Abstract
Developmental responses to nutritional variation represent one of the ecologically most important classes of adaptive plasticity. However, knowledge of genome-wide patterns of nutrition-responsive gene expression is limited. Here, we studied genome-wide transcriptional responses to nutritional variation and their dependency on trait and sex in the beetle Onthophagus taurus. We find that averaged across the transcriptome, nutrition contributes less to overall variation in gene expression than do sex or body region, but that for a modest subset of genes nutrition is by far the most important determinant of expression variation. Furthermore, our results reject the hypothesis that a common machinery may underlie nutrition-sensitive development across body regions. Instead, we find that magnitude (measured by number of differentially expressed contigs), composition (measured by functional enrichment) and evolutionary consequences (measured by patterns of sequence variation) are heavily dependent on exactly which body region is considered and the degree of sexual dimorphism observed on a morphological level. More generally, our findings illustrate that studies into the developmental mechanisms and evolutionary consequences of nutrition-biased gene expression must take into account the dynamics and complexities imposed by other sources of variation in gene expression such as sexual dimorphism and trait type.
Keywords: nutritional variation, polyphenism, condition, developmental plasticity, horned beetles, square combining table
1. Introduction
Differential gene expression is generally viewed as a central mechanism enabling context-specific development, such as tissue-specific growth, sexual differentiation or facultative responses to changes in external conditions, such as nutrient availability. Nutritional influences on gene expression are ubiquitous across taxa, genes and tissue types, and are thought to play a critical role in allowing organisms to adjust their development and physiology in response to environmental fluctuations [1–3]. Moreover, dysfunctional nutrition-dependent gene expression is thought to underlie many human diseases such as diabetes, obesity, Crohn's disease and possibly even cancer [4–8]. However, the transcriptome-wide magnitude and nature of nutritional plasticity in gene expression remain largely understudied for non-model systems, many of which exhibit remarkable nutrition-dependent developmental plasticity. Furthermore, the relative effects of nutrition-induced variation in gene expression, compared to the effects from other sources of variation such as tissue- or sex-based expression, are largely unclear for most organisms.
Obtaining such insights is especially important for understanding the evolution of morphological diversity in multicellular and sexually reproducing organisms. Here, individuals essentially constitute mosaics of body regions, organs and tissue types that differ, at times dramatically, in how their development is affected by nutrition [9–11]. While developmental genomic studies in social insects have yielded important insights into how genes involved in metabolism and reproduction have been co-opted in caste determination and differentiation [11–13], we still know little about how tissue- and sex-specific differences in nutritional responsiveness enable phenotypic diversification outside social insects and in organisms with more conventional forms of plasticity [14]. Lastly, tissue-, sex- and nutrition (environment)-specific gene expression all have important, but in part contrasting, evolutionary implications. On the one hand, tissue-specific expression is thought to limit pleiotropic constraints, resulting in higher rates of evolution and protein divergence owing to relaxed selective constraints [15–18]. On the other hand, genes whose expression is limited to a subset of individuals in the population are predicted to result in reduced selection intensity, hereon referred to as relaxed selection, relative to genes expressed in all individuals [19–21]. Genes with such environment-biased expression are predicted to harbour more genetic variation and fix more deleterious mutations over time. Thus, knowledge of the degree to which the nature and magnitude of different kinds of context-specific expression co-occurring in the same population are distinct or similar is critical for evaluating how they might combine to affect subsequent evolutionary trajectories [20].
Such knowledge is also central in evaluating nutrition's role in affecting individual condition, which can be defined as the ability to produce and maintain fitness-relevant traits [22–24]. Nutritional variation is one of the most significant sources of variation in condition, and condition-dependency is thought to impact diverse and fundamental evolutionary processes, including adaptation to novel environments and range expansions [25–28], the evolution of sex and sexual dimorphisms [29–31] or the maintenance of variation in sexually selected traits [22,23,30,32,33]. Theory predicts that sexually selected traits such as elaborate displays or weaponry should evolve condition-dependence if the fitness gains associated with trait exaggeration are greater for high—compared to low-condition individuals. In contrast, traits experiencing stabilizing or weak selection should exhibit minimal condition-dependence, because, in these cases, trait exaggeration should not result in increased fitness [24]. Morphological data across a wide range of taxa generally corroborate these predictions (e.g. birds: [34,35]; flies: [30,36]; beetles: [37]; fish: [38]; crustaceans: [39]), and at least one study on fruit flies has been able to demonstrated that high-condition populations are more sexually dimorphic in transcription than low-condition populations [40]. However, more precise knowledge of the transcriptomic underpinnings of condition-dependence is lacking [24]. Specifically, it remains unclear which developmental genetic mechanisms translate variation in nutrition into variation in condition, their similarities and differences across naturally and sexually selected traits or the overlap in developmental genetic mechanisms that are responsive to sex and condition.
In this study, we sought to address several fundamental questions regarding the nature, mechanisms and consequences of nutrition-dependent development in both males and females and across both naturally and sexually selected traits. Specifically, we sought to examine how nutrition-sensitive development is enabled, how it compares with other sources of variation in gene expression such as sex, how it evolves and diversifies and the evolutionary consequences of nutrition-dependent gene expression. In particular, we tested the hypotheses (i) that nutrition-specific gene expression is a significant source of population-wide variation in gene expression, (ii) that its magnitude and complexity parallel the diversity of nutrition-specific morphological development seen in different traits, (iii) that it is mediated by developmental pathways shared across different body regions and sexes, and (iv) that nutrition-specific gene expression results in relaxed selection relative to genes shared across nutritional contexts, leading to increased accumulation of genetic variation.
To test these hypotheses, we characterized the nutritional responses of four different body regions in males and females of the polyphenic beetle Onthophagus taurus (figure 1). We choose this species, because males and females differ substantially in body region- and tissue type-specific growth responses to nutritional variation, generating a remarkable sexual- as well as within-male dimorphism as a consequence [41,42]. Specifically, we focused our analysis on four body regions derived from epidermal tissue: abdominal epidermis, legs, thoracic horns and head horns (figure 1b). In females, all four body regions exhibit roughly proportional growth increases in response to increased nutrient availability, causing large, high nutrition females to represent proportionately enlarged versions of small, low-nutrition females. In contrast, in males, only abdominal epidermis shares the same growth response also seen in females, whereas male legs grow slightly—and male thoracic horns grow substantially—larger than their female counterparts when exposed to the same nutritional gradient. In this species, thoracic horns are only expressed in pupae, function as moulting organs to shed the larval head capsule during the larval–pupal moult, and are resorbed prior to the pupal–adult moult [43]. Lastly, male head horns, a pair of spectacular, sexually selected structures used in male–male competition [44] show the most extreme growth response and exhibit explosive, nonlinear growth after exceeding a specific nutrition threshold. Combined, even though males express the same range of nutritionally determined body sizes as females, the relative sizes of some of their body parts differ strikingly as a function of nutrition, so much so that alternative minor (‘hornless’) and major (‘horned’) male morphs were described initially as separate species (figure 1a).
Figure 1.
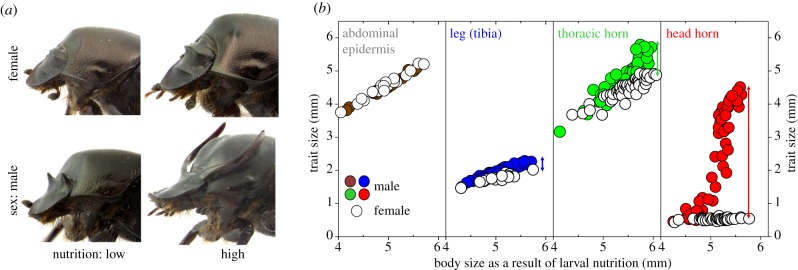
The effect of nutrition on growth in male and female O. taurus. (a) Representative female (top) and male (bottom) adults that were exposed to low (left) and high (right) levels of nutrition during larval development, respectively. (b) Scaling relationships between body size (as an estimate of larval nutrition; x-axis) and abdominal width (as an estimate of abdominal epithelium size), tibia length (as an estimate of leg size) and thoracic horn and head horn length, respectively. All plots are on the same scale to highlight relative differences in growth responses across body regions. Measurements from males are shown as solid circles, those from females as open circles to highlight the relative magnitude of sexual dimorphism for each focal trait (indicated by arrows for leg, thoracic and head horns).
In order to examine our basic hypotheses regarding the mechanisms, evolution and consequences of nutrition-dependent gene expression, we sought to answer four specific questions (i) how much variation in gene expression during development can be attributed to nutrition, body region, sex or the interactions among them? (ii) do body-region-specific responses in gene expression to nutrition covary with the overall magnitude of growth responses observed for those body regions? (iii) do homologous body regions in males and females differ in the number and/or composition of genes whose expression is affected by nutritional conditions? And (iv) what are the evolutionary consequences of nutrition-dependent gene expression with respect to sequence variability and divergence?
2. Material and methods
(a). Experimental design and statistical analysis
This study sought to characterize the nutritional responses of four different body regions in male and female O. taurus. Our experimental design therefore had to enable robust characterization of 16 conditions: two nutritional levels (large ‘L’, small ‘S’) × two sexes (male ‘M’, female ‘F’) × four body regions (abdominal epidermis ‘a’, leg ‘l’, thoracic horn ‘t’ and head horn ‘h’). We employed a microarray approach involving custom-made NimbleGen arrays (details below) developed for O. taurus based on a comprehensive 454-transcriptome [45] to estimate the effects of nutrition, sex and body region on gene expression. Exact design of our experiment, application of our ANOVA model and statistical procedures to estimate and compare nutrition-dependent variation in gene expression with other sources of variation are provided in the electronic supplementary material and in references [46,47].
(b). Animal husbandry
Beetles used in this study were collected and reared as described in references [48,49] and the electronic supplementary material. Nutrition consumed during larval development directly affects final, adult body size. We therefore used body mass, measured at the first day of pupal development, as an estimate of feeding conditions experienced during larval development. Pupae weighing less than 0.105 g were considered small, low-nutrition individuals. In males, this weight class uniformly matures into the minor, hornless morph. In contrast, pupae weighing more than 0.12 g were considered large, high nutrition individuals, which in males uniformly metamorphose into the major, horned morph. Pupae whose weight fell between 0.105 and 0.12 g were excluded from the experiment because their developmental fate is difficult to predict. We focused our analysis on the first 24 h of pupal development, because this stage allows us to capture the most important developmental processes underlying growth and differentiation of adult traits, because animals can be unambiguously sexed and phenotypes scored (e.g. horned and hornless) and because dissections are easy and fast, thus minimizing chances for contamination and degradation of RNA samples.
(c). Tissue harvesting and RNA extraction
Sixty-four O. taurus first-day pupae (within 24 h of pupation) were sacrificed and dissected for microarrays (16 each for large, horned male; small, horned male; large female and small female). Dissections were performed in RNase-free conditions and tissues were immediately ground in buffer RLT (RNeasy mini kit, Qiagen, CA), and flash frozen in liquid nitrogen. We dissected epidermal tissue of the developing head horns (the analogous area for females, which had no horns) and prothoracic horns, in addition to all six legs and a small section of the dorsal abdominal epidermis (with attached muscle gently scraped off), detailed further in references [50,51]. We then created biological replicates by pooling tissue of four individuals in the same tissue–sex–size category. Individuals were pooled, because we were more interested in overall patterns of gene expression that varied with sex, tissue and nutrition rather than individual-level variation in gene expression. We divided individuals into high nutrition (large) and low-nutrition (small) size classes based on mass as explained above, with identical cut-offs for both males (which metamorphose into discrete morphs) and females (which do not). The same individuals were used in batches across tissue types—for instance, small male replicate 3 included the same four small males for the thoracic horn, head horn, leg and abdominal tissue samples. A total of 64 batches of RNA were generated for microarray analyses (two sexes × two nutrition types × four body regions × four biological replicates). A subset of these samples were included as technical replicates (see array design details) where labelling and hybridization were repeated independently on the same RNA sample.
(d). Microarray platform and hybridization
We used a custom Nimblegen gene expression array (Roche Nimblegen, Inc., Madison, WI) as detailed in references [50,51]. For array hybridization, double-stranded cDNA (1.0 μg) was first labelled using dual-colour labelling kit (Roche NimbleGen, Inc., Madison, WI) in a reaction containing 1 OD of either CY3 or CY5 random nonomer primer, dNTP mix (1 mM each), and 100U Klenow fragment (3 > 5 exo-), followed by isopropanol precipitation with 50 mM EDTA and 500 mM sodium chloride and dual-colour hybridization and post-hybridization according to manufacturer's instructions (Roche NimbleGen, Inc., Madison, WI). Images were acquired using a GenePix 4200A scanner and GenePix v. 6.0 software (Molecular Devices, Sunnyvale, CA). Array data can be accessed under NCBI GEO accession number GSE58496.
(e). Enrichment analyses
We performed functional enrichment analyses using the eight t-statistic distributions that characterized the effect of nutrition for each tissue-by-sex gene expression set. Briefly, we characterized protein function based on homology with described genes in the Gene Ontology database [52] using the program Blast2GO [53] resulting in 1114 functional classes. Using the program ErmineJ [54], we tested for each functional class of proteins for an excess of differentially expressed genes relative to all other genes not in that category by using gene score resampling to generate a null distribution (see the electronic supplementary material, Methods for details).
(f). Cluster analyses
We used the TM4 microarray experiment viewer to quantify overall patterns of similarity in gene expression. We focused on patterns of differential gene expression with nutrition for each of the eight sex–tissue categories. We analysed all genes with a significant nutritional effect (false discovery rate (FDR) p < 0.01; n = 8303) for at least one of these categories. We used hierarchical clustering (with Pearson correlations as the distance metric) to test for patterns of similarity in expression and inspected pairwise Pearson correlations across these values. Confidence in the observed clustering was evaluated through a bootstrapping support tree (100 replicates).
(g). Single nucleotide polymorphism analyses
We analysed how gene expression was related to patterns of genetic variation using measures of single nucleotide polymorphisms (SNPs) described elsewhere [45]. We tested the prediction that nutritionally responsive genes (FDR of 0.05) were more genetically variable. For this analysis, we used an FDR cut-off of 0.05 to increase our sample size for comparing each tissue–sex category given that we had SNP information for only a subset of the contigs. We analysed zero and non-zero values separately (see the electronic supplementary material, Methods for details).
3. Results and discussion
In this study, we contrasted gene expression in four distinct body regions in males and females of high- and low-nutritional status. Using custom microarrays, we evaluated the differential expression of 42 010 contigs representing more than half of the genes annotated in the flour beetle Tribolium castaneum, the most closely related species with a fully sequenced genome [45]. Specifically, we sought to contrast nutrition-dependent changes in gene expression to two other sources of variation (sex, body region) as well as their respective interactions, characterize body region- and sex-specific responses to nutritional variation, and evaluate the evolutionary consequences of nutritional plasticity in gene expression.
(a). Nutrition-dependent variation in gene expression compared with other sources of variation
Of the 42 010 contigs that were initially investigated, 20 182 had R2-values that were significant at the 0.05 level. Adjusting for FDR at q = 0.05 reduced this number to 17 180 contigs whose R2-values ranged from 0.5157 to 0.9878. We then removed from further consideration the remaining 24 830 contigs (approx. 60%) for which the linear models failed to explain a significant amount of variation in expression levels, and compared the sums of squares owing to the three main effects only (body region, sex, nutrition). However, the sum of squares owing to body region involved three times the degrees of freedom (d.f.). To adjust for this difference in d.f., we considered the sum
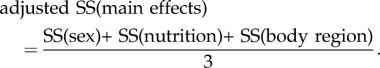 |
3.1 |
For most of the 17 180 contigs, either body region or sex explained the majority of the proportion of the adjusted-SS (table 1). For example, for 445 contigs, sex alone explained 100–75% of variation (75–50%: 2215 contigs; 50–25%: 4026 contigs), whereas body region alone explained 50–25% of variation in 4035 contigs. In contrast, nutrition explained 75–50% of the variation for only 37 of the 17 180 contigs (50–25%: 441 contigs). These results suggest that variation in gene expression contributed by nutrition is overall at least 1.5 orders of magnitude smaller when compared with the variance contributions of the other two main effects. However, for a subset of contigs nutritional variation appears to represent the major determinant of expression variation, at least at the particular developmental time point examined in this study (table 1). These findings suggest that the effect of nutritional variation on gene expression variation can be highly variable, and thus needs to be determined empirically, and ideally with reference to other sources of expression variation. In contrast, two- and three-way interactions accounted for a comparatively, and uniformly, small fraction of expression variation (table 1).
Table 1.
Effect of body region, sex, nutrition and two- and three-way interaction on variation in gene expression. Of 42 010 contigs initially examined in this study, 17 180 exhibited variation in expression attributable to the combined effects of nutrition, sex and body region, and their interactions.
| variation explained | 100–75% | 50–75% | 25–50% | 25–5% | <5% |
|---|---|---|---|---|---|
| body region | 0 | 0 | 4035 | 11 599 | 1546 |
| sex | 445 | 2215 | 4026 | 5768 | 4726 |
| nutrition | 0 | 37 | 441 | 4695 | 12 007 |
| two-way interactions | 0 | 0 | 0 | 1136 | 16 044 |
| three-way interactions | 0 | 0 | 0 | 362 | 16 818 |
(b). Nutritional responsiveness as a function of body region and sex
We then focused our attention on the nutritional responsiveness of each of our four focal body regions as a function of sex (figure 1). Recall that in females, all four body regions exhibit overall moderate and roughly proportional growth increases in response to increased nutrient availability. In contrast, male growth responses over the same nutritional gradient range from moderate (legs, abdominal epidermis) to accelerated (thoracic horns) to explosive and nonlinear (head horns). We found that different body regions within the same individuals, as well as the same body region across sexes, differed dramatically in the number of nutrition-responsive contigs, the magnitude of nutrition-induced differential expression, and the degree to which a given pool of nutrition-responsive contigs was unique to, or shared across, multiple body regions.
In males, nutrition-dependent growth of abdominal epidermis and leg tissue was associated with the differential expression of a modest 146 and 71 nutrition-responsive contigs, respectively (FDR p < 0.05; figure 2). Over the same nutritional gradient, and at the same FDR, thoracic horns revealed 1066 nutrition-responsive contigs, whereas the explosive growth response of head horn tissue was associated with the differential expression of 2093 contigs. A similar pattern was detected when we examined the magnitude of differential expression (electronic supplementary material, figures S1–S3). The degree of nutrition-dependent differential expression was lowest for male abdominal epidermis and legs, followed by thoracic horn tissue and by far highest for head horn tissue. In females, a largely corresponding pattern was detected. Abdominal epithelium, legs and head horn tissue, all of which exhibit moderate nutrition-dependent growth, also exhibited a moderate number of differentially expressed contigs (abdomen: 60; leg: 15; head horn: 22; FDR, p = 0.05). Furthermore, magnitude of differential expression was similar to that detected in male legs and abdominal epidermis. The only exception to this pattern was provided by female thoracic horn tissue. While female thoracic horns also show a moderate growth response to nutrition, this response was associated with the differential expression of 637 contigs and also exhibited the relatively highest magnitude of differential expression of any female body region (figure 2; electronic supplementary material, figures S1 and S2). Taken together, both the number of nutritionally responsive contigs and the average magnitude of nutrition-responsive expression therefore scaled positively with the degree of nutrition-responsive growth observed on a morphological level, and the extent of sexual dimorphism inherent in each body region.
Figure 2.
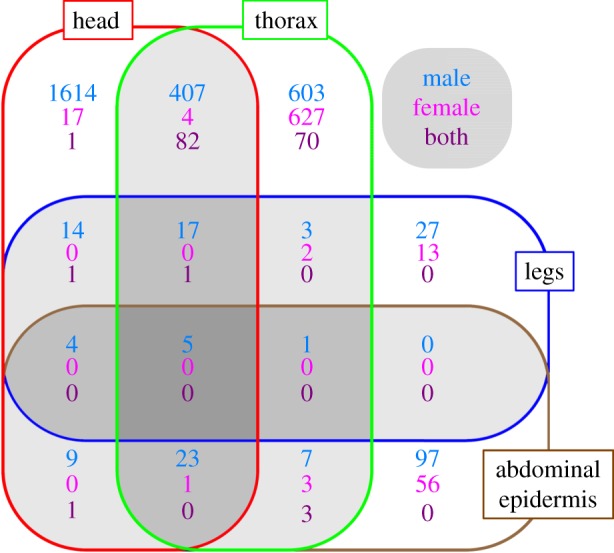
Number of nutrition-sensitive contigs unique to, or shared among, four different body regions. Different colours indicate numbers of contigs that exhibit nutrition sensitivity in males (blue), females (magenta) or both (purple), respectively.
Body regions also differed substantially in the degree to which their pool of nutrition-responsive contigs was unique to, or shared across, multiple body regions (figure 2). Within males, only head horn and thoracic horn epidermis overlapped appreciably in their nutrition-sensitive contig pool, whereas all other pairwise comparisons yielded essentially no overlap. Specifically, of 2707 contigs that exhibited nutrition-sensitive expression in either male head or thoracic horn tissue, 407 (15%) were shared between both body regions. In contrast, in all other pairwise comparisons shared contigs ranged from 0 to 14 (=0–0.3%). Three-way comparisons also yielded minimal to no overlap, and only five of a total of 2831 (0.18%) nutrition-responsive contigs were shared across all four male body regions. Within females, all two-way comparisons across body regions, including both horn types, yielded zero to near-zero overlap across nutrition-sensitive contig pools, as did all three-way comparisons, and 0 of a total of 723 nutrition-responsive contigs were shared across all four female body regions (figure 2).
Lastly, most pools of nutrition-responsive contigs were highly sex-specific for each individual body region or combinations thereof. For example, of the 153 contigs that exhibited nutrition-sensitive expression solely in male or female abdominal epidermis, none did so in both sexes. A similarly high degree of sex-specific nutritional responsiveness was found for all comparisons involving leg or abdominal epidermis, and any two- and three-way comparisons of both body regions with either horn type. The only exceptions to this pattern were provided by the contig pool specific to thoracic horns, and the pool shared between both horn types. In the former, of 1300 contigs that exhibited nutrition-responsive expression chiefly in either male or female thoracic horns, 70 (5.4%) did so in both sexes. In the latter, 82 contigs exhibited nutrition-responsive expression in at least one combination of male and/or female head and thoracic horns (figure 2).
Taken together, these data show that the magnitude of nutrition-responsive gene expression correlates closely with the magnitude of nutrition-responsive growth observed on a morphological level across different body regions and involves a contig repertoire that is, for the most part, specific to a given body region and sex. This suggests a surprising degree of modularity in nutrition-responsive gene expression, and one in contrast to the overall remarkable similarity in gene expression observed across horn types and legs, as documented by previous studies [50,51]. Combined, these results suggest that organisms may be using rather similar gene sets to build similar traits, but rely on very different, body-region- and sex-specific gene sets to facilitate their nutrition-responsiveness.
Our results are also in striking agreement with predictions derived from condition-dependence theory [22,23]. A long-standing challenge in sexual selection is to reconcile the predicted fixation of alleles beneficial under strong selection with the persistence of unexpectedly high levels of genetic variance in many sexually selected traits and the maintenance of mate choice benefits. This paradox is resolved by the genic capture hypothesis [22], which proposes that additive genetic variation in sexually selected traits reflects variance in condition. In other words, it posits that male secondary sexual traits are costly to produce and hence depend upon overall condition, which itself is affected by genes at a large number of loci [23]. One important prediction of this hypothesis is that condition-dependent exaggeration of secondary sexual traits should be associated with the differential expression of large numbers of genes compared with traits likely under weak directional or stabilizing selection. By extension, this hypothesis also predicts that the degree of sexual dimorphism attainable across traits should reflect, in part, the degree to which genic capture can evolve in males without correlated effects in females, thereby reducing intersexual genetic correlations and potential conflicts [30,31]. Both predictions are supported by our results: nutrition-responsive genes were by far most numerous in male head horns, which exhibit the largest degree of nutrition-dependent growth and most pronounced sexual dimorphism among the four body regions examined, whereas the overlap in nutrition-responsive contig repertoires between male and females head horns was much lower than that observed between the much less sexually dimorphic thoracic horns.
(c). Composition of nutritionally responsive transcriptomes
To assess the function of nutritionally responsive genes, we tested for the overrepresentation of differentially expressed genes in each of the 1114 biological process Gene Ontology categories represented in our dataset. Transcriptome-wide gene expression patterns of different body regions and sexes differed widely in the number and types of functional categories responding to nutrition (electronic supplementary material, figure S4). We found 497 categories enriched for response to nutrition in males (44.6% of the 1114 categories tested, FDR p < 0.05) and 338 categories enriched for response to nutrition in females (30.3% of the 1114 categories tested, FDR p < 0.05; electronic supplementary material, figure S4). The difference between sexes was more pronounced when considering functional categories that were nutritionally responsive exclusively in each sex; there were four times more categories responding to nutrition in males than in females (213 versus 57, FDR p < 0.05). These results parallel several previous studies that have identified males as the more nutritionally responsive sex across diverse biological contexts, from morphology [55] and behaviour [44,56, but see 57] to thermoregulation [58] and hormone physiology [59,60].
Across male body regions, we found the greatest number of nutritionally responsive functional categories in head horns (90), followed by thoracic horns (42), legs (32) and abdominal epidermis (16, electronic supplementary material, figure S4, FDR p < 0.05). This pattern was similar, as we would expect, to the pattern of nutritionally responsive contigs, suggesting that the genes responding to nutrition are associated with specific biological functions. In females, head horns, thoracic horns and legs exhibited similar numbers of categories responding to nutrition (45, 55, 55, respectively), followed by abdominal epidermis (15, electronic supplementary material, figure S4, FDR p < 0.05), again a pattern that parallels the number of nutritionally responsive contigs.
The types of functional categories responding to nutrition in male head horns involved functions that matched the explosive growth observed in this body region: 31 of 90 enriched categories in male head horns were related to metabolism or biosynthesis (34.4%, electronic supplementary material, table S2). Notably, these categories were not enriched in the other horn types that do not exhibit such explosive growth; electronic supplementary material, table S2). We then examined if head and thoracic horns shared nutritionally sensitive functional categories. We found that functional groups related to development, morphogenesis and response to hormone signalling were enriched for nutritional sensitivity in males and females (electronic supplementary material, table S3), complementing the results of several previous studies (reviewed in reference [61]). Finally, we found many more functional categories shared across all four body regions in males than expected based on the number of shared contigs that responded to nutrition. Specifically, while only five contigs (of 2831) exhibited significant differential expression owing to nutrition in all four body regions, 89 of the 497 functional categories showed significant enrichment across all four regions (=17.9%; compare central overlapping regions in figure 2 versus electronic supplementary material, figure S4). Similar to the categories overrepresented in male head horns, these categories served functions related to metabolism and biosynthesis. In contrast, in females, the degree of overlap among the four body regions was more modest, with 18 categories enriched out of the 338 represented (5.3%). These observations raise the possibility that males, but not females, use a broad set of developmental processes to enable nutritional responsiveness across male body regions but whose underlying specific genes, while acting in concert, did not individually show a strong enough response to nutrition to be detected as differentially expressed. Alternatively, different male body regions may employ the same broad set of developmental processes to enable different degrees of nutritional responsiveness, but use distinct subsets of genes and pathways to do so.
Lastly, our cluster analyses generated congruent results (electronic supplementary material, figure S5). We found that nutrition-sensitive gene expression (i) was most similar within each sex, (ii) exhibited the greatest similarity between thoracic and head horn epidermis, and (iii) was overall more similar across male than female body regions.
(d). Candidate genes and pathways underlying differential growth responses to nutrition
Our analysis identified diverse candidate genes and pathways possibly underlying the regulation of body region- and sex-specific responses to nutrition. These include several candidates identical, or functionally closely related, to genes and pathways identified previously as nutrition-sensitive through independent studies, such as genes associated with the regulation and execution of programmed cell death (PCD protein 7, apoptosis inhibitor 5, apoptosis-stimulating of p53 protein; see [62]), sexual differentiation (sex-lethal, doublesex; [63]); axis specification (pipsqueak, homothorax-like; [64]) and genes underlying epigenetic processes (e.g. dicer, methyltransferase-like protein 9, h3k9 methyltransferase; [65]).
In addition, we identified a large number of genes known to function in the regulation of growth and cell division, in particular in the nutritionally highly responsive head horns (e.g. ras-related/ras-associated genes, cyclin a, cdc6, cell division control protein, metastasis-associated protein, forkhead box proteins). Similarly, many genes related to cuticle synthesis and cuticular properties exhibited nutrition-sensitive expression in one or more body region and sex (e.g. cuticular proteins 47ef, 62bc, resilin).
Lastly, we detected nutrition-responsive expression in a series of genes or pathways whose function, in particular, biological contexts is very well understood, but whose differential expression in the context of nutrition-responsive growth was nevertheless unexpected. For instance, patched plays a key role in the Hedgehog-pathway and as such in the establishment of anterior/posterior polarity of a wide range of structures. Our study finds nutrition-responsive expression of patched in male head- and thoracic horns, but not legs or abdominal epidermis. Similarly, timeless, a central regulator of circadian rhythm, exhibits nutrition-responsive expression in male head- and thoracic horns. These pathways offer especially intriguing opportunities for future work.
(e). Evolutionary consequences of conditional gene expression
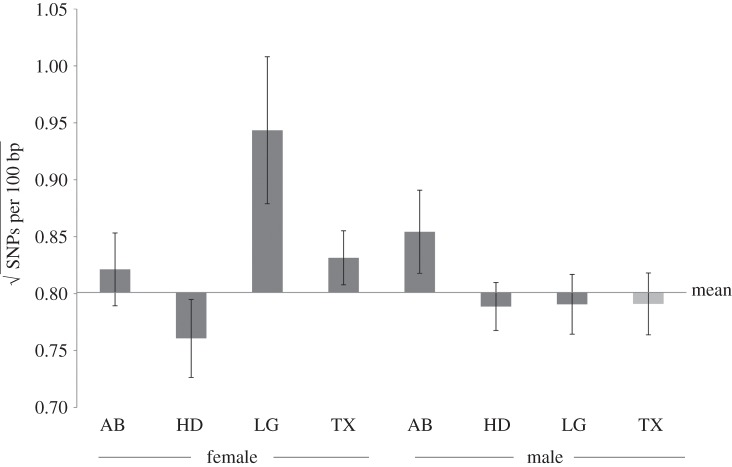
We sought to test the theoretical prediction that environment-specific gene expression results in relaxed selection and the accumulation of genetic variation, relative to genes expressed across all individuals. Previous work on this system has documented that nutrition-biased horn genes show relatively greater evolutionary divergence [20]; however, in this work, we were primarily interested in the theoretical prediction that these genes should also harbour more genetic variants. As our measure of genetic variation, we focused on the number of SNPs identified per 100 bp of contig, square root transformed for normality. We compared the distribution of genetic variation across all contigs with distributions of genetic variation for contigs specific in nutrition-biased expression to particular sex and tissue categories (5% FDR cut-off). Genes with nutrition-sensitive expression in male and female abdominal epidermis, female legs and female thoracic horns showed significantly more genetic variation than expected (figure 3 and the electronic supplementary material, table S4), consistent with relatively weaker selection intensity, or alternatively, balancing selection. However, genes expressed in female and male head horn primordial epidermis, and male legs exhibited less genetic variation than expected (figure 3 and the electronic supplementary, table S4), suggestive of purifying selection or a recent selective sweep. We analysed separately the frequency of ‘zero’ measures of genetic variation for contigs with sufficient read depth and length, thus ensuring that ‘zero’ was biologically relevant. Chi-squared tests revealed no significant difference in the frequency of zero values between our categories of interest relative to all contigs (electronic supplementary material, table S5).
Figure 3.
Patterns of genetic variation in nutrition-biased genes. Shown is the mean (and standard error) of the number of SNPs per 100 bp of contig (square root transformed for normality) for categories of genes with nutrition-biased expression. The horizontal line represents the mean value for all contigs. Tissue categories include abdominal epidermis (AB), head horn epidermis (HD), prothoracic horn epidermis (TX) and leg epidermis (LG). Patterns of genetic variation significantly different than the overall distribution for all genes are indicated with dark grey bars (see the electronic supplementary material, table S4 for statistics).
These results are partially consistent with the predictions of relaxed selection on genes with environment-biased expression across individuals, here, different nutritional morphs [20]. Relative to genes expressed across all individuals, environment-specific genes are subjected to weakened purifying and positive selection, resulting in the accumulation of genetic variation, much of which is thought to be deleterious [19,21]. Theory predicts that these genes should harbour greater genetic variation within species [21], a prediction that, to the best of our knowledge, has not previously been tested. Nutrition-responsive genes expressed in the abdominal epidermis (in both sexes), and in female leg and thoracic horn epidermis, are enriched for genetic variation. However, genes in several tissue regions, in particular, head horn tissue, showed less variation than expected. These results build on findings from other studies that show that morph-biased genes subjected to sexual selection (i.e. those in male head horns) show strong signatures of purifying and positive selection and greater evolutionary divergence relative to genes with less morph-bias [51,66]. These results emphasize that sexual selection intensity may interact with relaxed selection associated with environment-biased gene expression to result in different signatures of selection across traits. While models of relaxed selection assume that selection intensity is consistent across individuals and traits, it is likely that strong sexual selection on head horns predominates over relaxed selection on morph-biased genes, resulting in rapid evolution of head horn genes.
(f). Development and evolution of nutrition-responsiveness
Our findings illustrate that the developmental nature and evolutionary consequences of variation in gene expression owing to nutritional conditions are themselves heavily dependent on other conditions, in particular the body regions under consideration and the sex within which they occur. At the same time, we detect important and persistent patterns: first, body regions that morphologically respond more strongly to nutritional variation also exhibit a far more complex nutrition-responsive transcriptome. This observation rejects the hypothesis that a common machinery may orchestrate nutrition-responsive development, regardless of body region, and instead highlights an unexpected degree of modularity and developmental—and perhaps evolutionary—decoupling of body region and sex-specific nutrition-responsive genes and pathways. Here, male head horns—a recent evolutionary invention with striking nutrition-dependent growth—stand out dramatically as the most complex of the four body regions examined in this study, both in terms of numbers of nutrition-responsive contigs as well as enrichment of unique functional categories. Second, we observe that males, the nutritionally more responsive sex overall with respect to morphology, physiology and behaviour, engage a functionally more similar set of developmental processes (as measured by functional enrichment) across all four body regions than do nutritionally less-responsive females. While modest (5% of functional categories shared across body regions in females versus 18% in males), this difference nevertheless suggests that the degree of integration of nutrition-responsive transcriptomes across body regions may constitute an additional axis of diversification able to facilitate evolutionary changes in nutrition-dependent phenotype expression. Lastly, we observe that even though the degree of morphological responsiveness to nutrition may parallel the magnitude of transcriptional responsiveness for a given tissue and sex, it is a poor guide to predicting the degree to which nutrition-biased genes may be subjected to relaxed selection and mutation accumulation.
Acknowledgements
We thank John Colbourne, Justen Andrews, Brian Eads, Jacqueline Lopez and Amy Cash for their advice and guidance and two anonymous reviewers for their encouragement and constructive feedback.
Funding statement
This research was made possible through National Science Foundation grants IOS 0744585, 1120209 and 0820411 to A.P.M. E.C.S.R. was supported in part by NIH NRSA F32GM083830.
Data accessibility
Array data: NCBI GEO accession number GSE58496. SNP data: DRYAD accession number doi:10.5061/dryad.gr5n3.
References
- 1.Nijhout HF. 2003. Development and evolution of adaptive polyphenisms. Evol. Dev. 5, 9–18. ( 10.1046/j.1525-142X.2003.03003.x) [DOI] [PubMed] [Google Scholar]
- 2.Shingleton AW, Frankino WA, Flatt T, Nijhout HF, Emlen DJ. 2007. Size and shape: the developmental regulation of static allometry in insects. Bioessays 29, 536–548. ( 10.1002/bies.20584) [DOI] [PubMed] [Google Scholar]
- 3.Beldade P, Mateus A, Keller RA. 2011. Evolution and molecular mechanisms of adaptive developmental plasticity. Mol. Ecol. 20, 1347–1363. ( 10.1111/j.1365-294X.2011.05016.x) [DOI] [PubMed] [Google Scholar]
- 4.Gluckman PD, Hanson MA, Buklijas T, Low FM, Beedle AS. 2009. Epigenetic mechanisms that underpin metabolic and cardiovascular diseases. Nat. Rev. Endocrinol. 5, 401–408. ( 10.1038/nrendo.2009.102) [DOI] [PubMed] [Google Scholar]
- 5.Ferguson LR. 2012. Potential value of nutrigenomics in Crohn's disease. Nat. Rev. Gastroenterol. Hepatol. 9, 260–270. ( 10.1038/nrgastro.2012.41) [DOI] [PubMed] [Google Scholar]
- 6.Teegarden D, Romieu I, Lelievre SA. 2012. Redefining the impact of nutrition on breast cancer incidence: is epigenetics involved? Nutr. Res. Rev. 25, 68–95. ( 10.1017/S0954422411000199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duque-Guimaraes DE, Ozanne SE. 2013. Nutritional programming of insulin resistance: causes and consequences. Trends Endocrinol. Metab. 24, 525–535. ( 10.1016/j.tem.2013.05.006) [DOI] [PubMed] [Google Scholar]
- 8.Kirchner H, Osler ME, Krook A, Zierath JR. 2013. Epigenetic flexibility in metabolic regulation: disease cause and prevention? Trends Cell Biol. 23, 203–209. ( 10.1016/j.tcb.2012.11.008) [DOI] [PubMed] [Google Scholar]
- 9.Wheeler DE. 1986. Developmental and physiological determinants of caste in social Hymenoptera: evolutionary implications. Am. Nat. 128, 13–34. ( 10.1086/284536) [DOI] [Google Scholar]
- 10.Emlen DJ, Nijhout HF. 2000. The development and evolution of exaggerated morphologies in insects. Annu. Rev. Entomol. 45, 661–708. ( 10.1146/annurev.ento.45.1.661) [DOI] [PubMed] [Google Scholar]
- 11.Smith CR, Toth AL, Suarez AV, Robinson GE. 2008. Genetic and genomic analyses of the division of labour in insect societies. Nat. Rev. Genet. 9, 735–748. ( 10.1038/nrg2429) [DOI] [PubMed] [Google Scholar]
- 12.Toth AL, et al. 2007. Wasp gene expression supports an evolutionary link between maternal behavior and eusociality. Science 318, 441–444. ( 10.1126/science.1146647) [DOI] [PubMed] [Google Scholar]
- 13.Ament SA, Corona M, Pollock HS, Robinson GE. 2008. Insulin signaling is involved in the regulation of worker division of labor in honey bee colonies. Proc. Natl Acad. Sci. USA 105, 4226–4231. ( 10.1073/pnas.0800630105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aubin-Horth N, Renn SCP. 2009. Genomic reaction norms: using integrative biology to understand molecular mechanisms of phenotypic plasticity. Mol. Ecol. 18, 3763–3780. ( 10.1111/j.1365-294X.2009.04313.x) [DOI] [PubMed] [Google Scholar]
- 15.Liao BY, Scott NM, Zhang JZ. 2006. Impacts of gene essentiality, expression pattern, and gene compactness on the evolutionary rate of mammalian proteins. Mol. Biol. Evol. 23, 2072–2080. ( 10.1093/molbev/msl076) [DOI] [PubMed] [Google Scholar]
- 16.Pal C, Papp B, Lercher MJ. 2006. An integrated view of protein evolution. Nat. Rev. Genet. 7, 337–348. ( 10.1038/nrg1838) [DOI] [PubMed] [Google Scholar]
- 17.Larracuente AM, et al. 2008. Evolution of protein-coding genes in Drosophila. Trends Genet. 24, 114–123. ( 10.1016/j.tig.2007.12.001) [DOI] [PubMed] [Google Scholar]
- 18.Meisel RP. 2011. Towards a more nuanced understanding of the relationship between sex-biased gene expression and rates of protein-coding sequence evolution. Mol. Biol. Evol. 28, 1893–1900. ( 10.1093/molbev/msr010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitlock MC. 1996. The red queen beats the jack-of-all-trades: the limitations on the evolution of phenotypic plasticity and niche breadth. Am. Nat. 148, S65–S77. ( 10.1086/285902) [DOI] [Google Scholar]
- 20.Snell-Rood EC, VanDyken JD, Cruickshank TE, Wade MJ, Moczek AP. 2010. Toward a population genetic framework of developmental evolution: costs, limits, and consequences of phenotypic plasticity. Bioessays 32, 71–81. ( 10.1002/bies.200900132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Dyken JD, Wade MJ. 2010. The genetic signature of conditional expression. Genetics 184, 557–570. ( 10.1534/genetics.109.110163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rowe L, Houle D. 1996. The lek paradox and the capture of genetic variance by condition dependent traits. Proc. R. Soc. Lond. B 263, 1415–1421. ( 10.1098/rspb.1996.0207) [DOI] [Google Scholar]
- 23.Tomkins JL, Radwan J, Kotiaho JS, Tregenza T. 2004. Genic capture and resolving the lek paradox. Trends Ecol. Evol. 19, 323–328. ( 10.1016/j.tree.2004.03.029) [DOI] [PubMed] [Google Scholar]
- 24.Delcourt M, Rundle HD. 2011. Condition dependence of a multicomponent sexual display trait in Drosophila serrata. Am. Nat. 177, 812–823. ( 10.1086/659949) [DOI] [PubMed] [Google Scholar]
- 25.Proulx SR. 1999. Matings systems and the evolution of niche breadth. Am. Nat. 154, 89–98. ( 10.1086/303218) [DOI] [PubMed] [Google Scholar]
- 26.Proulx SR. 2002. Niche shifts and expansion due to sexual selection. Evol. Ecol. R 4, 351–369. [Google Scholar]
- 27.Lorch PD, Proulx S, Rowe L, Day T. 2003. Condition-dependent sexual selection can accelerate adaptation. Evol. Ecol. R 5, 867–881. [Google Scholar]
- 28.Whitlock MC, Agrawal AF. 2009. Purging the genome with sexual selection: reducing mutation load through selection on males. Evolution 63, 569–582. ( 10.1111/j.1558-5646.2008.00558.x) [DOI] [PubMed] [Google Scholar]
- 29.Agrawal AF. 2001. Sexual selection and the maintenance of sexual reproduction. Nature 411, 692–695. ( 10.1038/35079590) [DOI] [PubMed] [Google Scholar]
- 30.Bonduriansky R. 2007. The evolution of condition-dependent sexual dimorphism. Am. Nat. 169, 9–19. ( 10.1086/510214) [DOI] [PubMed] [Google Scholar]
- 31.Bonduriansky R. 2007. The genetic architecture of sexual dimorphism: the potential roles of genomic imprinting and condition-dependence. In Sex, size, and gender roles: evolutionary studies of sexual size dimorphism (eds Fairbairn D, Blanckenhorn J, Szekely T.), pp. 176–184. Oxford, UK: Oxford University Press. [Google Scholar]
- 32.Pomiankowski A, Moller AP. 1995. A resolution of the lek paradox. Proc. R. Soc. Lond. B 260, 21–29. ( 10.1098/rspb.1995.0054) [DOI] [Google Scholar]
- 33.Kotiaho JS, Simmons LW, Tomkins JL. 2001. Towards a resolution of the lek paradox. Nature 410, 684–686. ( 10.1038/35070557) [DOI] [PubMed] [Google Scholar]
- 34.Badyaev AV, Duckworth RA. 2003. Context-dependent sexual advertisement: plasticity in development of sexual ornamentation throughout the lifetime of a passerine bird. J. Evol. Biol. 16, 1065–1076. ( 10.1046/j.1420-9101.2003.00628.x) [DOI] [PubMed] [Google Scholar]
- 35.Bellamy L, et al. 2013. Sexual traits are sensitive to genetic stress and predict extinction risk in the stalk-eyed fly, Diasemopsis meigenii . Evolution 67, 2662–2673. ( 10.1111/evo.12135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cotton S, Fowler K, Pomiankowski A. 2004. Heightened condition dependence is not a general feature of male eyespan in stalk-eyed flies (Diptera: Diopsidae). J. Evol. Biol. 17, 1310–1316. ( 10.1111/j.1420-9101.2004.00754.x) [DOI] [PubMed] [Google Scholar]
- 37.Emlen DJ, et al. 2012. A mechanism of extreme growth and reliable signaling in sexually selected ornaments and weapons. Science 337, 860–864. ( 10.1126/science.1224286) [DOI] [PubMed] [Google Scholar]
- 38.Bakker T, Mundwiler B. 1999. Pectoral fin size in a fish species with paternal care: a condition-dependent sexual trait revealing infection status. Freshw. Biol. 41, 543–551. ( 10.1046/j.1365-2427.1999.00403.x) [DOI] [Google Scholar]
- 39.Cothran RD, Jeyasingh PD. 2010. Condition dependence of a sexually selected train in a crustacean species complex: importance of the ecological context. Evolution 64, 2535–2546. ( 10.1111/j.1558-5646.2010.00998.x) [DOI] [PubMed] [Google Scholar]
- 40.Wyman MJ, Agrawal AF, Rowe L. 2010. Condition-dependence of the sexually dimorphic transcriptome in Drosophila melanogaster. Evolution 62, 1836–1848. ( 10.1111/j.1558-5646.2009.00938.x) [DOI] [PubMed] [Google Scholar]
- 41.Moczek AP. 2006. Pupal remodeling and the development and evolution of sexual dimorphism in horned beetles. Am. Nat. 168, 711–729. ( 10.1086/509051) [DOI] [PubMed] [Google Scholar]
- 42.Moczek AP. 2007. Pupal remodeling and the evolution and development of alternative male morphologies in horned beetles. BMC Evol. Biol. 7, 151 ( 10.1186/1471-2148-7-151) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moczek AP, Cruickshank TE, Shelby JA. 2006. When ontogeny reveals what phylogeny hides: gain and loss of horns during development and evolution of horned beetles. Evolution 60, 2329–2341. ( 10.1111/j.0014-3820.2006.tb01868.x) [DOI] [PubMed] [Google Scholar]
- 44.Moczek AP, Emlen DJ. 2000. Male horn dimorphism in the scarab beetle Onthophagus taurus: do alternative reproductive tactics favor alternative phenotypes? Anim. Behav. 59, 459–466. ( 10.1006/anbe.1999.1342) [DOI] [PubMed] [Google Scholar]
- 45.Choi J-H, et al. 2010. Gene discovery in the horned beetle Onthophagus taurus. BMC Genomics 11, 703 ( 10.1186/1471-2164-11-703) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Godfrey K. 1985. Fitting by organized comparisons: the square combining table. In Exploring data tables, trends, and shapes (eds Hoaglin DC, Mosteller F, Tukey JW.), pp. 37–66. New York, NY: Wiley. [Google Scholar]
- 47.Moczek AP, Kijimoto T, Snell-Rood EC, Rocha G, Pespeni M, Kafadar K. 2014. Evolutionary and ecological genomics of plasticity: novel approaches and first insights from the study of horned beetles. In Ecology and the evolution of genes and genomes series: advances in experimental medicine and biology, vol. 781 (eds Landry, Christian R, Aubin-Horth, Nadia), VI, 359 p. 85 illus., 57 illus. in color. Springer. [DOI] [PubMed] [Google Scholar]
- 48.Moczek AP, Nagy LM. 2005. Diverse developmental mechanisms contribute to different levels of diversity in horned beetles. Evol. Dev. 7, 175–185. ( 10.1111/j.1525-142X.2005.05020.x) [DOI] [PubMed] [Google Scholar]
- 49.Shafiei M, Moczek AP, Nijhout HF. 2001. Food availability controls onset of metamorphosis in the dung beetle Onthophagus taurus (Coleoptera: Scarabaeidae). Physiol. Entomol. 26, 173–180. ( 10.1046/j.1365-3032.2001.00231.x) [DOI] [Google Scholar]
- 50.Kijimoto T, Costello J, Tang Z, Moczek AP, Andrews J. 2009. EST and microarray analysis of horn development in Onthophagus beetles. BMC Genomics 10, 504 ( 10.1186/1471-2164-10-504) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Snell-Rood EC, Cash A, Han MV, Kijimoto T, Andrews A, Moczek AP. 2011. Developmental decoupling of alternative phenotypes: insights from the transcriptomes of horn-polyphenic beetles. Evolution 65, 231–245. ( 10.1111/j.1558-5646.2010.01106.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ashburner M, et al. 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29. ( 10.1038/75556) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Conesa A, et al. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. ( 10.1093/bioinformatics/bti610) [DOI] [PubMed] [Google Scholar]
- 54.Lee H, Braynen W, Keshav K, Pavlidis P. 2005. ErmineJ: tool for functional analysis of gene expression data sets. BMC Bioinform. 6, 269 ( 10.1186/1471-2105-6-269) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moczek AP, Emlen DJ. 1999. Proximate determination of male horn dimorphism in the beetle Onthophagus taurus (Coleoptera: Scarabaeidae). J. Evol. Biol. 12, 27–37. ( 10.1046/j.1420-9101.1999.00004.x) [DOI] [Google Scholar]
- 56.Moczek AP. 1999. Facultative paternal investment in the polyphenic beetle Onthophagus taurus: the role of male morphology and social context. Behav. Ecol. 10, 641–647. ( 10.1093/beheco/10.6.641) [DOI] [Google Scholar]
- 57.Moczek AP, Cochrane J. 2006. Intraspecific female brood parasitism in the dung beetle Onthophagus taurus. Ecol. Entomol. 31, 1–6. ( 10.1111/j.1365-2311.2006.00773.x) [DOI] [Google Scholar]
- 58.Shepherd BL, Prange HD, Moczek AP. 2008. Some like it hot: body and weapon size affect thermoregulation in horned beetles. J. Insect Physiol. 54, 604–611. ( 10.1016/j.jinsphys.2007.12.007) [DOI] [PubMed] [Google Scholar]
- 59.Shelby JA, Madewell R, Moczek AP. 2007. Juvenile hormone mediates sexual dimorphism in horned beetles. J. Exp. Zool. B, Mol. Dev. Evol. 308B, 417–427. ( 10.1002/jez.b.21165) [DOI] [PubMed] [Google Scholar]
- 60.Snell-Rood EC, Moczek AP. 2012. Insulin signaling as a mechanism underlying developmental plasticity: the role of FOXO in a nutritional polyphenism. PLoS ONE 7, e34857 ( 10.1371/journal.pone.0034857) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kijimoto T, Andrews J, Moczek AP. 2010. Programmed cell death shapes the expression of horns within and between species of horned beetles. Evol. Dev. 12, 449–458. ( 10.1111/j.1525-142X.2010.00431.x) [DOI] [PubMed] [Google Scholar]
- 62.Kijimoto T, Pespeni M, Beckers O, Moczek AP. 2012. Beetle horns and horned beetles: emerging models in developmental evolution and ecology. WIREs Rev. Dev. Biol. 2, 405–418. ( 10.1002/wdev.81) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kijimoto T, Moczek AP, Andrews J. 2012. Diversification of doublesex function regulates morph-, sex-, and species-specific expression of beetle horns. Proc. Natl Acad. Sci. USA 109, 20 526–20 531. ( 10.1073/pnas.1118589109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moczek AP, Rose DJ. 2009. Differential recruitment of limb patterning genes during development and diversification of beetle horns. Proc. Natl Acad. Sci. USA 106, 8992–8997. ( 10.1073/pnas.0809668106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Snell-Rood EC, Troth A, Moczek AP. 2012. DNA Methylation as a mechanism of nutritional plasticity: insights from horned beetles. J. Exp. Zool. B, Mol. Dev. Evol. 9999B, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Warren IAJC, Vera A, Johns R, Zinna JH, Marden DJ, Emlen I, Lavine LC. 2014. Insights into the development and evolution of exaggerated traits using de novo transcriptomes of two species of horned scarab beetles. PLoS ONE 9, e88364 ( 10.1371/journal.pone.00883364) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Array data: NCBI GEO accession number GSE58496. SNP data: DRYAD accession number doi:10.5061/dryad.gr5n3.