Abstract
Individual differences in behaviour are often consistent across time and contexts, but it is not clear whether such consistency is reflected at the molecular level. We explored this issue by studying scouting in honeybees in two different behavioural and ecological contexts: finding new sources of floral food resources and finding a new nest site. Brain gene expression profiles in food-source and nest-site scouts showed a significant overlap, despite large expression differences associated with the two different contexts. Class prediction and ‘leave-one-out’ cross-validation analyses revealed that a bee's role as a scout in either context could be predicted with 92.5% success using 89 genes at minimum. We also found that genes related to four neurotransmitter systems were part of a shared brain molecular signature in both types of scouts, and the two types of scouts were more similar for genes related to glutamate and GABA than catecholamine or acetylcholine signalling. These results indicate that consistent behavioural tendencies across different ecological contexts involve a mixture of similarities and differences in brain gene expression.
Keywords: animal personality, gene expression, individual differences, novelty seeking, social insects
1. Introduction
Individual differences in behaviour are ubiquitous in nature. Many such differences are heritable, relatively stable over the lifetime of an animal, and consistent across distinct behavioural and ecological contexts, in both humans [1,2] and non-human animals [3–5]. This phenomenon exists in a wide range of species, from insects to primates [5–7], and also involves many different types of behaviour, including aggressiveness [8,9], fearfulness [10], risk-taking [11–13] and exploratory or novelty-seeking behaviour [14,15]. Relatively little is known about the molecular basis of consistent tendencies that are manifest across different behavioural and ecological contexts. Specifically, it is not known whether such consistent tendencies are associated with similar patterns of brain gene expression.
Scouting in colonies of the honeybee, Apis mellifera, provides an excellent system to study this question. Scout bees seek new resources for their colony, and they use a variety of similar and different behaviours to do so across two very different ecological contexts: foraging for floral resources and searching for a new nest site. Only a minority of a colony's adult worker bees act as food scouts and search for new food sources on their own; the majority of the foraging force wait to be ‘recruited’ and rely on information provided by scout bees to guide their foraging. The same is true for nest scouts; after colony reproduction (fission), a small group of bees search for new nest sites and then share information about these sites with the majority of the bees that have formed a temporary ‘swarm’, awaiting a move to a permanent location. Nest scouts recruit other individuals to investigate potential locations before the entire swarm moves [16]. The two forms of scouting behaviour also have very different search targets: they use different criteria to evaluate their targets and are influenced by different social environments [16]. The tendency to scout is influenced by both inherited and environmental factors [15,17–20]. Food scouting occurs daily and is strongly influenced by colony need and seasonal floral fluctuation, and food scouts search for new flower patches and evaluate how profitable they are as sources of sustenance. Nest scouting happens only when a colony reproduces and the swarm (reproductive propagule), seeking to establish a new colony, an event that happens once or twice a year [16]; nest scouts search for new locations, such as tree cavities, and evaluate how suitable they are as shelter [16].
Brain transcriptomic analysis is a powerful tool to quantify the dynamic nature of brain gene expression in relation to behavioural output, and has been used to identify neurogenomic profiles that are associated with distinct behavioural states [20]. Using this method, Liang et al. previously demonstrated [15] that food scouts show large differences in brain gene expression relative to non-scout foragers. Some of these differences suggested differences in the activity of certain neural signalling pathways in the scout brain that are known to be involved in vertebrate novelty seeking, and this information was used to demonstrate causal effects of these signalling pathways on food scouting probability [15]. However, Liang et al. [15] did not address the question of whether the two different types of scouts show similar or different patterns of brain gene expression.
We hypothesized that these two types of scouting behaviour share certain cross-contextual molecular signatures of novelty seeking, and tested our hypothesis by transcriptomic comparisons of the brains of food scouts and nest scouts relative to non-scout controls (recruit bees). There is a connection between two types of scouts; nest scouts are over three times more likely to act as food scouts than are other bees [15]. A link to novelty seeking has been shown for food scouts [15], but not yet for nest scouts.
Because of the big differences in ecological and behavioural contexts between the two types of scouts, we compared each type of scout to the relevant group of ‘recruits’, those bees who followed the information provided by the scouts in each context [21]. Recruits are similar to scouts in age, experience, search image and activity level during foraging and nest hunting, but differ significantly in their novelty-seeking tendency.
In this study, we predicted that food and nest scouts would show common patterns of brain gene expression to reflect common underlying novelty-seeking tendencies, despite the differences in behavioural and ecological contexts described above. We also predicted that food and nest scouts would show differences in brain gene expression to reflect the different behavioural and ecological contexts of the two types of scouting.
2. Material and methods
(a). Bees
Bees were collected from four colonies maintained at Liddell Laboratory, Cornell University, in Ithaca, NY. Each colony was headed by a queen that was inseminated by semen from a single drone (SDI colonies, drones were from unrelated colonies) to minimize the effects of genetic variation within each trial (queens were reared and instrumentally inseminated by Glenn Apiaries, Fallbrook, CA). Three colonies were used to collect food scouts and recruits, and two colonies were used to collect nest scouts and recruits (one colony was used in both food and nest scouting experiments). All bees were foraging age, and collected only in the morning to eliminate possible age and circadian effects on brain gene expression, respectively [22,23].
(b). Hive-moving assay to collect food scouts and recruits
Each colony that was used for identifying food scouts and recruits had its hive entrance closed and then was moved in the evening to a new location at least 4 km away; only bees showing scouting behaviour (independent search for floral resources) forage the next morning. We opened hive entrances the following morning at 08.00 h, and scouts were collected between 08.00 and 09.00 h. Scouts were identified as the first bees to leave the hive, collect food in the unfamiliar environment and return to their hive [18,18]. To prevent the scouts from activating recruits by performing waggle dances inside the hive, we installed a mesh-wired entrance tube at the hive entrance so foragers could leave but not re-enter [18]. We analysed each collected bee's foregut contents to verify her scout status [17,18]. Only bees that carried nectar (more than 1 µl) were identified as scouts (no pollen carriers were used). We collected recruits between 10.00 and 11.00 h the next morning, because by this time most of the foragers had been recruited to food sources by scouts [15]. We checked foregut contents to confirm that each bee was replete, which is typical for recruited foragers. In all experiments, we collected bees with soft forceps and immediately dropped them into liquid nitrogen to freeze brain gene expression at natural levels.
(c). Artificial swarm method to collect nest scouts and recruits
Artificial swarms were prepared according to standard procedures [24] during late May and early June. A different colony (each headed by an SDI queen) was used in each trial. We first located the colony's queen and put her in a small cage (3.2 × 10 × 1.6 cm). Using a large funnel, we then shook 1.5 kg (approx. 12 000) bees into a wooden ‘swarm box’ (15 × 25 × 35 cm) with screen wire sides and placed the queen cage inside it. To obtain both young bees and foragers, we shook bees off frames of comb located in both the upper and lower parts of the hive. The swarm box was then placed in a dark room and kept at room temperature for 3–5 days, during which time the screen sides were brushed with sucrose syrup (1 : 1 v/v) about five times a day, until hundreds of wax scales dropped off the bees, which occurs naturally in swarming bees. We took the artificial swarm outside in the morning at 08.00 h and installed it on a ‘swarm board’, a wooden board fixed vertically on a 1.7 m stake [25], as follows. First, we affixed the queen cage to the centre of the board, and placed two sugar water feeders on the swarm board so the bees would not need to forage. We then shook out the bees at the foot of the swarm board. Within about 1 h the bees had climbed onto the swarm board and were clustered around the queen, as in a natural swarm; nest-site scouting usually started 0.5–1 h after the cluster had formed. As soon as each nest scout identified herself by performing a waggle dance on the cluster, we collected her with a soft forceps before she could recruit bees to her site and dropped her into liquid nitrogen. Within 1 h, we collected 21–25 scouts; all were collected during the initial searching phase when each scout's waggle dance indicated a different location. The next morning, we identified a similar number of recruits as bees performing waggle dances during the ‘consensus’ phase [26], the period just prior to the swarm taking off, by which time all the dances were pointed in the same direction and most of the dancing bees were ones that had been recruited to the chosen nest site [26]. These recruits were also collected in liquid nitrogen. We collected both nest scouts and recruits in the morning between 9.00 and 12.00 to eliminate possible circadian effects on brain gene expression. (See the electronic supplementary material for description of standard brain dissection, RNA extraction and microarray analysis methods).
(d). Statistical analyses of gene expression
We analysed the microarray data with a linear mixed effects model implemented using restricted maximum likelihood (REML) to describe the normalized log2-transformed gene intensity values, including the effects of dye, behavioural group, bee and microarray (for loop design see electronic supplementary material, figure S2). We evaluated differences in mRNA abundance with an F-test statistic; F1-type false discovery rate p-values including multiple-test adjustment were used to generate lists of differentially expressed genes (DEG). A total of 10 001 of 11 886 were expressed on at least 75% of all the microarrays (104 of 138) and were used for further analysis. We excluded genes that were highly expressed in the hypopharyngeal glands owing to the risk of contamination during brain dissection [27]. Gene annotation was based on honeybee genome OGS 3.2 (updated July 2012). Six pairwise contrasts were tested with ANOVA across four groups, and four pairs were analysed further: scouts versus recruits within each context (FS versus FR and NS versus NR) and foragers versus nest seekers within each role (FS versus NS and FR versus NR). ‘Context’ refers to an involvement with either food sources or nest sites (both scouts and recruits), and ‘role’ refers to acting as either a scout or a recruit (for both food sources and nest sites). Two main factors, context and role, and a role × context interaction were tested separately in a mixed-model ANOVA on the same data. These relationships are illustrated in the electronic supplementary material, figure S1.
(e). Gene expression pattern analyses
We performed linear discriminant analyses with the ‘lda’ function in the ‘MASS’ package of R (v. 2.15.1). Hierarchal clustering and heatmaps were generated by using the ‘pheatmap’ package in R (v. 2.15.1). We performed class prediction analysis on 557 DEGs between scouts and recruits (regardless of context), using the uncorrelated centroid shrunken (USC) method [28] and identified a series of predictor gene sets. Using ‘leave-one-out’ cross-validation analysis with the supporting vector machine (SVM) method [29,30], we computed prediction results for each set and reported the one that had the strongest predictive value with the smallest number of predictor genes (for all tests and results see electronic supplementary material, table S3). We performed partially automated versions of these analyses with MultiExperiment Viewer (MeV, v. 4.8, developed and maintained by TM4.org: http://www.tm4.org/mev.html). We calculated a representation factor (RF) to determine whether the number of genes that overlapped on two gene lists was statistically significant. This RF factor is the number of observed overlapping genes divided by the expected number of overlapping genes. The denominator is calculated as the product of the number of oligos differentially expressed in each experiment divided by the total number of oligos analysed [27]. We also used RF analysis to determine whether the number of enriched gene ontology (GO) terms that overlapped on two gene lists was statistically significant. In general, RF = n(observed)/n(expected), where n(observed) is the number of the overlapped and n(expected) = n(list A) × n(list B)/n(background).
(f). Functional analysis of brain gene expression
We performed gene functional enrichment analysis with DAVID bioinformatics resources 6.7 (http://david.abcc.ncifcrf.gov) in order to identify the predominant molecular functions, biological processes and cellular components represented by a list of DEGs. These analyses used GO [31]. Differentially expressed honeybee orthologues of Drosophila melanogaster genes were analysed against a background set of genes, which are all the Drosophila orthologues (Drosophila genome v. Dmel r5.42) in the honeybee genome (Amel 4.5). Enrichment was determined by comparing overlapped GO terms between gene lists with an EASE score [31], which is a modified Fisher's Exact test primarily used by DAVID to determine significance levels in the gene enrichment analysis (http://david.abcc.ncifcrf.gov/helps/functional_annotation.html). EASE scores with a p-value cut-off (95% confidence) are generally more relaxed than Benjamini–Hochberg FDR with the same confidence. The results using both tests are summarized in the electronic supplementary material, table S2.
3. Results
(a). Similarities and differences in the brain gene expression profiles of food and nest scouts
There were extensive differences in brain gene expression between scouts and recruits, for both food and nest scouting (table 1, FDR < 0.05 with contrast p < 0.005). Over 1000 genes were differentially expressed between food scouts and food recruits (1003 for FS versus FR), and a similar amount between nest scouts and nest recruits (1032 for NS versus NR). This represents approximately 10% of all the transcripts analysed on the microarray (1032 of 9877 for food scouting and 1003 of 9889 for nest scouting) and is consistent with a previous brain transcriptomic study on food scout bees [15]. The brain gene expression profiles of food scouts in this study showed significant similarity with the profiles of food scouts in a previous study [15] (table 2, food 1 × food 2), despite the use of different behavioural assays and experimental settings to identify the scouts. The lists of DEGs that overlapped were significantly enriched (EASE score, p < 0.05) for genes with the GO terms ‘lipid particle’ (GO:0005811), ‘gland morphogenesis’ (GO:0022612) and ‘tissue death’ (GO:0016271).
Table 1.
Pairwise analyses of scouting behaviour across two ecological contexts: numbers of significantly DEGs are shown for each analysis (FDR < 0.05, contrast p < 0.001, except for interaction, which is FDR < 0.05, no contrast available). FS, food scouts; FR, food recruits; NS, nest scouts; NR, nest recruits.
| DEGs | FS versus FR | NS versus NR | FS versus NS | FR versus NR | FS versus NR | NS versus FR | context | role | interaction |
|---|---|---|---|---|---|---|---|---|---|
| total | 1003 | 1032 | 1682 | 1416 | 2368 | 1028 | 2251 | 1246 | 864 |
| up | 462↑ | 522↑ | 895↑ | 778↑ | 1206↑ | 441↑ | 1215↑ | 585↑ | |
| down | 541↓ | 510↓ | 787↓ | 638↓ | 1162↓ | 587↓ | 1036↓ | 661↓ |
Table 2.
Common patterns of DEGs and enriched GO terms across three scouting-related datasets. Representation factor (RF) analyses of food scouting, nest scouting (this study) and a previous study of food scouting [15]. RF = n(observed)/n(expected), where n(expected) = n(list A) × n(list B)/n(background). For DEGs, n = 9877 (background as total genes with Drosophlia homologues in this analysis); for enriched GO, n = 5195 (background as total GO terms available). Food 1: food scout versus recruits collected in the field experiments (this study); food 2: food scouts versus recruits collected in semi-natural enclosure [15]; nest: nest scouts versus nest recruits collected in the field experiments (this study).
| Experiments | differentially expressed genes |
GO terms |
||||||
|---|---|---|---|---|---|---|---|---|
| expected | observed | RF | p-value | expected | observed | RF | p-value | |
| food 1 × nest | 104.8 | 344 | 3.3 | <0.0001 | 0.54 | 10 | 18.6 | <0.0001 |
| food 1 × food 2 | 123.8 | 201 | 1.6 | <0.0001 | 0.76 | 8 | 10.5 | <0.0001 |
| nest × food 2 | 127.4 | 230 | 1.8 | <0.0001 | 1.05 | 26 | 24.8 | <0.0001 |
| food 1 × nest × food 2 | — | 84 | — | — | — | 8 | — | — |
Nest and food scouts showed strong similarities in brain gene expression profiles. The DEG list for NS versus NR overlapped significantly with the DEG list for FS versus FR, with 344 genes overlapping (table 2). Similar results were obtained when comparing nest scouts from this study and food scouts from a previous study [15] (table 2, nest × food 2). Moreover, the patterns of expression for these 344 overlapping genes showed significant concordance across scouting contexts (Pearson's correlation, R2 = 0.31, p < 0.00001), with 81% of them showing changes in the same direction. The set of 344 overlapping genes was significantly enriched for several GO terms (table 2, food 1 × nest, RF = 18.6, p < 0.0001), including genes related to the GO term ‘lipid particle’ (GO:0005811) and the GO term ‘protein folding’ (GO:0006457).
There also were strong brain gene expression differences between food scouts and nest scouts, and between food recruits and nest recruits, 1682 and 1416 genes, respectively (table 1). Among the differences, GO analysis revealed that genes related to glucose metabolism (GO:0006006) were enriched among the set of genes upregulated in the brains of food scouts but not nest scouts, perhaps related to differences in exposure to food for the two types of scouts.
Similar results were obtained with an independent factorial analysis that focused on brain gene expression patterns associated with ecological context and behavioural role. This is to complement the pairwise comparisons described above. ‘Context’ refers to an involvement with either food sources or nest sites (both scouts and recruits), and ‘role’ refers to acting as either a scout or a recruit (for both food sources and nest sites). There were over 2000 genes associated with differences in context, over 1000 associated with differences in role and almost 1000 associated with a role × context interaction (table 1 and figure 1a). Surprisingly, despite the large number of DEGs on the context DEG list, it was associated with relatively few significantly enriched GO terms. GO terms significantly enriched on the role DEG list and the role × context interaction list are indicated in figure 1b. Scouts showed more genes differentially expressed in the GO category of ‘lipid particle’ than did recruits. The role × context interaction DEG list contained several genes of particular interest. This includes several neurotransmitter receptor (figure 2a) and hormonal signalling (electronic supplementary material, table S3c) genes and the gene encoding the odourant binding protein obp4 (or asp4, GB53372), apparently unique to honeybees [32]. Obp4 was the most upregulated gene in the brains of nest scouts compared to nest recruits (FDR < 0.05, contrast p < 10−20), but was (modestly) downregulated in food scouts (FDR < 0.05, contrast p = 0.012; electronic supplementary material, figure S4a). Obp4 gene has been associated with a variety of foraging activities in previous microarray studies (electronic supplementary material, figure S4b).
Figure 1.
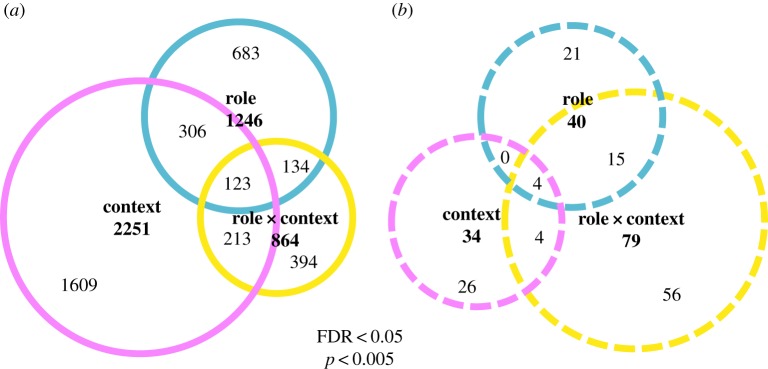
Factorial analyses of scouting across different behavioural and ecological contexts. (a) Numbers of DEG and their overlap as a function of role, context and their interaction, are shown in an area-proportional Venn diagram. ‘Context’ refers to an involvement with either food sources or nest sites (both scouts and recruits), and ‘role’ refers to acting as either a scout or a recruit (for food sources or nest sites). (b) Numbers of functional enrichments (based on GO terms and KEGG pathways) and their overlap as a function of role, context and their interaction (EASE score, p < 0.05) are shown in a similar diagram. Context was associated with the largest number of DEGs but smallest number of GO enrichment terms, with the opposite pattern for the role × context interaction.
Figure 2.
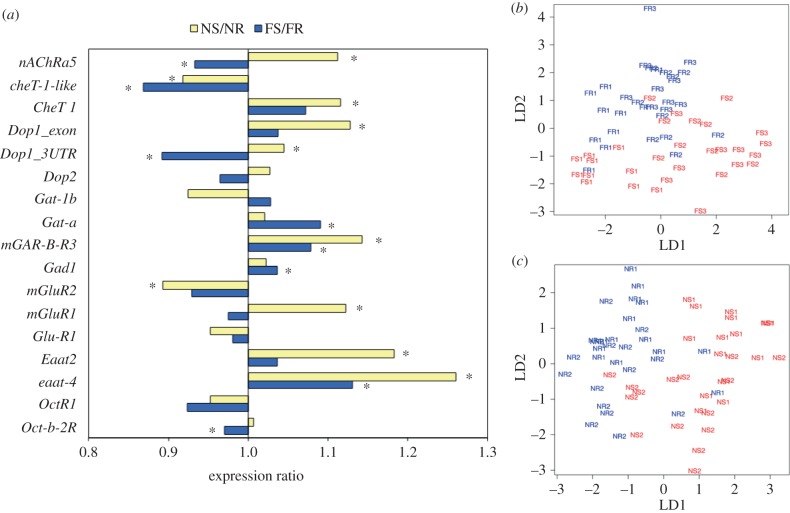
Brain transcriptomic analyses of 16 neurotransmitter-related genes in food scouting and nest scouting. (a) Expression ratio shows genes in five neurotransmitter systems are differentially expressed in both scouting behaviours (dopamine, glutamate, GABA, acetylcholine and octopamine systems). Yellow bars, expression ratios of nest scouts divided by nest recruits (NS/NR); blue bars, expression ratios of food scouts divided by food recruits (FS/FR). Asterisk (*) denotes FDR < 0.05 and contrast p < 0.005. Gene information: see electronic supplementary material, table S1. (b,c) Linear discriminant analysis using the expression profiles showed in (a) display a clear separation of scouts and recruits in 60 individual bees, plotted by food scouting (LD1 and LD2 accounted for 75.2% of the variation) and nest scouting (LD1 and LD2 accounted for 90.5% of the variation), respectively. FS, food scouts (red); FR, food recruits (blue); NS, nest scouts (red); NR, nest recruits (blue), with different colonies denoted as subscript 1, 2 and 3.
(b). Similarities and differences in the expression of hormone and neurotransmitter signalling genes in food and nest scouts
Nest scouts showed a stronger pattern of upregulation of genes related to hormone signalling than did food scouts (electronic supplementary material, table S3a–c). The DEG list for nest scouts versus recruits was enriched for genes associated with ‘response to ecdysone’ (GO:0035075) and ‘response to steroid hormone stimulus’ (GO:0048545). For example, two transcription factor genes, forkhead box P (foxP or fd85E) and ftz-transcription factor 1 (ftz-f1) showed consistent upregulation in the brains of both food scouts and nest scouts, but another closely related gene transcription factor gene, broad-complex (br), was upregulated only in the brains of nest scouts. Prior systems biology analysis predicted that these genes orchestrate important networks of behaviourally related gene expression in the honeybee brain [33].
Both nest and food scouts showed differential brain expression of several genes related to catecholamine, glutamate and GABA-related neural signalling relative to recruits. In addition to these three signalling systems, which also were found to be associated with scouting in a previous study [15], both nest and food scouts also showed differential expression of acetylcholine-related genes. Of a total of 16 genes related to dopamine, octopamine, glutamate, GABA-related and acetylcholine signalling (electronic supplementary material, table S1), food scouts showed significant expression differences in eight genes (relative to food recruits) and nest scouts showed significant expression differences in nine genes (relative to nest-site recruits; figure 1a).
There were also notable differences between nest and food scouts in the expression of neurotransmitter signalling genes. The two types of scouts were more similar for glutamate- and GABA-related genes, but more divergent for catecholamine- and acetylcholine-related genes. For example, while the upregulation of the genes encoding the excitatory amino acid transporter 4 (eaat-4) and the metabotropic GABA-B receptor subtype 3 (mgarb3) occurred in both types of scouts, dopamine receptor dopr1 and all three acetylcholine genes (nachra5, chet-1 and chet-1-like) were only upregulated in nest scouts, and downregulated or not significantly different in food scouts (figure 2a).
(c). Brain molecular signatures of scouting behaviour
Results from the context- and role-based factorial analysis presented above also were used to address the following question: are the individual differences consistent enough to allow for the classification of scouts and recruits solely on the basis of brain gene expression, regardless of ecological context? Based on the results of the factorial analysis we selected a set of 557 genes that varied only with role, not context or role × context. We then used class prediction to identify a series of ‘predictor gene sets’ out of these 557 genes, and used ‘leave-one-out’ cross-validation [29,30] to find the smallest predictor gene set with the best prediction results. A minimum set of 89 genes was able to predict whether a bee was a scout or a recruit 92.5% of the time (clustering and heatmap: electronic supplementary material, figure S5; selection: electronic supplementary material, table S4a, gene list: electronic supplementary material, table S5). A total of 111 of 120 bees were identified correctly as a scout or recruit (54 of 60 bees correct for scouts, 57 of 60 bees correct for recruits) using selected 89-gene expression profiles, compared with 78.3% success if all 557 genes were used without such selection (electronic supplementary material, table S3a). Among these 89 best predictor genes, the gene encoding the transcription factor Forkhead box P (foxP, GB40150) and tubulin β-1 (GB44133) were both consistently upregulated in nest scouts and in food scouts (consistent with the results in [15]). Similarly, the heat-shock protein gene lethal essential for life (l(2)efl-like, GB45906) showed consistent downregulation in both types of scouts. Metabotropic GABA receptor type B 3 (mgarb3) gene was upregulated in both food and nest scouts, which also is consistent with the results in [15].
We also used the same methods to ask whether predictor gene sets for nest scouts can correctly predict food scouts and vice versa. The best predictor gene set for nest scouts correctly classified food scouts and recruits significantly better than random (39 of 60, 65% success, binomial test, p < 0.05), but not vice versa (31 of 60, 58% success, p > 0.05; electronic supplementary material, table S4b). This result suggests that the neurogenomic signature of nest scouts may serve as a more stringent template for scouting behaviour than that of food scouts.
The 16 genes related to catecholamine, glutamate, GABA-related and acetylcholine signalling described above also provided a strong signature of scouting behaviour. Linear discriminant analysis (LDA) clearly separated scouts and recruits on the basis of this set of 16 genes (n = 30). For food scouting, LD1 and LD2 accounted for 75.2% of the variation between food scouts and recruits (figure 2b), and for nest scouting, LD1 and LD2 accounted for 90.5% of the variation between the nest scouts and recruits (figure 2c). By contrast, scouts did not show differences in expression for genes related to serotonin or tyramine signalling, and LDA using five genes involved in serotonin and tyramine signalling did not differentiate scouts from recruits (electronic supplementary material, figure S3).
4. Discussion
We predicted that consistent tendencies across different behavioural and ecological contexts involve common patterns of brain gene expression. In support of this hypothesis, we report that scouting behaviours, performed differently and expressed in two different ecological contexts, share a common core of genes. We suggest that this common core reflects a common underlying novelty-seeking tendency for the two types of scouting because some of the same neural signalling pathways that causally influence novelty seeking in food scouts [15] were also part of the brain molecular signature for nest scouts.
It is intriguing to find the same neurotransmitter systems involved in both types of scouting, regardless of the distinct social and natural environment in which each behaviour occurs. Dopamine and glutamate are known to be involved in novelty-seeking behaviour in vertebrates, including humans [34], and a causal relationship was found between increased glutamate signalling and increased scouting probabilities [15]. A weaker but still causal link has also been shown for dopamine and octopamine [15]. In this study, we found that nest scouts shared similar expression patterns with food scouts for glutamate- and GABA-related genes, but had different patterns for dopamine-, octopamine- and acetylcholine-related genes. Based on these findings, we speculate that the glutamate and GABA systems are part of a core mechanism that promotes novelty-seeking behaviour in bees across multiple contexts, while catecholamines influence scouting behaviour differently in different contexts.
Our results also suggest that in addition to neural signalling molecules, there is an important relationship between endocrine processes and scouting, because hormone-related regulator genes such as ftz-f1, br, usp and other ecdysone-related genes were consistently associated with scouting in both contexts. Suggestive links between steroid hormone signalling and cocaine-related novelty seeking have been reported recently in both mice and fruitflies [35,36]. We also found a robust link between genes related to the GO category ‘lipid particle’ and scouting behaviour—this might reflect unique aspects of lipid metabolism in the brains of scout bees, speculation that awaits functional analysis.
We also predicted that food and nest scouts would show differences in brain gene expression to reflect the different behavioural and ecological contexts of the two types of scouting. This prediction also was upheld. Food scouts search daily for brightly coloured flowers, whereas nest scouts search for dark tree holes on the infrequent occasions that colonies are homeless. Perhaps differences in the expression of genes such as odour-binding protein 4 (obp4) reflect some of the differences in the sensory aspects of these two types of scouting. The gene obp4 showed increased brain expression in nest scouts, but was downregulated in food scouts; it also was downregulated in a previous study of scouts [15] and downregulated in active versus inactive foragers [22]. These results suggest differences in responsiveness to olfactory stimuli in the foraging and swarming contexts.
It is possible that differences between scouts and recruits in brain gene expression are due, at least in part, to genetic differences. Previous research showed that both nest and food scouting tendencies have a heritable component [17–20]; certain genotypes are overrepresented among scouts, and a colony's scouting rate is affected by patriline diversity. It is also well known that scouting probabilities are regulated by the fluctuation of floral resource availability and colony needs for food and shelter over different seasons [23]. However, it is unclear what genetic elements contribute to the probability of becoming a scout or a recruit, and how such propensities interact with the environment inside and outside the hive to shape scouting probabilities. Moreover, previous research has demonstrated that it is possible to induce food scouting by manipulating some of the neurotransmitter systems implicated by the present and previous [15] transcriptomic analyses. These results make it likely that the overall patterns reported here reflect real similarities and differences in scouting behaviour across the two behavioural and ecological contexts.
Understanding both the mechanistic and evolutionary bases of consistent individual differences in behaviour are important challenges in the study of animal behaviour and animal personality. Progress on both challenges will require model systems similar to scouting in honeybees, in which similar tendencies across different behavioural and ecological contexts occur naturally and are amenable to mechanistic analysis.
Supplementary Material
Supplementary Material
Acknowledgements
We thank M. K. Wray, P. Date, M. Girard (Cornell University), M. K. Carr-Markell and J. Recchia-Rife (University of Illinois) for field assistance; M. R. Band, J. Drnevich, M. F. Majewski and K. K. Homann (Keck Center for Comparative and Functional Genomics, University of Illinois) for assistance with microarray experiments; and R. C. Fuller, J. S. Rhodes, C. W. Whitfield and members of the Robinson laboratory for reviewing the manuscript and providing valuable comments.
Data accessibility
Differential expressed genes (DEGs) and enriched GO categories were submitted as a part of the electronic supplementary material. Microarray data meet Minimum Information About Microarray Experiment (MIAME) standards and are available at ArrayExpress database (www.ebi.ac.uk/arrayexpress, accession number: E-MTAB-2918).
Funding statement
Supported by NSF Frontiers in Biological Research grant EF 0425852 (B. L. Schatz, PI, BeeSpace Project), NIH Director's Pioneer Award 1DP1OD006416 (G.E.R) and Cornell Agriculture Experiment Station grant NYC-191522 (T.D.S).
References
- 1.Bouchard TJ. 1994. Genes, environment, and personality. Science 264, 1700–1701. ( 10.1126/science.8209250) [DOI] [PubMed] [Google Scholar]
- 2.Plomin R, Owen MJ, McGuffin P. 1994. The genetic basis of complex human behaviors. Science 264, 1733–1739. ( 10.1126/science.8209254) [DOI] [PubMed] [Google Scholar]
- 3.Sih A, Bell A, Johnson JC. 2004. Behavioral syndromes: an ecological and evolutionary overview. Trends Ecol. Evol. 19, 372–378. ( 10.1016/j.tree.2004.04.009) [DOI] [PubMed] [Google Scholar]
- 4.Bell AM. 2007. Future directions in behavioural syndromes research. Proc. R. Soc. B 274, 755–761. ( 10.1098/rspb.2006.0199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sih A, Bell AM. 2008. Insights for behavioral ecology from behavioral syndromes. Adv. Study Behav. 38, 227–281. ( 10.1016/s0065-3454(08)00005-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gosling SD. 2001. From mice to men: what can we learn about personality from animal research? Psychol. Bull. 127, 45–86. ( 10.1037/0033-2909.127.1.45) [DOI] [PubMed] [Google Scholar]
- 7.Gosling SD. 2008. Personality in non-human animals. Soc. Pers. Psych. Compass 2, 985–1001. ( 10.1111/j.1751-9004.2008.00087.x) [DOI] [Google Scholar]
- 8.Huntingford FA. 1982. Do Inter- and intra-specifc aggression vary in relationship related to predation pressure in stickleback? Anim. Behav. 30, 909–916. ( 10.1016/S0003-3472(82)80165-6) [DOI] [Google Scholar]
- 9.Riechert SE, Hedrick AV. 1993. A test for correlations among fitness-linked behavioural traits in the spider Agelenopsis aperta (Araneae, Agelenidae). Anim. Behav. 46, 669–675. ( 10.1006/anbe.1993.1243) [DOI] [Google Scholar]
- 10.Boissy A. 1995. Fear and fearfulness in animals. Q. Rev. Biol. 70, 165–191. ( 10.1086/418981) [DOI] [PubMed] [Google Scholar]
- 11.Sloan Wilson D, Clark AB, Coleman K, Dearstyne T. 1994. Shyness and boldness in humans and other animals. Trends Ecol. Evol. 9, 442–446. ( 10.1016/0169-5347(94)90134-1) [DOI] [PubMed] [Google Scholar]
- 12.Fraser DF, Gilliam JF, Daley MJ, Le AN, Skalski GT. 2001. Explaining leptokurtic movement distributions: intrapopulation variation in boldness and exploration. Am. Nat. 158, 124–135. ( 10.1086/321307) [DOI] [PubMed] [Google Scholar]
- 13.van OERS K, DRENT PJ, de Goede P, van Noordwijk AJ. 2004. Realized heritability and repeatability of risk-taking behaviour in relation to avian personalities. Proc. R. Soc. Lond. B 271, 65–73. ( 10.1098/rspb.2003.2518) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dingemanse N. 2002. Repeatability and heritability of exploratory behaviour in great tits from the wild. Anim. Behav. 64, 929–938. ( 10.1006/anbe.2002.2006) [DOI] [Google Scholar]
- 15.Liang ZS, Nguyen T, Mattila HR, Rodriguez-Zas SL, Seeley TD, Robinson GE. 2012. Molecular determinants of scouting behavior in honey bees. Science 335, 1225–1228. ( 10.1126/science.1213962) [DOI] [PubMed] [Google Scholar]
- 16.Seeley TD. 2010. Honeybee democracy. Princeton, NJ: Princeton University Press. [Google Scholar]
- 17.Dreller C. 1998. Division of labor between scouts and recruits: genetic influence and mechanisms. Behav. Ecol. Sociobiol. 43, 191–196. ( 10.1007/s002650050480) [DOI] [Google Scholar]
- 18.Mattila HR, Seeley TD. 2010. Does a polyandrous honey bee queen improve through patriline diversity the activity of her colony's scouting foragers? Behav. Ecol. Sociobiol. 65, 799–811. ( 10.1007/s00265-010-1083-0) [DOI] [Google Scholar]
- 19.Robinson GE, Page RE., Jr 1989. Genetic determination of nectar foraging, pollen foraging, and nest-site scouting in honey bee colonies. Behav. Ecol. Sociobiol. 24, 317–323. ( 10.1007/BF00290908) [DOI] [Google Scholar]
- 20.Zayed A, Robinson GE. 2012. Understanding the relationship between brain gene expression and social behavior: lessons from the honey bee. Annu. Rev. Genet. 46, 591–615. ( 10.1146/annurev-genet-110711-155517) [DOI] [PubMed] [Google Scholar]
- 21.Biesmeijer JC, de Vries H. 2001. Exploration and exploitation of food sources by social insect colonies: a revision of the scout-recruit. Behav. Ecol. Sociobiol. 49, 89–99. ( 10.1007/s002650000289) [DOI] [Google Scholar]
- 22.Naeger NL, Van Nest BN, Johnson JN, Boyd SD, Southey BR, Rodriguez-Zas SL, Moore D, Robinson GE. 2011. Neurogenomic signatures of spatiotemporal memories in time-trained forager honey bees. J. Exp. Biol. 214, 979–987. ( 10.1242/jeb.053421) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez-Zas SL, Southey BR, Shemesh Y, Rubin EB, Cohen M, Robinson GE, Bloch G. 2012. Microarray analysis of natural socially regulated plasticity in circadian rhythms of honey bees. J. Biol. Rhythms 27, 12–24. ( 10.1177/0748730411431404) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seeley TD, Visscher PK. 2003. Choosing a home: how the scouts in a honey bee swarm perceive the completion of their group decision making. Behav. Ecol. Sociobiol. 54, 511–520. ( 10.1007/s00265-003-0664-6) [DOI] [Google Scholar]
- 25.Seeley TD, Buhrman SC. 1999. Group decision making in swarms of honey bees. Behav. Ecol. Sociobiol. 45, 19–31. ( 10.1007/s002650050536) [DOI] [Google Scholar]
- 26.Seeley TD, Visscher PK, Passino KM. 2006. Group decision making in honey bee swarms. Am. Sci. 94, 220–229. ( 10.1511/2006.59.993) [DOI] [Google Scholar]
- 27.Alaux C, et al. 2009. Honey bee aggression supports a link between gene regulation and behavioral evolution. Proc. Natl Acad. Sci. USA 106, 15 400–15 405. ( 10.1073/pnas.0907043106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown MP, Grundy WN, Lin D, Cristianini N, Sugnet CW, Furey TS, Ares M, Haussler D. 2000. Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc. Natl Acad. Sci. USA 97, 262–267. ( 10.1073/pnas.97.1.262) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitfield CW, Cziko AM, Robinson GE. 2003. Gene expression profiles in the brain predict behavior in individual honey bees. Science 302, 296–299. ( 10.1126/science.1086807) [DOI] [PubMed] [Google Scholar]
- 30.Yeung KY, Bumgarner RE. 2003. Multiclass classification of microarray data with repeated measurements: application to cancer. Genome Biol. 4, R83 ( 10.1186/gb-2003-4-12-r83) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. ( 10.1038/nprot.2008.211) [DOI] [PubMed] [Google Scholar]
- 32.Chandrasekaran S, Ament SA, Eddy JA, Rodriguez-Zas SL, Schatz BR, Price ND, Robinson GE. 2011. Behavior-specific changes in transcriptional modules lead to distinct and predictable neurogenomic states. Proc. Natl Acad. Sci.USA 108, 18 020–18 025. ( 10.1073/pnas.1114093108). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bardo MT, Donohew RL, Harrington NG. 1996. Psychobiology of novelty seeking and drug seeking behaviour. Behav. Brain Res. 77, 23–43. ( 10.1016/0166-4328(95)00203-0) [DOI] [PubMed] [Google Scholar]
- 34.Forêt S, Maleszka R. 2006. Function and evolution of a gene family encoding odorant binding-like proteins in a social insect, the honey bee (Apis mellifera). Genome Res. 16, 1404–1413. ( 10.1101/gr.5075706) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lasek AW, Gesch J, Giorgetti F, Kharazia V, Heberlein U. 2011. Alk is a transcriptional target of LMO4 and ERα that promotes cocaine sensitization and reward . J. Neurosci. 31, 14 134–14 141. ( 10.1523/JNEUROSCI.3415-11.2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai LT-Y, Bainton RJ, Blau J, Heberlein U. 2004. Lmo mutants reveal a novel role for circadian pacemaker neurons in cocaine-induced behaviors. PLoS Biol. 2, e408 ( 10.1371/journal.pbio.0020408) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Differential expressed genes (DEGs) and enriched GO categories were submitted as a part of the electronic supplementary material. Microarray data meet Minimum Information About Microarray Experiment (MIAME) standards and are available at ArrayExpress database (www.ebi.ac.uk/arrayexpress, accession number: E-MTAB-2918).