Abstract
Osteoprotegerin (OPG) is involved in bone homeostasis and tumor cell survival. Circulating OPG levels are also important biomarkers of various clinical traits, such as cancers and atherosclerosis. OPG levels were measured in serum or in plasma. In a meta-analysis of genome-wide association studies in up to 10 336 individuals from European and Asian origin, we discovered that variants >100 kb upstream of the TNFRSF11B gene encoding OPG and another new locus on chromosome 17q11.2 were significantly associated with OPG variation. We also identified a suggestive locus on chromosome 14q21.2 associated with the trait. Moreover, we estimated that over half of the heritability of OPG levels could be explained by all variants examined in our study. Our findings provide further insight into the genetic regulation of circulating OPG levels.
Osteoprotegerin (OPG), a secreted member of the tumor necrosis factor receptor superfamily, is a decoy receptor for receptor activator of nuclear factor κB ligand (RANKL) that inhibits osteoclastogenesis (1). OPG is found in multiple tissues, including bone, lung, heart, kidney, intestine and vascular tissues (2–4); notably, OPG knockout mice develop severe osteoporosis (5). In addition to its well-established role in bone homeostasis (6,7), OPG is known to be involved in tumor cell survival (8–11), normal B-cell function (12) and vascular calcification (13–19). Circulating OPG levels are also markers of bone metastases in prostate cancer patients (20), systemic lupus erythematosus (21) and atherosclerosis (22), as well as potential markers of osteoporosis (23), Paget's disease (24), colorectal cancer recurrence (25), intestinal inflammation (26), incident atrial fibrillation (27) and vascular disease (23,28).
Due to the clinical interest in OPG, several studies have investigated genetic and environmental influences on circulating OPG levels (29–31). Abrahamsen et al. (31) reported no significant genetic influence on OPG levels in Danish female twins, whereas Livshits et al. (29), Kwan et al. (30) and we found that the trait is highly heritable in Chuvashians (heritability estimate, h2 = 46%), Southern Chinese (h2 for age-adjusted OPG was 75% for females and 37% for males) and Framingham participants (h2 for age-adjusted OPG was 23% for females and 24% for males) (see Supplementary Material, Methods and Table S5), respectively. In addition, Vistoropsky et al. (32) reported that an intronic single-nucleotide polymorphism rs875525 in the ANKH gene is significantly associated with OPG levels in a candidate gene study. But whether genes other than ANKH contribute to the variation in circulating OPG levels remains to be clarified. Genome-wide association studies (GWAS) are now a standard approach to investigate the genetic architecture of human traits and diseases (33–36). To gain more insight into the genetic regulation of OPG levels, we conducted a meta-analysis of GWAS data from 10 336 individuals from five cohorts. We found that variants >100 kb upstream of the TNFRSF11B gene encoding OPG and a locus on chromosome 17q11.2 significantly contribute to OPG variation. We also identified a suggestive locus on chromosome 14q21.2 associated with OPG levels.
RESULTS
Figures 1 and 2 show, respectively, the Manhattan and QQ plots of association results from the fixed-effects inverse-variance weighted meta-analyses (37). The QQ plots of single-nucleotide polymorphisms (SNPs) for the sex-specific meta-analyses are provided in Supplementary Material, Figure S1. Summary association statistics of all SNPs are available at http://hpcf.cgs.hku.hk/gwas_pub/OPG. There was little evidence of inflation of the association results as the genomic inflation factors λ are 1.026, 1.023 and 1.018 for the sex-combined, female-specific and male-specific meta-analyses, respectively. Therefore, genomic control correction was not applied to any of the meta-analysis results. A sample-size weighted meta-analysis gave essentially the same results and significant loci.
Figure 1.
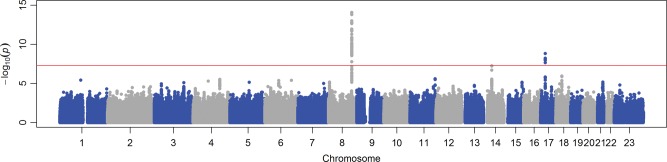
Manhattan plot of SNPs for the sex-combined meta-analysis. The X-axis indicates the chromosomal position of each SNP, whereas the Y-axis denotes the evidence of association shown as −log(P-value). The red line indicates genome-wide significance of association (P = 5 × 10−8).
Figure 2.
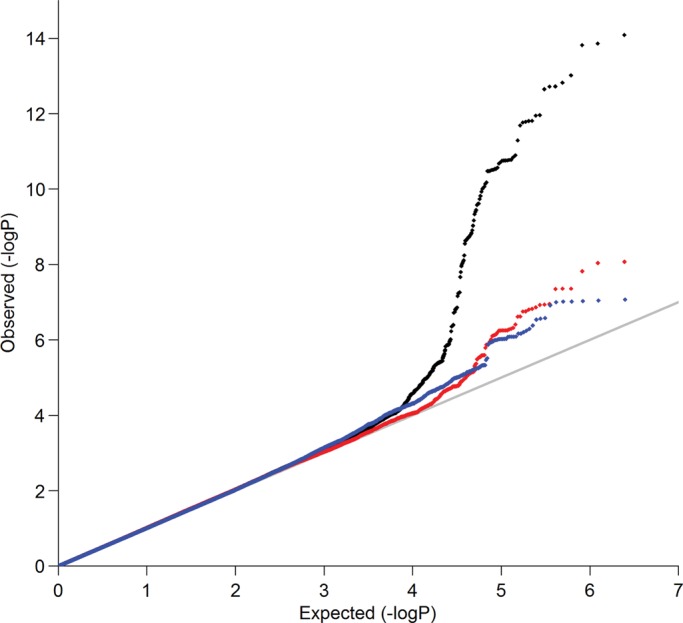
QQ plots of SNPs for sex-combined (in black), female-specific (in red) and male-specific (in blue) meta-analyses, respectively.
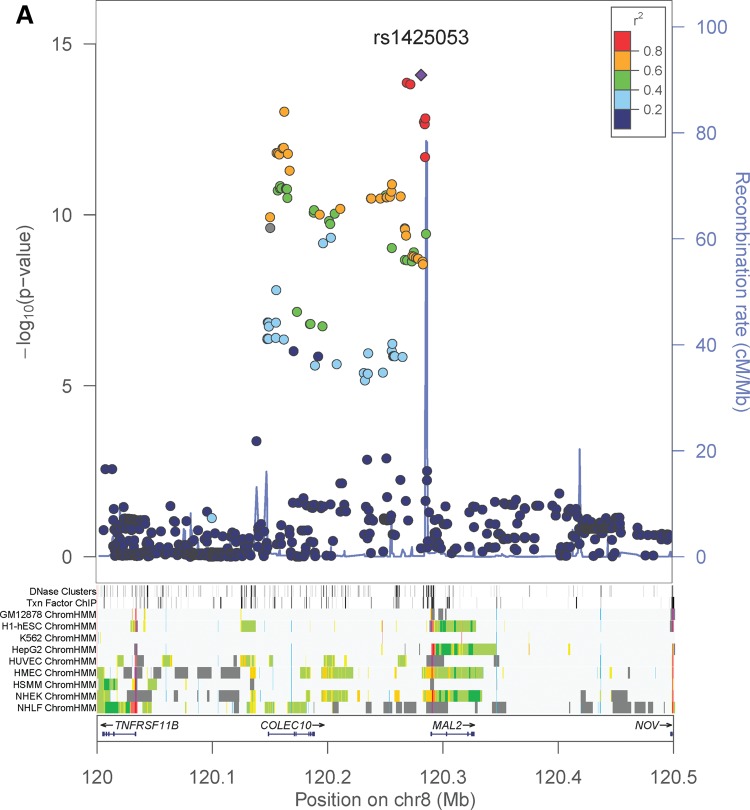
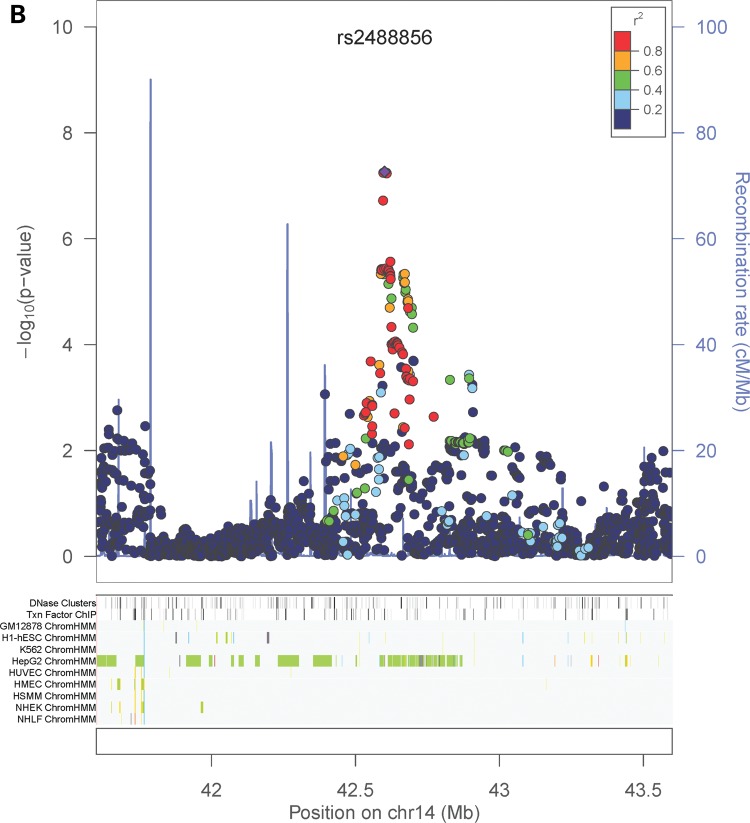
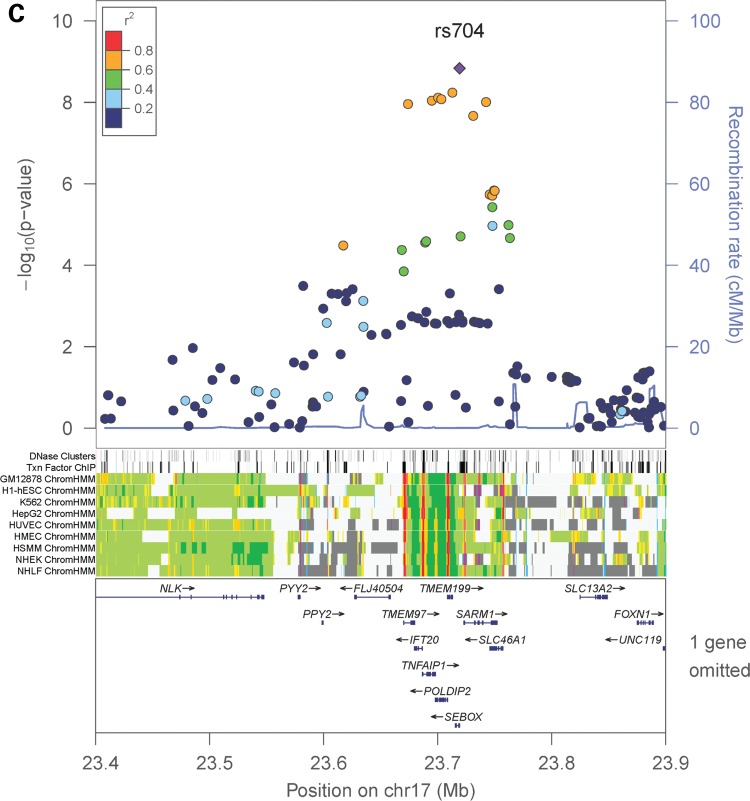
In the sex-combined meta-analysis, 72 SNPs in two loci had association P-values that reached genome-wide significance (P < 5 × 10−8): 8q23–q24.1 (smallest P = 8.15 × 10−15 near MAL2, COLEC10 and TNFRSF11B encoding OPG), and 17q11.2 (smallest P = 1.46 × 10−9 within a gene-rich region) (Table 1 and Fig. 3). The degree of heterogeneity was low (I2 < 0.5) across studies for the significant SNPs. In addition, four SNPs in a gene desert region at locus 14q21.2 almost reached genome-wide significance (smallest P = 5.45 × 10−8) (Table 1 and Fig. 3). In the female-specific meta-analysis, the chromosome 8 locus identified in the sex-combined meta-analysis remained significant (smallest P = 8.46 × 10−9) (Table 1; Supplementary Material, Fig. S2). None of the SNPs achieved genome-wide significance in the male-specific meta-analysis. Although we did not perform any replication study, the effect alleles of the top SNPs in the chromosome 8 and 17 loci showed consistent directions of effect in all participating studies (Table 1). Conditional analyses on the lead SNP in each significant locus found no strong evidence for additional independent signals reaching genome-wide significance. Pathway association analysis using a HYbrid Set-Based Test (HYST) (38) and hypergeometric test on 1320 canonical pathways revealed no significant pathways with genes moderately associated with OPG levels.
Table 1.
Meta-analysis results for loci that reached or marginally reached genome-wide significance (P < 5 × 10−8)
| Chr | Nearest gene(s) | Lead SNP | Positiona | EAFb | Effect allele | Other allele |
βc in individual study |
βc | SE | P | % Variance explained | I2 | Cochran's Q (P) | P for association with FNBMDd | P for association with LSBMDd | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FHS | GOOD | HKOS | MICROS | WHI | |||||||||||||||
| Sex-combined meta-analysis | |||||||||||||||||||
| 8 | COLEC10, MAL2, TNFRSF11B | rs1425053 | 120 282 171 | 0.607 (0.713) | A | T | 0.039 | 0.034 | 0.045 | 0.033 | 0.016 | 0.037 | 0.0048 | 8.15 × 10−15 | 0.58 | 0 | 0.80 | 0.047 | 0.24 |
| 17 | FLJ40504, POLDIP2, TMEM97, MIR4723, VTN, IFT20, SARM1, TNFAIP1, SLC46A1, TMEM199, SEBOX | rs704 | 23 718 988 | 0.494 (0.701) | A | G | −0.027 | −0.027 | −0.034 | −0.051 | −0.045 | −0.030 | 0.0049 | 1.46 × 10−9 | 0.35 | 0 | 0.74 | 7.07 × 10−3 | 0.025 |
| 14 | — | rs2488856 | 42 601 152 | 0.898 (NA) | A | C | −0.048 | −0.051 | NA | −0.027 | 0.023 | −0.044 | 0.0080 | 5.45 × 10−8 | 0.28 | 33 | 0.21 | 0.39 | 0.55 |
| Female-specific meta-analysis | |||||||||||||||||||
| 8 | COLEC10, MAL2, TNFRSF11B | rs2468186 | 120 269 883 | 0.623 (0.698) | T | C | 0.045 | – | 0.047 | 0.027 | 0.019 | 0.040 | 0.0070 | 8.46 × 10−9 | 0.59 | 0 | 0.56 | 0.45 | 0.45 |
Note: the LD r2 between rs1425053 and rs2468186 is >0.9 in both European and Chinese populations in HapMap.
Chr, chromosome.
aPositions are according to NCBI Build 36.
bEffect allele frequency (EAF) averaged from the samples, with the Chinese EAF specifically indicated in brackets.
cβ represents the change in the natural log-transformed serum OPG levels (pmol/l) per additional copy of the effect allele.
dP-value for association with BMD from a published meta-analysis (42).
Figure 3.
Regional association plots and ENCODE annotation of the loci that reached or marginally reached genome-wide significance (P < 5 × 10−8) in the sex-combined meta-analysis. The X-axis indicates the physical position of each SNP on the chromosome specified, whereas the Y-axis denotes the evidence of association shown as −log(P-value). ENCODE annotation are also provided, including transcription factor-binding sites (TFBS) from 95 cell lines, DNase I hypersensitivity sites (DNase HS) from 125 cell lines, and chromatin states in nine ENCODE cell lines (GM12878, H1-hSEC, K562, HepG2, HUVEC, HMEC, HSMM, NHEK and NHLF). The chromatin states are annotated as follows: strong enhancer (orange), weak enhancer (yellow), active promoter (red), poised promoter (pink), insulator (blue), transcribed (pale green), transcription transition (dark green), repressed (dark gray) and heterochromatin (pale gray).
The most significant association signal in our study came from 8q23–q24.1, located >100 kb upstream of the TNFRSF11B transcriptional start. The TNFRSF11B gene encodes OPG and so the region likely harbors important regulatory elements that control the expression of the OPG protein molecule. But our expression quantitative-trait loci (eQTL) analyses in various human tissues including bone cells found no association of the TNFRSF11B gene expression with the selected SNPs in the region (Table 2). Encyclopedia of DNA elements (ENCODE) annotation of the locus indicated that there are predicted insulators in 7 out of 9 cell lines examined within the COLEC10 gene (Fig. 3). HaploReg v2 (39) and RegulomeDB (40) indicated that some of the proxies (r2 > 0.8 in 1000 Genomes Pilot 1) of the top SNPs (i.e. rs1425053) lie at strong enhancers in the epithelial cell line HMEC (Supplementary Material, Table S6).
Table 2.
Results for e-QTL analyses
| Chr. | SNP | Transcript | GTEx whole blood (n = 144) | GTEx subcutaneous adipose (n = 111) | GTEx skeletal muscle (n = 138) | GTEx pancreas (n = 67) | Whole bone (n = 54) | Primary osteoblasts (n = 95) | Whole blood (n = 5311) |
|---|---|---|---|---|---|---|---|---|---|
| 8 | rs1425053 | TNFRSF11B | >0.1 | 0.05 | 0.03 | >0.1 | >0.1 | >0.1 | >0.1 |
| MAL2 | 0.002 | >0.1 | >0.1 | >0.1 | >0.1 | >0.1 | >0.1 | ||
| 8 | rs2468186 | TNFRSF11B | >0.1 | >0.1 | 0.02 | >0.1 | >0.1 | >0.1 | >0.1 |
| MAL2 | 0.002 | >0.1 | >0.1 | >0.1 | >0.1 | >0.1 | >0.1 | ||
| 17 | rs704 | VTN | >0.1 | >0.1 | >0.1 | >0.1 | >0.1 | >0.1 | >0.1 |
| TMEM199 | >0.1 | 0.03 | >0.1 | 4 × 10−4* | 0.004* | >0.1 | >0.1 | ||
| TMEM97 | >0.1 | 0.02 | >0.1 | >0.1 | >0.1 | >0.1 | 1.08 × 10−51* |
Significant P-values for association between the SNP and expression of the selected transcripts are marked with an asterisk.
The chromosome 17 locus identified in the sex-combined meta-analysis encompasses multiple genes, including FLJ40504, POLDIP2, TMEM97, MIR4723, VTN, IFT20, SARM1, TNFAIP1, SLC46A1, TMEM199, and SEBOX. The most significant SNP in this locus (rs704) encodes a possibly damaging (predicted by PolyPhen2) (41) missense mutation in the VTN gene. Moreover, eQTL analyses showed that the SNP was significantly associated with TMEM199 expression in pancreas and whole bone, as well as TMEM97 expression in whole blood (Table 2). The low RegulomeDB score of the SNP (i.e. 2b) also suggested that the SNP has important regulatory functions (Supplementary Material, Table S7).
Moreover, we examined the association of SNP rs875525 in the ANKH gene previously identified by Vistoropsky et al. (32) with OPG levels. However, it did not reach genome-wide significance in all three meta-analyses (P = 0.58, 0.52 and 0.88 in sex-combined, female-specific and male-specific meta-analyses, respectively). In addition, we interrogated published meta-analysis of BMD from the GEnetic Factors for OSteoporosis (GEFOS) Consortium (42) to see whether the lead SNPs in our study overlapped with any BMD-associated loci. However, no significant association between BMD and our lead SNPs was found (all P-values > 0.05 after Bonferroni correction for multiple testing) (Table 2).
We also estimated the proportion of age-adjusted OPG variation explained by all SNPs studied in all 10 336 samples using a density estimation method proposed by So et al. (43). We found that the ∼2.4 million SNPs examined in our study could explain ∼11% of age-adjusted OPG variation. Given that h2 for age-adjusted OPG levels was ∼20% in Framingham participants, which constitute the majority of our study samples (Supplementary Material, Methods and Table S5), over half of the h2 for age-adjusted OPG levels could be explained by all SNPs examined in our study alone.
DISCUSSION
Using a meta-analysis of GWAS from FIVE studies comprising >10 000 individuals, we identified two genome-wide significant loci (8q23-q24.1 and 17q11.2) and one locus on chromosome 14 associated with OPG levels with near genome-wide significance.
The association signal in the chromosome 8 resides in a linkage disequilibrium (LD) block containing the COLEC10 gene, whose functional relationship to OPG levels is unknown. Also, based on the GWAS catalog data (http://www.genome.gov/gwastudies/, accessed 7 January 2014) (36), no SNP in the LD block has been reported to be associated with any trait or disease. Although there existed predicted insulators generated by the ENCODE project in 7 out of 9 cell lines, the function of an insulator is to block the interaction between enhancers and promoters, and so the enhancer affected remains to be identified. Moreover, our eQTL analyses found no association of gene expression of TNFRSF11B, MAL2, VTN, TMEM199 and TMEM97with the top SNPs rs1425053 and rs2468186 or their proxies in strong LD. We acknowledge that the lack of eQTL associations may be due to small sample sizes except whole blood. So the functional roles of the genetic variants in the region remain to be elucidated. However, it is worth noting that HaploReg v2 (39) and RegulomeDB (40) indicated that some of the proxies (r2 > 0.8 in 1000 Genomes Pilot 1) of the top SNPs (i.e. rs1425053) lie at strong enhancers in the epithelial cell line HMEC and multiple studies suggested that OPG may promote the survival of endothelial cells (14,16,44).
The most significant SNP in the chromosome 17 hit (i.e. rs704) encodes a possibly damaging missense mutation in the VTN gene. The VTN protein encoded by the gene was previously found to bind to OPG (13), as well as to regulate osteogenesis in mesenchymal stem cells (45). Also, we found that the SNP was associated with TMEM199 expression in pancreas and whole bone as well as TMEM97 expression in whole blood. The biological function of TMEM199 in pancreas and whole bone tissues has not yet been established, but the gene was associated with cancer progression in breast tissues (46). TMEM97 plays a role in cholesterol and lipid metabolism (47) and is also involved in cancer development (48,49). But as pointed out by Edwards et al. (50), experiments are needed to elucidate the molecular mechanisms in detail.
We only identified two statistically significant regions and one suggestive region in which the potentially causal variants (i.e. the ones shown to be causal in influencing OPG levels that are in LD with other variants associated with OPG levels) reside. The next steps should be to fine-map these regions with a dense marker panel and a large sample size to identify candidate causal variants. Since the majority of participating studies in our meta-analysis were primarily of European ancestry, we recommend that fine-mapping studies be performed in populations of multiple ethnicities. First, the LD patterns differ between the different ethnic groups (Supplementary Material, Fig. S3), which may be able to reduce the number of candidate causal variants in the fine-mapping studies. Second, fine-mapping other racial groups may answer the question of whether the associations we found generalize to populations of non-European ancestry (51,52).
The results of our meta-analysis failed to confirm an association of a previously identified OPG-associated SNP rs875525 (32). As the study of Vistoropsky et al. was performed in Chuvashians, LD patterns between their subjects and ours (which were mostly European ancestry) should be similar. So the absence of such association in our study indicates either their association was a false positive or we had poor power to detect such association. Since Vistoropsky et al. did not provide the effect size of rs875525, we cannot estimate our power to discover such an effect.
The current study was limited by the measurement of circulating OPG. In addition, the studies varied by whether they measured OPG in serum versus plasma; however, we note that the βs and directions of association were similar across studies. Although monoclonal antibody was used, it is unclear whether the mono- or homodimeric form should be measured. The assay used by all studies detected both forms of OPG, and the RANKL-bound form. Also, one target action point of OPG is skeletal and there is no evidence that circulating OPG level reflects the amount of bioavailable OPG in the skeletal microenvironment. As OPG is produced locally in bone tissue by osteoblasts, another study design would be to examine genome-wide association of bone-specific OPG. A bone biopsy study in a large number of individuals is nonetheless impractical to resolve the matter.
In conclusion, we discovered that variants >100 kb upstream of the TNFRSF11B gene encoding OPG are associated with variation in circulating OPG levels and identified another new significant locus on chromosome 17q11.2 as well as a suggestive locus on chromosome 14q21.2 associated with the trait. Moreover, we estimated that over half of the heritability of age-adjusted OPG levels could be explained by all SNPs studied. Further work is needed to identify the causal variants and to understand how they influence the circulating OPG levels.
MATERIALS AND METHODS
Study populations
The meta-analysis comprised 10 336 individuals from five GWAS: Framingham Heart Study (FHS)—3223 men and 3685 women of European ancestry; Gothenburg Osteoporosis and Obesity Determinants (GOOD) study—935 men of European ancestry; Hong Kong Osteoporosis Study (HKOS)—702 women of Chinese ancestry; Microisolates in South Tyrol (MICROS) study—531 men and 692 women of European ancestry; and Women's Health Initiative (WHI)—568 women of European ancestry. Each participating study was approved by a research ethics committee and all participants provided written informed consent. OPG levels were measured in serum (except for FHS, which measured in plasma) by either ELISA or the Fluorescent Bead Immunoassay method. More details of all participating studies are provided in Supplementary Material, Tables S1–S3.
GWAS analysis
Genome-wide genotyping for SNPs and quality control were done independently by each study following standard protocols. Each study performed genotype imputation with reference to HapMap Phase II of the corresponding population. Detailed descriptions of the genotyping platforms, quality control and imputation procedures for each study are provided in the Supplementary Material, Table S4. Each study performed GWAS tests for natural log-transformed OPG level (in pmol/l) under an additive (per allele) genetic model (53), using either linear regression (for unrelated samples) or linear-mixed models (for related samples). Associations were adjusted for sex (if not sex-specific), age, age-squared, principal components of genetic ancestry as well as other study-specific covariates. Before meta-analysis, SNPs were removed in each study if they had a minor allele frequency <1%, an extreme deviation from Hardy–Weinberg equilibrium (P < 1 × 10−6), and/or an imputation quality score <0.3. In each study, we also ensured that the allele frequencies of the SNPs observed matched closely with those in the corresponding HapMap reference panel. We tested for population stratification of the observed associations using the genomic inflation (λ) method (54).
Meta-analysis
Summary statistics from the individual studies were combined using fixed-effects inverse-variance weighted meta-analysis using the METAL software (37). Since the approach requires the trait distribution to be identical across individual studies, we also combined the summary statistics using the sample-size weighted method implemented in the METAL software and compared the association results given by both methods. SNPs with missing data in more than one study were excluded. SNPs with P < 5 × 10−8 were considered genome-wide significant. We also performed a meta-analysis of sex-specific GWAS results. We examined between-study heterogeneity using I2 and Cochran's Q test. An association analysis conditioned on the lead SNP was also performed in each significant locus to search for additional independent signals (55).
Functional annotation
Significant loci found and their proxies (r2 > 0.8 in 1000 Genomes Pilot 1) were examined for regulatory functions generated by the ENCODE Project (56) using HaploReg v2 (39) (http://www.broadinstitute.org/mammals/haploreg/haploreg.php, accessed 14 July 2014) and RegulomeDB (40) (http://regulomedb.org, accessed 14 July 2014).
Pathway analysis
To examine whether any known biological pathway is associated with OPG levels, we applied HYST for GWAS (38). Briefly, each SNP was mapped to genes (including 10 kb upstream and downstream regions) according to hg18. An extended Simes' test was then applied to compute association P-value for each gene (57) and the P-values for the genes in a pathway were combined using a scaled χ2 test to yield an association P-value of the pathway (38). We analyzed 1320 canonical pathways from the pathway databases available in MSigDb v4.0 (58) and a pathway with P-value < 0.05/1320 was declared significant after Bonferroni correction for multiple testing. In order to remove pathways with statistical significance caused by only a few extreme gene-based P-values, we defined a set of candidate genes with a false discovery rate < 0.05 and tested for enrichment of this candidate gene set using a hypergeometric test. Any significant pathway with a nominal P-value < 0.001 in the hypergeometric test was filtered out.
eQTL analysis
We performed cis-eQTL analyses on the three lead SNPs (rs1425053, rs2468186 and rs704) or their proxies in strong LD (r2 ≥ 0.8) with selected transcripts within 500 kb of the SNP position. In each locus, linear regression was used to examine the association between the top-associated SNP and expression of the selected genes. The eQTL analyses were performed in several human tissues, including whole blood (n = 144), subcutaneous adipose (n = 111), skeletal muscle (n = 138) and pancreas tissues (n = 67) from postmortem donors in the Genotype Tissue Expression program (GTEx) (59) as well as transiliac bone biopsies (60), human primary osteoblast samples (obtained from 95 bone biopsies) (61) and peripheral blood samples (n = 5311) (62). The detailed methods of sample collection, DNA and RNA extraction, GWAS genotyping, gene expression profiling and RNA-sequence are described in the corresponding papers. Results were declared significant at P ≤ 0.05/2/5 = 0.005 after Bonferroni correction of the number of loci (i.e. two) and different tissues (i.e. five) tested.
Estimation of variance explained by all SNPs examined
To estimate the joint effect of all SNPs studied in explaining the variance of age-adjusted OPG levels, we applied the density estimation method proposed by So et al. (43). It required only summary association statistics and sample size for computation and was demonstrated to give estimates similar to those by GCTA (63).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Hong Kong Research Grant Council; the Small Project Funding HKU 201309176244 and 201109176063; the Bone Health Fund of HKU Foundation and Matching Grant; the CRCG Grant and the Osteoporosis Research Fund of The University of Hong Kong. The GOOD study was supported by the Swedish Research Council, the Swedish Foundation for Strategic Research, COMBINE, the ALF/LUA research grant from the Sahlgrenska University Hospital, the Lundberg Foundation, the Torsten and Ragnar Söderberǵs Foundation, the Novo Nordisk Foundation, the Gustav V and Queen Victoria Freemason Foundation and the European Commission Grant HEALTH-F2–2008-201865-GEFOS. The Framingham Heart Study of the NHLBI, NIH and Boston University School of Medicine were supported by the NHLBI's Framingham Heart Study (N01-HC-25195) and its contract with Affymetrix, Inc. for genotyping services (N02-HL-6-4278). The WHI work was supported in part by an allocation of computing time from the Ohio Supercomputer Center and the National Institute of General Medical Sciences (U01GM092655) as well as HHSN268201100002C, NO1-6H74316, U54RR0024384 and HHSN268200960002C. The MICROS study was supported by the Ministry of Health and Department for Promotion of Educational Policies, Universities and Research of the Autonomous Province of Bolzano, South Tyrol, the South Tyrolean Sparkasse Foundation, and the European Union Framework Program 6 EUROSPAN Project (contract no. LSHG-CT-2006-018947). In addition, D.P.K. received support from NIH (Grant R01 AR/AG 41398). E.J.B. received support from the FHS grants 1RO1HL64753, R01HL076784, 1 R01AG028321 and 2R01HL092577.
Supplementary Material
Acknowledgements
We thank all study participants for making this work possible. For the MICROS study, we also thank the primary care practitioners Raffaela Stocker, Stefan Waldner, Toni Pizzecco, Josef Plangger, Ugo Marcadent and the personnel of the Hospital of Silandro (Department of Laboratory Medicine) for their participation and collaboration in the research project.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Schoppet M., Preissner K.T., Hofbauer L.C. RANK ligand and osteoprotegerin – paracrine regulators of bone metabolism and vascular function. Arterioscl. Throm. Vas. 2002;22:549–553. doi: 10.1161/01.atv.0000012303.37971.da. [DOI] [PubMed] [Google Scholar]
- 2.Yun T.J., Chaudhary P.M., Shu G.L., Frazer J.K., Ewings M.K., Schwartz S.M., Pascual V., Hood L.E., Clark E.A. OPG/FDCR-1, a TNF receptor family member, is expressed in lymphoid cells and is up-regulated by ligating CD40. J. Immunol. 1998;161:6113–6121. [PubMed] [Google Scholar]
- 3.Tan K.B., Harrop J., Reddy M., Young P., Terrett J., Emery J., Moore G., Truneh A. Characterization of a novel TNF-like ligand and recently described TNF ligand and TNF receptor superfamily genes and their constitutive and inducible expression in hematopoietic and non-hematopoietic cells. Gene. 1997;204:35–46. doi: 10.1016/s0378-1119(97)00509-x. [DOI] [PubMed] [Google Scholar]
- 4.Simonet W.S., Lacey D.L., Dunstan C.R., Kelley M., Chang M.S., Luthy R., Nguyen H.Q., Wooden S., Bennett L., Boone T., et al. Osteoprotegerin: A novel secreted protein involved in the regulation of bone density. Cell. 1997;89:309–319. doi: 10.1016/s0092-8674(00)80209-3. [DOI] [PubMed] [Google Scholar]
- 5.Bucay N., Sarosi I., Dunstan C.R., Morony S., Tarpley J., Capparelli C., Scully S., Tan H.L., Xu W.L., Lacey D.L., et al. osteoprotegerin-deficient mice develop early onset osteoporosis and arterial calcification. Gene Dev. 1998;12:1260–1268. doi: 10.1101/gad.12.9.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Theoleyre S., Wittrant Y., Tat S.K., Fortun Y., Redini F., Heymann D. The molecular triad OPG/RANK/RANKL: involvement in the orchestration of pathophysiological bone remodeling. Cytokine Growth F. R. 2004;15:457–475. doi: 10.1016/j.cytogfr.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Sattler A.M., Schoppet M., Schaefer J.R., Hofbauer L.C. Novel aspects on RANK ligand and osteoprotegerin in osteoporosis and vascular disease. Calcified Tissue Int. 2004;74:103–106. doi: 10.1007/s00223-003-0011-y. [DOI] [PubMed] [Google Scholar]
- 8.Rachner T.D., Benad P., Rauner M., Goettsch C., Singh S.K., Schoppet M., Hofbauer L.C. Osteoprotegerin production by breast cancer cells is suppressed by dexamethasone and confers resistance against TRAIL-induced apoptosis. J. Cell Biochem. 2009;108:106–116. doi: 10.1002/jcb.22232. [DOI] [PubMed] [Google Scholar]
- 9.Neville-Webbe H.L., Cross N.A., Eaton C.L., Nyambo R., Evans C.A., Coleman R.E., Holen I. Osteoprotegerin (OPG) produced by bone marrow stromal cells protects breast cancer cells from TRAIL-induced apoptosis. Breast Cancer Res. Tr. 2004;86:269–279. doi: 10.1023/b:brea.0000036900.48763.b3. [DOI] [PubMed] [Google Scholar]
- 10.Holen I., Cross S.S., Neville-Webbe H.L., Cross N.A., Balasubramanian S.P., Croucher P.I., Evans C.A., Lippitt J.M., Coleman R.E., Eaton C.L. Osteoprotegerin (OPG) expression by breast cancer cells in vitro and breast tumours in vivo – a role in tumour cell survival? Breast Cancer Res. Tr. 2005;92:207–215. doi: 10.1007/s10549-005-2419-8. [DOI] [PubMed] [Google Scholar]
- 11.Fisher J.L., Thomas-Mudge R.J., Elliott J., Hards D.K., Sims N.A., Slavin J., Martin T.J., Gillespie M.T. Osteoprotegerin overexpression by breast cancer cells enhances orthotopic and osseous tumor growth and contrasts with that delivered therapeutically. Cancer Res. 2006;66:3620–3628. doi: 10.1158/0008-5472.CAN-05-3119. [DOI] [PubMed] [Google Scholar]
- 12.Yun T.J., Tallquist M.D., Aicher A., Rafferty K.L., Marshall A.J., Moon J.J., Ewings M.K., Mohaupt M., Herring S.W., Clark E.A. Osteoprotegerin, a crucial regulator of bone metabolism, also regulates B cell development and function. J. Immunol. 2001;166:1482–1491. doi: 10.4049/jimmunol.166.3.1482. [DOI] [PubMed] [Google Scholar]
- 13.Zannettino A.C.W., Holding C.A., Diamond P., Atkins G.J., Kostakis P., Farrugia A., Gamble J., To L.B., Findlay D.M., Haynes D.R. Osteoprotegerin (OPG) is localized to the Weibel-Palade bodies of human vascular endothelial cells and is physically associated with von Willebrand factor. J. Cell Physiol. 2005;204:714–723. doi: 10.1002/jcp.20354. [DOI] [PubMed] [Google Scholar]
- 14.Pritzker L.B., Scatena M., Giachelli C.M. The role of osteoprotegerin and tumor necrosis factor-related apoptosis-inducing ligand in human microvascular endothelial cell survival. Mol. Biol. Cell. 2004;15:2834–2841. doi: 10.1091/mbc.E04-01-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Min H., Morony S., Sarosi I., Dunstan C.R., Capparelli C., Scully S., Van G., Kaufman S., Kostenuik P.J., Lacey D.L., et al. Osteoprotegerin reverses osteoporosis by inhibiting endosteal osteoclasts and prevents vascular calcification by blocking a process resembling osteoclastogenesis. J. Exp. Med. 2000;192:463–474. doi: 10.1084/jem.192.4.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malyankar U.M., Scatena M., Suchland K.L., Yun T.J., Clark E.A., Giachelli C.M. Osteoprotegerin is an alpha(v)beta(3)-induced, NF-kappa B-dependent survival factor for endothelial cells. J. Biol. Chem. 2000;275:20959–20962. doi: 10.1074/jbc.C000290200. [DOI] [PubMed] [Google Scholar]
- 17.Hofbauer L.C., Schoppet M. Clinical implications of the osteoprotegerin/RANKL/RANK system for bone and vascular diseases. J. Am. Med. Assoc. 2004;292:490–495. doi: 10.1001/jama.292.4.490. [DOI] [PubMed] [Google Scholar]
- 18.Collin-Osdoby P. Regulation of vascular calcification by osteoclast regulatory factors RANKL and osteoprotegerin. Circ. Res. 2004;95:1046–1057. doi: 10.1161/01.RES.0000149165.99974.12. [DOI] [PubMed] [Google Scholar]
- 19.Brandstrom H., Stiger F., Lind L., Kahan T., Melhus H., Kindmark A. A single nucleotide polymorphism in the promoter region of the human gene for osteoprotegerin is related to vascular morphology and function. Biochem. Bioph. Res. Co. 2002;293:13–17. doi: 10.1016/S0006-291X(02)00137-7. [DOI] [PubMed] [Google Scholar]
- 20.Jung K., Lein M., Stephan C., Von-Hosslin K., Semjonow A., Sinha P., Loening S.A., Schnorr D. Comparison of 10 serum bone turnover markers in prostate carcinoma patients with bone metastatic spread: Diagnostic and prognostic implications. Int. J. Cancer. 2004;111:783–791. doi: 10.1002/ijc.20314. [DOI] [PubMed] [Google Scholar]
- 21.Shin Y., Kim H., Yoo S., Hur K., Choi J., Kim W., Cho C. Increased circulating levels of osteoprotegerin in patients with systemic lupus erythematous. Ann. Rheum. Dis. 2007;66:459–466. [Google Scholar]
- 22.Hosbond S.E., Poulsen T.S., Diederichsen A.C.P., Nybo M., Rasmussen L.M., Mickley H. Osteoprotegerin as a marker of atherosclerosis: a systematic update. Scand. Cardiovasc. J. 2012;46:203–211. doi: 10.3109/14017431.2012.685491. [DOI] [PubMed] [Google Scholar]
- 23.McFarlane S.I., Muniyappa R., Shin J.J., Bahtiyar G., Sowers J.R. Osteoporosis and cardiovascular disease – brittle bones and boned arteries, is there a link? Endocrine. 2004;23:1–10. doi: 10.1385/ENDO:23:1:01. [DOI] [PubMed] [Google Scholar]
- 24.Whyte M.P., Obrecht S.E., Finnegan P.M., Jones J.L., Podgornik M.N., McAlister W.H., Mumm S. Osteoprotegerin deficiency and juvenile Paget's disease. New Engl. J. Med. 2002;347:175–184. doi: 10.1056/NEJMoa013096. [DOI] [PubMed] [Google Scholar]
- 25.Tsukamoto S., Ishikawa T., Iida S., Ishiguro M., Mogushi K., Mizushima H., Uetake H., Tanaka H., Sugihara K. Clinical significance of osteoprotegerin expression in human colorectal cancer. Clin. Cancer Res. 2011;17:2444–2450. doi: 10.1158/1078-0432.CCR-10-2884. [DOI] [PubMed] [Google Scholar]
- 26.Galliera E., de Girolamo L., Dogliotti G., De Salvo C., Tosetti G., Pastorelli L. Circulating OPG levels are reduced following infliximab treatment and correlate with CRP levels: is serum OPG a potential marker of IBD disease activity? Inflamm. Bowel Dis. 2011;17:E59–E60. doi: 10.1002/ibd.21714. [DOI] [PubMed] [Google Scholar]
- 27.Schnabel R.B., Larson M.G., Yamamoto J.F., Kathiresan S., Rong J., Levy D., Keaney J.F., Wang T.J., Vasan R.S., Benjamin E.J. Relation of multiple inflammatory biomarkers to incident atrial fibrillation. Am. J. Cardiol. 2009;104:92–96. doi: 10.1016/j.amjcard.2009.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simonini G., Masi L., Giani T., Piscitelli E., Cimaz R., Vierucci S., Brandi M.L., Falcini F. Osteoprotegerin serum levels in Kawasaki disease: an additional potential marker in predicting children with coronary artery involvement. J. Rheumatol. 2005;32:2233–2238. [PubMed] [Google Scholar]
- 29.Livshits G., Pantsulaia I., Trofimov S., Kobyliansky E. Genetic influences on the circulating cytokines involved in osteoclastogenesis. J. Med. Genet. 2004;41:e76. doi: 10.1136/jmg.2003.014373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwan J.S.H., Xiao S.M., Bow C., Cheung C.L., Soong C., Lau K.S., Sham P.C., Kung A.W.C. Heritability of serum osteoprotegerin. Ann. Hum. Genet. 2011;75:584–588. doi: 10.1111/j.1469-1809.2011.00661.x. [DOI] [PubMed] [Google Scholar]
- 31.Abrahamsen B., Hjelmborg J.V., Kostenuik P., Stilgren L.S., Kyvik K., Adamu S., Brixen K., Langdahl B.L. Circulating amounts of osteoprotegerin and RANK ligand: Genetic influence and relationship with BMD assessed in female twins. Bone. 2005;36:727–735. doi: 10.1016/j.bone.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 32.Vistoropsky Y., Malkin I., Kobyliansky E., Livshits G. Osteoprotegerin plasma levels are strongly associated with polymorphisms in human homologue of the mouse progressive ankylosis (ANKH) gene. Ann. Hum. Genet. 2007;71:302–307. doi: 10.1111/j.1469-1809.2006.00331.x. [DOI] [PubMed] [Google Scholar]
- 33.Wang W.Y.S., Barratt B.J., Clayton D.G., Todd J.A. Genome-wide association studies: Theoretical and practical concerns. Nat. Rev. Genet. 2005;6:109–118. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- 34.McCarthy M.I., Abecasis G.R., Cardon L.R., Goldstein D.B., Little J., Ioannidis J.P.A., Hirschhorn J.N. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 35.Hirschhorn J.N., Daly M.J. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 36.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li M.X., Kwan J.S.H., Sham P.C. HYST: a HYbrid Set-Based Test for genome-wide association studies, with application to protein-protein interaction-based association analysis. Am. J. Hum. Genet. 2012;91:478–488. doi: 10.1016/j.ajhg.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ward L.D., Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S., et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Estrada K., Styrkarsdottir U., Evangelou E., Hsu Y.H., Duncan E.L., Ntzani E.E., Oei L., Albagha O.M.E., Amin N., Kemp J.P., et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat. Genet. 2012;44:491–501. doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.So H.C., Li M.X., Sham P.C. Uncovering the total heritability explained by all true susceptibility variants in a genome-wide association study. Genet. Epidemiol. 2011;35:447–456. doi: 10.1002/gepi.20593. [DOI] [PubMed] [Google Scholar]
- 44.Cross S.S., Yang Z.Y., Brown N.J., Balasubramanian S.P., Evans C.A., Woodward J.K., Neville-Webbe H.L., Lippitt J.M., Reed M.W.R., Coleman R.E., et al. Osteoprotegerin (OPG) – a potential new role in the regulation of endothelial cell phenotype and tumour angiogenesis? Int. J. Cancer. 2006;118:1901–1908. doi: 10.1002/ijc.21606. [DOI] [PubMed] [Google Scholar]
- 45.Kundu A.K., Putnam A.J. Vitronectin and collagen I differentially regulate osteogenesis in mesenchymal stem cells. Biochem. Bioph. Res. Co. 2006;347:347–357. doi: 10.1016/j.bbrc.2006.06.110. [DOI] [PubMed] [Google Scholar]
- 46.Grinchuk O.V., Motakis E., Kuznetsov V.A. Complex sense-antisense architecture of TNFAIP1/POLDIP2 on 17q11.2 represents a novel transcriptional structural-functional gene module involved in breast cancer progression. BMC Genomics. 2010;11 doi: 10.1186/1471-2164-11-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilcox C.B., Feddes G.O., Willett-Brozick J.E., Hsu L.C., DeLoia J.A., Baysal B.E. Coordinate up-regulation of TMEM97 and cholesterol biosynthesis genes in normal ovarian surface epithelial cells treated with progesterone: implications for pathogenesis of ovarian cancer. BMC Cancer. 2007;7:223. doi: 10.1186/1471-2407-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao Z.R., Zhang L.J., He X.Q., Zhang Z.Y., Zhang F., Li F., Pei Y.B., Hu Y.M., Wang M.W., Sun X.F. Significance of mRNA and protein expression of MAC30 in progression of colorectal cancer. Chemotherapy. 2011;57:394–401. doi: 10.1159/000331716. [DOI] [PubMed] [Google Scholar]
- 49.Moparthi S.B., Arbman G., Wallin A., Kayed H., Kleeff J., Zentgraf H., Sun X.F. Expression of MAC30 protein is related to survival and biological variables in primary and metastatic colorectal cancers. Int. J. Oncol. 2007;30:91–95. [PubMed] [Google Scholar]
- 50.Edwards S.L., Beesley J., French J.D., Dunning A.M. Beyond GWASs: illuminating the dark road from association to function. Am. J. Hum. Genet. 2013;93:779–797. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marigorta U.M., Navarro A. High trans-ethnic replicability of GWAS results implies common causal variants. PLos Genet. 2013;9 doi: 10.1371/journal.pgen.1003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carlson C.S., Matise T.C., North K.E., Haiman C.A., Fesinmeyer M.D., Buyske S., Schumacher F.R., Peters U., Franceschini N., Ritchie M.D., et al. Generalization and dilution of association results from European GWAS in populations of non-European ancestry: the PAGE Study. PLos Biol. 2013;11 doi: 10.1371/journal.pbio.1001661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Little J., Higgins J.P., Ioannidis J.P.A., Moher D., Gagnon F., von Elm E., Khoury M.J., Cohen B., Davey-Smith G., Grimshaw J., et al. Strengthening the reporting of genetic association studies (STREGA) – an extension of the STROBE statement. Eur. J. Clin. Invest. 2009;39:247–266. doi: 10.1111/j.1365-2362.2009.02125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 55.Yang J., Ferreira T., Morris A.P., Medland S.E., Madden P.A.F., Heath A.C., Martin N.G., Montgomery G.W., Weedon M.N., Loos R.J., et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 2012;44:369–375. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosenbloom K.R., Dreszer T.R., Long J.C., Malladi V.S., Sloan C.A., Raney B.J., Cline M.S., Karolchik D., Barber G.P., Clawson H., et al. ENCODE whole-genome data in the UCSC Genome Browser: update 2012. Nucleic Acids Res. 2012;40:D912–D917. doi: 10.1093/nar/gkr1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li M.X., Gui H.S., Kwan J.S.H., Sham P.C. GATES: a rapid and powerful gene-based association test using extended Simes procedure. Am. J. Hum. Genet. 2011;88:283–293. doi: 10.1016/j.ajhg.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N., et al. The genotype-tissue expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reppe S., Refvem H., Gautvik V.T., Olstad O.K., Hovring P.I., Reinholt F.P., Holden M., Frigessi A., Jemtland R., Gautvik K.M. Eight genes are highly associated with BMD variation in postmenopausal Caucasian women. Bone. 2010;46:604–612. doi: 10.1016/j.bone.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 61.Grundberg E., Kwan T., Ge B., Lam K.C.L., Koka V., Kindmark A., Mallmin H., Dias J., Verlaan D.J., Ouimet M., et al. Population genomics in a disease targeted primary cell model. Genome Res. 2009;19:1942–1952. doi: 10.1101/gr.095224.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Westra H.J., Peters M.J., Esko T., Yaghootkar H., Schurmann C., Kettunen J., Christiansen M.W., Fairfax B.P., Schramm K., Powell J.E., et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat .Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lubke G.H., Hottenga J.J., Walters R., Laurin C., de Geus E.J.C., Willemsen G., Smit J.H., Middeldorp C.M., Penninx B.W.J.H., Vink J.M., et al. Estimating the genetic variance of major depressive disorder due to all single nucleotide polymorphisms. Biol. Psychiat. 2012;72:707–709. doi: 10.1016/j.biopsych.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.