monitor and predict clinical outcomes. To this end, we will evaluate and test these biomarkers for their value in monitoring disease progression and response to therapies in DMD in longitudinal prospective studies.
MATERIALS AND METHODS
Animal experiments
Mouse strains used in this study included the spontaneous dystrophin-deficient allele (mdx-23; splice site mutation in exon 23) on C57BL/10 background (C57BL/10ScSn-Dmdmdx/J) and wild-type C57BL/10 mice, and the induced deletion of exon 52 allele (mdx-52) on the C57BL/6 background (65) and corresponding wild-type background control (C57BL/6). Mdx-23 and wild-type background strains were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). Mdx-52 mice were provided by Dr Shin'ichi Takeda, NCNP, Kokaira, Japan, and bred and maintained at Children's National Medical Center. All mice were handled under an approved protocol according to the Institutional Animal Care and Committee guidelines at the Children's National Medical Center.
Generating 13C6-Lys and 15N-labeled mouse colonies
A fully labeled 13C6-Lys BL6 mouse colony with more than 96% labeling efficiency was previously generated in our laboratory (34) using mouse-Express food containing ‘heavy’ L-lysine (13C6, 99%) from Cambridge Isotope Laboratories (Andover, MA, USA). Serum from these mice was collected, frozen at −80°C and used for subsequent experiments.
15N metabolic labeled mice were generated by feeding newly weaned 3-week-old wild-type C57/BL6 mice 15N (98% +) enriched Spirulina food (Cambridge Isotope Laboratories, Andover, MA, USA). The mice-tolerated Spurilina food well, showing no difference in body weight and overall health from mice that were kept on standard laboratory chow (Supplementary Material, Fig. S2A). Labeling efficiency was monitored via analysis of the urinary proteome every week from the start of the diet. Sufficient urinary proteome labeling with 15N (more than 95% labeling efficiency) was reached by 12 weeks of feeding (Supplementary Material, Fig. S2B). By 12 weeks, all organs and body fluids showed stable isotope labeling efficiencies of proteins greater than 90%. An example of labeling efficiency of the serum proteome is shown in Supplementary Material, Figure S2C. All identified serum proteins (n = 153) were enriched by 92–100% with 15N isotope. Sera collected from each 15N-labled mouse were pooled to make one 15N serum labeled stock which was aliquoted in small volumes and stored at −80°C for subsequent use.
Mouse serum collection
Each mouse used in this study was anesthetized by intraperitoneal injection of ketamine and xylazine (80 mg/kg ketamine and 10 mg/kg xylazine). Fully anesthetized animals (no response to toe or ear pinch, reduced heart rate and rate of breathing) were opened along the centerline of the chest, from the caudal end of the rib cage to the clavicle with small dissecting scissors. Ribs were then reflected laterally and stabilized with a retractor. Blood was immediately collected via heart puncture into an Eppendorf tube and allowed to clot at room temperature prior to centrifugation and serum collection. Typically 200 µl of blood could be collected per mouse aged from 3 weeks to 7 months, while only 10–20 µl of blood could be collected per mouse from 2 to 7 days old. This yielded about 80 µl of serum from each older mouse and about 10 µl of serum per young mouse. Serum samples including 15N and 13C6-Lys labeled sera were assayed for protein concentration using BCA assay (Thermo Scientific, Rockford, IL, USA). Each sample was then aliquoted in 10 µl volumes and stored at −80°C for proteome profiling as described below. After blood collection, all other organs were harvested and flash-frozen in liquid nitrogen-chilled isopentane for other studies.
Serum sample preparation and mass spectrometry analysis
The overall experimental workflow is shown in Supplementary Material, Figure S3. Initial biomarker discovery was performed on serum of 3-week-old mdx-52 mice (n = 3) and age-matched BL6 mice (n = 3). Serum aliquots containing 50 µg total protein from each of mouse were spiked with an equal amount of serum protein from SILAM 13C6-Lys labeled BL6 mouse serum stock.
Independent biomarker discovery was performed, using 15N labeled mouse strategy, on the naturally occurring mdx-23 mouse (n = 3) and its wild-type control BL10 mice (n = 3). Serum samples, containing 50 µg of total protein, from 3 months unlabeled mdx-23 mice (n = 3) and its age-matched wild-type BL10 mice (n = 3) were spiked with 15N labeled mouse serum stock. Unlike 13C6-Lys labeled mouse strategy which permits quantification of Lys containing peptides only, the 15N labeling strategy enables quantification of every tryptic peptide.
To test whether the serum levels of the identified biomarkers are associated with age-dependent muscle pathogenesis in mdx-23 mice, a longitudinal study was performed on serum samples of mdx-23 mice at 7 days, 3 weeks, 2 months, 4 months and 7 months of age (n = 3 per age group). Serum samples from 7-day-, 5-week- and 2-month-old BL10 mice were used as controls. Each serum sample containing 50 µg total protein was spiked with 15N-labeled BL6 serum aliquots containing 25 µg total proteins. We have chosen to spike at 2:1 ratio to minimize the usage of 15N labeled serum stock.
Each spiked sample was further fractionated by SDS–PAGE using 4–12% Bis Tris HCl pre-cast gels (Invitrogen Life Technologies, Grand Island, NY, USA). The gel was then stained with Bio-Safe Coomassie (Bio-Rad, Hercules, CA, USA) and each lane was cut into 20–30 bands. Each band was then in-gel digested using trypsin (Promega, Madison, WI, USA), and the resulting peptides were extracted and analyzed by nano-LC-MS/MS as described below.
Peptide mixtures from each band were resuspended in 10 µl of 0.1% formic acid and loaded (6 µl) via an auto-sampler onto a Symmetry C18 trap column (5 µm, 300 µm inner diameter × 23 mm, Waters Milford, MA, USA). The trapped peptides were washed for 10 min with 0.1% formic acid at a flow rate of 10 µl/min and then eluted on a C18 reversed-phase column (3.5 µm, 75 µm × 15 cm, LC Packings, Sunnyvale, CA, USA) at a flow rate of 250 nl/min using a Nano-HPLC system from Eksigent (Dublin, CA, USA) and a mobile phase consisting of water with 0.1% formic acid (A) and 90% acetonitrile (B). A 65-min linear gradient from 5 to 40% B was employed. Eluted peptides were introduced into the mass spectrometer via a 10 µm silica tip (New Objective Inc., Ringoes, NJ, USA) adapted to a nano-electrospray source (Thermo Fisher Scientific). The spray voltage was set at 1.2 kV, and the heated capillary at 200°C. The LTQ-Orbitrap-XL (ThermoFisher Scientific) was operated in the data-dependent mode with dynamic exclusion, in which one cycle of experiments consisted of a full MS survey scan in the Orbitrap (300–2000 m/z, at 30 000 resolution) and five subsequent MS/MS scans in the LTQ of the most intense peaks, using collision-induced dissociation with helium gas and normalized collision energy value set at 35%.
Database search and SILAC ratio measurement
Protein identification and quantification were performed using the IP2 software version 1.01 (Integrated Proteomics Applications, San Diego, CA, USA). Files from each lane were searched against the forward and reverse Uniprot mouse database (UniProt release 15.15, January 2013, 16 580 forward entries) for partially tryptic peptides, allowing two missed cleavages and the possible modifications of Met residue by oxidation (+15.99492 Da), of Lys residue (+6.0204 Da) in the case of 13C6-Lys spike-in strategy and nitrogen atom (+0.98 Da) in the case of 15N spike-in strategy. IP2 uses the Sequest search engine version 2010 (06 10 13 1836). The mass tolerance was set at ±30 ppm (13C6-Lys) or ±300 ppm (15N) for MS and 1.5 Da for MS/MS. Data were filtered by setting the protein false discovery rate at <1%. Only proteins that were identified by at least two unique peptides were retained for further quantitative analysis. Census software (version 1.77), built into the IP2 platform was used to determine the ratios of light to heavy peptide pairs using an extracted chromatogram approach. Quantitative data were filtered based on a determinant value of 0.5 and an outlier P-value of 0.1.
Label-free proteome profiling of serum samples of DMD and age-matched healthy controls
Serum samples from 4, 8, 12 and 15 years old DMD patients (n = 3 per age group) and from 3 healthy control subjects ranging in age from 6 to 15 years were collected through the Cooperative International Neuromuscular Research Group (CINRG) network according to an approved institutional IRB protocol and used for an independent biomarker discovery experiment for human subjects. Aliquots containing 100 µg total proteins from each serum sample were further separated by SDS–PAGE and in-gel digestion as described above. The resulting peptides were extracted and analyzed by nano-LC-MS/MS using ultra-high performance liquid chromatography system, connected to a Q-Exactive Hybrid Quadrupole Orbitrap mass spectrometer (Thermo Fisher Scientific) in data-dependent top 10 mode. The generated raw files were uploaded for protein identification using the Proteome Discoverer software version 1.4.0.288 (ThermoFisher Scientific) and searched against the Uniprot human database (UniProt release 2013_07), partially tryptic peptides, 2 missed cleavages, potential modification of oxidized methionine (+15.995 Da), peptide mass tolerance of ±10 ppm and fragment tolerance of ±0.02 Da. Results were filtered based on the following: false discovery rate <1%, peptide probability >0.01, Delta Cn >0.1 and Xcorr > 1.5, 2, 2.25, 2.5 for z 1, 2, 3, 4. Identified proteins and peptides were exported as PepXml format and directly uploaded into ProteoIQ software version 2.3.07 (NuSep, Bogart, GA, USA) for spectral count and label-free proteome profiling. Spectral counts were normalized based on total protein spectral count.
Western blot analysis
Aliquots (containing 30 µg total protein each) from same DMD and control serum samples above with an additional three controls were further separated by SDS–PAGE as described above. Replicate gels were run to test different antibodies. Separated proteins were transferred to nitrocellulose membranes (Millipore, Billerica, MA, USA) at 300 mA for 90 min at room temperature. Membranes were blocked in TBS-T (20 mm Tris, 500 mm NaCl, pH 7.5, with 0.1% Tween 20) supplemented with 5% non-fat dry milk (Bio-Rad) for 1 h at room temperature. The membranes were then incubated overnight at 4°C with rabbit antibodies raised against the human prophosphoglycerate mutase-2 (Origene, Rockville, MD, USA), glycogen phosphorylase (Origene), myomesin-3 (Sigma Aldrich, St Louis, MO, USA) and mouse antibodies raised against human skeletal muscle myosin (Sigma Aldrich) and human filamin-C (Origene).
Each antibody was diluted in TBS-T-5% milk according to the manufacturer instruction and used to react with membrane trans-blots. Membranes were washed three times (for 10 min each time) in TBS-T and incubated with goat anti-rabbit or rabbit anti-mouse secondary antibodies (Dako, Carpinteria, CA, USA) conjugated to horseradish peroxidase (1:3000 in TBS-T-5% milk) for 1 h at room temperature. The protein bands were revealed with ECL chemi-luminescence substrate (Amersham Biosciences). For quantification, the X-ray films were scanned, and densitometry analysis was carried out using a Bio-Rad GS-800 calibrated densitometer running Quantity One software (Bio-Rad). Ratios of the optical density of each specific protein to the total loaded proteins.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health (R01AR062380 to Y.H. and C.M.), partially by the following National Institutes of Health core grants (P50AR060836, R24HD050846, P30HD040677, UL1TR000075 to Y.H., E.H. and K.N.) and the following philanthropic funds (Board of Visitors foundation and the Lynn and Doug Parsons foundation).
Supplementary Material
ACKNOWLEDGEMENTS
Authors would like to thank Lauren Hache and Zoe Sund for their help coordinating initial patient's serum collection with CINRG sites and Dr Paula Clemens for her suggestions and guidance. Authors are also thankful to DMD patients and their families for their time and efforts.
REFERENCES
- 1.Danieli G.A., Mostacciuolo M.L., Bonfante A., Angelini C. Duchenne muscular dystrophy. A population study. Hum. Genet. 1977;35:225–231. doi: 10.1007/BF00393974. [DOI] [PubMed] [Google Scholar]
- 2.Bushby K., Finkel R., Birnkrant D.J., Case L.E., Clemens P.R., Cripe L., Kaul A., Kinnett K., McDonald C., Pandya S., et al. Diagnosis and management of Duchenne muscular dystrophy, part 1: diagnosis, and pharmacological and psychosocial management. Lancet Neurol. 2009;9:77–93. doi: 10.1016/S1474-4422(09)70271-6. [DOI] [PubMed] [Google Scholar]
- 3.Griggs R.C., Moxley R.T., III, Mendell J.R., Fenichel G.M., Brooke M.H., Pestronk A., Miller J.P. Prednisone in Duchenne dystrophy. A randomized, controlled trial defining the time course and dose response. Clinical Investigation of Duchenne Dystrophy Group. Arch. Neurol. 1991;48:383–388. doi: 10.1001/archneur.1991.00530160047012. [DOI] [PubMed] [Google Scholar]
- 4.Manzur A.Y., Kinali M., Muntoni F. Update on the management of Duchenne muscular dystrophy. Arch. Dis. Child. 2008;93:986–990. doi: 10.1136/adc.2007.118141. [DOI] [PubMed] [Google Scholar]
- 5.Manzur A.Y., Kuntzer T., Pike M., Swan A. Glucocorticoid corticosteroids for Duchenne muscular dystrophy. Cochrane Database Syst. Rev. 2008 doi: 10.1002/14651858.CD003725.pub3. CD003725. [DOI] [PubMed] [Google Scholar]
- 6.Angelini C., Pegoraro E., Turella E., Intino M.T., Pini A., Costa C. Deflazacort in Duchenne dystrophy: study of long-term effect. Muscle Nerve. 1994;17:386–391. doi: 10.1002/mus.880170405. [DOI] [PubMed] [Google Scholar]
- 7.DeSilva S., Drachman D.B., Mellits D., Kuncl R.W. Prednisone treatment in Duchenne muscular dystrophy. Long-term Benefit. Arch. Neurol. 1987;44:818–822. doi: 10.1001/archneur.1987.00520200022012. [DOI] [PubMed] [Google Scholar]
- 8.McAdam L.C., Mayo A.L., Alman B.A., Biggar W.D. The Canadian experience with long-term deflazacort treatment in Duchenne muscular dystrophy. Acta Myol. 2012;31:16–20. [PMC free article] [PubMed] [Google Scholar]
- 9.Ricotti V., Ridout D.A., Scott E., Quinlivan R., Robb S.A., Manzur A.Y., Muntoni F. Long-term benefits and adverse effects of intermittent versus daily glucocorticoids in boys with Duchenne muscular dystrophy. J. Neurol. Neurosurg. Psychiatry. 2013;84:698–705. doi: 10.1136/jnnp-2012-303902. [DOI] [PubMed] [Google Scholar]
- 10.Aartsma-Rus A., Bremmer-Bout M., Janson A.A., den Dunnen J.T., van Ommen G.J., van Deutekom J.C. Targeted exon skipping as a potential gene correction therapy for Duchenne muscular dystrophy. Neuromuscul. Disord. 2002;12(Suppl. 1):S71–S77. doi: 10.1016/s0960-8966(02)00086-x. [DOI] [PubMed] [Google Scholar]
- 11.Finkel R.S. Read-through strategies for suppression of nonsense mutations in Duchenne/Becker muscular dystrophy: aminoglycosides and ataluren (PTC124) J. Child. Neurol. 2010;25:1158–1164. doi: 10.1177/0883073810371129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson S.F., Crosbie R.H., Miceli M.C., Spencer M.J. Emerging genetic therapies to treat Duchenne muscular dystrophy. Curr. Opin. Neurol. 2009;22:532–538. doi: 10.1097/WCO.0b013e32832fd487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pichavant C., Aartsma-Rus A., Clemens P.R., Davies K.E., Dickson G., Takeda S., Wilton S.D., Wolff J.A., Wooddell C.I., Xiao X., et al. Current status of pharmaceutical and genetic therapeutic approaches to treat DMD. Mol. Ther. 2011;19:830–840. doi: 10.1038/mt.2011.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fairclough R.J., Wood M.J., Davies K.E. Therapy for Duchenne muscular dystrophy: renewed optimism from genetic approaches. Nat. Rev. Genet. 2013;14:373–378. doi: 10.1038/nrg3460. [DOI] [PubMed] [Google Scholar]
- 15.Jarmin S., Kymalainen H., Popplewell L., Dickson G. New developments in the use of gene therapy to treat Duchenne muscular dystrophy. Expert Opin. Biol. Ther. 2014;14:209–230. doi: 10.1517/14712598.2014.866087. [DOI] [PubMed] [Google Scholar]
- 16.Partridge T.A. Impending therapies for Duchenne muscular dystrophy. Curr. Opin. Neurol. 2011;24:415–422. doi: 10.1097/WCO.0b013e32834aa3f1. [DOI] [PubMed] [Google Scholar]
- 17.Cirak S., Arechavala-Gomeza V., Guglieri M., Feng L., Torelli S., Anthony K., Abbs S., Garralda M.E., Bourke J., Wells D.J., et al. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study. Lancet. 2011;378:595–605. doi: 10.1016/S0140-6736(11)60756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goemans N.M., Tulinius M., van den Akker J.T., Burm B.E., Ekhart P.F., Heuvelmans N., Holling T., Janson A.A., Platenburg G.J., Sipkens J.A., et al. Systemic administration of PRO051 in Duchenne's muscular dystrophy. N. Engl. J. Med. 2011;364:1513–1522. doi: 10.1056/NEJMoa1011367. [DOI] [PubMed] [Google Scholar]
- 19.Mendell J.R., Rodino-Klapac L.R., Sahenk Z., Roush K., Bird L., Lowes L.P., Alfano L., Gomez A.M., Lewis S., Kota J., et al. Eteplirsen for the treatment of Duchenne muscular dystrophy. Ann. Neurol. 2013;74:637–647. doi: 10.1002/ana.23982. [DOI] [PubMed] [Google Scholar]
- 20.Mendell J.R., Rodino-Klapac L., Sahenk Z., Malik V., Kaspar B.K., Walker C.M., Clark K.R. Gene therapy for muscular dystrophy: lessons learned and path forward. Neurosci. Lett. 2007;527:90–99. doi: 10.1016/j.neulet.2012.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heier C.R., Damsker J.M., Yu Q., Dillingham B.C., Huynh T., Van der Meulen J.H., Sali A., Miller B.K., Phadke A., Scheffer L., et al. VBP15, a novel anti-inflammatory and membrane-stabilizer, improves muscular dystrophy without side effects. EMBO Mol. Med. 2013;5:1569–1585. doi: 10.1002/emmm.201302621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDonald C.M., Henricson E.K., Han J.J., Abresch R.T., Nicorici A., Elfring G.L., Atkinson L., Reha A., Hirawat S., Miller L.L. The 6-minute walk test as a new outcome measure in Duchenne muscular dystrophy. Muscle Nerve. 2009;41:500–510. doi: 10.1002/mus.21544. [DOI] [PubMed] [Google Scholar]
- 23.Mazzone E.S., Messina S., Vasco G., Main M., Eagle M., D'Amico A., Doglio L., Politano L., Cavallaro F., Frosini S., et al. Reliability of the North Star Ambulatory Assessment in a multicentric setting. Neuromuscul. Disord. 2009;19:458–461. doi: 10.1016/j.nmd.2009.06.368. [DOI] [PubMed] [Google Scholar]
- 24.Scott E., Eagle M., Mayhew A., Freeman J., Main M., Sheehan J., Manzur A., Muntoni F. Development of a functional assessment scale for ambulatory boys with Duchenne muscular dystrophy. Physiother. Res. Int. 2012;17:101–109. doi: 10.1002/pri.520. [DOI] [PubMed] [Google Scholar]
- 25.Escolar D.M., Hache L.P., Clemens P.R., Cnaan A., McDonald C.M., Viswanathan V., Kornberg A.J., Bertorini T.E., Nevo Y., Lotze T., et al. Randomized, blinded trial of weekend vs daily prednisone in Duchenne muscular dystrophy. Neurology. 2011;77:444–452. doi: 10.1212/WNL.0b013e318227b164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okinaka S., Kumagai H., Ebashi S., Sugita H., Momoi H., Toyokura Y., Fujie Y. Serum creatine phosphokinase. Activity in progressive muscular dystrophy and neuromuscular diseases. Arch. Neurol. 1961;4:520–525. doi: 10.1001/archneur.1961.00450110050006. [DOI] [PubMed] [Google Scholar]
- 27.Ohlendieck K. Proteomic identification of biomarkers of skeletal muscle disorders. Biomark. Med. 2013;7:169–186. doi: 10.2217/bmm.12.96. [DOI] [PubMed] [Google Scholar]
- 28.Cacchiarelli D., Legnini I., Martone J., Cazzella V., D'Amico A., Bertini E., Bozzoni I. miRNAs as serum biomarkers for Duchenne muscular dystrophy. EMBO Mol. Med. 2011;3:258–265. doi: 10.1002/emmm.201100133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizuno H., Nakamura A., Aoki Y., Ito N., Kishi S., Yamamoto K., Sekiguchi M., Takeda S., Hashido K. Identification of muscle-specific microRNAs in serum of muscular dystrophy animal models: promising novel blood-based markers for muscular dystrophy. PLoS ONE. 2011;6:e18388. doi: 10.1371/journal.pone.0018388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaharieva I.T., Calissano M., Scoto M., Preston M., Cirak S., Feng L., Collins J., Kole R., Guglieri M., Straub V., et al. Dystromirs as serum biomarkers for monitoring the disease severity in Duchenne muscular Dystrophy. PLoS ONE. 2013;8:e80263. doi: 10.1371/journal.pone.0080263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nadarajah V.D., van Putten M., Chaouch A., Garrood P., Straub V., Lochmuller H., Ginjaar H.B., Aartsma-Rus A.M., van Ommen G.J., den Dunnen J.T., et al. Serum matrix metalloproteinase-9 (MMP-9) as a biomarker for monitoring disease progression in Duchenne muscular dystrophy (DMD) Neuromuscul. Disord. 2011;21:569–578. doi: 10.1016/j.nmd.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 32.Cynthia Martin F., Hiller M., Spitali P., Oonk S., Dalebout H., Palmblad M., Chaouch A., Guglieri M., Straub V., Lochmuller H., et al. Fibronectin is a serum biomarker for Duchenne muscular dystrophy. Proteomics Clin. Appl. 2014;8:269–278. doi: 10.1002/prca.201300072. [DOI] [PubMed] [Google Scholar]
- 33.McClatchy D.B., Liao L., Park S.K., Xu T., Lu B., Yates Iii J.R. Differential proteomic analysis of mammalian tissues using SILAM. PLoS ONE. 2011;6:e16039. doi: 10.1371/journal.pone.0016039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rayavarapu S., Coley W., Cakir E., Jahnke V., Takeda S., Aoki Y., Grodish-Dressman H., Jaiswal J.K., Hoffman E.P., Brown K.J., et al. Identification of disease specific pathways using in vivo SILAC proteomics in dystrophin deficient mdx mouse. Mol. Cell Proteomics. 2013;12:1061–1073. doi: 10.1074/mcp.M112.023127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cullen M.J., Jaros E. Ultrastructure of the skeletal muscle in the X chromosome-linked dystrophic (mdx) mouse. Comparison with Duchenne muscular dystrophy. Acta Neuropathol. 1988;77:69–81. doi: 10.1007/BF00688245. [DOI] [PubMed] [Google Scholar]
- 36.Nicholson L.V. Serum myoglobin in muscular dystrophy and carrier detection. J. Neurol. Sci. 1981;51:411–426. doi: 10.1016/0022-510x(81)90118-0. [DOI] [PubMed] [Google Scholar]
- 37.Ohta M., Itagaki Y., Itoh N., Hayashi K., Nishitani H., Ohta K. Carbonic anhydrase III in serum in muscular dystrophy and other neurological disorders: relationship with creatine kinase. Clin. Chem. 1991;37:36–39. [PubMed] [Google Scholar]
- 38.McArdle A., Edwards R.H., Jackson M.J. Time course of changes in plasma membrane permeability in the dystrophin-deficient mdx mouse. Muscle Nerve. 1994;17:1378–1384. doi: 10.1002/mus.880171206. [DOI] [PubMed] [Google Scholar]
- 39.Matsumura T., Saito T., Fujimura H., Shinno S. Cardiac troponin I for accurate evaluation of cardiac status in myopathic patients. Brain Dev. 2007;29:496–501. doi: 10.1016/j.braindev.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 40.Saito T., Takenaka M., Miyai I., Yamamoto Y., Matsumura T., Nozaki S., Kang J. Coagulation and fibrinolysis disorder in muscular dystrophy. Muscle Nerve. 2001;24:399–402. doi: 10.1002/1097-4598(200103)24:3<399::aid-mus1012>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 41.Colussi C., Banfi C., Brioschi M., Tremoli E., Straino S., Spallota F., Mai A., Rotili D., Capogrossi M.C., Gaetano C. Proteomic profile of differentially expressed plasma proteins from dystrophic mice and following suberoylanilide hydroxamic acid treatment. Proteomics Clin. Appl. 2010;4:71–83. doi: 10.1002/prca.200900116. [DOI] [PubMed] [Google Scholar]
- 42.Davalos D., Akassoglou K. Fibrinogen as a key regulator of inflammation in disease. Semin. Immunopathol. 2012;34:43–62. doi: 10.1007/s00281-011-0290-8. [DOI] [PubMed] [Google Scholar]
- 43.Vidal B., Ardite E., Suelves M., Ruiz-Bonilla V., Janue A., Flick M.J., Degen J.L., Serrano A.L., Munoz-Canoves P. Amelioration of Duchenne muscular dystrophy in mdx mice by elimination of matrix-associated fibrin-driven inflammation coupled to the alphaMbeta2 leukocyte integrin receptor. Hum. Mol. Genet. 2012;21:1989–2004. doi: 10.1093/hmg/dds012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lo A.S., Liew C.T., Ngai S.M., Tsui S.K., Fung K.P., Lee C.Y., Waye M.M. Developmental regulation and cellular distribution of human cytosolic malate dehydrogenase (MDH1) J. Cell. Biochem. 2005;94:763–773. doi: 10.1002/jcb.20343. [DOI] [PubMed] [Google Scholar]
- 45.Minarik P., Tomaskova N., Kollarova M., Antalik M. Malate dehydrogenases—structure and function. Gen. Physiol. Biophys. 2002;21:257–265. [PubMed] [Google Scholar]
- 46.Chen Y.W., Zhao P., Borup R., Hoffman E.P. Expression profiling in the muscular dystrophies: identification of novel aspects of molecular pathophysiology. J. Cell. Biol. 2000;151:1321–1336. doi: 10.1083/jcb.151.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frolova E.G., Sopko N., Blech L., Popovic Z.B., Li J., Vasanji A., Drumm C., Krukovets I., Jain M.K., Penn M.S., et al. Thrombospondin-4 regulates fibrosis and remodeling of the myocardium in response to pressure overload. FASEB J. 2012;26:2363–2373. doi: 10.1096/fj.11-190728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diez J.J., Iglesias P. The role of the novel adipocyte-derived hormone adiponectin in human disease. Eur. J. Endocrinol. 2003;148:293–300. doi: 10.1530/eje.0.1480293. [DOI] [PubMed] [Google Scholar]
- 49.Hollingsworth K.G., Garrood P., Eagle M., Bushby K., Straub V. Magnetic resonance imaging in Duchenne muscular dystrophy: longitudinal assessment of natural history over 18 months. Muscle Nerve. 2013;48:586–588. doi: 10.1002/mus.23879. [DOI] [PubMed] [Google Scholar]
- 50.Hindi S.M., Shin J., Ogura Y., Li H., Kumar A. Matrix metalloproteinase-9 inhibition improves proliferation and engraftment of myogenic cells in dystrophic muscle of mdx mice. PLoS ONE. 2013;8:e72121. doi: 10.1371/journal.pone.0072121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vandooren J., Van den Steen P.E., Opdenakker G. Biochemistry and molecular biology of gelatinase B or matrix metalloproteinase-9 (MMP-9): the next decade. Crit. Rev. Biochem. Mol. Biol. 2013;48:222–272. doi: 10.3109/10409238.2013.770819. [DOI] [PubMed] [Google Scholar]
- 52.Alagaratnam S., Mertens B.J., Dalebout J.C., Deelder A.M., van Ommen G.J., den Dunnen J.T., 't Hoen P.A. Serum protein profiling in mice: identification of Factor XIIIa as a potential biomarker for muscular dystrophy. Proteomics. 2008;8:1552–1563. doi: 10.1002/pmic.200700857. [DOI] [PubMed] [Google Scholar]
- 53.Chown P.J., Barnard E.A., Barnard P.J., Liu P.K., Carter N.D. Plasma phosphoglycerate mutase as a marker of muscular dystrophy. J. Neurol. Sci. 1984;65:201–210. doi: 10.1016/0022-510x(84)90084-4. [DOI] [PubMed] [Google Scholar]
- 54.Pearson C.M., Chowdhury S.R., Fowler W.M., Jr, Jones M.H., Griffith W.H. Studies of enzymes in serum in muscular dystrophy. II. Diagnostic and prognostic significance in relatives of dystrophic persons. Pediatrics. 1961;28:962–970. [PubMed] [Google Scholar]
- 55.Cohen P., Nimmo G.A., Burchell A., Antoniw J.F. The substrate specificity and regulation of the protein phosphatases involved in the control of glycogen metabolism in mammalian skeletal muscle. Adv. Enzyme Regul. 1977;16:97–119. doi: 10.1016/0065-2571(78)90069-9. [DOI] [PubMed] [Google Scholar]
- 56.Carberry S., Brinkmeier H., Zhang Y., Winkler C.K., Ohlendieck K. Comparative proteomic profiling of soleus, extensor digitorum longus, flexor digitorum brevis and interosseus muscles from the mdx mouse model of Duchenne muscular dystrophy. Int. J. Mol. Med. 2013;32:544–556. doi: 10.3892/ijmm.2013.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Infante J.P., Huszagh V.A. Mechanisms of resistance to pathogenesis in muscular dystrophies. Mol. Cell. Biochem. 1999;195:155–167. doi: 10.1023/a:1006972315739. [DOI] [PubMed] [Google Scholar]
- 58.Schoenauer R., Lange S., Hirschy A., Ehler E., Perriard J.C., Agarkova I. Myomesin 3, a novel structural component of the M-band in striated muscle. J. Mol. Biol. 2008;376:338–351. doi: 10.1016/j.jmb.2007.11.048. [DOI] [PubMed] [Google Scholar]
- 59.Fujita M., Mitsuhashi H., Isogai S., Nakata T., Kawakami A., Nonaka I., Noguchi S., Hayashi Y.K., Nishino I., Kudo A. Filamin C plays an essential role in the maintenance of the structural integrity of cardiac and skeletal muscles, revealed by the medaka mutant zacro. Dev. Biol. 2012;361:79–89. doi: 10.1016/j.ydbio.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 60.Bucciolini Di Sagni M.G., Vannelli Gori G., Oriolo R.A. Structural and ultrastructural changes in the skeletal muscles of patients in the early stages of Duchenne muscular dystrophy and "possible" carriers. Boll. Soc. Ital. Biol. Sper. 1982;58:632–638. [PubMed] [Google Scholar]
- 61.Chen Y.W., Nagaraju K., Bakay M., McIntyre O., Rawat R., Shi R., Hoffman E.P. Early onset of inflammation and later involvement of TGFbeta in Duchenne muscular dystrophy. Neurology. 2005;65:826–834. doi: 10.1212/01.wnl.0000173836.09176.c4. [DOI] [PubMed] [Google Scholar]
- 62.Matsumura K., Shimizu T., Sunada Y., Mannen T., Nonaka I., Kimura S., Maruyama K. Degradation of connectin (titin) in Fukuyama type congenital muscular dystrophy: immunochemical study with monoclonal antibodies. J. Neurol. Sci. 1990;98:155–162. doi: 10.1016/0022-510x(90)90256-m. [DOI] [PubMed] [Google Scholar]
- 63.Rouillon J., Zocevic A., Leger T., Garcia C., Camadro J.M., Udd B., Wong B., Servais L., Voit T., Svinartchouk F. Proteomics profiling of urine reveals specific titin fragments as biomarkers of Duchenne muscular dystrophy. Neuromuscul. Disord. 2014 doi: 10.1016/j.nmd.2014.03.012. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 64.Duguez S., Duddy W., Johnston H., Laine J., Le Bihan M.C., Brown K.J., Bigot A., Hathout Y., Butler-Browne G., Partridge T. Dystrophin deficiency leads to disturbance of LAMP1-vesicle-associated protein secretion. Cell. Mol. Life Sci. 2013;70:2159–2174. doi: 10.1007/s00018-012-1248-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Araki E., Nakamura K., Nakao K., Kameya S., Kobayashi O., Nonaka I., Kobayashi T., Katsuki M. Targeted disruption of exon 52 in the mouse dystrophin gene induced muscle degeneration similar to that observed in Duchenne muscular dystrophy. Biochem. Biophys. Res. Commun. 1997;238:492–497. doi: 10.1006/bbrc.1997.7328. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.