Abstract
Purpose
Neuroblastoma is an embryonal tumor with contrasting clinical courses. Despite elaborate stratification strategies, precise clinical risk assessment still remains a challenge. The purpose of this study was to develop a PCR-based predictor model to improve clinical risk assessment of patients with neuroblastoma.
Experimental Design
The model was developed using real-time PCR gene expression data from 96 samples and tested on separate expression data sets obtained from real-time PCR and microarray studies comprising 362 patients.
Results
On the basis of our prior study of differentially expressed genes in favorable and unfavorable neuroblastoma subgroups, we identified three genes, CHD5, PAFAH1B1, and NME1, strongly associated with patient outcome. The expression pattern of these genes was used to develop a PCR-based single-score predictor model. The model discriminated patients into two groups with significantly different clinical outcome [set 1: 5-year overall survival (OS): 0.93 ± 0.03 vs. 0.53 ± 0.06, 5-year event-free survival (EFS): 0.85 ± 0.04 vs. 0.042 ± 0.06, both P < 0.001; set 2 OS: 0.97 ± 0.02 vs. 0.61 ± 0.1, P = 0.005, EFS: 0.91 ± 0.8 vs. 0.56 ± 0.1, P = 0.005; and set 3 OS: 0.99 ± 0.01 vs. 0.56 ± 0.06, EFS: 0.96 ± 0.02 vs. 0.43 ± 0.05, both P < 0.001]. Multivariate analysis showed that the model was an independent marker for survival (P < 0.001, for all). In comparison with accepted risk stratification systems, the model robustly classified patients in the total cohort and in different clinically relevant risk subgroups.
Conclusion
We propose for the first time in neuroblastoma, a technically simple PCR-based predictor model that could help refine current risk stratification systems.
Introduction
Neuroblastoma, the most common solid extracranial pediatric tumor, is heterogeneous in terms of its biologic, genetic, and morphologic characteristics. Such tumors exhibit diverse clinical behaviors whereby patients with similar clinicopathologic features can have radically different outcomes. Since treatment strategies vary from surgery alone to intensive multimodal regimens, precise risk assessment is critical for therapeutic decisions. Currently, several clinical, biologic, and morphologic parameters, such as age at diagnosis, tumor stage, genomic amplification of MYCN oncogene, copy number alterations of the chromosomal regions 1p, 11q, and 17q, ploidy, and histologic features, are considered as markers of neuroblastoma outcome (1–4). However, despite elaborate risk stratification strategies, there remain cases where these markers have shown limited clinical use.
Microarray gene expression studies have contributed to identify sets of genes of prognostic importance in numerous neoplasias, including neuroblastoma (5–13). Nevertheless, the use of microarrays in clinical practice is limited by the large sets of genes identified and the need for complex statistical analyses required to extract informative patterns from raw microarray data. Furthermore, there has been little overlap in the prognostic gene sets identified by different groups. To translate these profiles into clinically applicable tests, it is essential to reduce the number of genes and create profiles that can be analyzed with a conventional assay such as quantitative real-time PCR (qRT-PCR).
In our previous study, we identified differentially expressed genes capable of discriminating between subgroups of neuroblastoma with radically different clinical course; namely, near-triploid (favorable prognosis) and near-diploid/tetraploid tumors (unfavorable prognosis). A substantial portion of these genes mapped to chromosomes 1 and 17, chromosomal regions found to be recurrently altered in neuroblastoma (14). Tumor cell ploidy has recently been included in the International Neuroblastoma Risk Group (INRG) classification system as a significant neuroblastoma biomarker together with MYCN status and 11q23 allelic status (3). Gene expression profiles associated with different neuroblastoma ploidy status may contribute to the identification of genes that are predictive of outcome. In this study, we investigated whether the identified gene expression profiling data allowed for the development of an outcome predictor model that could both accurately predict neuroblastoma outcome and be technically simple and applicable for routine clinical use. To this end, we examined the prognostic significance of 11 high-ranked, differentially expressed genes located on the chromosomes 1 and 17, chosen on the basis of our previous study (14). The results of a qRT-PCR–based analysis enabled the development of a single-score predictor model based on the expression pattern of 3 genes, CHD5, PAFAH1B1, and NME1, strongly associated with patient outcome.
Methods
Patients and samples
Tumor specimens from 96 patients with neuroblastoma were obtained at the time of diagnosis from different institutions [Memorial Sloan-Kettering Cancer Center (MSKCC), NY (n = 42), Hospital Sant Joan de Déu (HSJD), Barcelona, Spain (n = 32); Hospital Niño Jesús, Madrid, Spain (n = 11); Hospital La Paz, Madrid, Spain (n = 7); and Hospital La Fe, Valencia, Spain (n = 4)] and represented the training set of this study (Table 1 and Supplementary Table S1).
Table 1.
Clinical and biologic characteristics of the training and validation cohorts
| Training |
Validation |
|||
|---|---|---|---|---|
| Characteristics | Training set (N = 96) | Set 1 (N = 120) | Set 2 (N = 101) | Set 3 (N = 251) |
| Age, mo | ||||
| Median | 18.94 | 26.9 | 16.3 | 15.03 |
| Range | 0–216 | 0–299 | 0–156 | 0–276 |
| INSS, n (%) | ||||
| Stage I–III | 53 (56) | 43 (36) | 51 (50) | 153 (61) |
| Stage IV | 34 (35) | 52 (43) | 50 (50) | 67 (27) |
| Stage IVS | 9 (9) | 25 (21) | — | 31 (12) |
| MYCN status, n (%) | ||||
| Amplified | 20 (21) | 24 (20) | 20 (20) | 30 (12) |
| Nonamplified | 76 (79) | 96 (80) | 81 (80) | 220 (88) |
| Undetermined | — | — | — | 1 |
| 1p status, n (%) | ||||
| LOH | 11 (25) | 31 (26) | 28 (28) | 52 (21) |
| No LOH | 33 (75) | 88 (73) | 72 (72) | 194 (79) |
| Undetermined | 52 | 1 | 1 | 5 |
| Follow-up, mo | ||||
| Median | 48.22 | 89.21 | 11.73 | 64.46 |
The first 36 unselected cases of the training cohort (36 of 96 neuroblastoma) were used to develop the gene expression–based model; the remaining 60 primary neuroblastoma tumors were used as preliminary testing cohort. The complete training cohort (n = 96) was subsequently used to refine the obtained model (Table 1 and Supplementary Table S1).
A separate set of 120 primary neuroblastoma tumors (hereafter referred as set 1) of patients diagnosed and treated at the Department of Pediatric Oncology, University Children’s Hospital of Cologne, Cologne, Germany, was used as an independent, blinded set of RNA samples to validate the model by qRT-PCR.
Tumors were assessed by a pathologist, only tumors with more than 70% viable tumor cell content were included in the study. Risk assessment was defined by the International Staging System (INSS; ref. 1). Neuroblastoma stages I–IV were treated without use of cytotoxic therapy, when possible. Patients with stage IV neuroblastoma were treated with the combination of intensive chemotherapy (including high-dose therapy and autologous stem cell rescue), radiotherapy, and surgery. Written informed consent was obtained from each subject or from his or her guardian before collection of samples. This study was approved by the Institutional Review Boards.
Gene expression data
Two published gene expression data sets from different platforms comprising 352 patients in total were used as validations sets. Expression data were downloaded from the National Centre of Biotechnology Information Gene Expression Omnibus (GSE3960; ref. 15) and from the European Bioinformatics Institute ArrayExpress database (E-TABM-38; ref. 9). Updated clinical data were obtained from the authors. The validation sets will be hereafter referred to as set 2 [n = 101 (ref. 15)] and set 3 [n = 251 (ref. 9)]. Sets 2 and 3 were used to assess the prediction performance of the model across different neuroblastoma expression data sets. Set 3 includes expression data of 110 patients comprised in set 1. This set of overlapping samples (n = 110) was used to estimate the prediction performance of the model in neuroblastoma tumors analyzed by different expression methods (qRT-PCR and microarray analyses). Set 3 was used for further analyses aimed to test the classification performance of the model as compared with current risk stratification systems. Set 3 was selected for these analyses as it comprises a full representation of the neuroblastoma tumor spectra and a median follow-up time of more than 5 years (Table 1 and Supplementary Table S5).
The analyses were done blinded of clinical and biologic data and sample identification.
RNA isolation and qRT-PCR
Total RNA was isolated from the 96 snap-frozen samples using Tri Reagent (Sigma), following manufacturers’ protocols. The cDNA was synthesized from 1 µg total RNA as previously described (14). Gene expression was quantified with TaqMan Gene Expression Assays (Supplementary Table S2) products on an ABI PRISM 7000 Sequence Detection System, using the ΔΔCT relative quantification method. All experiments included no-template controls and were carried out in duplicate and repeated twice independently. Transcript levels were measured relative to 3 normal tissue samples (adrenal gland, lymph node, and bone marrow) and normalized to TATA-box–binding protein (TBP), hypoxanthine phosphoribosyltransferase 1 (HPRT1), and succinate dehydrogenase complex, subunit A (SDHA) expression values as previously described by Vandesompele and colleagues (16).
To control for possible variations among PCR runs conducted on different days, the expression of the reference genes was assessed in all tumor samples and control specimens at commencement, halfway through, and on completion of all the analyses. The assays were highly reproducible, with coefficient of variation (CV) less than 0.05 (TBP = 0.035, SDHA = 0.049, and HPRT1 = 0.040 in tumor samples and TBP = 0.029, SDHA = 0.045, and HPRT1 = 0.022 for control specimens).
Statistical analysis
Gene expression values were normalized with a z-score transformation. From the 96 neuroblastoma cases of the training set, the first 36 unselected cases were used for the selection of genes strongly associated, as independent factors, with overall (OS) and event-free (EFS) using Cox regression models. Relapse, progression, and death from disease were considered as events. Genes significantly associated with OS and EFS were further tested using principal components analysis (PCA; multivariable unsupervised method) with the VARIMAX rotation method and tested using Cox regression models. This procedure was conducted by means of a stepwise backward selection approach. Briefly, at each step of the backward selection procedure, one gene was removed from the set of genes. To select which gene was discarded (not independent predictor or the least statistically significant), all the possible combinations of genes were tested by removing and then reintroducing one by one each gene following an iterative procedure. Each gene set combination was thus analyzed using PCA and tested with Cox regression models to assess the performance and the association with OS and EFS. This backward selection procedure concluded when the combination with the minimum set of genes statistically significantly associated with OS and EFS is found. After testing all the combinations, genes that were not independent predictors and the least statistically significant genes were discarded.
The outcome predictor model was developed using a linear combination of the z-score transformed expression value of each of the selected genes, weighted by the regression coefficients (α) of components with Eigenvalue > 1. The model was tested using the remaining 60 neuroblastoma cases of the training set. The entire training cohort (n = 96) was used to refine the model. To validate the final model, we applied it to different expression data sets obtained by qRT-PCR (set 1) and microarray gene expression studies (sets 2 and 3). The performance of the model was examined with multivariable Cox regression models using a stepwise variable selection procedure. Survival curves were described using the Kaplan–Meier method and compared by means of the log-rank test. The area under the curve (AUC) of the receiver operating characteristic (ROC) test was used for quantitative assessment of the final model for the testing set (set 3). Predictive values (positive and negative), sensitivity, specificity, and the accuracy were calculated and expressed along with known prognostic factors (age, INSS, MYCN status, chromosome 11q and 1p alterations, and histologic features). Data were analyzed with SPSS program (version 15.0; SPSS, Inc.). All statistical tests were 2-sided. P values ≤0.05 were considered statistically significant.
Results
Selection of genes for the outcome predictor score model
In our previous study, we found specific transcriptional profiles associated with neuroblastoma with different ploidy (14). A statistically significant number of genes found to be differentially expressed mapped to chromosomes 1 (P = 0.01) and 17 (P < 0.0001), chromosomes with aberrations reported to be consistently associated with outcome in neuroblastoma (2). For this study, we selected 11 high-ranked, differentially expressed genes located on the chromosomes 1 and 17, some of these known to play a role in neuroblastoma pathogenesis. The expression levels of the 11 genes were quantified using qRT-PCR in 36 neuroblastoma tumors (Supplementary Table S1). To assess the association with OS and EFS, qRT-PCR expression levels were normalized by z-score transformation and analyzed using univariable Cox regression models (Supplementary Table S3). High expression of 6 genes was found to be significantly associated (P < 0.05) as independent factors with outcome: 5 (RERE, PTPRF, GNB1, CHD5, and PAFAH1B1) were associated with longer OS and EFS (HR < 1 for all) and one gene, NME1, was associated with worse clinical outcome (HR > 1). These 6 genes were considered potential prognostic markers and were further studied. We conducted PCA and univariable Cox regression analyses applying a stepwise backward selection approach. This selection procedure identified a predominant gene expression pattern. Three genes, CHD5 (chromodomain, helicase DNA-binding protein 5), PAFAH1B1 (platelet-activating factor acetylhydrolase, isoform 1B), and NME1/nm23-H1 (nonmetastatic cells 1, protein expressed in), had a principal component with Eigenvalue > 1 describing more than 60% of the expression variability and were found to be strongly associated with OS and EFS (P = 0.001, for both). These 3 genes were selected for the model as the strongest prognostic markers. We thus developed a gene expression–based outcome predictor score model using z-transformed qRT-PCR data of each of the 3 genes weighted by the regression coefficients from the PCA (Supplementary Table S4), as described in the equation:
where, (Y36) is the outcome prediction score, equation developed using 36 neuroblastomas; αn is the weighted value of each gene to the definition of (Y);α1 = 0.418; α2 = 0.430; α3 = −0.374; and (Gene) is the z-transformed gene expression data of the sample analyzed.
The negative weighting value of NME1 indicates that the contribution of this gene is inversely correlated with the (Y36) score; thus, high expression levels of NME1 correlate with low (Y36) values. Patients were ranked according to their (Y36) score. Low (Y36) values were associated with shorter OS and EFS; conversely, an increase of the (Y36) score was associated with longer survival. Gene expression transformation to a z-distribution (mean, 0; SD, 1) allowed the cutoff point to divide neuroblastoma tumors with high or low scores to be set at the value (Y36) = 0. Kaplan–Meier estimates showed how the model could clearly separate the patients into groups with divergent clinical course (Supplementary Fig. S1A and S1B).
The first approximation of the model was tested using an independent set of 60 primary neuroblastoma tumors (Supplementary Table S1). For each tumor, qRT-PCR expression levels of the 3 genes were z-transformed and inserted into the equation (Y36). Patients were ranked and divided into 2 groups, Y36 < 0 or >0. The outcome predictor score could separate patients with significant differences in OS [HR, 9.3; 95% confidence interval (CI), 1.1–79.7; P = 0.013] and EFS (HR, 3.1; 95% CI, 1.2–8.03; P = 0.014; Supplementary Fig. S1C and S1D).
To improve the performance of the predictor model, we proceeded to reestimate the component coefficient scores of the 3 genes using the entire training cohort of 96 cases (Supplementary Table S1). The component coefficient score values obtained were comparable with those of the (Y36) equation (Supplementary Table S4). Kaplan–Meier analyses with log-rank estimates of the 96 training samples analyzed with the (Y96) model confirmed the strong association of the (Y96) model with OS and EFS (P < 0.001, for both; Supplementary Fig. S1E and S1F). The (Y96) model was tested on an independent, blinded set of 120 neuroblastoma tumors (set 1) by qRT-PCR. Patients were separated into 2 groups with significantly diverse clinical outcome (5-year OS: 0.93 ± 0.03 vs. 0.53 ± 0.06, 5-year EFS: 0.85 ± 0.04 vs. 0.047 ± 0.06, both P < 0.001; OS: HR, 7.5; 95% CI, 2.9–19.5; and EFS: HR, 4.7; 95% CI, 2.3–9.5; Supplementary Fig. S1G and S1H).
Validation of outcome predictor model using independent microarray data
To assess whether the (Y96) model was able to predict prognosis across different neuroblastoma data sets, we applied it to 2 published gene expression databases from different microarray platforms: set 2 (n = 101; ref. 15) and set 3 (n = 251; ref. 9).
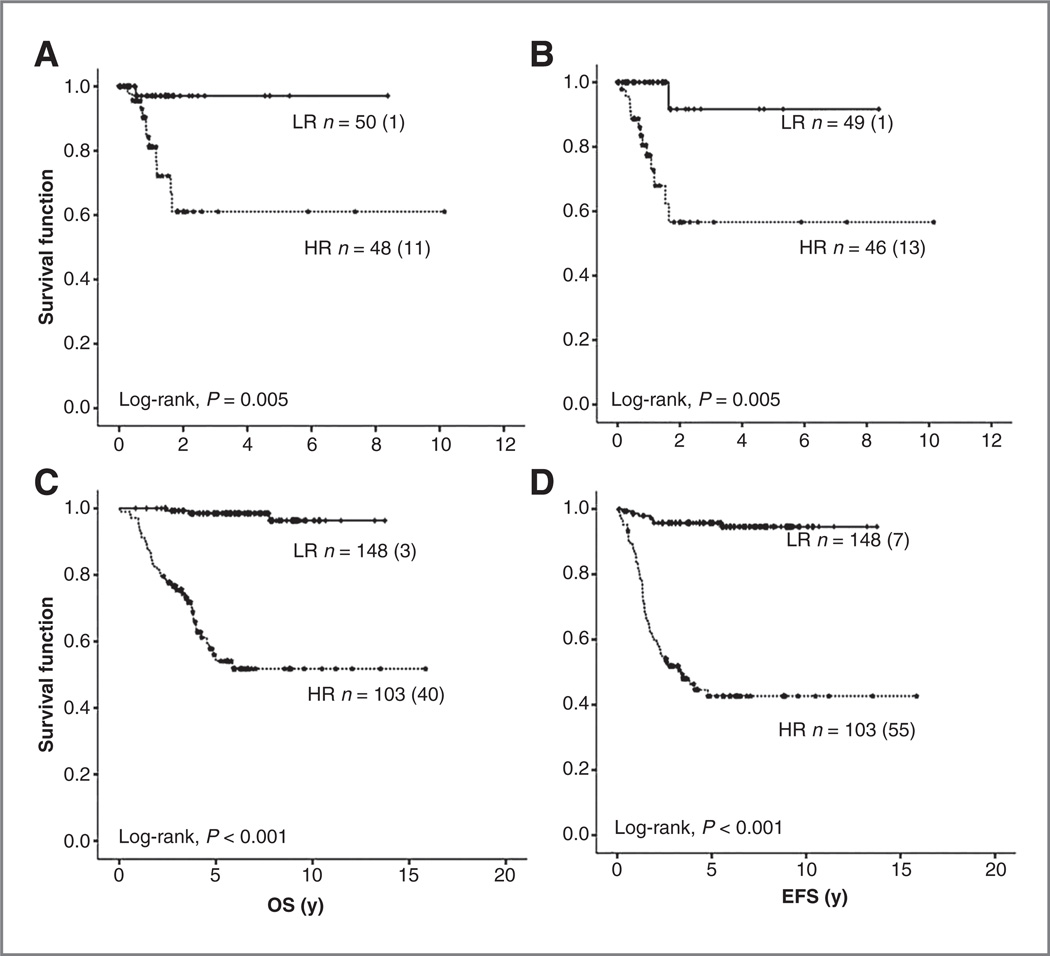
For each patient, microarray expression data of the 3 genes were z-transformed and inserted into the equation (Y96). Kaplan–Meier estimation with log-rank test and univariable Cox regression models showed that for both data sets, the (Y96) score could clearly separate patients into 2 groups with different OS and EFS [set 2: 5-year OS: 0.97 ± 0.02 vs. 0.61 ± 0.1; HR, 10.5; 95% CI, 1.3–80.7; 5-year EFS: 0.91 ± 0.8 vs. 0.56 ± 0.1; HR, 13.3; 95% CI, 1.7–101.6, both P = 0.005; set 3 5-year OS: 0.99 ± 0.01 vs. 0.56 ± 0.06; HR, 28.1; 95% CI, 8.7–91.2; 5-year EFS: 0.96 ± 0.02 vs. 0.43 ± 0.05; HR, 15.6; 95% CI, 7.1–34.3, both P < 0.001; (Fig. 1)]. For set 2, only one [stage III MYCN nonamplified (NA) neuroblastoma] of the 50 patients (2%) assigned to the low-risk group by the (Y96) score, had a fatal disease progression, and was thus misclassified. For set 3, 7 of the 148 patients (4.7%) assigned to the low-risk group by the (Y96) model had an event. Of them, 3 (2%; stage IV, >18 months, MYCN NA) died of disease progression.
Figure 1.
Kaplan–Meier analyses with log-rank estimates for OS and EFS of set 2 (A and B) and set 3 (C and D) classified according to the prediction (Y96) model. HR, high-risk; LR, low-risk.
The estimation of the performance of the (Y96) model for prediction of outcome showed an AUC for the ROC test of 0.87 (95% CI, 0.83–9.21) for OS and 0.89 (95% CI, 0.84– 0.93) for EFS. Prediction accuracies for the (Y96) model and established neuroblastoma prognostic parameters (age, INSS, MYCN amplification, and chromosome 1p status) are shown in Table 2. Furthermore, in a multivariable Cox regression analysis evaluating the prognostic parameters, the model remained significantly associated with OS and EFS (P < 0.001 for all), showing thus to be independent of other prognostic markers (Table 3).
Table 2.
Estimation of the performance of the (Y96) model for OS and EFS of set 3 (n = 251) in comparison with clinical and biologic parameters
| OS |
EFS |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alive | DOD | Sensitivity % |
Specificity % |
PPV % |
PNV % |
Accuracy % |
No event |
Event | Sensitivity % |
Specificity % |
PPV % |
PNV % |
Accuracy % |
|
| MYCN | ||||||||||||||
| No amplification | 195 | 25 | 41.9 | 93.8 | 60.0 | 88.6 | 84.9 | 177 | 43 | 30.6 | 93.7 | 63.3 | 80.5 | 78.1 |
| Amplification | 12 | 18 | 11 | 19 | ||||||||||
| INSS stage | ||||||||||||||
| I–III, IVs | 173 | 11 | 74.4 | 83.2 | 47.8 | 94.0 | 81.7 | 162 | 22 | 64.5 | 85.7 | 59.7 | 88.0 | 80.5 |
| IV | 35 | 32 | 27 | 40 | ||||||||||
| Age, mo | ||||||||||||||
| <18 | 160 | 8 | 81.4 | 76.9 | 42.2 | 95.2 | 77.7 | 149 | 19 | 69.4 | 78.8 | 51.8 | 88.7 | 76.5 |
| >18 | 48 | 35 | 40 | 43 | ||||||||||
| 1p | ||||||||||||||
| No | 181 | 18 | 58.1 | 87.0 | 48.1 | 91.0 | 82.1 | 165 | 34 | 45.2 | 87.3 | 53.8 | 82.9 | 76.9 |
| Del/imb | 27 | 25 | 24 | 28 | ||||||||||
| Model_Y96 | ||||||||||||||
| Low-risk | 145 | 3 | 93.0 | 69.7 | 38.8 | 98.0 | 73.7 | 141 | 7 | 88.7 | 74.6 | 53.4 | 95.3 | 78.1 |
| High-risk | 63 | 40 | 48 | 55 | ||||||||||
NOTE: The performance of the (Y96) model measured as the AUC for the ROC test was 0.87 (95% CI, 0.83–0.92) for OS and 0.89 (95% CI, 0.84–0.93) for EFS.
Abbreviations: Del/Imb, deletion/imbalance; DOD, dead of disease; PNV, predictive negative value; PPV, predictive positive value.
Table 3.
Multivariate Cox regression model for OS and EFS considering the (Y96) model and clinical and biologically relevant parameters for risk stratification of set 3 patients
| Survival |
Event |
|||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| Univariate model | ||||
| Model (Y96) | 28.09 (8.65–91.19) | <0.001 | 15.59 (7.07–34.34) | <0.001 |
| Age | 11.68 (5.40–25.27) | <0.001 | 5.44 (3.16–9.36) | <0.001 |
| Model (Y96) | 15.85 (4.75–52.91) | <0.001 | 11.07 (4.89–25.07) | <0.001 |
| Age | 5.41 (2.46–11.95) | <0.001 | 2.61 (1.49–4.59) | 0.001 |
| INSS stage | 10.14 (5.1–20.16) | <0.001 | 6.27 (3.72–10.57) | <0.001 |
| Model (Y96) | 15.13 (4.41–51.97) | <0.001 | 10.32 (4.46–23.86) | <0.001 |
| INSS stage | 3.65 (1.78–7.5) | <0.001 | 2.48 (1.42–4.31) | 0.001 |
| MYCN | 8.7 (4.7–16.1) | <0.001 | 4.7 (2.73–8.1) | <0.001 |
| Model (Y96) | 20.2 (6.02–67.73) | <0.001 | 13.53 (6–30.5) | <0.001 |
| MYCN | 2.74 (1.46–5.12) | 0.002 | 1.59 (0.91–2.77) | 0.105 |
| 1p LOH | 7.48 (4.06–13.77) | <0.001 | 4.25 (2.57–7.03) | <0.001 |
| Model (Y96) | 18.92 (5.63–63.52) | <0.001 | 12.83 (5.67–29.01) | <0.001 |
| 1p LOH | 2.93 (1.56–5.47) | 0.001 | 1.76 (1.05–2.95) | 0.033 |
| Multivariate model | ||||
| Model (Y96) | 9.9 (2.8–35.3) | <0.001 | 836 (3.6–30.4) | <0.001 |
| MYCW | 3.2 (1.7–6.0) | <0.001 | 1.7 (0.99–3.0) | 0.053 |
| INSS stage | 4.2 (2.0–8.4) | <0.001 | 2.6 (1.5–4.5) | 0.001 |
NOTE: The analysis was conducted sequentially, adding one variable at each step, to assess how the presence of each variable influences the performance of the (Y96) model. The (Y96) model was statistically significantly associated with OS and EFS in univariate and multivariate analyses.
Abbreviation: NE, "non evaluable" P value due to "no cases" in a combination of the 2 factors.
To further assess the model across different data sets, the prediction performance of samples analyzed by different expression methods (qRT-PCR and microarray analyses), included in sets 1 and 3 (n = 110), was compared. Equivalent results were obtained (data not shown), showing that the classification capacity of the model is independent of the type and origin of the expression data used.
Classification performance of the (Y96) model with respect to current risk stratification systems
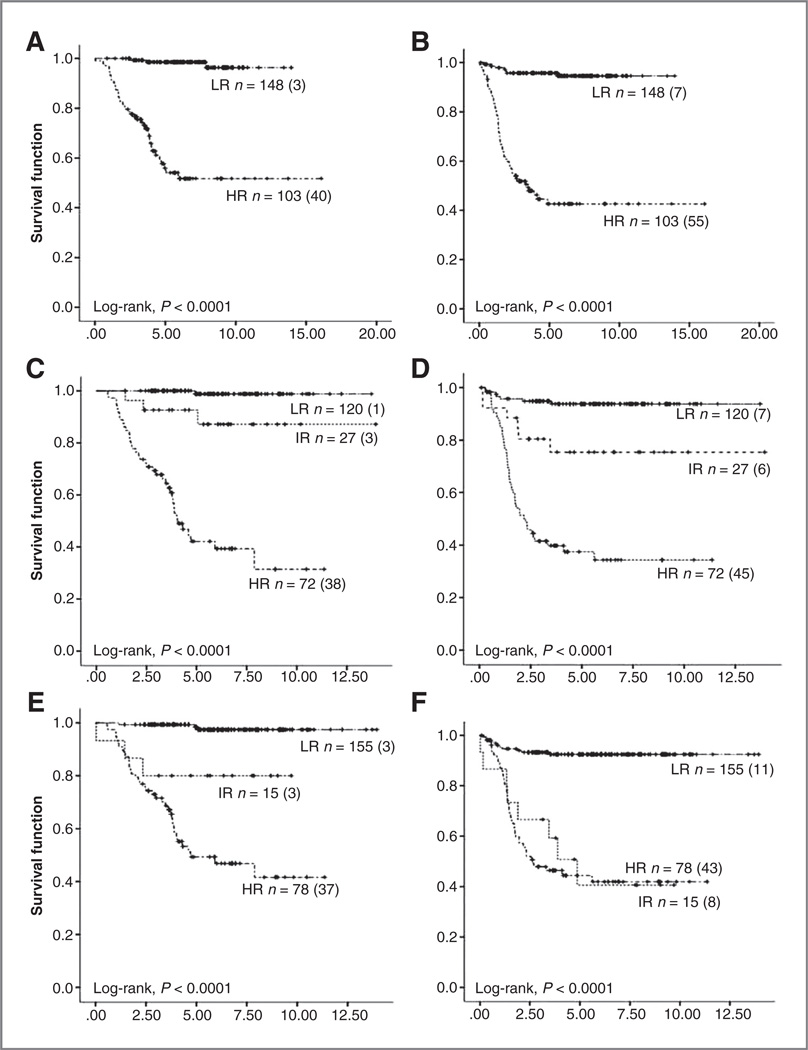
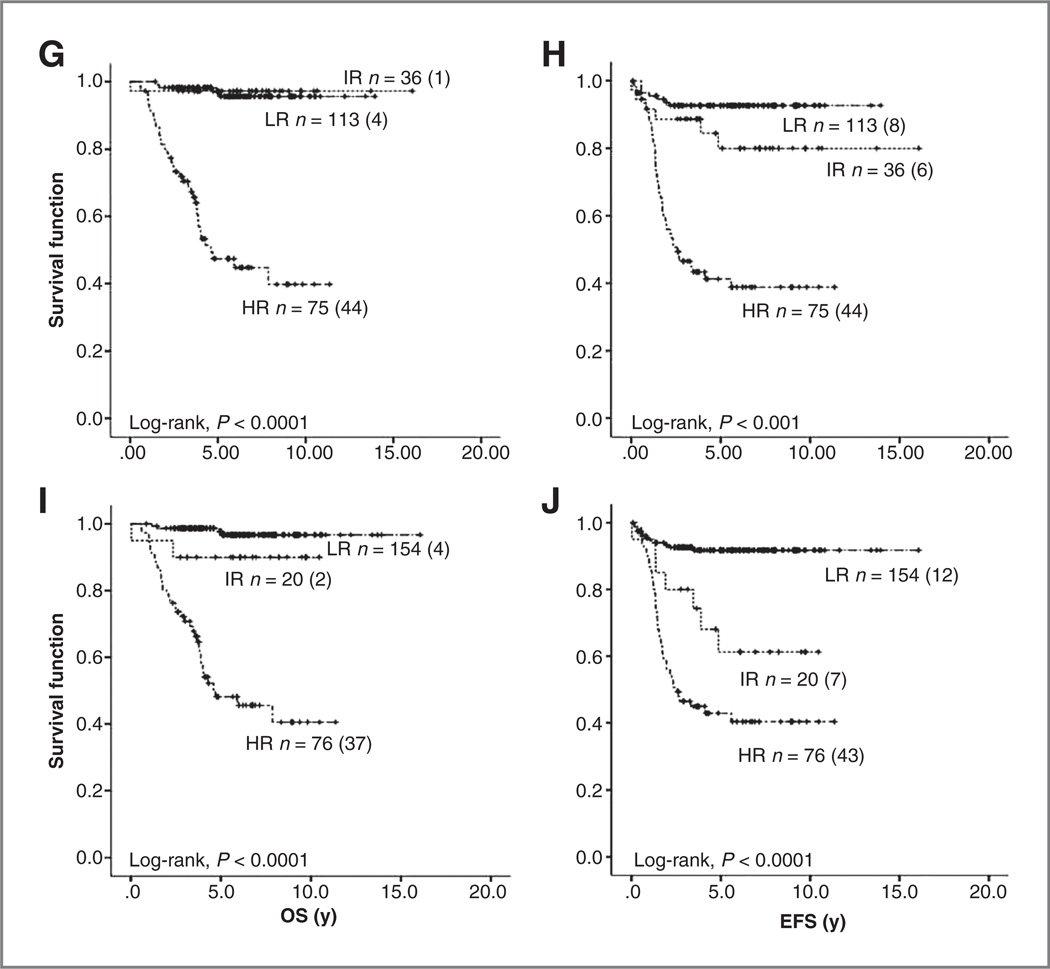
To evaluate the prediction performance of the (Y96) model, risk stratification systems of international neuroblastoma trials were applied to set 3; this set of data has a full representation of the neuroblastoma tumor spectra and a median follow-up time of more than 5 years (Table 1). According to the criteria of the INRG and of clinical trials from Germany [German Society for Pediatric Hematology/ Oncology (GPOH) NB2004], the United States [Children’s Oncology Group (COG)], and Japan [Japanese Advanced Neuroblastoma Study Group (JANB)], 219, 248, 224, and 250 patients could be classified, respectively (Supplementary Table S5). All classification systems separated patients into groups with significant differences in OS and EFS (Fig. 2, Table 4). In comparison, the classification prediction of the (Y96) model differed in a subset of patients (Fig. 2, Table 4 and Supplementary Table S5). Specifically, 5 of the patients favorably classified had a fatal outcome (INRG, n = 1 of 120; NB2004, n = 3 of 155; COG, n = 4 of 113; and JANB, n = 4 of 154; 4 cases matched), notably, these patients were classified in the high-risk group by the (Y96) model. Similarly, the 4 patients with a fatal outcome assigned to the intermediate-risk group (INRG, n = 3 of 27; NB2004, n = 3 of 15; COG, n = 1 of 36; and JANB, n = 2 of 20; 3 case matched) were classified as high-risk patients by the (Y96) model. Conversely, 3 cases assigned to the low-risk group according to the (Y96) model and unfavorably classified by the risk stratification systems, died of disease.
Figure 2.
Kaplan-Meier analyses and log-rank estimates for OS and EFS of the validation cohort set 3 classified according to the (Y96) model (A and B), INRG (C and D), GPOH NB2004 (E and F).
COG (G and H), and JANB (I and J). HR, high-risk; IR, intermediate-risk; LR, low-risk.
Table 4.
Summary of the Kaplan–Meier estimates shown in Fig. 2
| Low-risk |
High-risk |
Intermediate-risk |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Risk classification system |
No. of patients |
Survival probability |
SE | No. of patients |
Survival probability |
SE | No. of patients |
Survival probability |
SE |
| Y96 model (n = 251) | |||||||||
| EFS (3 y) | 148 | 0.96 | 0.02 | 103 | 0.52 | 0.05 | |||
| OS (3 y) | 0.99 | 0.01 | 0.77 | 0.04 | |||||
| EFS (5 y) | 0.96 | 0.02 | 0.43 | 0.05 | |||||
| OS (5 y) | 0.99 | 0.01 | 0.56 | 0.06 | |||||
| INRG (n = 219) | |||||||||
| EFS (3 y) | 120 | 0.95 | 0.02 | 72 | 0.41 | 0.06 | 27 | 0.8 | 0.08 |
| OS (3 y) | 1.00 | 0.69 | 0.05 | 0.93 | 0.05 | ||||
| EFS (5 y) | 0.94 | 0.02 | 0.37 | 0.06 | 0.75 | 0.09 | |||
| OS (5 y) | 0.99 | 0.01 | 0.42 | 0.07 | 0.93 | 0.05 | |||
| NB2004 (n = 248) | |||||||||
| EFS (3 y) | 155 | 0.93 | 0.02 | 78 | 0.48 | 0.06 | 15 | 0.67 | 0.12 |
| OS (3 y) | 0.99 | 0.01 | 0.73 | 0.05 | 0.8 | 0.1 | |||
| EFS (5 y) | 0.92 | 0.02 | 0.44 | 0.06 | 0.67 | 0.12 | |||
| OS (5 y) | 0.98 | 0.01 | 0.49 | 0.06 | 0.8 | 0.02 | |||
| COG (n = 224) | |||||||||
| EFS (3 y) | 113 | 0.93 | 0.03 | 75 | 0.47 | 0.06 | 36 | 0.89 | 0.05 |
| OS (3 y) | 0.98 | 0.01 | 0.72 | 0.05 | 0.97 | 0.03 | |||
| EFS (5 y) | 0.93 | 0.03 | 0.41 | 0.06 | 0.8 | 0.08 | |||
| OS (5 y) | 0.97 | 0.02 | 0.47 | 0.06 | 0.97 | 0.03 | |||
| JANB (n = 250) | |||||||||
| EFS (3 y) | 154 | 0.93 | 0.02 | 76 | 0.47 | 0.06 | 20 | 0.8 | 0.09 |
| OS (3 y) | 0.99 | 0.01 | 0.72 | 0.05 | 0.9 | 0.07 | |||
| EFS (5 y) | 0.92 | 0.02 | 0.43 | 0.06 | 0.61 | 0.12 | |||
| OS (5 y) | 0.98 | 0.01 | 0.48 | 0.06 | 0.9 | 0.07 | |||
(Y96) model classification of patient subgroups defined by current prognostic markers
Performance of the (Y96) model was assessed within patient subgroups of set 3 defined by currently used prognostic markers and compared with international neuroblastoma risk stratification systems. The (Y96) model correctly classified patients with fatal outcome in the subgroup of patients younger than 18 months (5-year OS: 1.0 vs. 0.80 ± 0.65, P < 0.0001; 5-year EFS: 0.98 ± 0.01 vs. 0.58 ± 0.83, P< 0.0001) and significantly separated patients with different OS and EFS within the cohort of age above 18 months (5-year OS: 0.93 ± 0.05 vs. 0.32 ± 0.06, P < 0.0001; 5-year EFS: 0.87 ± 0.06 vs. 0.34 ± 0.07, P < 0.0001; Supplementary Figs. S2 and S3). Furthermore, the model accurately classified all patients with localized neuroblastoma: stage I to III MYCN not amplified (5-year OS: 1.0 vs. 0.78 ± 0.1, P < 0.0001; 5-year EFS: 0.98 ± 0.13 vs. 0.64 ± 0.10, P < 0.0001), stage I to III age above 18 months (5-year OS: 1.0 vs. 0.44 ± 0.14, P < 0.001; 5-year EFS: 0.95 ± 0.44 vs. 0.43 ±0.11, P < 0.001), as well as patients older than 18 months with stage I to III MYCN not amplified disease (5-year OS: 1.0 vs. 0.56 ± 0.19, P < 0.001; 5-year EFS: 0.95 ± 0.41 vs. 0.33 ± 0.18, P < 0.0001), showing a higher classification accuracy than the risk stratification systems (Supplementary Figs. S4–S6). A significant classification capacity was observed for patients stage IV (5-year OS: 0.85 ± 0.1 vs. 0.41 ± 0.07, P = 0.02; 5-year EFS: 0.76 ± 0.1 vs. 0.27 ± 0.06, P = 0.01; Supplementary Fig. S7). Within stage IV neuroblastoma, the performance of the model was less robust for stage IV with MYCN nonamplified disease (5-year OS: 0.85 ± 0.1 vs. 0.52 ± 0.09, P = 0.095; 5-year EFS: 0.77 ±0.11 vs. 0.33 ± 0.09, P = 0.04) or stage IV above 18 months MYCN nonamplified tumors (5-year OS: 0.77 ± 0.13 vs. 0.33 ± 0.1, P = 0.096; 5-year EFS: 0.66 ± 0.15 vs. 0.25 ± 0.1, P = 0.125; Supplementary Figs. S8 and S9). However, the model showed the capacity to classify correctly all MYCN amplified neuroblastoma (stage I–IV) as high-risk tumors (Supplementary Fig. S10).
Validation of the robustness of the three-gene signature and the statistical procedure
To show that the 3 selected genes are highly prognostic independent of the test set used to build the predictive model, we developed for each validation set independently, an equation using exclusively microarray data. CHD5, PAFAH1B1, and NME1 microarray expression data were z-transformed and processed following the same statistical methodology mentioned above. The principal component coefficient scores obtained for each equation, (Yset2) and (Yset3), were similar to those of the (Y96) model (Supplementary Table S4). Set 2 and 3 data were analyzed applying these equations. For both data sets, Kaplan–Meier estimates and univariable Cox regression models showed equivalent results to those obtained with the (Y96) model (Supplementary Fig. S11 and Table S5). These results show the robustness of the gene expression signature and of the statistical methodology used to develop the model.
Discussion
In this study, we developed a PCR-based predictor model using the expression pattern of just 3 genes strongly associated with outcome of patients with neuroblastoma. CHD5, PAFAH1B1, and NME1 were identified through the analysis of the prognostic significance of 11 genes found differentially expressed in our previous study (14). The results of a qRT-PCR–based evaluation of the expression pattern of the selected genes in a training set of 96 primary neuroblastoma tumors enabled the development of a single-score predictor model. Its validity was assessed using different expression data sets obtained from qRT-PCR and microarray gene expression studies comprising a total of 362 patients (9, 15). Our scoring method reliably stratified patients into neuroblastoma groups with markedly divergent clinical course. Multivariable Cox models showed that the developed scoring model was an independent predictor marker for survival. Moreover, the prediction performance of the 3-gene model was found to be as robust as the international neuroblastoma risk stratification systems based on a combination of clinical and biologic parameters. Our classifier also significantly discriminated between patients in most subgroups defined by currently used prognostic markers. Interestingly, a significantly higher prediction performance than current risk stratification systems was observed in clinically relevant subgroups such as children with localized disease MYCN nonamplified. This is a relevant finding, as outcome prediction in this biologically broad subgroup of localized nonamplified neuroblastoma tumors still remains a challenge, as suggested by the low-and intermediate-risk patients experiencing adverse disease. The (Y96) model could, thus, help to better stratify these patients into subgroups with different clinical course and treatment regimes.
The genes that comprise the model have been previously reported to be involved in neuroblastoma biology. CHD5 is a tumor suppressor gene located on chromosome 1p36.31, region recurrently lost in high-risk neuroblastoma (17–21). Expression of this ATP-dependent chromatin remodeling helicase has been found to be restricted to neuronal-derived tissues and absent in neuroblastoma cell lines and neuroblastoma primary tumors with high-risk features, undifferentiated neuroblasts, MYCN amplification, advanced stage, and 1p monosomy (18, 21). Association between CHD5 expression and favorable prognosis in neuroblastoma has been reported previously in microarray gene expression studies (8, 11, 14, 22). PAFAH1B1, located on chromosome 17p13.3, encodes an acetylhydrolase involved in cerebral cortex development, neuronal migration, and axonal growth (23). Mutations of PAFAH1B1 have been related to the Miller–Dieker lissencephaly syndrome (23, 24). We identified PAFAH1B1 expression in low-risk neuroblastoma tumors, whereas in high-risk tumors, expression is significantly diminished. PAFAH1B1 has been reported previously associated with favorable prognosis in neuroblastoma (7). In our study, high expression of both CHD5 and PAFAH1B1 correlated with prolonged survival. In contrast, NME1 displayed high expression levels in high-risk neuroblastoma tumors, as reported previously (25, 26). NME1, located on a region frequently gained in clinically aggressive neuroblastoma 17q21.3 (27, 28), has been reported in several microarray analyses to be associated with high-risk neuroblastoma (6, 12, 13). The product of the NME1 gene is a nucleoside diphosphate kinase involved in cell proliferation, normal development, and cell differentiation (29).
Prognostic models, similar to ours, and gene signatures based on small sets of genes, have been reported previously to predict clinical outcome in diffuse large B-cell lymphoma (6 genes; ref. 30), clear cell renal carcinoma (3 genes; ref. 31), soft tissue sarcoma (3 genes; ref. 32), and non-small cell lung cancer (5 genes; ref. 33). In neuroblastoma, although diverse studies have proposed robust microarray multigene–based classification systems, to date, these gene signatures are still extensive and difficult to implement to clinical routine; that is, 429 genes (10), 144 genes (7, 9), 59 genes (8), 55 genes (5), 42 genes (6), 39 genes (11), or 19 genes (12). We have reduced the classifier complexity to 3 genes strongly associated with patient outcome and created a prognostic model that can be applied in routine laboratories using conventional qRT-PCR assay. Our preliminary results provide evidence of a prognostic model that can accurately define neuroblastoma clinical risk groups and could thus assist therapeutic decisions in patients with neuroblastoma.
In conclusion, we propose a robust and technically simple PCR-based one-score predictor model that requires only minimal amount of mRNA, easy to interpret, reproducible, and cost-effective for most laboratories. These features make the model a potentially practical classifier for neuroblastoma risk stratification that could help refine current risk stratification systems. The potential of this prototype model remains to be fully validated using qRT-PCR in a large, prospective, and independent cohort of patients with neuroblastoma samples.
Supplementary Material
Translational Relevance.
Neuroblastoma risk stratification is perhaps the most advanced of all pediatric solid tumors. International classification systems are based on various clinicopath-ologic and genetic parameters. Nevertheless, there remain cases where these elaborated stratification strategies have shown limited clinical use. In this study, a three-gene PCR-based single-score model was developed and tested using quantitative real-time PCR and micro-array expression data from four sets including 458 patients with primary neuroblastoma. The scoring method reliably stratified patients with significantly different outcome in all the test sets and in different neuroblastoma risk subgroups, showing to be robust and highly reproducible. Furthermore, multivariate analysis showed that the model is an independent prognostic factor for survival. The proposed model is a technically simple classifier that requires minimal amount of mRNA, easy to interpret, reproducible, and cost-effective. These results provide evidence of a practical prognostic classifier that could help refine pretreatment risk assessment in patients with neuroblastoma.
Acknowledgments
The Authors thank Drs. J. Alonso and P. Garcia-Miguel (Hospital La Paz, Madrid, Spain), Drs. R. Noguera, V. Castel, and S. Navarro (Hospital La Fe, Valencia, Spain), and Drs. A. Pérez-Martínez and I. de Prada Vicente (Hospital Niño Jesús, Madrid, Spain) for annotated neuroblastoma specimens. They also thank Yvonne Kahlert (Department of Pediatric Oncology, Children’s Hospital of Cologne, Cologne, Germany) for technical support.
Grant Support
This work has been supported by: the Spanish Ministry of Health (Instituto de Salud Carlos III, FIS PI070286); the Spanish Society against Cancer (AECC, 2007); the Catalan government (AGAUR, Generalitat de Catalunya, 2005SGR00605), NIH grant CA039771, and the Margarita del Pozo Fund donation.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interests were disclosed.
Authors’ Contributions
Conception and design: I. Garcia, G. Mayol, J. Rios, G. Domenech, N.-K. V. Cheung, C. de Torres, J. Mora, C. Lavarino.
Development of methodology: I. Garcia, G. Mayol, J. Rios, G. Domenech, M. Suñol, C. de Torres, J. Mora, C. Lavarino.
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): I. Garcia, G. Domenech, N.-K. V. Cheung, A. Oberthuer, M. Fischer, J.M. Maris, B. Hero, M. Suñol, C. de Torres, J. Mora, C. Lavarino.
Analysis and interpretation of data (e.g., statistical analysis, biosta-tistics, computational analysis): I. Garcia, G. Mayol, J. Rios, G. Domenech, A. Oberthuer, C. de Torres, J.M. Maris, J. Mora, C. Lavarino.
Writing, review, and/or revision of the manuscript: I. Garcia, G. Mayol, J. Rios, G. Domenech, N.-K. V. Cheung, A. Oberthuer, M. Fischer, G.M. Brodeur, M. Suñol, C. de Torres, J. Mora, C. Lavarino.
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): I. Garcia, J. Rios, G. Domenech, M. Fischer, P. Galvan, J. Mora, C. Lavarino.
Study supervision: I. Garcia, J. Rios, G. Domenech, J. Mora, C. Lavarino
Laboratory analyses: I. Garcia, G. Mayol, C. de Torres, P. Galvan, C. Lavarino.
Tissue banking and provision of the samples: E. Rodriguez, P. Galvan, N. K V. Cheung, M. Suñol.
Pathology review: M. Suñol.
All authors read and approved the final manuscript.
References
- 1.Brodeur GM, Pritchard J, Berthold F, Carlsen NL, Castel V, Castelberry RP, et al. Revisions of the international criteria for neuroblastoma diagnosis, staging, and response to treatment. J Clin Oncol. 1993;11:1466–1477. doi: 10.1200/JCO.1993.11.8.1466. [DOI] [PubMed] [Google Scholar]
- 2.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 3.Ambros PF, Ambros IM, Brodeur GM, Haber M, Khan J, Nakagawara A, et al. International consensus for neuroblastoma molecular diagnostics: report from the International Neuroblastoma Risk Group (INRG) Biology Committee. Br J Cancer. 2009;100:1471–1482. doi: 10.1038/sj.bjc.6605014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimada H, Ambros IM, Dehner LP, Hata J, Joshi W, Roald B, et al. The International Neuroblastoma Pathology Classification (the Shimada System) Cancer. 1999;86:364–372. [PubMed] [Google Scholar]
- 5.Asgharzadeh S, Pique-Regi R, Sposto R, Wang H, Yang Y, Shimada H, et al. Prognostic significance of gene expression profiles of metastatic neuroblastomas lacking MYCN gene amplification. J Natl Cancer Inst. 2006;98:1193–1203. doi: 10.1093/jnci/djj330. [DOI] [PubMed] [Google Scholar]
- 6.De Preter K, Vermeulen J, Brors B, Delattre O, Eggert A, Fischer M, et al. Accurate outcome prediction in neuroblastoma across independent data sets using a multigene signature. Clin Cancer Res. 2010;16:1532–1541. doi: 10.1158/1078-0432.CCR-09-2607. [DOI] [PubMed] [Google Scholar]
- 7.Oberthuer A, Hero B, Berthold F, Juraeva D, Faldum A, Kahlert Y, et al. Prognostic impact of gene expression-based classification for neuroblastoma. J Clin Oncol. 2010;28:3506–3515. doi: 10.1200/JCO.2009.27.3367. [DOI] [PubMed] [Google Scholar]
- 8.Vermeulen J, De Preter K, Naranjo A, Vercruysse L, Van Roy N, Hellemans J, et al. Predicting outcomes for children with neuroblastoma using a multigene-expression signature: a retrospective SIO-PEN/COG/GPOH study. Lancet Oncol. 2009;10:663–671. doi: 10.1016/S1470-2045(09)70154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oberthuer A, Berthold F, Warnat P, Hera B, Kahlert Y, Spitz R, et al. Customized oligonucleotide microarray gene expression-based classification of neuroblastoma patients outperforms current clinical risk stratification. J Clin Oncol. 2006;24:5070–5078. doi: 10.1200/JCO.2006.06.1879. [DOI] [PubMed] [Google Scholar]
- 10.Fischer M, Oberthuer A, Brors B, Kahlert Y, Skowron M, Voth H, et al. Differential expression of neuronal gene defines subtypes of disseminated neuroblastoma with favorable and unfavorable outcome. Clin Cancer Res. 2006;12:5118–5128. doi: 10.1158/1078-0432.CCR-06-0985. [DOI] [PubMed] [Google Scholar]
- 11.Schramm A, Schulte JH, Klein-Hitpass L, Havers W, Sieverts H, Berwanger B, et al. Prediction of clinical outcome and biological characterization of neuroblastoma by expression profiling. Oncogene. 2005;24:7902–7912. doi: 10.1038/sj.onc.1208936. [DOI] [PubMed] [Google Scholar]
- 12.Wei JS, Greer BT, Westermann F, Steinberg SM, Son CG, Chen QR, et al. Prediction of clinical outcome using gene expression profiling and artificial neural networks for patients with neuroblastoma. Cancer Res. 2004;64:6883–6891. doi: 10.1158/0008-5472.CAN-04-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohira M, Oba S, Nakamura Y, Isogai E, Kaneko S, Nakagawa A, et al. Expression profiling using a tumor-specific cDNA microarray predicts the prognosis of intermediate risk neuroblastomas. Cancer Cell. 2005;7:337–350. doi: 10.1016/j.ccr.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 14.Lavarino C, Garcia I, Mackintosh C, Cheung NK, Domenech G, Rios J, et al. Differential expression of genes mapping to recurrently abnormal chromosomal regions characterize neuroblastic tumours with distinct ploidy status. BMC Med Genomics. 2008;13:36. doi: 10.1186/1755-8794-1-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Q, Diskin S, Rappaport E, Attiyeh E, Mosse Y, Shue D, et al. Integrative genomics identifies distinct molecular classes of neuroblastoma and shows that multiple genes are targeted by regional alterations in DNA copy number. Cancer Res. 2006;66:6050–6062. doi: 10.1158/0008-5472.CAN-05-4618. [DOI] [PubMed] [Google Scholar]
- 16.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. Research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson PM, Gotoh T, Kok M, White PS, Brodeur GM. CHD5, a new member of the chromodomain gene family, is preferentially expressed in the nervous system. Oncogene. 2003;22:1002–1011. doi: 10.1038/sj.onc.1206211. [DOI] [PubMed] [Google Scholar]
- 18.Okawa ER, Gotoh T, Manne J, Igarashi J, Fujita T, Silverman KA, et al. Expression and sequence analysis of candidates for the 1p36.31 tumor suppressor gene deleted in neuroblastomas. Oncogene. 2007;27:803–8010. doi: 10.1038/sj.onc.1210675. [DOI] [PubMed] [Google Scholar]
- 19.Bagchi A, Papazoglu C, Wu Y, Capurso D, Brodt M, Francis D, et al. CHD5 is a tumor suppressor at human 1p36. Cell. 2007;128:459–475. doi: 10.1016/j.cell.2006.11.052. [DOI] [PubMed] [Google Scholar]
- 20.Fujita T, Igarashi J, Okawa ER, Gotoh T, Manne J, Kolla V, et al. CHD5, a tumor suppressor gene deleted from 1p36.31 in neuroblastomas. J Natl Cancer Inst. 2008;100:940–949. doi: 10.1093/jnci/djn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia I, Mayol G, Rodríguez E, Suñol M, Gershon TR, Ríos J, et al. Expression of the neuron-specific protein CHD5 is an independent marker of outcome in neuroblastoma. Mol Cancer. 2010;9:277. doi: 10.1186/1476-4598-9-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavarino C, Cheung NK, Garcia I, Domenech G, de Torres C, Alaminos M, et al. Specific gene expression profiles and chromosomal abnormalities are associated with infant disseminated neuroblastoma. BMC Cancer. 2009;9:44. doi: 10.1186/1471-2407-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gambello MJ, Darling DL, Yingling J, Tanaka T, Gleeson JG, Wynshaw-Boris A. Multiple dose-dependent effects of Lis1 on cerebral cortical development. J Neurosci. 2003;23:1719–1729. doi: 10.1523/JNEUROSCI.23-05-01719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cardoso C, Leventer RJ, Dowling JJ, Ward HL, Chung J, Petras KS, et al. Clinical and molecular basis of classical lissencephaly: mutations in the LIS1 gene (PAFAH1B1) Hum Mutat. 2002;19:4–15. doi: 10.1002/humu.10028. [DOI] [PubMed] [Google Scholar]
- 25.Hailat N, Keim DR, Melhem RF, Zhu XX, Eckerskorn C, Brodeur GM, et al. High levels of p19/nm23 protein in neuroblastoma are associated with advanced stage disease and with N-myc gene amplification. J Clin Invest. 1991;88:341–345. doi: 10.1172/JCI115299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leone A, Seeger RC, Hong CM, Hu YY, Arboleda MJ, Brodeur GM, et al. Evidence for nm23 overexpression, DNA amplification and mutation in aggressive childhood neuroblastomas. Oncogene. 1993;8:855–865. [PubMed] [Google Scholar]
- 27.Bown N, Cotterill S, Lastwska M, O'Neill S, Pearson AD, Plantza D, et al. Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. N Engl J Med. 1999;340:1954–1961. doi: 10.1056/NEJM199906243402504. [DOI] [PubMed] [Google Scholar]
- 28.Plantaz D, Mohapatra G, Matthay KK, Pellarin M, Seeger RC, Feuer-stein BG. Gain of chromosome 17 is the most frequent abnormality detected in neuroblastoma by comparative genomic hybridization. Am J Pathol. 1997;150:81–89. [PMC free article] [PubMed] [Google Scholar]
- 29.Lombardi D, Lacombe ML, Paggi MG. Nm23: unraveling its biological function in cell development. J Cell Physiol. 2000;182:144–149. doi: 10.1002/(SICI)1097-4652(200002)182:2<144::AID-JCP2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 30.Lossos IS, Czerwinski DK, Alizadeh AA, Wechser MA, Tibshirani R, Botstein D, et al. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N Engl J Med. 2004;350:1828–1837. doi: 10.1056/NEJMoa032520. [DOI] [PubMed] [Google Scholar]
- 31.Yao M, Huang Y, Shioi K, Hattori K, Murakami T, Sano F, et al. A three-gene expression signature model to predict clinical outcome of clear cell renal carcinoma. Int J Cancer. 2008;123:1126–1132. doi: 10.1002/ijc.23641. [DOI] [PubMed] [Google Scholar]
- 32.Hoffman AC, Danenberg KD, Taubert H, Danenberg PV, Wuerl P. A three-gene signature for outcome in soft tissue sarcoma. Clin Cancer Res. 2009;15:5191–5198. doi: 10.1158/1078-0432.CCR-08-2534. [DOI] [PubMed] [Google Scholar]
- 33.Chen HY, Yu SL, Chen CH, Chang GC, Chen CY, Yuan A, et al. A fivegene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med. 2007;356:11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.