Abstract
Herein, we described the development of two virtual screens to identify new vitamin D receptor (VDR) antagonists among nuclear receptor (NR) ligands. Therefore, a database of 14330 nuclear receptor ligands and their NR affinities was assembled using the online available “Binding Database”. Two different virtual screens were carried out in conjunction with a reported VDR crystal structure applying a stringent and less stringent pharmacophore model to filter docked NR ligand conformations. The pharmacophore models were based on the spatial orientation of the hydroxyl functionalities of VDR’s natural ligands 1,25(OH2)D3 and 25(OH2)D3. The first virtual screen identified 32 NR ligands with a calculate free energy of VDR binding of more than −6.0 kJ/mol. All but nordihydroguaiaretic acid (NDGA) are VDR ligands, which inhibited the interaction between VDR and coactivator peptide SRC2-3 with an IC50 value of 15.8 µM. The second screen identified 162 NR ligands with a calculate free energy of VDR binding of more than −6.0 kJ/mol. More than half of these ligands were developed to bind VDR followed by ERα/β ligands (26%), TRα/β ligands (7%) and LxRα/β ligands (7%). The binding between VDR and ERα ligand H6036 as well as TRα/β ligand triiodothyronine and a homoserine analog thereof was confirmed by fluorescence polarization.
Keywords: Virtual screening, nuclear receptor, vitamin D receptor, thyroid receptor, estrogen receptor
INTRODUCTION
Nuclear receptors (NR) are one of the most important drug targets today.[1] During the last decades, thousands of small molecules have been developed to selectively bind nuclear receptors. This development was supported by high throughput screening (HTS) and rational drug design. Although the activity of NR ligands is very important in terms of dosage and suppression of side effects, the selectivity of ligands towards a particular NR is crucial for specific pharmacological effects. Usually NR ligands are investigated in respect to their NR isoform-selectivity. For instance, estrogen receptor (ER) ligands are evaluated for their selectivity towards ERα and ERβ, which are distributed tissue-selectively in the human body.[2] Once a promising ligand has been identified, further analysis in respect to other closely related NR is conducted based on phylogenetic distance or NR sequence similarity.[3] Schapira et al. introduced an alternative concept of NR similarity based on the likelihood that two NRs share a common ligand.[4] Therefore, sixteen NR crystal structures and 78 NR ligands were used in a computational approach to determine the cross-reactivity of NR ligands. Herein, we present an alternative approach by using a large library of NR ligands and one receptor, the vitamin D receptor (VDR). Among 14330 compounds, we identified four new VDR antagonists that were originally developed as ligands for other nuclear receptors. Thus, virtual screening represents a useful tool to identify those NRs that are likely to interact with a newly synthesized NR ligand.
MATERIALS AND METHODS
Reagents
Virtual Screens
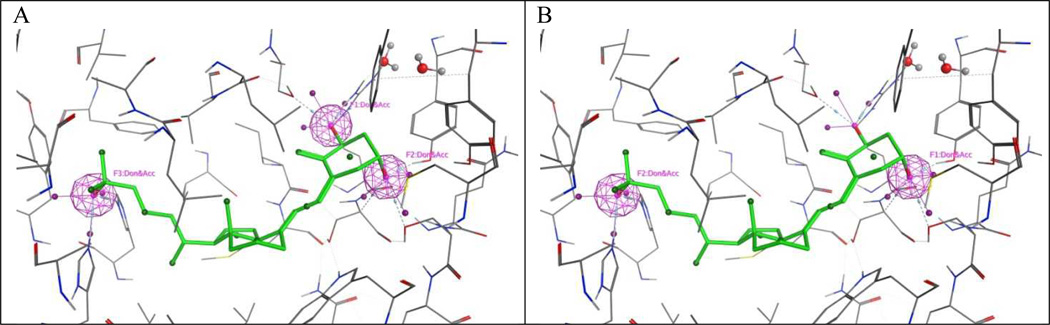
A library of nuclear receptor ligands were assembled using “the Binding Database”. The database included 14330 compound structures and their nuclear receptor binding data (EC50, IC50, or KD). Weakly active ligands that had estimated binding data (e.g. >5000 µM) or inactive compounds (e.g. no binding observed) were assigned a zero activity. Compounds that were not tested were assigned an empty field. The database only included compounds that with one or more nuclear receptors. For racemic compounds only one representative stereoisomer was used for the screen. All compounds were minimized using a MMFF94x force field. Energy minimization was terminated when the root mean square gradient fell below 0.1. The root mean square gradient is the norm of the gradient times the square root of the number of atoms. The ionization state of functional groups was adjusted to pH 7. Molecule conformations were generated from a single 3D conformer by applying a collection of preferred torsion angles to the rotatable bond during the virtual screen. The crystal structure of VDR bound to 1,25(OH)2D3 (PDB ID 1DB1)[6] was prepare for docking using the MOE structure preparation function to repair any structural defects in the pdb file. In addition, a protonation 3D function was used to optimize the hydrogen bond network and hydrogen positions. Finally unbound water molecules were removed. The virtual screen was carried out by selecting VDR-bound 1,25(OH)2D3 as binding site and a triangle matcher for the placement of compounds. The triangle matcher function generated poses by superposition of ligand atom triplets and triplets of receptor site points. The receptor site points were alpha sphere centers which represent locations of tight packing. At each iteration, a random triplet of ligand atoms and a random triplet of alpha sphere centers were used to determine the pose. The poses were scored using affinity London ΔG scoring that estimated the free energy of binding from each given pose given in kJ/mol. Compound conformations that did not satisfy the pharmacophore model 1 or 2 depicted in Figure 4 were eliminated.
Figure 4.
Two different pharmacophore models for VDR ligands.a
aPharmacophore models were established using MOE.
Labeled Coactivator Peptides
The peptide SRC2-3 (CLQEKHRILHKLLQNGNSPA),[7] was purchased and labeled with the cysteine-reactive fluorophore (Alexa Fluor 647 maleimide) in a 50:50 DMF/PBS mixture. After purification by high performance liquid chromatography, the corresponding labeled peptide was dissolved in DMSO and stored at −20°C.
Protein Expression and Purification
The VDR-LBDmt DNA was kindly provided by D. Moras[6] and cloned into the pMAL-c2X vector (New England Biolabs). A detailed expression and purification protocol for VDR-LBD was reported previously.[7]
Fluorescence Polarization Assay with VDR−SRC2-3
Agonistic and antagonistic activity was studied using a FP assay. This assay was conducted in 384-well black polystyrene plates (Corning) using a buffer [25 mM PIPES (pH 6.75) 50 mM NaCl, 0.01% NP-40, 2% DMSO], VDR-LBD protein (0.1 µM), LG190178 (3 µM), and Alexa Fluor 647-labeled SRC2-3 (5 nM). Small molecule transfer into a 20 µL assay solution was accomplished using a stainless steel pin tool (V&P Scientific), delivering 100 nL of the serially diluted compound solution. Fluorescence polarization was detected after 1 hour at excitation and emission wavelengths of 650 nm and 665 nm, respectively. Three independent experiments were conducted in quadruplicate. The data were analyzed using nonlinear regression with a variable slope (GraphPadPrism).
RESULTS
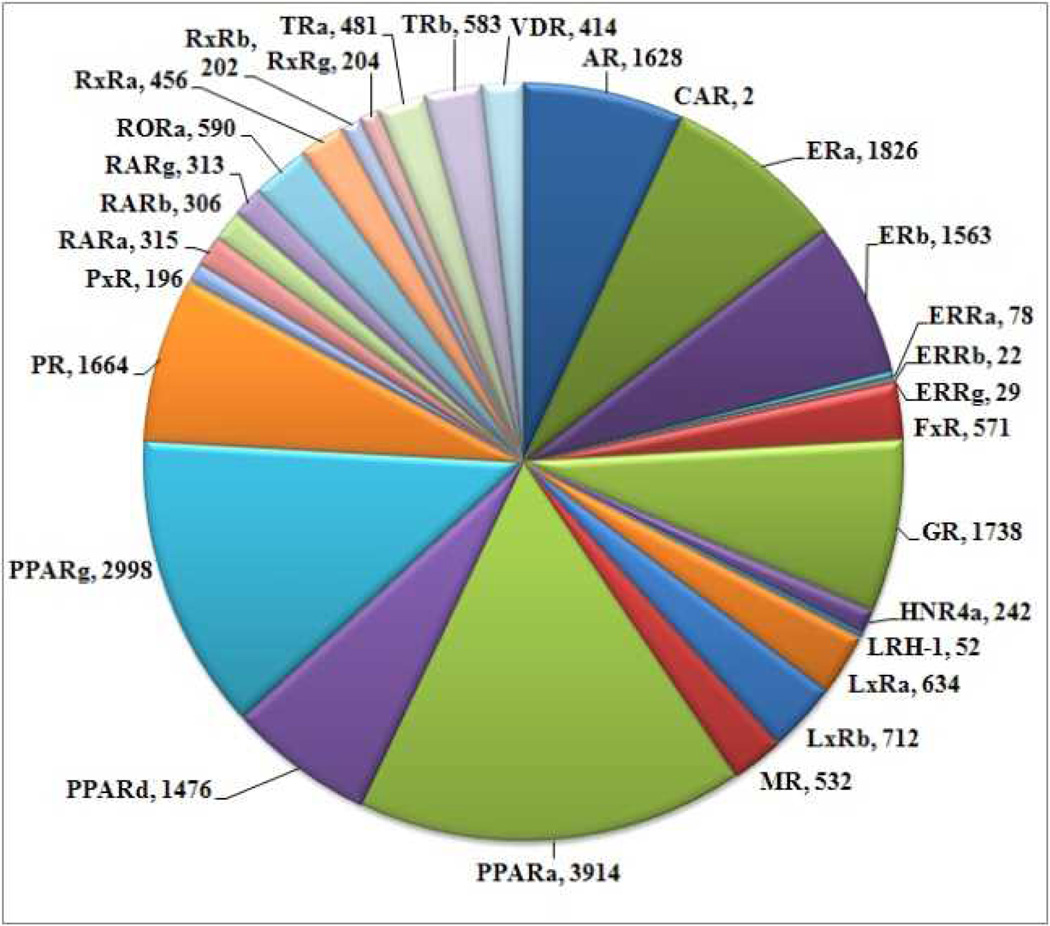
A library of 14330 NR ligands were compiled using “The Binding Database.org”.[8] The sets of NR ligands were downloaded individually and merged as a virtual small molecule library using MOE (molecular operating environment). The number of ligands downloaded per NR is given in Figure 1.
Figure 1.
Number of NR ligands deposited with “The Binding Database”.
The NRs with the largest ligand databases are the peroxisome proliferator-activated receptor (PPAR)γ, PPARδ, PPARα, the progesterone receptor (PR), the androgen receptor (AR), the ERα, the ERβ, and the glucocorticoid receptor (GR) with more than a thousand ligands each. Overall, 30 NRs are represented by their ligands in “The Binding Database” with a total of 14330 unique NR ligands. Many of these ligands were investigated in regards to multiple NRs and some of them exhibited a significant affinity towards more than one NR.
The analysis to determine the global selectivity of ligands among nuclear receptors is restricted by the fact that limited data are available. We were surprised that only one NR was investigated for the majority of NR ligands. 4006 ligands out of 14330, thus a quarter of the ligands, were investigated with two different NRs as illustrate in Figure 2.
Figure 2.
Number of NR ligands that bind to multiple NRs.
Most of NR ligands tested with two NRs were able to bind two NR isoforms with different affinity, such as NRα and NRβ, which applies to the liver × receptor (LXR), ER, and TR. Ligands that were developed for NRs having three isoforms such as the PPAR, the estrogen related receptor (ERR), the retinoid × receptor (RXR), and the retinoic acid receptor (RAR) represent almost half of the NR library members (Figure 1). However, only a fraction of these ligands (1853) were evaluated with more than two NRs (Figure 2). Some examples of ligands that bind multiple NRs, although with different affinity, are depicted in Figure 3.
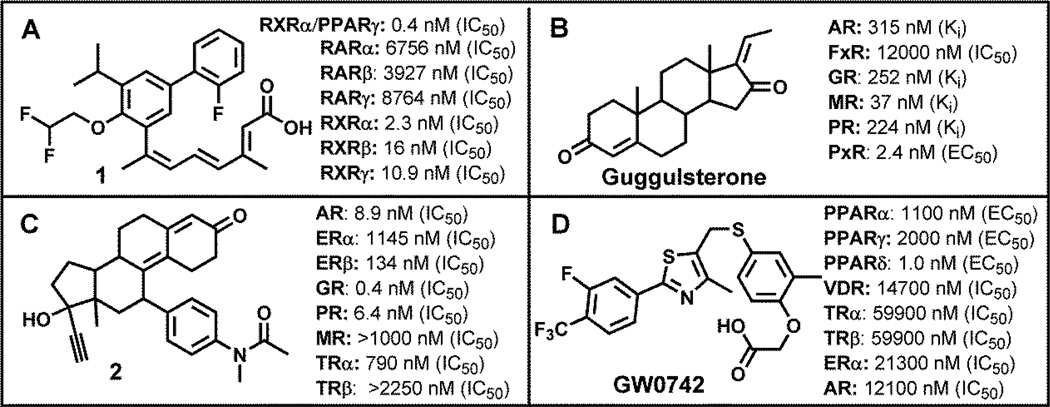
Figure 3.
NR ligands that were evaluated towards multiple nuclear receptors.
Compound 1 (Figure 3, A) was developed by Ligand Pharmaceuticals as RXRα antagonist.[9] Although the selectivity in respect to RARs is very high, there is a moderate selectivity toward other RXR subtypes. In addition, a synergistic activation of transcription was observed when cotransfected with PPARγ in the presence of selective PPARγ ligand. Guggulsterone (Figure 3, B) was predominately evaluated with steroid hormone receptors.[10] The compound has a strong affinity for the mineralocorticoid receptor (MR) and the pregnane × receptor (PXR).[11] Compound 2 (Figure 3, C) is a very potent GR antagonist, which still has a significant activity towards PR and AR.[12] Finally, GW0742 (Figure 3, D) was developed by GlaxoSmithKline as highly a selective agonist for the PPARδ.[13] The evaluation of GW0742, in respect to NR-mediated inhibition of transcription, identified this compound as antagonist for AR and VDR.[14]
Nevertheless, the exhaustive characterization of NR ligands is limited by the sheer number of different NRs resulting in a cost and time-intensive analysis for research labs and the pharmaceutical industry. Therefore, a prediction of NR-selectivity of new ligands using computational approaches might enable a selection of a smaller pool of NRs to be considered for evaluation. In addition, this approach might also identify groups of NRs that bind similar ligands, thus introducing a new relationship between NRs that is different from phylogenetic distance or NR sequence similarity. In order to test this hypothesis, we used the library of NR ligands and carried out two virtual screens applying the first crystal structure of liganded VDR.[6] For each screen, we applied a different pharmacophore model to filter all molecule conformations. The two different pharmacophore models are depicted in Figure 4.
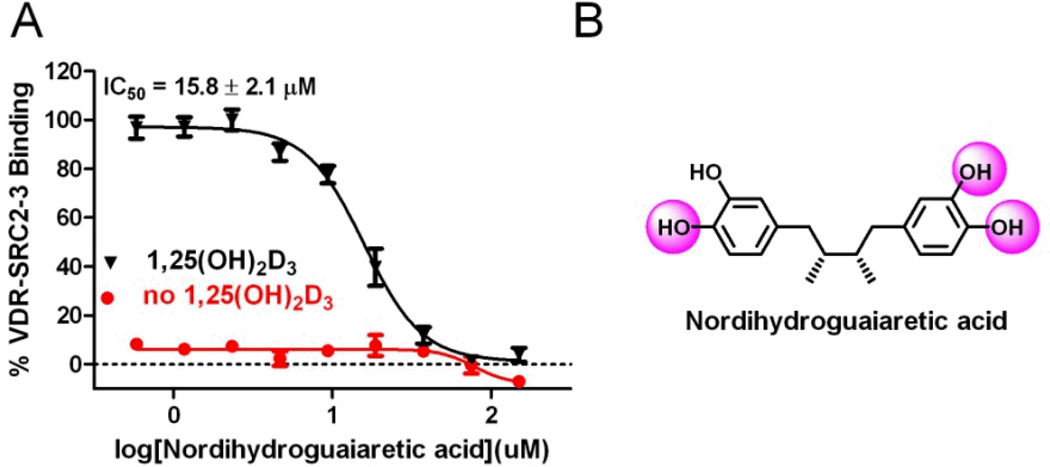
The virtual screen 1 was carried out with a pharmacophore model that specifies three electron donor/acceptor elements depicted as purple spheres (Figure 4, A). These three elements represent the spatial configuration of three hydroxyl groups of the most active endogenous VDR ligand 1,25-dihydroxy vitamin D3 (1,25(OH)2D3).[15] 1,25(OH)2D3 is a metabolic product of vitamin D3 formed from 25(OH)D3 by 1α-hydroxylase.[16] The binding affinity of 1,25(OH)2D3 is 0.1–1 nM, whereas 25(OH)D3 binds with a moderate affinity of 1420 nM towards VDR.[17] The virtual screen of 14330 compounds using pharmacophore model A (Figure 4) identified 64 compounds. 32 of the 64 compounds had a significant calculate free energy of VDR binding of more than −6.0 kJ/mol. Nordihydroguaiaretic acid (NDGA) was the only non-VDR ligand identified with a calculate ΔG of −11.1 kJ/mol (Figure 5).
Figure 5.
Nordihydroguaiaretic acid is inhibiting the interaction between VDR and coactivator peptide SRC2-3.
NDGA is a bioactive compound that inhibits lipoxygenases, functions as an antioxidant, and has shown promising anti-cancer activities.[18] The compound has also been reported to weakly interact with the androgen receptor by binding to a new BF3 binding site.[19] Herein, we confirmed the activity NDGA towards VDR using a fluorescence polarization assay. In the presence of 1,25(OH)2D3, NDGA was able to inhibit the interaction between VDR and coactivator peptide SRC2-3 with an IC50 values of 15.8 ± 2.1 µM. In the absence of 1,25(OH)2D3, NDGA was not able to promote the recruitment of coactivator towards VDR. Because of the fact that the interactions between VDR and coactivators are essential for VDR-mediated transcription we identified NDGA as novel VDR antagonist.
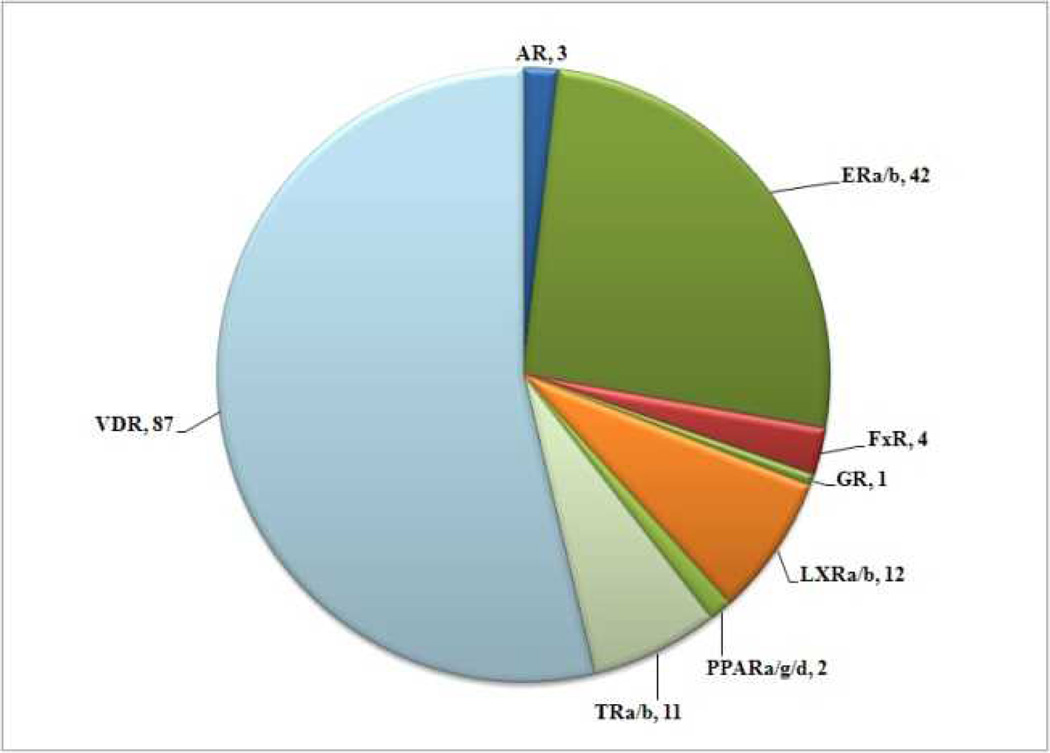
Virtual screen 2 was carried out using a less stringent pharmacophore model depicted in Figure 4, B bearing two acceptor/donor groups representative of VDR ligand 25(OH)D3. Among the 14330 molecules, 397 compounds were identified and 162 compounds exhibited a free energy of binding of more than −6.0 kJ/mol (Figure 6).
Figure 6.
Number and affiliation of NR ligands identified by virtual screen 2 using the pharmacophore model depicted in Figure 4, B.
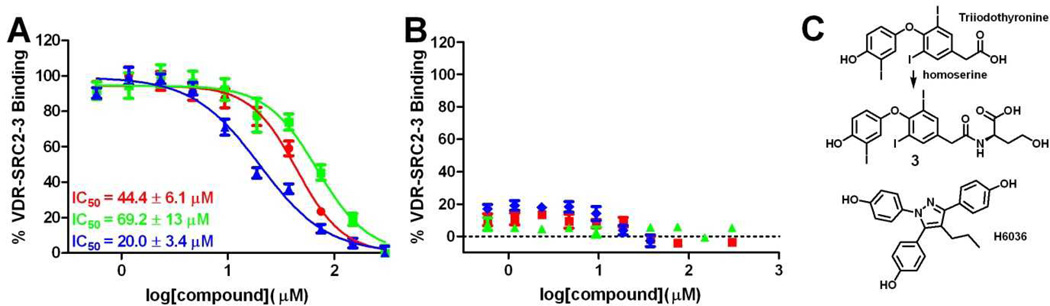
Among the hit compounds of virtual screen 2, the majority of molecules were developed as ligands for VDR. Ligands developed for LxRα/β, TRα/β, and ERα/β ligands were among the most frequent ligands that potentially interact with VDR. We picked one TRα ligand (3) and one ERα ligand (H6036) in order to confirm the activity in regard to VDR (Figure 6).
Both compounds (1 and H6036), identified by virtual screen 2, were able to inhibit the interaction between VDR and SRC2-3 with IC50 values of 44.5 ± 6.1 µM and 20.0 ± 3.4 µM, respectively (Figure 6, A). The original docking score for these compounds was −7.9 kJ/mol (3) and −9.1 kJ/mol (H6036), respectively. The thyroid receptor ligand triiodothyronine was not scored by virtual screen 2 because it did not satisfy the pharmacophore model. However, we observed that triiodothyronine did inhibit the interaction between VDR–SRC2-3, although at higher concentrations. In the absence of VDR ligand LG190178 no recruitment of SRC2-3 to VDR was observed in the presence of any of these ligands (Figure 6, B).
Figure 6.
Interaction between virtual screen hit compounds ( H6036,
H6036,  compound 3,
compound 3,  triiodothyronine) and VDR. A) Hit compound inhibition of the interaction between SRC2-3 and VDR in the presence of VDR agonist LG190178; B) Association of VDR-LBD and SRC2-3 in the presence of hit compounds; C) Structure and generation of NR ligands.
triiodothyronine) and VDR. A) Hit compound inhibition of the interaction between SRC2-3 and VDR in the presence of VDR agonist LG190178; B) Association of VDR-LBD and SRC2-3 in the presence of hit compounds; C) Structure and generation of NR ligands.
DISCUSSION
We showed that databases such as the “Binding database” can function as starting point for virtual screening. During the compilation of this focused library we only identified a small number of developed NR ligands that have been evaluated with other NRs. The main reason for this lack of investigation is the size of the NR superfamily in addition to the existing agonism and antagonism that would make an exhaustive evaluation with a panel of NRs very time intensive and costly. An alternative approach to predict NR selectivity conveyed herein is virtual screening. Using the first published VDR crystal structure and pharmacophore models representing the essential features of VDR ligands, we identified VDR ligands among a large library of nuclear receptor ligands using virtual screening. The essential features of VDR ligand 1,25(OH)2D3 are three hydroxyl functions that interact with VDR via hydrogen bonding. In addition, 1,25(OH)2D3 has a large hydrophobic surface area. Using a pharmacophore model based on 1,25(OH)2D3 that defines the spatial orientation of three hydroxyl groups as filter, we identify compounds that interact with VDR with 100% accuracy during virtual screen 1 using a cutoff of −6 kJ/mol for the calculated free energy of VDR binding. 31 out of the 32 hit compounds were VDR ligands such as agonists 2MD[20] or antagonist 4[21]. Both compounds have been shown to bind VDR, however 2MD promoted the recruitment of coactivators, whereas antagonist 4 inhibited the interaction between VDR and coactivator. Thus, virtual screen 1 identified VDR ligands but did not differentiate between VDR agonists and antagonists.
Virtual screen 2 applied a less stringent pharmacophore model based on VDR ligand 25(OH)D3, which is at least 1000-fold less potent than 1,25(OH)D3. As expected, we identified more and different NR ligands that are likely to interact with VDR. In total we found 162 compounds with a calculated free energy of VDR binding of more than −6.0 kJ/mol. 54%, thus 87 ligands were developed for VDR. We have to point out that 187 compounds of the 414 VDR binders are VDR–coactivator inhibitors developed by Mita et al.[22] and by us.[23] These compounds do not bind VDR at the 1,25(OH)2D3 binding site. Furthermore, VDR ligands change the three-dimensional structure of VDR upon binding.[24] The application of a pharmacophore model based on the 1,25(OH)2D3 binding mode will favor molecules that can bind VDR in a similar way and down-score those ligands that change the conformation of VDR. Nevertheless, it was reported that those ligand have a very strong VDR affinity.[24] As a result, we have 196 false negative molecules for virtual screen 1 and 140 false negatives for virtual screen 2 using a cutoff of −6 kJ/mol for the calculated free energy of VDR binding.
ERα and ERβ ligands made up 26% of the compounds identified by virtual screen 2 followed by TR ligands (7%) and LxR ligands (7%). Thus, there is a relationship between VDR, TR and ER that is beyond the phylogenetic distance or NR sequence similarity. Two compounds were picked that are commercially available or in case of 3 easy to synthesize. Both compounds inhibited the interaction between VDR and coactivator peptide SRC2-3 although at different concentrations. The calculated free energy of VDR binding correlated with the IC50 values observed for the newly identified VDR antagonists. NDGA exhibited the best VDR affinity and largest free energy of VDR binding followed by H6036 and 3, respectively. Interestingly, triiodothyronine was not among the hit compounds because it failed to satisfy the pharmacophore used for virtual screen 2. Nevertheless, we confirmed triiodothyronine as a weak VDR antagonist highlighting the fact again that other pharmacophore models exist to identify VDR ligands. An alternative approach to develop a new pharmacophore for a virtual screen could include the application of a VDR crystal structure bound to a VDR ligand with a low calculated free energy of VDR binding for the virtual screen 1 but an excellent reported affinity for VDR. Another approach could include the optimization of our current pharmacophore model by changing the volume of the donor/acceptor elements or by adding additional pharmacophore elements.
Overall, the development of NR-specific pharmacophore models is important because it can assist in the choice of NRs that should be evaluated in order to determine NR-selectivity of novel NR ligands. Although this approach can drastically decrease the cost and time to determine NR-selectivity of newly synthesized ligands it is not a full substitute for an exhaustive investigation of a comprehensive panel of NRs in respect to agonism and antagonism. In addition, NR-specific pharmacophore models can be used to identify new NR ligands. We demonstrate the utility of this approach using a library of NR ligands to identify new VDR antagonists. The application of larger virtual compound libraries such as the “Zinc Library” might result in the identification of more compounds that interact with VDR. Overall, it can be concluded that virtual screening can support both the identification of new NR ligands as well as the identification of NRs that are likely to interact with NR ligands in order to accelerate the determination of NR selectivity.
ACKNOWLEDGEMENT
This work was supported by the University of Wisconsin-Milwaukee [LAA], the UWM Research Growth Initiative (RGI grant 2012) [LAA], the NIH R03DA031090 [LAA], the UWM Research Foundation (Catalyst grant), the Lynde and Harry Bradley Foundation [LAA], the Richard and Ethel Herzfeld Foundation [LAA]. Add RO1 AR050023 [DDB] so I can get some credit on my grant.
REFERENCES
- 1.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5(12):993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 2.Kuiper GG, Carlsson B, Grandien K, Enmark E, Haggblad J, Nilsson S, Gustafsson JA. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors alpha and beta. Endocrinology. 1997;138(3):863–870. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- 3.(a) Escriva H, Safi R, Hanni C, Langlois MC, Saumitou-Laprade P, Stehelin D, Capron A, Pierce R, Laudet V. Ligand binding was acquired during evolution of nuclear receptors. Proc Natl Acad Sci U S A. 1997;94(13):6803–6808. doi: 10.1073/pnas.94.13.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Garcia-Vallve S, Palau J. Nuclear receptors, nuclear-receptor factors, and nuclear-receptor-like orphans form a large paralog cluster in Homo sapiens. Mol Biol Evol. 1998;15(6):665–682. doi: 10.1093/oxfordjournals.molbev.a025970. [DOI] [PubMed] [Google Scholar]
- 4.Schapira M, Abagyan R, Totrov M. Nuclear hormone receptor targeted virtual screening. J Med Chem. 2003;46(14):3045–3059. doi: 10.1021/jm0300173. [DOI] [PubMed] [Google Scholar]
- 5.Boehm MF, Fitzgerald P, Zou A, Elgort MG, Bischoff ED, Mere L, Mais DE, Bissonnette RP, Heyman RA, Nadzan AM, Reichman M, Allegretto EA. Novel nonsecosteroidal vitamin D mimics exert VDR-modulating activities with less calcium mobilization than 1,25-dihydroxyvitamin D3. Chem Biol. 1999;6(5):265–275. doi: 10.1016/S1074-5521(99)80072-6. [DOI] [PubMed] [Google Scholar]
- 6.Rochel N, Wurtz JM, Mitschler A, Klaholz B, Moras D. The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand. Mol Cell. 2000;5(1):173–179. doi: 10.1016/s1097-2765(00)80413-x. [DOI] [PubMed] [Google Scholar]
- 7.Teichert A, Arnold LA, Otieno S, Oda Y, Augustinaite I, Geistlinger TR, Kriwacki RW, Guy RK, Bikle DD. Quantification of the vitamin D receptor-coregulator interaction. Biochemistry. 2009;48(7):1454–1461. doi: 10.1021/bi801874n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. http://www.bindingdb.org/bind/index.jsp. [Google Scholar]
- 9.Michellys PY, Boehm MF, Chen JH, Grese TA, Karanewsky DS, Leibowitz MD, Liu S, Mais DA, Mapes CM, Reifel-Miller A, Ogilvie KM, Rungta D, Thompson AW, Tyhonas JS, Yumibe N, Ardecky RJ. Design and synthesis of novel RXR-selective modulators with improved pharmacological profile. Bioorg Med Chem Lett. 2003;13(22):4071–4075. doi: 10.1016/j.bmcl.2003.08.048. [DOI] [PubMed] [Google Scholar]
- 10.Burris TP, Montrose C, Houck KA, Osborne HE, Bocchinfuso WP, Yaden BC, Cheng CC, Zink RW, Barr RJ, Hepler CD, Krishnan V, Bullock HA, Burris LL, Galvin RJ, Bramlett K, Stayrook KR. The hypolipidemic natural product guggulsterone is a promiscuous steroid receptor ligand. Mol Pharmacol. 2005;67(3):948–954. doi: 10.1124/mol.104.007054. [DOI] [PubMed] [Google Scholar]
- 11.Owsley E, Chiang JY. Guggulsterone antagonizes farnesoid X receptor induction of bile salt export pump but activates pregnane X receptor to inhibit cholesterol 7alpha-hydroxylase gene. Biochem Biophys Res Commun. 2003;304(1):191–195. doi: 10.1016/s0006-291x(03)00551-5. [DOI] [PubMed] [Google Scholar]
- 12.von Geldern TW, Tu N, Kym PR, Link JT, Jae HS, Lai C, Apelqvist T, Rhonnstad P, Hagberg L, Koehler K, Grynfarb M, Goos-Nilsson A, Sandberg J, Osterlund M, Barkhem T, Hoglund M, Wang J, Fung S, Wilcox D, Nguyen P, Jakob C, Hutchins C, Farnegardh M, Kauppi B, Ohman L, Jacobson PB. Liver-selective glucocorticoid antagonists: a novel treatment for type 2 diabetes. J Med Chem. 2004;47(17):4213–4230. doi: 10.1021/jm0400045. [DOI] [PubMed] [Google Scholar]
- 13.Sznaidman ML, Haffner CD, Maloney PR, Fivush A, Chao E, Goreham D, Sierra ML, LeGrumelec C, Xu HE, Montana VG, Lambert MH, Willson TM, Oliver WR, Jr, Sternbach DD. Novel selective small molecule agonists for peroxisome proliferator-activated receptor delta (PPARdelta)--synthesis and biological activity. Bioorg Med Chem Lett. 2003;13(9):1517–1521. doi: 10.1016/s0960-894x(03)00207-5. [DOI] [PubMed] [Google Scholar]
- 14.Nandhikonda P, Yasgar A, Baranowski AM, Sidhu PS, McCallum MM, Pawlak AJ, Teske K, Feleke B, Yuan NY, Kevin C, Bikle DD, Ayers SD, Webb P, Rai G, Simeonov A, Jadhav A, Maloney D, Arnold LA. Peroxisome proliferation-activated receptor delta agonist GW0742 interacts weakly with multiple nuclear receptors, including the vitamin D receptor. Biochemistry. 2013;52(24):4193–4203. doi: 10.1021/bi400321p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holick MF, Schnoes HK, DeLuca HF, Suda T, Cousins RJ. Isolation and identification of 1,25-dihydroxycholecalciferol. A metabolite of vitamin D active in intestine. Biochemistry. 1971;10(14):2799–2804. doi: 10.1021/bi00790a023. [DOI] [PubMed] [Google Scholar]
- 16.Norman AW. Evidence for a new kidney-produced hormone, 1,25-dihydroxycholecalciferol, the proposed biologically active form of vitamin D. Am J Clin Nutr. 1971;24(11):1346–1351. doi: 10.1093/ajcn/24.11.1346. [DOI] [PubMed] [Google Scholar]
- 17.Barycki R, Sicinski RR, Plum LA, Grzywacz P, Clagett-Dame M, Deluca HF. Removal of the 20-methyl group from 2-methylene-19-nor-(20S)-1alpha,25-dihydroxyvitamin D(3) (2MD) selectively eliminates bone calcium mobilization activity. Bioorg Med Chem. 2009;17(22):7658–7669. doi: 10.1016/j.bmc.2009.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu JM, Nurko J, Weakley SM, Jiang J, Kougias P, Lin PH, Yao Q, Chen C. Molecular mechanisms and clinical applications of nordihydroguaiaretic acid (NDGA) and its derivatives: an update. Med Sci Monit. 2010;16(5):RA93–RA100. [PMC free article] [PubMed] [Google Scholar]
- 19.Lack NA, Axerio-Cilies P, Tavassoli P, Han FQ, Chan KH, Feau C, LeBlanc E, Guns ET, Guy RK, Rennie PS, Cherkasov A. Targeting the binding function 3 (BF3) site of the human androgen receptor through virtual screening. J Med Chem. 2011;54(24):8563–8573. doi: 10.1021/jm201098n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sicinski RR, Prahl JM, Smith CM, DeLuca HF. New 1alpha,25-dihydroxy-19-norvitamin D3 compounds of high biological activity: synthesis and biological evaluation of 2-hydroxymethyl, 2-methyl, and 2-methylene analogues. J Med Chem. 1998;41(23):4662–4674. doi: 10.1021/jm9802618. [DOI] [PubMed] [Google Scholar]
- 21.Inaba Y, Yoshimoto N, Sakamaki Y, Nakabayashi M, Ikura T, Tamamura H, Ito N, Shimizu M, Yamamoto K. A new class of vitamin D analogues that induce structural rearrangement of the ligand-binding pocket of the receptor. J Med Chem. 2009;52(5):1438–1449. doi: 10.1021/jm8014348. [DOI] [PubMed] [Google Scholar]
- 22.(a) Mita Y, Dodo K, Noguchi-Yachide T, Hashimoto Y, Ishikawa M. Structure-activity relationship of benzodiazepine derivatives as LXXLL peptide mimetics that inhibit the interaction of vitamin D receptor with coactivators. Bioorg Med Chem. 2013;21(4):993–1005. doi: 10.1016/j.bmc.2012.11.042. [DOI] [PubMed] [Google Scholar]; (b) Mita Y, Dodo K, Noguchi-Yachide T, Miyachi H, Makishima M, Hashimoto Y, Ishikawa M. LXXLL peptide mimetics as inhibitors of the interaction of vitamin D receptor with coactivators. Bioorg Med Chem Lett. 2010;20(5):1712–1717. doi: 10.1016/j.bmcl.2010.01.079. [DOI] [PubMed] [Google Scholar]
- 23.(a) Nandhikonda P, Lynt WZ, McCallum MM, Ara T, Baranowski AM, Yuan NY, Pearson D, Bikle DD, Guy RK, Arnold LA. Discovery of the first irreversible small molecule inhibitors of the interaction between the vitamin D receptor and coactivators. J Med Chem. 2012;55(10):4640–4651. doi: 10.1021/jm300460c. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sidhu PS, Nassif N, McCallum MM, Teske K, Feleke B, Yuan NY, Nandhikonda P, Cook JM, Singh RK, Bikle DD, Arnold LA. Development of Novel Vitamin D Receptor–Coactivator Inhibitors. ACS Med. Chem. Lett. 2014;5(2):199–204. doi: 10.1021/ml400462j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlberg C, Molnar F, Mourino A. Vitamin D receptor ligands: the impact of crystal structures. Expert Opin Ther Pat. 2012;22(4):417–435. doi: 10.1517/13543776.2012.673590. [DOI] [PubMed] [Google Scholar]