Abstract
Autophagy, the major lysosomal pathway for the turnover of intracellular organelles is markedly impaired in neurons in Alzheimer’s disease and Alzheimer mouse models. We have previously reported that severe lysosomal and amyloid neuropathology and associated cognitive deficits in the TgCRND8 Alzheimer mouse model can be ameliorated by restoring lysosomal proteolytic capacity and autophagy flux via genetic deletion of the lysosomal protease inhibitor, cystatin B. Here we present evidence that macroautophagy is a significant pathway for lipid turnover, which is defective in TgCRND8 brain where lipids accumulate as membranous structures and lipid droplets within giant neuronal autolysosomes. Levels of multiple lipid species including several sphingolipids (ceramide, ganglioside GM3, GM2, GM1, GD3 and GD1a), cardiolipin, cholesterol and cholesteryl esters are elevated in autophagic vacuole fractions and lysosomes isolated from TgCRND8 brain. Lipids are localized in autophagosomes and autolysosomes by double immunofluorescence analyses in wild-type mice and colocalization is increased in TgCRND8 mice where abnormally abundant GM2 ganglioside-positive granules are detected in neuronal lysosomes. Cystatin B deletion in TgCRND8 significantly reduces the number of GM2-positive granules and lowers the levels of GM2 and GM3 in lysosomes, decreases lipofuscin-related autofluorescence, and eliminates giant lipid-containing autolysosomes while increasing numbers of normal-sized autolysosomes/lysosomes with reduced content of undigested components. These findings have identified macroautophagy as a previously unappreciated route for delivering membrane lipids to lysosomes for turnover, a function that has so far been considered to be mediated exclusively through the endocytic pathway, and revealed that autophagic-lysosomal dysfunction in TgCRND8 brain impedes lysosomal turnover of lipids as well as proteins. The amelioration of lipid accumulation in TgCRND8 by removing cystatin B inhibition on lysosomal proteases suggests that enhancing lysosomal proteolysis improves the overall environment of the lysosome and its clearance functions, which may be possibly relevant to a broader range of lysosomal disorders beyond Alzheimer’s disease.
Keywords: autophagy, lysosome, lipids, Alzheimer’s disease, TgCRND8
The role of autophagy in lipid turnover is largely unknown. Yang et al. reveal macroautophagy to be a route for delivering membrane lipids to lysosomes. They demonstrate defective lysosomal lipid clearance in an Alzheimer’s mouse model leading to lipid accumulation in autolysosomes, which can be prevented by enhancing lysosomal proteolysis.
Introduction
Neurofibrillary tangles of tau protein and neuritic plaques containing dystrophic neurites associated with extracellular deposits of amyloid-β are the neuropathological hallmarks of Alzheimer’s disease (Selkoe, 2001; Foley, 2010). In his original descriptions of the disease, however, Alzheimer also reported granular lipid inclusions in multiple cell types within the brain and vasculature (Foley, 2010). Since their description, the compositions of such lipid deposits, the mechanisms underlying the deposition, and the role of lipid deposits in Alzheimer’s disease development remain unclear, although emerging evidence supports strong links between lipid dyshomeostasis and Alzheimer’s disease pathogenesis (Garner, 2010; Di Paolo and Kim, 2011; Hicks et al., 2012). Besides the well-established role of apolipoprotein E4 as a major risk factor for late-onset Alzheimer’s disease, other lipid species, including cholesterol, phospholipids, sphingolipids (e.g. gangliosides), have also been implicated in Alzheimer’s disease (Wolozin et al., 2006; Ariga et al., 2011; Frisardi et al., 2011; van Echten-Deckert and Walter, 2012). Collectively, the reported alterations in levels of specific lipid species in Alzheimer’s disease still present a complex picture, with certain ones increasing, others decreasing, and others showing opposite changes in different studies (Crino et al., 1989; Kalanj et al., 1991; Han, 2005; Molander-Melin et al., 2005; Barrier et al., 2007; Gylys et al., 2007; Ariga et al., 2010; Chan et al., 2012; Pernber et al., 2012). These observations possibly reflect differences in the brain regions analysed, the degree of neurodegeneration and/or gliosis, or the analytical techniques applied.
The endosomal-lysosomal pathway has long been considered the primary pathway for degrading endocytosed extracellular lipoproteins and membrane lipid components of intraluminal vesicles within multivesicular bodies/late endosomes (Kolter and Sandhoff, 2005). Only recently has autophagy also been implicated in lipid degradation based on observations that lipid droplets primarily storing cholesterol and triglycerides can be turned over by a form of autophagy, termed lipophagy, whereby a portion of, or small whole lipid droplets are sequestered in autophagosomes and delivered to lysosomes for degradation; and inhibition of autophagy induction increases lipid storage in lipid droplets (Singh et al., 2009). Sphingolipids, phospholipids and cholesterol are constituents of membrane structures/organelles (Andreyev et al., 2010; van Meer and de Kroon, 2011) that can be both substrates of autophagy and components of the autophagy machinery (e.g. autophagosomes) (Knaevelsrud and Simonsen, 2012; Lamb et al., 2013). It is therefore likely that this fraction of the cellular lipid pool is delivered to lysosomes through the route of autophagy (Kolter and Sandhoff, 2010) although this has not been previously investigated. Whether by an endocytic or autophagic route, such lipids become substrates for lysosomal hydrolases, the importance of which for lipid catabolism is amply illustrated by the lysosomal lipid storage disorders where undigested lipids accumulate (Walkley and Vanier, 2009; Lloyd-Evans and Platt, 2010; Schulze and Sandhoff, 2011; Lieberman et al., 2012; Platt et al., 2012; Nixon, 2013).
Dysfunction of the global lysosomal system, involving both endocytic and autophagic pathways, is a prominent and early neuropathological feature of Alzheimer’s disease and several other neurodegenerative diseases (Nixon, 2005; Harris and Rubinsztein, 2011; Nixon and Yang, 2011; Nixon, 2013). TgCRND8 mice overexpressing mutant human amyloid precursor protein (APP), a model of Alzheimer’s disease-related amyloidosis, also exhibit severe autophagy-related pathology in neurons, which is characterized by grossly enlarged autolysosomes, depressed cathepsin activity, and impaired lysosomal turnover of proteins (Yang et al., 2011). In the present study, we detected for the first time a range of membrane lipids in autophagic vacuole fractions isolated from brains of wild-type mice consistent with autophagy being a degradative pathway for membrane lipids. Moreover, we observed accentuated accumulation of lipids in autolysosomal/lysosomal compartments in TgCRND8 mice indicating that autophagic clearance of lipids is impaired. Finally, we could ameliorate lipid accumulation within lysosomes by restoring cathepsin activity and lysosomal protein degradation in TgCRND8 mice, which was achieved by deleting cystatin B (CSTB), a key endogenous lysosomal inhibitor of cysteine-active site cathepsins (Yang et al., 2011). Our findings, therefore, support a role for autophagy in the clearance of lipids in brain and demonstrate that enhancing lysosomal proteolytic function in a mouse model of Alzheimer’s disease-related amyloidosis has additional benefits in facilitating the clearance of stored lipids, which may contribute to the ameliorative effects of this intervention on pathological and functional features of this disease model (Yang et al., 2011).
Materials and methods
Animals and tissue preparation
All animal procedures were performed following the National Institutes of Health Guidelines for the Humane Treatment of Animals, with approval from the Institutional Animal Care and Use Committee at the Nathan Kline Institute for Psychiatric Research. Animals of both sexes were used in this study. TgCRND8 mice, expressing mutant human APP, Swedish (K670N/M671L) plus Indiana (V717F) mutations, were created on a 129S6/SvEvTac (129S6) strain background by Dr David Westaway (Chishti et al., 2001). CSTB knockout (CBKO) mice were initially from Dr Richard M. Myers (Pennacchio et al., 1998), which were on a 129X1/SvJ (129X1) strain background. Sequential breeding steps were performed for crossing CBKO with TgCRND8 to generate four main types of experimental animals: wild-type, CBKO, TgCRND8 and CBKO/TgCRND8 (i.e. the CBKO−/−/APP+/−); all on a 129X1 × 129S6 strain background, as described previously (Yang et al., 2011). All mice were genotyped by PCR. In experiments in which only TgCRND8 and wild-type were studied, the mice were used at indicated different ages ranging from 6- to 22 months old, whereas in experiments involving four types of mice in the CBKO × TgCRND8 cross colony (i.e. wild-type, CBKO, TgCRND8 and CBKO/TgCRND8), the mice were studied at 6 months old unless otherwise stated. All efforts have been made to minimize animal suffering and the numbers of animals used.
To obtain tissues for experiments, animals were anaesthetized with a mixture of ketamine (100 mg/kg body weight) and xylazine (10 mg/kg body weight). Mice for light microscopic analyses were usually fixed by cardiac perfusion using 4% paraformaldehyde (PFA) in 0.1 M sodium cacodylate buffer (pH 7.4, EMS). After perfusion fixation, the brains were immersion-fixed in the same fixative overnight at 4°C. For transmission electron microscopic study, 4% PFA was supplemented with 2% glutaraldehyde (EMS). For biochemical analyses, including western blotting and thin-layer chromatography, the brains were quickly frozen on dry ice and stored at −70°C. In some cases when both morphological and biochemical analyses were to be performed on the same brain, the brain was removed after brief perfusion with saline. One hemisphere was frozen at −70°C and the other half was immersion-fixed in 4% PFA for 3 days at 4°C.
Subcellular fractionation for isolation of autophagic-lysosomal compartments from brain tissue
For each mouse genotype, cerebral cortices from three or more fresh brains were pooled. Using a modified protocol (Marzella et al., 1982; Yu et al., 2005), the samples were homogenized and subjected to differential centrifugation to pellet first the nuclear fraction and any unbroken cells and then a fraction enriched in autophagic vacuoles, lysosomes and mitochondria (Cuervo et al., 1995; Singh et al., 2009). The different organelles in this fraction were isolated by floatation in a discontinuous gradient of Nycodenz® (50, 26, 24, 20 and 10%). A fraction containing mainly autophagosomes was recovered in the 10–20% interface, a fraction enriched in autolysosomes was isolated at the 20–24% interface, and a lysosome-enriched fraction was recovered in the 24–26% interface. These fractions were washed by centrifugation in 0.25 M sucrose and the pellets were treated differently depending on the type of analyses: frozen for later immunoblotting, resuspended in PBS and frozen for later lipid extraction, or immersed in 4% PFA and 2% glutaraldehyde in 0.1 M sodium cacodylate buffer supplemented with 0.25 M sucrose for electron microscopy. The cytosolic fraction was prepared by centrifugation at 100 000g for 1 h of the supernatant fraction resulting from the initial differential centrifugation steps. The cytosolic fraction was recovered in the supernatant whereas the pellet was enriched in endoplasmic reticulum vesicles (microsomes).
Antibodies for immunocytochemistry and immunofluorescence
NeuroTrace® fluorescent Nissl stain was purchased from Life Technologies (N-21480). The following primary antibodies were used in this study: homemade GM2 ganglioside monoclonal antibody (mouse IgM, cell culture supernatant, produced from the 10–11 hybridoma line provided by Progenics Pharmaceuticals; diluted 1:200 for immunocytochemistry, immunofluorescence, or thin layer chromatography-overlay immunostaining) (Micsenyi et al., 2009; Saito et al., 2012); GM3 ganglioside monoclonal antibody (mouse IgM, Clone M2590, Cosmo Bio Co., #NBT-M101, diluted 1:200 for immunofluorescence); ceramide monoclonal antibody (mouse IgM, Clone MID 15B4, Enzo Life Sci./Axxora, #ALX-804-196-T050, diluted 1:10 for immunofluorescence); LC3 polyclonal antibody (Novus biological, #NB100-2220 diluted 1:250 for immunofluorescence); cathepsin D (CTSD or CatD) rabbit polyclonal antibody directed against mouse CTSD (RU2; generated in-house diluted 1:5000–10 000 for immunofluorescence); GFAP rabbit polyclonal antibody directed against human GFAP (Sigma, G9269; diluted 1:400 for immunofluorescence); IBA1 rabbit polyclonal antibody directed against the C-terminus of the microglia/macrophage-specific protein IBA1 (Waco Chemicals, 019-19741; diluted 1:500 for immunofluorescence); neuron-specific enolase (NSE) mouse monoclonal antibody directed against human ENO2 (IgG1, kappa, Dako, M0873; diluted 1:100 for immunofluorescence). The following secondary antibodies and reagents for immunoperoxidase labelling were purchased from Vector Laboratories: biotinylated goat anti-mouse IgM (BA-2020), M.O.M.™ Immunodetection Kit (BMK-2202), Vectastain ABC kit (PK-4000), and DAB Peroxidase Substrate Kit (SK-4100). The following secondary antibodies for immunofluorescence were purchased from Molecular Probes/Life Technologies: Alexa Fluor® 488-conjugated goat anti-mouse IgM (µ chain, A21042), Alexa Fluor® 568-conjugated goat anti-mouse IgM (µ chain, A21043), Alexa Fluor® 488-conjugated goat anti-mouse IgG3 (A21151), Alexa Fluor® 568-conjugated goat anti-mouse IgG (A11031), Alexa Fluor® 488-conjugated goat anti-rabbit IgG (A11034), and Alexa Fluor® 568-conjugated goat anti-rabbit IgG (A11036).
Lipid extraction, thin layer chromatography and thin layer chromatography-overlay immunodetection
The total brain homogenates, autophagosome, autolysosome and lysosome fractions were lyophilized. The resulting powders were extracted twice with a mixture of methanol and methyl tert-butyl ether (MTBE) (1:1, V/V) containing 0.2 % butylated hydroxytoluene (BHT). The extracts were evaporated to dryness and suspended in 200 µl of water. The water suspensions were dialyzed extensively against distilled water. The dialyzed water suspensions were evaporated to dryness and partitioned to water phase and organic phase by the method described by Matyash et al. (2008). The water phase fractions contained almost all gangliosides and some polar materials. To reduce the polar materials that disturb high performance thin layer chromatography (HPTLC) of gangliosides, these fractions were evaporated to dryness and treated with 0.4 M potassium hydroxide at 23°C for 60 min. After the treatment, reaction mixtures were neutralized by concentrated HCl and dialyzed extensively against distilled water. The dialyzed ganglioside fractions were evaporated to dryness and analysed on HPTLC as described by Yu and Ledeen (1972). The plates were stained with an orcinol ferric chloride spray reagent (Bial’s reagent) (Manzi and Esko, 2001). The stained HPTLC plates including a GM2 standard purified from Tay-Sachs brains were scanned with the Odyssey infrared imaging system (LI-COR Biosciences). GM3, GM2, GM1, and GD1a gangliosides on the scanned images were quantified by Multi Gauge ver.2.0 (Fujifilm USA Medical Systems) using various amounts (0.31, 0.63, 1.25, 2.5. 5.0 μg) of standard GM2. It was assumed that the density of the band on HPTLC stained with an orcinol reagent was proportional to the sialic acid concentration and the molecular weights of GM3, GM1 and GD1a were 85%, 112%, and 134% of GM2, respectively. Alternatively, GM2 was detected by a thin layer chromatography overlay method (Yu and Ariga, 2000) using a mouse monoclonal anti-GM2 antibody. Anti-GM2 antibody bound to HPTLC sheets was visualized using Avidin-Biotin Complex (ABC) reagents (Vectastain ABC Kit, Vector Labs) and a peroxidase substrate (DAB) kit (Vector Labs) following the manufacturer’s instructions.
The organic phase separated by the method described above contains all lipids except for some polar lipids. To analyse simple and neutral lipids, the organic phases were evaporated to dryness and dissolved in a mixture of MTBE, methanol and water (30/60/8, v/v/v). These lipid solutions were passed through small (200 µl) DEAE Sephadex® columns to separate them from acidic lipids as described by Macala et al. (1983). The effluents were evaporated to dryness and applied on HPTLC plates. The HPTLC plates were developed first in the upper phase of a mixture of hexane, ethylacetate, water and acetic acid (30:40:50:20, v/v/v/v) until 4 cm from the origin, and then in the mixture of hexane, MTBE, and acetic acid (65:35:2, V/V/V) until 6.5 cm, and finally in the mixture of hexane, MTBE, and acetic acid (98:2:1) until 8.5 cm. The plates were air dried after every development. The HPTLC plates were stained with fluorescent dye primuline and analysed as described (van Echten-Deckert, 2000).
Ultrastructural analyses
For electron microscopy, vibratome sections of brain, or the pellets of the isolated brain autophagosome, autolysosome and lysosome fractions, were post-fixed in 1% osmium tetroxide. After alcohol dehydration, sections were embedded in Epon (EMS). One-micrometre thick sections were stained with toluidine blue for light microscopic examination and ultrathin sections prepared and stained with uranyl acetate and lead citrate. Material was viewed with a Philips CM 10 electron microscope equipped with a digital camera (Hamamatsu, model C4742-95) aided by AMT Image Capture Engine software (version 5.42.443a). For quantification of small autolysosome/lysosome profiles in TgCRND8 versus CBKO/TgCRND8, 30 electron microscopy images (×19 000) containing autophagic-lysosomal compartments were taken from the neuronal cytoplasm of the hippocampal CA1 sector of each mouse (n = 4 per genotype). The number of small autolysosomes/lysosomes, defined as <1 µm in diameter, round or oval in shape, single membrane-bound and containing none or only a small amount of undigested components; their total covering area did not exceed a half of the total area of a given profile, was counted, averaged as the number of profile per image, and expressed as the relative frequency.
Immunolabelling of brain sections
Immunocytochemistry and immunofluorescence were performed according to the protocols previously described (Yang et al., 2009) with two modifications: microwave antigen retrieval was performed and the time of detergent (0.2% Tween-20) treatment was limited to only 5 min. For quantitative analysis of GM2-positive puncta, four sagittal brain sections from each mouse (n = 5 TgCRND8 and n = 6 CBKO/TgCRND8), obtained with even space (400 µm) from the region between lateral 0.48 mm and 2.16 mm (Paxinos and Franklin, 2001), were immunostained with the anti-GM2 antibody and visualized with 3,3'-diaminobenzidine (DAB). Three ×40 digital images were taken from the CA1 area of each section and the GM-positive puncta were quantified with AutoMeasure software (Zeiss).
Results
Lipids are detected in autophagic vacuoles and accumulate in TgCRND8 mouse brain
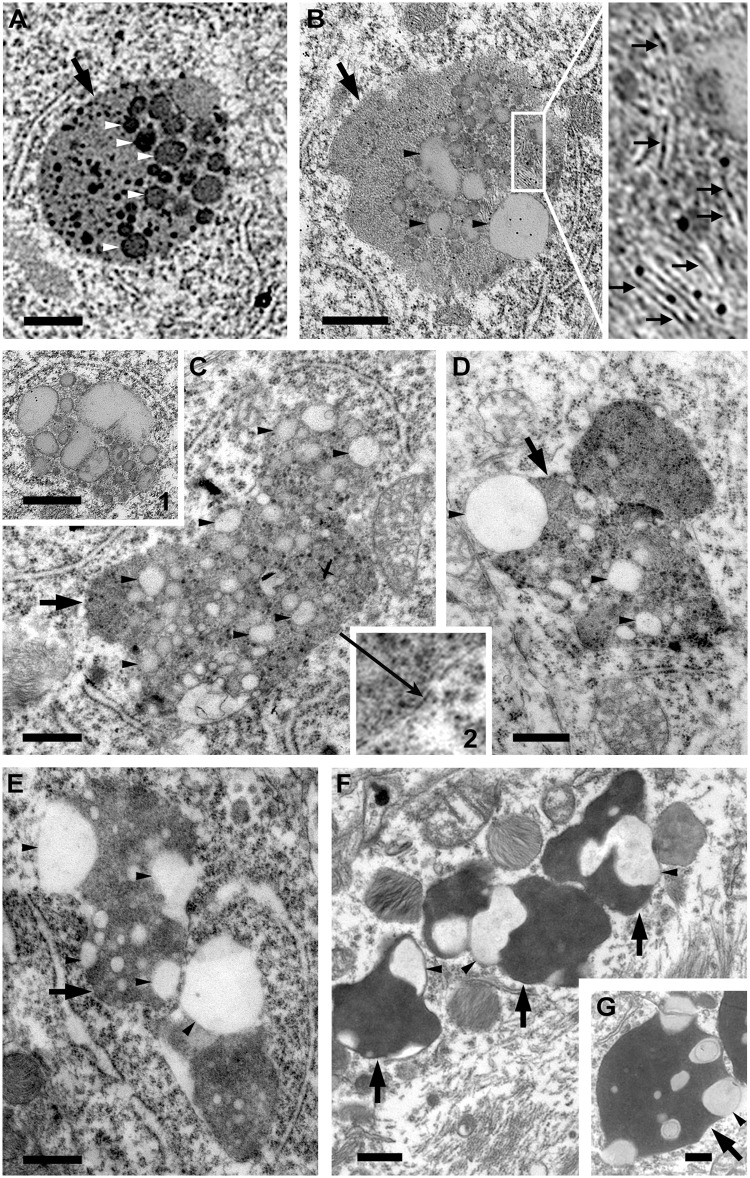
A characteristic feature of lysosomal pathology in the TgCRND8 brain is the presence of giant autolysosomes (usually >1.5 µm and up to 5 or 6 µm depending on ages, in diameter) containing varied compositions of undigested material (Yang et al., 2011). More extensive electron microscopy analyses of these structures (Fig. 1) reveal that a key feature of giant autolysosomes is abundant intraluminal membrane-bound vesicles, stacked membranes, and clear round structures corresponding to lipid droplets, similar to those previously described to appear transiently within lysosomes during lipophagy (Singh et al., 2009; Eid et al., 2013). Lipid droplets were also seen in the cytosol (Fig. 1C, inset 1) and their existence in both the cytosol and the single-membrane bound autolysosomes (Fig. 1C, inset 2) implied the competence of autophagic sequestration of lipid droplets and subsequent delivery to lysosomes in TgCRND8. Some of the lipid-containing giant autolysosomes (Fig. 1C–E) seemed to be derived from fusion of two or more autolysosomes, resulting in irregular shapes representing a possible transition to lipofuscin granules (also termed pigmented autophagic vacuoles) (Sulzer et al., 2008) that are commonly seen in Alzheimer’s disease brain (Terry et al., 1964) (Fig. 1F and G). A significant increase in such lipid-containing membranous structures and lipid droplets in neurons of TgCRND8 mice suggests that their clearance by the lysosomal system is impaired.
Figure 1.
Accumulation of membrane structures and lipid inclusions within autolysosomes in the brain of TgCRND8 suggesting impaired lipid degradation. (A–E) Electron microphotographs taken from the CA1 area of 6-month-old TgCRND8 showing a variety of giant autolysosomes (large arrows), which contain numerous stacked membranes (small arrows), round/oval vesicular structures (white arrowheads) and light-density lipid inclusions compatible with lipid droplets (black arrowheads). Some of the giant autolysosomes seem to be formed from fusion of two or more autolysosomes (C–E). The inset in B is the higher magnification view of the boxed area. Inset 1 in C shows a loosely packed cluster of lipid droplets found in the cytosol without limiting membrane, and Inset 2 in C demonstrates the limiting membrane (long arrow) of the giant autolysosome. (F and G) Images taken from biopsy samples from an Alzheimer’s disease brain, showing lipid inclusions (black arrowheads) in enlarged autolysosomes (lipofuscin granules) (large arrows). Scale bars = 500 nm.
Some of the membranous structures observed inside lysosomes could have their origin from the plasma membrane as part of the process of formation of multivesicular bodies in late endosomes and further delivery of these vesicles to lysosomes upon endosome/lysosome fusion (Schulze et al., 2009). It is also possible, however, that a significant fraction of lipid components of intracellular organelles and/or plasma membrane could be delivered to lysosomes via the autophagic pathway, especially under conditions of cellular stress. To investigate this hypothesis, we isolated autophagic-lysosomal compartments (i.e. autophagosome, autolysosome and lysosome) from brains of TgCRND8 and wild-type. These fractions exhibit enriched signals for the autophagosome/autolysosome protein marker MAP1LC3A (formerly known as LC3) and the autolysosome/lysosome protein marker CTSD (CatD) (Fig. 2). Their ultrastructure was examined by electron microscopy (Supplementary Fig. 1) and their lipid profiles were analysed by thin layer chromatography (Fig. 2A–D). As shown in Fig. 2A, a number of sphingolipids such as ceramide, GM2, GM1 and GD1a could be detected in the autophagosome and autolysosome fractions from wild-type brains, and the fact that their levels are considerably higher than those in the lysosome fraction indicates that even under normal conditions, at least a portion of these membrane lipids reach lysosomes by autophagy. Intriguingly, the levels of the majority of probed sphingolipids, including ceramide and ganglioside GM3, GM2, GM1, GD3, GD1a, were higher in the autophagic-lysosomal fractions from TgCRND8 compared to wild-type (Fig. 2A and Supplementary Fig. 2). Notably, these lipid species were highly abundant in the three autophagic-lysosomal fractions (Fig. 2A). By contrast, levels of these lipids in the endoplasmic reticulum or mitochondrial fractions were relatively low and were comparable in TgCRND8 and wild-type.
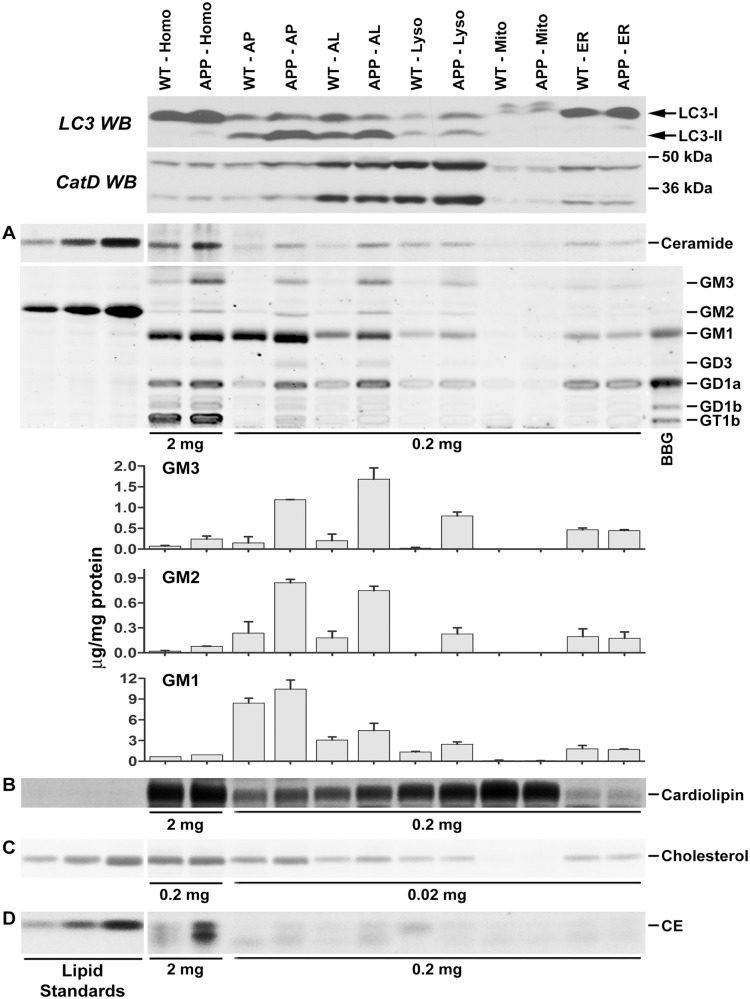
Figure 2.
Increased levels of lipids in autophagic-lysosomal fractions isolated from the brain of TgCRND8 detected by thin layer chromatography. Brain homogenates or subcellular fractions were prepared from 12-month-old TgCRND8 and wild-type mice (WT) and examined for relative enrichment of autophagic-lysosomal protein markers by western blotting (WB) with an anti-MAP1LC3A (LC3) or -CTSD antibody (CatD) (top two panels). Lipids, after extraction, were separated on thin layer chromatography plates for the detection of sphingolipids (A), cardiolipin (B), unesterified cholesterol (C) and cholesteryl esters (D). To achieve equal loading, we used equal total protein amounts (indicated at the bottom of thin layer chromatography panels), which were used for lipid extraction, to guide loading of samples. Note that 10-fold higher amounts of samples were used for ganglioside analyses of homogenates compared to those of other fractions. (A, bar graphs): the amounts of GM3, GM2 and GM1 obtained from two separate experiments (three to five pooled mouse brains per genotype per experiment) were calculated as μg/mg protein as described in the ‘Materials and methods’ section using various amounts (0.31, 0.63, 1.25, 2.5. 5.0 μg) of GM2 standard (In A, middle, only 1.25, 2.5. 5.0 μg of standard GM2 gangliosides are shown). Values are the mean ± SEM. (The quantitative data for ceramide, cardiolipin, unesterified cholesterol and cholesteryl esters are presented in Supplementary Fig. 2). Homo = homogenate; AP = autophagosome enriched fraction; AL = autolysosome enriched fraction; Lyso = lysosome; Mito = mitochondria; ER = endoplasmic reticulum-enriched; BBG = bovine brain gangliosides; CE = cholesteryl esters. The arrow indicates an unidentified lipid species. ‘APP’ = TgCRND8.
We also analysed a minor membrane phospholipid, cardiolipin (Scherer and Schmitz, 2011), which is derived from phosphatidylglycerol produced in the endoplasmic reticulum and primarily located in the inner membrane of mitochondria (Kolter and Sandhoff, 2010), which are degraded through autophagy. Because mitophagy is an exclusively autophagic process, cardiolipin may serve as an index for autophagic turnover of organellar membranes. As expected, cardiolipin was enriched in the mitochondrial fraction and its levels there were comparable in TgCRND8 and wild-type mice. Reflecting the activity of mitophagy in brain, we detected cardiolipin in autophagosomal/lysosomal fractions. The levels of cardiolipin in these fractions were higher in TgCRND8 than in wild-type mice (Fig. 2B and Supplementary Fig. 2), showing a similar trend to the alterations of the sphingolipids and, together, implying an impaired autophagic flux probably due to a defective degradative function in the late stage of the pathway.
Cholesterol, a membrane lipid especially enriched in lipid rafts of the plasma membrane, is believed to be delivered to endo/lysosomes through the endocytic pathway (Schulze et al., 2009); however, it was also detected in autophagosome and autolysosome fractions and was present at elevated levels in TgCRND8 compared to wild-type mice (Fig. 2C and Supplementary Fig. 2). Moreover, cholesteryl esters, which exist in lipid droplets as the storage form of cholesterol, have been previously shown to be elevated in brain samples from Alzheimer’s disease and Alzheimer’s disease mouse models (Chan et al., 2012). We also observed this lipid species in the autophagosome and autolysosome fractions and at increased levels in these fractions from TgCRND8 brain (Fig. 2D and Supplementary Fig. 2). The increased presence of these lipids in TgCRND8 mice is consistent with the electron microscopy observation of lipid droplets within the giant autolysosomes seen in these mutant mice (Fig. 1).
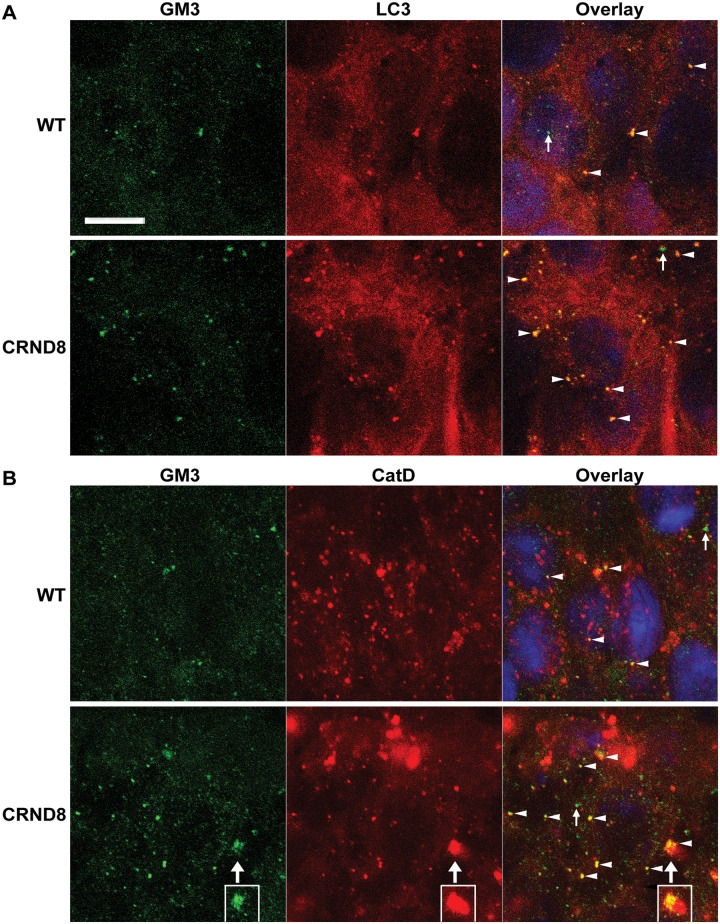
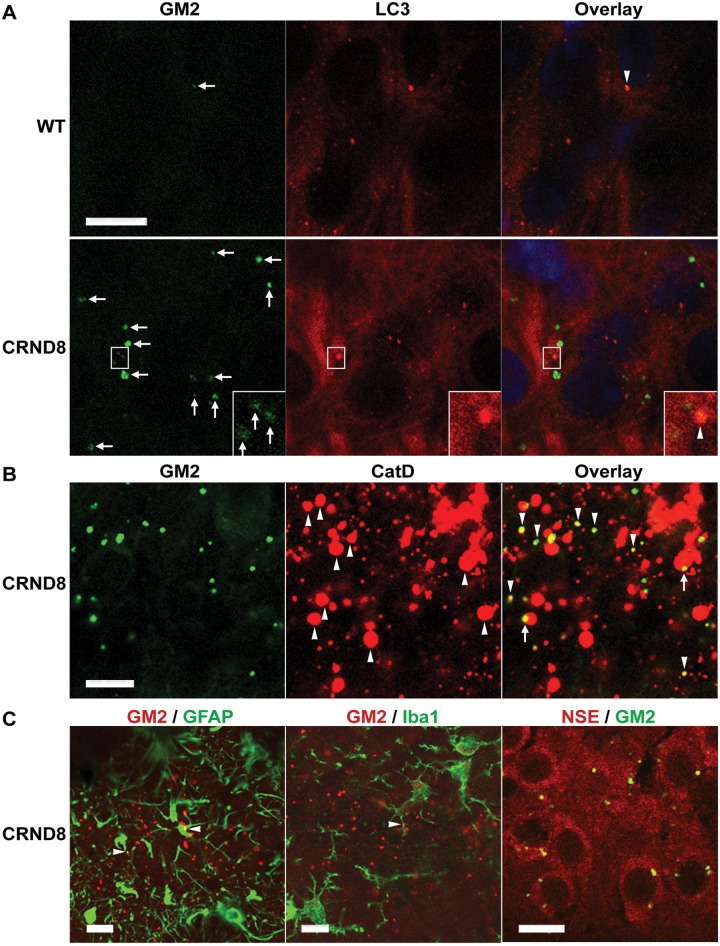
Consistent with the thin layer chromatography findings, double immunofluorescence studies with antibodies against GM3 and an autophagosome marker, MAP1LC3A (LC3), revealed existence of GM3 signals within MAP1LC3A-positive puncta in the hippocampal pyramidal cell layer of both wild-type and TgCRND8 (Fig. 3A). Double labelling of GM3 with an autolysosome/lysosome marker, CTSD, also detected co-localization of these two markers (Fig. 3B). Together, the results suggest a route of GM3 intracellular traveling from autophagosomes to autolysosomes. It is visible that there are more GM3-positve structures in TgCRND8 compared to wild-type (Fig. 3A and B) and that the giant autolysosomes in TgCRND8 can contain a large amount of GM3 (Fig. 3B), suggesting GM3 accumulation in the autolysosomes. In addition, immunofluorescence studies for GM2 detected little GM2 signal in the CA1 sector of wild-type mice, while many GM2 puncta were observed in the same area of TgCRND8, which exhibited a great variation in immunoreactive intensity (Fig. 4A). Interestingly, GM2 and MAP1LC3A co-localization was hardly detected and only some weakly-stained GM2 structures were found to be localized within MAP1LC3A puncta (Fig. 4A). Conversely, the strongly-stained GM2 puncta exhibited a high degree of co-localization with CTSD in TgCRND8 (Fig. 4B), even though such co-labelling was rarely found in wild-type sections (not shown) due to the nearly complete absence of GM2 signal in wild-type (Fig. 4A, also see Fig. 5D and Supplementary Fig. 4). These GM2 staining patterns therefore suggest that the levels of GM2 might be very low when GM2-containing structures are sequestered into autophagosomes, resulting in GM2-MAP1LC3A co-localization hardly detectable, but the GM2 levels are elevated in autolysosomes in TgCRND8 likely due to defective degradation, leading to easy detection of GM2-CatD co-localization.
Figure 3.
Co-localization of GM3 with autophagic-lysosomal markers in the hippocampus of wild-type and TgCRND8 brain. Vibratome brain sections from 12-month-old wild-type (WT) and TgCRND8 (n = 3/genotype) were double-stained for GM3 and an autophagosome marker, MAP1LC3A (LC3) (A) or an autolysosome/lysosome marker, CTSD (CatD) (B), followed by staining with Sudan Black B to block autofluorescence. Confocal digital single-plane (1 µm) images from z-stacks taken from the hippocampal pyramidal cell layer are shown. Examples showing co-localization of GM3 signal with MAP1LC3A or CTSD are indicated by arrowheads in the merged images. Small arrows depict profiles which are predominantly in green colour indicating that the green signals for GM3 are not due to carry-over from the red channel. The boxed areas in B are from a consecutive single-plane showing the same giant autolysosome (large arrows) with massive GM3 signal accumulation. Scale bars = 10 µm. ‘CRND8’ = TgCRND8.
Figure 4.
Co-localization of GM2 with autophagic-lysosomal or neuronal markers in the hippocampus of wild-type and TgCRND8 brain. (A–C) Vibratome brain sections from 12-month-old wild-type (WT) and TgCRND8 (n = 3/genotype) were double-stained for GM2 and other markers as indicated, followed by staining with Sudan Black B to block autofluorescence, and images taken from the hippocampal pyramidal cell layer are shown. (A) Double labelling of GM2 with the autophagosome marker MAP1LC3A (LC3). Examples showing co-localization of GM2 signal with MAP1LC3A are indicated by arrowheads in the merged images. Small arrows depict GM2-positive profiles with variable staining intensity. The insets are the enlarged views of the boxed areas showing that the weakly stained GM2 profile(s) can be observed within a strong MAP1LC3A-positive profile. (B) Double labelling of GM2 and CTSD (CatD). Upward arrowheads indicate numerous abnormal giant autolysosomes in TgCRND8 [Note: it seems that there is also a small CTSD-positive amyloid plaque(s) within the image, see top right corner]. The merged image depicts that the majority of GM2-signal is associated with CTSD-positive lysosomes, either having a high degree of co-localization with the CTSD signal (downward arrowheads) or occupying a portion of those CTSD-stained giant autolysosomes (arrows). (C) Double labelling of GM2 with a marker for either astrocytes (GFAP), microglia (IBA1) or neurons (neuron-specific enolase, NSE). Little GM2 signal is co-localized with GFAP or IBA1 staining, except the minimal areas indicated by arrowheads. GM2/NSE double labelling reveals overlapping of GM2 signal on NSE-stained neuronal cytoplasm. Scale bars = 10 µm. ‘CRND8’ = TgCRND8.
Figure 5.
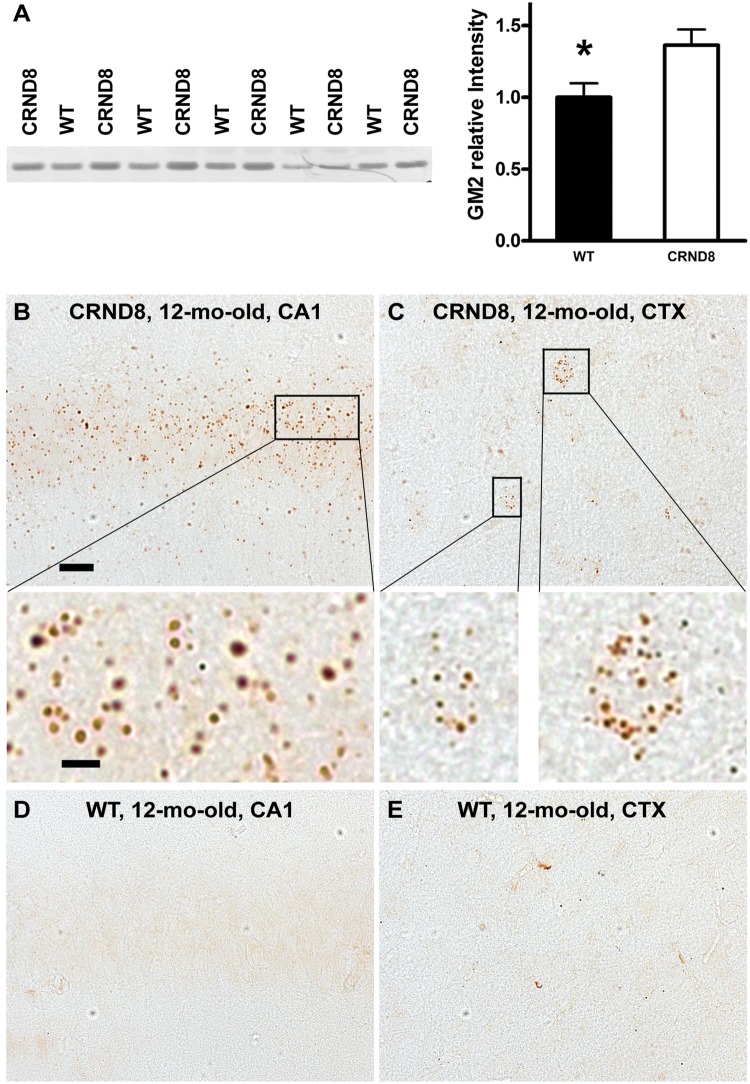
Accumulation of GM2 ganglioside in TgCRND8 brain revealed by immunodetection. (A) Thin layer chromatography immunostaining reveals increased GM2 levels in the brain of TgCRND8. Total lipid extracts were prepared from brain homogenates of 12-month-old TgCRND8 and wild-type (WT) mice (n = 5–6 mice per genotype) and loaded onto C18 Sep-Pak cartridges. The eluted fractions containing gangliosides were separated on thin layer chromatography plates and immunostained with a thin layer chromatography overlay method using an anti-GM2 antibody to reveal GM2 signal (left) which was scanned and the band relative densities were analysed (right). Values are the mean ± SEM for each group. Significant differences were analysed by two-tailed Student’s t-test. *P < 0.05. (B–E) Immunohistochemistry reveals abundance of GM2-positive granules in the brains of TgCRND8. Vibratome sections from TgCRND8 and age-matched wild-type mice (n = 4–6 mice per age per genotype) were immunostained with an anti-GM2 antibody. (B and C) Representative microphotograph images from the hippocampal CA1 sector (B) and the neocortex (CTX) (C) of a 12-month-old TgCRND8 mouse depict abundant GM2-positive granules in the CA1 pyramidal cell layer and in some cortical neurons. Magnified images of the boxed areas are shown below the low-power images. (D and E) Images of a brain section from a wild-type littermate exhibit little or no GM2-immunostaining in the CA1 sector (D) and the CTX (E). Scale bar in B = 20 µm for (B–E); bar for the enlarged images of (B and C) = 5 µm. ‘CRND8’ = TgCRND8.
Additional double immunofluorescence labelling with anti-GM2 antibody, coupled with markers for either neurons, astrocytes or microglia (Fig. 4C), revealed little co-localization between the GM2 immunoreactivity and the signals for GFAP (as a marker of astrocytes) or IBA1 (for microglia). By contrast, many GM2 puncta exhibited a close spatial relationship around the neuronal nuclei detected by a neuronal marker, NeuroTrace®, suggesting that the GM2 puncta are most abundant within neurons (not shown). Double labelling of GM2 with another neuronal marker, neuron-specific enolase (NSE), clearly demonstrated overlap of the GM2 and the NSE signals in neuron, indicating that the primary location for the GM2 puncta is neurons as expected, given the known enrichment of gangliosides in neurons.
Previous studies have consistently reported elevated levels of GM2 in brains from Alzheimer’s disease and several Alzheimer’s disease mouse models (see ‘Discussion’ section). These findings and the observation of remarkable difference in the immunofluorescent staining patterns of GM2 between wild-type and TgCRND8 in the current study (Fig. 4A) suggests pathological accumulation of GM2 in autolysosomes, which prompted us to carry out additional analyses on GM2. First, additional thin layer chromatography analysis using a thin layer chromatography-overlay method with the anti-GM2 antibody confirmed the elevation of GM2 in brain homogenates of TgCRND8 compared to wild-type (Fig. 5A). Next, we investigated the distribution of GM2 by immunoperoxidase labelling in mice at different ages ranging from 6- to 22-months-old. In TgCRND8 mice, we observed large numbers of GM2-positive punctate profiles in the pyramidal cell layer of the hippocampus (CA1 to CA3 sectors) and lower numbers in neocortex (CTX) (Fig. 5B and C). In wild-type mice, by contrast, GM2 puncta were rarely detected in these brain regions (Fig. 5D and E), except for a low level of immunoreactivity inconsistently seen in the CA3 sector (Supplementary Fig. 3). These differential patterns were similar in mice analysed at 6-, 12-, or 22-months of age (Fig. 5B–E and Supplementary Fig. 4).
Autophagic lipid turnover is rescued by enhancing lysosomal proteolysis
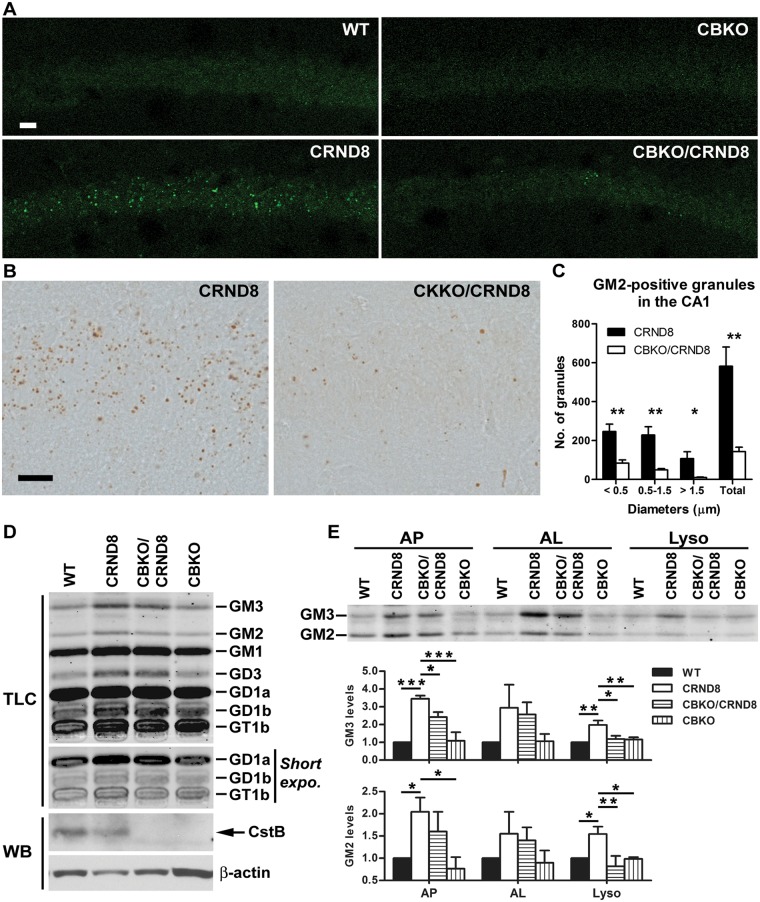
We previously demonstrated that lysosomal turnover of protein substrates is impeded in TgCRND8 brain and this impairment is ameliorated by improving lysosomal degradative capacity via genetic ablation of an inhibitor of lysosomal proteases, CSTB (Yang et al., 2011). We speculated that the accumulation of non-degraded material of any nature in the lysosome may disrupt the lysosomal intraluminal environment and interfere with the activity of all lysosomal hydrolases, including lipases and ganglioside degrading enzymes. If this is the case, we predicted that improved proteolytic function due to CSTB knockout (CBKO) might improve degradation not only of proteins but also lipids, particularly given that some of these exist as lipoproteins. As predicted, GM2 puncta numbers in the hippocampal CA1–3 sectors of CBKO/TgCRND8 mice were strikingly reduced compared to those in TgCRND8 mice and resembled the pattern seen in wild-type mice (Fig. 6A–C), indicating that CSTB deletion prevents GM2 accumulation within TgCRND8 lysosomes. Similarly, thin layer chromatography for gangliosides in isolated brain lysosome fractions also revealed reduced GM2 levels in 6-month-old CBKO/TgCRND8 mice compared to TgCRND8. GM3, like GM2, which are both degradative products of more complex gangliosides and most prone to accumulate under pathological conditions (e.g. lysosomal storage disorders), seemed to be decreased in CBKO/TgCRND8. Several additional gangliosides, which are major components of neurons and therefore their alterations, if subtle, might not be easily detected in total pool analysis, were unchanged in CBKO/TgCRND8 compared to TgCRND8 (Fig. 6D). Quantitative analyses of results from multiple experiments revealed significant differences in the levels of GM3 or GM2 in the lysosome fraction between TgCRND8 and wild-type or TgCRND8 and CBKO/TgCRND8. Similar and statistically significant changes were also observed in the autophagosome fraction, whereas a similar trend, while not significant, was seen in the autolysosome fraction (Fig. 6E). These results together indicate a restoration in the degradative function of lipids (e.g. GM3, GM2) in the late stage of the pathway in CBKO/TgCRND8, which leads to an overall improvement in autophagic flux.
Figure 6.
Effect of cystatin B deletion in TgCRND8 on the levels of gangliosides. (A–C) Reduction in the numbers of GM2-positive granules in the CA1 area of CBKO/TgCRND8. Vibratome brain sections from a group of 6-month-old wild-type (WT), CBKO, TgCRND8 and CBKO/TgCRND8 mice (strains: 129S6 × 129X1 for the four genotypes) were processed with the anti-GM2 antibody and visualized by either immunofluorescence (A) or DAB (B). Scale bars = 20 µm. (C) Sections visualized by DAB were quantified for the numbers of GM2-positive granules (grouped by diameters) in the CA1 area of either TgCRND8 or CBKO/TgCRND8. Values are the mean ± SEM for each group (n = 5 TgCRND8 or 6 CBKO/TgCRND8). Significant differences were analysed by two-tailed Student’s t-test. *P < 0.05, **P < 0.01. (D) Lipid extracts were prepared from the brain lysosome fraction from a group of 6-month-old wild-type, CBKO, TgCRND8 and CBKO/TgCRND8 mice and separated on a thin layer chromatography plate for the analysis of gangliosides. To achieve equal loading, we used equal total protein amounts [indicated at the bottom of thin layer chromatography (TLC) panels], which were used for lipid extraction, to guide loading of samples. Western blotting (WB) of CSTB in the lysosome fraction using a homemade anti-mouse CstB polyclonal antibody (Yang et al., 2011) depicts the presence of CSTB bands in samples from wild-type and TgCRND8 but the absence in samples from CBKO and CBKO/TgCRND8, confirming the genotypes of the mice used. Actin was used as a loading control. (E) The thin layer chromatography panel represents GM3 and GM2 profiles from lipid extracts of autophagosome, autolysosome and lysosome fractions, while the bar graphs depict the quantitative results from three separate experiments (n = 3; 4–6 pooled mouse brains per genotype per experiment). Values are mean ± SEM showing relative levels of GM3 or GM2 (normalized against the level of wild-type) in each fraction. Mean differences between genotypes were analysed by one way ANOVA followed by post hoc Bonferroni’s multiple comparison tests (‘selected pairs of columns’). *P < 0.05, **P < 0.01, ***P < 0.001. ‘CRND8’ = TgCRND8.
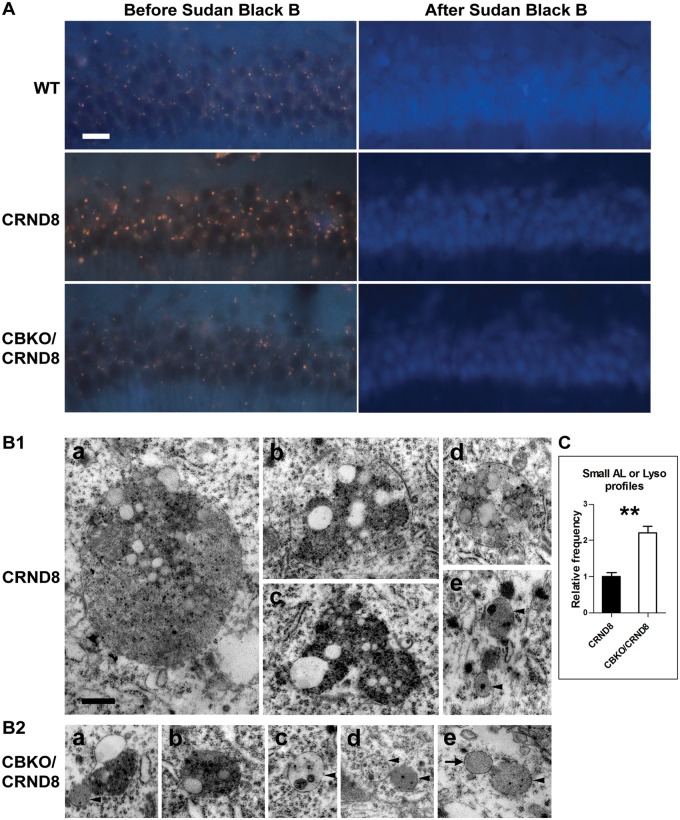
Autofluorescence in brain sections can be an indicator of lipofuscin, which contains a large amount of lipids (up to 50% by weight) (Yin, 1996; Warburton et al., 2005). TgCRND8 mice exhibited stronger and larger yellow-brown autofluorescent granules in the hippocampal pyramidal cell layer compared with age-matched wild-type mice (Fig. 7A), consistent with the electron microscopy observation of enlarged pigmented autolysosomes in TgCRND8 (Fig. 1). By contrast, autofluorescent granules in CBKO/TgCRND8 were smaller and weaker than TgCRND8, and comparable to the pattern seen in wild-type mice (Fig. 7A). The autofluorescent signals from all sections could be extinguished by Sudan Black B incubation (Fig. 7A), confirming that these granules are enriched in lipids (Romijn et al., 1999).
Figure 7.
Effect of cystatin B deletion in TgCRND8 on lipid accumulation. (A) Untreated vibratome brain sections from 6-month-old mice were examined under ultraviolet light and autofluorescence, presented as yellow-brown lipofuscin granules, in the CA1 area was captured (left). The sections were then incubated with Sudan Black B to block autofluorescence, and images from the same area were collected (right). Scale bars = 20 µm. (B) Comparison of electron microscopy images collected from the CA1 sector of TgCRND8 [B(1)] or CBKO/TgCRND8 [B(2)]. The bar graph shows the quantification of relative abundance of small autolysosomes/lysosomes (AL/Lyso), defined as being <1 µm in diameter, round or oval in shape, single membrane-bound and containing only a small amount (arrowheads) or none (arrow) of undigested components [i.e. those shown in B(1e) and B(2c–e)], in TgCRND8 (n = 4) or CBKO/TgCRND8 (n = 4). Significant differences were analysed by two-tailed Student’s t-test. **P < 0.01. Scale bar in B(1a) = 500 nm for all panels. ‘CRND8’ = TgCRND8.
Finally, electron microscopy examination of hippocampal CA1 samples from TgCRND8 and CBKO/TgCRND8 revealed more differences between these two genotypes. First, in contrast to TgCRND8 [Fig. 7B(1a–c)], lipid-containing giant autolysosomes (>1.5 µm in diameter) were rarely detected in CBKO/TgCRND8, consistent with evidence of rare giant autolysosomes detected by CTSD immunostaining (Yang et al., 2011; Fig. 6). Second, lipofuscin granules, when detected in CBKO/TgCRND8 [Fig. 7B(2a)], were much smaller than those in TgCRND8 [Fig. 7B(1c); also see Fig. 1]. Third, giant autolysosomes were replaced by a higher frequency of small autolysosomes/lysosomes (<1 µm in diameter) in CBKO/TgCRND8, which were devoid of lipid droplets and contained a small amount [Fig. 7B(2c–e)] or none [Fig. 7B(2e)] of undigested intraluminal components, suggesting improved lysosomal degradative capability. These small structures were fewer in TgCRND8 [Fig. 7B(1e)], consistent with CTSD immunocytochemical analyses (Yang et al., 2011; Fig. 6). Morphometric analyses of electron microscopy images indicated that autolysosomes/lysosomes of this type were 2.2-fold more in CBKO/TgCRND8 than in TgCRND8 mice (Fig. 7C), indicating a reduced total volume of lipid-containing components (intraluminal membranous structures and lipid droplets), consistent with the lowered autofluorescent signal (Fig. 7A).
Discussion
Autophagic pathway as a complementary route of lipid delivery to lysosomes
Our observations of lipid droplets in autolysosomes and elevated levels of cholesteryl esters in the isolated autophagosome and autolysosome fractions of wild-type mice indicate that lipophagy, an autophagic route of lipid droplet turnover well-studied in hepatocytes, macrophages and enterocytes (Singh et al., 2009; Ouimet et al., 2011; Khaldoun et al., 2014), is also constitutively active in neurons. This finding is consistent with previous observations that lipid droplets accumulate in the brains of patients with Huntington’s disease where the lipids accumulate in the neuronal cytosol due to problems in cargo recognition in the autophagic process (Martinez-Vicente et al., 2010). That lipid droplets are found in autolysosomes in addition to their presence in the cytosol implies that lipid droplet sequestration into autophagosomes and subsequent autophagosome fusion with lysosomes are not grossly impaired in TgCRND8. By contrast, the abnormal accumulation of lipid droplets within enlarged autolysosomes of TgCRND8 neural cells suggests that lysosomal turnover of these lipids is impaired.
Our data for the first time indicate that, beyond its role in lipophagy, autophagy also delivers membrane lipids to the lysosome for turnover under normal conditions, a function that has so far been demonstrated to be mediated through the endocytic pathway even though possible contributions by autophagy have been speculated (Schulze and Sandhoff, 2014). This conclusion is supported by observations that a number of membrane lipids are detectable in autophagic vacuole fractions isolated from brains of wild-type mice and in MAP1LC3A (LC3)-positive autophagosomes in wild-type brain sections. This sequestration is impeded in the brain of TgCRND8 as indicated by an increased abundance of these lipids in the autophagosomal/lysosomal compartments, suggesting defective autophagic flux as a result of decreased lysosomal degradative capacity.
Several possible mechanisms could be involved in the delivery of membrane lipids through the autophagic pathway to the lysosome. First, early or late endosomes/multivesicular bodies can fuse with autophagosomes to form amphisomes followed by a further fusion with lysosomes to form autolysosomes (Berg et al., 1998; Filimonenko et al., 2007; Razi et al., 2009). Amphisome formation is likely an entry point to the autophagic pathway for membrane lipids residing in the plasma membrane such as gangliosides. In addition, both plasma membrane and/or endosomes (e.g. recycling endosomes) can serve as membrane sources for the formation of phagophores and thus autophagosomes (Ravikumar et al., 2010; Puri et al., 2013), thus providing another entry point into the autophagic pathway. Moreover, the endoplasmic reticulum, which exhibits almost the same lipid species as those in plasma membrane and one-quarter to more than one-third abundance in total lipids, total sterol lipids or sphingolipids relevant to those in the plasma membrane (Andreyev et al., 2010), can contribute its membrane lipids to the autophagic pathway when serving as a membrane provider for phagophore/autophagosome formation (Axe et al., 2008; Ge et al., 2013; Hamasaki et al., 2013) and/or as substrates of autophagy during reticulophagy, the selective autophagic sequestration of endoplasmic reticulum membranes (Cebollero et al., 2012). Furthermore, the presence of the mitochondrial lipid, cardiolipin, in our autophagic vacuole fractions, suggests that mitophagy actively delivers additional membrane lipids into the autophagic pathway. Finally, compromised lysosomes can be degraded by the autophagic pathway in a process termed lysophagy or lysosomophagy (Klionsky et al., 2007; Hung et al., 2013; Maejima et al., 2013; Shen and Mizushima, 2014). Not only lipids of the lysosomal membrane, such as bis(monoacylglycero)phosphate (BMP) found by lipidomic analysis (Rodriguez-Navarro et al., 2012), but those delivered to lysosomes but not yet degraded will undergo a renewed round of degradation by becoming sequestered into autophagosomes. This form of organellar turnover may be particularly relevant to pathological conditions such as that in TgCRND8 mice where severely impaired lysosomal degradative capacity and the consequent damage incurred by oxidation of substrates may promote a futile cycle of lysosome sequestration and incomplete digestion leading to the formation of grossly enlarged autolysosomes.
Defective autophagic degradation of lipids in TgCRND8 brain
In the Alzheimer’s disease brain and in Alzheimer’s disease mouse models, the levels of various lipids are reported to be elevated, including ceramide (Han et al., 2002; Cutler et al., 2004), minor/simple gangliosides such as GM2 and/or GM3 (Kalanj et al., 1991; Molander-Melin et al., 2005; Barrier et al., 2007; Ariga et al., 2010; Chan et al., 2012), free cholesterol (Cutler et al., 2004) and cholesteryl esters (Chan et al., 2012). Consistent with these reports, we found elevated levels in these specific lipid species in TgCRND8 brain, which we observed to be accumulating particularly within autolysosomes, based on biochemical analyses of isolated autophagic-lysosomal fractions, immunohistochemical analyses revealing increased cathepsin and GM2-positive granules, and ultrastructural evidence of accumulated membrane structures and lipid droplets within enlarged autolysosomes.
Brain levels of major gangliosides (e.g. GM1, GD1a, GD1b, GT1b) have been reported to be decreased in Alzheimer’s disease brain and decreased or unchanged in Alzheimer’s disease mouse models (Crino et al., 1989; Kalanj et al., 1991; Barrier et al., 2007; Bernardo et al., 2009; Ariga et al., 2010), although several separate investigations of Alzheimer’s disease brain have revealed elevated GM1 levels (Molander-Melin et al., 2005; Pernber et al., 2012). In TgCRND8 mice, we observed elevated levels of GM1, GD1a, GD1b as well as GM2 and GM3. While previous studies involved analyses of brain homogenates, our analysis of isolated autophagic-lysosomal fractions enabled us to uncover lipid composition differences between TgCRND8 and wild-type more selective for lysosomal-related compartments.
Prevention of lysosomal lipid accumulation in TgCRND8 brain by specific enhancement of lysosomal proteolysis
We established for the first time by several approaches that CSTB deletion ameliorates lipid accumulation in the lysosomal compartments of TgCRND8 mice. First, autofluorescence signal in hippocampal neurons, which was greatly elevated in TgCRND8, was lowered in CBKO/TgCRND8 mice to levels comparable to those in wild-type mice, indicative of the markedly reduced amounts of lipofuscin. Second, numbers of abnormal GM2-positive granules were markedly diminished in brain sections in CBKO/TgCRND8 mice, consistent with a decrease in levels of minor/simple gangliosides (e.g. GM2 and GM3) in isolated lysosomes from these mice. Third, the giant autolysosomes that accumulated membrane structures, lipid droplets and other undigested materials in TgCRND8, were rarely detected in CBKO/TgCRND8 while the number of normal-sized autolysosomes/lysosomes containing only minimal amounts of undigested materials were restored to near normal levels.
Lysosomal lipid storage, as seen in lysosomal storage disorders, can inhibit lysosomal protease activity resulting in defective clearance of autophagosomes (Elrick et al., 2012) and impaired autophagic flux. In Alzheimer’s disease models, these deficits disrupt Alzheimer’s disease-related processes, such as the lysosome-dependent degradation of APP-CTFs (Tamboli et al., 2011) leading to the accumulation of APP-CTFs and amyloid-β, especially ganglioside-bound amyloid-β, within lysosomal compartments (Keilani et al., 2012) (for review, see van Echten-Deckert and Walter, 2012). Conversely, our findings indicate that the diminished proteolytic function in TgCRND8 brain (Yang et al., 2011) has similar adverse effects on the degradation of lipids by the lysosome. This is consistent with findings that not only chloroquine (Ivy et al., 1984), which raises lysosomal pH and thus can inhibit a wide range of lysosomal enzymes responsible for the degradation of proteins, lipids and other macromolecules, but leupeptin (Ivy et al., 1984, 1989; Nunomura and Miyagishi, 1993), an inhibitor of cysteine and some serine proteases, can induce formation of lipid-containing lipofuscin-like granules. Although the major component of leupeptin-induced lipofuscin is considered proteinaceous, membranous structures and lipid droplet-like vacuoles can also be detected within the lipofuscin-like granules (Nunomura and Miyagishi, 1993; Yang et al., unpublished data) and therefore indicate an effect of disrupting lysosomal proteolytic function on lipid accumulation. Similarly, the brains of mice deficient in CTSD (Koike et al., 2000, 2005; Jabs et al., 2008; Mutka et al., 2010), a lysosomal aspartic protease, exhibit accumulation of proteins (e.g. subunit c of mitochondrial ATP synthase and LC3-II), as well as signs of lipid storage. These include elevated levels of GM2, GM3, BMP and cholesteryl esters, autofluorescence, and accumulation of autophagic-lysosomal compartments filled with massive undigested membrane structures and ceroid-lipofuscin, the latter may contain a relatively high level of lipids (Jolly et al., 2002). Although CTSD plays a role in processing prosaposin to saposins involved in sphingolipid degradation (Hiraiwa et al., 1997), such a role may not completely explain all the observations of lipid accumulation in the CTSD-deficient mice. Rather, protein storage—as a general consequence of deficit in CTSD’s primary role as an aspartic protease—may also contribute to the lipid storage, and this assumption is supported by the similar phenotypes found in mice deficient in both cathepsin B and cathepsin L (Koike et al., 2005), which are lysosomal cysteine proteases without apparent roles in processing prosaposin.
Together, it is likely that storage material of any kind within autolysosomes and lysosomes can disturb the intraluminal environment altering lysosomal ion balance and pH that is critical for the constituent acidic hydrolases of the lysosome and causing defective autophagic flux (Platt et al., 2012; van Echten-Deckert and Walter, 2012), leading to further accumulation of multiple types of substrates. This can be particularly exemplified by lipofuscin, which is an advanced stage of lysosomal substrate accumulation, composed primarily of oxidatively modified proteins and lipids, along with some carbohydrates and metals, and capable of interfering with lysosomal functions in multiple ways. For example, components of lipofuscin can elevate lysosomal pH and inhibit lysosomal degradation of proteins, phospholipids and glycosaminoglycans (Eldred, 1995; Holz et al., 1999; Finnemann et al., 2002). Also, lipofuscin can compromise lysosomal degradation through diluting lysosomal hydrolases within the lysosome or diverting the distribution of hydrolases among lysosomal structures, and physically disturb the interactions between enzymes and their substrates, all resulting in defective substrate degradation and thus reduction of lysosomal release of amino acids and fatty acids into the cytosol for reuse. Additionally, lipofuscin may cause lysosomal leakage, which may further derange lysosomal acidic environment, and/or lysosomal membrane rupture (Eldred, 1995; Terman and Brunk, 2004; Kurz et al., 2007; Sulzer et al., 2008).
Considering the aforementioned intralysosomal interrelationships among substrates such as proteins and lipids, we therefore propose that improved lysosomal turnover of proteins after CSTB deletion (Yang et al., 2011) would have broader influences, directly or indirectly, on other lysosomal substrates besides proteins. Most likely, the overall intralysosomal environment has been improved which can facilitate/maintain the normal function of enzymes involved in lipid degradation, leading to prevention of lysosomal lipid storage.
Enhancing lysosomal degradative capacity versus enhancing autophagy induction
Our study demonstrates a strategy to improve lipid degradation through enhancing lysosomal proteolysis during the later stages of autophagy. The findings may have an implication that although enhancing autophagy at earlier stage(s) of the pathway (e.g. autophagy induction) might conceivably ameliorate lipid accumulation, it is increasingly accepted that stimulating autophagy induction may not have beneficial effects on autophagy efficiency (i.e. autophagy ‘flux’) if the later clearance stages of autophagy are blocked. This has already been shown in models of Niemann–Pick type C disease where inducing autophagy either worsened or had no effect on autolysosome substrate accumulation, whereas enhancing clearance did ameliorate the phenotype (Elrick et al., 2012; Meske et al., 2014). Additional studies have also demonstrated that enhancing early events in the autophagy pathway, e.g. overexpressing transcription factor EB (TFEB) (Settembre et al., 2013, Fig. 5) or treatment with rapamycin to induce autophagy (Sarkar et al., 2013, Fig. 5), could not rescue lipid accumulation since the later stage(s) was not functional.
Multiple lines of evidence obtained from the brain of Alzheimer’s disease and Alzheimer’s disease mouse models have indicated that autophagy dysfunction in Alzheimer’s disease is primarily represented by defective substrate degradation occurring at the later degradative stage of the pathway (i.e. within autolysosomes/lysosomes) (Nixon and Yang, 2011), which is believed to be the case for TgCRND8 (Yang et al., 2011).
Collectively, these foregoing findings imply that disruption in lysosomal clearance stages of autophagy in these diseases needs to be reversed in order to improve overall autophagy efficiency. Previous observations by us and others (Boland et al., 2008; Majumder et al., 2011; Yang et al., 2011; Elrick et al., 2012; Meske et al., 2014) support this notion and our current findings add further support. Our objective in this and the previous (Yang et al., 2011) studies was to enhance the efficiency of autophagy—a process that requires both the sequestration of substrates and their clearance/degradation by lysosomes. By enhancing lysosomal proteolysis, we achieved increased autophagic substrate turnover and thus enhanced overall autophagy efficiency in a manner relevant to possible disease modification in Alzheimer’s disease.
Possible relationship of lipid accumulation to Alzheimer’s disease pathogenesis
While it remains to be further explored in future studies how lipid accumulation fully relates to APP/amyloid-β pathobiology and influences pathogenesis of Alzheimer’s disease, there is evidence supporting both effects of autophagy disruption and lipid accumulation on APP/amyloid-β processing and clearance as well as possible negative influences of APP/amyloid-β on autophagy efficiency. Briefly, lipid accumulation has been found to exhibit effects on APP processing, APP-carboxy terminal fragments/amyloid-β clearance, amyloid-β aggregation etc. (Tamboli et al., 2011; Keilani et al., 2012; Maulik et al., 2012b) whereas some evidence implicates amyloid-β, and other Alzheimer’s disease-related causative factors in disrupting stages of autophagy (Nixon and Yang, 2011; van Echten-Deckert and Walter, 2012).
Regardless of whether lipid accumulation originates upstream or downstream of amyloid-β/APP metabolism, however, it is significant that the accumulation of lipids can be reduced by modulating lysosomal proteolysis, which likely contributes to amelioration of the phenotype(s) existing in this APP transgenic model (Yang et al., 2011), similar to previously observed beneficial effects of clearing lipids from lysosomes in other neurodegenerative diseases (Maulik et al., 2012a; Meske et al., 2014). Also, in an Alzheimer’s disease model carrying the same APP mutations as TgCRND8, manipulating lipids with beta-cyclodextrin, an agent commonly used to deplete lipids, has been shown to rescue aspects of the Alzheimer’s disease phenotype, including diminished amyloid-β plaque deposition, reduced tau immunoreactive dystrophic neurites and improved spatial learning and memory (Yao et al., 2012). Similarly, our preliminary studies in TgCRND8 have revealed that short-term cyclodextrin treatment (2-week intracerebroventricular infusion) lowers not only the number of GM2-positive granules in the hippocampal CA1 neurons, but also larger-size granules detected by antibody 4G8 that recognizes amyloid-β and/or other APP metabolites (Yang et al., unpublished results). Given these observations and others linking lipid accumulation in various neuropathological settings to pathogenesis (Schulze and Sandhoff, 2011; van Echten-Deckert and Walter, 2012), it is reasonable to propose that improvements in behavioural performance in TgCRND8 after CSTB deletion (Yang et al., 2011) reflect improved lysosomal degradative function related to increased clearance of both proteins and lipids. These findings raise the possibility that elevating cathepsin proteolysis to enhance lysosomal function may have applicability to a broader range of neurodegenerative diseases, including lysosomal storage disorders (Nixon et al., 2008; Nixon, 2013).
Acknowledgements
We are grateful to Nicole Gogel for assistance in article preparation. We are also grateful to Drs David Westaway (University of Alberta, Canada) and Richard M. Myers (HudsonAlpha Institute for Biotechnology, USA) for providing a breeding colony of TgCRND8 and CBKO mice, respectively, and to the Transgenic Mice Core in the Centre for Dementia at the Nathan Kline Institute, headed by Dr Efrat Levy, for providing some mice for this study. We thank Drs Kostantin Dobrenis (Albert Einstein College of Medicine, USA) and Marialuisa Melli (University of Bologna, Italy) for providing the anti-GM2 antibody and the rat cystatin B cDNA, respectively.
Glossary
Abbreviation
- CBKO
CSTB knockout
Funding
Supported by the Alzheimer’s Association: IIRG-08-90771 (D.-S.Y.), NIA: P01 AG017617 (R.A.N.), Takeda Pharmaceutical Company Limited (R.A.N.), NIAAA: AA015355 (M.S.), NIA: P30 AG038072 (A.M.C.) and NICHD: R01 HD045561 (S.U.W.). J.A.R. was supported by a Revson Postdoctoral fellowship and a Miguel Servet grant.
Supplementary material
Supplementary material is available at Brain online.
References
- Andreyev AY, Fahy E, Guan Z, Kelly S, Li X, McDonald JG, et al. Subcellular organelle lipidomics in TLR-4-activated macrophages. J Lipid Res. 2010;51:2785–97. doi: 10.1194/jlr.M008748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariga T, Wakade C, Yu RK. The pathological roles of ganglioside metabolism in Alzheimer's disease: effects of gangliosides on neurogenesis. Int J Alzheimers Dis. 2011;2011:193618. doi: 10.4061/2011/193618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariga T, Yanagisawa M, Wakade C, Ando S, Buccafusco JJ, McDonald MP, et al. Ganglioside metabolism in a transgenic mouse model of Alzheimer's disease: expression of Chol-1alpha antigens in the brain. ASN Neuro. 2010;2:e00044. doi: 10.1042/AN20100021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrier L, Ingrand S, Damjanac M, Rioux Bilan A, Hugon J, Page G. Genotype-related changes of ganglioside composition in brain regions of transgenic mouse models of Alzheimer's disease. Neurobiol Aging. 2007;28:1863–72. doi: 10.1016/j.neurobiolaging.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Berg TO, Fengsrud M, Stromhaug PE, Berg T, Seglen PO. Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J Biol Chem. 1998;273:21883–92. doi: 10.1074/jbc.273.34.21883. [DOI] [PubMed] [Google Scholar]
- Bernardo A, Harrison FE, McCord M, Zhao J, Bruchey A, Davies SS, et al. Elimination of GD3 synthase improves memory and reduces amyloid-beta plaque load in transgenic mice. Neurobiol Aging. 2009;30:1777–91. doi: 10.1016/j.neurobiolaging.2007.12.022. [DOI] [PubMed] [Google Scholar]
- Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, et al. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's disease. J Neurosci. 2008;28:6926–37. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebollero E, Reggiori F, Kraft C. Reticulophagy and ribophagy: regulated degradation of protein production factories. Int J Cell Biol. 2012;2012:182834. doi: 10.1155/2012/182834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan RB, Oliveira TG, Cortes EP, Honig LS, Duff KE, Small SA, et al. Comparative lipidomic analysis of mouse and human brain with Alzheimer disease. J Biol Chem. 2012;287:2678–88. doi: 10.1074/jbc.M111.274142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chishti MA, Yang DS, Janus C, Phinney AL, Horne P, Pearson J, et al. Early-onset amyloid deposition and cognitive deficits in transgenic mice expressing a double mutant form of amyloid precursor protein 695. J Biol Chem. 2001;276:21562–70. doi: 10.1074/jbc.M100710200. [DOI] [PubMed] [Google Scholar]
- Crino PB, Ullman MD, Vogt BA, Bird ED, Volicer L. Brain gangliosides in dementia of the Alzheimer type. Arch Neurol. 1989;46:398–401. doi: 10.1001/archneur.1989.00520400054019. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Palmer A, Rivett AJ, Knecht E. Degradation of proteasomes by lysosomes in rat liver. Eur J Biochem. 1995;227:792–800. doi: 10.1111/j.1432-1033.1995.tb20203.x. [DOI] [PubMed] [Google Scholar]
- Cutler RG, Kelly J, Storie K, Pedersen WA, Tammara A, Hatanpaa K, et al. Involvement of oxidative stress-induced abnormalities in ceramide and cholesterol metabolism in brain aging and Alzheimer's disease. Proc Natl Acad Sci USA. 2004;101:2070–5. doi: 10.1073/pnas.0305799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G, Kim TW. Linking lipids to Alzheimer's disease: cholesterol and beyond. Nat Rev Neurosci. 2011;12:284–96. doi: 10.1038/nrn3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid N, Ito Y, Maemura K, Otsuki Y. Elevated autophagic sequestration of mitochondria and lipid droplets in steatotic hepatocytes of chronic ethanol-treated rats: an immunohistochemical and electron microscopic study. J Mol Histol. 2013;44:311–26. doi: 10.1007/s10735-013-9483-x. [DOI] [PubMed] [Google Scholar]
- Eldred GE. Lipofuscin fluorophore inhibits lysosomal protein degradation and may cause early stages of macular degeneration. Gerontology. 1995;41(Suppl 2):15–28. doi: 10.1159/000213722. [DOI] [PubMed] [Google Scholar]
- Elrick MJ, Yu T, Chung C, Lieberman AP. Impaired proteolysis underlies autophagic dysfunction in Niemann-Pick type C disease. Hum Mol Genet. 2012;21:4876–87. doi: 10.1093/hmg/dds324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerod L, Fisher EMC, et al. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnemann SC, Leung LW, Rodriguez-Boulan E. The lipofuscin component A2E selectively inhibits phagolysosomal degradation of photoreceptor phospholipid by the retinal pigment epithelium. Proc Natl Acad Sci USA. 2002;99:3842–7. doi: 10.1073/pnas.052025899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley P. Lipids in Alzheimer's disease: a century-old story. Biochim Biophys Acta. 2010;1801:750–3. doi: 10.1016/j.bbalip.2010.05.004. [DOI] [PubMed] [Google Scholar]
- Frisardi V, Panza F, Seripa D, Farooqui T, Farooqui AA. Glycerophospholipids and glycerophospholipid-derived lipid mediators: a complex meshwork in Alzheimer's disease pathology. Prog Lipid Res. 2011;50:313–30. doi: 10.1016/j.plipres.2011.06.001. [DOI] [PubMed] [Google Scholar]
- Garner B. Lipids and Alzheimer's disease. Biochim Biophys Acta. 2010;1801:747–9. doi: 10.1016/j.bbalip.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Ge L, Melville D, Zhang M, Schekman R. The ER-golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife. 2013;2:e00947. doi: 10.7554/eLife.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gylys KH, Fein JA, Yang F, Miller CA, Cole GM. Increased cholesterol in Abeta-positive nerve terminals from Alzheimer's disease cortex. Neurobiol Aging. 2007;28:8–17. doi: 10.1016/j.neurobiolaging.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–93. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- Han X. Lipid alterations in the earliest clinically recognizable stage of Alzheimer's disease: implication of the role of lipids in the pathogenesis of Alzheimer's disease. Curr Alzheimer Res. 2005;2:65–77. doi: 10.2174/1567205052772786. [DOI] [PubMed] [Google Scholar]
- Han X, D MH, McKeel DW Jr, Kelley J, Morris JC. Substantial sulfatide deficiency and ceramide elevation in very early Alzheimer's disease: potential role in disease pathogenesis. J Neurochem 2002; 82: 809-18. [DOI] [PubMed]
- Harris H, Rubinsztein DC. Control of autophagy as a therapy for neurodegenerative disease. Nat Rev Neurol. 2011;8:108–17. doi: 10.1038/nrneurol.2011.200. [DOI] [PubMed] [Google Scholar]
- Hicks DA, Nalivaeva NN, Turner AJ. Lipid rafts and Alzheimer's disease: protein-lipid interactions and perturbation of signaling. Front Physiol. 2012;3:189. doi: 10.3389/fphys.2012.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraiwa M, Martin BM, Kishimoto Y, Conner GE, Tsuji S, O'Brien JS. Lysosomal proteolysis of prosaposin, the precursor of saposins (sphingolipid activator proteins): its mechanism and inhibition by ganglioside. Arch Biochem Biophys. 1997;341:17–24. doi: 10.1006/abbi.1997.9958. [DOI] [PubMed] [Google Scholar]
- Holz FG, Schutt F, Kopitz J, Eldred GE, Kruse FE, Volcker HE, et al. Inhibition of lysosomal degradative functions in RPE cells by a retinoid component of lipofuscin. Invest Ophthalmol Vis Sci. 1999;40:737–43. [PubMed] [Google Scholar]
- Hung YH, Chen LM, Yang JY, Yang WY. Spatiotemporally controlled induction of autophagy–mediated lysosome turnover. Nat Commun. 2013;4:2111. doi: 10.1038/ncomms3111. [DOI] [PubMed] [Google Scholar]
- Ivy GO, Kanai S, Ohta M, Smith G, Sato Y, Kobayashi M, et al. Lipofuscin-like substances accumulate rapidly in brain, retina and internal organs with cysteine protease inhibition. Adv Exp Med Biol. 1989;266:31–45. doi: 10.1007/978-1-4899-5339-1_3. [DOI] [PubMed] [Google Scholar]
- Ivy GO, Schottler F, Wenzel J, Baudry M, Lynch G. Inhibitors of lysosomal enzymes: accumulation of lipofuscin-like dense bodies in the brain. Science. 1984;226:985–7. doi: 10.1126/science.6505679. [DOI] [PubMed] [Google Scholar]
- Jabs S, Quitsch A, Kakela R, Koch B, Tyynela J, Brade H, et al. Accumulation of bis(monoacylglycero)phosphate and gangliosides in mouse models of neuronal ceroid lipofuscinosis. J Neurochem. 2008;106:1415–25. doi: 10.1111/j.1471-4159.2008.05497.x. [DOI] [PubMed] [Google Scholar]
- Jolly RD, Palmer DN, Dalefield RR. The analytical approach to the nature of lipofuscin (age pigment) Arch Gerontol Geriatr. 2002;34:205–17. doi: 10.1016/s0167-4943(01)00219-9. [DOI] [PubMed] [Google Scholar]
- Kalanj S, Kracun I, Rosner H, Cosovic C. Regional distribution of brain gangliosides in Alzheimer's disease. Neurol Croat. 1991;40:269–81. [PubMed] [Google Scholar]
- Keilani S, Lun Y, Stevens AC, Williams HN, Sjoberg ER, Khanna R, et al. Lysosomal dysfunction in a mouse model of Sandhoff disease leads to accumulation of ganglioside-bound amyloid-beta peptide. J Neurosci. 2012;32:5223–36. doi: 10.1523/JNEUROSCI.4860-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaldoun SA, Emond-Boisjoly MA, Chateau D, Carriere V, Lacasa M, Rousset M, et al. Autophagosomes contribute to intracellular lipid distribution in enterocytes. Mol Biol Cell. 2014;25:118–32. doi: 10.1091/mbc.E13-06-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Cuervo AM, Dunn WA, Jr, Levine B, van der Klei I, Seglen PO. How shall I eat thee? Autophagy. 2007;3:413–6. doi: 10.4161/auto.4377. [DOI] [PubMed] [Google Scholar]
- Knaevelsrud H, Simonsen A. Lipids in autophagy: constituents, signaling molecules and cargo with relevance to disease. Biochim Biophys Acta. 2012;1821:1133–45. doi: 10.1016/j.bbalip.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Koike M, Nakanishi H, Saftig P, Ezaki J, Isahara K, Ohsawa Y, et al. Cathepsin D deficiency induces lysosomal storage with ceroid lipofuscin in mouse CNS neurons. J Neurosci. 2000;20:6898–906. doi: 10.1523/JNEUROSCI.20-18-06898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike M, Shibata M, Waguri S, Yoshimura K, Tanida I, Kominami E, et al. Participation of autophagy in storage of lysosomes in neurons from mouse models of neuronal ceroid-lipofuscinoses (Batten disease) Am J Pathol. 2005;167:1713–28. doi: 10.1016/S0002-9440(10)61253-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter T, Sandhoff K. Principles of lysosomal membrane digestion: stimulation of sphingolipid degradation by sphingolipid activator proteins and anionic lysosomal lipids. Annu Rev Cell Dev Biol. 2005;21:81–103. doi: 10.1146/annurev.cellbio.21.122303.120013. [DOI] [PubMed] [Google Scholar]
- Kolter T, Sandhoff K. Lysosomal degradation of membrane lipids. FEBS Lett. 2010;584:1700–12. doi: 10.1016/j.febslet.2009.10.021. [DOI] [PubMed] [Google Scholar]
- Kurz T, Terman A, Brunk UT. Autophagy, ageing and apoptosis: the role of oxidative stress and lysosomal iron. Arch Biochem Biophys. 2007;462:220–30. doi: 10.1016/j.abb.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14:759–74. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- Lieberman AP, Puertollano R, Raben N, Slaugenhaupt S, Walkley SU, Ballabio A. Autophagy in lysosomal storage disorders. Autophagy. 2012;8:719–30. doi: 10.4161/auto.19469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Evans E, Platt FM. Lipids on trial: the search for the offending metabolite in Niemann-Pick type C disease. Traffic. 2010;11:419–28. doi: 10.1111/j.1600-0854.2010.01032.x. [DOI] [PubMed] [Google Scholar]
- Macala LJ, Yu RK, Ando S. Analysis of brain lipids by high performance thin-layer chromatography and densitometry. J Lipid Res. 1983;24:1243–50. [PubMed] [Google Scholar]
- Maejima I, Takahashi A, Omori H, Kimura T, Takabatake Y, Saitoh T, et al. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 2013;32:2336–47. doi: 10.1038/emboj.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder S, Richardson A, Strong R, Oddo S. Inducing autophagy by rapamycin before, but not after, the formation of plaques and tangles ameliorates cognitive deficits. PLoS One. 2011;6:e25416. doi: 10.1371/journal.pone.0025416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzi A, Esko J. Direct chemical analysis of glycoconjugates for carbohydrates. Curr Protoc Mol Biol. 2001 doi: 10.1002/0471142727.mb1709s32. Chapter 17: Unit17 9. [DOI] [PubMed] [Google Scholar]
- Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat Neurosci. 2010;13:567–76. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzella L, Ahlberg J, Glaumann H. Isolation of autophagic vacuoles from rat liver: morphological and biochemical characterization. J Cell Biol. 1982;93:144–54. doi: 10.1083/jcb.93.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res. 2008;49:1137–46. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maulik M, Ghoshal B, Kim J, Wang Y, Yang J, Westaway D, et al. Mutant human APP exacerbates pathology in a mouse model of NPC and its reversal by a beta-cyclodextrin. Hum Mol Genet. 2012a;21:4857–75. doi: 10.1093/hmg/dds322. [DOI] [PubMed] [Google Scholar]
- Maulik M, Westaway D, Jhamandas JH, Kar S. Role of cholesterol in APP metabolism and its significance in Alzheimer's disease pathogenesis. Mol Neurobiol. 2012b;47:37–63. doi: 10.1007/s12035-012-8337-y. [DOI] [PubMed] [Google Scholar]
- Meske V, Erz J, Priesnitz T, Ohm TG. The autophagic defect in Niemann-Pick disease type C neurons differs from somatic cells and reduces neuronal viability. Neurobiol Dis. 2014;64:88–97. doi: 10.1016/j.nbd.2013.12.018. [DOI] [PubMed] [Google Scholar]
- Micsenyi MC, Dobrenis K, Stephney G, Pickel J, Vanier MT, Slaugenhaupt SA, et al. Neuropathology of the Mcoln1(-/-) knockout mouse model of mucolipidosis type IV. J Neuropathol Exp Neurol. 2009;68:125–35. doi: 10.1097/NEN.0b013e3181942cf0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molander-Melin M, Blennow K, Bogdanovic N, Dellheden B, Mansson JE, Fredman P. Structural membrane alterations in Alzheimer brains found to be associated with regional disease development; increased density of gangliosides GM1 and GM2 and loss of cholesterol in detergent-resistant membrane domains. J Neurochem. 2005;92:171–82. doi: 10.1111/j.1471-4159.2004.02849.x. [DOI] [PubMed] [Google Scholar]
- Mutka AL, Haapanen A, Kakela R, Lindfors M, Wright AK, Inkinen T, et al. Murine cathepsin D deficiency is associated with dysmyelination/myelin disruption and accumulation of cholesteryl esters in the brain. J Neurochem. 2010;112:193–203. doi: 10.1111/j.1471-4159.2009.06440.x. [DOI] [PubMed] [Google Scholar]
- Nixon RA. Endosome function and dysfunction in Alzheimer's disease and other neurodegenerative diseases. Neurobiol Aging. 2005;26:373–82. doi: 10.1016/j.neurobiolaging.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19:983–97. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- Nixon RA, Yang DS. Autophagy failure in Alzheimer's disease-locating the primary defect. Neurobiol Dis. 2011;43:38–45. doi: 10.1016/j.nbd.2011.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon RA, Yang DS, Lee JH. Neurodegenerative lysosomal disorders: a continuum from development to late age. Autophagy. 2008;4:590–9. doi: 10.4161/auto.6259. [DOI] [PubMed] [Google Scholar]
- Nunomura A, Miyagishi T. Ultrastructural observations on neuronal lipofuscin (age pigment) and dense bodies induced by a proteinase inhibitor, leupeptin, in rat hippocampus. Acta Neuropathol. 1993;86:319–28. doi: 10.1007/BF00369443. [DOI] [PubMed] [Google Scholar]
- Ouimet M, Franklin V, Mak E, Liao X, Tabas I, Marcel YL. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 2011;13:655–67. doi: 10.1016/j.cmet.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. London: Academic Press; 2001. The mouse brain in streotaxic coordinates. [Google Scholar]
- Pennacchio LA, Bouley DM, Higgins KM, Scott MP, Noebels JL, Myers RM. Progressive ataxia, myoclonic epilepsy and cerebellar apoptosis in cystatin B-deficient mice. Nat Genet. 1998;20:251–8. doi: 10.1038/3059. [DOI] [PubMed] [Google Scholar]
- Pernber Z, Blennow K, Bogdanovic N, Mansson JE, Blomqvist M. Altered distribution of the gangliosides GM1 and GM2 in Alzheimer's disease. Dement Geriatr Cogn Disord. 2012;33:174–88. doi: 10.1159/000338181. [DOI] [PubMed] [Google Scholar]
- Platt FM, Boland B, van der Spoel AC. The cell biology of disease: lysosomal storage disorders: the cellular impact of lysosomal dysfunction. J Cell Biol. 2012;199:723–34. doi: 10.1083/jcb.201208152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri C, Renna M, Bento CF, Moreau K, Rubinsztein DC. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell. 2013;154:1285–99. doi: 10.1016/j.cell.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747–57. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razi M, Chan EY, Tooze SA. Early endosomes and endosomal coatomer are required for autophagy. J Cell Biol. 2009;185:305–21. doi: 10.1083/jcb.200810098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro JA, Kaushik S, Koga H, Dall'Armi C, Shui G, Wenk MR, et al. Inhibitory effect of dietary lipids on chaperone-mediated autophagy. Proc Natl Acad Sci U S A. 2012;109:E705–14. doi: 10.1073/pnas.1113036109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romijn HJ, van Uum JF, Breedijk I, Emmering J, Radu I, Pool CW. Double immunolabeling of neuropeptides in the human hypothalamus as analyzed by confocal laser scanning fluorescence microscopy. J Histochem Cytochem. 1999;47:229–36. doi: 10.1177/002215549904700211. [DOI] [PubMed] [Google Scholar]
- Saito M, Chakraborty G, Shah R, Mao RF, Kumar A, Yang DS, et al. Elevation of GM2 ganglioside during ethanol-induced apoptotic neurodegeneration in the developing mouse brain. J Neurochem. 2012;121:649–61. doi: 10.1111/j.1471-4159.2012.07710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Carroll B, Buganim Y, Maetzel D, Ng AH, Cassady JP, et al. Impaired autophagy in the lipid-storage disorder Niemann-Pick type C1 disease. Cell Rep. 2013;5:1302–15. doi: 10.1016/j.celrep.2013.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer M, Schmitz G. Metabolism, function and mass spectrometric analysis of bis(monoacylglycero)phosphate and cardiolipin. Chem Phys Lipids. 2011;164:556–62. doi: 10.1016/j.chemphyslip.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Schulze H, Kolter T, Sandhoff K. Principles of lysosomal membrane degradation: cellular topology and biochemistry of lysosomal lipid degradation. Biochim Biophys Acta. 2009;1793:674–83. doi: 10.1016/j.bbamcr.2008.09.020. [DOI] [PubMed] [Google Scholar]
- Schulze H, Sandhoff K. Lysosomal lipid storage diseases. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze H, Sandhoff K. Sphingolipids and lysosomal pathologies. Biochim Biophys Acta. 2014;1841:799–810. doi: 10.1016/j.bbalip.2013.10.015. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ. Alzheimer's disease: genes, proteins, and therapy. Physiol Rev. 2001;81:741–66. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013;15:647–58. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HM, Mizushima N. At the end of the autophagic road: an emerging understanding of lysosomal functions in autophagy. Trends Biochem Sci. 2014;39:61–71. doi: 10.1016/j.tibs.2013.12.001. [DOI] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–5. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulzer D, Mosharov E, Talloczy Z, Zucca FA, Simon JD, Zecca L. Neuronal pigmented autophagic vacuoles: lipofuscin, neuromelanin, and ceroid as macroautophagic responses during aging and disease. J Neurochem. 2008;106:24–36. doi: 10.1111/j.1471-4159.2008.05385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamboli IY, Hampel H, Tien NT, Tolksdorf K, Breiden B, Mathews PM, et al. Sphingolipid storage affects autophagic metabolism of the amyloid precursor protein and promotes Abeta generation. J Neurosci. 2011;31:1837–49. doi: 10.1523/JNEUROSCI.2954-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terman A, Brunk UT. Lipofuscin. Int J Biochem Cell Biol. 2004;36:1400–4. doi: 10.1016/j.biocel.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Terry RD, Gonatas NK, Weiss M. Ultrastructural studies in Alzheimer's Presenile Dementia. Am J Pathol. 1964;44:269–97. [PMC free article] [PubMed] [Google Scholar]
- van Echten-Deckert G. Sphingolipid extraction and analysis by thin-layer chromatography. Methods Enzymol. 2000;312:64–79. doi: 10.1016/s0076-6879(00)12900-3. [DOI] [PubMed] [Google Scholar]
- van Echten-Deckert G, Walter J. Sphingolipids: critical players in Alzheimer's disease. Prog Lipid Res. 2012;51:378–93. doi: 10.1016/j.plipres.2012.07.001. [DOI] [PubMed] [Google Scholar]
- van Meer G, de Kroon AI. Lipid map of the mammalian cell. J Cell Sci. 2011;124:5–8. doi: 10.1242/jcs.071233. [DOI] [PubMed] [Google Scholar]
- Walkley SU, Vanier MT. Secondary lipid accumulation in lysosomal disease. Biochim Biophys Acta. 2009;1793:726–36. doi: 10.1016/j.bbamcr.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton S, Southwick K, Hardman RM, Secrest AM, Grow RK, Xin H, et al. Examining the proteins of functional retinal lipofuscin using proteomic analysis as a guide for understanding its origin. Mol Vis. 2005;11:1122–34. [PubMed] [Google Scholar]
- Wolozin B, Manger J, Bryant R, Cordy J, Green RC, McKee A. Re-assessing the relationship between cholesterol, statins and Alzheimer's disease. Acta Neurol Scand Suppl. 2006;185:63–70. doi: 10.1111/j.1600-0404.2006.00687.x. [DOI] [PubMed] [Google Scholar]
- Yang DS, Lee JH, Nixon RA. Monitoring autophagy in Alzheimer's disease and related neurodegenerative diseases. Methods Enzymol. 2009;453:111–44. doi: 10.1016/S0076-6879(08)04006-8. [DOI] [PubMed] [Google Scholar]
- Yang DS, Stavrides P, Mohan PS, Kaushik S, Kumar A, Ohno M, et al. Reversal of autophagy dysfunction in the TgCRND8 mouse model of Alzheimer's disease ameliorates amyloid pathologies and memory deficits. Brain. 2011;134:258–77. doi: 10.1093/brain/awq341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Ho D, Calingasan NY, Pipalia NH, Lin MT, Beal MF. Neuroprotection by cyclodextrin in cell and mouse models of Alzheimer disease. J Exp Med. 2012;209:2501–13. doi: 10.1084/jem.20121239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin D. Biochemical basis of lipofuscin, ceroid, and age pigment-like fluorophores. Free Radic Biol Med. 1996;21:871–88. doi: 10.1016/0891-5849(96)00175-x. [DOI] [PubMed] [Google Scholar]
- Yu RK, Ariga T. Ganglioside analysis by high-performance thin-layer chromatography. Methods Enzymol. 2000;312:115–34. doi: 10.1016/s0076-6879(00)12903-9. [DOI] [PubMed] [Google Scholar]
- Yu RK, Ledeen RW. Gangliosides of human, bovine, and rabbit plasma. J Lipid Res. 1972;13:680–6. [PubMed] [Google Scholar]
- Yu WH, Cuervo AM, Kumar A, Peterhoff CM, Schmidt SD, Lee J-H, et al. Macroautophagy–a novel β-amyloid peptide-generating pathway activated in Alzheimer's disease. J Cell Biol. 2005;171:87–98. doi: 10.1083/jcb.200505082. [DOI] [PMC free article] [PubMed] [Google Scholar]