Abstract
Lethal congenital contracture syndrome (LCCS) is a lethal autosomal recessive form of arthrogryposis multiplex congenita (AMC). LCCS is genetically heterogeneous with mutations in five genes identified to date, all with a role in the innervation or contractile apparatus of skeletal muscles. In a consanguineous Saudi family with multiple stillbirths presenting with LCCS, we excluded linkage to all known LCCS loci and combined autozygome analysis and whole-exome sequencing to identify a novel homozygous variant in ZBTB42, which had been shown to be enriched in skeletal muscles, especially at the neuromuscular junction. Knockdown experiments of zbtb42 in zebrafish consistently resulted in grossly abnormal skeletal muscle development and myofibrillar disorganization at the microscopic level. This severe muscular phenotype is successfully rescued with overexpression of the human wild-type ZBTB42 gene, but not with the mutant form of ZBTB42 that models the human missense change. Our data assign a novel muscular developmental phenotype to ZBTB42 in vertebrates and establish a new LCCS6 type caused by ZBTB42 mutation.
INTRODUCTION
Arthrogryposis multiplex congenita (AMC) is a malformation syndrome characterized by non-progressive congenital contracture in two or more body areas with evidence of muscle wasting (1,2). This highly morbid condition occurs at a frequency that ranges widely between different populations (1 : 3000–1 : 56 000) and can be lethal in severe cases (3,4). One of the fatal forms of AMC is the lethal congenital contracture syndrome (LCCS). Although some advocate a strict definition of LCCS that limits its application to fetal deaths that occur on or prior to 32 weeks of gestation with evidence of anterior horn cell involvement, more recent literature has adopted a more permissive definition that includes cases of AMC that die at or shortly after birth even when evidence of anterior horn cell involvement is lacking (5).
Lack of fetal movement across the joints that are involved in AMC can be attributed to factors that impair the contractility or innervation of skeletal muscle, in addition to those that exert mechanical restriction on muscle movement internally (e.g. connective tissue defects), or externally (e.g. uterine fibroids) (6). To date, five genes have been implicated in the pathogenesis of LCCS and define five subtypes (LCCS1-5), and the proteins they encode play a role in the innervation or the contractile apparatus of skeletal muscles. LCCS1 is caused by mutations in GLE1 (MIM 603371), which encodes an mRNA export mediator that is thought to be involved in the survival of the anterior horn cell neurons (7). Interestingly, the genes linked to LCCS2 and LCCS3 [ERBB3 (MIM 190151) and PIP5K1C (MIM 611369), respectively] are involved in the synthesis of inositol hexakisphosphate, which is a binding partner of GLE1 (8,9). On the other hand, LCCS4 and LCCS5 are caused by homozygous mutations in genes [MYBPC1 (MIM 160794) and DNM2 (MIM 602378), respectively] that had previously been linked to a relatively mild form of arthrogryposis known as distal arthrogryposis in an autosomal dominant fashion. Both MYBCP1 and DNM2 were linked to LCCS based on a single mutation each (10,11). Unfortunately, there are no data on the relative contribution of each of five LCCS genes to the overall mutation spectrum of this lethal disorder, but it appears likely that its genetic heterogeneity is not limited to these five loci.
In this study, we show that a consanguineous family with multiple stillbirths owing to LCCS does not map to any of the above-mentioned loci and that it likely defines a novel LCCS6 subtype caused by a mutation in ZBTB42, which we show to be essential for normal muscle development in zebrafish.
RESULTS
Clinical report
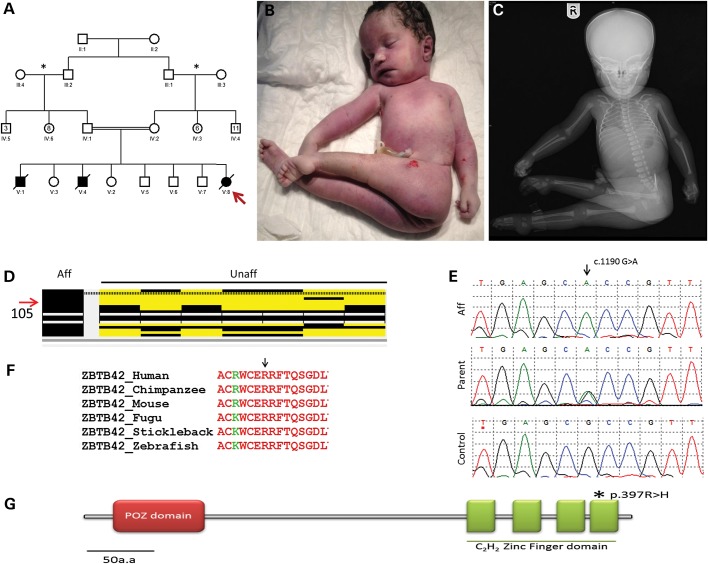
The parents are first cousins with five healthy children and history of male and female stillbirth neonates with arthrogryposis (Fig. 1A). The mother was 35 years old at the time of delivery of the index case. Pregnancy was complicated by reduced fetal movements as confirmed by antenatal ultrasounds, which also showed severe polyhydramnios, absent stomach, and multiple contracture deformities, but there were no major renal or CNS malformations. Multiple sessions of reductive amniocentesis were necessitated owing to the severity of the polyhydramnios. Karyotype was normal on the amniocentesis sample. Delivery was vaginal at 35 weeks of gestation. The female newborn had a birth weight of 2.31 kg, length of 53 cm and OFC of 36 cm. She had no respiratory effort and was announced dead (parents requested no resuscitation). There was no major craniofacial dysmorphism apart from macrocephaly. She had fixed–flexed hips, severely adducted lower limbs, fixed extension deformity of the knees symmetrically, fixed flexion of both hands, and dorsiflexion of the feet. No apparent digital defects were noted. Babygram showed no gross bony abnormality (Fig. 1B and C). Muscle biopsy and autopsy were denied by the family.
Figure 1.
(A) Pedigree for a consanguineous Saudi family with LCCS (asterisk indicates families from the same region; red arrow indicates index patient). (B and C) Clinical and X-ray images of stillborn baby showing fixed extension of limbs. (D) ZBTB42 is within a homozygous region found in affected individual in comparison with unaffected members of the family. (E) Chromatograms of the missense mutation found in ZBTB42. (F) Strong conservation of Arg397, which is mutated in the patient. (G) Protein schematic of ZBTB42 indicating the zinc finger domain in which the mutation lies.
Exclusion of known LCCS loci
Because of the consanguineous nature of the parents and the recurrence among male and female siblings, we hypothesized that LCCS in this family is caused by autozygosity of an ancestral mutation along with its surrounding haplotype in a pattern that can be tracked by autozygome analysis using runs of homozygosity as surrogates (12). Our analysis of the index's autozygome revealed that it covered 391 Mb of her genome, but only 65 Mb of the autozygome was unique to the index (i.e. not shared by any of the five unaffected siblings), spanning a total of 455 annotated OMIM genes (Supplementary Material, Table S1). Because none of the LCCS genes reside within the autozygome, we concluded that this family probably defines a novel LCCS gene (LCCS6) that maps to the autozygome of the index. DNA was not available from the two previous stillbirths.
WES identifies a novel ZBTB42 mutation
WES of the index generated 71 164 variants (Supplementary Material, Table S2). By only considering homozygous coding/splicing variants which were novel or reported in dbSNPs with low frequency up to 0.005 and within the autozygome of the index, the list of candidate variants was reduced to only two. One missense variant was found in ZBTB42 (ZBTB42:NM_001137601:exon2:c.1190G>A:p.R397H), a poorly characterized zinc finger protein-encoding gene (Fig. 1E). Both parents and two healthy siblings were found to be carriers for this variant whereas the other three healthy siblings were found to be homozygous wild type. This variant is absent in 485 Saudi exomes, 300 normal Saudi controls by Sanger sequencing, 1000 Genomes and in the Exome Variant Server. Interestingly, this variant replaces a highly conserved amino acid residue in the fourth zinc finger-binding domain of the protein (Fig. 1F and G). The second missense variant was found in SCN4A (MIM 603967) (SCN4A:NM_000334:exon9:c.1298T>G:p.L433R), a gene linked to hyperkalemic periodic paralysis and paramyotonia congenita. Because SCN4A encodes one of the major voltage-gated sodium channels in skeletal muscle and its associated pathologies manifest as weakness and myotonia, both ZBTB42 and SCN4A variants were regarded as potential disease-causing candidates in our subsequent functional analyses.
ZBTB42 is necessary for normal muscle development
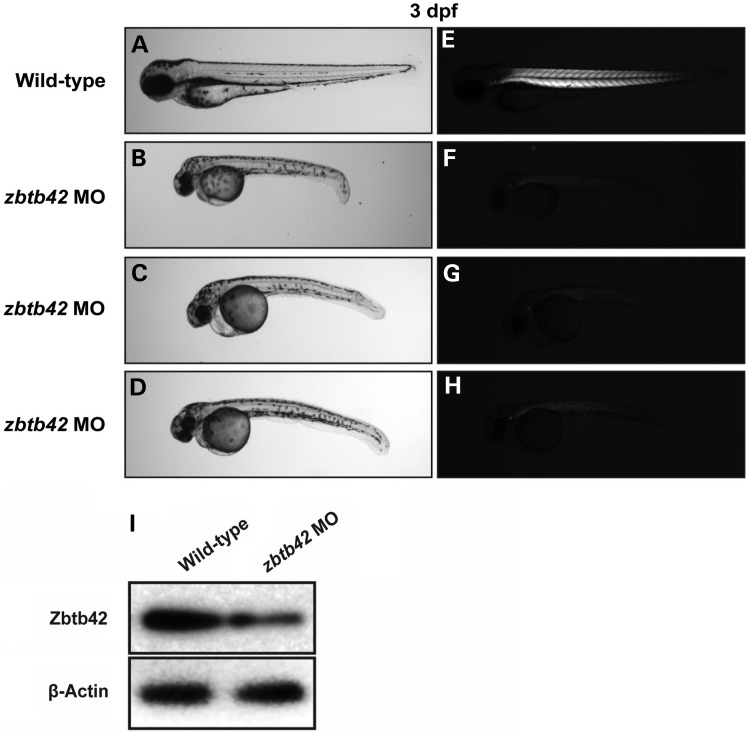
To understand the in vivo function of our lead candidate, ZBTB42, we used zebrafish as a model system. The zebrafish zbtb42 gene (NM_001137601) encodes a protein of 511 amino acids that is 43% identical to the human protein with even higher amino acid sequence identity (∼80%) in the conserved N-terminal BTB domain and C-terminal zinc finger domains. To ablate the function of zbtb42 in zebrafish, two independent morpholinos were designed to target the translational start site (ATG-MO) and exon1–intron1 (Ex1-MO) splice site. A standard morpholino directed against human β-globin that is absent in the zebrafish genome was used in all experiments to control for injection-related nonspecific effects in wild-type embryos. Knockdown using either the translation (ATG-MO) or splice site (Ex1-MO) blocking morpholinos resulted in similar phenotypes at relatively low concentrations (3–4 ng), suggesting the specificity of their action (Fig. 2A–H). For consistency, the ATG-MO was used for all later characterization studies of our zebrafish model. Efficacy of the ATG-MO to block translation of zbtb42 transcripts was investigated by western blot analysis of protein extracts from 3 dpf embryos. Highly reduced levels of Zbtb42 were observed in the morphant zebrafish in comparison with control-injected embryos (Fig. 2I).
Figure 2.
Knockdown of zbtb42 results in morphological defects and skeletal muscle disorganization in zebrafish. Representative zebrafish embryos injected with 3–5 ng of zbtb42 ATG-MO are hypotonic and shorter in body length (B–D) than wild-type controls (A) at 3 dpf. Zbtb42 morphants have a bent body axis, small eyes and head, in addition to decreased overall body girth. Imaging under normal light reveals cardiac edema and moderate bradycardia in MO-injected embryos, quantified as ∼75% of the heart rate of wild-type controls. Imaging under polarized light indicates highly reduced birefringence in zbtb42 morphants (F–H) compared with wild type (E), suggesting disruptions to normal skeletal muscle organization. (I) Western blot demonstrates that knockdown of zbtb42 (3 ng of ATG-MO) results in significantly decreased levels of Zbtb42 protein, with a 46% reduction in MO-injected morphants compared with wild-type controls at 3 dpf. β-Actin was used as a loading control, and band quantification was performed using ImageJ.
Zbtb42 morphants are leaner and smaller in size compared with controls and exhibited a dorsal curvature through the back and tail, instead of the normal flat dorsum, similar to other zebrafish models of myopathy, including klhl41 (nemaline myopathy with fetal arthrogryposis) and bin1 (centronuclear myopathy) (Fig. 2A–H) (13,14). Zbtb42 morphant fish also exhibited a mild bradycardia. Skeletal muscles of zebrafish embryos were examined using a birefringence assay that involves examination of axial skeletal muscles of live zebrafish embryos using polarized filter microscopy (15). Skeletal muscles of zbtb42 morphant zebrafish showed highly reduced birefringence in comparison with the controls suggesting a defect in myofiber organization in somites (Fig. 2A–H).
As the hallmark feature of AMC patients is the joint contractures associated with reduced mobility, the effect of Zbtb42 deficiency was examined on the motility behaviors of zebrafish. The first recognizable muscle-dependent motor activity in zebrafish is spontaneous embryo coiling, detectable between 17 and 22 hours post-fertilization (hpf). On average, wild-type embryos coil 31.3 ± 4.4 times per minute, whereas zbtb42 morphants coil only 9.6 ± 1.6 times per minute (P < 0.001). This suggests an early muscle weakness in the zbtb42 morphant zebrafish. Zebrafish embryos swim in response to touch by 26 hpf, and therefore, the swimming behaviors were analyzed in the control and zbtb42 morphant fish at 3 dpf. Zbtb42 morphant fish were largely immotile, and their touch-evoked response was blunted. Instead of rapidly swimming out of field of view like control fish (7.48 ± 2.1 cm/0.1 s), they twitched and only moved several lengths (1.59 ± 0.69 cm/0.1 s) on administration of touch stimuli (P < 0.001; Supplementary Material, Movies S1 and S2). The decreased spontaneous coiling and diminished touch-evoked escape behaviors are indicative of reduced muscle function and overall muscle weakness.
Zbtb42 deficiency results in sarcomeric disorganization and neuromuscular junction abnormalities in zebrafish
Skeletal muscle organization was examined in zbtb42 morphant zebrafish to understand whether the reduced birefringence and impaired motor function is a direct consequence of structural abnormalities in myofibers. Skeletal muscle histology was observed using toluidine blue-stained longitudinal sections of control and morphant zebrafish at 3 dpf. Histological survey of morphant fish revealed disorganized myofibers with fewer myofibers in somites. Moreover, large areas that completely lacked the sarcomeric organization were present in morphant zebrafish (Fig. 3A and B). To visualize the skeletal muscle abnormalities in Zbtb42 deficiency, whole-mount immunostaining was performed with phalloidin to stain for actin thin filaments. Phalloidin staining in wild-type zebrafish revealed that adjacent myofibers were positioned in close proximity to each other, whereas zbtb42 morphants exhibited thinner myofibers with large gaps between adjacent myofibers. Zbtb42-deficient myofibers also showed areas that lacked well-organized myofibrillar striations and appeared to contain aggregates of sarcomeric proteins (Fig. 3C–F). ZBTB42 is highly expressed in subsynpatic nuclei in skeletal muscles underlying the neuromuscular junctions (NMJs) (16). Therefore, the effect of Zbtb42 deficiency on NMJs was also evaluated in zebrafish. Immunofluorescence analysis with α-bungarotoxin, which binds to acetylcholine receptors on the postsynaptic side of NMJs, showed fewer and fragmented NMJs in zbtb42 morphants compared with the densely branching NMJs seen in normal controls (Fig. 3G and H).
Figure 3.
Abnormal histology of Zbtb42-deficient zebrafish. (A and B) Longitudinal semi-thin sections of 3 dpf zebrafish skeletal muscles stained with toluidine blue. Wild-type muscles exhibit well-differentiated myofibers with oblong nuclei localized to the fiber periphery, whereas zbtb42 morphant muscles appear highly disorganized, with pronounced spacing between misaligned fibers and evidence of wasting/atrophy in these areas (black arrow). (C–F) Whole-mount phalloidin staining of 3 dpf zebrafish embryos demonstrates widespread myofibrillar disarray in zbtb42 morphants (D and F) compared with wild-type (C and E). Higher magnifications emphasize the presence of sarcomeric aggregates in the myofibers (white arrows). Nuclei are stained with DAPI. (G and H) Staining of postsynaptic acetylcholine receptors using α-bungarotoxin shows smaller and fragmented NMJs (arrows) in zbtb42 morphants. Scale bars: 2 μm.
To identify ultrastructural defects in subcellular compartments of skeletal muscle, transmission electron microscopy was performed at 3 dpf. Longitudinal view of the skeletal muscle in zbtb42 morphants showed significant myofibrillar disarray lacking normal actin-myosin organization (arrows in Fig. 4B and D) in comparison with highly organized myofibrillar structure in wild-type zebrafish. Notably, skeletal muscle of Zbtb42-deficient zebrafish lacked the clearly defined H-zone that is present in wild-type muscles (arrowheads in Fig. 4C and D).
Figure 4.
Zbtb42 deficiency in zebrafish disrupts skeletal muscle ultrastructure. (A–D) Transmission electron micrographs of 3 dpf zebrafish embryos show extensive sarcomeric disorganization in zbtb42 morphants (B and D) in comparison with wild-type controls (A and C). In wild-type fish, myofibers are parallel to one another (A) whereas myofibrils present in morphant fish appear randomly oriented and disintegrating (B, arrow). Higher magnifications reveal the organization of thick and thin filaments to be disrupted in morphant myofibers (D, arrow). Discrete H bands characteristic of wild-type skeletal muscle are absent in zbtb42 morphants (C–D, arrowheads). Scale bars: A–B = 2 μm; C–D = 500 nm.
Human ZBTB42 mutation results in a null phenotype in zebrafish
In vivo consequence of the human p.Arg397His mutation was studied by modeling this change in our zebrafish model of Zbtb42 deficiency (Supplementary Material, Fig. S1). Human Arg397 is located in the highly conserved C-terminal zinc finger domain of ZBTB42 protein, and this amino acid residue is highly conserved in all vertebrate species. Human wild-type or mutant ZBTB42 mRNA transcripts were overexpressed and evaluated for their ability to rescue the skeletal muscle abnormalities observed in zbtb42 morphants. Overexpression of wild-type ZBTB42 resulted in restoration of muscle structure to normal in 57.2 ± 7.7% of morphant fish (Fig. 5A, B and D). A subset of these rescued fish had pericardial edema, as seen in morphant fish, which could be a nonspecific phenotype associated with morpholinos. In contrast, overexpression of mutant ZBTB42 (p.Arg397His) was neither able to rescue the morphology nor birefringence of zbtb42 morphant zebrafish (Fig. 5A, C and D). Clutches injected with both MO and mutant ZBTB42 showed only 5.9 ± 2.0% of normal fish, a percentage statistically insignificant from clutches injected with MO alone (P < 0.001). These data show that Arg397 is crucial for normal function of ZBTB42 protein in vertebrates.
Figure 5.
Skeletal muscle phenotype of zbtb42 morphants is rescued with wild-type but not mutant human ZBTB42 transcripts. (A and B) Polarized light microscopy of 4 dpf live embryos shows that overexpression of wild-type human ZBTB42 mRNA significantly increases birefringence and corrects the morphological abnormalities of zbtb42 morphant embryos. The phenotype indexed as ‘mild’ (asterisked in B) was considered a partial but not complete rescue, as the cardiac edema and reduced heartbeat frequently observed in traditional zbtb42 morphants (arrows) was not always restored alongside vast improvements in skeletal muscle architecture. (C) Overexpression of human ZBTB42 mRNA containing the Arg397His mutation fails to rescue the zbtb42 morphant phenotype in either a full or partial manner. (D) Quantification of phenotypic changes observed with mRNA overexpression in wild-type and zbtb42 morphants at 4 dpf. Wild-type embryos injected with only mRNA (no ATG-MO) are included as controls. Rescue experiments were performed in triplicate.
SCN4A p.Leu433Arg missense change is nonpathogenic variant
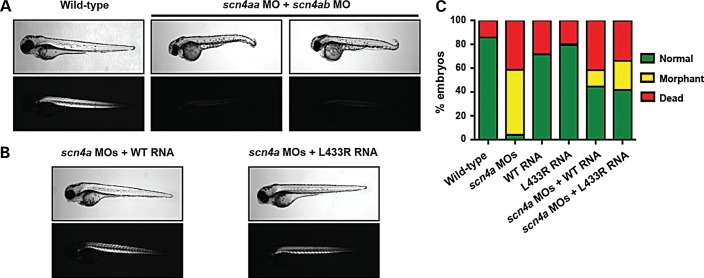
To determine whether the second missense variant detected by WES was pathogenic in our subject family, the human SCN4A p.Leu433Arg mutation was examined using the same mRNA rescue method. Because the zebrafish scn4a gene is duplicated (scn4aa and scn4ab isoforms), we first established the individual knockdown phenotypes of embryos injected with scn4aa MO and with scn4ab MO alone (Supplementary Material, Fig. S2). Each MO resulted in abnormal morphologies and reduced birefringence, with the scn4aa gene knockdown consistently being the more severe of the two. Morphants exhibited smaller, shorter trunks as well as bent tails. Because SCN4A is actively expressed in zebrafish skeletal muscle and encodes a major voltage-gated sodium channel in this tissue (17), such abnormalities were anticipated and are characteristic of several zebrafish models of neuromuscular disease (13,14,18). To achieve complete knockdown for rescue studies, a combination of both MOs was used (Fig. 6A). Human wild-type or mutant SCN4A transcripts were overexpressed in scn4a double morphant embryos (Fig. 6B and C). Overexpression of wild-type and mutant SCN4A transcripts were both found to rescue scn4a double morphants, with no statistically significant difference between the two (WT RNA: 73.5 ± 20.2% normal fish; p.Leu433Arg RNA: 67.1 ± 20.9% normal fish; P = 0.68). Both wild-type and mutant SCN4A rescues were statistically significant compared with clutches injected with the two MOs alone in terms of percentage of normal fish (MOs: 6.3 ± 6.2% normal fish; P < 0.001). These data serve as compelling in vivo evidence that SCN4A is not the causative gene of LCCS6 in the subject family.
Figure 6.
Skeletal muscle phenotype resulting from scn4aa and scn4ab double knockdown in zebrafish embryos is rescued with both wild-type and mutant human SCN4A transcripts. (A) Polarized light microscopy of 4 dpf live embryos show severely reduced birefringence in scn4a double morphants compared with wild-type siblings. (B) Wild-type human SCN4A mRNA and human SCN4A mRNA containing the Leu433Arg mutation each rescue the scn4a double morphant phenotype. Both mRNAs correct the morphological defects of double morphant embryos and restore skeletal muscle birefringence to near wild-type levels. (C) Quantification of phenotypic changes observed with mRNA overexpression in wild-type and scn4a morphants at 4 dpf. Wild-type embryos injected with only mRNA (no 4A- and 4B-MOs) are included as controls. Rescue experiments were performed in triplicate.
DISCUSSION
In this study, we expand the genetic heterogeneity of LCCS by describing a loss of function mutation in ZBTB42 as the likely cause of LCCS6 in a consanguineous family.
The initial characterization of ZBTB42 was triggered by the identification of an SNP therein that forms part of a small haplotype associated with increased risk of insulin resistance and the development of the metabolic syndrome (19). Although this haplotype involves the first exon and proximal upstream region of AKT1, a gene known to be involved in the pathophysiology of the metabolic syndrome, Devaney and colleagues decided to extend their investigation to the other gene contained within the haplotype (i.e. ZBTB42) (16). They found that ZBTB42 is highly expressed in skeletal muscles and ovaries and weakly expressed in other tissues. Immunohistochemical analysis confirmed a highly specific localization of ZBTB42 to the nucleoplasm of myofibrils, especially in nuclei at the NMJ.
The knockdown and rescue experiments we conducted on zebrafish revealed a previously unknown role of ZBTB42 in muscle development. Remarkably, the phenotype we observed in morphants is comparable with the severe phenotype we observed in morphants for established myopathy disease genes. It is interesting to note that the human phenotype of severe forms of these myopathies overlaps greatly with LCCS, as we showed previously for KLHL41-linked nemaline myopathy (14). Moreover, NMJ defects observed in Zbtb42-deficient zebrafish are highly similar to abnormalities as seen in mutations in ECEL1, a recently identified cause of distal arthrogryposis (20). Failure of the ZBTB42 mRNA that harbors the human mutation to rescue this phenotype is highly indicative that the mutation is at least hypomorphic and probably nullimorphic in nature. Lack of phenotype in the carrier parents suggests haplosufficiency for the encoded protein.
The mechanism through which ZBTB42 exerts its action on muscle development remains unclear. Although zinc finger domains are best known for their role in DNA binding by transcription factors, they can also facilitate binding to RNA and other protein molecules (21). The BTB domain is usually present as a single copy and implicated in protein–protein interactions. The presence of both of these domains is crucial for protein function as DNA-binding zinc finger domains determine the sequence specificity, whereas the BTB domain promotes oligomerization and the recruitment of transcriptional regulators (22). Transcriptional regulation by BTB-ZF proteins is often required for normal neurological development, skeletal morphogenesis, regulation of fertility and lymphocyte development (23–26). Therefore, while it is tempting to speculate that ZBTB42 with its nuclear localization and four fingers may play a role as a transcription factor that regulates the expression of important muscle developmental genes, we caution that future research is needed to test whether this is indeed the case, and we are interested in pursuing this line of research in the near future.
MATERIALS AND METHODS
Human subjects
The index, siblings, and parents were recruited with informed consent using an IRB-approved protocol (KFSHRC RAC#2080006). Venous blood was collected in EDTA for genomic DNA extraction and downstream molecular studies.
Fish and embryo maintenance
Fish were bred and maintained as described previously (27). Wild-type control embryos were obtained from the Oregon AB line and were staged by hours (hpf) or days (dpf) post-fertilization at 28.5°C. All animal work was performed with approval from the Boston Children's Hospital Animal Care and Use Committee (11-05-1955R).
Autozygome analysis
The full set of autozygous intervals in the genome of the index (autozygome) was determined by first performing genome-wide SNP genotyping on the Axiom SNP Chip platform (Affymetrix, Santa Clara, CA, USA) followed by mapping homozygous intervals that are >2 Mb in length as surrogates of autozygosity using the software AutoSNPa (28). Exclusion of linkage to known LCCS genes was as described before (29).
Whole-exome sequencing
Standard whole-exome sequencing (WES) was performed using TruSeq Exome Enrichment kit (Illumina) followed by preparation of an Illumina sequencing library, with enrichment for the desired target using the Illumina Exome Enrichment protocol. The captured libraries were sequenced using Illumina HiSeq 2000 Sequencer. The reads were mapped against UCSC hg19 by BWA. The SNPs and Indels were detected by SAMTOOLS. The resulting variants were filtered as described previously (30).
Morpholino knockdown and mRNA rescue
Two antisense morpholinos, one targeting the translational start site (ATG-MO) and one targeting the exon 1–intron1 splice site (Ex1-MO), were designed to knockdown the zebrafish zbtb42 transcript (GeneTools LLC, Philomath, OR, USA). Two translational morpholinos were also designed to knockdown both isoforms of zebrafish scn4a, scn4aa (4A-MO) and scn4ab (4B-MO). The morpholino sequences were zbtb42 ATG-MO: 5′ AACTCCATTCTTATGGACGAAAACC 3′; zbtb42 Ex1-MO: 5′ TTTCGGCCACATAATTGACACCGTT 3′; scn4aa 4A-MO: 5′ CCATCTTGGCATCGCGAAGAGACAC 3′ and scn4ab 4B-MO: 5′ GAGCAGACGCGCCATCTTGACATTC 3′. Morpholinos were dissolved in 1× Danieau buffer with 0.1% phenol red and 3–5 ng injected into the yolk of 1-cell-stage embryos. For rescue experiments, full-length human ZBTB42 (NM_001137601) and SCN4A (NM_000334) cDNAs were cloned into pCS2 + destination vectors (a gift from Nathan Lawson) using Gateway technology (Invitrogen, Carlsbad, CA, USA). The p.Arg397His (c.1190G>A) and p.Leu433Arg (c.1298T>G) substitutions were incorporated into ZBTB42 and SCN4A cDNAs, respectively, using GENEART site-directed mutagenesis (Invitrogen). The mutagenesis primer sequences were as follows: 5′ GCCGGTGGTGTGAGCACCGTTTCACGCAGTC 3′ (ZBTB42 forward); 5′ GACTGCGTGAAACGGTGCTCACACCACCGGC 3′ (ZBTB42 reverse); 5′ TGGTCATCATCTTCCGGGGCTCTTTCTACCT 3′ (SCN4A forward) and 5′ AGGTAGAAAGAGCCCCGGAAGATGATGACCA 3′ (SCN4A reverse). mRNA for all constructs was synthesized in vitro using mMessage kits (Ambion, Austin, TX, USA). 100–200 pg of mRNA was injected into embryos at the 1-cell stage independently or in combination with ATG-MO (for zbtb42 rescues) or 4A-MO and 4B-MO (for scn4a rescues). Phenotypic analyses performed through 4 dpf (15).
Western blotting and immunofluorescence
Zebrafish embryos (3 dpf) were homogenized in buffer containing Tris–Cl (20 mm, pH 7.6), NaCl (50 mm), EDTA (1 mm), NP-40 (0.1%) and complete protease inhibitor cocktail (Roche Applied Sciences, Indianapolis, IN, USA). Following centrifugation at 11 000 g at 4°C for 15 min, protein concentration in supernatants was determined by BCA protein assay (Pierce, Rockford, IL, USA). Total protein (25 μg) was separated by electrophoresis on 4–12% gradient Tris–glycine gels (Invitrogen) and transferred onto polyvinylidene difluoride membrane (Invitrogen). Membranes were blocked in PBS containing 5% casein/0.1% Tween 20 and then incubated with either rabbit polyclonal anti-ZBTB42 (1 : 500, ab100919, Abcam, Cambridge, MA, USA) or mouse monoclonal anti-β-actin (1 : 1000, A5441, Sigma) primary antibodies. After washing, membranes were incubated with horseradish peroxidase-conjugated anti-rabbit (1 : 2500, 170–6515) or anti-mouse (1 : 5000, 170–6516) IgG secondary antibodies (BioRad, Hercules, CA, USA). Proteins were detected using the SuperSignal chemiluminescent substrate kit (Pierce). Whole-mount direct immunofluorescence was performed with Alexa Fluor 546 phalloidin (1 : 50, A22283, Invitrogen) or α-bungarotoxin (Invitrogen, B13423, 1 μg/ml) on 3 dpf embryos as described previously (14). Embryos were mounted in 70% glycerol and visualized using an Ultra VIEW VoX spinning disc confocal microscope (Perkin Elmer).
Touch-evoked escape response assay
Mechanosensory stimuli were delivered to 3 dpf embryos by touching the yolk sac or tail with an insect pin as described previously (15). Motor behaviors were recorded using a SPOT RT3 digital camera system (SPOT Imaging Solutions, Diagnostic Instruments, Inc., Sterling Heights, MI, USA) mounted on a Nikon SMZ1500 stereomicroscope. Video frame capture (30 Hz) was performed using ImageJ (NIH).
Histopathology
For electron microscopy, 3 dpf zebrafish embryos were fixed in formaldehyde–glutaraldehyde–picric acid in cacodylate buffer overnight at 4°C, followed by osmication and uranyl acetate staining. Subsequently, embryos were dehydrated in a series of ethanol washes and embedded in TAAB Epon (Marivac Ltd, Halifax, NS, Canada). Sections (95 nm) were cut with a Leica UltraCut microtome, picked up on 100-μm Formvar-coated copper grids and stained with 0.2% lead citrate. Sections were viewed and imaged under a Philips Tecnai BioTwin Spirit electron microscope (Philips, Amsterdam, the Netherlands) at the Harvard Medical School Electron Microscopy Core.
Quantification and statistical analysis
Data were statistically analyzed by parametric student's t-test (two-tailed) and were considered significant when P < 0.001. All data analyses were performed using GraphPad Prism 6 software (GraphPad Software, Inc., La Jolla, CA, USA).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by a DHFMR Collaborative Research Grant (to F.S.A.), KACST 13-BIO1113-20 (to F.S.A.), the National Institute of Arthritis and Musculoskeletal and Skin Diseases of National Institutes of Health (K01 AR062601) and the Charles H. Hood Foundation Research Award (to V.A.G.) and the National Institute of Neurological Disorders and Stroke (F31 NS081928 to L.L.S.). Microscopy imaging was performed at the Boston Children′s Hospital IDDRC Cellular Imaging Core, and DNA sequencing was done through the Boston Children′s Hospital IDDRC Molecular Genetics Core, both supported by National Institutes of Health (NIH-P30-HD-18655).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the family for their enthusiastic participation in this study. We also thank the Genotyping and Sequencing Core Facilities at KFSHRC for their technical help and Ms. Mais Hashem for her help as a clinical coordinator. Thanks are also due to Laura Crowley for helping with molecular cloning.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Stern W.G. Arthrogryposis multiplex congenita. JAMA. 1923;81:1507–1510. [Google Scholar]
- 2.Fisher R.L., Johnstone W.T., Fisher W.H., Jr, Goldkamp O.G. Arthrogryposis multiplex congenita: a clinical investigation. J. Pediatr. 1970;76:255–261. doi: 10.1016/s0022-3476(70)80171-8. [DOI] [PubMed] [Google Scholar]
- 3.Laitinen O., Hirvensalo M. Arthrogryposis multiplex congenita. Ann. Paediatr. Fenn. 1966;12:133–138. [PubMed] [Google Scholar]
- 4.Wynne-Davies R., Lloyd-Roberts G.C. Arthrogryposis multiplex congenita. Search for prenatal factors in 66 sporadic cases. Arch. Dis. Child. 1976;51:618–623. doi: 10.1136/adc.51.8.618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vuopala K., Herva R. Lethal congenital contracture syndrome: further delineation and genetic aspects. J. Med. Genet. 1994;31:521–527. doi: 10.1136/jmg.31.7.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall J.G. Arthrogryposis multiplex congenita: etiology, genetics, classification, diagnostic approach, and general aspects. J. Pediatr. Orthop. B. 1997;6:159–166. [PubMed] [Google Scholar]
- 7.Nousiainen H.O., Kestila M., Pakkasjarvi N., Honkala H., Kuure S., Tallila J., Vuopala K., Ignatius J., Herva R., Peltonen L. Mutations in mRNA export mediator GLE1 result in a fetal motoneuron disease. Nat. Genet. 2008;40:155–157. doi: 10.1038/ng.2007.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narkis G., Ofir R., Manor E., Landau D., Elbedour K., Birk O.S. Lethal congenital contractural syndrome type 2 (LCCS2) is caused by a mutation in ERBB3(Her3), a modulator of the phosphatidylinositol-3-kinase/Akt pathway. Am. J. Hum. Genet. 2007;81:589–595. doi: 10.1086/520770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narkis G., Ofir R., Landau D., Manor E., Volokita M., Hershkowitz R., Elbedour K., Birk O.S. Lethal contractural syndrome type 3 (LCCS3) is caused by a mutation in PIP5K1C, which encodes PIPKI gamma of the phophatidylinsitol pathway. Am. J. Hum. Genet. 2007;81:530–539. doi: 10.1086/520771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Markus B., Narkis G., Landau D., Birk R.Z., Cohen I., Birk O.S. Autosomal recessive lethal congenital contractural syndrome type 4 (LCCS4) caused by a mutation in MYBPC1. Hum. Mutat. 2012;33:1435–1438. doi: 10.1002/humu.22122. [DOI] [PubMed] [Google Scholar]
- 11.Koutsopoulos O.S., Kretz C., Weller C.M., Roux A., Mojzisova H., Bohm J., Koch C., Toussaint A., Heckel E., Stemkens D., et al. Dynamin 2 homozygous mutation in humans with a lethal congenital syndrome. Eur. J. Hum. Genet. 2013;21:637–642. doi: 10.1038/ejhg.2012.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alkuraya F.S. Autozygome decoded. Genet. Med. 2010;12:765–771. doi: 10.1097/GIM.0b013e3181fbfcc4. [DOI] [PubMed] [Google Scholar]
- 13.Smith L.L., Gupta V.A., Beggs A.H. Bridging integrator 1 (Bin1) deficiency in zebrafish results in centronuclear myopathy. Hum. Mol. Genet. 2014;23:3566–3578. doi: 10.1093/hmg/ddu067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta V.A., Ravenscroft G., Shaheen R., Todd E.J., Swanson L.C., Shiina M., Ogata K., Hsu C., Clarke N.F., Darras B.T. Identification of KLHL41 mutations implicates BTB-Kelch-mediated ubiquitination as an alternate pathway to myofibrillar disruption in nemaline myopathy. Am. J. Hum. Genet. 2013;93:1108–1117. doi: 10.1016/j.ajhg.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith L.L., Beggs A.H., Gupta V.A. Analysis of skeletal muscle defects in larval zebrafish by birefringence and touch-evoke escape response assays. J. Vis. Exp. 2013;82:e50925. doi: 10.3791/50925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devaney S.A., Mate S.E., Devaney J.M., Hoffman E.P. Characterization of the ZBTB42 gene in humans and mice. Hum. Genet. 2011;129:433–441. doi: 10.1007/s00439-010-0940-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Novak A.E., Taylor A.D., Pineda R.H., Lasda E.L., Wright M.A., Ribera A.B. Embryonic and larval expression of zebrafish voltage-gated sodium channel alpha-subunit genes. Dev. Dyn. 2006;235:1962–1973. doi: 10.1002/dvdy.20811. [DOI] [PubMed] [Google Scholar]
- 18.Telfer W.R., Nelson D.D., Waugh T., Brooks S.V., Dowling J.J. Neb: a zebrafish model of nemaline myopathy due to nebulin mutation. Dis. Model. Mech. 2012;5:389–396. doi: 10.1242/dmm.008631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devaney J.M., Gordish-Dressman H., Harmon B.T., Bradbury M.K., Devaney S.A., Harris T.B., Thompson P.D., Clarkson P.M., Price T.B., Angelopoulos T.J., et al. AKT1 polymorphisms are associated with risk for metabolic syndrome. Hum. Genet. 2011;129:129–139. doi: 10.1007/s00439-010-0910-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaheen R., Al-Owain M., Khan A.O., Zaki M., Hossni H., Al-Tassan R., Eyaid W., Alkuraya F.S. Identification of three novel ECEL1 mutations in three families with distal arthrogryposis type 5D. Clin. Genet. 2014;85:568–572. doi: 10.1111/cge.12226. [DOI] [PubMed] [Google Scholar]
- 21.Matthews J.M., Sunde M. Zinc fingers – folds for many occasions. IUBMB. Life. 2002;54:351–355. doi: 10.1080/15216540216035. [DOI] [PubMed] [Google Scholar]
- 22.Perez-Torrado R., Yamada D., Defossez P.A. Born to bind: the BTB protein-protein interaction domain. Bioessays. 2006;28:1194–1202. doi: 10.1002/bies.20500. [DOI] [PubMed] [Google Scholar]
- 23.Kovalovsky D., Uche O.U., Eladad S., Hobbs R.M., Yi W., Alonzo E., Chua K., Eidson M., Kim H.J., Im J.S., et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat. Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barna M., Hawe N., Niswander L., Pandolfi P.P. Plzf regulates limb and axial skeletal patterning. Nat. Genet. 2000;25:166–172. doi: 10.1038/76014. [DOI] [PubMed] [Google Scholar]
- 25.Yatsenko A.N., Roy A., Chen R., Ma L., Murthy L.J., Yan W., Lamb D.J., Matzuk M.M. Non-invasive genetic diagnosis of male infertility using spermatozoal RNA: KLHL10mutations in oligozoospermic patients impair homodimerization. Hum. Mol. Genet. 2006;15:3411–3419. doi: 10.1093/hmg/ddl417. [DOI] [PubMed] [Google Scholar]
- 26.Bomont P., Cavalier L., Blondeau F., Hamida C.B., Belal S., Tazir M., Demir E., Topaloglu H., Korinthenberg R., Tüysüz B. The gene encoding gigaxonin, a new member of the cytoskeletal BTB/kelch repeat family, is mutated in giant axonal neuropathy. Nat. Genet. 2000;26:370–374. doi: 10.1038/81701. [DOI] [PubMed] [Google Scholar]
- 27.Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio Rerio) 5th edn. Univ. of Oregon Press, Eugene; 2007. [Google Scholar]
- 28.Carr I.M., Flintoff K.J., Taylor G.R., Markham A.F., Bonthron D.T. Interactive visual analysis of SNP data for rapid autozygosity mapping in consanguineous families. Hum. Mutat. 2006;27:1041–1046. doi: 10.1002/humu.20383. [DOI] [PubMed] [Google Scholar]
- 29.Alkuraya F.S. Homozygosity mapping: one more tool in the clinical geneticist's toolbox. Genet. Med. 2010;12:236–239. doi: 10.1097/GIM.0b013e3181ceb95d. [DOI] [PubMed] [Google Scholar]
- 30.Alkuraya F.S. The application of next-generation sequencing in the autozygosity mapping of human recessive diseases. Hum. Genet. 2013;132:1197–1211. doi: 10.1007/s00439-013-1344-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.