Abstract
Importance
Autosomal dominant Alzheimer disease (ADAD) is caused by rare genetic mutations in three specific genes, in contrast to late-onset Alzheimer Disease (LOAD), which has a more polygenetic risk profile.
Design, Setting, and Participants
We analyzed functional connectivity in multiple brain resting state networks (RSNs) in a cross-sectional cohort of ADAD (N=79) and LOAD (N=444) human participants using resting state functional connectivity MRI (rs-fcMRI) at multiple international academic sites.
Main Outcomes and Measures
For both types of AD, we quantified and compared functional connectivity changes in RSNs as a function of dementia severity as measured by clinical dementia rating (CDR). In ADAD, we qualitatively investigated functional connectivity changes with respect to estimated years from onset of symptoms within five RSNs.
Results
Functional connectivity decreases with increasing CDR were similar for both LOAD and ADAD in multiple RSNs. Ordinal logistic regression models constructed in each type of AD accurately predicted CDR stage in the other, further demonstrating similarity of functional connectivity loss in each disease type. Among ADAD participants, functional connectivity in multiple RSNs appeared qualitatively lower in asymptomatic mutation carriers near their anticipated age of symptom onset compared to asymptomatic mutation non-carriers.
Conclusions and Relevance
rs-fcMRI changes with progressing AD severity are similar between ADAD and LOAD. Rs-fcMRI may be a useful endpoint for LOAD and ADAD therapy trials. ADAD disease process may be an effective model for LOAD disease process.
Keywords: Resting-state functional connectivity, autosomal dominant Alzheimer's disease, late-onset Alzheimer's disease, default mode network, apolipoprotein E (APOE)
Introduction
Late-onset Alzheimer disease (LOAD) is the leading cause of dementia worldwide, currently affecting more than 18 million people1. AD is defined by pathological accumulation of tau neurofibrillary tangles and amyloid beta (Aβ) plaques2. While AD is typically late-onset and polygenetic (LOAD), in a small subset of individuals AD is inherited as an autosomal dominant trait (autosomal dominant AD or ADAD), which is typically early-onset and caused by monogenetic mutations in the genes encoding presenilin 1, presenilin 2, or amyloid precursor protein. These mutations are ~100% penetrant and cause AD by affecting Aβ cleavage and folding3.
Discovery of ADAD mutations has enabled researchers to develop transgenic mouse models and cell lines expressing these mutations4. These experimental models have enabled the preclinical testing of potential anti-amyloid AD therapies5. By studying ADAD individuals who will develop dementia at a predictable age, researchers can identify the temporal dynamics of changes in biomarker profiles before the development of clinical symptoms6. However, questions remain concerning the extent to which findings in ADAD translate to LOAD.
Converging evidence from cerebrospinal fluid (CSF), amyloid imaging, and brain volumetric studies7,8 suggests that ADAD and LOAD are similar disease processes. However, biomarker differences exist between LOAD and ADAD. Specifically, ADAD individuals may have greater amyloid plaque deposition in the basal ganglia compared to LOAD individuals9. Additionally, increased levels of CSF Aβ1−42 have been observed very early in ADAD but not in LOAD8.
One biomarker of interest in LOAD that is relatively unestablished in ADAD is resting state functional connectivity MRI (rs-fcMRI)10,11. Functional connectivity measures the correlation structure of blood oxygen-level dependent (BOLD) signals between regions of interest (ROI), collections of which form resting state networks (RSNs)12,13. In LOAD, reduced functional connectivity has been observed with progressing clinical status [measured by clinical dementia rating (CDR)]14 within the default mode network (DMN), a RSN comprised of regions known to harbor Aβ15 and tau16 pathology. DMN functional connectivity decreases have also been noted in presymptomatic individuals genetically at risk for LOAD17. Recently, abnormalities in functional connectivity have been observed in the dorsal attention (DAN); executive-control (CON); salience (SAL); and sensorimotor (SMN) networks that parallel deteriorating cognitive status18.
We measured functional connectivity in a cross-sectional cohort of asymptomatic and symptomatic ADAD participants [mutation positive (M+; n=54) and mutation negative (M–; n=25)] and a cross-sectional cohort of LOAD individuals (n=74 very mild AD dementia, n=27 mild AD dementia, and n=343 cognitively normal older adults). We show that functional connectivity changes with respect to CDR are similar for both types of AD (i.e., ADAD and LOAD)18.
Materials and Methods
Patient characteristics
The ADAD cohort was drawn from the international Dominantly Inherited Alzheimer Network (DIAN) and consisted of participants from ADAD families, both individuals with mutations (M+) and individuals lacking mutations (M–) (Table 1). We excluded from the analysis 26 M+ individuals and 7 M–individuals who were scanned with inconsistent sequence parameters. We removed one additional M– participant with questionable clinical status. Cross-sectional data available as of February 2012 were included in this analysis19. Only subjects that passed quality control (described below) were included in the final analysis.
Table 1.
Demographics for autosomal dominant Alzheimer's disease (ADAD) and late-onset AD (LOAD) participants. Five participants from the ADAD CDR ≥ 1 cohort had more advanced disease (n=4 CDR 2; n=1 CDR 3). In both cohorts, the CDR ≥ 1 groups tended to be older and less educated. CDR= clinical dementia rating scale, MMSE= mini-mental status exam [range: 0 to 30 with higher score reflecting healthier cognition], APOE= apolipoprotein E. For some participants LOAD APOE4 status was not obtained.
| Autosomal Dominant AD (ADAD) | Late-onset AD (LOAD) | ||||||
|---|---|---|---|---|---|---|---|
| M– CDR0 | M+ CDR0 | M+ CDR0.5 | M+ CDR>1 | CDR0 | CDR0.5 | CDR1 | |
| N | 25 | 31 | 15 | 8 | 343 | 74 | 27 |
| Age (yrs) (sd) | 30.9 (10.0) | 33.9 (8.5) | 41.4 (10.4) | 49.4 (8.7) | 68.7 (9.5) | 74.0 (7.7) | 70.1 (11.4) |
| Sex (% male) | 40% | 39°% | 33% | 63% | 34% | 58% | 37% |
| Education (yrs) (sd) | 14.6 (2.0) | 14.7 (2.3) | 13.9 (2.1) | 11.6 (1.1) | 15.8 (2.6) | 15.0 (2.6) | 14.3 (2.5) |
| MMSE (sd) | 29.5 (0.71) | 28.7 (3.6) | 26.5 (2.7) | 14.1 (8.1) | 28.9 (1.3) | 26.8 (2.9) | 21.2 (4.1) |
| % APOE4 | 20% | 16% | 27% | 25% | 29% | 49% | 42% |
| % Frame Rej | 5.3% (7.3%) | 6.6% (8.9%) | 7.1% (8.9%) | 12.2% (14.1%) | 9.4% (12.1%) | 12.2% (8.4%) | 13.2% (7.1%) |
A separate cohort of participants was enrolled in a longitudinal study at the Knight Alzheimer's Disease Research Center (ADRC) at Washington University in St. Louis (WUSTL) designed to track individuals at risk for LOAD through the stages of cognitive decline18. All participants from both cohorts provided informed consent according to institutional review board procedures at their respective institutions. Each participant completed a general physical (including neurologic) examination, health and medication history, and clinical assessment for dementia20. We used independent general linear mixed models to assess group differences in demographics.
Clinical Dementia Rating (CDR)
Experienced clinicians conducted semi-structured interviews of each participant and a knowledgeable collateral source. The clinical dementia rating scale (CDR) was used to determine and stage dementia severity14. CDR0 indicates cognitive normality, CDR0.5 corresponds to very mild dementia, and CDR≥1 specifies mild and moderate dementia. In other studies, certain CDR0.5 participants may be classified as having mild cognitive impairment (MCI) due to AD, depending on the staging criteria21. Five participants from the ADAD CDR≥1 cohort had more advanced disease [CDR2 (n=4); CDR3 (n=1)]. All CDR > 0 participants had a clinical diagnosis of AD dementia in accordance with standard criteria.22 Disease biomarkers such as PiB PET imaging23 and CSF measures24 were not explicitly taken into account for the diagnosis of LOAD, but when available were used to exclude participants with profiles inconsistent with AD.
Estimated years from onset (EYO)
Within the DIAN cohort, parent age at symptomatic onset was determined from semi-structured interviews with the participant, a knowledgeable collateral source, and/or other informants familiar with the parental history of disease. The age at onset of the affected parent was determined by estimating the time of onset of symptoms (e.g., memory/cognition, motor or behavior). The anticipated age at symptomatic onset (AAO) for each individual was indexed to the AAO for that individual's affected parent. The estimated years from symptom onset (EYO) for each DIAN individual was defined as [(age at testing) – AAO]5.
Apolipoprotein Eε4 (APOE4) allele determination
DNA was extracted from peripheral blood and apolipoprotein E (APOE) genotyping was conducted according to previously-published methods25. Individuals were defined to be APOE4 positive if they had at least one ε4 allele.
MRI data acquisition
For both cohorts, neuroimaging was performed using 3T Siemens Tim Trio scanners (Erlangen, Germany) equipped with the standard 12-channel head coil using previously-described methods (see supplemental; also see Table 2). Structural images were acquired to allow alignment of rs-fcMRI images to atlas space18.
Table 2.
MRI imaging parameters for autosomal dominant AD (ADAD) and late-onset AD (LOAD). Scanning parameter differences between the two cohorts are in bold.
| ADAD | LOAD | |
|---|---|---|
| MPRAGE (T1) | TE = 16 msec, TR = 2,400 msec, TI = 1,000 msec, flip angle = 8°, 256 × 256 acquisition matrix, 1 × 1 × 1 mm voxels) | TE = 16 msec, TR = 2,400 msec, TI = 1,000 msec, flip angle = 8°, 256 × 256 acquisition matrix, 1 × 1 × 1 mm voxels) |
| FSE (T2) | No FSE available. | TE = 86.0 msec, TR = 6150.0 msec, 256 × 256 acquisition matrix, 1 acquisition, 1 × 1 × 4 mm voxels, flip angle = 120° |
| rs-fcMRI | TE = 27 msec, TR = 2200 msec, field of view = 256 mm, flip angle = 90° | TE = 30 msec, TR = 2200 msec, field of view = 256 mm, flip angle = 90° |
| rs-fcMRI length | 1×(140) frames | 2×(164) frames |
TE: echo time
TR: repetition time
TI: inversion time
MPRAGE: magnetization-prepared rapid gradient echo
FSE: fast spin echo
Pre-processing of all rs-fcMRI
Initial preprocessing of all rs-fcMRI data (both ADAD and LOAD) followed conventional methods as previously described18,26 which were modified to correct for non-optimal order of operations27 (see supplemental). Spurious variance was reduced by regression of nuisance time-series derived from head motion correction and extraction of BOLD activity from white matter, CSF regions, as well as the BOLD time-series averaged over the whole brain (or global signal)28.
Quality Assurance (QA) of rs-fcMRI
rs-fcMRI analyses and quality control procedures for ADAD and LOAD participants followed previously-described methods (see supplemental)29. Subjects with either outlier rms movement or excessive frame removal (>40%) were excluded from further analysis.
Resting-state network (RSN) composite correlation
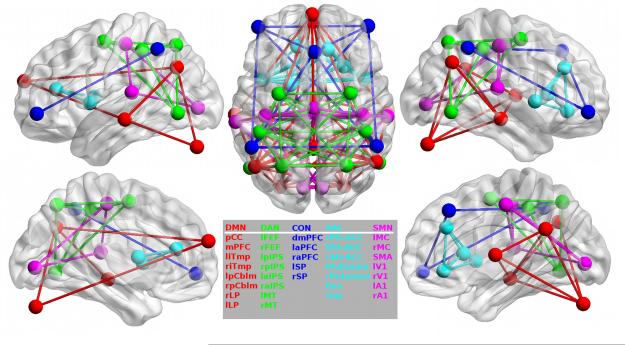
For all participants, we extracted time-series data from thirty-five 6-mm radius spherical brain regions of interest (ROIs) distributed throughout 5 functionally-defined RSNs including the DMN†, DAN, CON, SAL, and SMN (Figure 1). Briefly, intra-network composite scores were obtained by averaging BOLD correlation values computed between ROIs belonging to a particular RSN and inter-network composite scores were obtained by averaging correlations from ROIs belonging to separate RSNs. Using a composite score for intra and inter-network comparisons serves to reduce the amount of data while reducing the potential impact of sampling error. We analyzed composite scores for 5 intra-network (DMN, DAN, CON, SAL, SMN) and 3 inter-network (DMN:DAN, DMN:SMN, CON:SMN) composites which we have previously shown to be affected by LOAD18.
Figure 1.
Regions of Interest. Individual regions of interest are displayed on brain surfaces along with intra-network connections within each of the five networks analyzed in the current study: DMN=default mode, DAN=dorsal attention, CON= executive-control, SAL=salience, SMN=sensorimotor networks.
Statistical analysis
Generalized linear mixed models were used for each RSN composite to assess the fixed effects of CDR and AD type as well as their interaction. For ADAD, this model did not include CDR0 M– group in order to preserve the balance of the model between LOAD and ADAD. Differences between CDR0 M+ and CDR0 M– were assessed using the model that incorporates EYO described below. However, we include the CDR 0 M– group in each figure for comparison purposes. We also included ADAD family as a random effect because it is likely that functional connectivity measures are correlated for members of a common family. The AD type factor is a single fixed factor accounting for differences in average age and scanner acquisition parameters between the LOAD and ADAD groups. We assessed significant pair-wise effects (e.g., between CDR0 M+ and CDR0.5 M+) by extracting individual contrasts from the omnibus model. We compared the pair-wise effect size for different CDR stages between groups (e.g., CDR0.5 – CDR1 ADAD vs. CDR0.5 – CDR1 LOAD) using the Q test for effect size heterogeneity. We subsequently re-fit the preceding models adding factors in a stepwise fashion to account for the random effect of scanner and fixed effects of age and APOE ε4 status.
To analyze the effect of EYO on functional connectivity in the ADAD cohort, generalized linear mixed models were constructed for each RSN with EYO, quadratic effect of EYO (EYO2) and mutation status, as well as interactions among these factors. ADAD family membership was included as a random effect. Changes in RSN strength with respect to EYO were displayed using a locally weighted scatterplot smoothing (LOESS)7. To protect the confidentiality of participants’ mutation status, individual data points were not displayed.
To qualitatively assess whole-brain changes in DMN-associated functional connectivity with respect to EYO in the M+ ADAD group, we computed voxel-wise correlations between a 6 mm ROI in the posterior cingulate cortex (PCC; an important node of the DMN) and each voxel in the brain for each subject. We then used a LOESS model to predict PCC functional connectivity at each value of EYO in the range [-25,10] at 0.1 year increments for M+ individuals and displayed these predicted values using a movie. Each frame of the movie shows the predicted whole-brain average PCC-seed functional connectivity for a specific EYO value. Warm regions represent positive average within-DMN functional connectivity; cool regions represent negative between-network functional connectivity.
Cross-regression Analysis
We used ordinal logistic regression to perform a cross-regression analysis that further elucidated similarities between ADAD and LOAD. We fit a regression to predict CDR using the 5 intra-network and 3 inter-network composite values. We fit a separate model in each AD type and used this to predict CDR values for participants in the other AD type. We used Spearman rank correlations to assess the similarity between actual and predicted CDR values.
Results
Intra-network functional connectivity in LOAD and ADAD
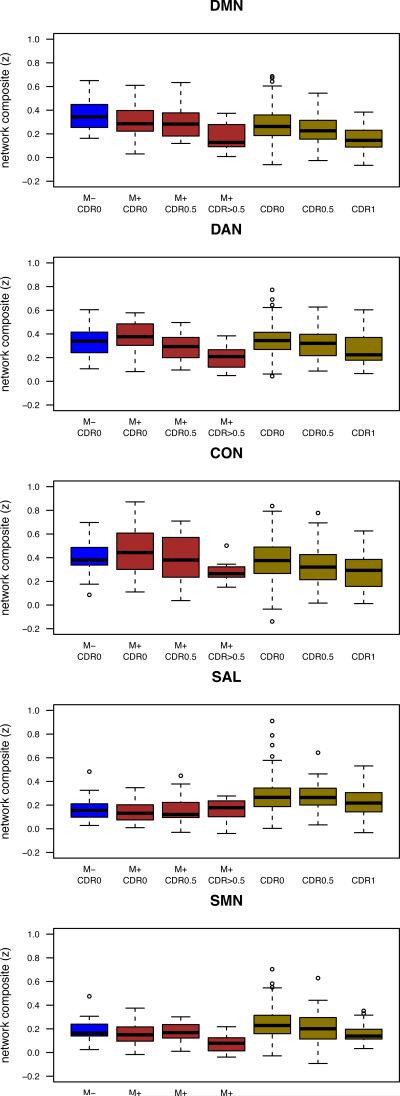
Initially, we combined both cohorts to test for the main effect of CDR stage on intra-network functional connectivity (Figure 2). A mixed model (corrected for mean age and acquisition differences between cohorts as well as a random effect of ADAD family membership) showed a significant main effect of CDR for multiple RSNs including the DMN, DAN, and CON (col. 1 Table 3). Only the SAL and SMN networks did not show a significant effect of CDR. In general, pairwise comparison between CDR stages showed that functional connectivity was lower in a step-wise fashion for the LOAD cohort (col. 2-4 Table 4). A similar pattern was observed for ADAD, although individual pair-wise differences (e.g., from CDR0 to CDR0.5) were generally not significant (cols. 2-4 Table 4). Stepwise inclusion of additional factors that assessed fixed effects of age as a continuous covariate and APOE ε4 status as well as a random effect of ADAD acquisition site reduced the observed effect sizes, but did not remove them (cols. 1-3 Table 3).
Figure 2.
Similar within RSN changes in LOAD and ADAD. Changes in intra-network resting state functional connectivity magnetic resonance imaging (rs-fcMRI) composite scores for participants for autosomal dominant Alzheimer disease (ADAD) and late-onset Alzheimer's disease (LOAD) participants as a function of clinical dementia rating (CDR). For both ADAD and LOAD, a stepwise loss of functional connectivity was seen for most resting state networks (RSNs) with increasing CDR. '*' denotes p<0.05, ‘**’ denotes p<0.005. Whiskers extend to 1.5 x interquartile range. DMN= default mode network, DAN= dorsal attention, CON= executive-control, SAL=salience, and SMN= sensorimotor.
Table 3.
Left columns) Results of independent omnibus mixed models that assessed the fixed effects of CDR and CDR by AD type interaction for both cohorts with a random effect of ADAD family; Right Columns) stepwise inclusion of additional factors (scanner, age as a continuous variable, and apolipoprotein ε4 (APOE) genotype) reduced power to observe a CDR effect but did not completely eliminate observed changes
| CDR | AD Type | CDR × AD Type | Step 1 | Step 2 | Step 3 | |||
|---|---|---|---|---|---|---|---|---|
| t(31) | t(6) | t(429) | Scanner | Age | APOE | |||
| DMN | t | –2.18 | 2.54 | 1.17 | CDR | 0.03* | 0.21 | 0.21 |
| p | 0.036* | 0.011 | 0.24 | CDRxAD type | 0.28 | 0.15 | 0.54 | |
| Age | 0.0001** | 0.0001** | ||||||
| APOE | 0.72 | |||||||
| DAN | t | –2.90 | 0.097 | 0.16 | CDR | 0.0086** | 0.04 | 0.043 |
| p | 0.0064* | 0.92 | 0.87 | CDRxAD type | 0.92 | 0.8 | 0.7 | |
| Age | 0.0014** | 0.0013** | ||||||
| APOE | 0.93 | |||||||
| CON | t | –2.31 | 2.41 | 0.29 | CDR | 0.034* | 0.19 | 0.21 |
| p | 0.027* | 0.016* | 0.77 | CDRxAD type | 0.72 | 0.47 | 0.89 | |
| Age | 0.0001** | 0.0001** | ||||||
| APOE | 0.62 | |||||||
| SAL | t | –0.38 | 6.91 | 0.63 | CDR | 0.71 | 0.96 | 0.96 |
| p | 0.71 | 0.001** | 0.52 | CDRxAD type | 0.52 | 0.5 | 0.54 | |
| Age | 0.086t | 0.05t | ||||||
| APOE | 0.15 | |||||||
| SMN | t | –1.36 | 4.95 | 1.44 | CDR | 0.17 | 0.25 | 0.29 |
| p | 0.18 | 0.001** | 0.15 | CDRxAD type | 0.16 | 0.14 | 0.15 | |
| Age | 0.33 | 0.17 | ||||||
| APOE | 0.55 | |||||||
| DMN:DAN | t | 3.16 | 3.11 | 0.35 | CDR | 0.0032** | 0.032* | 0.038* |
| p | 0.0032** | 0.002** | 0.73 | CDRxAD type | 0.7 | 0.98 | 0.64 | |
| Age | 0.0001** | 0.0001** | ||||||
| APOE | 0.63 | |||||||
| DMN:SMN | t | 0.96 | 1.12 | 1.09 | CDR | 0.34 | 0.59 | 0.65 |
| p | 0.34 | 0.26 | 0.28 | CDRxAD type | 0.27 | 0.17 | 0.39 | |
| Age | 0.037* | 0.015* | ||||||
| APOE | 0.9 | |||||||
| CON:SMN | t | –0.17 | 5.36 | 2.24 | CDR | 0.78 | 0.45 | 0.45 |
| p | 0.86 | 0.001** | 0.026* | CDRxAD type | 0.021* | 0.011* | 0.012* | |
| Age | 0.013* | 0.019* | ||||||
| APOE | 0.79 |
denotes trend p<0.1
denotes p<0.05
denotes p<0.005.
Table 4.
Pair-wise contrasts between each CDR group were extracted from the appropriate model. Contrasts comparing M+/– CDR 0 in ADAD are extracted from the model incorporating EYO. All other results were extracted from the omnibus mixed model. Effect sizes (Cohen's d) tend to be larger in ADAD, suggesting that loss of functional connectivity with respect to CDR is greater for this group. Shading indicate significantly larger effect size in ADAD as assessed using Q test for effect size heterogeneity.
| ADAD Cohort |
M– CDR0 v M+ CDR0 |
M+CDR0 v M+CDR0.5 |
M+CDR0.5 v M+ CDR1 |
M_ CDR0 v M+ CDR1 |
LOAD Cohort |
APOE4+ CDR0 v APOE4– CDR0 |
CDR0 v CDR0.5 |
CDR0.5 v CDR1 |
CDR0 v CDR1 |
|---|---|---|---|---|---|---|---|---|---|
| DMN | t=1.66, p=0.10t | t=0.16, p=0.88, d=0.05 | t=2.80, p=0.019,* d=1.22 | t=2.96, p=0.014,* d=1.18 | t=1.81, p=0.082t | t=2.33, p=0.02*, d=0.3 | t=3.00, p=0.003**, d=0.68 | t=4.87, p<0.001**, d=0.97 | |
| DAN | t=1.31, p=0.19 | t=2.21, p=0.051,t d=0.7 | t=1.64, p=0.132, d=0.72 | t=3.60, p=0.005,** d=1.42 | t=0.45, p=0.65 | t=1.18, p=0.24, d=0.15 | t=2.36, p=0.018*, d=0.53 | t=3.46, p<0.001**, d=0.68 | |
| CON | t=1.17, p=0.25 | t=0.60, p=0.36, d=0.3 | t=1.92, p=0.083,t d=0.84 | t=2.89, p=0.016,* d=1.14 | t=0.14, p=0.89 | t=1.80, p=0.073t, d=0.23 | t=1.64, p=0.10, d=0.37 | t=2.99, p=0.003**, d=0.6 | |
| SAL | t=1.40, p=0.17 | t=1.15, p=0.28, d=0.36 | t=0.45, p=0.67, d=0.2 | t=0.422, p=0.68, d=0.17 | t=1.70, p=0.089t | t=1.74, p=0.082t, d=0.22 | t=2.09, p=0.037*, d=0.47 | t=1.24, p=0.22, d=0.25 | |
| SMN | t=1.01, p=0.32 | t=0.38, p=0.71, d=0.12 | t=1.19, p=0.26, d=0.52 | t=1.62, p=0.14, d=0.64 | t=0.24, p=0.81 | t=2.85, p=0.005**, d=0.37 | t=2.64, p=0.008, d=0.59 | t=4.80, p<0.001**, d=0.96 | |
| DMN:DAN | t=0.81, p=0.42 | t=1.31, p=0.22, d=0.41 | t=3.32, p=0.008,* d=1.46 | t=4.69, t=0.001,** d=1.86 | t=1.53, p=0.13 | t=1.90, p=0.058t, d=0.24 | t=2.13, p=0.034, d=0.48 | t=3.61, p<0.001**, d=0.72 | |
| DMN:SMN | t=1.75, p=0.091t | t=1.26, p=0.26, d=0.4 | t=2.91, p=0.016,* d=1.27 | t=2.21, p=0.052,t d=0.88 | t=1.18, p=0.24 | t=0.78, p=0.44, d=0.1 | t=2.96, p=0.003, d=0.67 | t=3.83, p<0.001**, d=0.77 | |
| CON:SMN | t=1.17, p=0.25 | t=1.61, p=0.14, d=0.51 | t=0.50, p=0.63, d=0.22 | t=0.73, p=0.48, d=0.29 | t=0.097, p=0.92 | t=1.17, p=0.24, d=0.15 | t=2.32, p=0.021, d=0.52 | t=3.35, p=0.001**, d=0.67 |
indicates trends p<0.1
p<0.05
p<0.01
***p<0.001.
Although the general patterns of intra-network functional connectivity changes seen for ADAD and LOAD were similar, subtle differences were observed. When pair-wise effect sizes (Cohen's d) differed between ADAD and LOAD, the CDR effect was in general greater in ADAD compared to LOAD.
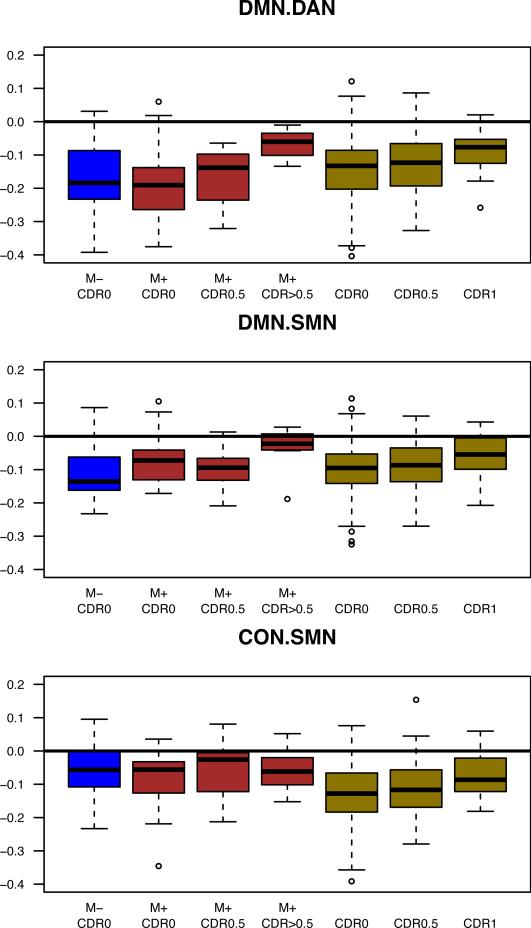
Inter-network functional connectivity in LOAD and ADAD
Inter-network functional connectivity was also decreased in magnitude with respect to CDR in both LOAD and ADAD (cols. 2-7 Figure 3). Inter-network (e.g., DMN:DAN) BOLD correlations typically are negative in sign (i.e., anti-correlations) in data preprocessed using global signal regression28. As previously reported, LOAD cross-network anti-correlations were diminished (i.e., closer to zero) with advancing CDR18. A similar finding was observed in ADAD (cols. 2-4 Figure 2), where decreased anti-correlation magnitude was observed for DMN:DAN but not DMN:SMN or CON:SMN (col. 1 Table 3). Stepwise inclusion of additional factors testing for fixed effects of age as a continuous covariate, APOE ε4 status, and the random effect of ADAD acquisition site reduced the effects, but did not remove them (Table 3b).
Figure 3.
Similar between RSN changes in LOAD and ADAD. Changes in inter-network composite scores for ADAD and LOAD participants as a function of CDR status. A loss of between-network functional connectivity was seen for the DMN:DAN and DMN:SMN with increasing CDR, though for CON:SMN, this pattern was only present in LOAD. ‘*’ denotes p<0.05, ‘**’ denotes p<0.005. Whiskers extend to 1.5 x interquartile range.
Cross-regression analysis
In order to further characterize the similarity between AD types, we fit ordinal logistic regression models (see Methods) in ADAD and used these to predict CDR levels in LOAD (and vice versa). The model fit in ADAD was able to predict LOAD CDR levels much better than chance (t(d.f.=442)=5.11; p<0.0001). The inverse process also allowed us to predict ADAD CDR levels based on LOAD data better than chance (t(d.f.=52)=4.51,p<0.0001). Cross-AD type classification was unsuccessful for predicting genetic risk in the absence of clinical symptoms.
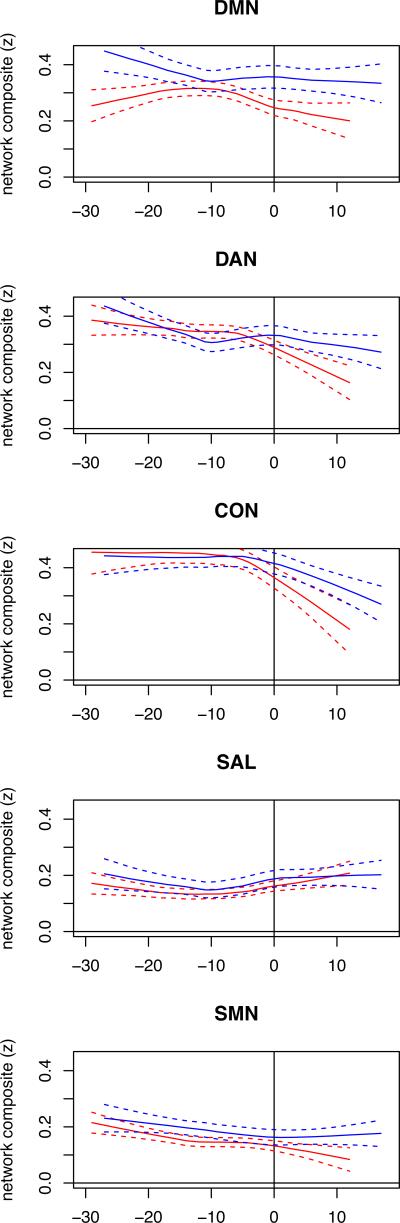
Functional connectivity in ADAD is lower in individuals closer to AAO
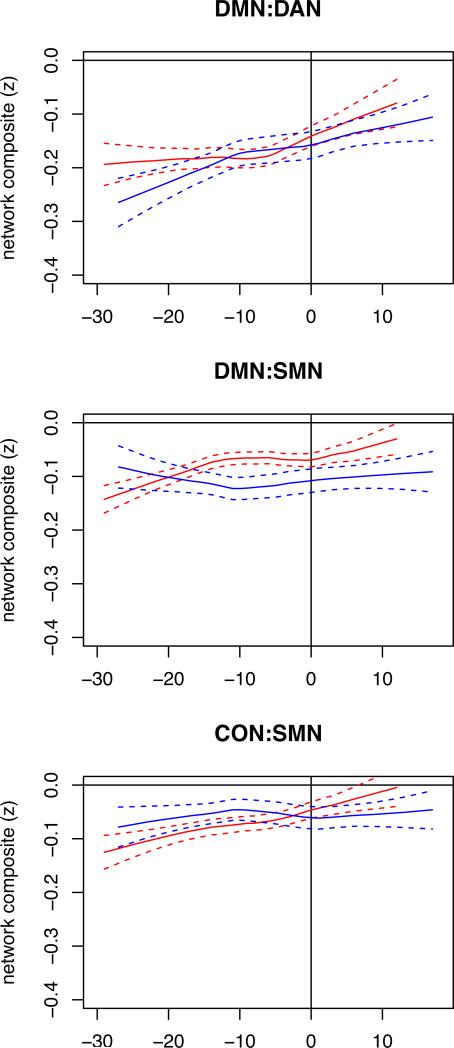
For ADAD, we show how functional connectivity changes occur relative to expected years from onset of symptoms (EYO) in all M+ individuals including individuals destined to develop cognitive impairment and those already symptomatic. Figure 4 presents LOESS plots of RSN composites scores against EYO and demonstrates a qualitative decrease in the DMN several years prior to expected symptom onset. Figure 5 presents the same analysis for the between RSN data. The limited size of this cohort spread over many decades of EYO precludes statistical demonstration of this effect but suggests that functional connectivity may slightly precede cognitive symptoms.
Figure 4.
EYO modulates within RSN FC in ADAD. Intra-network functional connectivity (and standard error bands) as function of estimated years from symptom onset (EYO) for all M+ (red) and M– (blue) ADAD individuals.
Figure 5.
EYO modulates between RSN FC in ADAD. Inter-network functional connectivity (and standard error bands) as function of estimated years from symptom onset (EYO) for all M+ (red) and M– (blue) ADAD participants.
We constructed a movie that demonstrates progressive loss of intra- and inter-network functional connectivity in the M+ group using the PCC as a seed. The fitted model predicted qualitative changes in functional connectivity in M+ participants prior to anticipated age of onset (AAO) (Movie 1).
Discussion
ADAD and LOAD manifest similar functional connectivity changes with respect to CDR. Moreover, regression models constructed in one cohort distinguished CDR stages in the other. This result demonstrates that functional connectivity changes manifest similarly in both types of AD. However, some differences exist between AD types in functional connectivity. A modestly greater effect of disease severity was seen for ADAD compared to LOAD. The available data suggest that ADAD may serve as an effective model to study LOAD pathophysiology, albeit with some reservations.
The first studies to investigate LOAD using rs-fcMRI detected changes in the DMN30. More recent work from our group has reported decreased functional connectivity in a wider set of intra and inter-network relationships18. These results are recapitulated in our current study, where we show similar effects of CDR on RSN connectivity in LOAD and ADAD. Similarities were also evident between ADAD and LOAD when a regression model was fit in each group using all analyzed RSNs as features and used these models to predict CDR levels in the other. Our success fitting CDR models in the ADAD cohort and predicting CDR status for the LOAD cohort (and vice versa) further suggests similar widespread RSN changes in both AD types.
However, analysis of the certain RSN composites suggested a slightly more pronounced decline for ADAD compared to LOAD. The greater loss in functional connectivity seen in ADAD in certain networks may suggest that ADAD is a more aggressive process than LOAD31,32. We previously hypothesized that inter-network correlations may reflect a mechanism by which pathology spreads from one functional network to the next in a cascading disease process33. There may be a more rapid and dramatic accumulation of Aβ and tau neurofibrillary tangle (NFT) pathology in ADAD compared to LOAD34. Hence, the observed rapid decline both within and between certain RSNs possibly reflects a faster spread of pathology from the DMN across diseased connections in ADAD.
Biomarker profiles accrue with age along distinct intra-individual trajectories in LOAD and ADAD6,35. In ADAD, we show evidence suggesting that functional connectivity decreases with EYO only in the M+ group. In M+ individuals, intra-individual changes in BOLD correlations within and between networks may serve as an effective biomarker of disease progression. Functional connectivity is a potentially useful biomarker in ADAD. However, we have only demonstrated qualitative differences between M+ and M– groups temporally proximate to the anticipated age of onset, suggesting that gross changes in intra-network functional connectivity likely occur later than changes in metabolism, hippocampal volume, and CSF Aβ and tau. Observed changes in BOLD correlations may reflect downstream pathophysiological processes7. Ongoing longitudinal studies will assess the usefulness of functional connectivity in tracking pre-clinical AD.
Our results differ from previous results on three points. First, individual RSN composite scores were not significantly different for asymptomatic participants with genetic risk factors in either cohort. This conflicts with previous studies of LOAD that showed DMN functional connectivity changes within network in asymptomatic individuals with Aβ plaque deposits36 or a family history of LOAD37. Second, we did not observe a transient increase in functional connectivity in the SAL for ADAD participants as was previously observed for LOAD18,38. This suggests another possible difference between LOAD and ADAD. Finally, in contrast to a recent study from Chhatwal et al.,11 we were unable to demonstrate at a statistically significant divergence between the M+ and M– individuals prior to symptom onset, though qualitatively our data are consistent with that finding. This difference possibly reflects the fact that Chhatwal et al analyzed ROI-level changes whereas here we analyzed network-level changes. We were able to demonstrate voxel-level DMN functional connectivity changes using a LOESS movie. This qualitatively confirmed the Chhatwal et al. results using a ROI. Indeed, the ADAD cohort reported by Chhatwal et al., is the same cohort reported here although we excluded several additional participants due to scan parameter issues.
This study made use of network composite scores as a measure of functional connectivity strength18 which have several strengths but also make two assumptions. First, composite scores are a data reduction strategy, reducing the burden of multiple comparisons. Second, they reduce sampling error of observing any single functional connectivity pair within an RSN. However, they assume that each functional connectivity pair in an RSN behaves similarly. This has been previously been shown to be valid in LOAD but may obscure focal changes such as those previously seen in ADAD11. In addition, composite scores assume that an ROI's RSN membership does not change with disease, which could bias the measurement.
Several limitations arose from the design of this study. First, there were scanning differences between cohorts. This complicates demonstration of average differences between cohorts, but this does not impact our ability to demonstrate similarities between AD types. Second, our LOAD cohort was significantly older than our ADAD cohort. This is an unavoidable confound in any study comparing LOAD to early-onset ADAD. We addressed this issue by correcting for age differences between the two cohorts. Finally, it has been argued that EYO might not be the best estimate of disease progression in CDR0.5 ADAD participants. However, because CDR0.5 individuals are difficult to stage precisely, EYO is the most practical measure in a cross-sectional study. Larger longitudinal studies will be able to more fully characterize ADAD and LOAD functional connectivity changes and place them in temporal relation to other biomarkers (especially CSF tau, Aβ, positron emission tomography (PET), volumetrics, and amyloid imaging). Volumetric comparisons are particularly important to this study since atrophy may influence the measured BOLD signal. Future studies directly comparing these two measures will be important.
Finally, this study made use of the global signal regression (GSR) preprocessing step. This procedure is controversial28,39. It is algebraically true that GSR forces the mean of correlations across the brain to be zero and can make negative correlations more apparent. However, correlations following GSR are essentially first-order partial correlations accounting for widely shared variance while correlations without GSR are canonical correlations. This makes correlations with and without GSR two fundamentally different statistical quantities reflecting different types of relationships. It is likely that some of the removed signal is of neural origin40 however a large fraction of the global signal is related to residual effects of head motion29 and fluctuations in pCO241. Thus, we viewed GSR as a necessary step for noise reduction in this cross-scanner, multi-site study. Beyond its noise reduction properties42, GSR has been shown to increase the concordance between BOLD correlation mapping and electrocorticography, particularly for negative correlations43, indicating an important relationship to neurobiology.
Supplementary Material
Acknowledgements
This work was funded by National Institute of Aging-U19-AG032438 (DIAN; JCM), National Institute of Mental Health-K23MH081786 (BMA), National Institute of Nursing Research-R01NR014449 (BMA), R01NR012657 (BMA), R01NR012907 (BMA), National Institute of Mental Health – R21MH099979 (BMA), National Institute of Neurological Disorder and Stroke P30NS048056 (DM, AZS), P50 AG05681 (JCM); P01 AG03991 (JCM), P01 AG026276 (JCM), Medical Research Council G0601846 (NCF). Supported by the NIHR Queen Square Dementia Biomedical Research Unit and the Washington University in St. Louis ADRC Genetics Core. BMA had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. The funding organizations had no role in the design and conduct of the study; collection, management, analysis, and interpretations of the data; and preparation, review, or approval of the manuscript; and the decision to submit the manuscript for publication.
Appendix
Financial Disclosure Information
Beau Ances has no disclosures
Randall Bateman receives funding from Fidelity Non-Profit Management Foundation. He is a board member for EvVivo Scientific Advisory Board and consults with C2N Diagnostics, Eisai Scientific Advisory Panel, Medtronics Scientific Advisory Panel and Novartis. Bateman is also funded or pending funding by the NIH, Alzheimers’ Association, Glenn Foundation for Medical Research, AstraZeneca LP, Merck & Company Inc., Eli Lilly and Co., Pharma Consortium (Biogen Idec, Elan Pharmaceuticals Inc., Eli Lilly and Co., Hoffman La-Roche Inc., Genentech nc., Janssen Alzheimer Immunotherapy, Mithridion Inc., Novartis Pharma AG, Pfizer Biotherapeutics R and D, Sanofi-Aventi), The Ruth K. Broad Biomedical, Elan and Astra Zeneca, Washington University, and C2N Diagnostics.
Tammie Benzinger has received research funding from Avid Radiopharmaceuticals, a wholly
owned subsidiary of Eli Lilly.
Tyler Blazey has no disclosures.
Matthew Brier has no disclosures.
Virginia Buckles has no disclosures.
Nigel Cairns has no disclosures.
Jasmeer Chhatwal has no disclosures
Anne Fagan receives funding from the NIH and is a member of the Scientific Advisory Boards for IBL International and Roche.
Nick Fox has received payment for consultancy or for conducting studies from AVID, Bristol-Myers Squibb, Elan Pharmaceuticals, Eisai, Lilly Research Laboratories, GE Healthcare, IXICO, Janssen Alzheimer Immunotherapy, Johnson & Johnson, Janssen-Cilig, Lundbeck, Neurochem Inc, Novartis Pharma AG, Pfizer Inc, Sanofi-Aventis and Wyeth Pharmaceuticals. Professor Fox has an NIHR Senior Investigator award and receives support from the Wolfson Foundation; NIHR Biomedical Research Unit (Dementia) at UCL; the EPSRC; Alzheimer's Research UK and the NIA. NCF receives no personal compensation for the activities mentioned above.
Bernardino Ghetti consults for Piramal imaging.
Alison Goate receives funding from AstraZeneca, Genentech, Pfizer, Tau Consortium and the DIAN Pharma-Consortium: DIAN Pharmaceutical-Consortium: Biogen Idec, Eisai, Inc., Elan Pharmaceuticals, Inc., Eli Lilly and Company, FORUM Pharmaceuticals, Hoffman La-Roche, Inc., Genetech, Inc., Janssen Alzheimer Immunotherapy, Mithridion, Inc., Novartis Pharm AG, Pfizer Biotherapeutics Research & Development, Sanofi-Aventis. Goate consults for Amgen and Cognition Therapeutics and received payments for lectures including service on speakers bureaus from Pfizer, AstraZeneca, and Genentech, she receives royalties for IP licensed to Taconic.
David Holtzman receives research funding from the NIH, AstraZeneca, C2N Diagnostics, Cure Alzheimer's Fund, and Tau Consortium. Holtzman serves on the scientific advisory board of C2N Diagnostics and consults for Bristol-Myers Squibb, Eli Lilly, and Genentech. Holtzman holds US patents; 7,892,845; 7,892,545; 7,771,722; 7,195,761; 7,015,044; 6,465,195. Russ Hornbeck has no disclosures.
Clifford Jack has provided consulting services for Janssen Research & Development, LLC, and Eli Lily. He receives research funding from the National Institutes of Health ((R01-AG011378, U01-HL096917, U01-AG024904, RO1 AG041851, R01 AG37551, R01AG043392, U01-AG06786)), and the Alexander Family Alzheimer's Disease Research Professorship of the Mayo Foundation.
Robert A Koeppe has no disclosures.
Daniel Marcus consults with Avid Radiopharmaceuticals, has stock options with Radiologics, Inc. He receives travel/accommodations/meeting expenses unrelated to activities listed from BlueArc, Inc.
Ralph NW Martins has no disclosures.
Colin Masters has advisory roles with Prana Biotechnology and Eli Lilly Company.
Chester A Mathis has no disclosures.
Richard Mayeux has no disclosures.
Eric McDade has no disclosures.
Dr. John C. Morris reports disclosures: Neither Dr. Morris nor his family owns stock or has equity interest (outside of mutual funds or other externally directed accounts) in any pharmaceutical or biotechnology company. Dr. Morris has participated or is currently participating in clinical trials of antidementia drugs sponsored by the following companies: Janssen Immunotherapy, and Pfizer. Dr. Morris has served as a consultant for Lilly USA. He receives research support from Eli Lilly/Avid Radiopharmaceuticals and is funded by NIH grants # P50AG005681; P01AG003991; P01AG026276 and U19AG032438”
Krista Moulder has no disclosures.
Angela Oliver has no disclosures.
Sébastien Ourselin receives funding from the Engineering and Physical Sciences Research Council (EPSRC, UK) (EP/H046410/1, EP/J020990/1, EP/K005278), the Medical Research Council (MRC,UK) (MR/J01107X/1), the EUFP7 project VPH-DARE@IT (FP7-ICT-2011-9-601055), the National Institute for Health Research University College London Hospitals Biomedical Research Centre (NIHR BRC UCLH/UCL High Impact Initiative), GE Healthcare, Siemens, and MIRADA Medical.
Dr. Ringman has received personal compensation for serving on scientific advisory boards for Takeda Pharmaceuticals and StemCells, Inc. and has received research support from Janssen, Pfizer, Accera, Bristol Myers Squibb, and Wyeth Pharmaceuticals.
Marcus Raichle has no disclosures.
Martin Rossor consults for Pfizer/Janssen.
Dr. Salloway is a consultant to Janssen AI, Astra-Zeneca, Avid-Lilly, GE, Baxter, Pfizer, Athena, BMS, Biogen, and Merck, all less than $5,000. His hospital receives research support for clinical trials from Janssen AI, Baxter, BMS, Pfizer, Genentech, Bayer, GE, Avid, Roche, Merck, Lilly, Functional Neuromodulation. He reports no conflicts with the current work.
Peter Schofield receives funding from the Australian NHMRC, ARC and Beyondblue. He is a board member of Neuroscience Research Australia, the Neuroscience Research Australia Foundation, The Health Science Alliance and is an employee of Neuroscience Research Australia. He has received speaker fees from Janssen Pharmaceuticals Australia. Aaron P Schultz served on an advisory board for Janssen Pharmaceuticals. Reisa Sperling consults for Pfizer, Janssen, Eisai, Roche, and Bristol-Myers Squibb. Sperling receives payment for lectures including service on speakers bureaus from Pfizer, Janssen, and Eli Lilly.
Abraham Snyder has no disclosures.
Jewell Thomas has no disclosures.
Paul M Thompson has no disclosures.
Michael W Weiner has served on the Scientific Advisory Boards for Pfizer, BOLT International, Neurotrope Bioscience, and Eli Lilly. He has provided consulting to Synarc, Pfizer, Janssen, KLJ Associates, Easton Associates, Harvard University, University of California, Los Angeles (UCLA), Alzheimer's Drug Discovery Foundation (ADDF), Avid Radiopharmaceuticals, Clearview Healthcare Partners, Perceptive Informatics, Smartfish AS, Decision Resources, Inc., Araclon, Merck, Defined Health, and Genentech. The following entities have provided funding for travel; Pfizer, Paul Sabatier University, MCI Group France, Travel eDreams, Inc., Neuroscience School of Advanced Studies (NSAS), Danone Trading, BV, CTAD Ant Congres, Kenes, Intl., ADRC, UCLA, UCSD, Sanofi-Aventis Groupe, University Center Hospital, Toulouse, Araclon, AC Immune, Eli Lilly, New York Academy of Sciences (NYAS), and National Brain Research Center, India for Johns Hopkins Medicine. He served on the Editorial Boards for Alzheimer's & Dementia and MRI. He received honoraria from Pfizer, Tohoku University, and Danone Trading, BV. He received research support from Merck, Avid, the Veterans Administration (VA) and Department of Defense (DOD).
Chengjie Xiong has no disclosures.
Footnotes
The DMN has previously included a thalamic ROI. However, this ROI was not included in this analysis because it has at best weak correlations with the DMN.
References
- 1.Blennow K, de Leon MJ, Zetterberg H. Alzheimer's disease. The Lancet. 2006;368(9533):387–403. doi: 10.1016/S0140-6736(06)69113-7. [DOI] [PubMed] [Google Scholar]
- 2.Hansson O, Zetterberg H, Buchhave P, Londos E, Blennow K, Minthon L. Association between CSF biomarkers and incipient Alzheimer's disease in patients with mild cognitive impairment: a follow-up study. Lancet neurology. 2006 Mar;5(3):228–234. doi: 10.1016/S1474-4422(06)70355-6. [DOI] [PubMed] [Google Scholar]
- 3.Ryan NS, Rossor MN. Correlating familial Alzheimer's disease gene mutations with clinical phenotype. Biomarkers in medicine. 2010 Feb;4(1):99–112. doi: 10.2217/bmm.09.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yagi T, Ito D, Okada Y, et al. Modeling familial Alzheimer's disease with induced pluripotent stem cells. Human molecular genetics. 2011 Dec 1;20(23):4530–4539. doi: 10.1093/hmg/ddr394. [DOI] [PubMed] [Google Scholar]
- 5.Bateman RJ, Aisen PS, De Strooper B, et al. Autosomal-dominant Alzheimer's disease: a review and proposal for the prevention of Alzheimer's disease. Alzheimer's research & therapy. 2011;3(1):1. doi: 10.1186/alzrt59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fleisher AS, Chen K, Quiroz YT, et al. Florbetapir PET analysis of amyloid-beta deposition in the presenilin 1 E280A autosomal dominant Alzheimer's disease kindred: a cross-sectional study. Lancet neurology. 2012 Dec;11(12):1057–1065. doi: 10.1016/S1474-4422(12)70227-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bateman RJ, Xiong C, Benzinger TL, et al. Clinical and biomarker changes in dominantly inherited Alzheimer's disease. N Engl J Med. 2012 Aug 30;367(9):795–804. doi: 10.1056/NEJMoa1202753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reiman EM, Quiroz YT, Fleisher AS, et al. Brain imaging and fluid biomarker analysis in young adults at genetic risk for autosomal dominant Alzheimer's disease in the presenilin 1 E280A kindred: a case-control study. The Lancet Neurology. 2012;11(12):1048–1056. doi: 10.1016/S1474-4422(12)70228-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villemagne VL, Ataka S, Mizuno T, et al. High striatal amyloid beta-peptide deposition across different autosomal Alzheimer disease mutation types. Archives of neurology. 2009 Dec;66(12):1537–1544. doi: 10.1001/archneurol.2009.285. [DOI] [PubMed] [Google Scholar]
- 10.Dickerson BC, Sperling RA. Large-scale functional brain network abnormalities in Alzheimer's disease: insights from functional neuroimaging. Behavioural neurology. 2009;21(1):63–75. doi: 10.3233/BEN-2009-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chhatwal JP, Schultz AP, Johnson K, et al. Impaired default network functional connectivity in autosomal dominant Alzheimer disease. Neurology. 2013 Aug 20;81(8):736–744. doi: 10.1212/WNL.0b013e3182a1aafe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biswal B, Yetkin Z, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magnetic Resonance in Medicine. 1995;34(4):537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- 13.Biswal BB, Mennes M, Zuo XN, et al. Toward discovery science of human brain function. Proceedings of the National Academy of Sciences of the United States of America. 2010 Mar 9;107(10):4734–4739. doi: 10.1073/pnas.0911855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris JC. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 1993 Nov;43(11):2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- 15.Buckner RL, Snyder AZ, Shannon BJ, et al. Molecular, structural, and functional characterization of Alzheimer's disease: evidence for a relationship between default activity, amyloid, and memory. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005 Aug 24;25(34):7709–7717. doi: 10.1523/JNEUROSCI.2177-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braak H, Thal DR, Ghebremedhin E, Tredici KD. Stages of the pathological process in Alzheimer disease: Age categories from 1 to 100 years. J Neuropathol Exp Neurol. 2011;70(11):960–969. doi: 10.1097/NEN.0b013e318232a379. [DOI] [PubMed] [Google Scholar]
- 17.Machulda MM, Jones DT, Vemuri P, et al. Effect of APOE epsilon4 status on intrinsic network connectivity in cognitively normal elderly subjects. Archives of neurology. 2011 Sep;68(9):1131–1136. doi: 10.1001/archneurol.2011.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brier MR, Thomas JB, Snyder AZ, et al. Loss of intranetwork and internetwork resting state functional connections with Alzheimer's disease progression. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012 Jun 27;32(26):8890–8899. doi: 10.1523/JNEUROSCI.5698-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moulder KL, Snider BJ, Mills SL, et al. Dominantly Inherited Alzheimer Network: facilitating research and clinical trials. Alzheimer's research & therapy. 2013 Oct 17;5(5):48. doi: 10.1186/alzrt213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris JC, Weintraub S, Chui HC, et al. The Uniform Data Set (UDS): clinical and cognitive variables and descriptive data from Alzheimer Disease Centers. Alzheimer disease and associated disorders. 2006 Oct-Dec;20(4):210–216. doi: 10.1097/01.wad.0000213865.09806.92. [DOI] [PubMed] [Google Scholar]
- 21.Morris JC, Blennow K, Froelich L, et al. Harmonized diagnostic criteria for Alzheimer's disease: recommendations. Journal of internal medicine. 2014 Mar;275(3):204–213. doi: 10.1111/joim.12199. [DOI] [PubMed] [Google Scholar]
- 22.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984 Jul;34(7):939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 23.Klunk WE, Engler H, Nordberg A, et al. Imaging brain amyloid in Alzheimer's disease with Pittsburgh Compound-B. Annals of neurology. 2004 Mar;55(3):306–319. doi: 10.1002/ana.20009. [DOI] [PubMed] [Google Scholar]
- 24.Fagan AM, Holtzman DM. Cerebrospinal fluid biomarkers of Alzheimer's disease. Biomarkers in medicine. 2010 Feb;4(1):51–63. doi: 10.2217/BMM.09.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pastor P, Roe CM, Villegas A, et al. Apolipoprotein Ee4 modifies Alzheimer's disease onset in an E280A PS1 kindred. Annals of neurology. 2003;54:163–169. doi: 10.1002/ana.10636. [DOI] [PubMed] [Google Scholar]
- 26.Shulman GL, Pope DL, Astafiev SV, McAvoy MP, Snyder AZ, Corbetta M. Right hemisphere dominance during spatial selective attention and target detection occurs outside the dorsal frontoparietal network. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010 Mar 10;30(10):3640–3651. doi: 10.1523/JNEUROSCI.4085-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hallquist MN, Hwang K, Luna B. The nuisance of nuisance regression: spectral misspecification in a common approach to resting-state fMRI preprocessing reintroduces noise and obscures functional connectivity. NeuroImage. 2013 Nov 15;82:208–225. doi: 10.1016/j.neuroimage.2013.05.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fox MD, Zhang D, Snyder AZ, Raichle ME. The global signal and observed anticorrelated resting state brain networks. Journal of neurophysiology. 2009 Jun;101(6):3270–3283. doi: 10.1152/jn.90777.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage. 2012 Feb 1;59(3):2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greicius MD, Menon V. Default-mode activity during a passive sensory task: uncoupled from deactivation but impacting activation. J Cogn Neurosci. 2004 Nov;16(9):1484–1492. doi: 10.1162/0898929042568532. [DOI] [PubMed] [Google Scholar]
- 31.Gregory GC, Macdonald V, Schofield PR, Kril JJ, Halliday GM. Differences in regional brain atrophy in genetic forms of Alzheimer's disease. Neurobiol Aging. 2006 Mar;27(3):387–393. doi: 10.1016/j.neurobiolaging.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 32.Ringman JM, Medina LD, Rodriguez-Agudelo Y, Chavez M, Lu P, Cummings JL. Current concepts of mild cognitive impairment and their applicability to persons at-risk for familial Alzheimer's disease. Current Alzheimer research. 2009 Aug;6(4):341–346. doi: 10.2174/156720509788929336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kfoury N, Holmes BB, Jiang H, Holtzman DM, Diamond MI. Trans-cellular propagation of Tau aggregation by fibrillar species. The Journal of biological chemistry. 2012 Jun 1;287(23):19440–19451. doi: 10.1074/jbc.M112.346072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shepherd C, McCann H, Halliday GM. Variations in the neuropathology of familial Alzheimer's disease. Acta neuropathologica. 2009 Jul;118(1):37–52. doi: 10.1007/s00401-009-0521-4. [DOI] [PubMed] [Google Scholar]
- 35.Jack CR, Jr., Vemuri P, Wiste HJ, et al. Shapes of the trajectories of 5 major biomarkers of Alzheimer disease. Archives of neurology. 2012 Jul;69(7):856–867. doi: 10.1001/archneurol.2011.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheline YI, Raichle ME, Snyder AZ, et al. Amyloid plaques disrupt resting state default mode network connectivity in cognitively normal elderly. Biol Psychiatry. 2010 Mar 15;67(6):584–587. doi: 10.1016/j.biopsych.2009.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L, Roe CM, Snyder AZ, et al. Alzheimer disease family history impacts resting state functional connectivity. Annals of neurology. 2012 Oct;72(4):571–577. doi: 10.1002/ana.23643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seeley WW, Menon V, Schatzberg AF, et al. Dissociable intrinsic connectivity networks for salience processing and executive control. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007 Feb 28;27(9):2349–2356. doi: 10.1523/JNEUROSCI.5587-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy K, Birn RM, Handwerker DA, Jones TB, Bandettini PA. The impact of global signal regression on resting state correlations: are anti-correlated networks introduced? NeuroImage. 2009 Feb 1;44(3):893–905. doi: 10.1016/j.neuroimage.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scholvinck ML, Maier A, Ye FQ, Duyn JH, Leopold DA. Neural basis of global resting- state fMRI activity. Proceedings of the National Academy of Sciences of the United States of America. 2010 Jun 1;107(22):10238–10243. doi: 10.1073/pnas.0913110107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Birn RM, Diamond JB, Smith MA, Bandettini PA. Separating respiratory-variation- related fluctuations from neuronal-activity-related fluctuations in fMRI. NeuroImage. 2006 Jul 15;31(4):1536–1548. doi: 10.1016/j.neuroimage.2006.02.048. [DOI] [PubMed] [Google Scholar]
- 42.Power JD, Mitra A, Laumann TO, Snyder AZ, Schlaggar BL, Petersen SE. Methods to detect, characterize, and remove motion artifact in resting state fMRI. NeuroImage. 2013 Aug 29;84C:320–341. doi: 10.1016/j.neuroimage.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keller CJ, Bickel S, Honey CJ, et al. Neurophysiological investigation of spontaneous correlated and anticorrelated fluctuations of the BOLD signal. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013 Apr 10;33(15):6333–6342. doi: 10.1523/JNEUROSCI.4837-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.