Abstract
Maternal inheritance of mitochondria creates a sex-specific selective sieve with implications for male longevity, disease susceptibility and infertility. Because males are an evolutionary dead end for mitochondria, mitochondrial mutations that are harmful or beneficial to males but not females cannot respond directly to selection. Although the importance of this male/female asymmetry in evolutionary response depends on the extent to which mitochondrial mutations exert antagonistic effects on male and female fitness, few studies have documented sex-specific selection acting on mitochondria. Here, we exploited the discovery of two highly divergent mitochondrial haplogroups (A and B2) in central Panamanian populations of the pseudoscorpion Cordylochernes scorpioides. Next-generation sequencing and phylogenetic analyses suggest that selection on the ND4 and ND4L mitochondrial genes may partially explain sexually antagonistic mitochondrial effects on reproduction. Males carrying the rare B2 mitochondrial haplogroup enjoy a marked advantage in sperm competition, but B2 females are significantly less sexually receptive at second mating than A females. This reduced propensity for polyandry is likely to significantly reduce female lifetime reproductive success, thereby limiting the spread of the male beneficial B2 haplogroup. Our findings suggest that maternal inheritance of mitochondria and sexually antagonistic selection can constrain male adaptation and sexual selection in nature.
Keywords: mitochondrial haplotype, sexually antagonistic selection, sperm competition, maternal inheritance, polyandry, Cordylochernes scorpioides
1. Introduction
At the cellular level, animals are fundamentally chimaeras, with a diploid genome in the nucleus inherited from both parents, and a haploid mitochondrial genome in the cytoplasm that replicates independently and is transmitted only by their mothers. Because males do not transmit their mitochondria to offspring, mutations in the mitochondrial genome that are either beneficial or detrimental to males cannot respond directly to selection ([1–3], but see [4]). The importance of this male/female asymmetry in evolutionary response depends critically on the extent to which mitochondrial mutations exert antagonistic effects on male and female fitness [1,5,6].
Until recently, animal mitochondria were considered minor players in evolution, with the small size of their genomes providing little scope for exerting significant effects on complex, fitness-related traits [7]. Over the past decade, this conventional view has been largely overturned, with accumulating evidence for strong mitochondrial haplotype effects on nervous system development, cognitive ability, disease susceptibility and sexual differences in longevity [8–15]. Nonetheless, the significance of the sex-biased, mitochondrial selective sieve as a contributor to variation in male fertility and sperm traits remains controversial and inadequately investigated. Several studies have reported marked effects of mitochondrial haplogroup on sperm motility in humans and other taxa [16–21], while others have failed to find an association [22,23]. Even less well understood are the contributions of mitochondrial DNA (mtDNA) variation to female re-mating behaviour and sperm competitive ability (but see [24–26]).
The coexistence of two sympatric but highly divergent mitochondrial haplogroups in the neotropical pseudoscorpion, Cordylochernes scorpioides [27], provides a unique opportunity to assess the influence of mitochondrial variation on sperm competitive ability and female sexual receptivity. In these pseudoscorpions, mating involves a sequence of stereotypical behaviours in which the male grasps the female while he constructs and deposits a spermatophore on the substrate (figure 1). The male then manoeuvres the female into a position in which the sperm packet directly contacts her genital aperture, and successful sperm transfer is associated with a pronounced abdominal flexure by the female [28]. Matings can be interrupted immediately following spermatophore deposition and the sperm packet collected for assessment of sperm number and viability [29]. External spermatophore deposition and diagnostic female behaviour facilitate unambiguous assessment of female sexual receptivity and success of sperm transfer. Non-invasive monitoring of female reproductive status and embryological development is made possible by C. scorpioides' ‘external womb’ mode of viviparity, in which females nourish developing embryos in an external, transparent brood sac overlying their genital aperture [27]. In the wild, C. scorpioides females produce mixed-paternity broods sired by up to four males [30], and sperm competitive ability is therefore likely to be an important component of male fitness in this pseudoscorpion.
Figure 1.
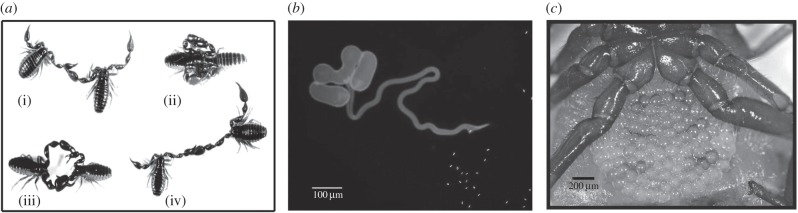
Reproductive biology of Cordylochernes scorpioides. (a) Mating sequence in which the male (i) grasps the female, (ii) holds her in a stationary position while he deposits a spermatophore, (iii) reverses and pulls the female over the spermatophore and (iv) maintains grasp on the female during the sperm uptake phase. (b) Sperm packet with the everted tube and evacuated sperm. (c) Ventral view of gravid female carrying a brood sac containing early stage embryos.
Here, we report the results of a study in which mitochondrial genome sequencing identified two highly divergent mitochondrial haplogroups, Clades A and B2, coexisting in populations of C. scorpioides in central Panamá. Although A- and B2-haplogroup males did not differ in sperm number and sperm viability, B2 males enjoyed a significant sperm competitive advantage, siring more than twice as many offspring as A males in a two-male sperm competition experiment. However, the B2 haplogroup had an adverse effect on female sexual receptivity at second mating, with B2 females significantly less likely than A females to accept sperm from a second male. The reduced propensity of B2 females to engage in polyandry is likely to incur major fitness costs, since mating with multiple males has been shown to significantly increase female lifetime reproductive success in C. scorpioides [31].
Taken together, these findings suggest that, despite its highly advantageous effect on sperm competitive ability, the B2 haplogroup is constrained to low frequency in central Panamá at least in part because of its negative consequences for females, in terms of reduced propensity for adaptive polyandrous behaviour in B2 females.
2. Material and methods
(a). Experimental pseudoscorpions
Individuals for this study were drawn from a large laboratory population established from 350 C. scorpioides males and females collected in 2006 and 2008 from six locations spanning a 60 km region in central Panamá [27]. In our laboratory population, pseudoscorpions are reared and maintained in individual vials to ensure virginity, and matings are staged to maintain the number of field-collected matrilines. No matings are carried out between full siblings, half siblings or first cousins to minimize inbreeding. Within these constraints, mating pairs for each generation are chosen randomly, and haplotype-based assortative mating, as might occur in mass-reared laboratory populations of Drosophila, is avoided.
(b). Mitochondrial genome sequencing
Previous sequencing of the C. scorpioides mitochondrial cytochrome oxidase 1 (COX1) gene from populations in central and western Panamá identified three highly divergent lineages: one clade consisting predominantly of individuals from central Panamá (Clade A), and two sister clades (B1 and B2), which appeared to be restricted to western Panamá [32]. However, subsequent sequencing of the mitochondrial NADH dehydrogenase 2 (ND2) gene from 66 C. scorpioides matrilines yielded an estimated frequency of the A and B2 haplogroups in central Panamá of 88 and 12%, respectively ([27]; D. Zeh 2011, unpublished data). Here, in order to characterize differences across the entire genome, we performed next-generation sequencing (NGS) of the mitochondrial genomes of five A-haplogroup and six B2-haplogroup individuals from our central Panamá laboratory population, as well as two B2- and two B1-haplogroup individuals previously collected from the Bocas del Toro province in extreme western Panamá [32].
(i). Sequencing of the reference Cordylochernes scorpioides mitochondrial genome
A reference C. scorpioides mtDNA genome for assembly of NGS sequences was generated using conventional sequencing (Applied Biosystems Prizm 3730 DNA Analyser, Life Technologies, Grand Island, NY, USA) and primer walking, an iterative process of forward and reverse sequencing of the two ends of successively shorter PCR products (for primers and PCR conditions, see the electronic supplementary material, table S1). Geneious Pro (Biomatters Ltd, Auckland, New Zealand) was used to assemble and annotate the reference genome, using the ORF-finder tool, BLAST searches of putative genes, proteins and rRNA sequences and the programs ARWEN [33] and tRNAscan-SE [34] for identifying the tRNAs.
(c). Next-generation sequencing of 15 mtDNA genomes
DNA templates for NGS were prepared by amplifying each individual's entire mitochondrial genome in three overlapping segments and pooling PCR products for each individual (electronic supplementary material, table S1). Library preparation and sequencing were performed by the Nevada Genomics Center at the University of Nevada, Reno. Starting with 100 ng of amplified mtDNA, libraries were prepared, according to the manufacturer's instructions, using an Ion Xpress Plus Fragment Library Kit (Life Technologies). Amplicons were sheared with the Ion Shear enzyme, barcoded with Ion Xpress Barcode Adapters, and size selected, using the E-Gel system (Life Technologies). Library size was verified and quantitation carried out, using the Agilent High Sensitivity DNA Kit (Agilent Technologies, Inc., Santa Clara, CA, USA). Templated Ion Sphere Particles were prepared, using the Ion PGM Template OT2 400 Kit, following the manufacturer's instructions. Sequencing was carried out on a Life Technologies Ion Torrent PGM Sequencer, using the Ion PGM Sequencing 400 Kit and the Ion 314v2 Chip. Across the 15 mtDNA genomes, mean read length was 150 bp and mean read number was 25 199, yielding a mean fold coverage per genome of 239 (range, 139–695).
(d). Phylogenetic analyses and tests of adaptive evolution
Sequences were assembled in Geneious Pro v. 7.1, aligned using MAFFT v. 7.017 [35], and edited to include the 13 protein-coding genes, all transfer RNAs, and both the small and large subunit ribosomal RNA genes. Phylogenetic reconstruction was carried out, using Bayesian estimation [36]. To identify functionally significant sequence divergence between the A and B2 haplogroups, adaptive evolution analyses were performed separately for each of the 13 protein-coding genes, using three methods. First, a codon-based test of neutrality for differences between sequences, dN−dS, where N and S represent non-synonymous and synonymous substitutions, respectively, was performed using MEGA6 [37]. Second, Bayesian estimates of the number of codons experiencing negative selection, neutrality and positive selection for each gene were obtained using MrBayes [36]. Third, TreeSAAP [38], a program that compares the distribution of observed changes inferred from a phylogenetic tree with neutral expectations, was used to evaluate potential selection on physico-chemical properties of amino acids (see the electronic supplementary material for details).
(e). Haplogroup effects on sperm number and sperm viability
Haplogroup effects on ejaculate traits were assessed by pairing experimental A and B2 males with virgin females, interrupting the matings and collecting sperm packets, as described elsewhere [29]. Sperm packets were ruptured in phosphate-buffered saline and stained with SYBR 14 and propidium iodide (Invitrogen Live/Dead Sperm Viability Kit, Life Technologies) to distinguish between live and dead sperm. Each 11 µl stained sample was pipetted onto a haemocytometer, and viewed under an Olympus BX51 fluorescence microscope equipped with an EM510 dual band pass fluorescence filter cube. Total number of sperm was estimated by multiplying the number of sperm counted in a 0.9 µl volume of the sample by a factor of 12.2 (11/0.9). The proportion of live sperm in the sample provided an estimate of sperm viability.
(f). Haplogroup effects on sperm competitive ability
To assess mitochondrial haplogroup effects on sperm competitive ability, initially virgin females were each mated to two males, one carrying A-haplotype mitochondria and the other B2-haplotype mitochondria, with no matings between siblings or first cousins, 48 h between matings and mating order randomized across male haplogroups. Given the low level of B2 female sexual receptivity at second mating (see below), only A-haplogroup females were used in this experiment. Each replication was initiated by placing a virgin female with a virgin male in a 28 mm diameter mating arena. Interactions were observed under red, fibre-optics illumination, using an Olympus SZ6145TR stereomicroscope, for 45 min or until the female accepted a sperm packet from the male. Only replicates in which the female unambiguously accepted a sperm packet from both males were retained for subsequent paternity analysis. Following their second mating, females were maintained in individual vials in a dark incubator at 28.5°C and 80% humidity, and monitored until they gave birth. Protonymphs were then removed from the brood nest, counted and frozen at −80°C, along with the dam and the two putative sires. DNA was extracted from the adults and PCR amplification of alleles at the cCscMS23 minisatellite locus (heterozygosity = 0.99) [39,40] was used to assign paternity (see the electronic supplementary material for methods). For each replication, PCR products from the mother, the two putative sires and an average of 22 offspring were run on a 1.5% agarose gel and stained with ethidium bromide to visualize alleles. Paternity was assigned based on the presence of unique paternal alleles in offspring.
(g). Haplogroup effects on female reproductive function
Haplotype effects on female reproduction were analysed by mating virgin females to virgin males in all four possible combinations: A♀ × A♂ (N = 21), A♀ × B2♂ (N = 22), B2♀ × A♂ (N = 21), B2♀ × B2♂ (N = 22). Female fecundity and reproductive success were assessed based on the number of early stage embryos produced and the number of protonymphs born to each female, respectively. Matings were carried out, as described elsewhere [27], and females were then monitored until they either gave birth to protonymphs, spontaneously aborted their brood of embryos or failed to become gravid within 30 days. Gravid females were carefully removed from their vials as soon as individual embryos became clearly discernable, and digital images of their brood sacs were recorded for embryo counting, as described elsewhere [41]. Females were then monitored until they gave birth or spontaneously aborted the brood. Females remain in a silken nest constructed on the vial wall throughout gestation, and embryonic development could therefore be observed without further disturbance to the female [31]. Within 24 h of birth, nymphs were removed from the nest and counted.
(h). Haplogroup effects on level of polyandry
For each haplogroup, we assessed female sexual receptivity across two mating opportunities, and used the proportion of females that were sexually receptive at second mating to quantify level of polyandry. Virgin A-haplogroup (N = 26) and B2-haplogroup (N = 32) females were paired with a randomly selected, unrelated male (Male A) of either the same or the alternative mitochondrial haplogroup, and matings were observed, as described above. After 48 h, each female that had accepted a sperm packet from a ‘same-haplogroup’ Male A was given the opportunity to mate with a ‘different-haplogroup’ Male B, and vice versa. Females were scored as sexually receptive if they remained stationary during construction of the spermatophore, allowed themselves to be pulled over the spermatophore and performed the abdominal flexure diagnostic of successful sperm transfer. Sexually unreceptive females either: (i) broke free from the male, (ii) refused to remain stationary during spermatophore construction or (iii) resisted being pulled over the spermatophore.
(i). Statistical analyses
Haplotype effects on female reproductive traits, male ejaculate traits and sperm competitive ability were analysed, using a general linear mixed model (GLMM), as performed in PROC GLIMMIX in SAS, v. 9.3 [42]. To avoid pseudoreplication, full-sibling family identity was included in the models as a random effect. Because total sperm number was approximately normally distributed, the GLMM for analysing this variable incorporated a Gaussian distribution, an identity link function, a Laplace maximum-likelihood approximation, and the SAS containment method for determining degrees of freedom [42]. Embryo and protonymph count data were square root transformed and analysed as above, with cephalothorax length included in the model as a covariate to control for and assess female size effects. Sperm viability was evaluated as the proportion of live sperm in an ejaculate and was not normally distributed. This response variable was therefore analysed using the GLIMMIX logit link function to fit a binomial response variable [42]. To analyse the effect of mitochondrial haplogroup on sperm competitive ability, male haplogroup and mating order were included as fixed factors, and male chela hand depth (HD) and dam identity as random factors in the model. The number of protonymphs sired by males was analysed using a GLMM that incorporated a log link function, a Gauss–Hermite Quadrature maximum-likelihood approximation, and a generalized Poisson mixed model for overdispersed count data to accommodate non-normality and overdispersion in the number of protonymphs sired by males of the two haplogroups (see [42], pp. 3123–3124). The Akaike Information Criterion corrected for finite sample size (AICc) was used to determine the best-fit model among the set of all possible models.
3. Results
(a). Mitochondrial genome sequencing
Sequencing of the 13 protein-coding genes of the oxidative phosphorylation (OXPHOS) pathway, together with the 22 transfer RNAs, and the small and large subunit ribosomal RNA genes, revealed substantial divergence between haplogroups, with a mean of 949 nucleotide substitutions between the A and B2 haplogroups (electronic supplementary material, table S2; figure 2). By contrast, within-haplogroup variation was low, and ranged from 11 to 47 nucleotide substitutions. Gene length was conserved for nine of the 13 OXPHOS genes but varied between haplogroups for ND2, ATP6 (ATP synthase 6), ND4 and CYTB (cytochrome B) (electronic supplementary material, table S3), with total genome size, excluding the control region, ranging from 13 704 to 13 736 bp. The two B2-haplogroup sequences from western Panamá were indistinguishable from the six B2-haplogroup sequences from our central Panamanian laboratory population. With a mean within-haplogroup divergence of only 0.04%, B2 is the least variable of the three clades (electronic supplementary material, table S2).
Figure 2.
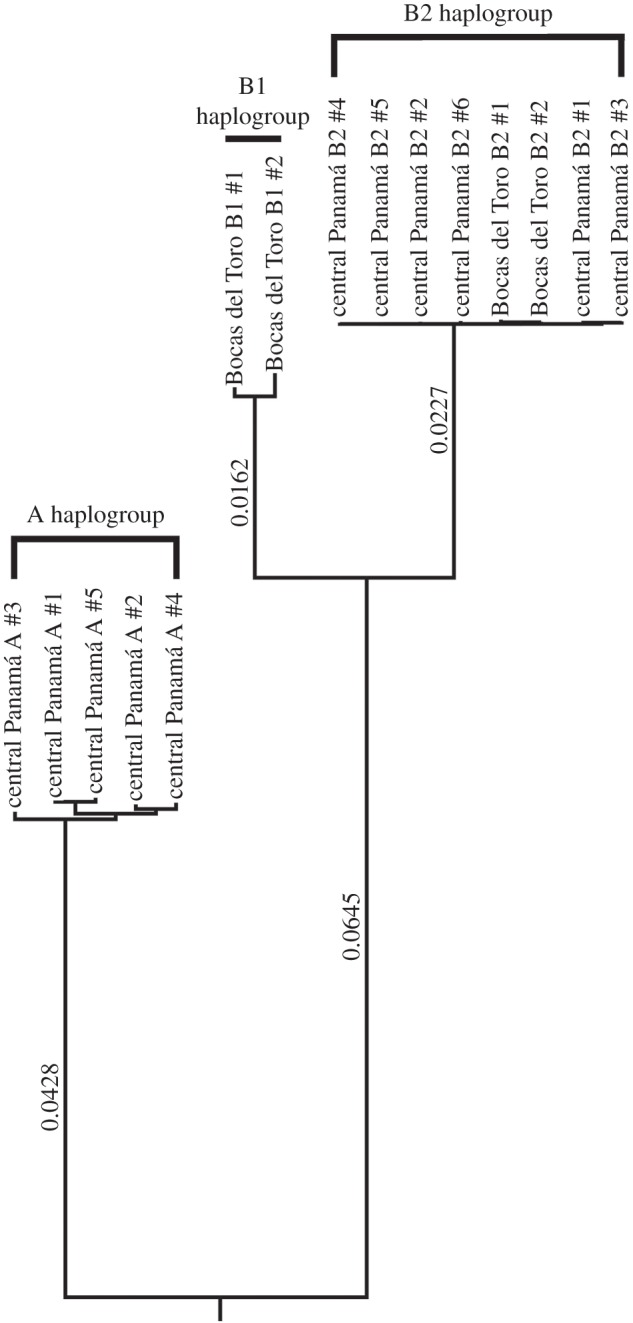
Bayesian phylogram of Cordylochernes scorpioides from Panamá based on mitochondrial genome sequencing. Central Panamá A- and B2-haplogroup individuals are representatives of laboratory matrilines that were used in the sperm competition experiment and assays of male and female reproductive traits. Bocas del Toro B1- and B2-haplogroup individuals were previously collected from Bocas del Toro province in extreme western Panamá [32]. The Bayesian phylogeny reconstruction incorporated a GTR substitution model with a gamma distribution variation and a Markov chain Monte Carlo length of 3 000 000. Bayesian estimated lengths between nodes are indicated to the left of each major branch.
(b). Adaptive evolution analyses of mitochondrial gene sequence
The percentage of codons exhibiting non-synonymous substitutions varied extensively between genes, with a minimum of 1.72% for COX2 (cytochrome oxidase 2) and a maximum of 17.70% for ND3. Across all 13 genes, 206 of 4568 codons were affected by non-synonymous substitutions. For 11 of the 13 genes, codon-based Z tests of neutrality for differences between sequences [42] revealed significant adaptive divergence for all pairwise sequence comparisons of A- versus B2-haplogroup sequences (p < 0.00179) when corrected for multiple comparisons using the sequential Bonferroni method [43]. Only ATP8 and ND3 showed no adaptive divergence between A and B2. For all genes, within-haplogroup sequence divergence was consistent with neutral expectations, when p-values were corrected for multiple comparisons.
Bayesian analyses of the overall frequency of neutral, negatively and positively selected sites within each gene [36] indicate that a high proportion of sites within most genes have been subjected to negative selection, with the frequency of negatively selected sites generally exceeding 90% (electronic supplementary material, table S3). Exceptions include ATP8, ND3, ND4L and ND1. Positive selection has been most pervasive on ND4L and ATP8, with positively selected sites identified across 28 and 24% of their gene regions, respectively.
TreeSAAP [38] analyses identified seven statistically and functionally significant amino acid substitutions, and all but one of these substitutions involved divergence between the A and B clades in NADH dehydrogenase genes (ND4 and ND4L) in Complex I of the electron transport chain (ETC; table 1). The other significant amino acid substitution involved divergence between all three haplogroups in ATP8, a Complex V gene.
Table 1.
Significant amino acid substitutions (p < 0.001) identified by TreeSAAP. ATP, ATP synthase gene; ND, NADH dehydrogenase gene.
| gene/codon | functional property | OXPHOS complex | A-haplogroup amino acid | B1-haplogroup amino acid | B2-haplogroup amino acid |
|---|---|---|---|---|---|
| ATP8/41 | polar requirement | V | serine | alanine | aspartic acid |
| ND3/20 | solvent accessible reduction ratio | I | valine | methionine | valine |
| ND4/24 | equilibrium constant (ionization of COOH) | I | isoleucine | no codon present | methionine |
| ND4/72 | equilibrium constant (ionization of COOH) | I | isoleucine | valine | valine |
| ND4/102 | equilibrium constant (ionization of COOH) | I | isoleucine | valine | valine |
| ND4L/2 | solvent accessible reduction ratio | I | valine | threonine | threonine |
| ND4L/38 | solvent accessible reduction ratio | I | valine | threonine | threonine |
(c). Haplogroup effects on sperm number and sperm viability
The mean number of sperm in the sperm packets of A-haplogroup males (N = 80) and B2-haplogroup males (N = 82) did not differ significantly (F1,39 = 0.08, p = 0.78), with A-haplogroup males producing a mean (±s.e.) of 1386 sperm ± 100 and B2-haplogroup males a mean of 1345 ± 99. Similarly, the proportion of viable sperm did not differ between A and B2 males (mean ± s.e.: A♂ = 97.40 ± 1.35% versus B2♂ = 96.36 ± 1.35%; F1,37 = 0.02, P = 0.88). Neither the sexually dimorphic trait, chela HD, nor male age had a significant impact on sperm number (HD: F1,39 = 2.62, p = 0.11; age: F1,39 = 1.89, p = 0.18) or sperm viability (HD: F1,37 = 0.00, p = 0.96; age: F1,37 = 0.48, p = 0.49).
(d). Haplogroup effects on sperm competitive ability
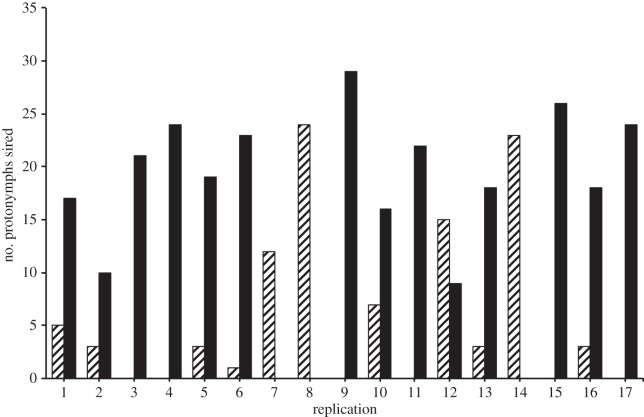
PCR amplification of the ND2 locus followed by ClaI digestion established that A and B2 putative sires from all successful replicates of the sperm competition experiment possessed the correct haplogroup sequence (see the electronic supplementary material for methods). In the full model analysis, male mitochondrial haplogroup was found to exert a strong and significant effect (F1,14 = 5.69, p = 0.03) on the number of protonymphs sired, with B2 males siring 2.47 times more offspring (mean ± s.e.: 15.71 ± 2.03) than A-haplogroup males (6.35 ± 2.03). The effects of mating order (F1,14 = 1.68, p = 0.22) and male chela HD (F1,15 = 0.00, p = 0.99) on paternity were not significant. According to the AICc, the best-fit model incorporated only male haplogroup (F1,16 = 7.32, p = 0.02). In eight replications, paternity was mixed and B2-haplogroup males outcompeted A-haplogroup males in all but one of those replicates (figure 3).
Figure 3.
Sperm competitive ability and male mitochondrial haplogroup. The striped bar indicates the number of protonymphs sired by the A-haplogroup male and the black bar the number sired by the B2-haplogroup male in each of the 17 replications of the sperm competition experiment.
(e). Haplogroup effects on female reproductive function
The proportion of females that became gravid and produced early stage embryos did not differ significantly between mating cross types (likelihood ratio χ2 = 1.403, p = 0.70). Mean number of early stage embryos produced was not affected by male haplogroup (F1,31 = 0.00, p = 0.97), female haplogroup (F1,31 = 1.00, p = 0.33) or by interaction between male and female haplogroups (F1,31 = 0.19, p = 0.66; table 2). Qualitatively similar results were obtained when data from females that failed to become gravid were excluded from the analysis (male haplogroup: F1,15 = 0.77, p = 0.39; female haplogroup: F1,15 = 0.24, p = 0.63; male × female haplogroup: F1,15 = 1.02, p = 0.33). Likewise, neither male nor female haplogroup significantly affected the number of protonymphs born, irrespective of whether non-gravid females were included (male haplogroup: F1,31 = 0.46, p = 0.50; female haplogroup: F1,31 = 0.84, p = 0.37; male × female haplogroup: F1,31 = 0.12, p = 0.73) or excluded from the analyses (male haplogroup: F1,14 = 0.65, p = 0.43; female haplogroup: F1,14 = 0.15, p = 0.70; male × female haplogroup: F1,14 = 0.46, p = 0.51; table 2). Finally, female cephalothorax length had only marginally significant effects on embryo number (F1,31 = 3.38, p = 0.08) and the number of protonymphs born (F1,31 = 3.76, p = 0.06). Taken together, these results demonstrate that, among singly mated females, reproductive success is not influenced by male or female haplogroup, or by mating pair haplogroup combination.
Table 2.
Male, female and male × female haplogroup effects on female fecundity and female reproductive success. A, A-haplogroup mitochondria; B2, B2-haplogroup mitochondria.
| cross type | early stage embryos produced (mean ± s.e.)a | number of protonymphs born (mean ± s.e.)a |
|---|---|---|
| A♀ × A♂ | 70.53 ± 8.43 | 42.45 ± 6.83 |
| A♀ × B2♂ | 69.12 ± 8.38 | 43.37 ± 6.83 |
| B2♀ × A♂ | 60.59 ± 8.36 | 34.91 ± 6.67 |
| B2♀ × B2♂ | 57.14 ± 8.46 | 40.27 ± 6.66 |
aMeans incorporate zero value observations in which females failed to become gravid or aborted the entire brood of embryos.
(f). Haplogroup effects on level of polyandry
Females carrying A-haplogroup mitochondria did not differ significantly from B2-haplogroup females in sexual receptivity at first mating (Fisher's exact test, p = 1.00), with nearly all A-haplogroup (97%) and B2-haplogroup females (96%) accepting a sperm packet. By contrast, females differed significantly in their propensity for polyandry, with only 44% of B2-haplogroup females accepting a sperm packet from a second male, compared with 71% of A-haplogroup females (Fisher's exact test, p = 0.05; electronic supplementary material, figure S1). This marked difference in level of polyandry could not be attributed to the mitochondrial haplogroup of the female's first (Fisher's exact test, p = 1.00) or second mate (Fisher's exact test, p = 1.00).
4. Discussion
The co-occurrence of highly divergent mitochondrial haplogroups in central Panamanian populations of the harlequin beetle riding pseudoscorpion provided a unique opportunity to investigate the mechanisms through which maternal inheritance of mitochondria and sexually antagonistic selection constrain male adaptation and the operation of sexual selection. This study demonstrates that natural variation in mtDNA has significant and opposing effects on components of reproductive success in C. scorpioides males and females. The pseudoscorpions used in this study were drawn from matrilines that have been maintained in the laboratory for a minimum of 14 generations through matings that were random with respect to mitochondrial haplogroup. Consequently, the A and B2 mitochondrial clades in our C. scorpioides laboratory population are nearly identical with respect to nuclear genetic background, greatly increasing the likelihood that systematic differences between haplogroups in sperm competitive ability and female sexual receptivity are causally related to mitochondrial sequence divergence.
In our two-male sperm competition experiment, B2-haplogroup males sired 2.47 times more offspring than A-haplogroup males, and this striking discrepancy could not be explained by differences in sperm number, sperm viability or mating order. Whether or not the sperm competitive superiority of the B2 haplogroup is intrinsic or dependent on female haplogroup remains to be determined, since only A-haplogroup females were used in this experiment. Given the evidence of male and female genotype effects on sperm competition in other species [44,45], it is possible that sperm competitive ability in C. scorpioides may be influenced by female mitochondrial haplogroup. While haplogroup-dependent cryptic female choice favouring B2 sperm could theoretically account for the B2 male advantage in fertilization, this hypothesis is not consistent with the results of our assessment of possible haplotype effects on female reproductive function. In a non-competitive context, neither male haplogroup, female haplogroup nor the interaction between male and female haplogroup exerted an effect on the proportion of females that became gravid, the number of early stage embryos produced and the number of nymphs born. Furthermore, because the B2 haplogroup is rare in central Panamá, the fitness of B2-carrying males must largely be mediated through matings with A-haplogroup females.
The sequence and phylogenetic analyses performed in this study suggest that adaptive evolution, involving, in particular, selection on peptides encoded by ND4 and ND4L, has played a critical role in divergence between C. scorpioides mitochondrial haplogroups. The ETC embedded in the mitochondrion's inner membrane is responsible for establishing the voltage differential that drives eukaryotic metabolism and generates as much as 90% of cellular energy [12]. Four of the five complexes that compose the ETC include polypeptides encoded by mitochondria, as well as nuclear-encoded proteins. OXPHOS begins with NADH, the essential link in the ETC. NADH is oxidized by complex I, with the mitochondrial genome encoding seven (ND 1–6 and 4L) of the approximately 45 polypeptides of this complex [9]. Given the fundamental role of mitochondria in sperm metabolism and motility, it seems likely that the functionally significant amino acid substitutions identified here may at least partially account for the profound difference in sperm competitive ability between males carrying A- and B2-haplogroup mitochondria. Interestingly, a recent RNA sequencing study performed in our laboratory [46] has revealed that ND4 and ND4L, the two mitochondrial genes identified by TreeSAAP as exhibiting functionally significant amino acid substitutions between the A and B2 haplogroups, are the two genes most differentially expressed by mitochondrial haplogroups in testicular tissue. These findings indicate that the divergent mitochondrial haplogroups differ not only in genetic architecture and protein structure, but also in levels of expression in male gonadal tissue.
In contrast to its beneficial effect on male reproductive success, the B2 mitochondrial haplogroup was associated with maladaptive mating behaviour by females. Sexual receptivity at second mating was significantly lower in females carrying B2-haplogroup mitochondria, with 38% fewer B2 females than A females accepting a sperm packet from a second male. This reduced propensity for polyandry is likely to incur significant costs for B2 females, given the evidence for substantial fitness benefits of polyandry in this pseudoscorpion. In a previous study, in which twice-mated females received either one sperm packet from each of two different males or two sperm packets from a single male, C. scorpioides females mated to two males gave birth to 32% more offspring over their lifetime than did females restricted to mating with a single male [31].
According to theory, virgin females should mate with the first male they encounter in order to ensure fertilization [47]. However, once mated, females may become less receptive in subsequent matings, either as a result of male manipulation via products in the seminal fluid that inhibit re-mating [48] or because females become more discriminating after securing an adequate supply of sperm [49–51]. In the context of the reproductive biology of C. scorpioides females, male suppression of female re-mating would be highly adaptive even in the absence of last male sperm precedence. In this viviparous species, females produce an external brood sac of embryos approximately 3 days after mating [52]. Successful inhibition of polyandry during this 3-day period would represent a high reproductive benefit to males. Once gravid, females are invariably sexually unreceptive until the nymphs are born approximately 16 days later. Since the brood sac of developing embryos overlies the female's genital aperture, complete brood abortion would be required for females to accept sperm. Of course, mechanisms other male manipulation, such as effects of females' mitochondria on fertilization rate and oogenesis, could account for the haplogroup differences in female re-mating frequency observed in this study. However, we found no evidence for such effects, as the two haplogroups do not differ in the interval between mating and brood sac production.
In the study reported here, although all females became less sexually receptive at second mating, B2-haplogroup females were significantly less likely to re-mate than A-haplogroup females, regardless of the haplogroup of the females' first mate. This suggests that female mitochondrial haplogroup, rather than differences between A and B2 males in their seminal fluid products, is responsible for the divergent re-mating behaviour in females, and that B2 females may be more susceptible to male manipulation. Mitochondrial DNA sequence variation is known to have significant effects on neurodevelopment and cognition, with the expression of over 200 nuclear genes in brain tissue modified by haplotype [8]. Maternally inherited mitochondria suffer the direct fitness costs of manipulation by seminal products, such as sex peptides that reduce female re-mating frequency and longevity [48], but receive no indirect benefits from the production of competitively superior sons [2]. Female susceptibility to male manipulation may therefore be influenced by variation in mtDNA sequence, and we hypothesize that the low re-mating frequency of C. scorpioides B2 females is mediated by haplogroup effects on mitochondrial/nuclear crosstalk in the female brain.
It is instructive to consider our findings in the context of the sexually selected sperm hypothesis, which argues that postcopulatory sexual selection favours male traits that increase sperm competitive ability and female propensity for polyandry, since polyandrous females benefit most from producing sons with superior sperm competitive ability [53]. As recognized by Pizzari & Birkhead [6], two factors act to constrain this Fisherian-like process: (i) sex-biased inheritance of traits that enhance male success in fertilization and (ii) sexual antagonism. Both of these constraints apply in the case of the divergent mitochondrial haplogroups in C. scorpioides. Exclusively maternal inheritance of mitochondria is an extreme case of sex-biased inheritance that prevents a direct response to selection acting on males, while the opposing effects of mtDNA haplogroup on level of polyandry and sperm competitive ability undermine the Fisherian process. Because B2 females are less polyandrous, if conditions were to favour the spread of B2 females, an increase in their frequency would erode the conditions required for B2 males to outcompete A males through postcopulatory sexual selection.
Maternal inheritance of mitochondria creates male/female asymmetries in response to selection that have potentially important implications for evolutionary processes ranging from sexual differences in longevity and disease susceptibility to population viability and speciation [13,54]. Paradoxically, in the field of sexual selection, where maternal inheritance of mitochondria may well have its greatest impact, female-limited response to selection remains inadequately investigated. Our findings suggest that mitochondrial haplotype effects on components of male and female reproductive success may often go undetected in non-competitive contexts. In C. scorpioides, mitochondrial haplogroup had no discernable effects on sperm number or sperm viability, and essentially all females were sexually receptive at first mating. Only when females were given the opportunity to mate with a second male did the sexually antagonistic effects of the rare haplogroup on sperm competitive ability and level of polyandry become apparent. Similar mitochondrial effects on competitive but not non-competitive male fertility have recently been demonstrated in Drosophila melanogaster [26]. Such cryptic effects of mtDNA sequence variation suggest that mitochondria may play a more important role in postcopulatory sexual selection than is currently appreciated.
Although more comprehensive sampling in western Panamá is required, the relative frequencies of the three C. scorpioides mitochondrial haplogroups vary considerably between western and central Panamá [32]. In the Chiriquí and Boca del Toro provinces of extreme western Panamá, the frequencies of the A, B1 and B2 haplogroups are 47, 23 and 30%, respectively [32]. As reported above, the corresponding frequencies in central Panamá are 88%, 0 and 12%, suggesting gene flow from western Panamá as the source of the B2 haplogroup in populations of C. scorpioides in central Panamá.
This is one of the first studies to show opposing effects of naturally occurring mitochondrial genetic variation on reproductive success in males and females. Cordylochernes scorpioides males benefit from B2 mitochondria that they inherit from their mothers but this rare mitochondrial haplogroup which enhances sperm competitive ability appears to be constrained to low frequency in the lowland rainforests of central Panamá by its adverse effect in the physiological environment of their sisters.
Supplementary Material
Acknowledgements
We thank La Autoridad Nacional del Ambiente for permission to collect pseudoscorpions in Panamá, the Smithsonian Tropical Research Institute for logistical support, Craig Osborne and Kris Kruse (Nevada Genomics Center) for performing the mitochondrial genome sequencing, Eleanor Su and Rachel Anderson for help with sperm data collection, and two anonymous referees for their constructive criticisms of an earlier draft of this manuscript.
Data accessibility
All data recorded were made available online on Dryad at the provisional DOI: doi:10.5061/dryad.66v6q.
Funding statement
This research was supported by a grant from the National Science Foundation to J.A.Z. and D.W.Z.
References
- 1.Frank SA, Hurst LD. 1996. Mitochondria and male disease. Nature 383, 224 ( 10.1038/383224a0) [DOI] [PubMed] [Google Scholar]
- 2.Zeh JA, Zeh DW. 2005. Maternal inheritance, sexual conflict and the maladapted male. Trends Genet. 21, 281–286. ( 10.1016/j.tig.2005.03.006) [DOI] [PubMed] [Google Scholar]
- 3.Hedrick PW. 2012. Reversing mother's curse revisited. Evolution 66, 612–616. ( 10.1111/j.1558-5646.2011.01465.x) [DOI] [PubMed] [Google Scholar]
- 4.Wade MJ, Brandvain Y. 2009. Reversing mother's curse: selection on male mitochondrial fitness effects. Evolution 63, 1084–1089. ( 10.1111/j.1558-5646.2009.00614.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Innocenti P, Morrow EH, Dowling DK. 2011. Experimental evidence supports a sex-specific selective sieve in mitochondrial genome evolution. Science 332, 845–848. ( 10.1126/science.1201157) [DOI] [PubMed] [Google Scholar]
- 6.Pizzari T, Birkhead TR. 2002. The sexually-selected sperm hypothesis: sex-biased inheritance and sexual antagonism. Biol. Rev. 77, 183–209. ( 10.1017/S1464793101005863) [DOI] [PubMed] [Google Scholar]
- 7.Hurst LD. 1993. The incidences, mechanisms and evolution of cytoplasmic sex ratio distorters in animals. Biol. Rev. 68, 121–193. ( 10.1111/j.1469-185X.1993.tb00733.x) [DOI] [Google Scholar]
- 8.Roubertoux PL, et al. 2003. Mitochondrial DNA modifies cognition in interaction with the nuclear genome and age in mice. Nat. Genet. 35, 65–69. ( 10.1038/ng1230) [DOI] [PubMed] [Google Scholar]
- 9.Wallace DC. 2005. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 39, 359–407. ( 10.1146/annurev.genet.39.110304.095751) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camus MF, Clancy DJ, Dowling DK. 2012. Mitochondria, maternal inheritance, and male aging. Curr. Biol. 22, 1717–1721. ( 10.1016/j.cub.2012.07.018) [DOI] [PubMed] [Google Scholar]
- 11.Maklakov AA, Lummaa V. 2013. Evolution of sex differences in lifespan and aging: causes and constraints. Bioessays 35, 717–724. ( 10.1002/bies.201300021) [DOI] [PubMed] [Google Scholar]
- 12.Wallace DC. 2013. Bioenergenetics in human evolution and disease: implications for the origin of biological complexity and the missing genetic variation of common diseases. Phil. Trans. R. Soc. B 368, 20120267 ( 10.1098/rstb.2012.0267) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolff JN, Gemmell NJ. 2013. Mitochondria, maternal inheritance, and asymmetric fitness: why males die younger. Bioessays 352, 93–99. ( 10.1002/bies.201200141) [DOI] [PubMed] [Google Scholar]
- 14.Ballard JWO, Pichaud N. 2014. Mitochondrial DNA: more than an evolutionary bystander. Funct. Ecol. 28, 218–231. ( 10.1111/1365-2435.12177) [DOI] [Google Scholar]
- 15.Dowling DK. 2014. Evolutionary perspectives on the links between mitochondrial genotype and disease phenotype. Biochim. Biophys. Acta 1840, 1393–1403. ( 10.1016/j.bbagen.2013.11.013) [DOI] [PubMed] [Google Scholar]
- 16.Ruiz-Pesini E, et al. 2000. Human mtDNA haplogroups associated with high or reduced spermatozoa motility. Am. J. Human Genet. 67, 682–696. ( 10.1086/303040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.May-Panloup P, Chrétien M-F, Savagner F, Vasseur C, Jean M, Malthièry Y, Reynier P. 2003. Increased sperm mitochondrial DNA content in male infertility. Hum. Reprod. 18, 550–556. ( 10.1093/humrep/deg096) [DOI] [PubMed] [Google Scholar]
- 18.Froman DP, Kirby JD. 2005. Sperm mobility: phenotype in roosters Gallus domesticus determined by mitochondrial function. Biol. Reprod. 72, 562–567. ( 10.1095/biolreprod.104.035113) [DOI] [PubMed] [Google Scholar]
- 19.Montiel-Sosa F, Ruiz-Pesini E, Enriquez JA, Marcuello A, Diez-Sánchez C, Montoya J, Wallace DC, López-Pérez MJ. 2006. Differences of sperm motility in mitochondrial DNA haplogroup U sublineages. Gene 368, 21–27. ( 10.1016/j.gene.2005.09.015) [DOI] [PubMed] [Google Scholar]
- 20.Smith S, Turbill C, Suchentrunk F. 2010. Introducing mother's curse: low male fertility associated with an imported mtDNA haplotype in a captive colony of brown hares. Mol. Ecol. 19, 36–43. ( 10.1111/j.1365-294X.2009.04444.x) [DOI] [PubMed] [Google Scholar]
- 21.Feng GF, Zhang J, Feng LM, Shen NX, Li LJ, Zhu YM. 2013. Mitochondrial DNA haplogroup associated with sperm motility in the Han population. Asian J. Androl. 15, 630–633. ( 10.1038/aja.2013.83) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mossman JA, Slate J, Birkhead TR, Moore HD, Pacey AA. 2010. Mitochondrial haplotype does not influence sperm motility in a UK population of men. Human Reprod. 27, 641–651. ( 10.1093/humrep/der438) [DOI] [PubMed] [Google Scholar]
- 23.Mossman JA, Slate J, Birkhead TR. 2012. Mitochondrial haplotype does not affect sperm velocity in the zebra finch Taeniopygia guttata. J. Evol. Biol. 23, 422–432. ( 10.1111/j.1420-9101.2009.01913.x) [DOI] [PubMed] [Google Scholar]
- 24.Dowling DK, Friberg U, Arnqvist G. 2007. A comparison of nuclear and cytoplasmic genetic effects on sperm competitiveness and female remating in a seed beetle . J. Evol. Biol. 20, 2113–2125. ( 10.1111/j.1420-9101.2007.01433.x) [DOI] [PubMed] [Google Scholar]
- 25.Friberg U, Dowling DK. 2008. No evidence of mitochondrial genetic variation for sperm competition within a population of Drosophila melanogaster. J. Evol. Biol. 21, 1798–1807. ( 10.1111/j.1420-9101.2008.01581.x) [DOI] [PubMed] [Google Scholar]
- 26.Yee WKW, Sutton KL, Dowling DK. 2013. In vivo male fertility is affected by naturally occurring mitochondrial haplotypes. Curr. Biol. 23, R55–R56. ( 10.1016/j.cub.2012.12.002) [DOI] [PubMed] [Google Scholar]
- 27.Zeh JA, Bonilla MM, Su EJ, Padua MV, Anderson RV, Kaur D, Yang D-S, Zeh DW. 2012. Degrees of disruption: projected temperature increase has catastrophic consequences for reproduction in a tropical ectotherm. Global Change Biol. 18, 1833–1842. ( 10.1111/j.1365-2486.2012.02640.x) [DOI] [Google Scholar]
- 28.Zeh JA, Newcomer SD, Zeh DW. 1998. Polyandrous females discriminate against previous mates. Proc. Natl Acad. Sci. USA 95, 13 732–13 736. ( 10.1073/pnas.95.23.13732) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonilla MM, Zeh DW, White AM, Zeh JA. 2011. Discriminating males and unpredictable females: males bias sperm allocation in favor of virgin females. Ethology 117, 740–748. ( 10.1111/j.1439-0310.2011.01928.x) [DOI] [Google Scholar]
- 30.Zeh DW, Zeh JA, Bermingham E. 1997. Polyandrous, sperm-storing females: carriers of male genotypes through episodes of adverse selection. Proc. R. Soc. Lond. B 264, 119–125. ( 10.1098/rspb.1997.0018) [DOI] [Google Scholar]
- 31.Newcomer SD, Zeh JA, Zeh DW. 1999. Genetic benefits enhance the reproductive success of polyandrous females. Proc. Natl Acad. Sci. USA 96, 10 236–10 241. ( 10.1073/pnas.96.18.10236) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeh JA, Zeh DW, Bonilla MM. 2003. Phylogeography of the harlequin beetle-riding pseudoscorpion and the rise of the Isthmus of Panamá. Mol. Ecol. 12, 2759–2769. ( 10.1046/j.1365-294X.2003.01914.x) [DOI] [PubMed] [Google Scholar]
- 33.Laslett D, Canbäck B. 2008. ARWEN, a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 24, 172–175. ( 10.1093/bioinformatics/btm573) [DOI] [PubMed] [Google Scholar]
- 34.Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33, W686–W689. ( 10.1093/nar/gki366) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 15, 3059–3066. ( 10.1093/nar/gkf436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. ( 10.1093/sysbio/sys029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. ( 10.1093/molbev/mst197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woolley S, Johnson J, Smith MJ, Crandall KA, McClellan DA. 2003. TreeSAAP: Selection on amino acid properties using phylogenetic trees. Bioinformatics 19, 671–672. ( 10.1093/bioinformatics/btg043) [DOI] [PubMed] [Google Scholar]
- 39.Zeh DW, Zeh JA, May CA. 1994. Charomid cloning vectors meet the pedipalpal chelae: single-locus minisatellite DNA probes for paternity assignment in the beetle-riding pseudoscorpion. Mol. Ecol. 3, 517–522. ( 10.1111/j.1365-294X.1994.tb00130.x) [DOI] [PubMed] [Google Scholar]
- 40.Zeh JA, Zeh DW. 2006. Outbred embryos rescue inbred half-siblings in mixed-paternity broods of live-bearing females. Nature 439, 201–203. ( 10.1038/nature04260) [DOI] [PubMed] [Google Scholar]
- 41.Koop JL, Zeh DW, Bonilla MM, Zeh JA. 2009. Reproductive compensation favours male-killing Wolbachia in a live-bearing host. Proc. R. Soc. B 276, 4021–4028. ( 10.1098/rspb.2009.1230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.SAS Institute Inc. 2011. SAS/STAT® 9.3 user‘s guide. Cary, NC: SAS Institute Inc. [Google Scholar]
- 43.Holm S. 1979. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 6, 65–70. [Google Scholar]
- 44.Clark AG, Begun DJ, Prout T. 1999. Female×male interactions in Drosophila sperm competition. Science 283, 217–220. ( 10.1126/science.283.5399.217) [DOI] [PubMed] [Google Scholar]
- 45.Lüpold S, Pitnick S, Berben KS, Blengini CS, Belote JM, Manier MK. 2013. Female mediation of competitive fertilization success in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 110, 10 693–10 698. ( 10.1073/pnas.1300954110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Su E. 2014. Simulated climate warming and mitochondrial haplogroup modulate small non-coding RNA expression in the neotropical pseudoscorpion, Cordylochernes scorpioides. MS thesis, University of Nevada, Reno. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kokko H, Maples J. 2005. Sexual selection when fertilization is not guaranteed. Evolution 59, 1876–1885. ( 10.1111/j.0014-3820.2005.tb01058.x) [DOI] [PubMed] [Google Scholar]
- 48.Chapman T, Bangham J, Vinti G, Seifried B, Lung O, Wolfner MF, Smith HK, Partridge L. 2003. The sex peptide of Drosophila melanogaster: female post-mating responses analyzed by using RNA interference. Proc. Natl Acad. Sci. USA 100, 9923–9928. ( 10.12073/pnas.1631635100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gabor CR, Halliday TR. 1997. Sequential mate choice by multiply mating smooth newts: females become more choosy. Behav. Ecol. 8, 162–166. ( 10.1093/beheco/8.2.162) [DOI] [Google Scholar]
- 50.Pitcher TE, Neff BD, Rodd FH, Rowe L. 2003. Multiple mating and sequential mate choice in guppies: females trade up. Proc. R. Soc. Lond. B 270, 1623–1629. ( 10.1098/rspb.2002.2280) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeh JA, Zeh DW. 2007. Mate choice by non-virgin females contributes to reproductive isolation between populations of the harlequin beetle-riding pseudoscorpion. Ethology 113, 1202–1211. ( 10.1111/j.1439-0310.2007.01432.x) [DOI] [Google Scholar]
- 52.Zeh JA, Zeh DW. 2006. Male-killing Wolbachia in a live-bearing arthropod: brood abortion as a constraint on the spread of a selfish microbe. J. Invert. Pathol. 92, 33–38. ( 10.1016/j.jip.2006.02.004) [DOI] [PubMed] [Google Scholar]
- 53.Keller L, Reeve HK. 1995. Why do females mate with more than one male? The sexually selected sperm hypothesis. Adv. Stud. Behav. 24, 291–315. ( 10.1016/s0065-3454(08)60397-6) [DOI] [Google Scholar]
- 54.Gemmell NJ, Metcalf VJ, Allendorf FW. 2004. Mother's curse: the effect of mtDNA on individual fitness and population viability. Trends Ecol. Evol. 19, 238–244. ( 10.1016/j.tree.2004.02.002) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data recorded were made available online on Dryad at the provisional DOI: doi:10.5061/dryad.66v6q.