Abstract
Sexual selection and sexual conflict are considered important drivers of speciation, based on both theoretical models and empirical correlations between sexually selected traits and diversification. However, whether reproductive isolation between species evolves directly as a consequence of intrapopulation sexual dynamics remains empirically unresolved, in part because knowledge of the genetic mechanisms (if any) connecting these processes is limited. Here, we provide evidence of a direct mechanistic link between intraspecies sexual selection and reproductive isolation. We examined genes with known roles in intraspecific sperm competition (ISC) in D. melanogaster and assayed their impact on conspecific sperm precedence (CSP). We found that two such genes (Acp36DE and CG9997) contribute to both offensive sperm competition and CSP; null/knockdown lines both had lower competitive ability against D. melanogaster conspecifics and were no longer able to displace heterospecific D. simulans sperm in competitive matings. In comparison, Sex Peptide (Acp70A)—another locus essential for ISC—does not contribute to CSP. These data indicate that two loci important for sperm competitive interactions have an additional role in similar interactions that enforce post-mating reproductive isolation between species, and show that sexual selection and sexual isolation can act on the same molecular targets in a gene-specific manner.
Keywords: sexual selection, reproductive isolation, sperm competition
1. Introduction
Substantial evidence suggests that sexual selection and conflict could be powerful drivers of speciation. Empirically, sister species are often differentiated by traits thought to evolve by sexual selection or conflict, and these traits are correlated with different rates of macroevolutionary diversification in a broad range of organisms [1–3]. Theoretically, models of speciation by sexual selection and sexual conflict suggest that traits involved in intrapopulation sexual dynamics can ultimately contribute to reproductive isolation [4,5]. Nonetheless, for intraspecific sexual selection to directly drive speciation requires that these two processes are mechanistically linked, but evidence of shared genetic mechanisms connecting these processes is limited [6]. Recent studies have uncovered quantitative trait loci (QTL) that control intraspecific mating differences as well as interspecies mating cues [7,8], but without knowledge of the underlying genes, the specific connection between these intraspecific mating traits and interspecific isolating barriers is unclear. As a result, whether reproductive isolation between species evolves directly as a consequence of intrapopulation sexual dynamics remains unresolved [9,10]. Here, we examine evidence for a direct mechanistic connection between intraspecific sperm competition (ISC) and post-copulatory interspecific reproductive isolation, via shared underlying genes.
Seminal fluid proteins associated with male success in sperm competition (hereafter called ‘intraspecific sperm competition’ or ‘ISC’ genes) in Drosophila melanogaster are among the best-characterized examples of molecules involved in sexual selection and sexual conflict. Their underlying genes are typically expressed in the male accessory gland (though other expression patterns have been observed; see [11,12]); however, they are known to influence female oviposition rate, female remating rate and female lifespan [13], and evidence for male × female genetic interactions at these loci is consistent with coevolution between the sexes [14]. Moreover, many of these seminal fluid proteins evolve rapidly, suggesting strong selection imposed by sexual selection and/or sexual conflict [15,16]. Rapidly evolving genes could result in elevated protein divergence between species, leading to reduced effectiveness of these proteins in heterospecific matings. These genes would then have larger relative roles in mediating reproductive isolation. Alternatively, rapid evolution could also represent strong selection to increase efficiency of sperm competition within a species, resulting in these genes having greater effects on outcompeting other sperm, regardless of whether they are conspecific or heterospecific. Given the role of sperm competition genes in intraspecies sexual interactions and their frequently rapid evolutionary rates, these genes might also be important for the development and expression of reproductive barriers between populations [17], particularly post-mating pre-zygotic reproductive barriers that are mediated by sperm competition and/or male–female interactions.
One such barrier is conspecific sperm precedence (CSP). CSP in Drosophila is the observation that when a female is inseminated by both conspecific and heterospecific males, the conspecific male sires the majority of the progeny, regardless of whether he is the first or second male to mate [18]. Therefore, CSP acts as a species barrier where post-mating interactions reduce the production of heterospecific (hybrid) individuals in favour of conspecific offspring. CSP is broadly observed among Drosophila species and other insects [19], and we confirmed its operation in crosses between D. melanogaster and D. simulans here. However, unlike genes involved in intraspecific sexual interactions, no specific genes have been identified for CSP.
Our goal in this study was to evaluate whether known ISC genes have a mechanistic role in CSP. Although there are hundreds of seminal fluid proteins, only a handful have been phenotypically characterized. We focused on three of the best-characterized genes: Sex Peptide (Acp70A) has been called a master regulator of female reproduction [20]; it is necessary for the efficient release of sperm from storage [21] and is responsible for the long-term mating response in females (i.e. reduced female receptivity, increased sperm release and oviposition [22,23]). CG9997 is required for the transfer of three other accessory gland proteins that localize and bind Sex Peptide to sperm, and when lacking also results in improper release of sperm from storage [24,25]. Acp36DE is known to contribute to male reproductive success via effects on correct sperm storage [26,27]. We confirmed the effects of these loci on ISC and then tested these genes for a role in CSP.
2. Material and methods
(a). Fly stocks and maintenance
All flies were maintained at room temperature under standard laboratory conditions. Flies were cultured in 8 dram glass vials on Bloomington media recipe food. Wild-type female D. melanogaster were from the Austria w132 line, originally collected by Christian Schlötterer and donated to us by Kristi Montooth (Indiana University). Green fluorescent protein (GFP)-marked D. melanogaster males and D. simulans males were ordered from the Bloomington Drosophila Stock Center (32170) and University of California Stock Center (14021-0251.263), respectively. GFP can be visualized in the pseudo-pupil and ocelli of both strains, and is a dominant marker. The creation of these lines is described by Holtzman et al. [28]. Null/knockdown lines for Sex Peptide, CG9997 and Acp36DE (and control Acp36DE) were generously provided by Mariana Wolfner (Cornell University). For Sex Peptide, null males and wild-type males were generated by crossing the SP null line (0325/TM3, Sb ry) to a deficiency line (Δ130/TM3, Sb ry) [22], resulting in null (0325/Δ130) and control (0325/TM3, Sb ry or Δ130/TM3, Sb ry) siblings. For Acp36DE, we used a null Acp36DE1 and control Acp36DE+. The null and control chromosomes are maintained by backcrossing every generation to CyO/Df(2L)H2O females (a deficiency that lacks the Acp36DE locus) [26]. This backcrossing generated null (Acp36DE1/Df(2L)H2O) and control (Acp36DE+/Df(2L)H2O) males. For CG9997, we crossed a sympUAST-CG9997 line to tubulin-GAL4/TM3, Sb flies to generate the knockdown males (tubulin-GAL4/UAS-CG9997-UAS) and wild-type males (TM3, Sb/UAS-CG9997-UAS) [25]. The sympUAST-CG9997 line was donated by Mariana Wolfner, and the tubulin-GAL4 line was ordered from the Bloomington Drosophila Stock Center (5138). The CG9997 RNAi construct was previously determined to be efficient [24,25]. Because our results reiterated the described phenotypic effect of RNAi (reduced sperm competitive ability in conspecific crosses, specifically in the RNAi line but not control line), we did not further test the efficacy of RNAi for this locus.
(b). Competitive mating experimental procedure
We completed a minimum of 15 replicates of each (conspecific-GFP or heterospecific-GFP) × (null/knockdown or control) combination for a total of 60 replicates for each locus. We focused on offensive sperm competitive ability for each locus because, after mating with a conspecific D. melanogaster male, D. melanogaster females strongly reject heterospecific D. simulans males; this behaviour precludes an analysis of crosses where D. simulans is the second male (i.e. defensive ability in D. melanogaster)
(i) Day 0: virgin D. melanogaster females were collected and maintained in groups of 5 to age for 3–5 days prior to the first mating. This ensures that they are reproductively mature and will readily mate, and represents the beginning of the experiment. Experimental null/knockdown males, control males and GFP males were also collected as virgins and stored in the groups of 5 for 3–5 days.
(ii) Day 1: five D. melanogaster females were transferred to a new vial with five GFP males (either D. melanogaster or D. simulans depending on whether the treatment was ISC or CSP, respectively) without anaesthesia and allowed to mate for 3 days. This design maximizes the number of heterospecifically mated females for use in the remaining experiment. In preliminary data collection, it was observed that mass matings resulted in a higher number of mated females compared with single-pair matings. In addition, shorter mating times resulted in very poor average mating success, owing to premating behavioural isolation between the species.
(iii) Day 4: females were transferred individually to a fresh vial of food and allowed to oviposit for 1 day. In general, D. melanogaster females refuse to remate within 24 h [29,30], so isolating females increases the chances that they will readily mate with the second male. Isolating females for longer periods of time can result in use of all the sperm from the first male, and the effect we would observe would not be due to sperm competition. Females that did not lay eggs when isolated for one day were discarded, as these females are assumed not to have mated with the GFP-marked male.
(iv) Day 5: females were transferred to a fresh vial, to which a single experimental or control male was added. (In preliminary data collection, we observed matings at this time point and found that all females had remated with the second male within 2 h, regardless of genotype; therefore, we did not continue to watch matings for the remainder of the experiment). These pairings were maintained and transferred daily until day 7, when all flies were discarded. Daily transfers reduce crowding in a given vial, so that hybrid progeny will not be absent owing to larval competition. After adults were removed, cotton substrate was added to day 5–7 vials to encourage pupation of hybrid individuals because D. simulans (and to some extent hybrids) make pupal mats rather than climb up the side of the glass vials.
(v) Scoring progeny: vials of days 4–7 were kept until all progeny had eclosed. As progeny began to emerge they were sexed and scored under UV light for the presence of the GFP (visible in pseudo-pupil and ocelli). The phenotype used in the data analysis is the proportion of GFP progeny in vials from day 5 to day 7. For conspecific matings we used the total number of progeny and for heterospecific matings, we restricted the proportion to the number of females, because male hybrids are inviable (results were not different if we used proportion of females for both CSP and ISC assays). We excluded any replicates where we could not ensure that the first male had mated (i.e. where no progeny were observed in the day 4 vial when the female was isolated after first mating) or that the second male had mated (i.e. where all progeny were GFP). Within these constraints, we were still able to maintain a sample size of 15 replicates per treatment. To confirm that CSP operates between D. melanogaster and D. simulans, we also performed this experiment (with the same replication and quality controls) using wild-type D. simulans as the first male and D. melanogaster-GFP as the second male.
(c). Statistical analysis
Differences in competitive ability between null and wild-type males were compared using a logit model in R:
The binomial response variable was the number of green-eyed progeny observed compared with the number of wild-type progeny observed, as this represents the magnitude of the sperm competitive ability of the first male versus the second male, regardless of whether the first male is conspecific or heterospecific. The variable x1 is a binary variable describing whether the second male to mate was null/knockdown or wild-type (x1 = 0 for wild-type males), and β1 is the log-odds describing the difference in competitive ability between null/knockdown and wild-type males. The variable x2 is a binary variable describing whether the GFP-marked male is conspecific or heterospecific (x2 = 0 for D. melanogaster-GFP males) and β2 is the log-odds for the difference in competing against conspecific versus heterospecific males. The last variable is also a binary variable, describing the interaction between the first two variables (x1x2 = 1 for null/knockdown males competing with D. simulans GFP males), and β3 describes the log-odds for the interaction term.
The differences in the total number of progeny produced between crosses that involved control males and crosses that involved null/knockdown males were compared using a linear model similar to the model above in R. In this model, the response variable was the total number of progeny produced after the second mating, and we were primarily interested in the significance of β1 (which we refer to as βnull to reduce redundancy). The differences in oviposition and total number of progeny produced after mating with only a control or null genotype for Acp36DE over 3 days (analogous to days 5–7 described above, but without a first mating having had occurred) were compared using a Wilcoxon rank-sum test in R. For these data, we used eight replicates for each genotype and counted eggs in each vial after the female and male pair were transferred without aspiration to a new vial.
(d). Measures of molecular evolution
Nucleotide alignments of the coding regions (CDS) for all genes identified as members of the Sex Peptide network [31] and Acp36DE were downloaded from UCSC genome browser. The species and species tree we used in the analysis were (D. melanogaster (D. simulans, D. sechellia)). A single dN/dS value was estimated for the entire unrooted tree using PAML (M0 model) [32]. Besides providing the phylogeny, all other options were left as default. Pairwise comparisons of the total number of non-synonymous and synonymous changes were made for all genes using the Nei–Gojobori method for just D. melanogaster and D. simulans. This was completed using MEGA v. 6 [33].
3. Results and discussion
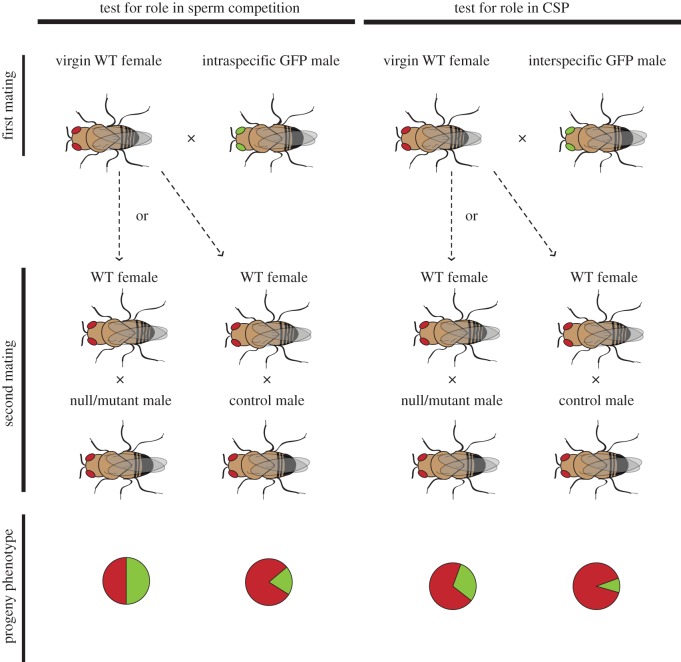
Using sperm-competition assays, we assessed the sperm competitive ability of null/knockdown males versus wild-type individuals at each locus (figure 1). Sperm competition assays involve mating a female sequentially to males of two different genotypes, to determine the ability of a given male genotype either to resist displacement by the second male (defensive ability) or to displace the first male's sperm (offensive ability); competitive ability is revealed in the relative siring success of each male. This same design can be used to assess both ISC and CSP, simply by varying whether the first mating involves conspecific or heterospecific males, respectively (figure 1). In our experiments, either null/knockdown or wild-type (control) males were the second males to mate following a first mating with either a wild-type (GFP) conspecific male (to confirm a role in ISC) or a wild-type heterospecific (GFP) male (to assess CSP). Our assays therefore assess the offensive ability of each null/knockdown male in comparison with conspecific wild-type males. Using GFP-marked first males allows us to quantify progeny siring rates directly in offspring using this dominant marker.
Figure 1.
Experimental design of the sperm competition assay. The same design is used to test for a role in ISC and CSP, by varying the identity of the first mated (GFP) male. The response phenotype is the proportion of progeny with green eyes; schematic expectations for each case are represented with pie charts. For genes involved in ISC (left-hand side), females are expected to produce proportionately more GFP offspring when mated to null/knockdown second males, compared with wild-type second males, as these individuals are less able to displace first male (GFP) sperm (owing to reduced offensive competitive ability). For genes involved in CSP (right-hand side), the same pattern is expected to hold, although the total proportions of green-eyed progeny are expected to be less than the respective crosses assessing ISC because of reduced compatibility in heterospecific crosses. (Online version in colour.)
We found that two ISC loci also had large and significant effects on the magnitude of CSP (figure 2). Acp36DE is known to contribute to both defensive and offensive ability; in our offensive assay, null males were less successful competitors than wild-type males when competing against conspecific males, confirming previous results (β1 = 0.9409, p < 0.0001; figure 2a). Importantly, null males were also less successful than wild-type males when competing against heterospecific males (β2 = −2.9463, p < 0.0001; figure 2a). The effect of Acp36DE on CSP was also greater than the effect on ISC (β3 = 0.7644, p < 0.0001); that is, the magnitude of the difference between null versus wild-type males is greater when measuring interspecific than ISC. Similar to Acp36DE, CG9997 contributed both to ISC and CSP. When competing against conspecific males, knockdown males were less successful at displacing sperm than wild-type males (β1 = 0.3465, p < 0.0001; figure 2b). This pattern was also seen for knockdown versus wild-type males when competing against heterospecific males (β2 = −1.7399, p < 0.0001), and again the magnitude of the effect is greater for CSP compared with ISC (β3 = 0.4205, p = 0.0064; figure 2b).
Figure 2.
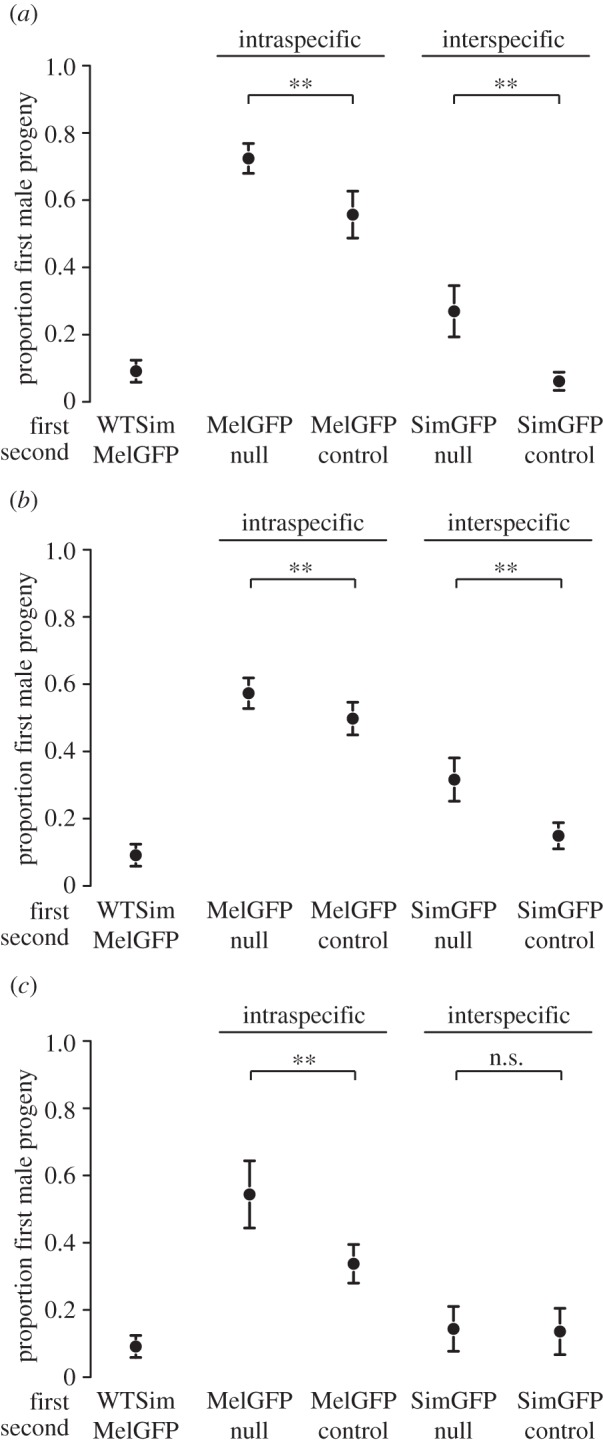
The ability of null/knockdown versus wild-type males to displace competitor sperm when the competitor is either conspecific or heterospecific for each locus we examined. (a) Acp36DE, (b) CG9997 and (c) Sex Peptide. Every panel depicts the control: wild-type D. simulans competing against GFP D. melanogaster. Each point represents mean ± s.e. proportion of 15 replicates. Asterisk designates a null/knockdown versus wild-type male comparison that is significant from the logistic regression. n.s. stands for not significant.
Unlike these two loci, Sex Peptide did not affect the magnitude of CSP, although it had an effect on sperm offensive ability among conspecifics (figure 2c). Compared with wild-type males, null Sex Peptide males were less successful at displacing conspecific sperm (β1 = 0.9556, p < 0.0001). By contrast, wild-type and null males both successfully displaced sperm from heterospecific males (β2 = −1.5036, p < 0.0001 and β3 = −0.9612, p < 0.0001, respectively; figure 2c). Although Sex Peptide is better known for its defensive role in conspecific matings [14,21], which we also confirmed here (electronic supplementary material, figure S1), our analysis shows that it also plays a clear role in offensive ISC, but does not contribute to CSP-mediated reproductive isolation through offensive ability.
Differences in fertility or in differences in mating frequency/copulation intensity between control and null/knockdown males could result in spurious observation of differences in competitive ability. We evaluated whether control and null/knockdown males differed in fecundity by comparing the total number of progeny produced for the competitive crosses containing control males versus null/knockdown males (electronic supplementary material, figures S2–S4). For CG9997 and Sex Peptide, there was no difference in the number of progeny produced between control and null crosses (βnull = 11.467; p = 0.307 for CG9997, and βnull = −1.867; p = 0.866 for Sex Peptide). For Acp36DE, crosses involving the null male did produce fewer progeny (βnull = −26.628; p = 0.0129), similar to previous reports that also analysed sperm competitive ability for this locus [26], but this reduction was observed regardless of whether this male was competing with a heterospecific or conspecific male. Because we could not disentangle fecundity effects from competitive effects for Acp36DE, we also looked at differences in oviposition and progeny production over 3 days after single (non-competitive) matings with either control or null genotypes for this locus. This time frame is identical to the time frame of our experimental design. We observed no difference between number of eggs laid over the 3 days (W = 31.5; p = 1.00) or progeny produced (W = 35; p = 0.751) between the null and control Acp36DE male genotypes (electronic supplementary material, figures S5–S6).
If mating frequency and intensity is correlated with progeny production, these data also suggest that there was no difference in mating frequency between null/knockdown and control males. Interestingly, previous studies have demonstrated that females that mate with Sex Peptide null males remate more quickly in comparison with females initially mated to control males [23]; remating rate might therefore be increased using null Sex Peptide males. Because CG9997 is similarly thought to play a role in remating rate and the long-term response, similar logic could also apply for this locus. In either case, this would result in increased matings by null/knockdown males compared with control males, which could not explain why null/knockdown males specifically performed poorly in CSP assays. We therefore infer that differences in fertility and mating frequency of control versus null/knockdown males are unlikely to explain the observed involvement of CG9997 and Acp36DE in CSP.
To examine the evolutionary dynamics of Acp36DE, CG9997 and Sex Peptide, we examined sequence data for each locus and for 11 additional genes known to be involved in the D. melanogaster ‘sex peptide network’ of which Sex Peptide and CG9997 (but not Acp36DE) are known members [31]. Of the 14 genes, our three focal loci had relatively high values of dN (the number of non-synonymous substitutions per non-synonymous site; table 1), reflective of high relative rates of protein-changing evolution. However, there was no obvious distinction between the values of dN observed at the two genes found to affect CSP (CG9997 and Acp36DE) and the one that did not (Sex Peptide; table 1). Likewise, although the values of dN/dS (which can provide evidence for positive selection between species when dN/dS is >1) were uniformly higher in all 14 loci compared with the D. melanogaster (D. simulans) genome-wide average of 0.07 (0.11) [34], these estimates of protein-coding evolution do not clearly differentiate CG9997 and Acp36DE from Sex Peptide (table 1).
Table 1.
Estimates of dN, dS, and dN/dS for genes in the sex peptide network and Acp36DE using D. melanogaster, D. simulans and D. sechellia, with the number of codons, non-synonymous changes (N) and synonymous changes (S) from a pairwise comparison of D. melanogaster and D. simulans. Asterisk denotes genes examined in this study.
| gene | chromosome position | dN | dS | dN/dS | codons | N | S |
|---|---|---|---|---|---|---|---|
| CG1652 | chr2R:5703015–5704105 | 0.1258 | 0.4172 | 0.3014 | 323 | 90.083 | 58.917 |
| CG3239 | chrX:5387007–5390082 | 0.0887 | 0.2618 | 0.3387 | 842 | 101 | 71 |
| Acp36DE* | chr2L:18356047–18359026 | 0.0831 | 0.1697 | 0.4895 | 913 | 95.5 | 79.5 |
| Sex Peptide* | chr3L:13294735–13295022 | 0.0599 | 0.2228 | 0.2686 | 56 | 4 | 4 |
| CG30488 | chr2R:8734353–8735509 | 0.0491 | 0.2205 | 0.2226 | 266 | 28.167 | 27.833 |
| CG12558 | chr3R:24616244–24617183 | 0.0422 | 0.1804 | 0.2338 | 299 | 17 | 25 |
| CG9997* | chr3R:24581206–24582701 | 0.0382 | 0.1196 | 0.3191 | 331 | 26 | 20 |
| Seminase | chr3L:20948855–20949850 | 0.0374 | 0.1135 | 0.3296 | 276 | 21 | 18 |
| CG5630 | chr3R:16740018–16754935 | 0.0330 | 0.1910 | 0.1727 | 378 | 26.5 | 31.5 |
| CG14061 | chr3R:24582721–24583956 | 0.0258 | 0.1841 | 0.1403 | 367 | 17 | 33 |
| CG1656 | chr2R:5704805–5706040 | 0.0177 | 0.1390 | 0.1272 | 329 | 10.5 | 17.5 |
| Esp | chr3R:20667657–20676134 | 0.0048 | 0.1530 | 0.0315 | 655 | 5 | 50 |
| CG17575 | chr2R:8733049–8734101 | 0.0040 | 0.1991 | 0.0219 | 292 | 1 | 21 |
| SPR | chrX:5340689–5388045 | 0.0039 | 0.3281 | 0.0117 | 436 | 4 | 59 |
Overall, our analyses support several new findings. First, they demonstrate that CG9997 and Sex Peptide play a role in offensive sperm competition (previous analyses of these loci only focused on defensive sperm competition). Second, they uncover two genes that are mechanistically involved in CSP. Although studies have previously mapped QTL for CSP [35,36], no genes have yet been identified for this widespread post-mating pre-zygotic reproductive isolating barrier. (Note that neither Acp36DE nor CG9997, nor any other ‘male gene’ members of the Sex Peptide network, co-localize with the described CSP QTL in Drosophila [36]). Second, because Acp36DE and CG9997 are known actors in sperm competition and sperm storage within D. melanogaster, their involvement in CSP provides evidence for a direct mechanistic connection between conspecific and heterospecific competitive sexual interactions, via the same causative loci. Previous studies have linked sexually selected traits with reproductive isolation by demonstrating that specific phenotypes are associated both with sexual selection and reproductive isolation [37,38], although the genetics of these traits remain unknown. Other studies have deduced that genomic regions (QTL) associated with reproductive isolation are also sexually selected; however, the contribution of sexual selection is inferred (generally via assortative mating), rather than demonstrated directly [39,40]. Nonetheless, for models of speciation via sexual selection to be plausible, at least one locus must be demonstrably involved in both sexual selection and reproductive isolation. Here, we have identified two such loci.
Although our data show that specific molecular players can mediate both ISC and CSP, not all ISC genes appear to be involved in CSP, at least via the mechanisms we probably observed here (sperm storage, retention and release from storage). (Note, however, that these genes could still contribute to CSP through their effects on female remating rate, particularly as remating sets the stage for CSP.) Differences among genes observed here could be explained by differences in their evolutionary rates of change, and/or in the magnitude of pleiotropy experienced by these genes. In the first case, genes that are more rapidly evolving could be more likely to contribute to CSP because they have increased functional divergence between species. However, we find no clear differences among our three focal genes in common measures of protein evolution (table 1), suggesting that differences in the rates of protein evolution per se are not responsible for differential involvement of these ISC genes in CSP. In the second case, genes with smaller pleiotropic effects might be less constrained, therefore more responsive to selection for increased reproductive isolation, and correspondingly more important for CSP. Although all three genes studied here are required for effective sperm competition, Sex Peptide is known to be involved in at least nine different phenotypes [20], whereas Acp36DE is only known to act on sperm accumulation [26], and CG9997 is only known to be required for the transfer of certain accessory gland proteins into females [25]. Therefore, current data suggest that the locus without an effect on CSP (Sex Peptide) is a highly pleiotropic gene, whereas the other two loci are less pleiotropic by comparison, possibly contributing to the differential involvement of these genes in CSP.
Finally, if the functional significance of some but not all post-mating pre-fertilization mechanisms differs in the context of sexual selection versus reproductive isolation, this could explain the differential involvement of specific sperm competition genes in CSP. Our observations that Acp36DE and CG9997 clearly contribute to both ISC and CSP indicate that their functional roles in proper sperm storage and utilization [24,26] are important mechanisms for both ISC and CSP. This inference is consistent with recent studies in D. melanogaster, D. simulans and D. mauritiana that suggest that siring success in both ISC and CSP is determined by female control of sperm use and displacement, although the specific patterns of use and displacement are different in the two reproductive contexts [41,42]. By contrast, our finding that Sex Peptide does not contribute to differences in siring success in CSP indicates that the specific functional roles of this critical ISC gene are less important in the altered reproductive context of CSP. Our findings might also suggest additional, previously undescribed functional roles for CG9997 and Acp36DE, specifically in the context of CSP. For example, the known role of CG9997 is to transfer other seminal fluid proteins into the reproductive tract, which then bind Sex Peptide to sperm [25]. Sex Peptide and CG9997 are therefore mechanistically linked, and they both show similar sperm release phenotypes in intraspecific matings [25,21], which is consistent with our ISC results. However, our finding that CG9997, but not Sex Peptide, has a phenotypic effect on CSP suggests that CG9997 or the proteins it helps transfer have additional roles not directly linked to the function of Sex Peptide, at least in the alternative reproductive context of interspecific matings. These inferences can be more directly tested with further functional data in the future.
4. Conclusion
Seminal fluid loci are strong a priori candidates for genes that could play dual roles in intraspecific sexual interactions and interspecific reproductive isolation. They are critical for sexual selection, frequently evolve rapidly and have functions that appear to be phenotypically and behaviourally similar to those operating during post-mating pre-zygotic sexual isolation. Although it remains to be seen how many other sperm competition proteins are similarly involved in species reproductive barriers, our data confirm that at least two known D. melanogaster seminal fluid proteins do have a second function in sexual isolation. Accordingly, these findings confirm that a necessary condition for sexual selection/conflict to drive sexual isolation—a shared genetic basis for these processes—is met for ISC and CSP. Further studies linking these two processes will allow us to better understand how reproductive isolation is facilitated or constrained by sexual selection, and which loci are unique versus shared between these processes. Although sexual selection and sexual conflict are often discussed as important drivers of speciation, this broader understanding of the prevalence of direct genetic connections between these processes is essential for evaluating their relative importance compared with other drivers of isolation and diversification.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
Null/knockdown lines for Sex Peptide, CG9997 and Acp36DE, and the Acp36DE control line, were generously provided by Mariana Wolfner (Cornell University). The wild-type (Austria w132) female D. melanogaster line was donated by Kristi Montooth (Indiana University). M. W. Hahn and two anonymous reviewers provided helpful comments on the manuscript.
Data accessibility
All data used in the study are available as an electronic supplementary material file.
Funding statement
Research was supported by Indiana University Department of Biology funding to L.C.M., and a National Science Foundation Graduate Research Fellowship to D.M.C.
References
- 1.Arnqvist G, Edvardson M, Friberg U, Nilsson T. 2000. Sexual conflict promotes speciation in insects. Proc. Natl Acad. Sci. USA 97, 10 460–10 464. ( 10.1073/pnas.97.19.10460) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Panhuis TM, Butlin R, Zuk M, Tregenza T. 2001. Sexual selection and speciation. Trends Ecol. Evol. 16, 364–371. ( 10.1016/S0169-5347(01)02160-7) [DOI] [PubMed] [Google Scholar]
- 3.Ritchie MG. 2007. Sexual selection and speciation. Annu. Rev. Ecol. Evol. Syst. 38, 79–102. ( 10.1146/annurev.ecolsys.38.091206.095733) [DOI] [Google Scholar]
- 4.Kirkpatrick M, Ravigne V. 2002. Speciation by natural and sexual selection: models and experiments. Am. Nat. 159, S22–S35. ( 10.1086/338370) [DOI] [PubMed] [Google Scholar]
- 5.Gavrilets S, Hayashi TI. 2005. Speciation and sexual conflict. Evol. Ecol. 19, 167–198. ( 10.1007/s10682-004-7916-4) [DOI] [Google Scholar]
- 6.Chenoweth SF, McGuigan K. 2010. The genetic basis of sexually selected variation. Annu. Rev. Ecol. Evol. Syst. 41, 81–101. ( 10.1146/annurev-ecolsys-102209-144657) [DOI] [Google Scholar]
- 7.Arbuthnott D. 2009. The genetic architechture of insect sourtship behavior and premating isolation. Heredity 103, 15–22. ( 10.1038/hdy.2009.22) [DOI] [PubMed] [Google Scholar]
- 8.Groot AT, et al. 2013. One quantitative trait locus for intra- and interspecific variation in a sex pheromone. Mol. Ecol. 22, 1065–1080. ( 10.1111/mec.12171) [DOI] [PubMed] [Google Scholar]
- 9.Ryan MJ, Rand AS. 1993. Species recognition and sexual selection as a unitary problem in animal communication. Evolution 47, 647–657. ( 10.2307/2410076) [DOI] [PubMed] [Google Scholar]
- 10.Bolnick DI, Kirkpatrick M. 2012. The relationship between intraspecific assortative mating and reproductive isolation between divergent populations. Curr. Zool. 58, 484–492. [Google Scholar]
- 11.Findlay GD, Yi X, MacCoss MJ, Swanson WJ. 2008. Proteomics reveals novel Drosophila seminal fluid proteins transferred at mating. PLoS Biol. 6, e178 ( 10.1371/journal.pbio.0060178) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Findlay GD, MacCoss MJ, Swanson WJ. 2009. Proteomic discovery of previously unannotated, rapidly evolving seminal fluid genes in Drosophila. Genome Res. 19, 886–896. ( 10.1101/gr.089391.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sirot LK, LaFlamme BA, Sitnik JL, Rubinstein CD, Avila FW, Chow CY, Wolfner MF. 2009. Molecular social interactions: Drosophila melanogaster seminal fluid proteins as a case study. In Socio-genetics (ed. Sokolowski MB.), pp. 23–56. Waltham, MA: Academic Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chow CY, Wolfner MF, Clark AG. 2001. The genetic basis for male x female interactions underlying variation in reproductive phenotypes of Drosophila. Genetics 186, 1355–1365. ( 10.1534/genetics.110.123174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Begun DJ, Whitley P, Todd BL, Waldrip-Dail HM, Clark AG. 2000. Molecular population genetics of male accessory gland proteins in Drosophila. Genetics 156, 1879–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong A, Turchin MC, Wolfner MF, Aquadro CF. 2008. Evidence for positive selection on Drosophila melanogaster seminal fluid protease homologs. Mol. Biol. Evol. 25, 497–506. ( 10.1093/molbev/msm270) [DOI] [PubMed] [Google Scholar]
- 17.Snook RR, Chapman T, Moore PJ, Wedell N, Crudgington HS. 2009. Interactions between the sexes: new perspectives on sexual selection and reproductive isolation. Evol. Ecol. 23, 71–91. ( 10.1007/s10682-007-9215-3) [DOI] [Google Scholar]
- 18.Price CSC. 1997. Conspecific sperm precedence in Drosophila. Nature 388, 663–666. ( 10.1038/41753) [DOI] [PubMed] [Google Scholar]
- 19.Howard DJ. 1999. Conspecific sperm and pollen precedence and speciation. Annu. Rev. Ecol. Evol. Syst. 30, 109–132. ( 10.1146/annurev.ecolsys.30.1.109) [DOI] [Google Scholar]
- 20.Gioti A, Wigby S, Wertheim B, Schuster E, Martinez P, Pennington CJ, Partridge L, Chapman T. 2012. Sex peptide of Drosophila melanogaster males is a global regulator of reproductive processes in females. Proc. R. Soc. B 279, 4423–4432. ( 10.1098/rspb.2012.1634) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Avila FW, Ravi Ram K, Qazi MCB, Wolfner MF. 2010. Sex peptide is required for the efficient release of stored sperm in mated Drosophila females. Genetics 186, 595–600. ( 10.1534/genetics.110.119735) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu H, Kubli E. 2003. Sex-peptide is the molecular basis of the sperm effect in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 100, 9929–9933. ( 10.1073/pnas.1631700100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chapman T, Bangham J, Vinti G, Seifried B, Lung O, Wolfner MF, Smith HK, Partridge L. 2003. The sex peptide of Drosophila melanogaster: female post-mating responses analyzed by using RNA interference. Proc. Natl Acad. Sci. USA 100, 9923–9928. ( 10.1073/pnas.1631635100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ravi Ram K, Wolfner MF. 2007. Sustained post-mating response in Drosophila melanogaster requires multiple seminal fluid proteins. PLoS Genet. 3, e238 ( 10.1371/journal.pgen.0030238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ravi Ram K, Wolfner MF. 2009. A network of interactions among seminal proteins underlies the long-term postmating response in Drosophila. Proc. Natl Acad. Sci. USA 106, 15 384–15 389. ( 10.1073/pnas.0902923106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neubaum DM, Wolfner MF. 1999. Mated Drosophila melanogaster females require a seminal fluid protein, Acp36DE, to store sperm efficiently. Genetics 153, 845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chapman T, Neubaum DM, Wolfner MF, Partridge L. 2000. The role of male accessory gland protein Acp36DE in sperm competition in Drosophila melanogaster. Proc. R. Soc. Lond. B 267, 1097–1105. ( 10.1098/rspb.2000.1114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holtzman S, Miller D, Eisman R, Kuwayama H, Niimi T, Kaufman T. 2010. Transgenic tools for members of the genus Drosophila with sequenced genomes. Fly 4, 349–362. ( 10.4161/fly.4.4.13304) [DOI] [PubMed] [Google Scholar]
- 29.Pyle DW, Gromko MH. 1981. Genetic basis for repeated mating in Drosophila melanogaster. Am. Nat. 117, 133–146. ( 10.1086/283694) [DOI] [Google Scholar]
- 30.Markow TA, O'Grady PM. 2005. Drosophila: a guide to species identification and use, 1st edn Waltham, MA: Academic Press. [Google Scholar]
- 31.Findlay GD, Sitnik JL, Wang W, Aquadro CF, Clark NL, Wolfner MF. 2014. Evolutionary rate covariation identifies new members of a protein network required for Drosophila melanogaster female post-mating responses. PLoS Genet. 10, e1004108 ( 10.1371/journal.pgen.1004108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. ( 10.1093/molbev/msm088) [DOI] [PubMed] [Google Scholar]
- 33.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. ( 10.1093/molbev/mst197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heger A, Ponting CP. 2007. Evolutionary rate analyses of orthologs and paralogs from 12 Drosophila genomes. Genome Res. 17, 1837–1849. ( 10.1101/gr.6249707) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Britch SC, Swartout EJ, Hampton DD, Draney ML, Chu J, Marshall JL, Howard DJ. 2007. Genetic architecture of conspecific sperm precedence in Allonemobius fasciatus and A. socius. Genetics 176, 1209–1222. ( 10.1534/genetics.106.064949) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levesque L, Brouwers B, Sundararajan V, Civetta A. 2010. Third chromosome candidate genes for conspecific sperm precedence between D. simulans and D. mauritiana. BMC Genet. 11, 12 ( 10.1186/1471-2156-11-21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiernasz DC, Kingsolver JG. 1991. Wing melanin pattern mediates intraspecific mate choice and species recognition in Pontia occidentalis. Anim. Behav. 42, 276–280. [Google Scholar]
- 38.Williams TH, Gumm JM, Mendelson TC. 2013. Sexual selection acting on a speciation trait in darters (Percidae: Etheostoma). Behav. Ecol. 24, 1407–1414. ( 10.1093/beheco/art080) [DOI] [Google Scholar]
- 39.Sæther SA, et al. 2007. Sex chromosome-linked species recognition and evolution of reproductive isolation in flycatchers. Science 318, 95–97. ( 10.1126/science.1141506) [DOI] [PubMed] [Google Scholar]
- 40.Parnell NF, Streelman JT. 2013. Genetic interactions controlling sex and color establish the potential for sexual conflict in Lake Malawi cichlid fishes. Heredity 110, 239–246. ( 10.1038/hdy.2012.73) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manier MK, Belote JM, Berben KS, Lüpold S, Ala-Honkola O, Collins WF, Pitnick S. 2013. Rapid diversification of sperm precedence traits and processes among three sibling Drosophila species. Evolution 67, 2348–2362. ( 10.1111/evo.12117) [DOI] [PubMed] [Google Scholar]
- 42.Manier MK, Lüpold S, Belote JM, Starmer WT, Berben KS, Ala-Honkola O, Collins WF, Pitnick S. 2013. Postcopulatory sexual selection generates speciation phenotypes in Drosophila. Curr. Biol. 23, 1853–1862. ( 10.1016/j.cub.2013.07.086) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in the study are available as an electronic supplementary material file.