Abstract
Background:
The previous studies showed that herpes human virus-6 (HHV-6) and HHV-7 exist in salivary glands. One of the important areas in oral and maxillofacial pathology field is tumors of the salivary glands. In this study, to declare the major sites of persistent infection with HHV-6 and HHV-7, the existence of HHV-6 and HHV-7 genomes in formalin-fixed paraffin embedded tissue samples of salivary gland tumors.
Methods:
This analytical study was performed in 60 paraffin blocks samples of malignant and benign neoplasms of both major and minor salivary glands. This study performed with highly sensitive real time PCR method.
Results:
Among 60 paraffin blocks salivary gland tumors with equal chances of presence of the HHV-7 and HHV-6 in the samples, 34% were positive for both HHV-7 and HHV-6 while 47.2% were only positive for HHV-7, 18.9% samples were positive for HHV-6. A relationship was noticed between HHV-7 and HHV-6 genomes.
Conclusion:
In conclusion, this study showed no relation between virus and diseases with P=0.953. Also it could be inferred that there is a relationship between HHV-6 and 7 in salivary glands neoplasms.
Keywords: Human herpes virus, HHV-6, HHV-7, Real time PCR, Salivary gland, Tumors
1. INTRODUCTION
Human herpes virus 6 (HHV-6) and human herpes virus 7 (HHV-7) during childhood are really widespread (1, 2), and the same as other herpesviruses, they are latent infections throughout life. Salivary glands act as major sites harbouring persistent HHV-6 infection when HHV-6 is frequently isolated from the saliva of healthy individuals (3, 4, 5). However, some studies represented that HHV-7, instead of HHV-6, is isolated from saliva frequently (6, 7, 8, 9), contradict previous reports. Tumours of the salivary glands are an important area in the field of oral and maxillofacial pathology.
The annual incidence of salivary gland tumours shows in Isfahan, Iran is far greater (1.13%) than the world incidence that is about 1 to 6.5 cases per 100,000 people (10). 20% of human malignancies are due to persistent viral infections, and tobacco is the second major risk factor for human carcinomas. Herpes viruses have recognized to be the reason of several malignant and benign oral lesions (11). In the present study, to declare the major sites of persistent infection with HHV-6 and HHV-7, the existence of HHV-6 and HHV-7 genomes in formalin-fixed paraffin embedded tissue samples of salivary gland tumours were examined through highly sensitive real time PCR method.
2. PATIENTS AND METHODS
This analytical, descriptive study was performed in 60 formalin-fixed paraffin embedded tissue samples of salivary gland tumours, 23 benign salivary gland tumours including pleomorphic adenoma. Monomorphic adenoma and 37 malignant salivary gland tumours including mucoepidermiod carcinoma and adenoid salivary gland specimens were obtained from surgery or autopsy from adult patients with oral cancer, sialoadenitis, pleomorphic adenoma of the salivary glands, and other diseases. First, 5-10 µm tissue sections of formalin fixed, paraffin wax embedded tissue blocks (depending on tissue size) were transferred into 1.5 mL eppendorf tubes for DNA isolation. To avoid cross-contamination, a new, sterile, and disposable microtome blade was used immediately before cutting each block for cleaning purposes.
Subsequently, DNA extraction was performed by using the procedures according to the manufacturer’s protocol described in the high pure nucleic acid extraction kit (Roche, Germany). First, the paraffin wax was dissolved in 300 µl of citrisoly (xylene substitute) and was washed with ethanol to remove the citrisoly. Then cell lysis buffer (200 µl) and 20 µl proteinase K solutions (20 mg/ml) were added to each sample, followed by an overnight incubation at 55°C. After the solution had cooled down to room temperature, 200 µl binding buffer was added and the high pure filter tube was combined with collection tube. They were centrifuged and the flow-through was discarded, then 500 µl Inhibitor removal buffer was added followed by centrifuging. In the next step, the DNA was washed with 500 µl washing buffer and eluted with the elution buffer.
The absorbance of a sample of DNA solution was measured for concentration at 260 nm using an ultraviolet spectrometer. The absorbance ratio at 260 and 280 nm (A260/280) was used to evaluate DNA purity. The procedure was carried out with some minor modifications, for instance, longer incubation times (overnight, approximately 16 hours) and doubling proteinase K concentration (12).
The Real-time PCR mixture contained 50 Mm KCl, 10 Mm Tris-Hcl (PH=8. 3) and 2. 25 M MgCl2 as a 1X reaction buffer, 200 mM of each dNTP, 10 µm of each of primers and 1 U Taq polymerase (Roche, Germany).
Real time PCR was performed with HHV-7 and HHV-6 primers including forward and reverse primer with labels probe for HHV-7 and HHV-6 (Table1). The PCR cycling temperatures for HHV-7 were 2 min of incubation at 50 °C then followed by 2 min at 95° C; the samples were subjected to 45 cycle’s for 20 sec at 95 °C followed by 1 min at 60° C (13).
Table 1.
Primer and probe for PCR. *Herpes virus 6- forward primer (HHV-6-F), Herpes virus 6- reverses primer (HHV-6-R); Herpes virus 7- forward primer (HHV-7-F), Herpes virus 7- reverses primer (HHV-7-R); Herpes virus 6-probe (HHV-6-P), Herpes virus7-probe (HHV-7-P).
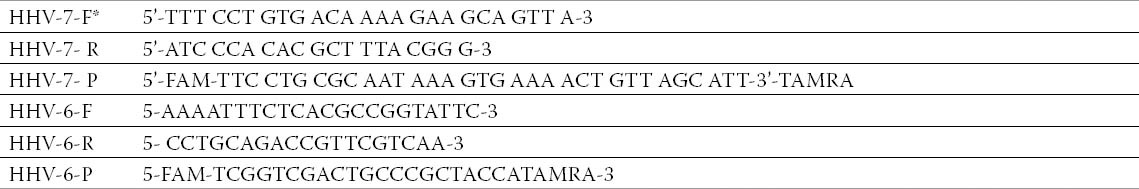
Amplification HHV-6 was carried out in a 25 µl volume reaction mixture by use 1,100 nM each primer, and 200 nM probe. The reaction mixtures were incubated at 95 °C for 10 min, followed by 45 cycles of 95 °C for 15 sec and 60 °C for 1 min (14).
3. STATISTICAL ANALYSIS
The relationship of two groups was analysed through Mc-Nemar test by SPSS software (version 11, Chicago, IL, USA). A p value less than 0.05 were considered significant.
4. RESULTS
The persistent infection with HHV-6 and HHV-7, the existence of HHV-6 and HHV-7 genomes in formalin-fixed paraffin embedded tissue samples of salivary gland tumors is shown in Tables 2, 3 4 and 5. Of the 60 paraffin blocks of malignant and benign neoplasms of both major and minor salivary gland with equal chances of presence of HHV-7 and HHV-6 in the samples were compared. Out of the 60 samples, 18 were positive for both HHV-7 and HHV-6 while 25 were only positive for HHV-7, 10 samples were positive for HHV-6 but negative for HHV-7, and 7 samples were reported negative for both HHV-6 and HHV-7. A relationship was noticed between HHV-7 and HHV-6 genomes. It can, therefore, be suggested that a relationship is likely to exist between these two viruses in salivary glands neoplasms.
Table 2.
Disease and HHV-6 and HHV-7 positive
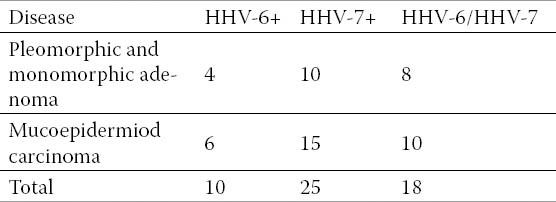
Table 3.
Case processing

Table 4.
Disease viruses cross tabulation
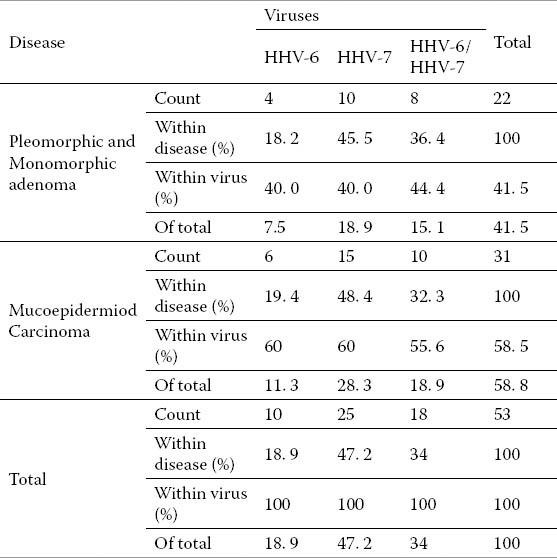
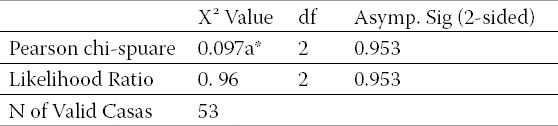
Table 5.
Chi-square tests. *a.1 cells (16.7) have expected count less than 5. The minimum expected count is 4.15.
5. DISCUSSION
In Maxillofacial pathology, salivary gland tumours are prominent, (14). Recent studies have focused on the role of viruses in salivary gland neoplasms. The HHV-6 and the HHV-7 have been identified as one of the most important oncogenic viruses capable of producing a number of oncogenic factors such as BCl2, BCl10. The HHV-6 has also been identified in salivary gland neoplasms (15). A number of studies have suggested the presence of numerous oncogenic zones in the genome of this virus. The role of this virus as an etiological agent in cancer requires further research for validation. Di-Luca at 1995 (7), reported the presence of HHV-6 genome in 63% of their healthy salivary glands and only in 3% of their saliva samples. Levy at 1997 (16), proposed HHV-6 as the agent for certain neoplasms such as lymphoma, leukemia, and cervical carcinoma. This is while HHV-6 has been recognized to have the capacity to activate other HHVs such as EBV and CMV as well as papilloma viruses. Zhou et al 2007 (17), reported a relationship between EBV and HHV-6 infections and the histological progress of angio-immunoblastic T-cell lymphoma. Chen and Hudnall (18), in 2006 examined 8 autopsy samples from all body parts including 4 males and 4 females in the presence of 8 types of herpes virus (EBV, CMV, VZV, HSV-2, HSV-1, HHV-6, HHV-7, and HHV-8) and only EBV, HHV-6, and HHV-7 in all their samples were found. Based on these findings, we decided to investigate, for the first time, the presence of HHV-7 and HHV-6 in salivary gland neoplasms in order to determine the relationships, if any, between these viruses and the related salivary gland tumours. The hypothesis of no correlation between HHV-7 and HHV-6 genomes in salivary gland neoplasms was refuted using the Mcnemar’s statistical test. Chi-square tests have shown in Table 5.
6. conclusion
This study shows no relation between virus and diseases.
Footnotes
CONFLICT OF INTEREST: NONE DECLARED.
REFERENCES
- 1.Wyatt LS, Rodriguez WJ, Balachandran N, et al. Human herpes virus 7 antigenic properties and prevalence in children and adults. J Virol. 1991;65:6260–6265. doi: 10.1128/jvi.65.11.6260-6265.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamanishi K. Human herpes virus 6. Microbiol Immunol. 1992;36:551–561. doi: 10.1111/j.1348-0421.1992.tb02055.x. [DOI] [PubMed] [Google Scholar]
- 3.Cone RW, Huang MLW, Ashley R, et al. Human herpesvirus 6 DNA in peripheral blood cells and saliva from immunocompetent individuals. J Clin Microbiol. 1993;31:1262–1267. doi: 10.1128/jcm.31.5.1262-1267.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fox JD, Briggs M, Ward PA, et al. Human herpesvirus 6 in salivary glands. Lancet. 1990;336:590–593. doi: 10.1016/0140-6736(90)93392-3. [DOI] [PubMed] [Google Scholar]
- 5.Harnett GB, Farr TJ, Pietroboni GR, et al. Frequent shedding of human herpesvirus 6 in saliva. J Med Virol. 1990;30:128–130. doi: 10.1002/jmv.1890300209. [DOI] [PubMed] [Google Scholar]
- 6.Black JB, Inoue N, Kite-Powell K, et al. Frequent isolation of human herpes virus 7 from saliva. Virus Res. 1993;29:91–98. doi: 10.1016/0168-1702(93)90128-a. [DOI] [PubMed] [Google Scholar]
- 7.Di-Luca D, Mirandola P, Ravaioli T, et al. Human herpesviruses 6 and 7 in salivary glands and shedding in saliva of healthy and human immunodeficiency virus positive individuals. J Med Virol. 1995;45:462–468. doi: 10.1002/jmv.1890450418. [DOI] [PubMed] [Google Scholar]
- 8.Hidaka Y, Liu Y, Yamamoto M, et al. Frequent isolation of human herpesvirus 7 from saliva samples. J Med Virol. 1993;40:343–346. doi: 10.1002/jmv.1890400416. [DOI] [PubMed] [Google Scholar]
- 9.Sada E, Yasukawa M, Ito C, et al. Detection of human herpesvirus 6 and human herpesvirus 7 in the submandibular gland, parotid gland, and lip salivary gland by PCR. J Clin Mic. 1996:2320–2321. doi: 10.1128/jcm.34.9.2320-2321.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghafghazi SH. Isfahan University of Medical Sciences Press; 1998. Studding the frequenc of patients with neoplastic salivary gland lesions in regard to sex and age; pp. 1981–1996. [Google Scholar]
- 11.Flaitz CM, Hicks MJ. Molecular piracy: the viral link to carcinogenesis. Oral Oncol. 1998;34:448–53. doi: 10.1016/s1368-8375(98)00057-8. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert MT, Haselkorn T, et al. The Isolation of Nucleic Acids from Fixed, Paraffin-Embedded Tissues – Which Methods Are Useful When? PLOS One. 2007;2:537. doi: 10.1371/journal.pone.0000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zerr DM, Huang ML, et al. Sensitive Method for Detection of Human Herpesviruses 6 and 7 in Saliva Collected in Field Studies. J Clin Microbiol. 2000;38:1981–1983. doi: 10.1128/jcm.38.5.1981-1983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tavakoli NP, Nattanmai S, Hull R, et al. Detection and typing of human herpesvirus 6 by molecular methods in specimens from patients diagnosed with encephalitis or meningitis. J Clin Microbiol. 2007;45:3972. doi: 10.1128/JCM.01692-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nevile BW, Damm DD, Allen CM, Bouquot JE. 2nd ed. New York: McGraw Hill; 2001. Oral and maxillofacial pathology; p. 406. [Google Scholar]
- 16.Levy JA. Three new human herpesviruses (HHV6, 7, and 8) Lancet. 1997;349:558–563. doi: 10.1016/S0140-6736(97)80119-5. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Y, Attygalle AD, Chuang SS, et al. Angioimmunoblastic T-cell lymphoma: histological progression associates with EBV and HHV6B viral load. Br J Haematol. 2007;138:44–53. doi: 10.1111/j.1365-2141.2007.06620.x. [DOI] [PubMed] [Google Scholar]
- 18.Chen T, Hudnall SD. Anatomical mapping of human herpesvirus reservoirs of infection. Mod Pathol. 2006;19:726–737. doi: 10.1038/modpathol.3800584. [DOI] [PubMed] [Google Scholar]


