Abstract
Many models of mutualisms show that mutualisms are unstable if hosts lack mechanisms enabling preferential associations with mutualistic symbiotic partners over exploitative partners. Despite the theoretical importance of mutualism-stabilizing mechanisms, we have little empirical evidence to infer their evolutionary dynamics in response to exploitation by non-beneficial partners. Using a model mutualism—the interaction between legumes and nitrogen-fixing soil symbionts—we tested for quantitative genetic variation in plant responses to mutualistic and exploitative symbiotic rhizobia in controlled greenhouse conditions. We found significant broad-sense heritability in a legume host's preferential association with mutualistic over exploitative symbionts and selection to reduce frequency of associations with exploitative partners. We failed to detect evidence that selection will favour the loss of mutualism-stabilizing mechanisms in the absence of exploitation, as we found no evidence for a fitness cost to the host trait or indirect selection on genetically correlated traits. Our results show that genetic variation in the ability to preferentially reduce associations with an exploitative partner exists within mutualisms and is under selection, indicating that micro-evolutionary responses in mutualism-stabilizing traits in the face of rapidly evolving mutualistic and exploitative symbiotic bacteria can occur in natural host populations.
Keywords: mutualism, genetic variation, partner choice, host sanctions, selection, Medicago lupulina
1. Introduction
Mutualistic interactions between two species occur when both partners exchange reciprocal fitness benefits [1,2]. Despite the ubiquity of mutualisms in nature, partners are known to vary in the fitness rewards they provide [1,3], and phenotypes or species that exploit the mutualism are common (reviewed in [3]). Exploiters, by definition, are thought to have a fitness advantage over their mutualistic competitors because they receive benefits while providing fewer or no rewards to their interacting partners [3,4]. Because of the apparent fitness benefits of exploitation, mutualisms are predicted to be susceptible to invasion and spread of species or phenotypes that exploit the interaction [5–10], which could lead to a breakdown in the mutualism [11]. Yet mutualisms persist through time, and very little breakdown or shift is observed in nature, suggesting that mechanisms exist that maintain their stability [11–18]. Despite the theoretical and empirical importance of mutualism-stabilizing mechanisms, there is little empirical evidence on whether genetic variation exists for these underlying mechanisms, a fundamental prerequisite for any evolutionary response that would either stabilize or destabilize mutualisms [19]. Here, we test for standing genetic variation in mutualism-stabilizing traits in populations of host plants that form mutualistic associations with symbiotic soil microbes, and whether these traits are subject to natural selection.
For hosts interacting with symbiotic partners, theory has demonstrated that mutualisms can remain stable through several mechanisms: partner fidelity feedback [5,15], partner choice [15] and host sanctions [16]. While there is debate on the origin and the maintenance of these mechanisms [20–23], their viability as an evolutionary stable state [15–17,24], and how we can distinguish between them in natural systems [20,22,25], all three mechanisms share a common outcome: the host increases the relative fitness of more beneficial partners and/or decreases the relative fitness of less beneficial or exploitative partners. Given the recurrent observation of exploitative partners [4,8,26], selection on stabilizing mechanisms is predicted to be strong because exploitative symbionts are assumed to have negative effects on host fitness [13,16,27–30] (but see [31]). If there is a cost to stabilizing mechanisms, in the absence of exploiters, selection will favour the loss of host stabilizing traits [17].
In the mutualism between legumes and rhizobia, both partners gain substantial fitness benefits from the interaction [32]. Rhizobia form infections on legume roots and fix atmospheric nitrogen into a plant available form in return for protection (i.e. residence inside a root nodule) and carbon metabolites from the host [32]. However, nodule production is energetically expensive [33,34], so a conflict of interest between the host and bacteria (from the host's perspective) limits the number of rhizobial infections a plant forms [8]. An individual legume host typically associates with multiple rhizobia strain genotypes and is also known to form biased infections with some rhizobia genotypes over others, leading to higher fitness rewards for rhizobial genotypes with higher nodule occupancy [35]. Non-beneficial symbionts co-occur with beneficial partners in both natural and agricultural systems [36–41], and selection favours less beneficial rhizobia in the absence of mutualism-stabilizing mechanisms [42]. Empirical evidence also suggests that legumes possess stabilizing traits that reduce overall infection by less beneficial or non-beneficial partners, which is expected to stabilize the mutualism by reducing fitness rewards to non-beneficial genotypes and prevent them from spreading to fixation in populations [27–30,43]. Although the microevolution of stabilizing mechanisms will directly affect mutualism stability or breakdown, we have little data on whether they are genetically variable or subject to natural selection, and how they evolve in contemporary populations.
In this study, we address two outstanding empirical issues that have critical implications for the evolutionary dynamics of mutualism stabilizing traits. First, we ask whether stabilizing traits in a natural host population meet the fundamental requirements for a response to natural selection. Second, we examine the potential evolutionary dynamics of mutualism-stabilizing traits in the absence of exploiters by testing for the presence of a cost to the trait, and whether they are likely to evolve due to selection on genetically correlated traits. Collectively, these forces could facilitate the loss or maintenance of stabilizing traits in the presence and absence of exploitative partners.
Using a quantitative genetic framework (cf. [44,45]), we exposed a large legume population to soil containing a mixture of mutualist and exploitative rhizobia strains, and estimated the degree of exploiter infection—a mutualism-stabilizing trait reflecting a host's ability to influence the frequency of non-beneficial rhizobia and prevent the spread of exploiters by increasing fitness rewards to beneficial rhizobia through biased infection frequency. We find significant genetic variation for differential association with exploitative partners and direct natural selection to reduce the percentage of symbiotic infections by exploitative partners. Our results suggest that mutualism-stabilizing traits in natural legume populations have potential to evolve in response to exploiters. However, we found no evidence for fitness costs of the trait or indirect selection on other correlated traits, suggesting that costs will not lead to the loss of stabilizing traits in the absence of exploitation.
2. Material and methods
(a). Natural history: plant populations and rhizobia strains
Medicago lupulina is a common exotic that grows in roadsides, fields and disturbed habitats in North America. It is largely selfing and typically forms facultative symbiotic associations with rhizobia in the Ensifer genus (previously Sinorhizobium [46]), especially Ensifer medicae and Ensifer meliloti in southern Ontario [47].
We collected plant lines from 11 sub-populations at the Koffler Scientific Reserve (www.ksr.utoronto.ca) from a range of habitats (old fields, recently disturbed fields, along paths and roads). Sites were at least 100 m apart and spanned an approximately 1.2 km2 zone on the reserve. Prior to the experiment, we selfed lines for one generation in a common greenhouse environment to equalize maternal effects and randomly selected 10 plant lines from each sub-population. Although the genetic variation measured in our experiment represents broad-sense variation (additive and non-additive), M. lupulina shows high selfing rates in natural populations (95.8% [48]), and hence selection will act on broad-sense variation [49].
We used two rhizobia strains: a mutualist rhizobia strain (RB7) collected as a wild isolate from a M. lupulina individual from the largest sub-population at the reserve, and an exploitative rhizobia strain (T173, obtained from [49]) that was isolated from Meliloti albus in a fallowed field in southern Ontario. Bromfield et al. [50] collected T173 from a field where M. albus and M. lupulina co-occur, and it thus represents a strain that M. lupulina can potentially encounter [51]. Previous phylogenetic analysis indicates that T173 nests within the Ensifer clade of mutualist rhizobia, including the mutualist rhizobia strain RB7 used in this study [31,51]. T173 forms non-fixing nodules on multiple legume hosts in the Medicago genus and its original host, Melilotus alba [51].
During preliminary inoculation trials, both strains formed nodules on M. lupulina; single-strain inoculations of T173 consistently formed many small non-fixing, white nodules (i.e. ineffective nodules), while RB7 tended to form large, pink nitrogen-fixing nodules in addition to a fraction of small white immature nodules, similar in appearance to nodules induced by T173 but occupied by RB7 (see the electronic supplementary material, appendix A) and is consistent with other Medicago–Ensifer studies reporting small numbers of white nodules in mutualist-only inoculations [52,53]. When T173 was inoculated as a single strain on M. lupulina, we observed faster plant death compared with uninoculated controls and 100% mortality when plants produced their third true leaf (on average), well before flowering [31]. When supplemented with high nitrogen fertilizer, the exploiter reduced host biomass and delayed flowering compared with uninoculated controls [31]. We confirmed strain identity on agar media with differing antibiotic resistance profiles: T173 can be distinguished from RB7 by high resistance to kanamycin and neomycin on agar plates [51]. In preliminary trials of strain culturing from nodules in plants grown in mixed inoculation treatment (n = 653), we found that visually scoring exploiter strain occupancy as the percentage of non-nitrogen-fixing nodules to the total number of nodules on the whole plant, based on the absence of pink coloration, was highly correlated with scoring occupancy using antibiotic resistance assays (r = 0.86). We found very few nodules infected by a mixture of strains (0.003% of cultured isolates).
(b). Experimental design
We grew replicates of 110 plant families (10 lines from nine sub-populations, and five from two sub-populations) in a randomized blocked design in the greenhouse. We exposed replicates of each line to one of three treatments: mutualist rhizobia [M], exploiter rhizobia [E] and a mixture of the mutualist and exploiter in a 3 : 1 ratio [M + E]. In total, we grew 110 plant lines, eight replicates/line for M and E treatments and 10 replicates/line for [M + E].
Seeds were germinated in sterile conditions to synchronize germination among lines, and pre-germinated radicles were planted in clean cell pack tray inserts containing sterilized low-nutrient charge soil (details in the electronic supplementary material, appendix A). We randomly assigned strain treatments to each tray within each block, randomized plant lines across trays within each strain treatment and inoculated seedlings with approximately 106 rhizobia cells of the assigned strain treatment, with little contamination observed during the experiment (see the electronic supplementary material, appendix A). For established plants, we recorded plant mortality throughout the experiment. Following 100 days of growth, we terminated the experiment and harvested all surviving plants.
(c). Data collection
We measured several plant traits indicative of performance, fitness components and the interaction with rhizobia: (i) mortality, (ii) dry shoot and root biomass, (ii) exploiter strain root occupancy, (iv) nodule density and (v) number of days until death. Mortality was scored during the experiment while all remaining traits were scored at harvest. Exploiter strain root occupancy was estimated as the frequency of nodules that were non-fixing nodules, which was scored based on nodule colour (nitrogen-fixing nodules have a pink hue). We analysed exploiter occupancy as a proportion rather than the absolute number of non-fixing nodules because it would reflect a better measure of host preference for either the mutualist or exploiter strain. Nodule density was measured as the number of nodules per gram dry root biomass and reflects host investment in the number of nodules per unit of root tissue.
Scoring exploiter strain occupancy required destructive harvesting, precluding estimates of reproductive fitness and strain occupancy on the same individual plant. Similarly, following experimental plants until plant senescence and fruit set (more than approx. 175 days) would have led to nodule senescence and precluded accurate estimates of nodule strain occupancy on the experimental plants. However, a parallel experiment conducted in the same conditions as the experimental plants shows that biomass is highly correlated with fruit set, suggesting that it is a reliable proxy of reproductive fitness (see the electronic supplementary material, appendix B, for details).
(d). Statistical analysis
(i). Testing for genetic variance in plant traits and calculating trait line means
We calculated broad-sense heritability, H2 = Vg/Vp [45], and the coefficient of genetic variation, CV = √Vg/mean [54], for each trait within each treatment separately, except in the mixed treatment, where broad-sense H2 and CV were also calculated on dead and live plants separately. Mortality in the mutualist treatment was too low to estimate genetic variation. Genetic variance components (Vg) for all traits were obtained from the covariance parameter estimate from the plant line term (nested within sub-population) in a mixed-model ANCOVA using Gaussian assumptions. We treated tray, population and plant line (nested within sub-population) as random effects, and block and harvest date as fixed effects. We tested for significant genetic variation among plant families and populations in all traits using a log-likelihood ratio test between full models and models without the plant line or population term, respectively [55]. As some non-fixing nodules may not contain the exploiter in the mixed inoculation, we recalculated CV and H2 but included line mean estimates of percentage non-fixing nodules from the mutualist treatment as a fixed covariate (see next paragraph for line mean estimation methods) to exclude the possibility that Vg in exploiter occupancy reflects a general propensity for plant lines to form non-fixing nodules, regardless of exploiter presence or absence. Using this, covariate did not affect statistical significance and lead to similar H2 and CV estimates, so we present results without the additional fixed covariate.
We estimated line means for shoot biomass, percentage survival, nodule density and exploiter strain occupancy (proportion of non-fixing nodules to total nodule number) from plants in the mutualist and mixed treatments. Trait means were estimated for each line in each treatment using least square mean estimates from an ANCOVA model containing block, rhizobia treatment, plant line and plant line × rhizobia treatment as fixed effects, and tray as a random effect. For shoot size and nodule density, we used a standard Gaussian distribution assumption and untransformed data; for shoot biomass, we log-transformed it to increase the normality of residuals (proc mixed, SAS v. 9.3). For survival, we used a binary distribution (proc glimmix, dist = binary). For the proportion of non-fixing nodules, we used a binomial distribution (proc glimmix, dist = binomial, total number of non-fixing nodules/total nodule number as the response). We removed four highly influential outliers (two and two observations in mixed and mutualist treatment, respectively) with nodule density estimates more than 10 s.d. from the mean.
(ii). Estimating selection on plant traits and the cost of excluding non-mutualists
Using biomass and mortality as separate fitness measures, we tested for selection on exploiter strain occupancy and nodule density by regressing line means of biomass and survival against both traits following standard methods in Lande & Arnold [44] and Rausher [56]. For biomass selection, we used line means calculated from plants that survived the duration of the experiment. For mortality selection, we used line means calculated from all established experimental plants in the mixed and mutualist treatments.
Selection gradients were calculated using raw unstandardized data (β and γ refer to linear and nonlinear selection gradients, respectively). To determine whether selection on non-fixing nodules or nodule density significantly differed between rhizobia treatments, we used an ANCOVA model containing the treatment, linear and second-order terms (i.e. quadratic and cross-product terms: treatment × trait and treatment × trait × trait). Second-order terms (i.e. trait1 × trait1, trait1 × trait2 terms) were non-significant for biomass selection, suggesting no evidence of stabilizing, disruptive or correlational selection on surviving individuals. Because a fraction of non-fixing nodules in the mixed treatment could be immature or ineffective nodules occupied by the mutualist strain, we compared the magnitude of selection in the mixed and mutualist treatments. If we found significantly stronger, negative selection on non-fixing nodules in the mixed treatment, and either zero or positive selection in the mutualist treatment (i.e. a significant trait × treatment interaction), this would strongly suggest that selection on the proportion of non-fixing nodules is due specifically to the presence of the exploiter, and not due to a general propensity for host lines to produce more or less immature nodules occupied by the mutualist. We repeated the ANCOVA analysis, including line mean estimates of root : shoot ratios as a covariate to exclude the inference that associations between nodule density or non-fixing nodules with shoot size reflect differences in allocation to roots. Inclusion of the root: shoot ratios in the selection analysis did not change the significance or direction of selection for either nodulation trait.
We evaluated potential costs of excluding exploiters by testing for a positive correlation between plant fitness in the absence of exploiters (shoot size in the mutualist treatment) and exploiter occupancy (proportion of non-fixing nodules in the mixed treatment) (cf. [57,58]). However, a neutral or negative correlation does not conclusively eliminate the possibility of a cost.
3. Results
(a). Exploiters reduce host fitness
The presence of the exploitative strain had significant negative impacts on plant performance. As we expected, plant biomass was highest in the mutualist treatment, at intermediate values in the mixed treatment and lowest in the exploiter treatment (electronic supplementary material, table S1). All plants exposed to single inoculations of the exploiter strain died during the course of the experiment (electronic supplementary material, table S1). Plants exposed to mixed inoculations suffered 36.7% mortality, compared with mortality of 5.25% in the mutualist treatment (F1,1723 = 184.48, p < 0.0001; electronic supplementary material, tables S1 and S2), and were 65.1% smaller than plants in the mutualist treatment (F2,1651 = 163.40, p < 0.0001; electronic supplementary material, tables S1 and S2). We estimated exploiter occupancy in the mixed treatment by measuring the proportion of non-nitrogen-fixing root infections, determined by nodule colour; nodule colour is highly correlated (r = 0.86) with strain occupancy of nodules, based on antibiotic and plating assays (see Material and methods). Hosts that died in the mixed inoculation had 116% greater percentage of non-fixing nodules compared with plants that survived the mixed inoculation (85.89 ± 01.36% and 39.75 ± 0.67%, respectively; F1,779 = 1201.35, p < 0.0001; electronic supplementary material, table S3). While hosts produced some non-fixing nodules in the absence of the exploiter, plants in the mixed treatment had 185.6% higher proportion of non-fixing nodules on roots compared with plants in the mutualist treatment (F1,1574 = 214.10, p < 0.0001; electronic supplementary material, tables S1 and S2), suggesting that decreased survivorship and performance in mixed inoculation was likely to be due to higher infection rates of the exploitative strains. These data indicate that the exploiter was harmful for plant performance and fitness.
(b). Exploiter strain occupancy in host nodules is genetically variable
We consistently detected genetic variance among plant families for almost all traits across all three rhizobia inoculation treatments. Traits related to plant performance (i.e. shoot and root mass, nodule number, time until death and mortality) in the mutualist and mixed inoculation treatment showed significant genetic variation, with the exception of days until death in the mutualist treatment (electronic supplementary material, table S4). We found significant genetic variation for the proportion of non-fixing nodules in the mixed inoculation treatment, based on a significant effect of plant line in mixed models (χ2 = 19.6, p < 0.00001; H2 = 0.1334, CV = 0.1522; electronic supplementary material, table S4). These data indicate that the plant lines used in the experiment showed significant genetic variation in mutualism-stabilizing traits: their propensity for associating with partners of variable quality.
(c). Selection acts to reduce exploiter occupancy
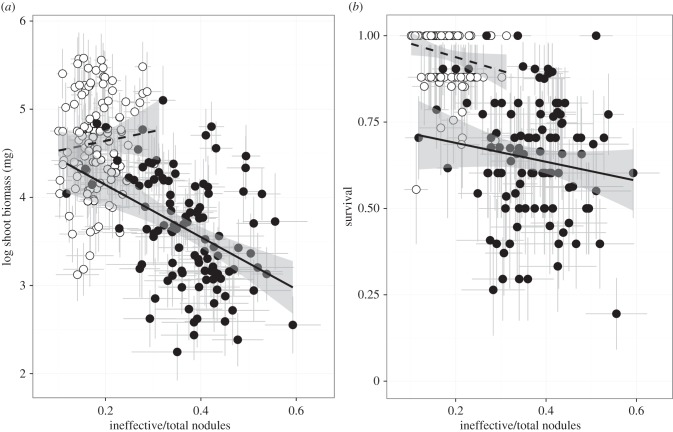
For plants that survived in the mixed treatment, we found strong linear biomass selection to decrease non-fixing nodules (figure 1a); parallel experiments show that biomass significantly increases with fruit set, suggesting that it accurately reflects fecundity selection (see Material and methods). While some non-fixing nodules may also contain mutualist or no symbionts in the mixed and mutualist treatment, our analysis demonstrates that selection was acting to explicitly reduce exploiter occupancy, as selection to reduce non-fixing nodules only occurred in the mixed treatment (treatment × proportion non-fixing nodules; F1,213 = 8.96, p = 0.0031; electronic supplementary material, table S5), and no main effect of the proportion of non-fixing nodules was detected across either treatment (F1,213 = 0.02, p = 0.8925; electronic supplementary material, table S5). Selection gradients calculated within each rhizobia treatment confirmed stronger negative selection in the mixed treatment compared with the mutualist treatment on the proportion of non-fixing nodules (β = −0.6272 ± 0.1858 compared with β = 0.4550 ± 0.2817; electronic supplementary material, table S6). Mortality selection on the proportion of non-fixing nodules was not significant (electronic supplementary material, table S5), but was consistent with biomass selection, showing negative selection on the trait the mixed treatment (β = −0.4447 ± 0.3378; figure 1b; electronic supplementary material, table S6).
Figure 1.
Genotypic relationship between the proportion of non-fixing nodules (ineffective nodules) and (a) shoot biomass and (b) mortality in the mutualist (dashed line, open circles) and mixed treatment (solid line, closed circles). Each dot is a line mean (n = 110 plant lines/treatment). Grey error bars are ±1 standard error and the 95% CI of the fitted line is shaded in grey. The proportion of non-fixing nodules gives an estimate of exploiter strain occupancy in the mixed rhizobia treatment, which contains both mixed and mutualist rhizobia strains in the soil.
(d). There is no cost to excluding exploitative partners
We evaluated whether there was a potential cost to excluding exploiters by determining whether host families that have a low proportion of non-fixing nodules in the presence of the exploiter (i.e. greater association with mutualist partners) also have lower relative fitness in the absence of the exploiter (cf. [57,58]). However, we found the opposite (albeit non-significant) trend, where lines with a low proportion of non-fixing nodules showed higher relative biomass in the mutualist treatment (r = −0.12676, p = 0.1890; figure 2), indicating that no cost to filtering exploitative rhizobia partners was detected despite a large quantitative genetic design (n = 110 plant lines). These data preliminarily suggest that costs of mutualism-stabilizing traits are either absent or weak, at least for the genetic variation expressed in our experiment.
Figure 2.
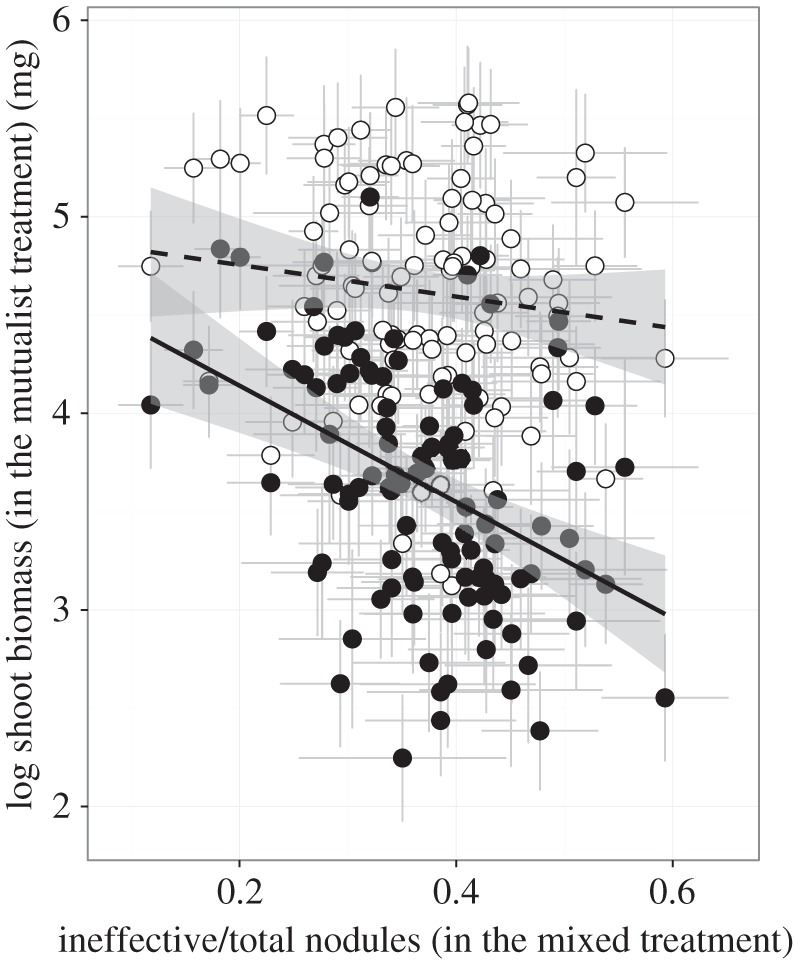
Relationship between the proportion of ineffective nodules and shoot biomass in the mixed and mutualist treatment, demonstrating the cost of assortative preference with mutualist rhizobia. Each dot is a line mean. Line means for exploiter occupancy (estimated in the mixed treatment) are correlated with the corresponding line means for plant biomass in the absence of exploitation (mutualist treatment, dashed line). A positive correlation shows a cost to increasing associations with beneficial partners, but our data indicate no significant positive or negative associations with these traits (dashed line, open circles). The relationship between shoot biomass in the mixed treatment and exploiter occupancy (solid line, closed circles) is shown for comparison.
(e). Selection to reduce investment in symbiotic associations does not affect selection to reduce exploiter occupancy
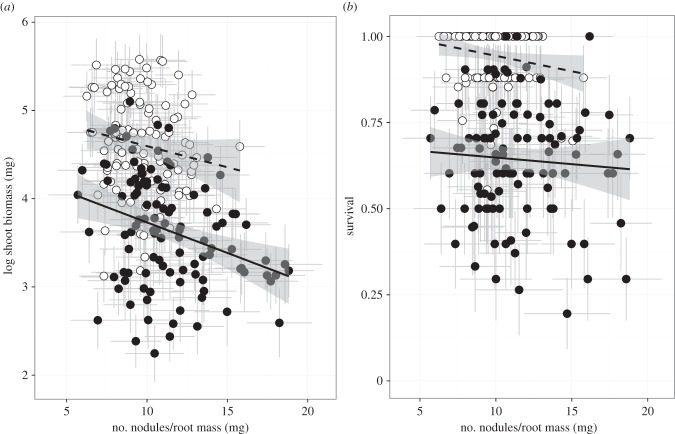
In addition to measuring the proportion of non-fixing nodules, we measured the total nodule number per unit of root mass (nodule density), which characterizes host investment in symbiotic associations. In both the mutualist and mixed treatment, we detected linear selection for hosts to decrease nodule density (figure 3a; β = −0.01204 ± 0.0063 and β = −0.0094 ± 0.0056, respectively; electronic supplementary material, table S6). We found no significant difference in the strength of selection on nodule density between mutualist or mixed treatments, as we detected no significant nodule density × treatment interaction (F1,213 = 0.35, p = 0.5535), and only a main effect for nodule density (F1,213 = 6.35, p = 0.0124; electronic supplementary material, table S5). We failed to detect significant correlational selection between proportion of non-fixing nodules and nodule density across mixed and mutualist treatments (non-fixing nodule × nodule density interaction: F1,212 = 0.17, p = 0.6837), suggesting that selection is acting independently on nodule density and the ability to exclude exploiters. We found no significant mortality selection on nodule density that varied by treatment (F1,213 = 0.49, p = 0.4562; figure 3b; electronic supplementary material, table S5), which is consistent with patterns of selection found with biomass selection.
Figure 3.
Genotypic relationship between nodule density and (a) shoot biomass and (b) mortality in the mutualist (dashed line, open circles) and mixed treatment (solid line, closed circles). Each dot is a line mean (n = 110 plant lines/treatment). Grey error bars are ±1 standard error and the 95% CI of the fitted line is shaded in grey.
Despite evidence that nodule density is under selection, we found no evidence that this trait will affect selection on the ability to exclude exploiters when exploiters are absent from the environment, as we found no genetic association between exploiter strain occupancy in the mixed treatment and nodule density in the mutualist treatment (partial r = 0.13801, p = 0.1543; after accounting for the proportion of non-fixing nodules in the mutualist treatment).
4. Discussion
Mutualism theory has shown that partner choice, host sanctions or partner fidelity feedback can stabilize the mutualism by preventing exploitative genotypes from spreading to fixation in the population [15,16]. Although evidence consistent with host stabilizing traits has been found in legumes [27–30,43], we have little empirical data to infer potential micro-evolutionary dynamics of these traits in natural host populations [19]. Our study has filled several important empirical gaps that are critical to understand any potential evolutionary dynamics of host stabilizing traits, by showing the following. (i) Natural populations exhibit genetic variation for the ability to exclude exploitative partners, and selection will favour hosts with higher partner filtering abilities. (ii) There is no evidence to suggest that selection will favour a loss in the ability to exclude exploitative partners in the absence of exploitation, as indicated by a lack evidence of fitness cost in the trait and the lack of potential correlated evolutionary responses with investment in total symbiotic associations.
Our experiment also demonstrates selection to reduce investment in total symbiotic associations. However, the similarity in selection on nodule density between environments where an exploiter is present or absent suggests that reducing investment in the total number of rhizobia infections, per unit of root growth, is not a general mechanism to reduce association with non-beneficial hosts. We discuss these results with respect to the potential (co)evolutionary dynamics of host stabilizing traits in the presence and absence of exploitative partners.
(a). Evolution of stabilizing traits in the presence and absence of exploitative partners
The various mechanisms that might stabilize mutualisms are hotly debated [20,22–25,59]. However, there is remarkably little evidence that these mechanisms are genetically variable in natural populations, and could indeed evolve in a direction that would either stabilize or destabilize mutualistic interactions. To our knowledge, evidence is limited to a study by Heath & Tiffin [35], who demonstrated genetic variation in M. truncatula consistent with partner choice mechanisms using a suite of variably beneficial strains. Partner choice is usually defined as pre-interaction mechanisms to reduce infection by a potential exploiter, and several studies have found genetic variation in loci involved in partner signalling in the host [60–63]. Thus, evidence of genetic variation has been limited to stabilizing mechanisms either among partners that are variably beneficial or in pre-infection signalling mechanisms that determine symbiotic or non-symbiotic properties of rhizobia strains. Theoretical models developed to explain mutualism stability, however, have primarily modelled symbiotic (i.e. compatible) yet binary symbiotic partner quality, where non-beneficial partners are assumed to be harmful to its host—that is, mutualist versus exploitative partners that are able to infect hosts. Our experiment is able to link the empirical and theoretical gap in the mutualism literature by using an exploiter strain that has either evolved methods of invading plant roots and inducing nodule formation or lost its ability to fix nitrogen [51]. The mere existence of a harmful yet compatible exploiter strain, combined with the fact that bacteria can reproduce (and hence respond to selection) much more quickly than their host, suggests that there will probably be strong selective pressure for the host to reduce post-infection feedback benefits by aborting or inactivating nodules [64]. Post-infection stabilizing mechanisms, such as host sanction mechanisms (as defined by Denison [8]) and partner fidelity feedbacks (as defined by Weyl et al. [22]) will probably play a key role in reducing infection by exploitative or inefficient rhizobia partners that possess compatible traits to invade and form root nodules. Since coevolutionary responses on post-infection traits are currently intensely debated [20,22,24,25,59], more data on genetic variation in symbiosis-related traits (e.g. nodule size [65], energy storage in rhizobia [66]) and post-infection stabilizing traits, as well as the strength of natural selection acting on them, would clarify their role in stabilizing or destabilizing mutualisms [19]. Similarly, identifying trait mismatches between species due to differing coevolutionary histories (cf. [67]) would be helpful. Recent evidence by Regus et al. [43] suggests that partner choice is not plastic to environmental alterations in the costs and benefits of the mutualism. Consistent with partner choice and host sanctions, our data indicate that M. lupulina expresses significant genetic variation for the ability to filter out compatible exploitative partners and that selection will favour hosts with higher partner filtering, thus favouring a mutualism-stabilizing trait that reduces fitness rewards to exploiters. More generally, our study provides the empirical (as opposed to theoretical) possibility that hosts have the required standing genetic variation required to rapidly respond to selection imposed by invasion of exploiters.
Our study also gives empirical insight into an additional, yet important evolutionary dynamic of host stabilizing traits that has not previously been considered: the maintenance of stabilizing traits in the absence of exploitative partners. Foster & Kokko [17] showed that the presence of exploitative partners was critical to maintain partner choice. An important feature of their model was a built-in assumption that partner choice is costly for hosts, which leads to it being selectively removed in the absence of exploiters. We did not detect evidence that excluding non-beneficial partners is costly for host fitness, implying that selection would not eliminate mutualism-stabilizing mechanisms in the absence of exploitation. Nor did we find strong evidence that selection would facilitate a loss in stabilizing mechanisms through a correlated evolutionary response with investment in total symbiotic associations in the absence of exploiters. However, further studies will be required to confirm these suggestive data.
(b). Alternative interpretations for genetic variation in stabilizing mechanisms
We found that plant lines that were poor at excluding exploitative partners generally had much lower performance (figure 1a). An alternative interpretation of the results we observed could be genetic variation in compatibility with the specific mutualist strain we used. If some plant lines in our experiment have reduced compatibility with the mutualist partner, it could result in reduced nitrogen acquisition, which could subsequently affect the ability to exclude exploiters. In other words, the observed genetic variation in strain occupancy could be due to host responses to the mutualist, rather than the exploiter specifically. Genetic variation in mutualist compatibility would change our interpretation on what putative agent of selection is acting on host traits to reduce exploiter occupation. If variation in compatibility was causing the observed selection patterns, the putative agent of selection acting on the host originates from the mutualist, not the exploiter. While we cannot completely exclude the compatibility hypothesis, several lines of evidence suggest that it is unlikely. First, if differential compatibility explained our results, we would expect that plants that did poorly in the mutualist treatment (a potential sign of incompatibility) would also have lower filtering abilities in the mixed treatment. However, we found no significant genetic correlation between plant fitness in the mutualist treatment and host filtering abilities in the mixed treatment (r = −0.12468, p = 0.189; figure 2). Second, the mutualist rhizobia strain we used for our experiment came from the same locality as the host plants used in this study. Preliminary experiments using six other candidate mutualist rhizobia strains inoculated on a subset of plant lines used in this study showed no differences in host biomass for all strains tested (electronic supplementary material, figure S1) and consistent phenotypic selection to reduce nodule density across all beneficial strains (electronic supplementary material, figure S2). These data indicate that the mutualist strain used in this study did not substantially vary in mutualistic behaviour from other rhizobia strains from the same locality. Based on this evidence, it is less likely that poor partner filtering abilities are due to decreased nitrogen acquisition from beneficial, yet less compatible partners.
We also detected a positive, non-significant trend to increase the proportion of non-fixing nodules in the mutualist treatment (figure 1a), which could be interpreted as selective pressure to increase the number of immature nodules. While we found little evidence of contamination, if low-level contamination by the exploiter caused the positive trend, it suggests that rhizobia (exploiter, mutualist or both) have complex frequency-dependent behaviour that enables the exploiter to reverse its negative fitness effects to become positive when it is at low frequencies.
(c). Selection to reduce investment in symbiotic associations is consistent with mutualism theory
Our results show that host populations will be under selection to reduce investment in total symbiotic associations, regardless of the quality of microbial partners belowground, indicating that host fitness is reduced if rhizobia infections are too prolific (figure 3a). As a mutualism is defined as a positive fitness feedback between reciprocating partners [5], hosts that form no or few nodules with mutualist rhizobia are expected to have low fitness. Our data show low fitness at high nodule investment, suggesting that nodule investment is under some form of stabilizing selection. We suggest that two factors explain why we observed linear rather than stabilizing selection in our experiments. First, genetic variants with zero or low nodule investment are likely to be rare in natural populations with selection acting strongly against them, given the fitness advantages of the mutualism. Mutation accumulation or other experimental designs that minimize the strength of selection might be required to identify and study variants with zero or low nodule investment. Second, the direction of selection could be explained by density of rhizobial partners: under low rhizobial densities, higher nodule investment is likely to be favoured. However, under rhizobial densities exceeding available host infection sites, genotypes with the lowest investment will generally be favoured because of the costs of nodule production causing negative linear selection.
As reported cell densities vary in field soil (more than 10–107 cells g−1 soil [47]), follow-up experiments measuring other aspects of symbiotic association (e.g. nodule size, energy storage in rhizobia cells) and manipulating symbiotic bacteria densities, such that rhizobia availability is lower than the expected total number of infection sites on the host, could be used to test this hypothesis. Generally, our result of negative linear selection on nodule investment is consistent with the view in mutualism theory that a conflict of interest can occur between the hosts and symbionts due to the differing costs and benefits of the interaction, even when the interaction is mutually beneficial and lacks pure exploiters [68–70].
5. Conclusion
The ability for host plants to control infection from beneficial and exploitative symbionts is a critical trait that can maintain the stability of the mutualisms. We found that lines of M. lupulina, when exposed to an exploitative symbiont, showed significant genetic variation for the ability to associate differentially between mutualist and exploitative symbionts. As a proof of principle, our study shows that microevolutionary responses in mutualism-stabilizing traits mediated by the host are possible. Follow-up studies quantifying variation among multiple populations, and in the ability to regulate nodule production, as well as further investigation of any potential costs of the trait or genetic correlations with other traits, will provide useful information for predicting the evolution of host stabilizing traits.
Supplementary Material
Supplementary Material
Acknowledgements
We are grateful for all the graduate and undergraduate students that helped in the greenhouse: Charise Currier, Adriana Salcedo, Lorena Cannon, Emily Lo, Amanda Gorton, Amanda Stock, Russell Dinnage, Linda Qu, Rufina Kim and Alex Jung. We thank Eden Bromfield at Agriculture and Agri-Foods Canada (AAFC) for kindly sending us several rhizobia strains. Comments by Sharon Strauss, Art Weis, James Thomson, Megan Frederickson and two anonymous reviewers improved the manuscript. A.K.S. designed and performed the experiment and all statistical analyses; A.K.S. and J.R.S. wrote the manuscript. All authors discussed the analysis and results, and contributed to editing the manuscript.
Data accessibility
Data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.j2063. DNA sequences for bacterial strains used in this study: GenBank accession no. KF898182 (RB7) and KF898184 (T173).
Funding statement
Grants from NSERC Canada (A.K.S. and J.R.S.) and CFI made this study possible.
References
- 1.Bronstein JL. 1994. Conditional outcomes in mutualistic interactions. Trends Ecol. Evol. 9, 214–217. ( 10.1016/0169-5347(94)90246-1) [DOI] [PubMed] [Google Scholar]
- 2.Boucher DH, James S, Keeler KH. 1982. The ecology of mutualism. Annu. Rev. Ecol. Syst. 13, 315–347. ( 10.2307/2097071) [DOI] [Google Scholar]
- 3.Bronstein JL. 2001. The exploitation of mutualisms. Ecol. Lett. 4, 277–287. ( 10.1046/j.1461-0248.2001.00218.x) [DOI] [Google Scholar]
- 4.Sachs JL, Simms EL. 2008. The origins of uncooperative rhizobia. Oikos 117, 961–966. ( 10.1111/j.0030-1299.2008.16606.x) [DOI] [Google Scholar]
- 5.Trivers RL. 1971. The evolution of reciprocal altruism. Q. Rev. Biol. 46, 35–57. ( 10.1086/406755) [DOI] [Google Scholar]
- 6.Doebeli M, Knowlton N. 1998. The evolution of interspecific mutualisms. Proc. Natl Acad. Sci. USA 95, 8676–8680. ( 10.1073/pnas.95.15.8676) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herre EA, Knowlton N, Mueller UG, Rehner SA. 1999. The evolution of mutualisms: exploring the paths between conflict and cooperation. Trends Ecol. Evol. 14, 49–53. ( 10.1016/s0169-5347(98)01529-8) [DOI] [PubMed] [Google Scholar]
- 8.Denison RF. 2000. Legume sanctions and the evolution of symbiotic cooperation by rhizobia. Am. Nat. 156, 567–576. ( 10.1086/316994) [DOI] [PubMed] [Google Scholar]
- 9.Pellmyr O, Huth CJ. 1994. Evolutionary stability of mutualism between yuccas and yucca moths. Nature 372, 257–260. ( 10.1038/372257a0) [DOI] [Google Scholar]
- 10.Soberon Mainero J, Martinez del Rio C. 1985. Cheating and taking advantage in mutualistic associations. In The biology of mutualism (ed. DH Boucher), pp. 192–216. New York, NY: Oxford. [Google Scholar]
- 11.Sachs JL, Simms EL. 2006. Pathways to mutualism breakdown. Trends Ecol. Evol. 21, 585–592. ( 10.1016/j.tree.2006.06.018) [DOI] [PubMed] [Google Scholar]
- 12.Ferriere R, Bronstein JL, Rinaldi S, Law R, Gauduchon M. 2002. Cheating and the evolutionary stability of mutualisms. Proc. R. Soc. Lond. B 269, 773–780. ( 10.1098/rspb.2001.1900) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simms EL, Taylor DL. 2002. Partner choice in nitrogen-fixation mutualisms of legumes and rhizobia. Integr. Comp. Biol. 42, 369–380. ( 10.1093/icb/42.2.369) [DOI] [PubMed] [Google Scholar]
- 14.Foster KR, Wenseleers T. 2006. A general model for the evolution of mutualisms. J. Evol. Biol. 19, 1283–1293. ( 10.1111/j.1420-9101.2005.01073.x) [DOI] [PubMed] [Google Scholar]
- 15.Bull JJ, Rice WR. 1991. Distinguishing mechanisms for the evolution of co-operation. J. Theor. Biol. 149, 63–74. ( 10.1016/S0022-5193(05)80072-4) [DOI] [PubMed] [Google Scholar]
- 16.West SA, Kiers ET, Simms EL, Denison RF. 2002. Sanctions and mutualism stability: why do rhizobia fix nitrogen? Proc. R. Soc. Lond. B 269, 685–694. ( 10.1098/rspb.2001.1878) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foster KR, Kokko H. 2006. Cheating can stabilize cooperation in mutualisms. Proc. R. Soc. B 273, 2233–2239. ( 10.1098/rspb.2006.3571) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janzen DH. 1975. Pseudomyrmex nigropilosa: a parasite of a mutualism. Science 188, 936–937. ( 10.1126/science.188.4191.936) [DOI] [PubMed] [Google Scholar]
- 19.Heath KD, Stinchcombe JR. 2013. Explaining mutualism variation: a new evolutionary paradox? Evolution 68, 309–317. ( 10.1111/evo.12292) [DOI] [PubMed] [Google Scholar]
- 20.Archetti M, Scheuring I, Hoffman M, Frederickson ME, Pierce NE, Yu DW. 2011. Economic game theory for mutualism and cooperation. Ecol. Lett. 14, 1300–1312. ( 10.1111/j.1461-0248.2011.01697.x) [DOI] [PubMed] [Google Scholar]
- 21.Richter KS, Weis AE. 1995. Differential abortion in the yucca. Nature 376, 557–558. ( 10.1038/376557b0)7637801 [DOI] [Google Scholar]
- 22.Weyl EG, Frederickson ME, Yu DW, Pierce NE. 2010. Economic contract theory tests models of mutualism. Proc. Natl Acad. Sci. USA 107, 15 712–15 716. ( 10.1073/pnas.1005294107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frederickson ME. 2013. Rethinking mutualism stability: cheaters and the evolution of sanctions. Q. Rev. Biol. 88, 269–295. ( 10.1086/673757) [DOI] [PubMed] [Google Scholar]
- 24.Kiers ET, Denison RF, Kawakita A, Herre EA. 2011. The biological reality of host sanctions and partner fidelity. Proc. Natl Acad. Sci. USA 108, E7 ( 10.1073/pnas.1014546108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Archetti M, Úbeda F, Fudenberg D, Green J, Pierce Naomi E, Yu Douglas W. 2011. Let the right one in: a microeconomic approach to partner choice in mutualisms. Am. Nat. 177, 75–85. ( 10.1086/657622) [DOI] [PubMed] [Google Scholar]
- 26.Sachs JL, Skophammer RG, Regus JU. 2011. Evolutionary transitions in bacterial symbiosis. Proc. Natl Acad. Sci. USA 108(Suppl. 2), 10 800–10 807. ( 10.1073/pnas.1100304108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiers ET, Rousseau RA, West SA, Denison RF. 2003. Host sanctions and the legume–rhizobium mutualism. Nature 425, 78–81. ( 10.1038/nature01931) [DOI] [PubMed] [Google Scholar]
- 28.Simms EL, Taylor DL, Povich J, Shefferson RP, Sachs JL, Urbina M, Tausczik Y. 2006. An empirical test of partner choice mechanisms in a wild legume–rhizobium interaction. Proc. R. Soc. B 273, 77–81. ( 10.1098/rspb.2005.3292) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gubry-Rangin C, Garcia M, Béna G. 2010. Partner choice in Medicago truncatula–Sinorhizobium symbiosis. Proc. R. Soc. B 277, 1947–1951. ( 10.1098/rspb.2009.2072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oono R, Anderson CG, Denison RF. 2011. Failure to fix nitrogen by non-reproductive symbiotic rhizobia triggers host sanctions that reduce fitness of their reproductive clonemates. Proc. R. Soc. B 278, 2698–2703. ( 10.1098/rspb.2010.2193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simonsen AK, Stinchcombe JR. 2014. Herbivory eliminates fitness costs of mutualism exploiters. New Phytol. 202, 651–661. ( 10.1111/nph.12668) [DOI] [PubMed] [Google Scholar]
- 32.Sprent JI, Sutherland JM, Faria SMD, Dilworth MJ, Corby HDL, Becking JH, Materon LA, Drozd JW. 1987. Some aspects of the biology of nitrogen-fixing organisms [and discussion]. Phil. Trans. R. Soc. Lond. B 317, 111–129. ( 10.2307/2396530) [DOI] [Google Scholar]
- 33.Layzell DB, Pate JS, Atkins CA, Canvin DT. 1981. Partitioning of carbon and nitrogen and the nutrition of root and shoot apex in a nodulated legume. Plant Physiol. 67, 30–36. ( 10.1104/pp.67.1.30) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pate JS, Layzell DB, Atkins CA. 1979. Economy of carbon and nitrogen in a nodulated and nonnodulated (NO(3)-grown) legume. Plant Physiol. 64, 1083–1088. ( 10.1104/pp.64.6.1083) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heath KD, Tiffin P. 2009. Stabilizing mechanisms in a legume-rhizobium mutualism. Evolution 63, 652–662. ( 10.1111/j.1558-5646.2008.00582.x) [DOI] [PubMed] [Google Scholar]
- 36.Friesen ML. 2012. Widespread fitness alignment in the legume–rhizobium symbiosis. New Phytol. 194, 1096–1111. ( 10.1111/j.1469-8137.2012.04099.x) [DOI] [PubMed] [Google Scholar]
- 37.Thrall PH, Burdon JJ, Woods MJ. 2000. Variation in the effectiveness of symbiotic associations between native rhizobia and temperate Australian legumes: interactions within and between genera. J. Appl. Ecol. 37, 52–65. ( 10.1046/j.1365-2664.2000.00470.x) [DOI] [Google Scholar]
- 38.Sachs JL, Russell JE, Lii YE, Black KC, Lopez G, Patil AS. 2010. Host control over infection and proliferation of a cheater symbiont. J. Evol. Biol. 23, 1919–1927. ( 10.1111/j.1420-9101.2010.02056.x) [DOI] [PubMed] [Google Scholar]
- 39.Singleton PW, Stockinger KR. 1983. Compensation against ineffective nodulation in soybean. Crop Sci. 23, 69–72. ( 10.2135/cropsci1983.0011183X002300010019x) [DOI] [Google Scholar]
- 40.Burdon JJ, Gibson AH, Searle SD, Woods MJ, Brockwell J. 1999. Variation in the effectiveness of symbiotic associations between native rhizobia and temperate Australian Acacia: within-species interactions. J. Appl. Ecol. 36, 398–408. ( 10.1046/j.1365-2664.1999.00409.x) [DOI] [Google Scholar]
- 41.Denton MD, Coventry DR, Bellotti WD, Howieson JG. 2000. Distribution, abundance and symbiotic effectiveness of Rhizobium leguminosarum bv. trifolii from alkaline pasture soils in South Australia. Aust. J. Exp. Agric. 40, 25–35. ( 10.1071/EA99035) [DOI] [Google Scholar]
- 42.Porter SS, Simms EL. 2014. Selection for cheating across disparate environments in the legume–rhizobium mutualism. Ecol. Lett. 17, 1121–1129. ( 10.1111/ele.12318) [DOI] [PubMed] [Google Scholar]
- 43.Regus JU, Gano KA, Hollowell AC, Sachs JL. 2014. Efficiency of partner choice and sanctions in Lotus is not altered by nitrogen fertilization. Proc. R. Soc. B 281, 20132587 ( 10.1098/rspb.2013.2587) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lande R, Arnold SJ. 1983. The measurement of selection on correlated characters. Evolution 37, 1210–1226. ( 10.2307/2408842) [DOI] [PubMed] [Google Scholar]
- 45.Lynch M, Walsh JB. 1998. Genetics and analysis of quantitative traits. Sunderland, MA: Sinauer Assocs Inc. [Google Scholar]
- 46.Young JM. 2010. Sinorhizobium versus Ensifer: may a taxonomy subcommittee of the ICSP contradict the judicial commission? Int. J. Syst. Evol. Microbiol. 60, 1711–1713. ( 10.1099/ijs.0.025163-0) [DOI] [PubMed] [Google Scholar]
- 47.Prévost D, Bromfield ES. 2003. Diversity of symbiotic rhizobia resident in Canadian soils. Can. J. Soil Sci. 83, 311–319. ( 10.4141/s01-066) [DOI] [Google Scholar]
- 48.Yan J, Chu H-J, Wang H-C, Li J-Q, Sang T. 2009. Population genetic structure of two Medicago species shaped by distinct life form, mating system and seed dispersal. Ann. Bot. 103, 825–834. ( 10.1093/aob/mcp006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roughgarden J. 1979. Theory of population genetics and evolutionary ecology. New York, NY: Macmillan. [Google Scholar]
- 50.Bromfield ESP, Butler GW, Barran LR. 2001. Temporal effects on the composition of a population of Sinorhizobium meliloti associated with Medicago sativa and Melilotus alba. Can. J. Microbiol. 47, 567–573. ( 10.1139/w01-034) [DOI] [PubMed] [Google Scholar]
- 51.Bromfield ESP, Tambong JT, Cloutier S, Prévost D, Laguerre G, van Berkum P, Thi TVT, Assabgui R, Barran LR. 2010. Ensifer, Phyllobacterium and Rhizobium species occupy nodules of Medicago sativa (alfalfa) and Melilotus alba (sweet clover) grown at a Canadian site without a history of cultivation. Microbiology 156, 505–520. ( 10.1099/mic.0.034058-0) [DOI] [PubMed] [Google Scholar]
- 52.Campbell GR, Taga ME, Mistry K, Lloret J, Anderson PJ, Roth JR, Walker GC. 2006. Sinorhizobium meliloti bluB is necessary for production of 5, 6-dimethylbenzimidazole, the lower ligand of B12. Proc. Natl Acad. Sci. USA 103, 4634–4639. ( 10.1073/pnas.0509384103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marketon MM, Gronquist MR, Eberhard A, González JE. 2002. Characterization of the Sinorhizobium meliloti sinR/sinI locus and the production of novel N-acyl homoserine lactones. J. Bacteriol. 184, 5686–5695. ( 10.1128/JB.184.20.5686-5695.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Houle D. 1992. Comparing evolvability and variability of quantitative traits. Genetics 130, 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Littell RC, Milliken GA, Stroup WW, Wolfinger RD. 1996. SAS system for mixed models. Cary, NC: SAS Institute. [Google Scholar]
- 56.Rausher MD. 1992. The measurement of selection on quantitative traits: biases due to environmental covariances between traits and fitness. Evolution 46, 616–626. ( 10.2307/2409632) [DOI] [PubMed] [Google Scholar]
- 57.Carr DE, Murphy JF, Eubanks MD. 2005. Genetic variation and covariation for resistance and tolerance to Cucumber mosaic virus in Mimulus guttatus (Phrymaceae): a test for costs and constraints. Heredity 96, 29–38. [DOI] [PubMed] [Google Scholar]
- 58.Simms EL, Rausher MD. 1987. Costs and benefits of plant resistance to herbivory. Am. Nat. 130, 570–581. ( 10.2307/2461704) [DOI] [Google Scholar]
- 59.Weyl EG, Frederickson ME, Yu DW, Pierce NE. 2011. Reply to Kiers et al.: economic and biological clarity in the theory of mutualism. Proc. Natl Acad. Sci. USA 108, E8 ( 10.1073/pnas.1015734108) [DOI] [Google Scholar]
- 60.De Mita S, Ronfort J, McKhann HI, Poncet C, El Malki R, Bataillon T. 2007. Investigation of the demographic and selective forces shaping the nucleotide diversity of genes involved in nod factor signaling in Medicago truncatula. Genetics 177, 2123–2133. ( 10.1534/genetics.107.076943) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Mita S, Santoni S, Hochu I, Ronfort J, Bataillon T. 2006. Molecular evolution and positive selection of the symbiotic gene NORK in Medicago truncatula. J. Mol. Evol. 62, 234–244. ( 10.1007/s00239-004-0367-2) [DOI] [PubMed] [Google Scholar]
- 62.Liu J, Yang S, Zheng Q, Zhu H. 2014. Identification of a dominant gene in Medicago truncatula that restricts nodulation by Sinorhizobium meliloti strain Rm41. BMC Plant Biol. 14, 167 ( 10.1186/1471-2229-14-167) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Broghammer A, et al. 2012. Legume receptors perceive the Rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc. Natl Acad. Sci. USA 109, 13 859–13 864. ( 10.1073/pnas.1205171109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kiers ET, Denison RF. 2008. Sanctions, cooperation, and the stability of plant–rhizosphere mutualisms. Ann. Rev. Ecol. Evol. Syst. 39, 215–236. ( 10.1146/annurev.ecolsys.39.110707.173423) [DOI] [Google Scholar]
- 65.Heath KD, Stock AJ, Stinchcombe JR. 2010. Mutualism variation in the nodulation response to nitrate. J. Evol. Biol. 23, 2494–2500. ( 10.1111/j.1420-9101.2010.02092.x) [DOI] [PubMed] [Google Scholar]
- 66.Ratcliff WC, Kadam SV, Denison RF. 2008. Poly-3-hydroxybutyrate (PHB) supports survival and reproduction in starving rhizobia. FEMS Microbiol. Ecol. 65, 391–399. ( 10.1111/j.1574-6941.2008.00544.x) [DOI] [PubMed] [Google Scholar]
- 67.Thompson JN. 1999. Specific hypotheses on the geographic mosaic of coevolution. Am. Nat. 153, S1–S14. ( 10.1086/303208) [DOI] [Google Scholar]
- 68.Grman E. 2012. Plant species differ in their ability to reduce allocation to non-beneficial Arbuscular mycorrhizal fungi. Ecology 93, 711–718. ( 10.1890/11-1358.1) [DOI] [PubMed] [Google Scholar]
- 69.Noë R, Hammerstein P. 1995. Biological markets. Trends Ecol. Evol. 10, 336–339. ( 10.1016/S0169-5347(00)89123-5) [DOI] [PubMed] [Google Scholar]
- 70.Akçay E, Roughgarden J. 2007. Negotiation of mutualism: rhizobia and legumes. Proc. R. Soc. B 274, 25–32. ( 10.1098/rspb.2006.3689) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.j2063. DNA sequences for bacterial strains used in this study: GenBank accession no. KF898182 (RB7) and KF898184 (T173).