Abstract
Protein palmitoylation is a widespread lipid modification in which one or more cysteine thiols on a substrate protein are modified to form a thioester with a palmitoyl group. This lipid modification is readily reversible; a feature of protein palmitoylation that allows for rapid regulation of the function of many cellular proteins. Mutations in palmitoyltransferases (PATs), the enzymes that catalyze the formation of this modification, are associated with a number of neurological diseases and cancer progression. This review summarizes the crucial role of palmitoylation in biological systems, the discovery of the DHHC protein family that catalyzes protein palmitoylation, and the development of methods for investigating the catalytic mechanism of PATs.
Keywords: protein palmitoylation, palmitoyltransferase (PAT), DHHC protein family, catalytic mechanism, palmitoylation assay
1 Introduction
As many as 25–40% of eukaryotic cellular proteins are estimated to be membrane-associated proteins (1). Proteins can interact with the hydrophobic lipid bilayer of cell membranes via a hydrophobic surface formed by structures such as α-helices or β-sheets. In addition, a protein can be modified with a lipid anchor under enzymatic control that interacts with the lipid bilayer and localizes proteins to the membrane surface. Lipid modifications are increasingly recognized as important mechanisms for both targeting proteins to the membrane and subcellular protein trafficking. To date hundreds of lipid-modified proteins have been identified and, in many cases, the modification has been demonstrated to be important for the cellular function of these proteins. For example, the lipidated forms of receptors, monomeric and trimeric G-proteins (2–5) and protein tyrosine kinases (6–11) play crucial roles in a variety of cell signaling events.
Palmitoylation (12, 13), prenylation (14–17), and myristoylation (18, 19) represent three types of common lipid modifications that occur in the cell (Table 1). During the past decade, the enzymes responsible for prenylation and myristoylation have been extensively studied, and the target sequences for these modifications have been identified. In recent years, palmitoylation, which involves the formation of a thioester bond between a cysteine thiol side chain and a saturated 16-carbon fatty acid (20–28), has attracted great attention. Compared to the other lipid modifications, palmitoylation is readily reversible due to the lability of the thioester bond. Therefore rapid cycles of palmitoylation and depalmitoylation allow proteins to be facilely shuttled between the plasma membrane and the Golgi apparatus to regulate many cellular functions (29–35). Palmitoylation is catalyzed by protein palmitoyltransferases (PATs). PATs were first identified in yeast more than a decade ago using a genetic screen against palmitoylation-dependent Ras proteins (36). Since then many PATs have been identified using sequence homology; however the recognition motifs in the substrate proteins are still poorly defined. The structures, recognition signals, and the corresponding enzymes that catalyze lipid modifications are summarized in Table 1. Studies conducted over the past decade have substantially advanced our understanding of not only the molecular mechanisms of PATs and the functional consequences of protein palmitoylation, but have also begun to disclose important roles of these enzymes in human health. This review focuses on recent discoveries demonstrating the biological significance of protein palmitoylation and PATs and the development of methods to investigate the catalytic mechanism of these enzymes.
Table 1. Structures, signals, and corresponding enzymes that catalyze three types of lipid modifications.
Common recognition signals in substrate proteins for both prenylation and N-myristoylation have been identified, while a consensus signal for palmitoylation has not yet been defined. In the signal sequences of prenylation and N-myristoylation, “a” refers to any aliphatic amino acid, and “X” could be a subset of amino acids.
| Lipid Modifications | Signals | Enzymes | |
|---|---|---|---|
| Prenylation |
|
–CaaX | Farnesyltransferase Geranylgeranyltransferase 1 |
|
|
–CC or –CXC (Rab proteins only) | Geranylgeranyltransferase II | |
| N-myristoylation |
|
MGxxS— | N-myristoyltransferase |
| Palmitoylation |
|
Poorly defined | Palmitoyltransferase |
2 Biological Significance of Protein Palmitoylation
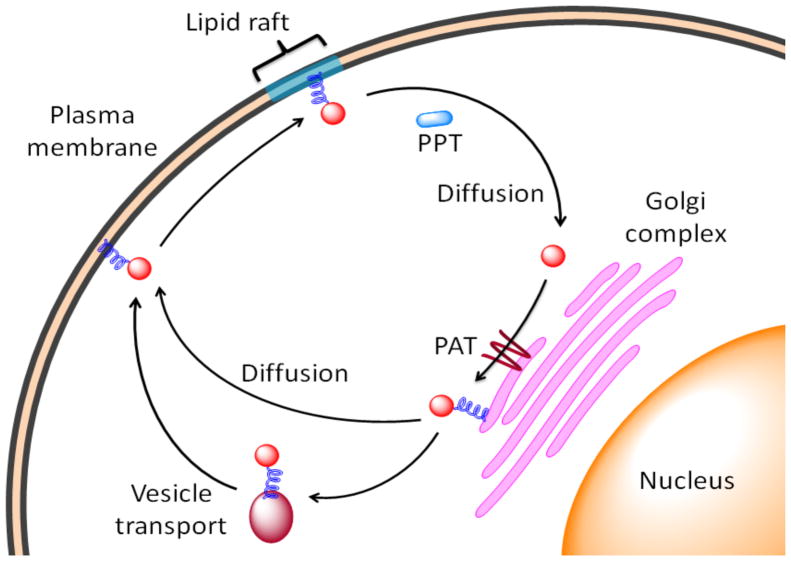
Protein palmitoylation is an important reversible lipid modification (29, 30, 32–35). However, in the cell palmitoylation does not function solely as a lipid anchor to localize proteins to the membrane. The reversible cycles of palmitoylation and depalmitoylation play an important function in shuttling modified proteins between cellular compartments, allowing relocalization of the protein in the cell or within different regions of the membrane (13). A scheme of this process is shown in Figure 1. Therefore, in addition to enhancing the membrane association of proteins, palmitoylation also helps target proteins in neurons, recruit proteins into lipid rafts, regulate cell signaling processes, etc. (32–34, 37). Identification of the enzymes responsible for catalyzing the formation and hydrolysis of specific palmitoylated proteins is a key step towards understanding the biological significance and function of this modification.
Figure 1. Inter-compartment protein shuttling through palmitoylation-depalmitoylation cycles.
Palmitoylated proteins (shown as a red circle with an attached blue fatty acid chain) is trafficked to the plasma membrane though either diffusion or vesicle transport, and is then targeted to lipid rafts. Depalmitoylation is catalyzed by a putative palmitoyl protein thioesterase (PPT, shown as a blue oval), leading to a palmitoylated half-time estimated at 0.3 to 2 h. The depalmitoylated protein diffuses back to the Golgi complex where it can be palmitoylated for another cycle.
One major function of protein palmitoylation is to promote protein membrane association. Proteins can either contain a single palmitoyl modification or be dually modified with one or more palmitoyl groups and either a prenyl group or a myristoyl group. Palmitoylated proteins can be divided into 4 distinct types (38–44), shown in Table 2, including (1) a single palmitoyl modification frequently near the end of a protein; (2) palmitoylation proximal to a transmembrane domain; (3) dual palmitoylation and prenylation; and (4) dual palmitoylation and myristoylation.
Table 2. Four different types of substrate proteins that are palmitoylated.
Although many palmitoylated proteins have been identified, the recognition signals in these proteins remain poorly defined. In addition to the palmitoyl modification, proteins may contain other membrane interactions, such as a transmembrane domain or a second lipid modification (prenyl or myristoyl group). Palmitoylation sites are identified with a *.
| palmitoylation type | substrate proteins |
|---|---|
| palmitoylation + prenylation −CXC*aaX | Ras proteins, Rho Proteins, Gγ subunits of S. cerevisiae |
| palmitoylation + myristoylation MGC*− | Gα subunits, Src family kinases (Yes, Fyn, Lck) |
| proximal to TMDs | GPCRs, viral-envelope proteins (HIV, influenza) |
| palmitoylation alone | Gα subunits, GRKs, RGS, SNAP-25, PSD-95 |
Examples of proteins that are only modified with a palmitoyl group include the G-protein α subunits. These subunits are palmitoylated to enhance the interaction with Gβγ subunits, an essential step in cell signaling cycles (27). Additionally, regulators of G-protein signaling (RGS) are also palmitoylated, regulating membrane localization and inactivation of G proteins to turn off G protein-coupled receptor signaling pathways (27, 35, 45). In contrast, mammalian Ras proteins may be dually lipidated. Ras proteins are modified with a farnesyl moiety to target the protein to the cell membrane. However, additional interactions are needed to enhance membrane localization; H-Ras and N-Ras contain both farnesyl and palmitoyl modifications, while K-Ras membrane association is enhanced by the electrostatic interaction between a positively charged basic sequence and the negatively charged plasma membrane (46–49). For dually lipid modified proteins, the first modification (e.g. prenylation or myristoylation) provides substrate proteins with weak membrane interaction, while the subsequent palmitoylation generates sufficient hydrophobicity for strong membrane affinity (50, 51).
A large number of neuronal proteins are palmitoylated, including synaptic scaffolding proteins, G-protein-coupled receptors (GPCRs) and synaptic vesicle proteins (35). Palmitoylation of these proteins is vital for regulating proper neuronal development and function, including neuronal differentiation, neurotransmission, and neurotransmitter release (52–54). One of the first neuronal proteins that was demonstrated to be palmitoylated was rhodopsin, in which two cysteines proximal to the seventh transmembrane domain are modified with palmitoyl groups (55). Similarly, many GPCRs are palmitoylated on analogous cysteines (53). A second example is the dendritic postsynaptic density protein 95 (PSD-95) that requires palmitoylation to enhance clustering of AMPA receptors at excitatory synapses; furthermore, palmitate cycling of PSD-95 also regulates synaptic plasticity (56–59). Palmitoylation of the SNARE protein (soluble N-ethylmaleimide-sensitive fusion protein-attachment protein receptor) SNAP-25 (25 kDa synaptosome-associated protein) is also important for neuronal function (60). SNAP-25 is located at presynaptic axon terminals, and is essential for neurotransmitter release (61). Palmitoylation of SNAP-25 is proposed to help regulate aspects of synaptic vesicle fusion, and to target synaptic vesicles to the site of transmitter release (62).
Another important function of palmitoylation is to target modified proteins to lipid rafts. Lipid rafts are dynamic nanoscale assemblies in the cell membrane that are enriched in sphingolipid, cholesterol, GPI-anchored proteins and palmitoylated proteins (63, 64). When palmitoylation is blocked by either mutagenesis or inhibition of PATs, the unmodified proteins no longer localize to lipid rafts, implicating the palmitoyl modification as an important localization determinant (65–69). Localization to lipid rafts is especially important for signaling proteins. The best-studied examples are nonreceptor tyrosine kinases (NRTKs) and linker for activation of T cells (LAT) which are involved in T cell signaling events; blocking palmitoylation in these proteins by mutation of the palmitoylated cysteine residue disrupts the lipid raft structure, and leads to attenuated T-cell responses (70–72). These studies assert the important role that palmitoylation plays in localizing proteins to functional membrane subdomains, such as lipid rafts.
3 DHHC Protein Family and Palmitoyltransferases (PATs)
Protein thiols can be palmitoylated nonenzymatically in vitro upon incubation with palmitoyl-CoA within a few hours at neutral pH (73). However, in a cell protein palmitoylation occurs much more rapidly and is catalyzed by palmitoylating enzymes, namely, protein palmitoyltransferases (PATs) (47). PATs catalyze the thioesterification of cysteines in the substrate protein with a palmitoyl group using palmitoyl-CoA as a co-substrate. One of the first identified PATs was the Saccharomyces cerevisiae Ras palmitoyltransferase Erf2p/Erf4p (effectors of Ras function) (74). After the CaaX sequence in Ras is farnesylated to enhance localization to membranes, the Erf2p/Erf4p PAT catalyzes palmitoylation of an upstream cysteine to further stabilize membrane interactions. The two proteins in the Erf2p/Erf4p complex co-purify and are both required for Ras palmitoylation activity. Deletion of either the ERF2 or ERF4 gene in yeast leads to a decrease in palmitoylation and mislocalization of Ras in the cell (36). A second palmitoyltransferase, Akr1p (ankyrin-repeat-containing protein 1), was identified in yeast by phenotypic analysis (75). This enzyme catalyzes palmitoylation of Yck2p (yeast casein kinase 2) both in vivo and in vitro, as well as other proteins (76). In the AKR1Δ strain, Yck2p localizes diffusely in the cytosol rather than associating with the membrane (77).
Both Erf2p and Akr1p are integral membrane proteins that contain several transmembrane domains (TMDs), and share a common DHH/YC-CRD (Asp-His-His/Tyr-Cys-cysteine rich domain) positioned between two TMDs (47, 78). The majority of PATs contain a consensus DHHC sequence and a conserved cysteine-rich domain (CRD), although Akr1p contains a DHYC sequence and an attenuated CRD with fewer cysteines conserved (47). For both PATs, mutations in the DHH(Y)C motif abolish the palmitoyltransferase activity (77, 79), indicating that this motif is important for palmitoylation activity.
Based on the prediction that the DHHC motif may be the active site of PATs, a search of the yeast genome for DHHC-containing proteins led to the discovery of five other putative PATs, including Akr2p, Swf1p, Pfa3p, Pfa4p, and Pfa5p (41, 76, 80, 81). All of these proteins catalyze palmitoylation with varying substrate recognition, confirming the importance of the DHHC-CRD sequence for PAT activity. Similarly, 23 mammalian DHHC proteins have been identified and most possess palmitoyltransferase activity (47). In addition, 15 genes from Caenorhabditis elegans and 22 genes from Drosophila melanogaster are also predicted to have PAT activity from their conserved DHHC sequence (82).
Among the 23 mammalian PATs, several are suggested to have biomedical relevance as their palmitoylated substrates are involved in the pathogenesis of neurological disorders, such as Huntington’s disease (DHHC17) (83), schizophrenia (DHHC8) (84), and X-linked mental retardation (DHHC9 and DHHC15) (85). One of the most notable examples is DHHC17, also known as HIP14 (huntingtin interacting protein 14) (83, 86, 87), which catalyzes the palmitoylation of several neuronal proteins, including huntingtin, PSD-95 (56, 57, 88, 89), and SNAP-25 (60). Furthermore, HIP14 activity regulates the subcellular trafficking and function of huntingtin, leading to the suggestion that it is involved in the pathogenesis of Huntington’s disease (87). Consistent with this, studies have indicated that the N-terminal polyglutamine (polyQ) expansion of the huntingtin protein prevents interaction with HIP14, and therefore leads to a decrease in palmitoylation and enhanced aggregation (87). Further studies on the palmitoylation mechanism of HIP14 as well as other DHHC proteins may disclose their exact pathophysiological roles in neurological disorders (90).
4 Akr1p as a Protein Palmitoyltransferase
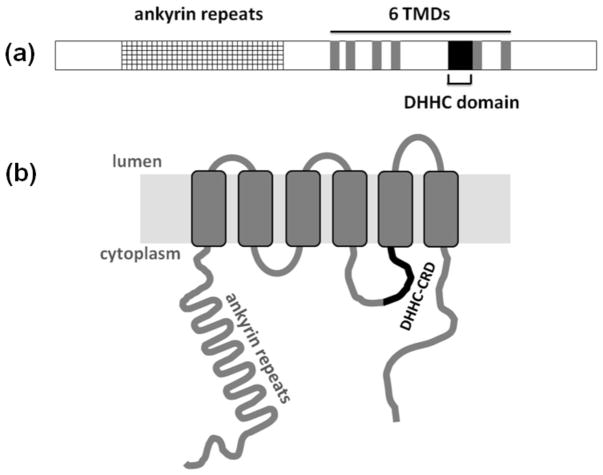
Akr1p, the yeast homologue of HIP14, is an 86-kDa integral membrane protein with six transmembrane domains. The N-terminal hydrophilic domain contains six ankyrin repeat sequences. The topology of Akr1p in the membrane indicates that the conserved DHHC-CRD sequence is located on the cytosolic side of the membrane between transmembrane domains four and five (91). A scheme of the Akr1p domains and topology is shown in Figure 2.
Figure 2.
(a) Schematic representation of the identified domains in the Akr1p sequence. Ankyrin repeats are shown in the grid region; the six transmembrane domains are indicated in grey and the DHHC domain is highlighted in black; (b) Schematic representation of Akr1p membrane topology. Both ankyrin repeats and the DHHC-cysteine rich domain (CRD) are positioned on the cytoplasmic side of the membrane.
Davis and colleagues have carried out multiple studies demonstrating that Akr1p is a palmitoyltransferase that catalyzes the palmitoylation of yeast casein kinase (Yck2p) both in vivo and in vitro (77). In wild-type (AKR1+) yeast cells Yck2p is exclusively trafficked to the cell membrane, whereas in akr1Δ cells Yck2p is diffusely localized in the cytoplasm (77). This finding is consistent with palmitoylation playing an important role in tethering proteins to the membrane. In addition, the C-terminus of Yck2p contains two cysteines that are hypothesized as the palmitoylation site(s). Consistent with this, mutation of the two C-terminal cysteine residues (-CC) to serine (-SS), leads to the relocalization of Yck2p from the membrane to the cytoplasm, as observed for WT Yck2p in akr1Δ cells (77). Furthermore, the DHYC motif of Akr1p is important for catalytic activity, as also observed for Erf2p (79), mutation of DH to AA, or C to A in Akr1p disables catalysis of Yck2p palmitoylation both in vivo and in vitro (77).
To search for palmitoylated proteins in yeast, the Davis lab carried out a global analysis of yeast membrane proteins using MudPIT (multi-dimensional protein identification technology) tandem-MS-based proteomic methodology coupled with methods to enrich palmitoylated proteins (76). Forty-seven palmitoylated proteins, including Akr1p, were identified by MS/MS data. To identify the substrate proteins that are palmitoylated by each of the DHHC-containing palmitoyltransferases, this proteomic method was applied to mutant yeast strains deficient in one of more DHHC proteins. Decreased representation in the MudPIT analysis of proteins isolated from the akr1Δ strain led to the identification of six yeast proteins as potential Akr1p substrates: Yck1p, Yck2p, Akr1p, Ypl199c, Ykl047w, and Meh1p.
Akr1p is palmitoylated upon incubation with palmitoyl-CoA, which is known as auto-palmitoylation (77). Interestingly, this property is shared with many other DHHC protein palmitoyltransferases, including Erf2p (79). In Akr1p alanine mutations for DH or C in the conserved DHYC sequence abolish both the auto-palmitoylation and transpalmitoylation activity both in vivo and in vitro (77). Furthermore, mutations in this conserved motif in other PATs, including C203S in Erf2p (79) and C468S in HIP14 (83), also abolish both palmitoylation activities. These data led to the hypothesis that auto-palmitoylation is enzyme-catalyzed and, further, that the auto-palmitoylated enzyme could be a covalent intermediate that occurs during the catalytic cycle. This palmitoylated intermediate is cleaved upon addition of hydroxylamine, suggesting a thioester linkage (75). In this proposed mechanism (Figure 3), a cysteine thiol at the active site of the PAT reacts with palmitoyl-CoA to form a covalent palmitoyl thioester enzyme intermediate. In a second step, the palmitoyl group is transferred from the enzyme to the substrate protein. Support for this mechanism is provided by kinetic measurements of the Erf2p/Erf4p PAT that show a “burst” of CoA formation, consistent with the formation of a reaction intermediate (92). In comparison to the activation of thiol nucleophiles in thiol proteases (93) and N-acetyltransferases (94–96), the conserved Asp and/or His in the DHHC domain may function to stabilize a nucleophilic thiolate anion in both steps of the reaction.
Figure 3. Scheme of the proposed two-step palmitoylation mechanism of PATs.
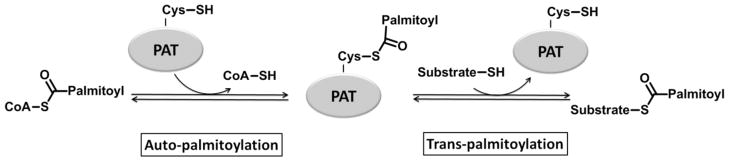
A covalent palmitoyl thioester enzyme intermediate is formed in the first auto-palmitoylation step, followed by the transfer of the palmitoyl moiety from PAT to the substrate protein in the second trans-palmitoylation step.
5 Methods Used for Studying PATs
Palmitoyltransferases are integral membrane proteins that contain several transmembrane domains. Therefore these proteins are difficult to overexpress and purify in large quantities, presenting a significant challenge to the characterization of PATs. Recently methods have been developed to enhance the expression, purification, and characterization of yeast PATs which are reviewed in the next section.
5.1 Expression and purification of yeast PATs
The largest quantities of active yeast PATs have been obtained using galactose inducible yeast expression systems. No high recombinant expression of a PAT in E. coli has yet been described, although small amounts of active HIP14 (DHHC17) (97) and Erf2p/Erf4p (79) were observed. This difficulty is likely due to both the problems associated with expression of eukaryotic membrane proteins in bacteria and the lack of necessary posttranslational modifications.
Deschenes and colleagues have optimized the expression of yeast PATs, including Erf2p, Akr1p, Pfa3p, Pfa4p, and Pfa5p (98) while the Davis lab has developed methods to overexpress Akr1p (77). The FLAG-tagged and/or His-tagged versions of these protein genes encoded on a yeast plasmid were transformed into yeast strain, and the cells were grown on selective media. The culture was then induced by addition of galactose, harvested by centrifugation, lysed by vigorous vortexing with glass beads and the membrane fractions were collected using ultracentifugation. The PAT activity in these membrane fractions was sufficient to catalyze palmitoylation of their corresponding protein substrates.
The Erf2p/Erf4p purification method serves as the prototypical method for other PATs. The first step is solubilization of the membrane fractions by addition of 0.75% (w/v) detergent. Dodecylmaltoside (DDM), deoxycholic acid (DCA), and Triton X-100 were all efficient at solubilizing the PAT Erf2p/Erf4p; however, DDM was most effective in maintaining the palmitoylation activity (98). Following protein solubilization, His6-Erf2p/Flag-Erf4p was then purified using nickel affinity and anion exchange chromatography, with DDM present in all wash and elution buffers. This procedure provides purified active protein based on absorbance, SDS-PAGE analysis, and PAT activity assay.
5.2 Palmitoylation assays
To study catalysis of palmitoylation and obtain a better understanding of its biological significance, a robust palmitoylation assay is required. Several palmitoylation assays have been developed and used to study PATs; each assay has both advantages and disadvantages (99).
The first developed and most frequently used assay is the in vivo metabolic labeling of proteins using radiolabeled palmitate, most commonly [3H]palmitate (57, 74, 100–102). The radiolabeled palmitate is added to the cell culture during growth, where the palmitoyl moiety is incorporated onto target proteins. The labeling of proteins is evaluated by fractionation of the proteins by SDS-PAGE of the cell lysate followed by exposing the dried gel to high-sensitivity films. This method provides an efficient way to identify palmitoylated proteins and to assess the palmitoylation level of a certain protein in living cells. In a pulse-chase format this assay can also be used to measure both the palmitoylation and depalmitoylation rates. Disadvantages of this method are: low sensitivity requiring long hours of labeling and exposure time for detection of the radioactive signal and difficulty with accurately quantifying the result, since the in vivo labeling process is affected by many cellular factors. Therefore, this method is useful for the initial identification of PATs but is not generally suitable for further mechanistic characterization of the enzymes.
A similar assay labels palmitoylated proteins with ω-azido-fatty acids. In this method synthetic ω-azido-fatty acid is added to the cell culture during growth, so the target proteins are labeled with a palmitoyl group containing an azido group at the end of the fatty acid chain (103). Following in vivo labeling, proteins can be extracted and subjected to click chemistry (104) to add a detectable chemical probe, such as biotin or a fluorophore, to quantify the palmitoylated proteins. This method is both safer and more sensitive than labeling with radiolabeled palmitate; however, it requires multiple steps and presents challenges for quantification and measurement of the time-dependence of the reactions.
Another commonly used assay is the in vitro palmitoylation assay, in which PATs catalyze palmitoylation of a substrate (either whole proteins or peptides mimicking the protein palmitoylation motif) upon addition of exogenous palmitoyl-CoA (105). When using peptides as substrates, fluorescent labeling enhances the sensitivity of the assay and the palmitoylated peptides can be separated from the non-palmitoylated peptides by high performance liquid chromatography (HPLC) (106). However, not all PATs react readily with peptide substrates. When using whole proteins as substrates, radiolabeled palmitoyl-CoA is used so that palmitoylation is detected by autoradiography after separation by SDS-PAGE. This method is more quantitative, requires less reaction handling for signal detection, and can be used to measure time-dependent reactions. This assay has also been applied in a cell-based assay format, in which intact cells are incubated with fluorescently-labeled peptides and endogenous palmitoyl-CoA (105). This assay has the advantage of speed and simplicity and can be used to study the intracellular trafficking of the palmitoylated peptides. The in vitro palmitoylation assay has been widely used to study the catalytic mechanism of PATs.
In a recent kinetic study of Erf2p/Erf4p, the Ras PAT, Deschenes and colleagues introduced a coupled fluorescence-based assay measuring NADH production to monitor the rate of CoASH production from palmitoyl-CoA in the palmitoylation reaction (92). This assay does not require using radioactive material, and can be performed in a 96-well plate making it useful for high-throughput inhibitor screening. However, this assay is not as sensitive as the radioactive assay, and therefore larger quantities of active enzyme and substrate protein are required to generate a detectable signal.
Besides the traditional biochemical assays, mass spectrometry has become an important tool for studying the posttranslational modification of proteins, including lipidation (102, 107, 108). In this case, the palmitoylated proteins are subjected to protease digestion and the resulting peptides are separated by chromatography. By comparing the sample spectrum with the control (no modification), the peptides with the palmitoyl group are identified by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) based on the 272 Da mass shift for the palmitoyl group (109). This method is not quantitative but is a versatile tool to identify palmitoylated proteins. However, this method has some limitations. For example, the thioester linkage could be cleaved during sample preparation, especially at high or low pH in the presence of exogenous thiols. In addition, peptides containing a hydrophobic fatty acyl chain, like palmitoyl, have low solubility, can stick to the separation column, and may have poor ionization efficiency. Based on these difficulties, this method was optimized by performing an additional step, called fatty acyl exchange chemistry, before mass spectral analysis to replace the labile thioester bond with a more stable thioether linkage (62, 76, 110–114). In this method first all of the unlabeled free cysteine thiols are blocked by reaction with N-ethylmaleimide (NEM) after reduction of disulfide bonds by incubation with tris(2-carboxyethyl)phosphine (TCEP). Next, the thioester bond with the palmitoyl group is cleaved by the addition of hydroxylamine, exposing the palmitoylated cysteine. Last, the exposed cysteine thiols are reacted with thiol-specific reagents with detectable functional groups, such as Btn-BMCC (1-biotinamido-4-[4′-(maleimidomethyl) cyclohexanecarboxamido] butane) and biotin-HPDP (N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)propionami de). Finally, the labeled peptide is purified and analyzed by mass spectrometry. This optimized method has been successfully used to identify palmitoylated proteins in several biological systems. For example, Davis and colleagues identified a host of palmitoylated proteins in yeast (76), and Freeman and colleagues identified palmitoylated proteins and their sites of modification in lipid raft-enriched and non-raft membranes of mammalian cells (112). This method can generate false positives, due to the non-specific nature of the reactions; therefore, careful controls and confirmation of the results using other biological tools are essential. Overall, this method has great advantages for discovering novel palmitoylated proteins but may not be suitable for detailed mechanistic studies of the enzymatic reactions.
To sum up, there are a number of assays that have been developed to study protein palmitoylation, with each having their own advantages and disadvantages. Using these assays, many novel palmitoylated proteins have been identified, some of which have been further mechanistically characterized. However, to obtain an in-depth picture of the palmitoylation mechanism, faster, more sensitive, and higher-throughput assays are desired.
6 Conclusions and Perspectives
This review summarized recent advances in the analysis of protein palmitoylation and PATs, from the roles of protein palmitoylation in nature and the discovery of the conserved residues in PATs, to the development of methods to characterize protein palmitoylation. To unravel the catalytic mechanism of PATs, a number of useful methods have been developed, including approaches for expressing and purifying PATs and assays for measuring palmitoylation activity both in vitro and in vivo. The discovery of the large DHHC protein family has led to the prediction that the DHHC motif is the active site, providing insight into the catalytic mechanism of PATs.
Even though progress has been made toward understanding various aspects of protein palmitoylation, several critical issues still remain unclear, including the detailed catalytic mechanism of protein palmitoylation, substrate specificity of PATs, and the development of possible inhibitors. Furthermore, the current data suggest that protein palmitoylation proceeds with two steps: auto-palmitoylation of the enzyme followed by trans-palmitoylation of the substrate protein. However, mechanistic information about these steps remains to be obtained, including identification of the site of auto-palmitoylation in these enzymes. Although both trans- and auto-palmitoylation disappear when mutations are made in the DHHC domain of PATs, there is not yet any direct evidence that the DHHC cysteine is the site of auto-palmitoylation. Furthermore, even though a number of palmitoylated proteins have been discovered both in vivo and in vitro, there is no consensus signal for protein palmitoylation. Hence, analysis of the substrate specificity determinants of PATs will provide important information about molecular recognition of the enzymes, and will help produce reagents that will facilitate mechanistic biochemistry on PATs. These results will eventually lead to the development of robust high-throughput screens for PAT inhibitors and, potentially, specific PAT inhibitors that have the potential to be used to treat a variety of diseases.
Acknowledgments
This work was financially supported by the NIH R01 grant GM040602 (CAF).
List of Abbreviations
- Biotin-HPDP
N-[6-(biotinamido)hexyl]-3′-(2′-pyridyl dithio)propionamide
- Btn-BMCC
1-biotinamido-4-[4′-(maleimidomethyl) cyclohexanecarboxamido] butane
- DCA
deoxycholic acid
- DDM
dodecylmaltoside
- DHHC-CRD
Asp-His-His-Cys cysteine rich domain
- GPCR
G protein-coupled receptor
- GRK
G protein-coupled receptor kinase
- HIP14
huntingtin interacting protein 14
- LAT
linker for activation of T cells
- MudPIT
multi-dimensional protein identification technology
- NEM
N-ethylmaleimide
- NRTK
nonreceptor tyrosine kinase
- PAT
palmitoyl acyltransferase
- PPT
palmitoyl protein thioesterase
- PSD-95
dendritic postsynaptic density protein 95
- RGS
regulator of G protein signaling
- SNAP-25
25 kDa synaptosome-associated protein
- SNARE
soluble N-ethylmaleimide-sensitive fusion protein-attachment protein receptor
- TCEP
tris(2-carboxyethyl)phosphine
- TMD
transmembrane domain
References
- 1.Stevens TJ, Arkin IT. Do more complex organisms have a greater proportion of membrane proteins in their genomes? Proteins: Struct Funct Genet. 2000;39:417–420. doi: 10.1002/(sici)1097-0134(20000601)39:4<417::aid-prot140>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 2.Terry KL, Casey PJ, Beese LS. Conversion of protein farnesyl-transferase to a geranylgeranyltransferase. Biochemistry. 2006;45:9746–9755. doi: 10.1021/bi060295e. [DOI] [PubMed] [Google Scholar]
- 3.Fields TA, Casey PJ. Signalling functions and biochemical properties of pertussis toxin-resistant G-proteins. Biochem J. 1997;321:561–571. doi: 10.1042/bj3210561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Higgins JB, Casey PJ. The role of prenylation in G-protein assembly and function. Cell Signal. 1996;8:433–437. doi: 10.1016/s0898-6568(96)00071-x. [DOI] [PubMed] [Google Scholar]
- 5.Thomason PA, James SR, Casey PJ, Downes CP. A G-protein beta gamma-subunit-responsive phosphoinositide 3-kinase activity in human platelet cytosol. J Biol Chem. 1994;269:16525–16528. [PubMed] [Google Scholar]
- 6.Kurosaki T, Hikida M. Tyrosine kinases and their substrates in B lymphocytes. Immunol Rev. 2009;228:132–148. doi: 10.1111/j.1600-065X.2008.00748.x. [DOI] [PubMed] [Google Scholar]
- 7.Chiarugi P, Fiaschi T. Redox signalling in anchorage-dependent cell growth. Cell Signalling. 2007;19:672–682. doi: 10.1016/j.cellsig.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 8.Latour S, Veillette A. Proximal protein tyrosine kinases in immunoreceptor signaling. Curr Opin Immunol. 2001;13:299–306. doi: 10.1016/s0952-7915(00)00219-3. [DOI] [PubMed] [Google Scholar]
- 9.Ellery JM, Kempshall SJ, Nicholls PJ. Activation of the interleukin 2 receptor: a possible role for tyrosine phosphatases. Cell Signalling. 2000;12:367–373. doi: 10.1016/s0898-6568(00)00085-1. [DOI] [PubMed] [Google Scholar]
- 10.Kamata H, Hirata H. Redox regulation of cellular signaling. Cell Signalling. 1998;11:1–14. doi: 10.1016/s0898-6568(98)00037-0. [DOI] [PubMed] [Google Scholar]
- 11.Neel BG, Tonks NK. Protein tyrosine phosphatases in signal transduction. Curr Opin Cell Biol. 1997;9:193–204. doi: 10.1016/s0955-0674(97)80063-4. [DOI] [PubMed] [Google Scholar]
- 12.Charollais J, Van DGFG. Palmitoylation of membrane proteins. Mol Membr Biol. 2009;26:55–66. doi: 10.1080/09687680802620369. [DOI] [PubMed] [Google Scholar]
- 13.Fukata Y, Fukata M. Protein palmitoylation in neuronal development and synaptic plasticity. Nat Rev Neurosci. 2010;11:161–175. doi: 10.1038/nrn2788. [DOI] [PubMed] [Google Scholar]
- 14.Zhang FL, Casey PJ. Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 15.Winter-Vann AM, Casey PJ. Opinion: Post-prenylation-processing enzymes as new targets in oncogenesis. Nat Rev Cancer. 2005;5:405–412. doi: 10.1038/nrc1612. [DOI] [PubMed] [Google Scholar]
- 16.Sebti SM, Hamilton AD. Farnesyltransferase and geranylgeranyl-transferase I inhibitors and cancer therapy: lessons from mechanism and bench-to-bedside translational studies. Oncogene. 2000;19:6584–6593. doi: 10.1038/sj.onc.1204146. [DOI] [PubMed] [Google Scholar]
- 17.Magee T, Seabra MC. Fatty acylation and prenylation of proteins: what’s hot in fat. Curr Opin Cell Biol. 2005;17:190–196. doi: 10.1016/j.ceb.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Johnson DR, Bhatnagar RS, Knoll LJ, Gordon JI. Genetic and biochemical studies of protein N-myristoylation. Annu Rev Biochem. 1994;63:869–914. doi: 10.1146/annurev.bi.63.070194.004253. [DOI] [PubMed] [Google Scholar]
- 19.Farazi TA, Waksman G, Gordon JI. The biology and enzymology of protein N-myristoylation. J Biol Chem. 2001;276:39501–39504. doi: 10.1074/jbc.R100042200. [DOI] [PubMed] [Google Scholar]
- 20.Greaves J, Chamberlain LH. DHHC palmitoyl transferases: substrate interactions and (patho)physiology. Trends Biochem Sci. 2011;36:245–253. doi: 10.1016/j.tibs.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 21.Planey SL, Zacharias DA. Palmitoyl acyltransferases, their substrates, and novel assays to connect them. Mol Membr Biol. 2009;26:14–31. doi: 10.1080/09687680802646703. [DOI] [PubMed] [Google Scholar]
- 22.Baekkeskov S, Kanaani J. Palmitoylation cycles and regulation of protein function. Mol Membr Biol. 2009;26:42–54. doi: 10.1080/09687680802680108. [DOI] [PubMed] [Google Scholar]
- 23.Wan J, Roth AF, Bailey AO, Davis NG. Palmitoylated proteins: purification and identification. Nat Protoc. 2007;2:1573–1584. doi: 10.1038/nprot.2007.225. [DOI] [PubMed] [Google Scholar]
- 24.Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- 25.Chakrabandhu K, Herincs Z, Huault S, Dost B, Peng L, Con-chonaud F, Marguet D, He H-T, Hueber A-O. Palmitoylation is required for efficient Fas cell death signaling. EMBO J. 2007;26:209–220. doi: 10.1038/sj.emboj.7601456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smotrys JE, Linder ME. Palmitoylation of intracellular signaling proteins: Regulation and function. Annu Rev Biochem. 2004;73:559–587. doi: 10.1146/annurev.biochem.73.011303.073954. [DOI] [PubMed] [Google Scholar]
- 27.Osterhout JL, Waheed AA, Hiol A, Ward RJ, Davey PC, Nini L, Wang J, Milligan G, Jones TLZ, Druey KM. Palmitoylation Regulates Regulator of G-protein Signaling (RGS) 16 Function: II. Palmitoylation of a cysteine residue in the RGS box is critical for RGS16 GTPase accelerating activity and regulation of Gi-coupled signaling. J Biol Chem. 2003;278:19309–19316. doi: 10.1074/jbc.M210124200. [DOI] [PubMed] [Google Scholar]
- 28.Mumby SM, Kleuss C, Gilman AG. Receptor regulation of G-protein palmitoylation. Proc Natl Acad Sci U S A. 1994;91:2800–2804. doi: 10.1073/pnas.91.7.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greaves J, Carmichael JA, Chamberlain LH. The palmitoyl transferase DHHC2 targets a dynamic membrane cycling pathway: regulation by a C-terminal domain. Mol Biol Cell. 2011;22:1887–1895. doi: 10.1091/mbc.E10-11-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang MM, Tsou LK, Charron G, Raghavan AS, Hang HC. Tandem fluorescence imaging of dynamic S-acylation and protein turnover. Proc Natl Acad Sci U S A. 2010;107:8627–8632. S8627/8621–S8627/8626. doi: 10.1073/pnas.0912306107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salaun C, Greaves J, Chamberlain LH. The intracellular dynamic of protein palmitoylation. J Cell Biol. 2010;191:1229–1238. doi: 10.1083/jcb.201008160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conibear E, Davis NG. Palmitoylation and depalmitoylation dynamics at a glance. J Cell Sci. 2010;123:4007–4010. doi: 10.1242/jcs.059287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwanaga T, Tsutsumi R, Noritake J, Fukata Y, Fukata M. Dynamic protein palmitoylation in cellular signaling. Prog Lipid Res. 2009;48:117–127. doi: 10.1016/j.plipres.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 34.Basu J. Protein palmitoylation and dynamic modulation of protein function. Curr Sci. 2004;87:212–217. [Google Scholar]
- 35.Qanbar R, Bouvier M. Role of palmitoylation/depalmitoylation reactions in G-protein-coupled receptor function. Pharmacol Ther. 2003;97:1–33. doi: 10.1016/s0163-7258(02)00300-5. [DOI] [PubMed] [Google Scholar]
- 36.Bartels DJ, Mitchell DA, Dong X, Deschenes RJ. Erf2, a novel gene product that affects the localization and palmitoylation of Ras2 in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6775–6787. doi: 10.1128/mcb.19.10.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian L, Jeffries O, McClafferty H, Molyvdas A, Rowe ICM, Saleem F, Chen L, Greaves J, Chamberlain LH, Knaus H-G, Ruth P, Shipston MJ. Palmitoylation gates phosphorylation-dependent regulation of BK potassium channels. Proc Natl Acad Sci U S A. 2008;105:21006–21011. doi: 10.1073/pnas.0806700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsutsumi R, Fukata Y, Noritake J, Iwanaga T, Perez F, Fukata M. Identification of G protein α subunit-palmitoylating enzyme. Mol Cell Biol. 2009;29:435–447. doi: 10.1128/MCB.01144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masuda K, Itoh H, Sakihama T, Akiyama C, Takahashi K, Fukuda R, Yokomizo T, Shimizu T, Kodama T, Hamakubo T. A combinatorial G protein-coupled receptor reconstitution system on budded baculovirus. Evidence for Gαi and Gαo coupling to a human leukotriene B4 receptor. J Biol Chem. 2003;278:24552–24562. doi: 10.1074/jbc.M302801200. [DOI] [PubMed] [Google Scholar]
- 40.Sandilands E, Brunton VG, Frame MC. The membrane targeting and spatial activation of Src, Yes and Fyn is influenced by palmitoylation and distinct RhoB/RhoD endosome requirements. J Cell Sci. 2007;120:2555–2564. doi: 10.1242/jcs.003657. [DOI] [PubMed] [Google Scholar]
- 41.Dighe SA, Kozminski KG. Swf1p, a member of the DHHC-CRD family of palmitoyltransferases, regulates the actin cytoskeleton and polarized secretion independently of its DHHC motif. Mol Biol Cell. 2008;19:4454–4468. doi: 10.1091/mbc.E08-03-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang D-A, Sebti SM. Palmitoylated cysteine 192 is required for RhoB tumor-suppressive and apoptotic activities. J Biol Chem. 2005;280:19243–19249. doi: 10.1074/jbc.M411472200. [DOI] [PubMed] [Google Scholar]
- 43.Bhattacharyya R, Wedegaertner PB. Gα13 requires palmitoylation for plasma membrane localization, Rho-dependent signaling, and promotion of p115-RhoGEF membrane binding. J Biol Chem. 2000;275:14992–14999. doi: 10.1074/jbc.M000415200. [DOI] [PubMed] [Google Scholar]
- 44.Patterson SI, Skene JHP. A shift in protein S-palmitoylation, with persistence of growth-associated substrates, marks a critical period for synaptic plasticity in developing brain. J Neurobiol. 1999;39:423–437. doi: 10.1002/(sici)1097-4695(19990605)39:3<423::aid-neu8>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 45.Wang J, Xie Y, Wolff DW, Abel PW, Tu Y. DHHC protein-dependent palmitoylation protects regulator of G-protein signaling 4 from proteasome degradation. FEBS Lett. 2010;584:4570–4574. doi: 10.1016/j.febslet.2010.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wright LP, Philips MR. CAAX modification and membrane targeting of Ras. J Lipid Res. 2006;47:883–891. doi: 10.1194/jlr.R600004-JLR200. [DOI] [PubMed] [Google Scholar]
- 47.Mitchell DA, Vasudevan A, Linder ME, Deschenes RJ. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J Lipid Res. 2006;47:1118–1127. doi: 10.1194/jlr.R600007-JLR200. [DOI] [PubMed] [Google Scholar]
- 48.Pechlivanis M, Kuhlmann J. Hydrophobic modifications of Ras proteins by isoprenoid groups and fatty acids--More than just membrane anchoring. Biochim Biophys Acta. 2006;1764:1914–1931. doi: 10.1016/j.bbapap.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 49.Resh MD. Membrane targeting of lipid modified signal transduction proteins. Subcell Biochem. 2004;37:217–232. doi: 10.1007/978-1-4757-5806-1_6. [DOI] [PubMed] [Google Scholar]
- 50.Peitzsch RM, McLaughlin S. Binding of acylated peptides and fatty acids to phospholipid vesicles: Pertinence to myristoylated proteins. Biochemistry. 1993;32:10436–10443. doi: 10.1021/bi00090a020. [DOI] [PubMed] [Google Scholar]
- 51.Shahinian S, Silvius JR. Doubly-lipid-modified protein sequence motifs exhibit long-lived anchorage to lipid bilayer membranes. Biochemistry. 1995;34:3813–3822. doi: 10.1021/bi00011a039. [DOI] [PubMed] [Google Scholar]
- 52.Morello JP, Bouvier M. Palmitoylation: a post-translational modification that regulates signalling from G-protein coupled receptors. Biochem Cell Biol. 1996;74:449–457. doi: 10.1139/o96-049. [DOI] [PubMed] [Google Scholar]
- 53.Bouvier M, Moffett S, Loisel TP, Mouillac B, Hebert T, Chidiac P. Palmitoylation of G-protein-coupled receptors: a dynamic modification with functional consequences. Biochem Soc Trans. 1995;23:116–120. doi: 10.1042/bst0230116. [DOI] [PubMed] [Google Scholar]
- 54.Bouvier M, Chidiac P, Hebert TE, Loisel TP, Moffett S, Mouillac B. Dynamic palmitoylation of G-protein-coupled receptors in eukaryotic cells. Methods Enzymol. 1995;250:300–314. doi: 10.1016/0076-6879(95)50080-4. [DOI] [PubMed] [Google Scholar]
- 55.O’Brien PJ, Zatz M. Acylation of bovine rhodopsin by [3H]palmitic acid. 1984. pp. 5054–5057. [PubMed] [Google Scholar]
- 56.Noritake J, Fukata Y, Iwanaga T, Hosomi N, Tsutsumi R, Matsuda N, Tani H, Iwanari H, Mochizuki Y, Kodama T, Matsuura Y, Bredt DS, Hamakubo T, Fukata M. Mobile DHHC palmitoylating enzyme mediates activity-sensitive synaptic targeting of PSD-95. J Cell Biol. 2009;186:147–160. doi: 10.1083/jcb.200903101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fukata M, Fukata Y, Adesnik H, Nicoll RA, Bredt DS. Identification of PSD-95 Palmitoylating enzymes. Neuron. 2004;44:987–996. doi: 10.1016/j.neuron.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 58.Topinka JR, Bredt DS. N-terminal palmitoylation of PSD-95 regulates association with cell membranes and interaction with K+ channel Kv1.4. Neuron. 1998;20:125–134. doi: 10.1016/s0896-6273(00)80440-7. [DOI] [PubMed] [Google Scholar]
- 59.Ho GH, Selvakumar B, Mukai J, Hester L, Wang Y, Gogos J, Snyder S. S-Nitrosylation and S-Palmitoylation Reciprocally Regulate Synaptic Targeting of PSD-95. Neuron. 2011;71:131–141. doi: 10.1016/j.neuron.2011.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gonzalo S, Greentree WK, Linder ME. SNAP-25 is targeted to the plasma membrane through a novel membrane-binding domain. J Biol Chem. 1999;274:21313–21318. doi: 10.1074/jbc.274.30.21313. [DOI] [PubMed] [Google Scholar]
- 61.Greaves J, Gorleku OA, Salaun C, Chamberlain LH. Palmitoylation of the SNAP25 protein family: specificity and regulation by DHHC palmitoyl transferases. J Biol Chem. 2010;285:24629–24638. doi: 10.1074/jbc.M110.119289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kang R, Wan J, Arstikaitis P, Takahashi H, Huang K, Bailey AO, Thompson JX, Roth AF, Drisdel RC, Mastro R, Green WN, Yates JR, III, Davis NG, El-Husseini A. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature. 2008;456:904–909. doi: 10.1038/nature07605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edidin M. The state of lipid rafts: From model membranes to cells. Annu Rev Biophys Biomol Struct. 2003;32:257–283. doi: 10.1146/annurev.biophys.32.110601.142439. [DOI] [PubMed] [Google Scholar]
- 64.Brown DA, London E. Functions of lipid rafts in biological membranes. Annu Rev Cell Dev Biol. 1998;14:111–136. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- 65.Shenoy-Scaria AM, Dietzen DJ, Kwong J, Link DC, Lublin DM. Cysteine3 of Src family protein tyrosine kinase determines palmitoylation and localization in caveolae. J Cell Biol. 1994;126:353–363. doi: 10.1083/jcb.126.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robbins SM, Quintrell NA, Bishop JM. Myristoylation and differential palmitoylation of the HCK protein-tyrosine kinases govern their attachment to membranes and association with caveolae. Mol Cell Biol. 1995;15:3507–3515. doi: 10.1128/mcb.15.7.3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guzzi F, Zanchetta D, Chini B, Parenti M. Thioacylation is required for targeting G-protein subunit Go1α to detergent-insoluble caveolin-containing membrane domains. Biochem J. 2001;355:323–331. doi: 10.1042/0264-6021:3550323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arni S, Keilbaugh SA, Ostermeyer AG, Brown DA. Association of GAP-43 with detergent-resistant membranes requires two palmitoylated cysteine residues. J Biol Chem. 1998;273:28478–28485. doi: 10.1074/jbc.273.43.28478. [DOI] [PubMed] [Google Scholar]
- 69.Webb Y, Hermida-Matsumoto L, Resh MD. Inhibition of protein palmitoylation, raft localization, and T cell signaling by 2-bromopalmitate and polyunsaturated fatty acids. J Biol Chem. 2000;275:261–270. doi: 10.1074/jbc.275.1.261. [DOI] [PubMed] [Google Scholar]
- 70.Kabouridis PS, Magee AI, Ley SC. S-acylation of LCK protein tyrosine kinase is essential for its signaling function in T lymphocytes. EMBO J. 1997;16:4983–4998. doi: 10.1093/emboj/16.16.4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang W, Trible RP, Samelson LE. LAT palmitoylation: its essential role in membrane microdomain targeting and tyrosine phosphorylation during T cell activation. Immunity. 1998;9:239–246. doi: 10.1016/s1074-7613(00)80606-8. [DOI] [PubMed] [Google Scholar]
- 72.Lin J, Weiss A, Finco TS. Localization of LAT in glycolipid-enriched microdomains is required for T cell activation. J Biol Chem. 1999;274:28861–28864. doi: 10.1074/jbc.274.41.28861. [DOI] [PubMed] [Google Scholar]
- 73.Duncan JA, Gilman AG. Autoacylation of G protein subunits. J Biol Chem. 1996;271:23594–23600. doi: 10.1074/jbc.271.38.23594. [DOI] [PubMed] [Google Scholar]
- 74.Lobo S, Greentree WK, Linder ME, Deschenes RJ. Identification of a Ras Palmitoyltransferase in Saccharomyces cerevisiae. J Biol Chem. 2002;277:41268–41273. doi: 10.1074/jbc.M206573200. [DOI] [PubMed] [Google Scholar]
- 75.Feng Y, Davis NG. Akr1p and the type I casein kinases act prior to the ubiquitination step of yeast endocytosis: Akr1p is required for kinase localization to the plasma membrane. Mol Cell Biol. 2000;20:5350–5359. doi: 10.1128/mcb.20.14.5350-5359.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roth AF, Wan J, Bailey AO, Sun B, Kuchar JA, Green WN, Phinney BS, Yates JR, III, Davis NG. Global analysis of protein palmitoylation in yeast. Cell. 2006;125:1003–1013. doi: 10.1016/j.cell.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roth AF, Feng Y, Chen L, Davis NG. The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J Cell Biol. 2002;159:23–28. doi: 10.1083/jcb.200206120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Greaves J, Chamberlain LH. S-acylation by the DHHC protein family. Biochem Soc Trans. 2010;38:522–524. doi: 10.1042/BST0380522. [DOI] [PubMed] [Google Scholar]
- 79.Lobo S, Greentree WK, Linder ME, Deschenes RJ. Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J Biol Chem. 2002;277:41268–41273. doi: 10.1074/jbc.M206573200. [DOI] [PubMed] [Google Scholar]
- 80.Smotrys JE, Schoenfish MJ, Stutz MA, Linder ME. The vacuolar DHHC-CRD protein Pfa3p is a protein acyltransferase for Vac8p. J Cell Biol. 2005;170:1091–1099. doi: 10.1083/jcb.200507048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hou H, John Peter AT, Meiringer C, Subramanian K, Ungermann C. Analysis of DHHC acyltransferases implies overlapping substrate specificity and a two-step reaction mechanism. Traffic. 2009;10:1061–1073. doi: 10.1111/j.1600-0854.2009.00925.x. [DOI] [PubMed] [Google Scholar]
- 82.Bannan BA, Van Etten J, Kohler JA, Tsoi Y, Hansen NM, Sigmon S, Fowler E, Buff H, Williams TS, Ault JG, Glaser RL, Korey CA. The Drosophila protein palmitoylome: characterizing palmitoyl-thioesterases and DHHC palmitoyl-transferases. Fly. 2008;2:198–214. doi: 10.4161/fly.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ducker CE, Stettler EM, French KJ, Upson JJ, Smith CD. Huntingtin interacting protein 14 is an oncogenic human protein: palmitoyl acyltransferase. Oncogene. 2004;23:9230–9237. doi: 10.1038/sj.onc.1208171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mukai J, Dhilla A, Drew LJ, Stark KL, Cao L, MacDermott AB, Karayiorgou M, Gogos JA. Palmitoylation-dependent neurodevelopmental deficits in a mouse model of 22q11 microdeletion. Nat Neurosci. 2008;11:1302–1310. doi: 10.1038/nn.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Raymond FL, Tarpey PS, Edkins S, Tofts C, O’Meara S, Teague J, Butler A, Stevens C, Barthorpe S, Buck G, Cole J, Dicks E, Gray K, Halliday K, Hills K, Hinton J, Jones D, Menzies A, Perry J, Raine K, Shepherd R, Small A, Varian J, Widaa S, Mallya U, Moon J, Luo Y, Shaw M, Boyle J, Kerr B, Turner G, Quarrell O, Cole T, Easton DF, Wooster R, Bobrow M, Schwartz CE, Gecz J, Stratton MR, Futreal PA. Mutations in ZDHHC9, which encodes a palmitoyltransferase of NRAS and HRAS, cause X-linked mental retardation associated with a marfanoid habitus. Am J Hum Genet. 2007;80:982–987. doi: 10.1086/513609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ohyama T, Verstreken P, Ly CV, Rosenmund T, Rajan A, Tien A-C, Haueter C, Schulze KL, Bellen HJ. Huntingtin-interacting protein 14, a palmitoyl transferase required for exocytosis and targeting of CSP to synaptic vesicles. J Cell Biol. 2007;179:1481–1496. doi: 10.1083/jcb.200710061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yanai A, Huang K, Kang R, Singaraja RR, Arstikaitis P, Gan L, Orban PC, Mullard A, Cowan CM, Raymond LA, Drisdel RC, Green WN, Ravikumar B, Rubinsztein DC, El-Husseini A, Hayden MR. Palmitoylation of huntingtin by HIP14 is essential for its trafficking and function. Nat Neurosci. 2006;9:824–831. doi: 10.1038/nn1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.El-Husseini AE-D, Schnell E, Dakoji S, Sweeney N, Zhou Q, Prange O, Gauthier-Campbell C, Aguilera-Moreno A, Nicoll RA, Bredt DS. Synaptic strength regulated by palmitate cycling on PSD-95. Cell. 2002;108:849–863. doi: 10.1016/s0092-8674(02)00683-9. [DOI] [PubMed] [Google Scholar]
- 89.El-Husseini AE, Craven SE, Chetkovich DM, Firestein BL, Schnell E, Aoki C, Bredt DS. Dual palmitoylation of PSD-95 mediates its vesiculotubular sorting, postsynaptic targeting, and ion channel clustering. J Cell Biol. 2000;148:159–171. doi: 10.1083/jcb.148.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Singaraja RR, Hadano S, Metzler M, Givan S, Wellington CL, Warby S, Yanai A, Gutekunst C-A, Leavitt BR, Yi H, Fichter K, Gan L, McCutcheon K, Chopra V, Michel J, Hersch SM, Ikeda J-E, Hayden MR. HIP14, a novel ankyrin domain-containing protein, links huntingtin to intracellular trafficking and endocytosis. Hum Mol Genet. 2002;11:2815–2828. doi: 10.1093/hmg/11.23.2815. [DOI] [PubMed] [Google Scholar]
- 91.Politis EG, Roth AF, Davis NG. Transmembrane topology of the protein palmitoyl transferase Akr1. J Biol Chem. 2005;280:10156–10163. doi: 10.1074/jbc.M411946200. [DOI] [PubMed] [Google Scholar]
- 92.Mitchell DA, Mitchell G, Ling Y, Budde C, Deschenes RJ. Mutational analysis of Saccharomyces cerevisiae Erf2 reveals a two-step reaction mechanism for protein palmitoylation by DHHC enzymes. J Biol Chem. 2010;285:38104–38114. doi: 10.1074/jbc.M110.169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Storer AC, Menard R. Catalytic mechanism in papain family of cysteine peptidases. Methods Enzymol. 1994;244:486–500. doi: 10.1016/0076-6879(94)44035-2. [DOI] [PubMed] [Google Scholar]
- 94.Zhou X, Zhang N, Liu L, Walters KJ, Hanna PE, Wagner CR. Probing the catalytic potential of the hamster arylamine N-acetyltransferase 2 catalytic triad by site-directed mutagenesis of the proximal conserved residue, Tyr190. FEBS Journal. 2009;276:6928–6941. doi: 10.1111/j.1742-4658.2009.07389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang H, Liu L, Hanna PE, Wagner CR. Catalytic Mechanism of Hamster Arylamine N-Acetyltransferase 2. Biochemistry. 2005;44:11295–11306. doi: 10.1021/bi047564q. [DOI] [PubMed] [Google Scholar]
- 96.Sandy J, Mushtaq A, Holton SJ, Schartau P, Noble MEM, Sim E. Investigation of the catalytic triad of arylamine N-acetyltransferases: essential residues required for acetyl transfer to arylamines. Biochem J. 2005;390:115–123. doi: 10.1042/BJ20050277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huang K, Yanai A, Kang R, Arstikaitis P, Singaraja RR, Metzler M, Mullard A, Haigh B, Gauthier-Campbell C, Gutekunst C-A, Hayden MR, El-Husseini A. Huntingtin-interacting protein HIP14 is a palmitoyl transferase involved in palmitoylation and trafficking of multiple neuronal proteins. Neuron. 2004;44:977–986. doi: 10.1016/j.neuron.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 98.Budde C, Schoenfish MJ, Linder ME, Deschenes RJ. Purification and characterization of recombinant protein acyltransferases. Methods. 2006;40:143–150. doi: 10.1016/j.ymeth.2006.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Draper JM, Smith CD. Palmitoyl acyltransferase assays and inhibitors. Mol Membr Biol. 2009;26:5–13. doi: 10.1080/09687680802683839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fernandez-Hernando C, Fukata M, Bernatchez PN, Fukata Y, Lin MI, Bredt DS, Sessa WC. Identification of Golgi-localized acyl transferases that palmitoylate and regulate endothelial nitric oxide synthase. J Cell Biol. 2006;174:369–377. doi: 10.1083/jcb.200601051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Resh MD. Palmitoylation of ligands, receptors, and intracellular signaling molecules. Sci STKE. 2006:re14. doi: 10.1126/stke.3592006re14. [DOI] [PubMed] [Google Scholar]
- 102.Drisdel RC, Alexander JK, Sayeed A, Green WN. Assays of protein palmitoylation. Methods. 2006;40:127–134. doi: 10.1016/j.ymeth.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 103.Hang HC, Geutjes E-J, Grotenbreg G, Pollington AM, Bijlmakers MJ, Ploegh HL. Chemical Probes for the Rapid Detection of Fatty-Acylated Proteins in Mammalian Cells. J Am Chem Soc. 2007;129:2744–2745. doi: 10.1021/ja0685001. [DOI] [PubMed] [Google Scholar]
- 104.Martin BR, Cravatt BF. Large-scale profiling of protein palmitoylation in mammalian cells. Nat Methods. 2009;6:135–138. doi: 10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Draper JM, Xia Z, Smith CD. Cellular palmitoylation and trafficking of lipidated peptides. J Lipid Res. 2007;48:1873–1884. doi: 10.1194/jlr.M700179-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Varner AS, De Vos ML, Creaser SP, Peterson BR, Smith CD. A fluorescence-based high performance liquid chromatographic method for the characterization of palmitoyl acyl transferase activity. Analytical Biochemistry. 2002;308:160–167. doi: 10.1016/s0003-2697(02)00212-9. [DOI] [PubMed] [Google Scholar]
- 107.Hensel J, Hintz M, Karas M, Linder D, Stahl B, Geyer R. Localization of the palmitoylation site in the transmembrane protein p12E of Friend murine leukemia virus. Eur J Biochem. 1995;232:373–380. doi: 10.1111/j.1432-1033.1995.373zz.x. [DOI] [PubMed] [Google Scholar]
- 108.Liang X, Nazarian A, Erdjument-Bromage H, Bornmann W, Tempst P, Resh MD. Heterogeneous fatty acylation of Src family kinases with polyunsaturated fatty acids regulates raft localization and signal transduction. J Biol Chem. 2001;276:30987–30994. doi: 10.1074/jbc.M104018200. [DOI] [PubMed] [Google Scholar]
- 109.Hoffman MD, Kast J. Mass spectrometric characterization of lipid-modified peptides for the analysis of acylated proteins. J Mass Spectrom. 2006;41:229–241. doi: 10.1002/jms.981. [DOI] [PubMed] [Google Scholar]
- 110.Zhao Z, Hou J, Xie Z, Deng J, Wang X, Chen D, Yang F, Gong W. Acyl-biotinyl exchange chemistry and mass spectrometry-based analysis of palmitoylation sites of in vitro palmitoylated rat brain tubulin. Protein J. 2010;29:531–537. doi: 10.1007/s10930-010-9285-x. [DOI] [PubMed] [Google Scholar]
- 111.Drisdel RC, Green WN. Labeling and quantifying sites of protein palmitoylation. BioTechniques. 2004;36:276–282. 284–285. doi: 10.2144/04362RR02. [DOI] [PubMed] [Google Scholar]
- 112.Yang W, Di Vizio D, Kirchner M, Steen H, Freeman MR. Proteome scale characterization of human S-acylated proteins in lipid raft-enriched and non-raft membranes. Mol Cell Proteomics. 2010;9:54–70. doi: 10.1074/mcp.M800448-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yount JS, Moltedo B, Yang Y-Y, Charron G, Moran TM, Lopez CB, Hang HC. Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat Chem Biol. 2010;6:610–614. doi: 10.1038/nchembio.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Merrick BA, Dhungana S, Williams JG, Aloor JJ, Peddada S, Tomer KB, Fessler MB. Proteomic profiling of S-acylated macrophage proteins identifies a role for palmitoylation in mitochondrial targeting of phospholipid scramblase. :3. doi: 10.1074/mcp.M110.006007. [DOI] [PMC free article] [PubMed] [Google Scholar]