Abstract
Cerebrospinal fluid amyloid-beta 1–42 (Aβ1–42) and phosphorylated Tau at position 181 (pTau181) are biomarkers of Alzheimer's disease (AD). We performed an analysis and meta-analysis of genome-wide association study data on Aβ1–42 and pTau181 in AD dementia patients followed by independent replication. An association was found between Aβ1–42 level and a single-nucleotide polymorphism in SUCLG2 (rs62256378) (P = 2.5×10−12). An interaction between APOE genotype and rs62256378 was detected (P = 9.5 × 10−5), with the strongest effect being observed in APOE-ε4 noncarriers. Clinically, rs62256378 was associated with rate of cognitive decline in AD dementia patients (P = 3.1 × 10−3). Functional microglia experiments showed that SUCLG2 was involved in clearance of Aβ1–42.
INTRODUCTION
Alzheimer's disease (AD) is a complex disorder in which several pathways contribute to pathology and clinical phenotype. Delineation of each pathological pathway and identification of the factors which modulate them are crucial for the development of effective treatment. Information on individual pathological pathways is provided by biomarkers. In AD, cerebrospinal fluid (CSF) levels of Aβ1–42 and phosphorylated Tau (pTau) reflect cerebral amyloid deposition and tau-related neurodegeneration, respectively. However, many of the biological factors that influence these core AD pathways remain elusive.
Research has shown that AD risk genes contribute to CSF marker variance (1–4). However, genes may also exist that affect CSF markers without conferring disease susceptibility. These genes may be promising candidates for modulation of the disease process.
The aim of the present study was to identify genetic factors related to heterogeneity in pathological pathways involved in amyloid deposition and tau-related neurodegeneration—as manifested through CSF Aβ1–42 and pTau levels—and which might influence the clinical course of AD. To achieve this, we performed a four-step investigation involving analysis and meta-analysis of data from genome-wide association studies (GWASs) of these two CSF biomarkers; replication of our top findings in an independent sample; and specific analyses and functional experiments to follow-up promising findings. In contrast to previous GWAS of CSF biomarkers (1,4,5), the present GWAS was restricted to patients with dementia secondary to AD only in order to enrich for genetic effects occurring after disease onset.
RESULTS
Study design
We analyzed GWAS data on CSF biomarkers Aβ1–42 and Tau phosphorilated at position 181 (pTau181) from three independent AD cohorts (Bonn1CSF, n = 113; Bonn2CSF, n = 167; ADNI, n = 83). The resulting data were then combined in a meta-analysis. The three GWAS cohorts are described in detail in Materials and Methods and Table 1. In all three cohorts, the CSF biomarkers Aβ1–42 and pTau181 showed a normal distribution (Supplementary Material, Figs S1 and S2).
Table 1.
Description of the samples used in the present study
| Bonn1CSF | Bonn2CSF | ADNI | ADC | AD dementia patients |
Healthy controls |
|||
|---|---|---|---|---|---|---|---|---|
| Bonn1 : 438 | Bonn2 : 641 | Bonn1 : 1179 | Bonn2 : 1096 | |||||
| N | 113 | 167 | 83 | 515 | 1079 | 2275 | ||
| Age (years ± SD) | 71.6 (±7.4) | 71.8 (±7.7) | 74.8 (±7.5) | 66.4 (±8.8) | 73.3 (±8.8) | 56.7 (±13.8) | ||
| F : M ratio | 1.26 | 1.19 | 1 | 1.05 | 1.7 | 1.3 | ||
| Aβ1–42 (±SD) | 478.3 (±186.5) | 529.2 (±238.1) | 149.4 (±51.1) | 462.5 (±147.9) | NA | NA | NA | NA |
| pTau181 (±SD) | 87.5 (±36.5) | 88.6 (±45.2) | 39.5 (±20.1) | 88.2 (±38.8) | NA | NA | NA | NA |
| Genotype platform | Illumina 610 | Illumina 1M-quad | Illumina 610 | Sequenom | Bonn1 : 438 | Bonn2 : 641 | Bonn1 : 1179 | Bonn2 : 1096 |
| Illumina 610 | Illumina 1M-quad | Illumina HumanHap550k | Illumina 1M-quad | |||||
F : M ratio, female : male ratio; SD, standard deviation. Aβ1–42, amyloid-beta 1–42; pTau181, phosphorylated Tau at position 181; AD, Alzheimer's disease; CSF, cerebrospinal fluid; ADNI, Alzheimer's Disease Neuroimaging Initiative cohort; ADC, Amsterdam Dementia Cohort.
For the replication step, we selected single-nucleotide polymorphisms (SNPs) that showed consistent allele direction across all three GWAS, which modulated either Aβ1–42 or pTau181 with P<5 × 10−6, and which showed consistent allele direction across all studies. These 30 SNPs, together with 10 SNPs representing known AD susceptibility genes, were then genotyped in an independent sample of 515 AD dementia patients (Amsterdam Dementia Cohort, ADC). These 40 SNPs represented a total of 39 loci. Two SNPs showing association with CSF levels of Aβ1–42 in the GWAS meta-analysis were replicated in the ADC, i.e. rs429358 at the APOE locus and rs62256378 at a novel locus containing the SUCLG2 gene. The ADC sample is described in detail in Materials and Methods and Table 1. In the ADC cohort, the CSF biomarkers Aβ1–42 and pTau181 showed a normal distribution (Supplementary Material, Figs. S1 and S2).
Since these two replicated loci both modulated Aβ1–42 levels in the CSF, we explored a possible genetic interaction, since this might suggest a biological link between the two loci. We also investigated whether rs62256378 (i) confers AD risk, using a case–control sample and (ii) has a possible effect on cognitive decline in AD dementia patients, using prospective data on Mini-Mental State Examination (MMSE) scores. The samples used in these two analyses are described in detail in Materials and Methods.
Functional experiments were then performed to elucidate the functional role of SUCGL2 in Aβ1–42 homeostasis. First, the cellular distribution of SUCLG2 was examined in postmortem human brain tissue from individuals who had died from non-neurological disease. Secondly, to follow-up our finding concerning genetic interaction with APOE, the effect of APOE-ε4 carrier status on the protein expression of SUCLG2 was investigated using protein extracts from human AD and control postmortem frontal cortex. Thirdly, the role of SUCLG2 in microglial Aβ1–42 phagocytosis was analyzed by reducing SUCLG2 expression through siRNA approaches.
GWAS meta-analysis
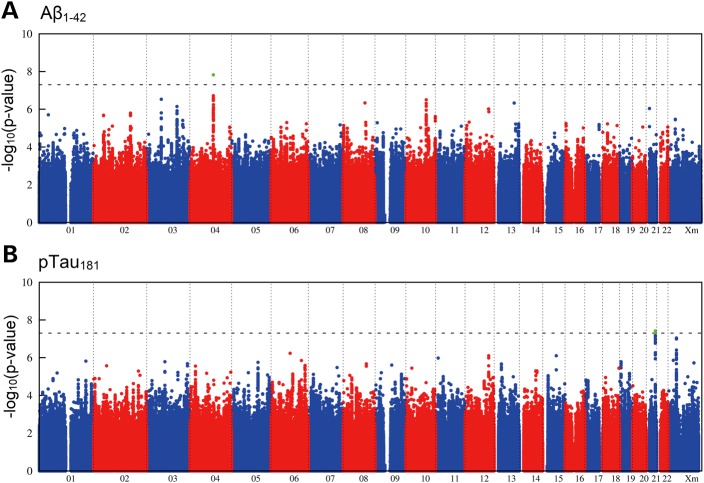
Meta-analysis of the three GWAS yielded genome-wide inflation factors of λ = 1.05 for Aβ1–42 and λ = 1.03 for pTau181 (Supplementary Material, Fig. S3). After λ-adjustment, a genome-wide significant association was observed between APOE-ε4 (rs429358) and Aβ1–42 (P = 1.8 × 10−10), thus confirming published data (Table 2) (1,4,5). A total of 335 SNPs were below the threshold of genome-wide significance but had a promising P-value of <5 × 10−6 (Fig. 1). Of these, 163 showed consistent allele direction across all three GWAS.
Table 2.
Summary of GWAS, replication and overall meta-analysis for SNPs with P < 5 × 10−6 and the same effect direction in all three GWAS samples
| GWAS |
Replication |
Combined analysis |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | rsa | chrb | bpc | EAd/OAe | βf | SEg | dirh | P-adji | rsa | EAd/OAe | MAFj | βk | SEg | Pl | EAd/OAe | dirm | βn | SEo | Pp |
| Aβ1–42 | 2-45978647 | 2 | 45 978 642 | G/GT | −0.382 | 0.08 | −−− | 2.1 × 10−6 | rs6753292q | T/C | 0.251 | 0.01 | 0.08 | 0.91 | G/GT | +− | −0.18 | 0.05 | 1.1 × 10−3 |
| rs374159788 | 2 | 172 759 319 | A/AGT | 0.608 | 0.12 | +++ | 1.6 × 10−6 | rs34737109q | A/G | 0.383 | −0.08 | 0.07 | 0.23 | A/AGT | +− | 0.07 | 0.06 | 0.21 | |
| rs62256378 | 3 | 67 457 033 | A/G | 0.89 | 0.17 | +++ | 3.0 × 10−7 | rs62256378 | A/G | 0.061 | 0.61 | 0.13 | 1.9 × 10−6 | A/G | −−− | 0.71 | 0.10 | 2.5 × 10−12 | |
| rs893703 | 3 | 139 250 649 | G/A | 0.639 | 0.13 | +++ | 1.2 × 10−6 | rs4407373q | T/G | 0.099 | −0.05 | 0.11 | 0.68 | G/A | +− | 0.25 | 0.08 | 3.2 × 10−3 | |
| rs12493397 | 3 | 165 284 889 | A/G | −0.422 | 0.09 | −−− | 3.9 × 10−6 | rs2222464q | C/T | 0.274 | −0.02 | 0.07 | 0.76 | A/G | −−− | −0.18 | 0.06 | 1.2 × 10−3 | |
| rs388575 | 4 | 107 973 286 | C/T | −0.518 | 0.09 | −−− | 1.5 × 10−8 | rs388575 | C/T | 0.252 | 0.05 | 0.07 | 0.50 | C/T | −+ | −0.18 | 0.06 | 1.4 × 10−3 | |
| 6-63065777 | 6 | 63 065 777 | A/AAAT | 1.241 | 0.20 | +++ | 2.9 × 10−9 | rs12527308q | T/C | 0.15 | 0.09 | 0.09 | 0.31 | A/AAAT | ++ | 0.28 | 0.08 | 7.0 × 10−4 | |
| rs2258946 | 10 | 93 624 672 | A/G | 0.521 | 0.10 | +++ | 6.6 × 10−7 | rs2258946 | T/C | 0.145 | −0.10 | 0.09 | 0.29 | A/G | +− | 0.17 | 0.07 | 1.1 × 10−2 | |
| rs61023801 | 13 | 91 828 308 | G/A | 1.084 | 0.21 | +++ | 4.7 × 10−7 | rs74922937q | A/G | 0.026 | 0.02 | 0.20 | 0.92 | G/A | ++ | 0.52 | 0.14 | 3.0 × 10−4 | |
| APOE-ε4 (rs429358) | 19 | 45 411 941 | ε4/ε4(−) | −0.48 | 0.07 | −−− | 1.8 × 10−10 | APOE-ε4 (rs429358) | ε4/ε4(−) | 0.442 | −0.70 | 0.15 | 1.1 × 10−5 | ε4/ε4(-) | −− | −0.40 | 0.05 | 4.3 × 10−17 | |
| pTau181 | rs72804029 | 2 | 59 273 581 | T/C | −0.804 | 0.17 | −−− | 2.7 × 10−6 | rs72804029 | T/C | 0.089 | 0.04 | 0.13 | 0.73 | T/C | −+ | −0.26 | 0.10 | 1.0 × 10−2 |
| rs313541 | 4 | 25 263 478 | G/A | 0.501 | 0.11 | +++ | 2.8 × 10−6 | rs313561q | G/A | 0.192 | −0.06 | 0.09 | 0.50 | G/A | +− | 0.16 | 0.07 | 1.3 × 10−2 | |
| rs73276571 | 5 | 120 616 307 | A/G | 0.66 | 0.14 | +++ | 1.8 × 10−6 | rs73276571 | A/G | 0.082 | −0.20 | 0.13 | 0.13 | A/G | +− | 0.22 | 0.10 | 2.4 × 10−2 | |
| rs4896367 | 6 | 138 807 281 | C/T | 0.493 | 0.10 | +++ | 1.4 × 10−6 | rs4896367 | C/T | 0.268 | −0.04 | 0.08 | 0.62 | C/T | +− | 0.16 | 0.06 | 1.1 × 10−2 | |
| rs9397718 | 6 | 154 753 469 | C/T | 0.397 | 0.08 | +++ | 3.4 × 10−6 | rs6911312q | C/A | 0.356 | −0.01 | 0.07 | 0.90 | C/T | +− | 0.18 | 0.06 | 1.4 × 10−3 | |
| rs9397794 | 6 | 155 575 942 | A/G | 0.449 | 0.09 | +++ | 2.6 × 10−6 | rs9397794 | A/G | 0.245 | −0.01 | 0.08 | 0.88 | A/G | +− | 0.18 | 0.06 | 3.0 × 10−3 | |
| rs77131199 | 10 | 26 798 222 | A/G | 1.484 | 0.32 | +++ | 3.6 × 10−6 | rs77131199 | A/G | 0.037 | 0.01 | 0.18 | 0.95 | A/G | ++ | 0.37 | 0.16 | 1.7 × 10−2 | |
| rs12418935 | 11 | 11 863 410 | A/G | 0.615 | 0.12 | +++ | 1.1 × 10−6 | rs12418935 | A/G | 0.107 | −0.05 | 0.12 | 0.65 | A/G | +− | 0.26 | 0.09 | 2.1 × 10−3 | |
| rs7304039 | 12 | 108 736 352 | A/G | 0.414 | 0.08 | +++ | 7.9 × 10−7 | rs7304039 | A/G | 0.295 | −0.07 | 0.08 | 0.34 | A/G | +− | 0.15 | 0.06 | 6.9 × 10−3 | |
| rs4942417 | 13 | 32 887 389 | C/A | 0.441 | 0.09 | +++ | 2.1 × 10−6 | rs4942417 | C/A | 0.154 | −0.08 | 0.10 | 0.42 | C/A | +− | −0.19 | 0.07 | 3.6 × 10−3 | |
| 18-13887179 | 18 | 13 887 179 | GAA/G | −0.775 | 0.14 | −−− | 5.0 × 10−8 | rs7230126q | C/T | 0.317 | 0.03 | 0.07 | 0.67 | GAA/G | −+ | −0.14 | 0.06 | 3.1 × 10−2 | |
| rs3794851 | 18 | 74 709 351 | C/G | 1.050 | 0.22 | +++ | 3.4 × 10−6 | rs3794851 | C/G | 0.046 | −0.01 | 0.16 | 0.93 | C/G | +− | 0.36 | 0.13 | 6.7 × 10−3 | |
| rs149933701 | 19 | 6 636 839 | C/T | 10.678 | 0.22 | +++ | 1.6 × 10−6 | rs61111347q | A/G | 0.039 | −0.08 | 0.17 | 0.63 | C/T | +− | 0.36 | 0.14 | 8.0 × 10−3 | |
| rs11700591 | 21 | 41 507 695 | C/T | 0.723 | 0.13 | +++ | 9.9 × 10−8 | rs11700591 | C/T | 0.069 | −0.02 | 0.13 | 0.89 | C/T | +− | −0.35 | 0.09 | 2.3 × 10−4 | |
| X-118495317 | X | 118 495 317 | C/CT | 0.619 | 0.12 | +++ | 3.0 × 10−7 | rs217969q | T/A | 0.256 | 0.04 | 0.07 | 0.50 | C/CT | ++ | 0.18 | 0.06 | 2.0 × 10−3 | |
r2-values for proxy SNPs were computed on the basis of 1000 Genomes reference data, May 2012 release, CEU sample. GWAS, genome-wide association study.
ars number.
bChromosome.
cPhysical position in base pairs, according to dbSNP build 37.
dEffect allele: allele for which the effect estimate is reported.
eOther allele.
fEffect estimate (fixed effects meta-analysis) for Step 1 meta-analysis.
gCorresponding standard error.
hEffect directions in the three initial studies.
iLambda-adjusted P-value (fixed effects meta-analysis), GWAS meta-analysis.
jMinor allele frequency.
kEffect estimate from regression analysis. Represents changes in Aβ1–42 or pTau181 levels in standard deviations per minor allele.
lP-value of regression analysis.
mEffect directions for Step 1 (joint direction) and replication analysis.
nEffect estimate (fixed effects meta-analysis) GWAS meta-analysis plus replication.
oCorresponding standard error.
pP-value (fixed effects meta-analysis) GWAS meta-analysis plus replication.
qProxy SNP supporting the signal of the GWAS.
Figure 1.
Genome-wide signal intensity (Manhattan) plots. (A) Results for the association with CSF Aβ1–42. (B) Results for the association with CSF pTau181. Both plots show the individual P-values against genomic position. Chromosome number is shown on the x-axis. The results within each chromosome are plotted left to right, with left being the P-terminal end of the corresponding chromosome. Horizontal dashed lines indicate a P-value threshold of 5 × 10−8 (i.e. genome-wide significance). Green dots depict single-nucleotide polymorphisms with genome-wide significance in the genome-wide association study step.
Replication and meta-analysis of combined data
Thirty-five of the 40 SNPs included in the replication step were successfully genotyped (Supplementary Material, Table S1).
The replication step revealed a strong association between rs429358 (APOE-ε4) and Aβ1–42 levels (P = 1.1 × 10−5, β = −0.7 ± 0.015, Table 2). The units used to express the effect size of the SNPs (β ± SE) are standardized. This association became stronger in a meta-analysis of the combined GWAS and replication data (P = 4.3 × 10−17, β = −0.4 ± 0.05, Table 2).
The second strongest replication signal was for association between an SNP on chromosome 3p14.1 [rs62256378, minor allele frequency (MAF) = 6.1%], and Aβ1–42 levels (P = 1.9 × 10−6, β = 0.61 ± 0.13; Supplementary Material, Fig. S4). The meta-analysis of the GWAS and replication data generated a highly significant finding (P = 2.5 × 10−12, β = 0.71 ± 0.10), with the low-frequency allele being associated with higher Aβ1–42 levels and showing an effect size (standardized units for β ± SE) that was larger than that of APOE-ε4. Since rs62256378 was imputed, we genotyped this SNP in 264 individuals from the GWAS step for whom DNA was available. The genotyping of rs62256378 using Sequenom technology revealed a concordance between imputed and experimental genotypes of >99%, demonstrating the validity of the imputed rs62256378 genotypes in the GWAS step (the results of the statistical calculations that involved imputed and experimental genotypes are provided in Supplementary Material, Table S2).
With the exception of APOE, none of the known AD susceptibility genes showed genome-wide significant association with Aβ1–42 or pTau181 levels in the meta-analysis of the three GWAS and the replication sample (Table 3). Both CD2AP (rs9349407) and APOE (rs429358) showed a nominally significant association with increased pTau181 levels (P = 0.031 and 0.029, respectively).
Table 3.
Effect of known AD susceptibility genes on CSF levels of Aβ1–42 and pTau181
| Reference: case–control analysis (GWAS catalog) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Reported SNP | Risk allele frequency | OR | Minor allele | Major allele | Risk allele | ||||||
| Effect of susceptibility allele on Aβ1-42 levelsa | Consistencyb | P | Effect of susceptibility allele on pTau181 levelsa | Consistencyb | P | |||||||
| APOE | rs429358 (APOE-ε4) | 0.09 | 3–4 | C | T | C | −0.404 | Yes | 4.3 × 10−17 | 0.113 | Yes | 0.024 |
| ABCA7 | rs3764650 | 0.1 | 1.23 | G | T | G | −0.038 | Yes | 0.649 | 0.004 | Yes | 0.96 |
| CR1 | rs6656401 | 0.19 | 1.21 | A | G | A | −0.104 | Yes | 0.094 | 0.121 | Yes | 0.06 |
| PICALM | rs3851179 | 0.63 | 1.16 | T | C | C | 0.006 | No | 0.909 | 0.031 | Yes | 0.581 |
| CLU | rs11136000 | 0.6 | 1.16 | T | C | C | 0 | No | 0.995 | −0.007 | No | 0.909 |
| BIN1 | rs744373 | 0.29 | 1.15 | G | A | G | 0.007 | No | 0.895 | 0.078 | Yes | 0.17 |
| EPHA1 | rs11767557 | 0.79 | 1.11 | C | T | T | 0.003 | No | 0.964 | 0.015 | Yes | 0.818 |
| CD2AP | rs9349407 | 0.29 | 1.11 | C | G | C | −0.086 | Yes | 0.111 | 0.119 | Yes | 0.031 |
| CD33 | rs3865444 | 0.69 | 1.1 | A | C | C | 0.003 | No | 0.959 | 0.005 | Yes | 0.936 |
| MS4A | rs610932 | 0.58 | 1.1 | T | G | G | 0.024 | No | 0.652 | 0.009 | Yes | 0.871 |
GWAS, genome-wide association study; CSF, cerebrospinal fluid; OR, odds ratio.
aEffect estimate from regression analysis. Covariates used for the analysis are APOE, age and sex. Represents changes in Aβ1–42 or pTau181 levels in standard deviations per minor allele.
bConsistency in allele direction between the OR observed in the present study and the OR reported in the GWAS catalog.
Effect of rs62256378 on AD risk
In the case–control analysis, no association was found between rs62256378 and AD risk (P = 0.8, OR = 1.03, 95% CI 0.79–1.27). However, we cannot exclude the possibility that an AD association might be detected in a larger case–control sample. To test this, we obtained Stage I association data for rs62256378 from the case–control meta-analysis study conducted by the International Genomics of Alzheimer's Project (IGAP) (6). Here, the SNP rs62256378 showed no association with AD risk in either the complete Stage I analysis (P = 0.7, OR = 0.98, 95% CI 0.91–1.06) or the stratum of APOE-ε4 noncarrier (P = 0.7, OR = 1.02, 95% CI 0.91–1.13).
Genetic interaction between rs429358 and rs62256378
Both of the presently replicated loci modulated Aβ1–42 levels. APOE-ε4 was associated with lower Aβ1–42 levels, and the minor allele A of rs62256378 was associated with higher Aβ1–42 levels. Investigation of a possible interaction between the two association signals revealed strong evidence for allelic interaction (P = 9.5 × 10−5, Table 4). The effect of the minor allele A of rs62256378 varied substantially depending on APOE-ε4 carrier–noncarrier status (Table 5). The strongest effect of rs62256378-A in terms of elevation of Aβ1–42 levels was observed in APOE-ε4 noncarriers (P = 1.5 × 10−8, β = 0.97 ± 0.17, Table 5; Supplementary Material, Fig. S5). The units used to express the effect of the SNPs (β ± SE) are standardized. In heterozygous APOE-ε4 carriers, the effect remained significant, although the level of significance was reduced (P = 1.5 × 10−3, β = 0.35 ± 0.11, Table 5; Supplementary Material, Fig. S5). In APOE-ε4 homozygotes, no significant effect was detectable for rs62256378-A (P = 0.84). This finding was supported by the consistent pattern of interaction observed across all four sub-samples (Table 5). However, the study-specific effect estimates for rs62256378 (Table 5) indicate substantial heterogeneity (I2 = 72%, P = 9.9 × 10−4 with χ²-test for heterogeneity). This heterogeneity was driven by the Bonn1CSF sample, which had a markedly higher effect estimate than the other studies. Interestingly, our signal was not driven by this sub-study, since meta-analysis of the remaining three studies also yielded genome-wide significance for rs62256378 (P = 2.2 × 10−9, β = 0.64 ± 0.11).
Table 4.
Analysis of interaction between Aβ1–42 levels, rs492358 and rs62256378
| SNP1 | Alleles SNP1 |
SNP2 | Alleles SNP2 |
Bonn1CSF |
Bonn2CSF |
ADNI |
ADC |
Meta-analysis | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Minor | Major | Minor | Major | βa | SEb | Pc | βa | SEb | Pc | βa | SEb | Pc | βa | SEb | Pc | βa | SEb | Pc | ||
| rs62256378 | A | G | rs429358 | ε4 | ε4(−) | −1.32 | 0.58 | 0.02 | −0.55 | 0.31 | 0.08 | −0.09 | 0.40 | 0.82 | −0.56 | 0.18 | 2.0 × 10−3 | −0.55 | 0.14 | 9.5 × 10−5 |
CSF, cerebrospinal fluid; ADNI, Alzheimer's Disease Neuroimaging Initiative cohort; ADC, Amsterdam Dementia Cohort.
aEffect estimate for allelic interaction term x1x2. Represents changes in Aβ1–42 or pTau181 levels in standard deviations per minor allele.
bStandard error of effect estimate.
cP-value.
Table 5.
Association results for Aβ1–42 and rs62256378 according to sample and APOE strata
| Stratuma | rsb | chrc | bpd | Minore | Majorf | Bonn1CSF |
Bonn2CSF |
ADNI |
ADC |
Meta-analysis | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| βg | SEh | Pi | βg | SEh | βi | βg | SEh | Pi | βg | SEh | Pi | βg | SEh | Pi | ||||||
| ALL | rs429358 (e4) | 19 | 45 411 941 | ε4 | ε4(−) | −0.51 | 0.11 | 6.1 × 10−6 | −0.33 | 0.07 | 5.9 × 10−7 | −0.39 | 0.15 | 0.01 | −0.70 | 0.15 | 1.1 × 10−5 | −0.40 | 0.05 | 4.3 × 10−17 |
| ALL | rs62256378 | 3 | 67 457 033 | A | G | 2.13 | 0.46 | 2.5 × 10−5 | 0.82 | 0.24 | 6.0 × 10−4 | 0.50 | 0.29 | 0.09 | 0.61 | 0.13 | 1.9 × 10−6 | 0.71 | 0.10 | 2.5 × 10−12 |
| APOE-ε4 noncarriers | rs62256378 | 3 | 67 457 033 | A | G | 1.68 | 0.57 | 4.8 × 10−3 | 0.96 | 0.36 | 8.8 × 10−3 | 0.66 | 0.43 | 0.14 | 0.94 | 0.24 | 1.1 × 10−4 | 0.97 | 0.17 | 1.5 × 10−8 |
| APOE-ε4 heterozygous | rs62256378 | 3 | 67 457 033 | A | G | 0.24 | 0.30 | 0.44 | 0.45 | 0.22 | 0.04 | 0.48 | 0.49 | 0.33 | 0.32 | 0.15 | 0.03 | 0.35 | 0.11 | 1.5 × 10−3 |
| APOE-ε4 homozygous | rs62256378 | 3 | 67 457 033 | A | G | – | – | – | −0.13 | 0.33 | 0.71 | −0.10 | 0.76 | 0.90 | 0.01 | 0.25 | 0.96 | −0.04 | 0.19 | 0.83 |
aAnalysis of all patients and within strata defined by APOE-ε4 carrier status.
brs number.
cChromosome.
dPhysical position in base pairs, according to dbSNP build 37.
eMinor allele.
fMajor allele.
gEffect estimate from regression analysis. Represents increase in Aβ1–42 levels in standard deviations per minor allele.
hStandard error of effect estimate.
iP-value; ADNI, Alzheimer's Disease Neuroimaging Initiative cohort; ADC, Amsterdam Dementia Cohort.
Rs62256378 and cognitive decline
The SNP rs62256378-A significantly attenuated cognitive decline in AD dementia patients (P = 3.1 × 10−3, β = 0.32 ± 0.12, Table 6; Supplementary Material, Fig. S6). After stratification for APOE genotype, this effect was significant in APOE-ε4 noncarriers (P = 0.021, β = 0.32 ± 0.16, Table 6). A non-significant trend was observed in heterozygous APOE-ε4 carriers (P = 0.07, β = 0.31 ± 0.21). No evidence was found for interaction between SUCLG2 (rs62256378) and APOE genotype (P = 0.456).
Table 6.
Effect of rs62256378 on decline in MMSE score according to sample and APOE strata
| Stratuma | rsb | Allele |
Bonn1 |
Bonn2 |
ADNI |
ADC |
Meta-analysis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Minor | Major | βc | SEd | Pe | βc | SEd | Pe | βc | SEd | Pe | βc | SEd | Pe | βc | SEd | Pe | ||
| ALL | rs62256378 | A | G | 0.57 | 0.32 | 0.08 | 0.66 | 0.64 | 0.15 | 0.24 | 0.21 | 0.11 | 0.28 | 0.16 | 0.08 | 0.32 | 0.12 | 3.1 × 10−3 |
| APOE-ε4 noncarriers | rs62256378 | A | G | 0.57 | 0.46 | 0.23 | 0.26 | 0.93 | 0.39 | 0.01 | 0.23 | 0.50 | 0.61 | 0.25 | 0.02 | 0.32 | 0.16 | 0.021 |
| APOE-ε4 heterozygous | rs62256378 | A | G | 0.38 | 0.56 | 0.25 | 1.07 | 1.03 | 0.16 | 0.43 | 0.42 | 0.15 | 0.18 | 0.28 | 0.29 | 0.31 | 0.21 | 0.072 |
| APOE-ε4 homozygous | rs62256378 | A | G | −0.36 | 1.10 | 1.00 | 1.38 | 2.15 | 0.25 | 1.23 | 0.92 | 0.14 | 0.29 | 0.45 | 0.26 | 0.40 | 0.37 | 0.141 |
aAnalysis of all patients and within strata defined by APOE-e4 carrier status.
bPhysical position in base pairs, according to dbSNP build 37.
cEffect obtained from regression analysis. The β represents attenuation of memory decline as shown by standard deviations of MMSE score decline.
dStandard error of effect estimate.
eOne-sided P-value for rs62256378-A as the protective allele.
Conditional analysis of the SUCLG2 locus
This marker is located within a recombination hotspot. This hotspot defines two linkage disequilibrium blocks that span several exons of SUCLG2 on chromosome 3p14.1 (Supplementary Material, Fig. S4). Conditional analysis showed that all additional signals in this region were dependent on rs62256378 (Supplementary Material, Table S4).
Functional investigation of the role of SUCLG2
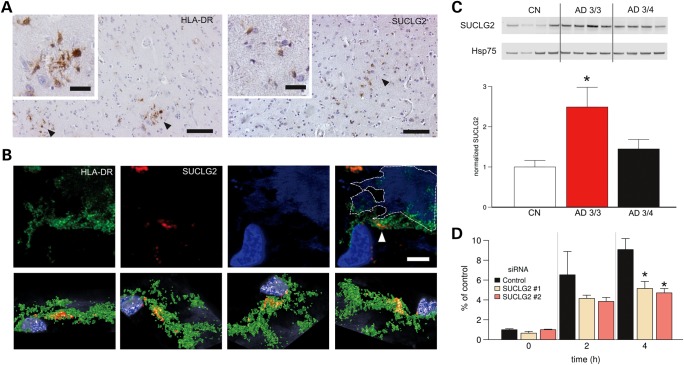
Immunohistochemistry revealed that SUCLG2 expression was mainly confined to microglial cells (7) in the postmortem brains of AD dementia patients and age-matched controls (Fig. 2A and B). The strongest association between rs62256378 and higher Aβ1–42 levels was observed in APOE-ε4 noncarriers. To determine whether APOE status influenced SUCLG2 levels, SUCLG2 protein levels were analyzed in human postmortem samples from the frontal cortex, a brain region affected at an early stage of AD. Here, ApoE3/3 carriers showed significantly higher SUCLG2 levels compared with ApoE3/4 carriers (Fig. 2C). Controls in turn showed the lowest levels, reinforcing the hypothesis that rs62256378 may influence the disease course in AD, rather than overall disease risk. SUCLG2 levels in microglial cells were then lowered using various siRNA approaches (Supplementary Material, Fig. S7). Decreasing microglial SUCLG2 compromised Aβ1–42 phagocytosis in a time-dependent manner (Fig. 2D).
Figure 2.
Functional studies of SUCLG2. (A and B) SUCLG2 localized to microglia in AD brains. (A) Representative DAB immunostaining with SUCLG2 and HLA-DR (as a human microglial marker) in brain cortex of AD subjects. DAB-positive cells with characteristic romboidal nuclei (typically microglial cells) are indicated by arrows. Scale bars are 100 and 20 µm on the main and small images, respectively. (B) High-resolution confocal image of an area of frontal cortex from an AD dementia patient immune labelled with HLA-DR (green) and SUCLG2 (red). Plaque autofluorescence is visible in the blue channel and it is highlighted with a discontinuous line. Most of the SUCLG2 is enclosed by the HLA-DR (arrow). The top panel shows several viewpoints of a 3D reconstruction of the same area, which is shown in the lower panel. (Scale is 10 μm.) (C) Increased levels of SUCLG2 in AD ApoE 3/3 brains. Soluble extracts from postmortem frontal cortex of AD dementia patients with ApoE 3/4 and ApoE 3/3 status plus age-matched healthy controls (CN) were probed with an antibody to SUCLG2. Expression levels were normalized to Hsp75 (CN, n = 6; AD, 3/4, n = 8; AD, 3/3, n = 4). Data are presented as mean ± SEM. Asterisk denotes P < 0.05. (D) SUCLG2 silencing is detrimental to extracellular Aβ1–42. FAM-Aβ1–42 phagocytosis was impaired in BV2 cells in which SUCLG2 was silenced by two distinct siRNA compared with negative control siRNA (n = 6). FAM-Aβ1–42 was added at a 500 nm final concentration and the amount of internalized Aβ1–42 was assessed at the indicated time points, showing that cells contain less Aβ1–42 in the absence of SUCLG2. Data are presented as mean ± SEM. Asterisk denotes P < 0.05.
DISCUSSION
The present study demonstrated that genetic variation in SUCLG2 modulated Aβ1–42 levels in AD dementia patients. Furthermore, we found that SUCLG2 was associated with AD clinical course, as measured by change in MMSE over time. Despite substantial evidence for heterogeneity in the effect of rs62256378, the stronger effect estimate observed in Bonn1CSF does not call the validity of our initial result for rs62256378 into question, since the overall signal was not determined by Bonn1CSF; the meta-analysis that excluded this sample also demonstrated genome-wide significance for rs62256378 (P = 2.2 × 10−9, β = 0.64 ± 0.11).
To our knowledge, no previous genetic or biological study has implicated SUCLG2 in AD. The negative result of our case–control analysis suggests that this gene does not confer AD susceptibility, which would explain why it has not been identified in previous case–control GWAS of AD. Possible reasons for the failure of previous GWAS of CSF markers to identify SUCLG2 are 3-fold. First, they included unaffected subjects and individuals with mild cognitive impairment (MCI). Secondly, they analyzed non-imputed datasets. In this regard, we computed r2 values for rs62256378 with SNPs from the 1000 Genomes project data, release May 2012. Only five SNPs (rs76856038, rs76856038, rs80028595, rs115298081 and rs74727963) showed r² values of >0.50 with rs62256378, and none of these are included in the Illumina 550K or Omni marker panels. Thirdly, APOE status was not used as a covariate in the analyses. In contrast, analysis in the present study was restricted to AD dementia patients only, genetic data were imputed, and APOE status was included as a covariate.
Our data also confirmed the previously reported association between the APOE locus and Aβ1–42 levels. Interestingly, the effect of SUCLG2 was mainly observed in APOE-ε4 noncarriers. This suggests that SUCLG2 and APOE converge in their effect on Aβ1–42 levels in some, as yet unidentified, common biological pathway.
The effect of SUCLG2 on Aβ1–42 levels was surprisingly high, since we did not expect to identify a novel gene with an effect comparable with APOE. SUCLG2 explained 10.7% of the variance in Aβ1–42 levels in APOE-ε4 noncarriers, which is more than the variance explained by APOE-ε4 in the overall sample (7.1%, Supplementary Material, Table S5).
None of the presently investigated SNPs showed genome-wide significant association with pTau181. To date, genome-wide significant association with total Tau/pTau181 levels has been reported for four loci, i.e. APOE, SNAR-I, GLIS3 and NCR2 (1,4,5). Besides APOE, a recent GWAS by Cruchaga et al. (5) identified the following three novel genome-wide significant findings for total Tau and pTau181 levels: (i) rs9877502 at SNAR-I, P = 4.89 × 10−9 and P = 1.68 × 10−7 for total Tau and pTau181, respectively; (ii) rs514716 at GLIS3, P = 1.07 × 10−8 and P = 3.22 × 10−9 for Tau and pTau181, respectively and (iii) rs6922617 at NCR2, P = 2.55 × 10−5 and P = 3.58 × 10−8 for total Tau and pTau181, respectively. Of these three SNPs, only rs9877502 and pTau181 showed a nominally significant association in the present GWAS meta-analysis of pTau181 (P = 9.21 × 10−4). Interestingly, we observed a non-significant trend for rs6922617 (P = 0.07). This locus received further support from the adjacent SNP rs11966476 (P = 0.04) (Supplementary Material, Table S3).
We observed a nominally significant association between the pTau181 level and three SNPs: rs429358 (APOE locus), rs9877502 (SNAR-I) and rs11966476 (NCR2). To our knowledge, our results for SNAR-I and NCR2 represent the first independent replications of these loci. A possible reason for our failure to replicate the finding for GLIS3 is that in addition to AD dementia patients, the study by Cruchaga et al. also included MCI patients and controls. If GLIS3 only contributes to pTau181 variance in unaffected individuals or during the preclinical stages of disease, this effect may not be present in our sample.
With the exception of APOE, none of the known AD susceptibility genes showed genome-wide significant association with either Aβ1–42 or pTau181 levels in the present study. Previous studies have reported associations between Aβ1–42 or pTau181 levels and the AD susceptibility genes PICALM, CLU, MS4A4A and TREM2 (2,3,5). Again, our failure to replicate these findings, even at a level of nominal significance, may have been due to the fact that our study focused on AD dementia patients only. The present study is the first to report an association between the AD susceptibility gene CD2AP (rs9349407) and pTau181 level (P = 0.031), although this finding was only nominally significant and requires replication. In the case of TREM2, a genuine association between the SNP originally associated with AD (rs75932628; p.R47H) and pTau181 cannot be ruled out, since none of the present SNPs in this region displayed sufficient linkage disequilibrium with rs75932628, and this SNP cannot be imputed adequately using available variant databases.
SUCLG2 encodes a GTP-specific β-subunit of the succinyl-CoA ligase (SUCL). The mitochondrial matrix enzyme SUCL is a heterodimer, which is formed by an invariable α-subunit (encoded by SUCLG1), and a variable β-subunit which confers nucleoside specificity. This β-subunit is encoded by either SUCLA2 or SUCLG2. In the case of SUCLG2, the SUCL enzyme catalyzes the reversible conversion of succinyl-CoA and GDP to succinate and GTP, respectively (8). Previous studies have identified mutations in SUCLG1 and SUCLA2 in patients with mitochondrial DNA (mtDNA) depletion syndromes (MDS). MDS are characterized by a decrease in mtDNA content and a reduction in the activity of mitochondrial respiratory chain complexes I, II, IV and V (9). Interestingly, both features have also been found, to a lesser extent, in AD (10). In line with this, Miller et al. demonstrated that reducing the expression of SUCLG2 led to a decrease in both mtDNA and the activity mitochondrial complex IV (11).
Our observation of higher SUCLG2 expression in APOE-ε4 noncarriers may be explained in terms of the amyloid cascade hypothesis. While hereditary AD is caused by an overproduction of Aβ1–42 secondary to autosomal dominant mutations in genes coding for APP and presenilin 1 and 2, sporadic AD is more likely to arise from limited clearance of Aβ1–42 (12). Limited clearance by microglia is likely to contribute to increased Aβ1–42 burden in the brains of sporadic AD dementia patients, as described recently for the CD33 risk allele rs3865444 (13). In terms of the interaction of APOE with SUCLG2, an improvement in the microglial clearance of Aβ1–42 may be of central importance in terms of explaining this genetic relationship. In this regard, ApoE is indeed tightly connected to the turnover of Aβ1–42, since it plays a critical role in the normal proteolytic clearance of Aβ1–42 (14). Increased levels of ApoE and lipidation of ApoE increase the intra- and extracellular microglial degradation of Aβ1–42 by increasing the interaction between ApoE and Aβ1–42 (15). Notably, ApoE-ε2 and ε3 isoforms have a higher lipidation status than the ApoE-ε4 isoform, and ApoE-ε4 is much less efficient in promoting soluble Aβ1–42 degradation by microglia (14). In the present study, the association between rs62256378 and Aβ1–42 CSF levels was modified by ApoE-ε4. The latter also decreased the level of SUCLG2 in microglia, and this in turn impaired Aβ1–42 phagocytosis. Conversely, in AD patients carrying the SUCLG2 A allele and being APOE-ε4 noncarriers, microglial Aβ1–42 clearance is probably improved through the combination of two mechanisms. First, an ApoE-dependent mechanism that delivers more efficiently Aβ1–42 to microglia. Second, microglia of these AD patients can more efficiently phagocyte Aβ1–42. Removal of Aβ1–42 by microglia is likely to decrease the propensity of Aβ1–42 to aggregate and deposit. In AD patients, the levels of Aβ1–42 in the CSF are lowered quite early after clinical disease onset or possibly even before (16). This is explained by the intrinsic tendency of Aβ1–42 to form aggregates ranging from soluble oligomers to ultimately insoluble senile plaques. Since this process is positively correlated with the concentration of Aβ1–42 in the brain parenchyma, decreased degradation of Aβ1–42 will increase the concentration of Aβ1–42 and concomitantly its tendency to form aggregates. As a consequence of this process, the amount of Aβ1–42 that is available to escape into the CSF is lowered. Therefore, stabilized SUCLG2 levels and concomitant microglial clearance may maintain higher Aβ1–42 CSF levels by reducing the tendency of Aβ1–42 to aggregate in the brain.
Furthermore, microglial function may also be of relevance to cognitive performance, as suggested by recent experimental data, which showed that improved microglial clearance was protective in terms of memory function (17). A plausible hypothesis is that improved microglial function may account for the stabilization of MMSE scores in rs62256378-A carriers. These findings are consistent with a number of previous reports, which have implicated innate immune activation and microglial clearance in sporadic AD.
Microglial phagocytosis of Aβ1–42 is highly energy dependent, and most of this energy is provided by mitochondria. Alternatively, the effect of SUCLG2 on microglial clearance of Aβ1–42 may be mediated by modulation of oxidative stress and inflammation. The latter is involved in Aβ1–42 clearance in the brain, in which microglia participate actively by internalizing and degrading soluble (18) and aggregated forms of Aβ1–42 (19). On the other hand, microglia have been identified as an important source of reactive oxygen species (ROS) (20). The mitochondrial respiratory chain is a major source of ROS. The association between inflammation and oxidative damage is mediated by the release of ROS during the inflammatory process. In old transgenic AD mice, microglial function is impaired, as shown by the decrease expression of Aβ1–42-binding receptors and Aβ1–42-degrading enzymes, whereas microglia maintain their ability to produce pro-inflammatory cytokines (21). Thus, a chronic inflammatory status is produced, which increases oxidative stress and the accumulation of Aβ1–42 in the brain, and reduces the clearance of Aβ1–42. In addition, reducing oxidative stress in transgenic AD mice has been shown to diminish amyloid plaque burden and microglial activation (22). The maintenance of mtDNA is crucial for mitochondrial function, since it encodes the key enzymes of mitochondrial respiration. Hence, the beneficial effect of the SUCLG2 A allele on the clearance of Aβ1–42 and on the clinical course of AD may be mediated by improved respiratory chain and mtDNA maintenance, a compensatory mechanism which enhances mitochondrial function by reducing microglial oxidative stress and pro-inflammatory activity. This in turn increases the clearance of Aβ1–42 and reduces the rate of neuronal death.
Interestingly, while the evidence for interaction between SUCLG2 and APOE was strong in terms of the effect on Aβ1–42 levels (P = 9.5 × 10−5), no such interaction was evident in the analysis of clinical course (P = 0.456). This may suggest that SUCLG2 contributes to Aβ1–42 and clinical course through (at least partially) independent pathways and that the pathway leading to variance in clinical course is not—or at least not strongly—influenced by APOE. This hypothesis is consistent with the lack of consistent evidence for an association between APOE genotype and clinical course, despite the very strong effect of APOE genotype on Aβ1–42 levels and on disease susceptibility.
Further studies are now warranted to disentangle the detailed molecular mechanisms underlying the influence of SUCLG2 on both Aβ1–42 levels and disease course.
MATERIALS AND METHODS
GWAS samples
GWAS data on the CSF biomarkers Aβ1–42 and phosphorylated pTau181 from three independent AD cohorts were analyzed (Bonn1CSF, n = 113; Bonn2CSF, n = 167; ADNI, n = 83). All AD dementia patients included in the GWAS analysis fulfilled NINCDS/ADRDA criteria for probable AD (23). The demographic data of the GWAS AD dementia patients are shown in Table 1.
The Bonn1CSF and Bonn2CSF GWAS samples were derived from a larger German GWAS cohort (n = 1079) which was recruited from the following three sources: (i) the German Dementia Competence Network (24) (DCN; n = 391); (ii) the German study on Aging, Cognition and Dementia in primary care patients (25) (AgeCoDe, n = 64); and (iii) the interdisciplinary Memory Clinic at the University Hospital of Bonn (n = 624). These German patients were classified as two separate samples (i.e. Bonn1, n = 438; and Bonn2, n = 641), as they were genotyped using two different platforms (see below). The Bonn1CSF and Bonn2CSF samples were comprised of AD dementia patients from Bonn1 and Bonn2, respectively, for whom CSF data were available. The Bonn1CSF included 61 patients from the Memory Clinic and 52 from the DCN. The Bonn2CSF consisted of 84 patients from the Memory Clinic and 83 from the DCN.
Between 2003 and 2005, the DCN recruited a large cohort of patients with MCI (n = 1095 patients) or early AD (n = 391). Baseline analysis comprised extensive neuropsychological tests including those of the Consortium to Establish a Registry for Alzheimer's Disease (CERAD), the MMSE and the Clinical Dementia rating (CDR-SB). Follow-up assessments were performed at 12 and 24 months (24). Whole blood, genomic DNA, plasma and CSF were collected from all participants.
AgeCoDe is a German multicenter prospective cohort study. The sample was drawn at random from general practice registries in six German cities, and the selected individuals were contacted by mail. Main inclusion criteria were age 75 years or above and the absence of dementia according to the clinical judgment of the respective general practitioner. A total of 3327 AgeCoDe participants were recruited between January 2002 and November 2003 (25,26). Each participant is assessed at baseline and at 18 months intervals thereafter. All assessments are performed at the participant's home by a trained study psychologist or physician. At all visits, assessment includes the Structured Interview for Diagnosis of Dementia of Alzheimer type, Multi-Infarct Dementia and Dementia of other etiology according to DSM-IV and ICD-10 (SIDAM) (27). The SIDAM comprises: (i) a 55-item neuropsychological test battery, including all 30 items of the MMSE and assessment of several cognitive domains (orientation, verbal and visual memory, intellectual abilities, verbal abilities/ calculation, visual–spatial constructional abilities and aphasia/ apraxia); (ii) a 14-item scale for the assessment of the activities of daily living (SIDAM-ADL-Scale) and (iii) the Hachinski Rosen Scale. Dementia was diagnosed according to DSM-IV criteria. The AgeCoDe study was approved by all of the respective ethics committees. Written informed consent was obtained from all study participants or their legal guardians. At the time of writing, four waves of follow-up have been completed in the AgeCoDe cohort.
The interdisciplinary Memory Clinic of the Department of Psychiatry and Department of Neurology at the University Hospital in Bonn provided further patients (n = 624). Diagnoses were assigned according the NINCDS/ADRDA criteria (23) and on the basis of clinical history, physical examination, neuropsychological testing (using the CERAD neuropsychological battery, including the MMSE), laboratory assessments and brain imaging.
The third GWAS sample was a subsample of the Alzheimer's Disease Neuroimaging Initiative (ADNI) cohort. This comprised 83 AD dementia patients of non-Hispanic Caucasian origin for whom CSF biomarker data were available. The ADNI is a $60 million, 5-year public-private partnership which was launched in 2003 by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, the Food and Drug Administration and various private pharmaceutical companies and non-profit making organizations. The Principal Investigator of this initiative is Michael W. Weiner, MD. ADNI is the result of the efforts of many coinvestigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the USA and Canada. The initial goal of ADNI was to recruit 800 adults aged 55–90 years, i.e. 200 cognitively normal individuals for 3-year follow-up, 400 individuals with MCI for 3-year follow-up and 200 individuals with early AD for 2-year follow-up. Up-to-date information on the cohort is available at www.adni-info.org. All AD dementia patients fulfilled NINCDS/ADRDA criteria (23), and their magnetic resonance imaging (MRI) data were consistent with this diagnosis. ADNI participants undergo evaluation at 6- to 12-month intervals through a battery of assessments including cognitive and neuropsychological testing, neuroimaging (MRI with or without positron emission tomography) and measurement of plasma and CSF biomarkers. Among other instruments, cognitive status is assessed using the MMSE score. The ADNI study is approved by the Institutional Review Board at each of the participating centers, and all participants provide written informed consent. Data used in the preparation of the present study were obtained from the ADNI database.
Replication sample
These patients were drawn from the ongoing memory-clinic-based ADC of the VU University Medical Center. All patients underwent a standardized 1-day assessment, comprising medical history, informant based history, physical and neurological examination, laboratory tests, neuropsychological assessment and brain MRI. Probable AD diagnoses were assigned according to both NINCDS/ADRDA criteria (23), and the core clinical NIA-AA criteria (28). Follow-up took place during routine clinical attendance at the memory clinic, and involved patient history, cognitive testing including the MMSE, and a general physical and neurological examination. Patients who failed to attend routine follow-up for at least 2 years after baseline were contacted by telephone, and their cognitive status was evaluated using a standardized interview that included questions concerning all cognitive domains, physical complaints and medical history, complemented by the Telephone Interview for Cognitive Status (29). In cases where the telephone interview suggested clinical progression, patients underwent a detailed evaluation at the outpatient clinic.
Investigation of rs62256378 in AD risk
A case–control sample was used to investigate whether rs62256378 confers AD risk. The AD dementia patients were derived from the Bonn1 and Bonn2 samples (n = 1079). The control sample was mainly comprised of individuals from three population-based studies: the Heinz Nixdorf Recall (HNR) study cohort (n = 1188) (30), the POPGEN biobank (n = 471) (31) and the KORA study (n = 458) (32). These controls were not screened for cognitive impairment. Furthermore, 287 healthy elderly individuals from the AgeCoDe cohort, in whom dementia and MCI had been excluded at the last follow-up visit, were also included. The demographic data for these samples are provided in Table 1. In addition, Stage I association results for rs62256378 were obtained from a case–control meta-analysis study conducted by the IGAP (6). In Stage I of this meta-analysis 17 008 AD cases and 37 154 controls were analyzed. In addition, APOE-ε4 carrier–noncarrier stratified association results were obtained from the IGAP.
Investigation of rs62256378 in cognitive decline
The effect of rs62256378 on cognitive decline in AD dementia patients was explored using longitudinal MMSE scores in AD dementia patients from: the Bonn1 (n = 80), Bonn2 (n = 110), ADNI (n = 140) and the ADC (n = 339). The effect of APOE-ε4 carrier–noncarrier status on the rs62256378 effect on MMSE progression was also analyzed.
CSF collection and analysis
For the Bonn1CSF and Bonn2CSF samples, diagnostic lumbar punctures were performed at the respective Department of Neurology or Psychiatry using a standardized technique. The subject was placed in a sitting or supine position, and a 22G atraumatic spinal needle was inserted. The CSF samples were kept on ice for a maximum of 1 h and then centrifuged for 10 min (2000 g at 4°C). Samples were aliquoted to 0.25 ml and stored in polypropylene tubes at −80°C until analysis. All CSF samples were sent to the Department of Psychiatry in Erlangen for quantification. Levels of Aβ1–42 and pTau181 were measured using commercially available ELISA immunoassays INNOTEST® β-amyloid(1–42) and INNOTEST® PhosphoTAU(181p) (Innogenetics), in accordance with the protocols described by Popp et al. (33).
In the ADNI cohort, lumbar puncture was performed using a 20G or 24G spinal needle, as described in the ADNI procedures manual. In brief, CSF was collected into collection tubes provided to each site. The CSF samples were then transferred into polypropylene transfer tubes and placed on dry ice within 1 h of collection. They were then shipped overnight to the ADNI Biomarker Core Laboratory at the University of Pennsylvania Medical Center on dry ice. After thawing for 1 h at room temperature and gentle mixing, aliquots (0.5 ml) of these samples were prepared. The aliquots were stored in bar code-labelled polypropylene vials at −80°C. CSF levels of Aβ1–42 and pTau181 were measured using the multiplex xMAP Luminex technology (Luminex Corp) and INNO-BIA AlzBio3 immunoassays (Innogenetics) according to protocols described elsewhere (1).
In the ADC cohort, lumbar puncture was performed using a 25G needle. The CSF was collected in polypropylene tubes and centrifuged at 1800 g for 10 min at 4°C. The CSF was immediately stored at −20°C until further analysis (maximum 2 months). CSF levels of Aβ1–42 and pTau181 were measured using similar commercial ELISA immunoassays to those used for Bonn1CSF and Bonn2CSF. Quantification was performed in the neurological laboratory of the VU University Medical Center in Amsterdam in accordance with the protocols described by Mulder et al. (34).
Standardization of CSF biomarker levels was necessary since values in the ADNI cohort differed substantially from those observed in the Bonn1CSF, Bonn2CSF and ADC cohorts, due to the different platforms used (regular ELISA versus Luminex). Individual CSF levels were standardized within each study using a standard normal transformation. In all four cohorts, the CSF biomarkers Aβ1–42 and pTau181 data showed a normal distribution (Supplementary Material, Figs S1 and S2).
Genotyping
The Bonn1CSF sample was genotyped on the Illumina 610-quad chip, and was part of the sample investigated by Harold et al. (35). The Bonn2CSF sample was genotyped using the Illumina Omni 1M-quad chip. The ADNI cohort was genotyped using the Illumina 610 chip.
The control sample of Bonn1 was genotyped using the Illumina HumanHap550k chip. The control sample of Bonn2 was genotyped with the Illumina 1M-quad chip.
Genotyping in the ADC sample was performed using the MassArray system and a Sequenom Compact MALDI-TOF device (Sequenom). SNPs were selected for the replication step if they modulated either Aβ1–42 or pTau181 with a P-value of <5 × 10−6 and showed consistent allele direction across all three GWAS. A total of 163 SNPs representing 36 novel loci met these criteria. Successful Sequenom assay design was achieved for 40 SNPs. These included rs429358, which characterizes APOE-ε4 versus APOE-ε4 noncarriers, and 30 SNPs representing 29 potentially novel loci. For seven of the 36 novel loci, design of a Sequenom assay failed due to technical reasons and a lack of additional proxies for the respective locus. SNPs representing the nine other known AD susceptibility genes were also included (Supplementary Material, Table S1) (35–38). Primer sequences and assay conditions are available upon request. Thus, a total of 40 SNPs were genotyped in the ADC cohort.
Since rs62256378 was imputed in the GWAS step, the SNP was genotyped directly in GWAS patients for whom DNA was available using Sequenom (Bonn1CSF, n = 112 and Bonn2CSF, n = 152).
Quality control
The Bonn1 sample was part of the sample investigated by Harold et al. (35) which comprised 19 000 samples prior to QC. Following stringent QC, Harold et al. eliminated 53 383 SNPs and 2850 subjects from their data analysis. No German subjects were excluded. For the present analyses, a check was made for deviations from Hardy–Weinberg equilibrium within the German subgroup, and 55 SNPs with an HWE P < 10−6 were excluded.
In the Bonn2 sample, seven individuals were excluded due to potential relatedness, as indicated by high genome-wide identity-by-state values (4 SDs greater than overall mean IBS). SNPs with a genotyping rate of <99% were excluded. Four individuals were also excluded due to a genotyping rate of <99%. Finally, 88 SNPs were excluded due to deviation from HWE (P < 10−6), and 80 000 SNPs due to low MAF (<1%). The QC criteria used for Bonn2 were applied to the entire ADNI cohort. In total, 24 individuals and 42 199 SNPs were excluded due to a low genotyping rate, 80 SNPs due to deviation from HWE, and 5961 SNPs due to low MAF.
The QC procedure applied to the population-based Bonn1 controls is described elsewhere (39). In this sample, 49 controls were removed due to a low genotyping rate. In addition, 80 controls were removed because they overlapped with the HNR control sample used in Bonn2.
In the ADC replication cohort, three samples were excluded due to a genotyping rate of <95%. In addition, 101 individuals were excluded because their covariate APOE status was unavailable. A total of five SNPs were excluded due to a low genotyping rate (<95%), strong deviation from HWE (P = 1 × 10−38) or monomorphic status (Supplementary Material, Table S1).
Statistical analyses
For each of the three GWAS (Bonn1CSF, Bonn2CSF and ADNI), genotype data were imputed using the 1000 Genomes (40) reference data, May 2012 release, and the IMPUTEv2 software package (41). Imputed SNPs with an information score of <0.4 or an MAF of <3% were excluded. Imputation accuracy was satisfactory, with info scores of 0.61, 0.67 and 0.75, in Bonn1CSF, Bonn2CSF and ADNI, respectively. After imputation, a total of 6 812 394 SNPs were included in the initial GWAS step and the meta-analysis of the three GWAS. A complete (genome wide) set of summary association statistics for both Aβ1–42 and pTau181 were deposited at http://intersnp.meb.uni-bonn.de/CSF_AD.html.
To identify possible stratification within each cohort, a multidimensional scaling analysis was performed using PLINK (42). The three leading principal components were retained as covariates.
In each cohort, CSF levels were analyzed as quantitative traits (QTs) using an additive allelic linear regression model, as implemented in PLINK. Age, sex and APOE status—as defined by the number of APOE-ε4 copies—were additional covariates.
The association results were combined in a meta-analysis using the fixed effects model (43), as implemented in YAMAS (44). In addition, heterogeneity measures (45) were computed and meta-analysis was performed under a random effects model (43). Significance of cross-study differences in the β estimates for rs62256378 was assessed using the χ²-test for heterogeneity.
GWAS generated genome-wide inflation factors for Aβ1–42 of λ = 1.012 in Bonn1CSF, λ = 1.005 in Bonn2CSF and λ = 1.05 in ADNI. For pTau181, GWAS produced genome-wide inflation factors of λ = 1.006 in Bonn1CSF, λ = 1.013 in Bonn2CSF and λ = 1.001 in ADNI. The meta-analysis of the three GWAS data yielded genome-wide inflation factors of λ = 1.05 and 1.03 for Aβ1–42 and pTau181, respectively (Supplementary Material, Fig. S3). Therefore, we decided to present λ-adjusted P-values for the genome-wide analysis instead, and to validate any particular potentially genome-wide significant findings via Monte-Carlo simulation.
The experimental genotypes of the replication ADC cohort were analyzed with INTERSNP (46), and combined with the GWAS results in the meta-analysis fashion described above.
The explained variance was determined using the coefficient of determination R² of the linear regression model without the covariates that were used for P-value computation.
Two SNPs showing association with Aβ1–42 level were replicated in the ADC, i.e. rs429358 at the APOE locus and rs62256378 at a novel locus containing the SUCLG2 gene. A possible genetic interaction between these two association signals was explored as an indicator of a possible biological link. For this, SNP–SNP interaction was analyzed using an allelic interaction test (one degree of freedom test, 1 d.f.). The standard genotype coding and definition of interaction parameters provided by Cordell and Clayton (47) were used, as implemented in INTERSNP.
Case–control analysis was performed using a standard logistic regression test (1 d.f.). Cognitive decline was computed as ΔMMSE/Δt, where ΔMMSE was the difference in the MMSE scores between time point A and time point B, and where Δt was the time in days between time points A and B. ΔMMSE/Δt was then analyzed as a QT. Educational status and MMSE at time point A were used as additional covariates.
Human brain tissue
Postmortem human brain tissue specimens from patients with histologically confirmed AD and from age-matched controls who had died from non-neurological disease (75 ± 10 years at the age of death) were obtained from the Neurological Tissue Bank of the Biobank from the Hospital Clinic-Institut d'Investigacions Biomèdiques August Pi i Sunyer in Barcelona. Postmortem duration varied from 1.5 to 5 h; neither age nor postmortem sampling time differed significantly between controls and AD dementia patients.
Immunohistochemistry
For colocalization of SUCLG2 in human brain cortex, tissue sections were subjected to paraffin removal at 60°C for 30 min, xylene treatment and rehydration through a graded ethanol series. Antigen unmasking was performed by heating the tissue sample to 100°C for 20 min in 10 mm sodium citrate buffer (pH = 6). Sections were incubated with rabbit anti-SUCLG2 antibody (Novus Biologicals, NBP1-32521) diluted to 1 : 400 and mouse anti-HLA-DR (Thermo scientific, MA1-35420) diluted to 1 : 100. Consecutive sections were further incubated with biotinylated secondary antibody, and signal amplification was achieved with HRP–streptavidin–biotin conjugates (Vectastain Elite ABC system). Diaminobenzidine (DAB)-stained sections were then counterstained with hematoxylin. Brightfield images were then obtained using an axioplan microscope (Zeiss). In parallel, sections were incubated with Alexa 488 and Alexa 546-conjugated secondary antibodies and mounted on a dapi-containing mounting media (Vectorlabs). An attempt was made to quench autofluorescence. Hi-Res confocal images were obtained using a Leica SP5 Laser Scanning Confocal Microscope (Leica). Following deconvolution, 3D reconstruction was performed using Huygens Scientific Volume Imaging.
Cell culture and transfection
BV2 murine microglial cells were obtained from the American Type Culture Collection. The cells were maintained at 37°C in 5% CO2 DMEM medium, supplemented with 10% fetal calf serum and 1% penicillin/streptomycin. The following were obtained from Invitrogen: three different Stealth siRNA directed to exons 4, 5 and 6–7 of SUCLG2 (5′-GCGGUUUGAAAGGAGGUGUUCAUU-3′, 5′-CCCUGGAUAUUUCUAGAGAAACAUA-3′ and 5′-ACCAGGCUGCAGAUCAGAUUACAAA-3′); the recommended Stealth negative control; and the positive (GAPDH) control siRNA duplexes. Transfections were performed using lipofectamine 2000 (Invitrogen) in accordance with the manufacturer's recommendations.
Western blot
Protein extracts from BV2 cells or frozen human brain tissue were homogenized in TNE buffer (10 mm Tris–HCl, pH 7.4; 150 mm NaCl, 5 mm EDTA; 0.5% NP-40; 1× complete protease inhibitor cocktail (Roche); 1× phosphatase inhibitor cocktail I and II (Sigma–Aldrich). Protein concentration was determined using the BCA method (Pierce Biotechnology). A total of 20 mg of protein was resolved by SDS–PAGE, transferred to PVDF and probed with rabbit anti-SUCLG2 antibody (Novus Biologicals, NBP1-32521) diluted to 1 : 500. Following overnight incubation, the membranes were rinsed three times in TTBS and incubated at 22°C in 800CW donkey anti-rabbit secondary antibody in 5% nonfat milk in TTBS, pH = 7.5. The blots were visualized using an LI-COR Biosciences Odyssey infrared imaging system (Lincoln). After visualization, the blots were stripped through rinsing with TBS. They were then incubated at 55°C for 30 min in a buffer containing 200 mm glycine, 2% SDS and 100 mm mercaptoethanol. Membranes were rinsed in TBS and then blocked and reprobed with mouse monoclonal anti-Hsp75 (UC David/NIH NeuroMab Facility, clone N52A/42) diluted to 1 : 1000; rinsed three times in TTBS; incubated at 22°C in 800CW donkey anti-mouse secondary antibody in 5% nonfat milk in TTBS, pH = 7.5; and visualized using the LI-COR system (Lincoln).
Phagocytosis of FAM-labeled Aβ1–42 (FAM-Aβ1–42)
Following incubation of Aβ1–42 at 37°C for 3 days, microglial phagocytosis of aggregated FAM-Aβ1–42 (Anaspec) was measured using a plate-based assay. Control or SUCLG2-silenced BV2 cells were plated at 50 000 cells per 100 μl in black 96-well plates, and FAM-Aβ1–42 was added to a final concentration of 500 nm and incubated for 0, 2 or 4 h. The FAM-Aβ1–42-containing medium was then removed and the remaining extracellular FAM-Aβ1–42 was quenched for 1 min with 100 μl 0.2% trypan blue in PBS, pH = 4.4. Following aspiration, fluorescence was measured at 485 nm excitation/535 nm emission using an Infinite 200 reader (Tecan). To normalize for cell numbers, 100 μl at 50 μg/ml Hoechst Dye 33342 in PBS was added and the sample was incubated for 30 min. Fluorescence was measured at 360 nm excitation/465 nm emission using the Infinite 200 reader (Tecan). In addition, microglial FAM-Aβ1–42 phagocytosis was verified by confocal microscopy using a BX61 microscope equipped with a disc-spinning unit (Olympus).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by grants from the Bundesministerium für Bildung und Forschung (BMBF) (KND grant 01 GI 0422 and KNDD grant 01GI 0711), as well as grants from the Deutsche Forschungsgemeinschaft (DFG) (BE 3828/3-2 and BE 3828/4-1 to T.B., HE 3350/8-2 (KFO177) to M.T.H.), T.B., M.T.H. and M.M.N. are members of the DFG cluster of excellence ImmunoSensation. This work was also supported by the BiomarkAPD Project of the Joint Programme—Neurodegenerative Disease Research by a grant from the BMBF (01ED1203D to P.L. and M.T.H.). Research of the VUmc Alzheimer center is part of the neurodegeneration research program of the Neuroscience Campus Amsterdam, the Netherlands. The VUmc Alzheimer Center is supported by Alzheimer Nederland and Stichting VUmc fonds. The clinical database structure was developed with funding from Stichting Dioraphte. Data used in preparation of this article were obtained from the ADNI database (http://adni.loni.ucla.edu/). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering and through generous contributions from the following: Alzheimer's Association; Alzheimer's Drug Discovery Foundation; BioClinica, Inc.; Biogen Idec, Inc.; Bristol-Myers Squibb Company; Eisai, Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd; Janssen Alzheimer Immunotherapy Research & Development, LLC; Johnson & Johnson Pharmaceutical Research & Development LLC; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC; NeuroRx Research; Novartis Pharmaceuticals Corporation; Pfizer, Inc.; Piramal Imaging; Servier; Synarc, Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by National Institute of Health (grants numbers P30 AG010129 and K01 AG030514).
AUTHORS’ CONTRIBUTIONS
Study concept and design: A.R., W.M.v.d.F., C.H., F.J., W.M., M.T.H., T.B., M.M.N. Acquisition data: A.R., W.M.v.d.F., C.H., D.R., S.H., P.L., J.P., A.L., D.D., P.H., R.H., L.B., Sherms, A.G., F.J., W.M., M.T.H., T.B., M.M.N. Sample contribution: W.M.v.d.F., D.R., J.P., E.L., C.T., H.H., J.K., O.P., R.H., L.F., M.H., H.K., P.S., M.M.B., E.R., J.W., F.J., W.M., M.T.H. Data analysis: A.R., W.M.v.d.F., C.H., D.R., S.H., P.L., A.L., D.D., E.L., M.P.K., C.C., P.H., L.B., Sherms, M.M.B., F.J., W.M., M.T.H., T.B., M.M.N. Statistical analysis and interpretation: A.R., W.M.v.d.F., C.H., D.R., A.L., D.D., E.L., M.P.K., L.B., Sherms, F.J., W.M., M.T.H., T.B., M.M.N. Drafting of the manuscript: A.R., W.M.v.d.F., C.H., D.R., S.H., P.L., J.P., A.L., D.D., E.L., M.P.K., C.C., P.H., C.T., H.H., J.K., O.P., R.H., L.F., M.H., L.B., Sherms, H.K., P.S., M.M.B., E.R., J.W., A.G., F.J., W.M., M.T.H., T.B., M.M.N.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Lara Buscemi from the Université de Lausanne, Switzerland for her assistance with confocal microscopy. We also thank Dr Ellen Gelpí from the Neurological Tissue Bank of the Biobank of the Hospital Clinic-Institut d'Investigacions Biomèdiques August Pi i Sunyer, Barcelona, Spain for providing postmortem human brain tissue from AD dementia patients and controls.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Kim S., Swaminathan S., Shen L., Risacher S.L., Nho K., Foroud T., Shaw L.M., Trojanowski J.Q., Potkin S.G., Huentelman M.J., et al. Genome-wide association study of CSF biomarkers Abeta1–42, t-tau, and p-tau181p in the ADNI cohort. Neurology. 2011;76:69–79. doi: 10.1212/WNL.0b013e318204a397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schjeide B.M., Schnack C., Lambert J.C., Lill C.M., Kirchheiner J., Tumani H., Otto M., Tanzi R.E., Lehrach H., Amouyel P., et al. The role of clusterin, complement receptor 1, and phosphatidylinositol binding clathrin assembly protein in Alzheimer disease risk and cerebrospinal fluid biomarker levels. Arch. Gen. Psychiatry. 2011;68:207–213. doi: 10.1001/archgenpsychiatry.2010.196. [DOI] [PubMed] [Google Scholar]
- 3.Elias-Sonnenschein L.S., Helisalmi S., Natunen T., Hall A., Paajanen T., Herukka S.K., Laitinen M., Remes A.M., Koivisto A.M., Mattila K.M., et al. Genetic loci associated with Alzheimer's disease and cerebrospinal fluid biomarkers in a Finnish case-control cohort. PLoS ONE. 2013;8:e59676. doi: 10.1371/journal.pone.0059676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han M.R., Schellenberg G.D., Wang L.S. Genome-wide association reveals genetic effects on human Abeta42 and tau protein levels in cerebrospinal fluids: a case control study. BMC Neurol. 2010;10:90. doi: 10.1186/1471-2377-10-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cruchaga C., Kauwe J.S., Harari O., Jin S.C., Cai Y., Karch C.M., Benitez B.A., Jeng A.T., Skorupa T., Carrell D., et al. GWAS of cerebrospinal fluid tau levels identifies risk variants for Alzheimer's disease. Neuron. 2013;78:256–268. doi: 10.1016/j.neuron.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C., DeStafano A.L., Bis J.C., Beecham G.W., Grenier-Boley B., et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat. Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mattiace L.A., Davies P., Dickson D.W. Detection of HLA-DR on microglia in the human brain is a function of both clinical and technical factors. Am. J. Pathol. 1990;136:1101–1114. [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson J.D., Mehus J.G., Tews K., Milavetz B.I., Lambeth D.O. Genetic evidence for the expression of ATP- and GTP-specific succinyl-CoA synthetases in multicellular eucaryotes. J. Biol. Chem. 1998;273:27580–27586. doi: 10.1074/jbc.273.42.27580. [DOI] [PubMed] [Google Scholar]
- 9.Ostergaard E. Disorders caused by deficiency of succinate-CoA ligase. J. Inherit. Metab. Dis. 2008;31:226–229. doi: 10.1007/s10545-008-0828-7. [DOI] [PubMed] [Google Scholar]
- 10.Silva D.F., Selfridge J.E., Lu J., E L., Cardoso S.M., Swerdlow R.H. Mitochondrial abnormalities in Alzheimer's disease: possible targets for therapeutic intervention. Adv. Pharmacol. 2012;64:83–126. doi: 10.1016/B978-0-12-394816-8.00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller C., Wang L., Ostergaard E., Dan P., Saada A. The interplay between SUCLA2, SUCLG2, and mitochondrial DNA depletion. Biochim. Biophys. Acta. 2011;1812:625–629. doi: 10.1016/j.bbadis.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 12.Mawuenyega K.G., Sigurdson W., Ovod V., Munsell L., Kasten T., Morris J.C., Yarasheski K.E., Bateman R.J. Decreased clearance of CNS beta-amyloid in Alzheimer's disease. Science. 2010;330:1774. doi: 10.1126/science.1197623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bradshaw E.M., Chibnik L.B., Keenan B.T., Ottoboni L., Raj T., Tang A., Rosenkrantz L.L., Imboywa S., Lee M., Von Korff A., et al. CD33 Alzheimer's disease locus: altered monocyte function and amyloid biology. Nat. Neurosci. 2013;16:848–850. doi: 10.1038/nn.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang Q., Lee C.Y., Mandrekar S., Wilkinson B., Cramer P., Zelcer N., Mann K., Lamb B., Willson T.M., Collins J.L., et al. ApoE promotes the proteolytic degradation of Abeta. Neuron. 2008;58:681–693. doi: 10.1016/j.neuron.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tokuda T., Calero M., Matsubara E., Vidal R., Kumar A., Permanne B., Zlokovic B., Smith J.D., Ladu M.J., Rostagno A., et al. Lipidation of apolipoprotein E influences its isoform-specific interaction with Alzheimer's amyloid beta peptides. Biochem. J. 2000;348:359–365. [PMC free article] [PubMed] [Google Scholar]
- 16.Tarawneh R., Holtzman D.M. Biomarkers in translational research of Alzheimer's disease. Neuropharmacology. 2010;59:310–322. doi: 10.1016/j.neuropharm.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heneka M.T., Kummer M.P., Stutz A., Delekate A., Schwartz S., Vieira-Saecker A., Griep A., Axt D., Remus A., Tzeng T.C., et al. NLRP3 is activated in Alzheimer's disease and contributes to pathology in APP/PS1 mice. Nature. 2013;493:674–678. doi: 10.1038/nature11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandrekar S., Jiang Q., Lee C.Y., Koenigsknecht-Talboo J., Holtzman D.M., Landreth G.E. Microglia mediate the clearance of soluble Abeta through fluid phase macropinocytosis. J. Neurosci. 2009;29:4252–4262. doi: 10.1523/JNEUROSCI.5572-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee C.Y., Landreth G.E. The role of microglia in amyloid clearance from the AD brain. J. Neural. Transm. 2010;117:949–960. doi: 10.1007/s00702-010-0433-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dumont M., Beal M.F. Neuroprotective strategies involving ROS in Alzheimer disease. Free Radic. Biol. Med. 2011;51:1014–1026. doi: 10.1016/j.freeradbiomed.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hickman S.E., Allison E.K., El Khoury J. Microglial dysfunction and defective beta-amyloid clearance pathways in aging Alzheimer's disease mice. J. Neurosci. 2008;28:8354–8360. doi: 10.1523/JNEUROSCI.0616-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilkinson B.L., Cramer P.E., Varvel N.H., Reed-Geaghan E., Jiang Q., Szabo A., Herrup K., Lamb B.T., Landreth G.E. Ibuprofen attenuates oxidative damage through NOX2 inhibition in Alzheimer's disease. Neurobiol. Aging. 2012;33:197. doi: 10.1016/j.neurobiolaging.2010.06.014. e121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKhann G., Drachman D., Folstein M., Katzman R., Price D., Stadlan E.M. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 24.Kornhuber J., Schmidtke K., Frolich L., Perneczky R., Wolf S., Hampel H., Jessen F., Heuser I., Peters O., Weih M., et al. Early and differential diagnosis of dementia and mild cognitive impairment: design and cohort baseline characteristics of the German Dementia Competence Network. Dement. Geriatr. Cogn. Disord. 2009;27:404–417. doi: 10.1159/000210388. [DOI] [PubMed] [Google Scholar]
- 25.Jessen F., Wiese B., Bickel H., Eifflander-Gorfer S., Fuchs A., Kaduszkiewicz H., Kohler M., Luck T., Mosch E., Pentzek M., et al. Prediction of dementia in primary care patients. PLoS ONE. 2011;6:e16852. doi: 10.1371/journal.pone.0016852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luck T., Riedel-Heller S.G., Kaduszkiewicz H., Bickel H., Jessen F., Pentzek M., Wiese B., Koelsch H., van den Bussche H., Abholz H.H., et al. Mild cognitive impairment in general practice: age-specific prevalence and correlate results from the German study on ageing, cognition and dementia in primary care patients (AgeCoDe) Dement. Geriatr. Cogn. Disord. 2007;24:307–316. doi: 10.1159/000108099. [DOI] [PubMed] [Google Scholar]
- 27.Zaudig M., Mittelhammer J., Hiller W., Pauls A., Thora C., Morinigo A., Mombour W. SIDAM – a structured interview for the diagnosis of dementia of the Alzheimer type, multi-infarct dementia and dementias of other aetiology according to ICD-10 and DSM-III-R. Psychol. Med. 1991;21:225–236. doi: 10.1017/s0033291700014811. [DOI] [PubMed] [Google Scholar]
- 28.McKhann G.M., Knopman D.S., Chertkow H., Hyman B.T., Jack C.R., Jr, Kawas C.H., Klunk W.E., Koroshetz W.J., Manly J.J., Mayeux R., et al. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kempen G.I., Meier A.J., Bouwens S.F., van Deursen J., Verhey F.R. [The psychometric properties of the Dutch version of the Telephone Interview Cognitive Status (TICS)] Tijdschr. Gerontol. Geriatr. 2007;38:38–45. [PubMed] [Google Scholar]
- 30.Schmermund A., Mohlenkamp S., Stang A., Gronemeyer D., Seibel R., Hirche H., Mann K., Siffert W., Lauterbach K., Siegrist J., et al. Assessment of clinically silent atherosclerotic disease and established and novel risk factors for predicting myocardial infarction and cardiac death in healthy middle-aged subjects: rationale and design of the Heinz Nixdorf RECALL Study. Risk Factors, Evaluation of Coronary Calcium and Lifestyle. Am. Heart J. 2002;144:212–218. doi: 10.1067/mhj.2002.123579. [DOI] [PubMed] [Google Scholar]
- 31.Krawczak M., Nikolaus S., von Eberstein H., Croucher P.J., El Mokhtari N.E., Schreiber S. PopGen: population-based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genet. 2006;9:55–61. doi: 10.1159/000090694. [DOI] [PubMed] [Google Scholar]
- 32.Wichmann H.E., Gieger C., Illig T. KORA-gen – resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen. 2005;67(Suppl. 1):S26–S30. doi: 10.1055/s-2005-858226. [DOI] [PubMed] [Google Scholar]
- 33.Popp J., Lewczuk P., Frommann I., Kolsch H., Kornhuber J., Maier W., Jessen F. Cerebrospinal fluid markers for Alzheimer's disease over the lifespan: effects of age and the APOEepsilon4 genotype. J. Alzheimers Dis. 2010;22:459–468. doi: 10.3233/JAD-2010-100561. [DOI] [PubMed] [Google Scholar]
- 34.Mulder C., Verwey N.A., van der Flier W.M., Bouwman F.H., Kok A., van Elk E.J., Scheltens P., Blankenstein M.A. Amyloid-beta(1–42), total tau, and phosphorylated tau as cerebrospinal fluid biomarkers for the diagnosis of Alzheimer disease. Clin. Chem. 2010;56:248–253. doi: 10.1373/clinchem.2009.130518. [DOI] [PubMed] [Google Scholar]
- 35.Harold D., Abraham R., Hollingworth P., Sims R., Gerrish A., Hamshere M.L., Pahwa J.S., Moskvina V., Dowzell K., Williams A., et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat. Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hollingworth P., Harold D., Sims R., Gerrish A., Lambert J.C., Carrasquillo M.M., Abraham R., Hamshere M.L., Pahwa J.S., Moskvina V., et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat. Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lambert J.C., Heath S., Even G., Campion D., Sleegers K., Hiltunen M., Combarros O., Zelenika D., Bullido M.J., Tavernier B., et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat. Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 38.Naj A.C., Jun G., Beecham G.W., Wang L.S., Vardarajan B.N., Buros J., Gallins P.J., Buxbaum J.D., Jarvik G.P., Crane P.K., et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat. Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mangold E., Ludwig K.U., Birnbaum S., Baluardo C., Ferrian M., Herms S., Reutter H., de Assis N.A., Chawa T.A., Mattheisen M., et al. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat. Genet. 2010;42:24–26. doi: 10.1038/ng.506. [DOI] [PubMed] [Google Scholar]
- 40.Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Bakker P.I., Ferreira M.A., Jia X., Neale B.M., Raychaudhuri S., Voight B.F. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meesters C., Leber M., Herold C., Angisch M., Mattheisen M., Drichel D., Lacour A., Becker T. Quick, "imputation-free" meta-analysis with proxy-SNPs. BMC Bioinformatics. 2012;13:231. doi: 10.1186/1471-2105-13-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huedo-Medina T.B., Sanchez-Meca J., Marin-Martinez F., Botella J. Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychol. Methods. 2006;11:193–206. doi: 10.1037/1082-989X.11.2.193. [DOI] [PubMed] [Google Scholar]
- 46.Herold C., Steffens M., Brockschmidt F.F., Baur M.P., Becker T. INTERSNP: genome-wide interaction analysis guided by a priori information. Bioinformatics. 2009;25:3275–3281. doi: 10.1093/bioinformatics/btp596. [DOI] [PubMed] [Google Scholar]
- 47.Cordell H.J., Clayton D.G. A unified stepwise regression procedure for evaluating the relative effects of polymorphisms within a gene using case/control or family data: application to HLA in type 1 diabetes. Am. J. Hum. Genet. 2002;70:124–141. doi: 10.1086/338007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.