Abstract
Long non-coding RNAs (lncRNAs) represent an emerging layer of cancer biology, contributing to tumor proliferation, invasion, and metastasis. Here, we describe a role for the oncogenic lncRNA PCAT-1 in prostate cancer proliferation through cMyc. We find that PCAT-1–mediated proliferation is dependent on cMyc protein stabilization, and using expression profiling, we observed that cMyc is required for a subset of PCAT-1–induced expression changes. The PCAT-1–cMyc relationship is mediated through the post-transcriptional activity of the MYC 3′ untranslated region, and we characterize a role for PCAT-1 in the disruption of MYC-targeting microRNAs. To further elucidate a role for post-transcriptional regulation, we demonstrate that targeting PCAT-1 with miR-3667-3p, which does not target MYC, is able to reverse the stabilization of cMyc by PCAT-1. This work establishes a basis for the oncogenic role of PCAT-1 in cancer cell proliferation and is the first study to implicate lncRNAs in the regulation of cMyc in prostate cancer.
Keywords: PCAT-1, lncRNA, cell proliferation, 3’UTR, cMyc
Introduction
The maturation and spread of high-throughput sequencing technologies has enabled increasingly complex analyses of the cellular transcriptome, with the nomination of numerous novel RNA species [1], [2]. Among these, long non-coding RNAs (lncRNAs) over 200 bp in length have been implicated as fundamental actors in numerous molecular processes, including cell differentiation, lineage specificity, neurologic disorders, and cancer [3], [4], [5].
In prostate cancer, the first prominent lncRNA, PCA3, was initially described as a novel biomarker of disease [6] and subsequently defined as a promising urine test for this disease [7]. Similarly, the lncRNA PCGEM1 has been implicated in prostate cancer as a regulator of apoptosis [8] and newer lncRNAs are being pursued through high-throughput technologies [9]. Using next-generation sequencing of a large cohort of prostate cancer tissues, we recently defined the landscape of lncRNAs in prostate cancer, including SChLAP1 as a regulator of metastasis [10], [11]. On the basis of this unbiased whole-transcriptome analysis, we have previously nominated PCAT-1 as the top-ranked, upregulated lncRNA in prostate cancer tissues [10], and independent studies have confirmed overexpression of PCAT-1 in prostate cancer [9] and implicated PCAT-1 as a prognostic biomarker for colorectal cancer metastasis and poor patient survival [12].
Here, we characterize the function of the lncRNA PCAT-1 in prostate cancer. We show that PCAT-1 promotes prostate cell proliferation and that this phenotype is mediated through up-regulation of the cMyc protein (encoded by the MYC gene). Antagonism of cMyc is able to reverse PCAT-1–mediated cell proliferation. We show that PCAT-1 regulates cMyc post-transcriptionally through the MYC 3′ untranslated region (UTR). Further, we find a protective effect of PCAT-1 on cMyc by interfering with the regulation of MYC by miR-34a. Conversely, microRNA-based targeting of PCAT-1 reverses its effect on cMyc. These studies elucidate a novel mechanism of cMyc stabilization in prostate cancer and describe a post-transcriptional function of PCAT-1 in the antagonism of microRNAs.
Material and Methods
Cell Lines and Reagents
All cell lines were purchased from the American Type Culture Collection (Manassas, VA) and maintained in standard media. PCAT-1 overexpression cell lines were generated using the pLenti6 vector (Invitrogen, Carlsbad, CA), and stably transfected cells were selected by 2.5 μg/ml blasticidin (Invitrogen). PCAT-1 stable knockdown LNCaP cell lines were transfected with appropriate shRNA lentiviral constructs and a green fluorescent protein (GFP)-positive cell fraction was selected by 1 μg/ml puromycin (Sigma, St Louis, MO). All cell lines were used for fewer than 6 months after resuscitation. Cell lines were confirmed by genotyping and were analyzed by the University of Michigan DNA Sequencing Core using the Profiler Plus PCR Amplification Kit (Applied Biosystems, Foster City, CA). Details on the generation of these isogenic cell lines have been previously published and can be found in reference [13] as well as in the Supplementary Materials.
Cell Proliferation Assays
Cell proliferation assays were performed according to standard protocols using trypan blue staining to enumerate the surviving fraction of cells (see Supplementary Materials).
siRNA Knockdown and MicroRNA Transfection Studies
siRNA knockdowns were performed according to standard protocols (see Supplementary Materials). siRNA sequences (in sense format) for PCAT-1 knockdown were given as follows: siRNA 3 AUACAUAAGACCAUGGAAAU; siRNA 4 GAACCUAACUGGACUUUAAUU. Commercial cMyc siRNAs were obtained from Dharmacon (Dharmacon, Lafayette, CO).
For microRNA transfection, respective microRNA mimics and negative controls were purchased from Ambion (Austin, TX), and transfections were performed with Oligofectamine (Invitrogen) as described previously [14].
Bromouridine Pulse-Chase Labeling Assay
Cells were plated in 100-mm dishes and cultured until the cell confluency reaches 75%. After 30 minutes of bromouridine labeling, followed by uridine chases for 0 and 6 hours, cells were washed with cold phosphate-buffered saline and harvested with TRIzol. Total cell RNA was isolated by a standard protocol, and nascent Bru-labeled mRNA was specifically isolated with anti-bromodeoxyuridine antibodies. Samples were subjected to quantitative polymerase chain reaction (qPCR), and the expression level of target gene was determined. The gene expression levels at 0 and 6 hours represent nascent mRNA synthesis and stability, respectively.
RNA Isolation, cDNA Synthesis, and qPCR Experiments
RNA isolation and cDNA synthesis were performed with TRIzol and Superscript III (Invitrogen), respectively, according to standard protocols (see Supplementary Materials). qPCR was performed with SYBR Green. The primer sequences are listed below: PCAT-1-Forward: TGAGAAGAGAAATCTATTGGAACC, PCAT-1-Reverse: GGTTTGTC-TCCGCTGCTTTA; MYC-Forward: GCTCGTCTCAGAGAAGCTGG, MYC-Reverse: GCTCAGATCCTGCAGGTACAA; ACTB-Forward: AAGGCCAACCGCGAGAAG, ACTB-Reverse: ACAGCCTGGATAGCAACGTACA. Additional primer pair sequences can be found in Table W9.
Immunoblot Analysis
With or without treatment, cells were harvested and lysed in RIPA lysis buffer (Sigma) supplemented with protease inhibitor cocktail (Roche, Basel, Switzerland). Western blot analysis was performed with standard protocols as described before [14], [15]. The protein band densitometry was determined using the image analysis software package Image J (version 1.42, available from the Research Services Branch of NIH).
Luciferase Reporter Assays
The cMyc 3′UTR was amplified using reverse transcription–PCR from LNCAP prostate cancer cells and subcloned into the pRL-TK vector. DU145 control, DU145–PCAT-1, RWPE-LacZ, RWPE–PCAT-1 pool, LNCaP sh–NT, LNCaP sh–PCAT-1–#1, and LNCaP sh–PCAT-1–#2 were plated onto 24-well plates and transfected with cMyc 3′UTR luciferase constructs as well as pRL-TK vector as internal control for luciferase activity. After 48 hours of incubation, luciferase reporter assay were conducted with the dual luciferase assay kit (Promega, Madison, WI).
Gene Expression Microarrays
Expression profiling was performed using the Agilent Whole Human Genome Oligo Microarray (Santa Clara, CA), according to previously published protocols. All samples were run in technical duplicates comparing RWPE and Du145 cells overexpressing PCAT-1 and treated with either non-targeting siRNAs or siRNAs targeting cMyc. Expression array data were processed to yield log-2 expression values. Genes that were at least > 1.5-fold changed in one cell line and at least > 0.5-fold changed in the second cell line were considered to be significantly upregulated or downregulated. All cell line replicates were considered. For gene ontology analysis, upregulated and downregulated probes were separated and probe lists were analyzed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatics platform with the functional annotation clustering tool. See the Supplementary Materials for additional details.
Statistical Analyses for Experimental Studies
All data are expressed as means ± SD or SEM. All experimental assays were performed in duplicate or triplicate. Statistical analysis was performed by two-sided Student’s t test and one-way analysis of variance (ANOVA) with a Student-Newman-Keuls follow-up test. Significance was designed as “*” when P < .05.
Results
cMyc Is Required for PCAT-1–Mediated Cell Proliferation
To establish a role for PCAT-1 in prostate cancer, we overexpressed PCAT-1 in Du145 prostate cancer and RWPE benign immortalized prostate cells (Figure W1A). We further used two shRNA constructs targeting PCAT-1 to deplete endogenous PCAT-1 levels in the LNCAP prostate cancer cell line (Figure W1A). Additional details on the characterization of these cell lines can be found in reference [13]. We found that overexpression of PCAT-1 substantially increased cell proliferation in Du145 and RWPE cells, whereas PCAT-1 knockdown in LNCAP cells reduced cell proliferation (Figure W1, B–D). We further confirmed that knockdown of PCAT-1 in Du145–PCAT-1 and RWPE–PCAT-1 cells reversed this cell proliferation phenotype, supporting the specificity of our overexpression models (Figure W2). Finally, we confirmed that our in vitro models expressed PCAT-1 at a level similar to endogenous expression by comparing our models with PCAT-1 expression from a previously published cohort [10] of prostate cancer tissues (Figure W3).
We next sought to describe a mechanistic basis for the role of PCAT-1 in cell proliferation. To this end, we noted that PCAT-1 is located on chr8q24, just 725 kb upstream of the MYC oncogene (Figure 1A, left). Given this close proximity, we hypothesized that PCAT-1 may regulate MYC to achieve its functions. Indeed, the cis-regulation of neighboring protein-coding genes is a common mechanism for numerous lncRNAs, including the well-studied lncRNAs such as H19 and Xist [5]. In cancer, the cis-regulation of neighboring protein-coding genes by lncRNAs has also been implicated in oncogenic processes such as cell invasion [16].
Figure 1.
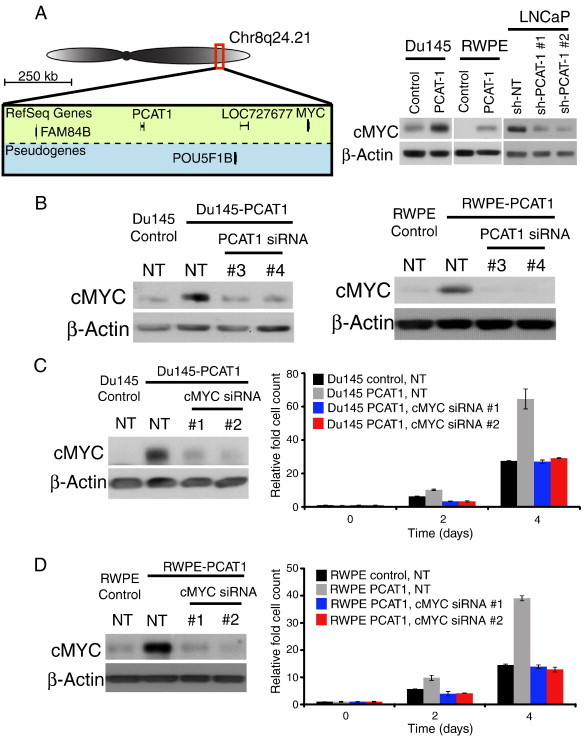
PCAT-1 mediates cell proliferation through up-regulation of cMyc. (A) Left: A schematic demonstrating the genomic location of MYC and PCAT-1 on chr8q24. PCAT-1 is approximately 750 kb upstream of MYC. Right: Western blot analysis correlating cMyc protein levels with PCAT-1 expression. cMyc is upregulated with PCAT-1 overexpression and downregulated with PCAT-1 knockdown. (B) Left: Knockdown of PCAT-1 in Du145–PCAT-1 cells decreases cMyc protein abundance. Right: Knockdown of PCAT-1 in RWPE–PCAT-1 cells decreases cMyc protein abundance. (C) Left: Western blot analysis demonstrating efficiency of cMyc knockdown in Du145–PCAT-1 cells. Right: Cell proliferation of Du145–PCAT-1 cells with and without cMyc knockdown. (D) Left: Western blot analysis demonstrating efficiency of cMyc knockdown in RWPE–PCAT-1 cells. Right: Cell proliferation of RWPE–PCAT-1 cells with and without cMyc knockdown. Error bars represent mean ± SEM. In this figure, NT stands for non-targeting control.
Surprisingly, we found that cMyc protein levels were dramatically increased by PCAT-1 overexpression in Du145 and RWPE (Figure 1A, right). Consistent with these observations, knockdown of PCAT-1 in LNCAP cells reduced cMyc protein (Figure 1A, right). To verify this relationship, we next performed siRNA knockdown of PCAT-1 in Du145–PCAT-1 and RWPE–PCAT-1 cells and observed a substantial down-regulation of the cMyc protein, confirming the specificity of PCAT-1–mediated up-regulation of cMyc (Figure 1B). To determine whether cMyc up-regulation played a role in PCAT-1–mediated pathogenesis, we performed siRNA knockdown of cMyc in PCAT-1–overexpressing cell lines. We found that cMyc knockdown fully abrogated the proliferative effect of PCAT-1 overexpression in Du145 and RWPE (Figure 1, C and D). PCAT-1–mediated cell proliferation is thus dependent on cMyc overexpression.
Identification of a PCAT-1–cMyc Regulatory Gene Expression Program
We next sought to define downstream effectors of PCAT-1 and cMyc through the analysis of gene expression changes following modulation of these factors. Specifically, to evaluate whether cMyc stabilization was necessary for PCAT-1–mediated gene expression signatures, we performed gene expression array analysis of Du145–PCAT-1 and RWPE–PCAT-1 cells treated with cMyc or non-targeting siRNAs and compared them to control RWPE and Du145 cells lacking PCAT-1. We confirmed the microarray reproducibility by running each sample in duplicate (Figure W4A). Next, we observed a significant overlap (P < .05 by Fisher exact test) for probes differentially regulated with PCAT-1 overexpression in RWPE and Du145 (Figure W4B). Using this set of probes, we defined a signature of 184 genes that were significantly upregulated or downregulated by PCAT-1 and whose expression change was abrogated by cMyc knockdown (Figure 2A and Table W1). Analysis of 91 genes downregulated by PCAT-1 and dependent on cMyc using the DAVID bioinformatics platform revealed enrichment for cytochrome P450 genes, which are known downregulated targets of cMyc (Table W2) [17], [18], [19]. Conversely, analysis of 93 genes upregulated by PCAT-1 and dependent on cMyc demonstrated an enrichment of genes involved in protein biosynthesis and transcriptional elongation, well-described functions of cMyc (Table W3) [20], [21]. This included both ribosomal subunits such as RPL9 and RPLP0 as well as the translation initiation factors EIF4B and EEF1A2. Interestingly, PCAT-1 also required cMyc for the down-regulation of several well-established tumor suppressor genes such as CDKN2B, a known cMyc target [22], [23], as well as TP73 and IKZF2. Together, these data support a role for cMyc in the control of cancer-specific gene expression signatures mediated through PCAT-1.
Figure 2.
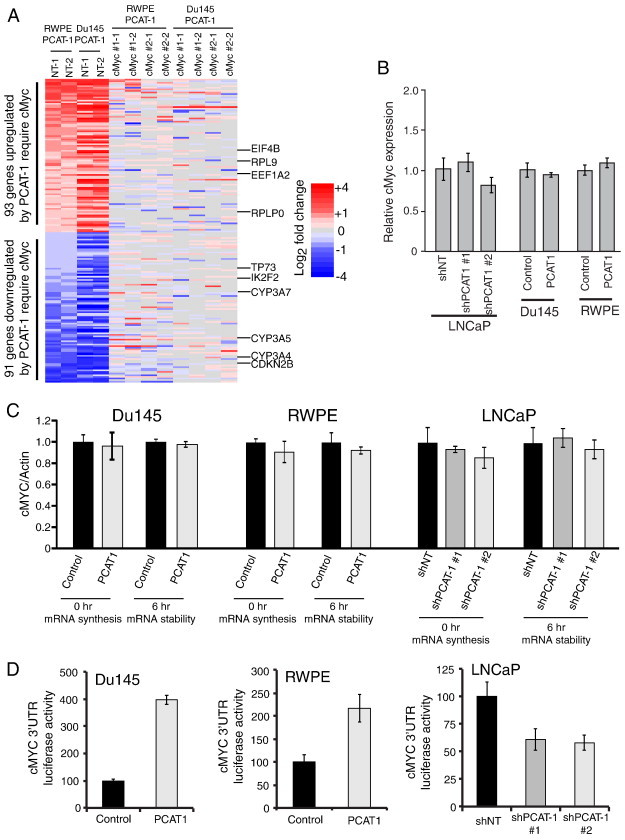
PCAT-1 regulates cMyc in a post-transcriptional manner by 3′UTR activation. (A) Gene expression analysis of Du145–PCAT-1 and RWPE–PCAT-1 cells treated with non-targeting or cMyc siRNAs reveals a gene signature of genes co-regulated by PCAT-1 and cMyc. In this figure, the columns on the far left show gene expression changes in RWPE–PCAT-1 and Du145–PCAT-1 cells treated with non-targeting siRNA compared to RWPE and Du145 control cells treated with non-targeting siRNA, respectively. On the right, RWPE–PCAT-1 and Du145–PCAT-1 cells were treated with two independent siRNAs for cMyc, and the indicated gene expression values of these cells were calculated relative to the RWPE and Du145 control cells treated with non-targeting siRNA. All experiments were analyzed with duplicate microarrays, as indicated at the top of the heat map. (B) MYC mRNA expression levels are unchanged in three in vitro model systems of PCAT-1. (C) MYC mRNA stability is unchanged in Du145–PCAT-1, RWPE–PCAT-1, or LNCaP sh–PCAT-1 cells. (D) Luciferase signal for the MYC 3′UTR activity is increased in Du145–PCAT-1 and RWPE–PCAT-1 cells but decreased in LNCaP sh–PCAT-1 cells. In this figure, NT stands for non-targeting control.
Post-Transcriptional Regulation of cMyc by PCAT-1
To establish a mechanistic basis for cMyc overexpression by PCAT-1, we next assessed MYC mRNA levels in our in vitro models. We found that MYC mRNA levels were unchanged in our LNCaP, Du145, and RWPE cell lines (Figure 2B). To further examine the regulation of cMyc, we performed bromouridine pulse-chase labeling assays to test the effect of PCAT-1 on the synthesis and stability of MYC mRNA. Similarly, we found no change in MYC RNA stability in our model systems (Figure 2C). Taken together, these data argue that the regulation of cMyc by PCAT-1 operates at the post-transcriptional level.
Post-transcriptional regulation of mRNAs is frequently coordinated through the activity of microRNAs binding to the 3′UTR [24], [25]. Specifically, increased activity of the 3′UTR can result in increased protein abundance and gene activation. To test whether PCAT-1 regulated MYC through its 3′UTR, we transfected MYC 3′UTR luciferase constructs into our isogenic cell lines. Supporting a role in post-transcriptional regulation, we found that PCAT-1 overexpression increased MYC 3′UTR activity, whereas knockdown of PCAT-1 decreased MYC 3′UTR activity (Figure 2D).
MicroRNA-Mediated Cross Talk between MYC and PCAT-1
Recently, several groups have argued that a subset of lncRNAs may operate as coordinators of microRNA activity by disrupting microRNA regulation of mRNA 3′UTRs [26], [27], [28]. lncRNAs may thus interfere with microRNA function. Conversely, microRNAs that target lncRNAs would be predicted to titrate away the effect of the lncRNAs. To apply this model to PCAT-1 regulation of cMyc, we employed four microRNA-target prediction algorithms (miRanda, miRBase, TargetScan, and PicTar) to nominate microRNAs that target MYC or PCAT-1 (Tables W4–W7). For MYC, we recovered known microRNA regulators such as the miR-34 family (including miR-34a, miR-34b, and miR-34c), which has been previously reported to downregulate MYC in multiple prostate cancer model systems [29], [30], [31]. For PCAT-1, we surprisingly found only one microRNA, miR-3667-3p, that was predicted to form high-confidence complementary base pairing with PCAT-1 (Figure 3A).
Figure 3.
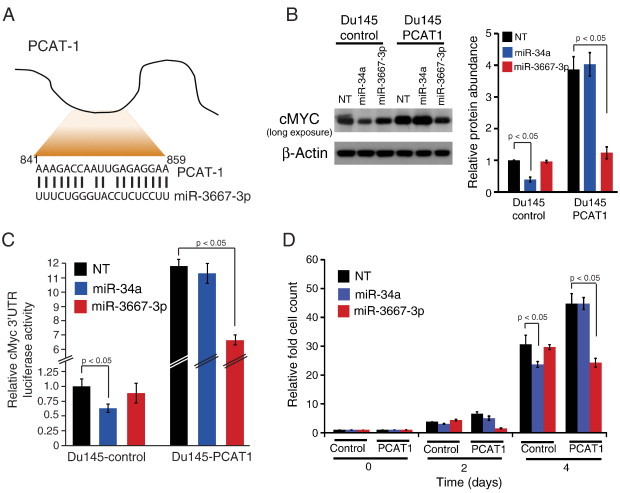
PCAT-1 interferes with microRNA networks regulating cMyc. (A) A schematic demonstrating base-pair complementation between PCAT-1 and miR-3667-3p. (B) Left: cMyc protein abundance in Du145 control and Du145–PCAT-1 cells following overexpression of miR-34a or miR-3667p. Note that a long exposure of the immunoblot is shown to illustrate relative protein abundance. Right: Quantification of cMyc protein abundance. Significance is determined by three independent experiments. (C) MYC 3′UTR luciferase signal following overexpression of miR-34a or miR-3667-3p in cells with and without PCAT-1 overexpression. (D) Cell proliferation of Du145 control or Du145–PCAT-1 cells following overexpression of miR-34a or miR-3667-3p. A non-targeting NT scrambled RNA serves as a control. Error bars represent mean ± SEM. Statistical significance is determined by Student's t test. In this figure, NT stands for non-targeting control.
To assess the effect of PCAT-1 on microRNA signaling, we overexpressed miR-34a, a known regulator of MYC[29], and miR-3667-3p, which was nominated as a PCAT-1 regulator, in Du145 control and Du145–PCAT-1 cells. We predicted that PCAT-1 should antagonize the effects of miR-34a, preventing its regulation of cMyc; conversely, miR-3667-3p should antagonize PCAT-1 and impact cMyc levels only in PCAT-1–overexpressing cells. When tested in our in vitro models, we observed that miR-34a appropriately decreased cMyc abundance in Du145 control cells but not in Du145–PCAT-1 cells (Figure 3B). This suggests that PCAT-1 expression exerts a protective effect on cMyc by interfering with microRNA-based regulation. By contrast, overexpression of miR-3667-3p decreased cMyc protein levels exclusively in Du145–PCAT-1 cells (Figure 3B). Quantification of cMyc protein abundance from these experiments confirmed the significance of these results (Figure 3B, right). We further confirmed these phenomena in RWPE PCAT-1 overexpression cells, where miR-34a and miR-3667-3p overexpression produced highly concordant results (Figure W5). We also assessed the endogenous levels of miR-34a and miR-3667-3p in both parental cell lines and overexpression lines (Figure W6).
To confirm that this regulation of cMyc was post-transcriptional, we verified that overexpression of miR-34a and miR-3667-3p specifically decreased the MYC 3′UTR activity in Du145 control and Du145–PCAT-1 cells, respectively (Figure 3C). Further, we reasoned that if miR-3667-3p affects cMyc protein abundance by targeting PCAT-1, then PCAT-1 mRNA levels should be depleted on overexpression of this microRNA. Indeed, we found that PCAT-1 mRNA levels decreased approximately 50% when miR-3667-3p was overexpressed in Du145–PCAT-1 cells (Figure W7). Finally, to demonstrate the functional impact of miR-34a and miR-3667-3p, we performed cell proliferation assays. Consistent with our model, miR-34a significantly decreased proliferation only in Du145 control cells, whereas miR-3667-3p decreased proliferation only in Du145–PCAT-1 cells (Figure 3D). Analyzing the data shown in Figure 3, B to D, for statistical significance using a one-way ANOVA test further confirmed a significant P value < .05 for all experiments (Table W8). Together, these data support a model in which PCAT-1 interferes with the regulation of MYC by microRNAs (e.g., miR-34a), thereby increasing cMyc protein levels and enhancing cell proliferation (Figure W8). Targeting of PCAT-1 by miR-3667-3p is able to reverse its effects by depleting PCAT-1 and preventing it from upregulating MYC 3′UTR activity and protein abundance.
Discussion
Taken together, this study represents the first analysis of lncRNA-based regulation of cMyc in prostate cancer. We find that PCAT-1 post-transcriptionally upregulates cMyc protein, leading to cell proliferation and specific gene expression programs, including increased expression of cMyc target programs such as protein translation. Moreover, we investigate a post-transcriptional regulatory web in which PCAT-1 disrupts microRNA-based regulation of cMyc. Specifically, we find that PCAT-1 is able to abrogate the down-regulation of cMyc by miR-34a. By contrast, targeting PCAT-1 with miR-3667-3p is able to reverse the stabilizing effect of PCAT-1 on cMyc protein levels. These findings correlate with the 3′UTR activity of MYC, which is also regulated by PCAT-1.
cMyc is a well-studied regulator of cell proliferation and is frequently dysregulated in prostate cancer, where amplification of the cMyc locus occurs in > 20% of patients with localized disease and > 50% of patients with metastatic disease [32]. cMyc is a ubiquitous transcription factor essential for cell cycle progression, and its oncogenic up-regulation results in wide-ranging consequences on cancer cell biology [33]. Indeed, it is estimated that up to one third of all human genes may be regulated by cMyc in certain contexts [34]. Numerous mechanisms of cMyc regulation have been described in cancer, including the post-transcriptional regulation of cMyc by microRNAs. In particular, miR-34a has been shown to target the 3′UTR of MYC, thereby degrading cMyc protein and strongly suppressing cancer cell proliferation and transformation [29], [35]. However, regulation of cMyc by prostate cancer lncRNAs has not been previously studied.
In addition, these data give insight into the complex web of post-transcriptional regulation of RNA and protein species in the cell cytoplasm. Here, our work derives from the theory of competing endogenous RNAs proposed by Pandolfi and colleagues [36]. According to this model, cytoplasmic RNA species are able to “titrate away” the presence of other regulatory entities, particularly microRNAs but also pseudogenes and other mRNAs [26], [27], [28]. This occurs through competitive disruption of microRNA binding to target genes. Characterization of the PTEN mRNA by Pandolfi and colleagues has led to an increased appreciation for the numerous cytoplasmic interactions that may occur at the RNA level and contribution to overall protein abundance and production [36]. We propose that PCAT-1 functions through a similar web in prostate cancer to coordinate increased cMyc abundance (Figure W8).
With respect to mechanism specifically, we have previously suggested that PCAT-1 may interact with the nuclear polycomb repressive complex 2 (PRC2) [10]. While a subset of PCAT-1 transcripts appear to localize to the nucleus, possibly engaging PRC2 for functional effects, we subsequently found that a majority of PCAT-1 transcripts are located in the cytoplasm [13] and thus likely operates independently of PRC2 in this cellular compartment. This current manuscript therefore supports a function of PCAT-1 in the cytoplasm but does not exclude potential mechanisms for gene expression regulation in the nucleus through PRC2 or other means. One additional notable point is that we previously showed that PCAT-1 expression does not correlate well with MYC mRNA expression in human prostate cancer tissues. Our current work would support that observation: we find no effect of PCAT-1 on MYC mRNA. In this regard, our work is consistent with evidence showing that microRNA-based regulation can occur primarily through translational inhibition in certain situations [37].
We have also independently shown that PCAT-1 downregulates BRCA2 [13], and a recent study in breast cancer cells demonstrated that cMyc could downregulate BRCA2 through genomic binding to, and up-regulation of, miR-1245 [38]. However, given the well-documented cell type–specific functions of both microRNAs and cMyc [39], [40], [41], [42], the relationship between miR-1245 and cMyc in prostate cells remains an unexplored area of research.
Further, we have observed that PCAT-1 is able to interfere with the regulation of MYC by miR-34a. However, we have been unable to locate a direct binding site for miR-34a in the PCAT-1 gene sequence. Our analyses are limited by the capabilities of microRNA target prediction software packages, which almost exclusively include protein-coding genes only. miRBase, in fact, was the only software that allowed us to search for microRNAs targeting PCAT-1 directly. Thus, the effect of PCAT-1 on miR-34a–mediated regulation may be indirect and may reflect a broader function of PCAT-1 in which PCAT-1 disrupts microRNA function indirectly. Here, we are intrigued by the presence of numerous repeat elements in PCAT-1, including retroviral long-terminal repeats and an Alu element (see [10]). Repeat elements are known to have effects on microRNA regulation and function [43], [44], [45], which may mean that PCAT-1 is able to impact microRNAs in this manner as well.
More broadly, our work gives insight into role of the 8q24 chromosomal locus in tumor aggressiveness. The 8q24 locus has been studied at length given its frequent amplification in cancer [46], including prostate cancer [32]. While MYC is a presumed target for this amplification, it is also clear that the function of 8q24 amplification in cancer is more expansive than simply MYC amplification. This is supported by the fact that 8q24 aberrations are typically very large or arm-level amplifications. Indeed, additional targets such as NCOA2 and PVT1 have been proposed [47], [48]. However, to date, no model has been able to fully elucidate the role of 8q24 amplification in cancer. Here, we suggest that PCAT-1 may also contribute to the role of 8q24 amplification by augmenting the function of cMyc. This function of PCAT-1 may support a role for other 8q24 ncRNAs in cancer pathogenesis.
Clinically, PCAT-1 may be an attractive candidate for biomarker development given its substantial overexpression in prostate cancer and functional role in cancer proliferation. To this end, PCAT-1 overexpression has been implicated in the poor prognosis of lethal colorectal cancers [12], and in situ hybridization assays have been suggested as a possible avenue for the clinical translation of lncRNAs [11]. While the direct therapeutic targeting of lncRNAs has not yet been proven to date, RNA-based therapies are increasingly investigated as potential therapeutic agents. Ultimately, disease-specific targeted therapies against both proteins and lncRNAs may improve the clinical management of refractory prostate cancer.
Acknowledgements
We thank Yi Sun, Dongping Wei, and Dan Hamstra for helpful discussions.
Footnotes
This article refers to supplementary materials, which are designated by Tables W1 to W9 and Figures W1 to W8 and are available online at www.neoplasia.com.
This work was supported in part by the Prostate Cancer Foundation (F.Y.F. and A.M.C.), the Prostate Cancer Foundation Young Investigator Award (J.R.P. and Q.C.), National Institutes of Health Prostate Specialized Program of Research Excellence grant P50CA69568, Department of Defense grants PC094231 (F.Y.F.) and PC100171 (A.M.C.), the Early Detection Research Network grant UO1 CA111275 (A.M.C.), the US National Institutes of Health R01CA132874-01A1 (A.M.C.), and the National Center for Functional Genomics support by the Department of Defense (to A.M.C.). A.M.C. is also supported by the Doris Duke Charitable Foundation Clinical Scientist Award and the Howard Hughes Medical Institute. A.M.C. is an American Cancer Society Research Professor and a Taubman Scholar of the University of Michigan. J.R.P., M.K.I, and Q.C. were supported by the Department of Defense Fellowship grants PC094290 (to J.R.P), BC100238 (to M.K.I.), and PC094725 (to Q.C.). The University of Michigan has filed a patent on PCAT-1 in which A.M.C., J.R.P., and M.K.I. are named as co-inventors. Wafergen, Inc has a non-exclusive license for creating commercial research assays for the detection of lncRNAs including PCAT-1. A.M.C. serves on the Scientific Advisory Board of Wafergen. Gen-Probe or Wafergen had no role in the design or experimentation of this study nor has it participated in the writing of the manuscript. Gene expression array data: Microarray data have been deposited at the NCBI Gene Expression Omnibus as GSE54872. Microarray data will be made available on publication. Author contributions: J.R.P., W.C., A.M.C., and F.Y.F. designed the project and directed the experimental studies. W.C., J.R.P., Q.C., M.T.P., V.K., S.H., J.R.E., and M.L. performed the experimental studies. M.K.I. performed the bioinformatics and statistical analyses. J.R.P. and M.K.I. performed the microRNA analyses. T.S.L., K.E.K., and M.L. facilitated the experiments and interpreted the data. J.R.P., W.C., and F.Y.F. interpreted the data and wrote the manuscript.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.neo.2014.09.001.
Appendix A. Supplementary data
Supplementary Materials.
Supplementary tables.
References
- 1.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19:R152–R161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Kok JB, Verhaegh GW, Roelofs RW, Hessels D, Kiemeney LA, Aalders TW, Swinkels DW, Schalken JA. DD3(PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res. 2002;62:2695–2698. [PubMed] [Google Scholar]
- 7.Tomlins SA, Aubin SM, Siddiqui J, Lonigro RJ, Sefton-Miller L, Miick S, Williamsen S, Hodge P, Meinke J, Blase A. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci Transl Med. 2011;3:94ra72. doi: 10.1126/scitranslmed.3001970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Srikantan V, Zou Z, Petrovics G, Xu L, Augustus M, Davis L, Livezey JR, Connell T, Sesterhenn IA, Yoshino K. PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer. Proc Natl Acad Sci U S A. 2000;97:12216–12221. doi: 10.1073/pnas.97.22.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M, Chen Y, Liu XS. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013;20:908–913. doi: 10.1038/nsmb.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ge X, Chen Y, Liao X, Liu D, Li F, Ruan H, Jia W. Overexpression of long noncoding RNA PCAT-1 is a novel biomarker of poor prognosis in patients with colorectal cancer. Med Oncol. 2013;30:588. doi: 10.1007/s12032-013-0588-6. [DOI] [PubMed] [Google Scholar]
- 13.Prensner JR, Chen W, Iyer MK, Cao Q, Ma T, Han S, Sahu A, Malik R, Wilder-Romans K, Navone N. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res. 2014;74:1651–1660. doi: 10.1158/0008-5472.CAN-13-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao Q, Mani RS, Ateeq B, Dhanasekaran SM, Asangani IA, Prensner JR, Kim JH, Brenner JC, Jing X, Cao X. Coordinated Regulation of Polycomb Group Complexes through microRNAs in Cancer. Cancer Cell. 2011;20:187–199. doi: 10.1016/j.ccr.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han S, Brenner JC, Sabolch A, Jackson W, Speers C, Wilder-Romans K, Knudsen KE, Lawrence TS, Chinnaiyan AM, Feng FY. Targeted radiosensitization of ETS fusion-positive prostate cancer through PARP1 inhibition. Neoplasia. 2013;15:1207–1217. doi: 10.1593/neo.131604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tinel M, Elkahwaji J, Robin MA, Fardel N, Descatoire V, Haouzi D, Berson A, Pessayre D. Interleukin-2 overexpresses c-myc and down-regulates cytochrome P-450 in rat hepatocytes. J Pharmacol Exp Ther. 1999;289:649–655. [PubMed] [Google Scholar]
- 18.Tinel M, Berson A, Elkahwaji J, Cresteil T, Beaune P, Pessayre D. Downregulation of cytochromes P450 in growth-stimulated rat hepatocytes: role of c-Myc induction and impaired C/EBP binding to DNA. J Hepatol. 2003;39:171–178. doi: 10.1016/s0168-8278(03)00238-1. [DOI] [PubMed] [Google Scholar]
- 19.Arora V, Knapp DC, Smith BL, Statdfield ML, Stein DA, Reddy MT, Weller DD, Iversen PL. c-Myc antisense limits rat liver regeneration and indicates role for c-Myc in regulating cytochrome P-450 3A activity. J Pharmacol Exp Ther. 2000;292:921–928. [PubMed] [Google Scholar]
- 20.Ruggero D. The role of Myc-induced protein synthesis in cancer. Cancer Res. 2009;69:8839–8843. doi: 10.1158/0008-5472.CAN-09-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt EV. The role of c-myc in regulation of translation initiation. Oncogene. 2004;23:3217–3221. doi: 10.1038/sj.onc.1207548. [DOI] [PubMed] [Google Scholar]
- 22.Seoane J, Pouponnot C, Staller P, Schader M, Eilers M, Massague J. TGFβ influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat Cell Biol. 2001;3:400–408. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- 23.Staller P, Peukert K, Kiermaier A, Seoane J, Lukas J, Karsunky H, Möröy T, Bartek J, Massagué J, Hänel F. Repression of p15INK4b expression by Myc through association with Miz-1. Nat Cell Biol. 2001;3:392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- 24.Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science. 2011;331:550–553. doi: 10.1126/science.1191138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prensner JR, Chen W, Iyer MK, Cao Q, Ma T, Han S, Sahu A, Malik R, Wilder-Romans K, Navone N. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res. 2014;74:1651–1660. doi: 10.1158/0008-5472.CAN-13-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamura S, Saini S, Majid S, Hirata H, Ueno K, Deng G, Dahiya R. MicroRNA-34a modulates c-Myc transcriptional complexes to suppress malignancy in human prostate cancer cells. PLoS One. 2012;7:e29722. doi: 10.1371/journal.pone.0029722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siemens H, Jackstadt R, Hunten S, Kaller M, Menssen A, Gotz U, Hermeking H. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10:4256–4271. doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
- 31.Benassi B, Flavin R, Marchionni L, Zanata S, Pan Y, Chowdhury D, Marani M, Strano S, Muti P, Blandino G. MYC is activated by USP2a-mediated modulation of microRNAs in prostate cancer. Cancer Discov. 2012;2:236–247. doi: 10.1158/2159-8290.CD-11-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim JH, Dhanasekaran SM, Mehra R, Tomlins SA, Gu W, Yu J, Kumar-Sinha C, Cao X, Dash A, Wang L. Integrative analysis of genomic aberrations associated with prostate cancer progression. Cancer Res. 2007;67:8229–8239. doi: 10.1158/0008-5472.CAN-07-1297. [DOI] [PubMed] [Google Scholar]
- 33.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 34.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamamura S, Saini S, Majid S, Hirata H, Ueno K, Chang I, Tanaka Y, Gupta A, Dahiya R. MicroRNA-34a suppresses malignant transformation by targeting c-Myc transcriptional complexes in human renal cell carcinoma. Carcinogenesis. 2012;33:294–300. doi: 10.1093/carcin/bgr286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meijer HA, Kong YW, Lu WT, Wilczynska A, Spriggs RV, Robinson SW, Godfrey JD, Willis AE, Bushell M. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science. 2013;340:82–85. doi: 10.1126/science.1231197. [DOI] [PubMed] [Google Scholar]
- 38.Song L, Dai T, Xie Y, Wang C, Lin C, Wu Z, Ying Z, Wu J, Li M, Li J. Up-regulation of miR-1245 by c-myc targets BRCA2 and impairs DNA repair. J Mol Cell Biol. 2012;4:108–117. doi: 10.1093/jmcb/mjr046. [DOI] [PubMed] [Google Scholar]
- 39.Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci U S A. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carroll AP, Tooney PA, Cairns MJ. Context-specific microRNA function in developmental complexity. J Mol Cell Biol. 2013;5:73–84. doi: 10.1093/jmcb/mjt004. [DOI] [PubMed] [Google Scholar]
- 41.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 42.Lee BK, Bhinge AA, Battenhouse A, McDaniell RM, Liu Z, Song L, Ni Y, Birney E, Lieb JD, Furey TS. Cell-type specific and combinatorial usage of diverse transcription factors revealed by genome-wide binding studies in multiple human cells. Genome Res. 2012;22:9–24. doi: 10.1101/gr.127597.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu TJ, Yi X, Zhao XW, Zhao Y, Yin JQ. Alu-directed transcriptional regulation of some novel miRNAs. BMC Genomics. 2009;10:563. doi: 10.1186/1471-2164-10-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smalheiser NR, Torvik VI. Alu elements within human mRNAs are probable microRNA targets. Trends Genet. 2006;22:532–536. doi: 10.1016/j.tig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 46.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guan Y, Kuo WL, Stilwell JL, Takano H, Lapuk AV, Fridlyand J, Mao JH, Yu M, Miller MA, Santos JL. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin Cancer Res. 2007;13:5745–5755. doi: 10.1158/1078-0432.CCR-06-2882. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials.
Supplementary tables.


