Abstract
Genetic diversity was measured by allozyme electrophoresis in eight natural populations of the threatened Canarian endemic Viola palmensis Webb & Berth. (Violaceae). Nineteen alleles corresponding to 11 gene loci were detected. High levels of genetic diversity were found, ranging from 36·3 to 45·4 % for the percentage of polymorphic loci (P), from 1·45 to 1·60 for the average number of alleles per locus (A) and from 0·128 to 0·200 for the expected heterozygosity (He). Between 85·5 and 96·6 % of genetic variability was apportioned within populations. As a whole, populations were not at Hardy–Weinberg equilibrium, with a deficit of heterozygous individuals attributable to the existence of genetic structuring in the populations analysed. The levels of interpopulation genetic differentiation were low (mean FST = 0·100), while genetic identity pair‐wise comparisons were high (mean I = 0·973) suggesting considerable levels of gene flow among populations. No relationship was detected between genetic differentiation and geographical distances between populations. An outcrossing insect‐mediated breeding system might contribute to pollen dispersion of this species. For conservation genetics we suggest in situ preservation areas are defined that are free of disturbance and that include populations with the highest genetic diversity.
Key words: Allozymes, Canary Islands, conservation genetics, gene flow, genetic diversity, Viola palmensis
INTRODUCTION
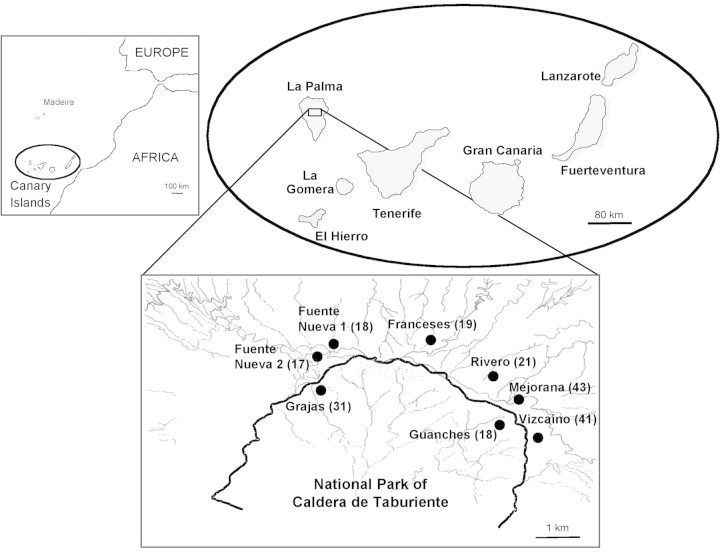
The Canary Islands consist of seven oceanic islands close to the African Continent (Fig. 1). They are reasonably accessible to continental colonizers and to multiple introductions by closely related taxonomic groups. As in other oceanic islands, the Canaries are characterized by a large number of endemic species as a result of apparent extensive speciation, putatively due to adaptive radiation and genetic drift (Francisco‐Ortega et al., 2000; Sosa, 2001). However, more than 20 % of the approx. 570 flowering plant species endemic to the Canaries (Santos‐Guerra, 1999) are endangered (Gómez‐Campo, 1996). The basic explanations offered for endangerment are habitat destruction or fragmentation (Young et al., 1996), the impact of non‐native animals and plants, and small and disjunct population sizes (García‐Casanova et al., 2001). On oceanic islands, however, negative human impact is considered to be the primary cause of extinction (Rieseberg and Swensen, 1996). In the Canaries, these factors have been acting together for the last four decades, resulting in degradation of insular habitats and reduction of population size of many endemic species (Francisco‐Ortega et al., 2000; Sosa, 2001; Bouza et al., 2002).
Fig. 1. Location of Viola palmensis populations studied. Number of individuals analysed in parentheses.
Theoretically, maintenance of sufficient genetic diversity within and among populations is crucial for the long‐term survival of most species. Thus, the loss of genetic variability associated with a reduction in the population size of the predominantly outcrossing plant groups in the Canaries could lead to both inbreeding depression and decreased fitness (Francisco‐Ortega et al., 2000). This could reduce the ability of endemic species to compete with alien species, to cope with habitat disturbance and to adapt to environmental change, thereby increasing the probability of extinction of small populations (Ellstrand and Elam, 1993).
Studies on genetic diversity of endangered species using molecular polymorphisms have increased in frequency in recent years (Batista et al., 2001; Freville et al., 2001; Bouza et al., 2002) because of their central importance in planning in situ and ex situ conservation efforts (Amos and Hoelzel, 1992; Francisco‐Ortega et al., 2000; Francisco‐Ortega and Santos, 2001; Sosa, 2001). Viola palmensis Webb & Berth. is one of four known endemics of the genus Viola in the Canaries; its populations are found near to the National Park of Caldera de Taburiente (Fig. 1). This species, commonly named the summit pansy due to its occurrence at high altitudes (1800–2400 m a.s.l.), was catalogued as vulnerable by the International Union for the Conservation of Nature (Walter and Guillet, 1998), and as sensitive to habitat alterations by the Regional Catalogue of Threatened Species from the Canaries (BOC, 2001), in recognition of its distribution being restricted to La Palma island. Viola palmensis is a herbaceous and long‐lived perennial species. Individuals of V. palmensis, like those of other Viola species (Schellner et al., 1982), can be maintained in a population for a long time as a result of the perennating root system (A. Palomares, pers. comm.).
The genus Viola has been studied by researchers in many disciplines. Although there are no published studies on V. palmensis, two species of the genus have been the subject of research in the Canaries. Calero and Santos (1988, 1993) assessed the reproductive biology of V. anagae and V. cheiranthifolia, finding moderate levels of compatibility for both species, but with allogamy and asexual reproduction by stolons in the former, and low levels of autogamy in the latter. In contrast, continental species of Viola have been much studied with a primary focus on their demography (Newell et al., 1981), spatial pattern of ramets and seedlings (Schellner et al., 1982), and plant size and fitness (Antlfinger et al., 1985). With respect to population genetics, allozyme electrophoresis (Kim et al., 1991; Marcussen and Nordal, 1998; Nordal and Jonsell, 1998; Marcussen and Borgen, 2000) and random amplified polymorphic DNA (RAPD) markers (Ko et al., 1998; Oh et al., 1998; Neuffer et al., 1999) have been used to analyse within‐species genetic variation and relationships between many continental Viola species. Other researchers have compared two different markers (allozyme and ISSR) to assess the population genetic structure of the studied species (Culley and Wolfe, 2001).
Of the molecular techniques available, allozyme electrophoresis was deemed adequate to estimate the amount of genetic variation that resides within and among plant populations. This technique is amenable for conservation genetic surveys because data can be obtained quickly for many individuals and it provides single‐gene molecular markers (alleles) that are biparentally inherited and that generally adjust to a codominant pattern of expression, which is readily checked from the band intensities in heterozygous individuals. There are many examples of allozymic variability assessments that have contributed to the conservation genetics (Williamson and Werth, 1999; Batista et al., 2001; Sosa, 2001) of different plant families.
The goal of the present work was to assess the genetic variation contained within and among populations of the Canarian endemic Viola palmensis using allozyme electrophoresis. Such information can be used to determine management strategies for the conservation genetics of this threatened species and to assess which populations are priorities for preservation measures.
MATERIALS AND METHODS
Plant material
Between 17 and 43 individuals from eight natural populations of Viola palmensis were collected from around the National Park of Caldera de Taburiente (Fig. 1). Due to the threatened status of V. palmensis, only a few young leaves were taken from each adult individual sampled. These were transported in a hand cooler to the laboratory, where they were stored at –80 °C until required. Populations analysed represent the regions in which V. palmensis is commonly found. Although some of the population‐level sample sizes appear small, these samples represent a large proportion (more than 50 %) of the total specimens in each locality.
Allozyme electrophoresis
Each leaf sample (approx. 1 cm2) was ground to a fine powder with liquid nitrogen in a mortar. Extraction buffer [0·1 m Tris, pH 7·5; 1 % ascorbic acid; 10 mm MgCl2; 20 mm diethyldithiocarbamic acid; 0·1 % mercaptoethanol; 5 % polyvinylpyrrolidone] (1 : 3 w/v) was then added. Extracts were adsorbed onto Whatman No. 1 filter papers and subjected to a horizontal 14 % starch gel (containing 3 % sucrose) electrophoresis that varied in duration depending on the buffer system used. After electrophoresis, gels were sliced into layers that underwent a specific staining procedure following the protocols reported by Wendel and Weeden (1989) with slight modifications. Twenty‐eight enzyme systems were assayed using 12 different gel and electrode buffers. The seven enzyme systems with the best resolution were obtained from the following buffer combinations: (1) Electrode buffer consisting of 40 mm citric acid adjusted to pH 6·5 with N‐(3‐aminopropyl)‐morpholine, diluted 1 : 19 as a gel buffer. This buffer system was utilized to reveal phosphoglucoisomerase (PGI, EC 3.4.11.1), isocitrate dehydrogenase (IDH, EC 1.1.1.42), peroxidase (PRX, EC 1.11.1.7) and shikimate dehydrogenase (SKD, EC 1.1.1.25); (2) Electrode buffer of 0·065 m l‐histidine and 0·019 m citric acid, pH 6·5, diluted 1 : 6 as a gel buffer. This system was used to resolve phosphoglucomutase (PGM, EC 2.7.5.1), phosphogluconate dehydrogenase (PGD, EC 1.1.1.44) and glucose‐6‐phosphate dehydrogenase (G6PDH, EC 1.1.1.49). The genetic interpretation of the relative mobility for the observed bands led us to assign an ‘a’ to the most anodal allele, and a letter following the alphabetic sequence to each subsequent (slower) cathodal allele. The isozymes of each given enzyme were also numbered sequentially following a similar methodology. The genotypes were inferred according to knowledge of quaternary structure and the number of loci per enzyme system reported for other plants (Weeden and Wendel, 1989).
Data analysis
Genstat‐PC 3.3 (Lewis, 1993) was used to calculate the basic descriptors of allozymic variability. FSTAT (Goudet, 2000) was utilized for estimating and testing Wright’s F‐statistics (Wright, 1965), and calculating intra‐ and interpopulation apportionment of allozymic variation (Nei, 1973). The FST pair‐wise values among populations and the value of FST overall (based on Weir and Cockerham, 1984) were estimated using GENEPOP (Raymond and Rousset, 1995). A Mantel–Cox test was performed between genetic differentiation and geographic distances among pairs of populations to determine a possible ‘isolation by distance’ pattern of differentiation. In addition, estimates of genetic similarity between populations were subjected to a principal component analysis (PCA) using the software SPSS 11.0 (SPSS, Chicago).
RESULTS
Allelic frequencies
Although the chromosome number and ploidy of Viola palmensis have not been described (Nadot et al., 2000), the banding patterns correspond to a diploid plant species, as described by Weeden and Wendel (1989). Eleven putative loci and 19 alleles (Table 1) were discerned from banding patterns. Two loci were identified for PGI, PGM, PRX and PGD, whereas SKD, IDH and G6PDH were coded by only one locus. Six loci (Pgi‐1, Pgm‐2, Skd, Idh, Pgd‐1 and G6pdh) were monomorphic in all populations. Patterns of allelic distribution and the number of alleles per locus varied depending on the population. For the polymorphic loci there was at least one common allele at high frequency in most populations analysed. No one population displayed all the allelic variation found in the species. Fifteen alleles (79 %) were found in all populations (with variable frequency), but two populations (Vizcaíno and Mejorana) had 18 of the 19 alleles observed. Thus, 94·7 % of the allelic diversity detected in Viola palmensis was located in these two populations. Finally, only one allele (Prx‐1c) was exclusive to one population (Grajas), but with low frequency (Table 1).
Table 1.
Allelic frequencies estimated at 11 loci in the eight populations of Viola palmensis studied
| Populations | |||||||||
| Locus | Allele | Grajas | Fuente Nueva 1 | Fuente Nueva 2 | Guanches | Rivero | Vizcaíno | Mejorana | Franceses |
| Pgi‐1 | 1·000 | 1·000 | 1·000 | 1·000 | 1·000 | 1·000 | 1·000 | 1·000 | |
| (31) | (17) | (18) | (18) | (21) | (41) | (43) | (19) | ||
| Pgi‐2 | a | 0·565 | 0·853 | 0·778 | 0·778 | 0·806 | 0·736 | 0·628 | 0·500 |
| b | 0·435 | 0·147 | 0·222 | 0·222 | 0·194 | 0·264 | 0·372 | 0·500 | |
| (31) | (17) | 18) | (18) | (18) | (36) | (39) | (19) | ||
| Pgm‐1 | a | 0·435 | 0·692 | 0·542 | 0·750 | 0·275 | 0·760 | 0·500 | 0·921 |
| b | 0·565 | 0·308 | 0·458 | 0·250 | 0·725 | 0·240 | 0·500 | 0·079 | |
| (23) | (13) | (12) | (8) | (20) | (25) | (23) | (19) | ||
| Pgm‐2 | 1·000 | 1·000 | 1·000 | 1·000 | 1·000 | 1·000 | 1·000 | 1·000 | |
| (31) | (17) | (18) | (18) | (21) | (41) | (43) | (19) | ||
| Prx‐1 | a | 0·000 | 0·000 | 0·028 | 0·063 | 0·206 | 0·167 | 0·067 | 0·000 |
| b | 0·964 | 1·000 | 0·972 | 0·938 | 0·794 | 0·833 | 0·933 | 1·000 | |
| c | 0·036 | 0·000 | 0·000 | 0·000 | 0·000 | 0·000 | 0·000 | 0·000 | |
| (28) | (15) | (18) | (16) | (17) | (39) | (30) | (19) | ||
| Prx‐2 | a | 0·000 | 0·033 | 0·000 | 0·000 | 0·000 | 0·103 | 0·033 | 0·000 |
| b | 0·321 | 0·633 | 0·333 | 0·281 | 0·382 | 0·487 | 0·533 | 0·447 | |
| c | 0·589 | 0·333 | 0·667 | 0·594 | 0·559 | 0·282 | 0·300 | 0·368 | |
| d | 0·089 | 0·000 | 0·000 | 0·125 | 0·059 | 0·128 | 0·133 | 0·184 | |
| (28) | (15) | (18) | (16) | (17) | (39) | (30) | (19) | ||
| Skd | 1·000 | 1·000 | 1·000 | 1·000 | 1·000 | 1·000 | 1·000 | 1·000 | |
| (31) | (17) | (18) | (18) | (21) | (41) | (43) | (19) | ||
| Idh | 1·000 | 1·000 | 1·000 | 1·000 | 1·000 | 1·000 | 1·000 | 1·000 | |
| (31) | (17) | (18) | (18) | (21) | (41) | (43) | (19) | ||
| Pgd‐1 | 1·000 | 1·000 | 1·000 | 1·000 | 1·000 | 1·000 | 1·000 | 1·000 | |
| (31) | (17) | (18) | (18) | (21) | (41) | (43) | (19) | ||
| Pgd‐2 | a | 0·103 | 0·292 | 0·222 | 0·028 | 0·190 | 0·597 | 0·395 | 0·053 |
| b | 0·897 | 0·708 | 0·778 | 0·972 | 0·810 | 0·403 | 0·605 | 0·947 | |
| (29) | (12) | (18) | (18) | (21) | (36) | (38) | (19) | ||
| G6pdh | 1·000 | 1·000 | 1·000 | 1·000 | 1·000 | 1·000 | 1·000 | 1·000 | |
| (31) | (17) | (18) | (18) | (21) | (41) | (43) | (19) |
Number of individuals analysed in parentheses.
Genetic diversity
High levels of allozyme diversity were detected. The average number of alleles per locus (A) ranged from 1·45 to 1·64 in the different populations (mean = 1·53, s.e. = 0·0268). The average expected heterozygosity (He) was 0·164 (s.e. = 0·0095), whereas the average percentage of polymorphic loci (P, 0·95 criterion) was 43·2 % (s.e. = 0·0149) (Table 2). The populations Vizcaíno and Mejorana were more genetically variable than the others; the lowest level of variability was found in the Franceses population.
Table 2.
Genetic diversity of Viola palmensis at the population level
| Population | n | H o | H e | P | A |
| Grajas | 29·5 | 0·129 | 0·165 | 45·4 | 1·54 |
| Fuente Nueva 1 | 15·8 | 0·054 | 0·149 | 36·3 | 1·45 |
| Fuente Nueva 2 | 17·5 | 0·093 | 0·158 | 45·4 | 1·45 |
| Guanches | 16·7 | 0·075 | 0·137 | 45·4 | 1·54 |
| Rivero | 19·9 | 0·074 | 0·176 | 45·4 | 1·54 |
| Vizcaíno | 38·3 | 0·115 | 0·201 | 45·4 | 1·64 |
| Mejorana | 38·0 | 0·141 | 0·201 | 45·4 | 1·64 |
| Franceses | 19·0 | 0·124 | 0·128 | 36·3 | 1·45 |
| Mean | 24·3 | 0·100 | 0·164 | 43·2 | 1·53 |
n, Mean sample size per locus; Ho, observed heterozygosity; He, expected heterozygosity; P, percentage of polymorphic loci; A, mean number of alleles per locus.
Genetic structure
The partitioning of total genetic variation was similar in all populations. Nearly all genetic variability detected was due to variation within populations, with little genetic differentiation among populations. On average, 91·7 % of the genetic variation was found within populations, ranging from 85·5 to 96·6 % depending on the locus.
Given the eight populations and five polymorphic loci, 38 valid tests were available to elucidate the possible deviation of the genotypic frequencies from Hardy–Weinberg expectations (Fis) (Table 3). Sixteen tests (42 %) indicated no significant differences from zero; 20 of the 22 statistically significant tests were positive (53 %), but only two (Pgi‐2 in the populations Mejorana and Franceses) were negative. On average, Fis showed a significant deficiency of heterozygotes (Fis > 0) for all populations. Overall, these results demonstrate that natural populations of Viola palmensis are not in Hardy–Weinberg equilibrium.
Table 3.
FIS values at polymorphic loci in natural populations of Viola palmensis
| Population | ||||||||
| Loci | Grajas | Fuente Nueva 1 | Fuente Nueva 2 | Guanches | Rivero | Vizcaíno | Mejorana | Franceses |
| Pgi‐2 | –0·231 | –0·143 | –0·259 | 0·064 | –0·214 | 0·227 | –0·361* | –0·561* |
| Pgm‐1 | –0·039 | 0·662* | 0·203 | 1·000* | 0·395 | 0·360 | 0·410 | –0·059 |
| Prx‐1 | 1·000* | – | 0·000 | –0·034 | 0·830** | 0·365* | –0·055 | – |
| Prx‐2 | 0·616*** | 0·872*** | 1·000*** | 0·460* | 0·793*** | 0·308** | 0·410*** | 0·356* |
| Pgd‐2 | 0·639* | 0·814* | 0·694* | 0·000 | 1·000*** | 0·831*** | 0·785*** | 1·000* |
| All | 0·221*** | 0·644*** | 0·418*** | 0·464** | 0·586*** | 0·426*** | 0·303*** | 0·032*** |
*** P < 0·001; ** P < 0·01; * P < 0·05.
–, Monomorphic locus.
Pair‐wise comparisons of the genetic differentiation coefficient (FST) among populations for all loci ranged from 0·006 (between the populations Fuente Nueva 2 and Guanches) to 0·241 (between Franceses and Rivero) (Table 4), with a mean value of FST = 0·100 for the species. These values suggest high levels of interpopulational genetic exchange.
Table 4.
Pair‐wise values for FST found between populations of Viola palmensis
| Grajas | Fuente Nueva 1 | Fuente Nueva 2 | Guanches | Rivero | Vizcaíno | Mejorana | |
| Fuente Nueva 1 | 0·106 | ||||||
| (0·208) | |||||||
| Fuente Nueva 2 | 0·012 | 0·026 | |||||
| (0·022) | (0·046) | ||||||
| Guanches | 0·051 | 0·057 | 0·006 | ||||
| (0·093) | (0·095) | (0·009) | |||||
| Rivero | 0·037 | 0·097 | 0·017 | 0·099 | |||
| (0·071) | (0·196) | (0·032) | (0·194) | ||||
| Vizcaíno | 0·171 | 0·039 | 0·105 | 0·145 | 0·156 | ||
| (0·424) | (0·084) | (0·242) | (0·339) | (0·393) | |||
| Mejorana | 0·054 | 0·019 | 0·040 | 0·099 | 0·056 | 0·037 | |
| (0·117) | (0·041) | (0·086) | (0·220) | (0·127) | (0·087) | ||
| Franceses | 0·127 | 0·123 | 0·146 | 0·065 | 0·241 | 0·159 | 0·125 |
| (0·239) | (0·208) | (0·265) | (0·098) | (0·539) | (0·366) | (0·273) |
Standard errors in parentheses.
In an ‘isolation by distance’ model, genetic similarities decrease when geographical distances between populations increase (Wright, 1943). However, the genetic resemblance and the geographical proximity did not appear significantly related in this study of Viola palmensis, as shown by the negative slope and non‐significance (r2 = 0·023) of the regression (data not shown). In fact, the lowest FST values (0·006–0·171) were detected among populations separated by more than 4 km, whereas populations separated by less than 2 km showed the highest levels of genetic differentiation (0·012–0·241; Table 4).
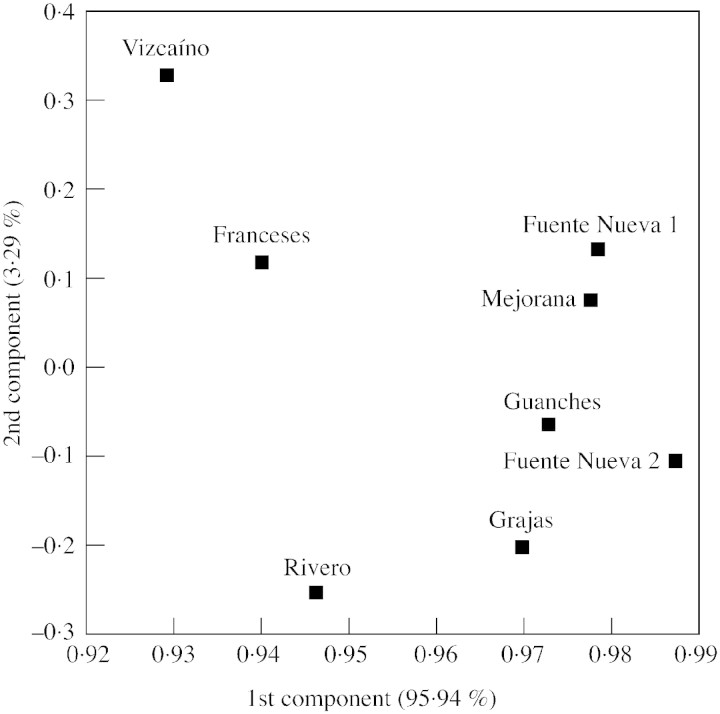
The PCA ordination based on allelic frequencies (Fig. 2) accounted for 95·94 % of genetic differentiation detected among populations. Most populations of Viola palmensis were located very close together, with only the Franceses population being slightly separate from the others.
Fig. 2. Principal component analysis of Viola palmensis populations. Percentages in parentheses indicate the proportion of total variation explained by each component.
DISCUSSION
Genetic diversity
Viola palmensis displays high levels of genetic diversity (P = 43·2; A = 1·53; He = 0·164) (Table 2). These values are similar to the average values reported by Hamrick and Godt (1989) for widespread species (P = 43·0; A = 1·72; He = 0·159). Thus, V. palmensis belongs to the subset of endemics possessing high levels of genetic variability (Lewis and Crawford, 1995; Smith and Pham, 1996).
It has been established that narrowly distributed endemic species have lower levels of genetic diversity than species with an extensive geographic distribution (Hamrick and Godt, 1989). So, considering the narrow distribution of Viola palmensis (in an area of about 15 km2 exclusively in La Palma island), the amounts of genetic variability detected are particularly notable. Culley and Wolfe (2001) found high levels of genetic variation in the continental species V. pubescens (P = 66·7 %; A = 2·6; He = 0·32), and argued that such levels may represent the effect of outcrossing through chasmogamous flowers. Although some authors (Kim et al., 1991; Marcussen and Nordal, 1998; Nordal and Jonsell, 1998; Marcussen and Borgen, 2000) have detected low levels of allozyme variation within other Viola species, V. palmensis seems to constitute a new example of the small set of instances in which genetic diversity in an endemic plant from the Macaronesian islands is greater than that detected in most species from oceanic archipelagos (Francisco‐Ortega et al., 2000; Batista et al., 2001).
Some authors have proposed recent speciation (Aradhya et al., 1991) or multiple origins for various oceanic species (Smith and Pham, 1996) as potential explanations for the high levels of genetic diversity detected within narrowly distributed endemic species. Francisco‐Ortega et al. (2000) proposed the occurrence of an old lineage refuge in the Canaries and a possible origin by multiple colonization from continental taxa to explain the high genetic diversity shown by some Canarian endemics. However, phylogenetic relationships in the pansies and related groups, discerned using internal transcribed spacer DNA sequences, suggest that the Section Melanium, to which Viola palmensis belongs, is probably very recently derived and has radiated very rapidly (H. Ballard, pers. comm.). There are other plausible explanations for the high levels of genetic variability found in Viola palmensis. This species may be considered to be perennial, maintaining individuals in a population for long periods through perennating roots. Also, evidence suggests that populations of V. palmensis have not experienced a recent bottleneck. Moreover, although the reproductive biology of V. palmensis has been not studied, the high levels of intraspecific variation detected could be an outcome of cross‐pollination, as found in other Viola species (Ko et al., 1998; Culley and Wolfe, 2001).
Genetic structure
In general, species that show self‐fertilization or that undergo outcrossing among genetically related individuals have an excess of homozygotes in their natural populations. In contrast, species with a strictly allogamous breeding system usually show a percentage of heterozygous individuals in agreement with the Hardy–Weinberg equilibrium (Hartl and Clark, 1997).
Viola palmensis exhibits highly significant heterozygote deficiency in most loci (FIS > 0; Table 3). These results cannot be explained by chance, and there are two main potential causes: first, the existence of genetic structure in the populations analysed (Wahlund effect); and secondly, a breeding system with a tendency to self‐pollination due to restricted pollinator movements or reduced pollinator activity. Outcomes suggest that populations may be genetically structured, i.e. they are composed of subpopulations within which mating is nearly random, but between which mating may be infrequent. Such structuring is frequently reported for insect‐pollinated herbs (Heywood, 1991; Williamson and Werth, 1999) and, in most rare species, large discrepancies exist between mean observed and mean expected heterozygosity values (Ellstrand and Ellam, 1993; Hedrick, 2000).
Only a hierarchical sampling of individuals or an exhaustive analysis of the reproductive biology of Viola palmensis could test the hypotheses proposed and specify the factors (Wahlund effect vs. inbreeding through restricted pollinator movement or reduced pollinator visitation) that determine the excess of homozygotes. Since low levels of autogamy have been described for another Canarian endemic (V. cheiranthifolia; Calero and Santos, 1993), we cannot ignore the fact that there may be some level of inbreeding in V. palmensis. On the other hand, cleistogamy has not been described for the Section Melanium (H. Ballard, pers. comm.). This, together with the high levels of intrapopulation genetic differentiation and the low levels of interpopulation genetic differentiation detected, suggests that a genetic structure is responsible for deviations in Hardy–Weinberg equilibrium gene frequencies.
Gene flow and genetic drift
The proportion of the total genetic diversity found among populations of Viola palmensis was similar to that described by Hamrick and Godt (1989) in species with a mixed, wind‐mediated breeding system, and in species with an outcrossing wind‐breeding system. The range of pair‐wise FST values (0·006–0·241; mean FST = 0·100) was small and showed weak differentiation among natural populations of V. palmensis (Table 4); Culley and Wolfe (2001) reported a higher value (FST = 0·290) for V. pubescens.
The estimate of gene flow, based on the mean FST value, suggests that substantial genetic exchange between natural populations of Viola palmensis is sufficient to counteract genetic drift (Slatkin, 1987) or, alternatively, that original genetic variation in the narrow populations has been maintained. High gene flow among populations is supported by the PCA ordination, which groups all populations together. Although we have no data on the pollination mechanism or pollinator(s) of V. palmensis, a possible insect‐mediated pollination involving Hymenoptera as pollinators (Calero and Santos, 1988) would result in relatively frequent long‐distance pollen dispersion, contributing to a low genetic differentiation among populations. However, gene flow is a collective term that not only represents the amount of pollen flow, but also includes movement of seeds and, more rarely in terrestrial plants, movement of individuals. As Slatkin (1987) noted, if the geographic distribution of a species remains the same and if local populations persist for a long time, as seems to occur in V. palmensis, then gene flow (if it actively takes place) occurs primarily through the movement of individual propagules (seeds) or pollen grains between established populations. However, in V. palmensis, as in other Viola species (Beattie, 1978), seeds are dispersed ballistically a few metres within populations. At this point, the action of an outcrossing breeding system and a possible insect‐mediated pollination could explain the low levels of genetic differentiation detected among V. palmensis populations. Alternatively, gradual habitat fragmentation and isolation of populations may not yet have impacted on differentiation of populations if within‐ and between‐population gene flow has occurred.
As noted in other studies (Young et al., 1996), there are some circumstances in which gene flow among populations can be increased by habitat fragmentation. Despite the presumed present action of gene flow, we cannot reject the possibility that the currently known populations of summit pansy are a set of fragmented populations or relics of a formerly extended population that have maintained their historic variation. The strong pressure exerted by man and introduced herbivores during the last few decades could have reduced that much larger population to the present distribution. Thus, the results obtained are also interpretable in this way: i.e. Viola palmensis still has low levels of genetic differentiation due to the maintenance of variation in the populations over a long period and to the lack of sufficient time since fragmentation for processes such as genetic drift and inbreeding to erode genetic variation or to generate genetic differentiation among populations.
In the case of the Franceses population, genetic drift has taken place in this last stand. The number of individuals in this population has decreased considerably since 1997. Therefore, the genetic study was performed on all remaining individuals in the population. In general, however, except for the Franceses population, it seems that the recent evolutionary history of the natural populations of V. palmensis has involved typical processes of population expansion, dispersal by seeds and pollen, and intermittent genetic exchange between populations, mostly without impacts from stochastic processes. However, it would be necessary to know the real population sizes, the possible migration pathways and potential gene flow amongst them to determine the influence of these factors and to ascertain which have affected distribution. Also, the largely random genetic relationship among geographically close or distant populations indicates that gene flow, if active, operates at multiple scales including the landscape level, and that pollen and seed dispersal probably occur at very different scales themselves. Finally, the presence of approx. 95 % of the allelic variation in the species being found in the Vizcaíno and Mejorana populations could be interpreted as evidence for these two sites serving as the original founder colonies for the others, or at least being remnants of the early centre of the range of the species prior to the onset of habitat fragmentation.
Considerations for conservation
Knowledge about genetic variation within a taxon is crucial for conservation purposes, when interpreted within a broader ecological and organismal context, because of its implications for the long‐term survival and continued evolution of a population or species (Young and Clarke, 2000; Sosa, 2001; Frankham et al., 2002). The information gained on the levels and distribution of allozymic variation in the threatened endemic Viola palmensis can be used to suggest appropriate management strategies. The low FST values detected among populations of this species indicate that the populations are not considerably differentiated, and that moderate or high interpopulational gene flow occurs, via pollen, seeds or both.
In terms of in situ conservation, preservation areas must be defined that are free of anthropogenic or herbivore disturbance, and that include the populations that have the greatest genetic diversity. Preserving two particular populations (Vizcaíno and Mejorana) would maintain about 95 % of the allelic variation found in Viola palmensis. Moreover, if the Grajas population was also included, the allelic variation preserved would be increased to 100 %. However, it is necessary to conserve more than just three populations. Although, theoretically, almost 95 % of the total allozyme diversity detected in the species can be conserved in just two populations, it is important to bear in mind that introduced herbivores could still eliminate these populations. In addition, it should be recognized that allozymes often underestimate levels of interpopulational differentiation for adaptive traits critical to the survival and reproduction of plants (Hamrick et al., 1991; Francisco‐Ortega et al., 2000).
For ex situ conservation, our results indicate a need to collect individual seeds from every population, both from individuals a few metres apart and from others a long distance apart. In this way, a representative sample of seeds from across the population as a whole and its possible subpopulations would be assured. Obviously, collecting seed from the populations with the highest genetic diversity would increase the probability of obtaining the greatest amount of genetic variation in the germplasm bank for this endemic species.
Gene flow by transplantation would entail some risks; it is not advisable to mix seeds obtained from distinct populations as a first option (Leberg, 1993): this could generate changes in the genetic composition of the populations, producing a decreased fitness through outbreeding depression and disruption of locally adapted gene combinations (Storfer, 1999). However, our results indicate that all of the populations analysed, with the exception of Franceses, may have enough genetic variation potentially to avoid inbreeding depression, and that they do not currently require reinforcement from other populations. Both the current population sizes and the high levels of genetic variation found seem sufficient to maintain the natural genetic variation in populations of Viola palmensis. Given a weak level of genetic differentiation among populations, the introduction or movement of individuals from one population to another should not impact or affect the existing genetic structure, composition and genetic balance of the recipient population.
Finally, the results also indicate additional lines of investigation for future research. One approach would be to expand the present sampling to include other Viola species from the Canaries and some of their mainland relatives. This would clarify the probable sequence of founding events that gave rise to this and other island species, and could explain why the level of genetic variation in V. palmensis is not noticeably depressed relative to that of other oceanic island endemics. This could be done in combination with a phylogenetic approach, perhaps involving other kinds of molecular data, to test relationships among Canarian and continental species. A promising second line of investigation would be to utilize other molecular markers, such as microsatellites and RAPDs, to re‐evaluate variation in populations. A third profitable line of inquiry would be to study pollen dispersal, the pollination mechanism and insect vectors, as well as the breeding system and seed dispersal strategies in this species. An integrated research programme that combines genetic analyses with studies of reproductive biology and demography would provide valuable additional data that would extend the present conclusions.
ACKNOWLEDGEMENTS
We thank Ángel Palomares (National Park of Caldera de Taburiente) for helping to locate populations and collect individual plants; Dr Juli Caujapé‐Castells (Jardín Botánico Canario Viera y Clavijo) and Dr Miguel Ángel González‐Pérez (Universidad de Las Palmas de Gran Canaria) for technical assistance. This work was financed by Transformaciones Agrarias (TRAGSA) and Organismo Autónomo de Parques Nacionales (Parque Nacional de la Caldera de Taburiente, Ministerio de Medio Ambiente), with the collaboration of Fundación Universitaria de Las Palmas. We also thank Dr H. Ballard and an anonymous referee for their comments.
Supplementary Material
Received: 1 May 2002; Returned for revision 24 June 2002; Accepted: 29 August 2002 Published electronically: 31 October 2002
References
- AmosB, Hoelzel AR.1992. Applications of molecular genetic techniques to the conservation of small populations. Biological Conservation 61: 133–144. [Google Scholar]
- AntlfingerAE, Curtis WF, Solbrig OT.1985. Environmental and genetic determinants of plant size in Viola sororia Evolution 39: 1053–1064. [DOI] [PubMed] [Google Scholar]
- AradhyaKM, Mueller‐Dombois D, Ranker TA.1991. Genetic evidence for recent and incipient speciation in the evolution of Hawaiian Metrosideros (Myrtaceae). Heredity 67: 129–138. [Google Scholar]
- BatistaF, Bañares A, Caujapé‐Castells J, Carqué E, Marrero‐Gómez M, Sosa PA.2001. Allozyme diversity in three endemic species of Cistus (Cistaceae) from the Canary Islands: Intraspecific and interspecific comparisons and implications for genetic con servation. American Journal of Botany 88: 1582–1592. [PubMed] [Google Scholar]
- BeattieA.1978. Plant‐animal interactions affecting gene flow in Viola In: Richards AJ, ed. The pollination of flowers by insects. Linnean Society Symposium Series, Number 6. London: Academic Press, 151–164. [Google Scholar]
- BOC.2001. Decreto 151, de 23 de julio, por el que se crea el catálogo de especies amenazadas de Canarias. Boletín Oficial de Canarias 97: 11101–11111. [Google Scholar]
- BouzaN, Caujapé‐Castells J, González‐Pérez MA, Batista F, Sosa PA.2002. Population structure and genetic diversity of two endangered endemic species of the Canarian laurel forest: Dorycnium spectabile (Fabaceae) and Isoplexis chalcantha (Scrophulariaceae). International Journal of Plant Sciences 163: 619–630. [Google Scholar]
- CaleroA, Santos A.1988. Biología reproductiva de especies amenazadas en la flora Canaria. Lagascalia 15 (suppl.): 661–664. [Google Scholar]
- CaleroA, Santos A.1993. Reproductive biology of the high altitude Canarian flora. In: Demiritz H, Zhatay O, eds. Proceedings of the 5th OPTIMA Meeting, 8–15 September 1986. Istanbul, Turkey: University of Istanbul. [Google Scholar]
- CulleyTM, Wolfe AD.2001. Population genetic structure of the cleistogamous plant species Viola pubescens Aiton (Violaceae), as indicated by allozyme and ISSR molecular markers. Heredity 86: 545–556. [DOI] [PubMed] [Google Scholar]
- EllstrandNC, Ellam DR.1993. Population genetic consequences of small population size: implications for plant conservation.Annual Review of Ecology and Systematics 24: 217–242. [Google Scholar]
- Francisco‐OrtegaJ, Santos‐Guerra A.2001. Genes y conservación de plantas vasculares. In: Martín‐Esquivel J, Fernández‐Palacios JM, eds. Naturaleza de las Islas Canarias Santa Cruz de Tenerife, Canary Islands: Editorial Turquesa, 357–365. [Google Scholar]
- Francisco‐OrtegaJ, Santos‐Guerra A, Kim SC, Crawford DJ.2000. Plant genetic diversity in the Canary Islands: a conservation perspective. American Journal of Botany 87: 909–919. [PubMed] [Google Scholar]
- FrankhamR, Ballou JD, Briscoe DA.2002. Introduction to conservation genetics. Cambridge: Cambridge University Press. [Google Scholar]
- FrevilleH, Justy F, Olivieri I.2001. Comparative allozyme and microsatellite population structure in a narrow endemic plant species, Centaurea corymbosa Pourret (Asteraceae). Molecular Ecology 10: 879–889. [DOI] [PubMed] [Google Scholar]
- García‐CasanovaJ, Rodríguez‐Luengo JL, Rodríguez‐Piñero C.2001. Especies amenazadas. In: Martín‐Esquivel J, Fernández‐Palacios JM, eds. Naturaleza de las Islas Canarias Santa Cruz de Tenerife, Canary Islands: Editorial Turquesa, 167–172. [Google Scholar]
- Gómez‐CampoC.1996. Libro rojo de especies vegetales amenazadas de las Islas Canarias. Santa Cruz de Tenerife, Canary Islands: Viceconsejería de Medio Ambiente del Gobierno de Canarias. [Google Scholar]
- GoudetJ.2000. FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.1). www.unil.ch/izea/softwares/fstat.html. [Google Scholar]
- HamrickJL, Godt MJW.1989. Allozyme diversity in plant species. In: Brown HD, Clegg MT, Kahler AL, Weir BS, eds. Plant population genetics, breeding, and genetic resources Sunderland, Massachusetts: Sinauer, 43–63. [Google Scholar]
- HamrickJL, Murawski DA, Loveless MD.1991. Correlations between species traits and allozyme diversity: Implications for conservation biology. In: Falk DA, Holsinger KE, eds. Genetics and conservation of rare plants New York: Oxford University Press, 75–86. [Google Scholar]
- HartlDL, Clark AG.1997. Principles of population genetics. Sunderland, Massachusetts: Sinauer. [Google Scholar]
- HedrickPW.2000. Genetics of populations. Sudbury, Massachusetts: Jones and Barlett. [Google Scholar]
- HeywoodJS.1991. Spatial analysis of genetic variation in plant populations. Annual Review of Ecology and Systematics 22: 335–355. [Google Scholar]
- KimKS, Sun BY, Whang SS, Chung GH.1991. Biosystematic study of the genus Viola in Korea: comparative morphology of the Viola albida complex. Korean Journal of Botany 34: 229–238. [Google Scholar]
- KoMK, Yang J, Jin YH, Lee CH, Oh J.1998. Genetic relationships of Viola species evaluated by random amplified polymorphic DNA analysis. Journal of Horticultural Science and Biotechnology 73: 601–605. [Google Scholar]
- LebergPL.1993. Strategies for population reintroduction: effects of genetic variability on population growth and size. Conservation Biology 7: 194–199. [Google Scholar]
- LewisPO.1993. Genestat‐pc 3.3. Raleigh: Department of Statistics, North Carolina State University. [Google Scholar]
- LewisPO, Crawford DJ.1995. Pleistocene refugium endemics exhibit greater allozymic diversity than widespread congeners in the genus Polygonella (Polygonaceae). American Journal of Botany 82: 141–149. [Google Scholar]
- MarcussenT, Borgen L.2000. Allozymic variation and relationships within Viola subsection Viola (Violaceae). Plant Systematics and Evolution 223: 29–57. [Google Scholar]
- MarcussenT, Nordal I.1998. Viola suavis, a new species in the Nordic flora, with analyses of the relation to other species in the subsection Viola (Violaceae). Nordic Journal of Botany 18: 221–237. [Google Scholar]
- NadotS, Ballard HE, Creach JB, Dajoz I.2000. The evolution of pollen heteromorphism in Viola: a phylogenetic approach.Plant Systematics and Evolution 223: 155–171. [Google Scholar]
- NeiM.1973. Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences of the USA 70: 3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NeufferB, Auge H, Mesch H, Amarell U, Brandl R.1999. Spread of violets in polluted pine forest: morphological and molecular evidence for the ecological importance of interspecific hybridization. Molecular Ecology 8: 365–377. [Google Scholar]
- NewellSJ, Solbrig OT, Kincaid DT.1981. Studies on the population biology of the genus Viola III. The demography of Viola blanda and Viola pallens. Journal of Ecology 69: 997–1016. [Google Scholar]
- NordalI, Jonsell B.1998. A phylogeographical analysis of Viola rupestris: three post‐glacial immigration routes into the Nordic area? Botanical Journal of the Linnean Society 128: 105–122. [Google Scholar]
- OhBJ, Ko MK, Lee CH,1998. Identification of the series‐specific random amplified polymorphic DNA markers of Viola species. Plant Breeding 117: 295–296. [Google Scholar]
- RaymondM, Rousset F.1995. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity 86: 248–249. [Google Scholar]
- RiesebergLH, Swensen SM.1996. Conservation genetics of endangered island plants. In: Avise JC, Hamrick JL, eds. Conservation genetics: case studies from nature. New York: Chapman and Hall. [Google Scholar]
- Santos‐GuerraA.1999. Origen y evolución de la flora Canaria. In: Fernández‐Palacios JM, Bacallado JJ, Belmonte JA, eds. Ecología y cultura en Canarias Santa Cruz de Tenerife, Canary Islands: Museo de la Ciencia, Cabildo Insular de Tenerife, 107–130. [Google Scholar]
- SchellnerRA, Newell SJ, Solbrig OT.1982. Studies on the population biology of the genus Viola IV. Spatial pattern of ramets and seedlings in three stoloniferous species. Journal of Ecology 70: 273–290. [Google Scholar]
- SlatkinM.1987. Gene flow and the geographic structure of natural populations. Science 236: 787–792. [DOI] [PubMed] [Google Scholar]
- SmithJF, Pham TV.1996. Genetic diversity of the narrow endemic Allium aaseae (Alliaceae). American Journal of Botany 83: 717–726. [Google Scholar]
- SosaPA.2001. Genes, poblaciones y especies. In: Martín‐Esquivel J, Fernández‐Palacios JM, eds. Naturaleza de las Islas Canarias Santa Cruz de Tenerife, Canary Islands: Editorial Turquesa, 151–155. [Google Scholar]
- StorferA.1999. Gene flow and endangered species translocations: a topic revisited. Biological Conservation 87: 173–180. [Google Scholar]
- SwoffordDL, Selander RB.1981. BIOSYS‐1: a FORTRAN program for the comprehensive analysis of electrophoretic data in population genetics and systematics. Journal of Heredity 72: 281–283. [Google Scholar]
- WalterKS, Gillett HJ.1998. 1997 Red list of threatened plants. Gland, Switzerland: The World Conservation Union. [Google Scholar]
- WeedenN, Wendel JF.1989. Genetics of plant isozymes. In: Soltis D, Soltis P, eds. Isozymes in plant biology Portland: Dioscorides Press, 46–72. [Google Scholar]
- WeirBS, Cockerham CC.1984. Estimating F‐statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- WendelJF, Weeden N.1989. Visualization and interpretation of plant isozymes. In: Soltis D, Soltis P, eds. Isozymes in plant biology Portland: Dioscorides Press, 5–45. [Google Scholar]
- WilliamsonPS, Werth CR.1999. Levels and patterns of genetic variation in the endangered species Abronia macrocarpa (Nyctaginaceae). American Journal of Botany 86: 293–301. [PubMed] [Google Scholar]
- WrightS.1943. Isolation by distance. Genetics 28: 114–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WrightS.1965. The interpretation of population structure by F‐statistics with special regard to systems of mating. Evolution 19: 395–420. [Google Scholar]
- YoungA, Boyle T, Brown T.1996. The population genetic consequences of habitat fragmentation for plants. Trends in Ecology and Evolution 11: 413–418. [DOI] [PubMed] [Google Scholar]
- YoungAG, Clarke GM.2000. Genetics, demography and viability of fragmented populations. Cambridge: Cambridge University Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.