Abstract
Sexual dimorphisms, which are phenotypic differences between males and females, are driven by sexual selection. Interestingly, sexually selected traits show geographical variations within species despite strong directional selective pressures. This paradox has eluded many evolutionary biologists for some time, and several models have been proposed (e.g. ‘indicator model’ and ‘trade-off model’). However, disentangling which of these theories explains empirical patterns remains difficult, because genetic polymorphisms that cause variation in sexual differences are still unknown. In this study, we show that polymorphisms in cytochrome P450 (CYP) 1B1, which encodes a xenobiotic-metabolizing enzyme, are associated with geographical differences in sexual dimorphism in the anal fin morphology of medaka fish (Oryzias latipes). Biochemical assays and genetic cross experiments show that high- and low-activity CYP1B1 alleles enhanced and declined sex differences in anal fin shapes, respectively. Behavioural and phylogenetic analyses suggest maintenance of the high-activity allele by sexual selection, whereas the low-activity allele possibly has experienced positive selection due to by-product effects of CYP1B1 in inferred ancestral populations. The present data can elucidate evolutionary mechanisms behind genetic variations in sexual dimorphism and indicate trade-off interactions between two distinct mechanisms acting on the two alleles with pleiotropic effects of xenobiotic-metabolizing enzymes.
Keywords: sexual dimorphism, sexual selection, cytochrome P450, genetic polymorphisms
1. Introduction
Charles R. Darwin proposed that sexual selection drives sexual dimorphisms and speciation according to the reproductive strategy of each species [1]. Under such strong directional selection, alleles that enhance sexually selected traits are predicted to spread and become fixed within populations [2], leading to increased sexual dimorphism and decreased genetic diversity [3] due to more successful mating of individuals with specific pronounced traits. However, phenotypic variations in the degree of sexual dimorphism are often found among wild populations within species. Evolutionary biologists have long discussed this paradox [4,5], and several explanations have been proposed. Firstly, sexually selected traits can be indicative of a male's quality and condition [6]. Under this ‘indicator model’, alleles that contribute to male qualities can influence sexually selected traits, resulting in corresponding phenotypic variations [7]. Secondly, ecology can influence the degree of sexual dimorphism [8], even in cases where sexual selection acts on male or female traits [9], leading to increased sexual differences and competitive mating that selects phenotypes that are advantageous for reproductive success but are disadvantageous for survival [10,11]. Under this ‘trade-off model’ between sexual and natural selection, it is predicted that genes related to sexual dimorphism have pleiotropic functions, and may underpin differences in sexual dimorphisms as by-products of environmental adaptation. As a result, sexually selected traits can stabilize at an equilibrium point for both pressures, varying with environmental differences between geographically local populations. Although recent studies have indicated which evolutionary mechanism explains empirical phenotypic variation [11,12], the exact mechanism by which related genes influence the variation remains unknown.
To unravel evolutionary and molecular mechanisms of this paradox, we focused on medaka fish (Oryzias latipes), which are prevalent throughout East Asia [13,14] (electronic supplementary material, figure S1), because medaka have been shown to be an excellent animal model for studies of phenotypic variation [15]. Specifically, this model organism allows functional genetic comparisons to be made among geographically local populations with abundant genetic diversity [16,17], according to geographical differences in sexual dimorphism of anal fin morphology [18,19]. This variation of anal fin morphology is observed in wild medaka populations, and also has been reproduced during their rearing in constant environments of artificial breeding systems in laboratories [18], indicating that some genetic polymorphisms between populations may cause geographical differences in sexual dimorphism. In addition to such wild populations, it is empirically known that males from Tanabe (TN), which is a local population in mainland Japan, have larger anal fins, which are shaped as parallelograms and show clear sexual dimorphism (figure 1a,b,e,f), whereas males from the Maegok (MG) population, which is another local population in the Korean peninsula, have smaller right-angled triangle-shaped anal fins showing little sexual dimorphism (figure 1c,d,g,h). Furthermore, an ecological study of medaka proposed that the anal fin was a sexually selected trait because the anal fin had a sexual function in that it was used to hold females during mating (electronic supplementary material, video S1) and its morphology difference was positively correlated with reproductive success [20]. This geographical difference in medaka fish sexual dimorphism may provide a unique opportunity to examine phenotypic variations that underlie sexually selected traits.
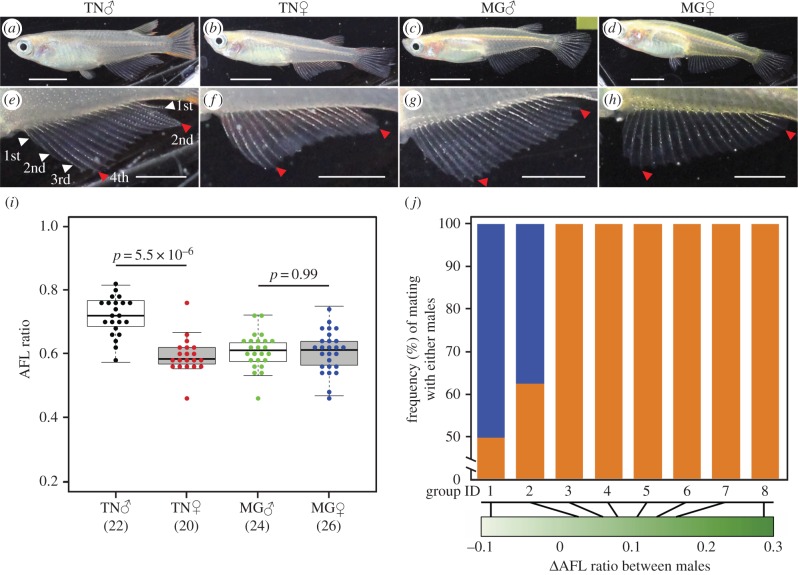
Figure 1.
Phenotypic diversity of anal fin shape between TN and MG populations. (a–h) Differences in anal fin shape between Tanabe (a,b,e,f) and Maegok (c,d,g,h). The photos (e–h) are the enlarged images of their anal fin (a–d), respectively. White and red arrowheads show the order of anal fin ray. Red arrowheads indicate the A-AFL and P-AFL defined as the lengths of the fourth and second anal fin rays from the anterior and posterior, respectively. White scale bars in (a–d) and (e–h) indicate 5 and 2.5 mm, respectively. (i) Sex-based variations in anal fin shape within Tanabe and Maegok. Boxplots represent five-number summary statistics for each group, with lower and upper error bars indicating minimum and maximum observations, the tops and bottoms of boxes represent the third and the first quartiles, respectively, and the middle bar represents the median. Numbers in parentheses represent the numbers of individual fish examined. Significant differences were identified using Kruskal–Wallis' test and Scheffe's type multiple tests. (j) Frequencies of successful mating events with male fish from Tanabe (orange bars) and Maegok (blue bars) are ordered by increasing differences (Δ) in AFL ratios between males (green gradient bar), which were calculated by subtracting AFL ratio of MG from of TN (TN-MG) (see also the electronic supplementary material, table S3).
Geographical differences in sexual dimorphism might be caused by polymorphisms of enzymes synthesizing/metabolizing sex hormones. A previous study discussed that geographical differences in anal fin morphology can correspond to differences in internal densities of androgen and oestrogen based on comparative data among local medaka populations [18]. Sex steroid hormones such as androgens and oestrogens often affect the development of sex differentiation in vertebrates [21]. In medaka, androgens and oestrogens are responsible for male- and female-specific characteristics of fin morphology, respectively [22,23]. Additionally, oestrogens are synthesized from androgens, a system that is highly conserved in vertebrates [24], and then are metabolized by xenobiotic-metabolizing enzymes [25]. Thus, it is just conceivable that the difference in the degree of sexual dimorphism between geographical populations can be caused by small imbalances between androgen and oestrogen internal concentrations, which attribute to some enzymatic variation in metabolizing the oestrogens.
Sex steroid hormones are synthesized by various vertebrate cytochrome P450 (CYP) enzymes (KEGG PATHWAY: map00140). CYP is known as a haem protein and shows a 450 nm spectral peak when it is reduced and bound to carbon monoxide [26]. CYPs compose a super-family exhibiting mono-oxygenase activity against their substrates and are classified according to amino acid sequence homology; their name starts with the abbreviation CYP, followed by a number for the gene family, a capital letter for the subfamily and the last number indicating the individual gene. These enzymatic functions have been well studied in humans because CYPs play an important role in xenobiotic metabolism and drug digestion [25]. Also in various fish species, the functions of CYPs have been well examined because their metabolites produced from environmental pollutants are known to influence sex determination or differentiation [27]. A balance between synthesis and excretion of sex steroids governs their internal levels in fish [28], indicating that CYP enzymes involved in such a process have a capability to alter the hormonal environment during a stage of development [27]. Thus, if CYP enzyme activity is changed according to amino acid substitution(s), then it might potentially cause sex reversal and/or offset of sex difference.
Considering the relationship between sex hormones and CYPs described above, variations in CYP activity metabolizing oestrogens can affect geographical differences in the degree of sexual dimorphism in medaka. Initial screening in our study exhibited high degrees of polymorphism among medaka CYPs and suggested that differing activities of CYP1A and CYP1B1, which metabolize xenobiotic and endogenous compounds [25], alleles contribute to differential regulation of sex steroid metabolism between medaka populations from various locations (electronic supplementary material, text S1 and figure S2). Therefore, we investigated the role of CYP polymorphisms and the ensuing molecular mechanisms that lead to geographical variations in medaka anal fin morphology and elucidate the evolution of sexual dimorphisms under CYP genes. To this end, we (i) test for the effect of geographical differences in sexual dimorphisms of anal fin morphology, (ii) measure differences in enzyme activity between CYP alleles using biochemical assays, and (iii) examine whether these alleles are associated with differences in degrees of sexual dimorphism and (iv) detect selective pressures acting on each allele of CYP1B1 by identifying mutations acquired from common ancestors.
2. Material and methods
All details of materials and methods are described in the electronic supplementary material. A brief description of our methodology is shown below.
(a). Samples
We initially examined 26 geographical populations and found two geographical populations originated from Tanabe in mainland Japan and Maegok in the Korean peninsula, which belong to S.JPN (Southern Japanese group) and W.KOR (Western Korean group) in mitochondrial genetic groups [14], respectively (electronic supplementary material, figure S1). In the subsequence experiments, we mainly used the two populations because those have candidate alleles concerning sexual dimorphism (see the electronic supplementary material, text S1 and figure S2).
(b). Morphological analysis of anal fins
Photos of Tanabe, Maegok and F2 individuals with body lengths more than 10 mm were taken using a digital camera (Eos Kiss, Canon) with a micro lens (Ultrasonic EF 100 mm, Canon). Standard body lengths (SL), anterior anal fin lengths (A-AFL) and posterior anal fin lengths (P-AFL) were then measured from photographs using Adobe Illustrator CS5 (Adobe). A-AFL and P-AFL were defined as the lengths of the fourth and second anal fin rays from the anterior and posterior, respectively (figure 1e). After the measurements, we performed CYP genotyping using genomic DNA extracted from the caudal fins, and then analysed the association between differences in anal fin shape and CYP genotypes using R programs.
(c). Mating experiment
To test whether males with a larger AFL ratio were more likely to mate with females, we placed two medaka males from Tanabe and Maegok, respectively, and one female from Tanabe or Maegok in a tank to observe competitive mating (electronic supplementary material, figure S3). We collected fertile eggs after each mating event (total of 198 eggs from 55 events) of eight groups (figure 1j and electronic supplementary material, table S3). Subsequently, we genotyped the LG22-1 (MF01SSA023H11) locus to determine whether a clutch of and/or abandoned eggs was fertilized by either males in each event, and then calculated the fraction of eggs fertilized by each male in each event in a group for the estimation of reproductive success.
(d). Measurement of cytochrome P450 enzyme activities
Previously, we established a cell-based assay for functional analyses of CYPs using Drosophila S2 cells [29]. Isolated full-length cDNAs of CYP1A and 1B1 were inserted into an expression vector. S2 cells were transfected with the vectors using Effectene transfection reagent (Qiagen) according to the manufacturer's protocol. Medaka CYP1A and CYP1B1 activities were quantified using P450-Glo CYP1A1 and P450-Glo CYP1B1 assay kits (Promega), respectively [30], and measured as d-luciferin luminescence using a microplate reader (PerkinElmer). The enzyme activities were calculated as luminescence ratios relative to that produced by the Tanabe allele.
(e). Estimation of dN/dS (ω) and assessment of ancestral CYP1B1 enzyme activities
We assumed a tree topology ((Tanabe, Kasumi), (Maegok, Shanghai), Luzon) and estimated ω (=dN/dS) values for each branch using maximum-likelihood calculations (figure 4a; electronic supplementary material, tables S8 and S9) using the program PAML [31]. To estimate the ancestral CYP1B1 amino acid sequences in O. latipes, we inferred the ancestral CYP1B1 sequences using the maximum-likelihood method [32] under the JTT matrix-based model [33] using the program MEGA5 [34]. Subsequently, we generated the putative ancestral CYP1B1 amino acid sequence using PCR-based site-direct mutagenesis and measured the activity of each CYP1B1.
Figure 4.
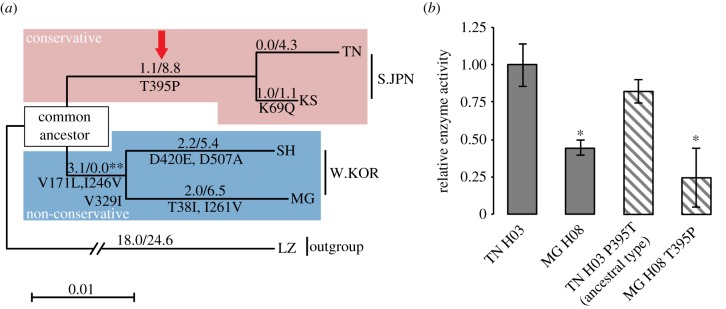
Reconstruction of ancestral CYP1B1. (a) Maximum-likelihood tree with synonymous and non-synonymous SNPs of Oryzias genus. LZ (O. lusonensis) from the Philippines was used as the out-group. The numbers on each branch show dN/dS (ω). Amino acid substitutions are indicated under each branch. The square on the tree indicates common ancestral sequence of S.JPN and W.KOR CYP1B1 inferred based on this tree; significant likelihood ratio test, **p < 0.02 (see also electronic supplementary material, table S10), are shown under the square. The red arrow shows the amino acids replaced in subsequent mutagenesis analyses. (b) Comparison of enzyme activities between wild-type and mutant enzymes. Constructs were generated using site-direct mutagenesis. This figure indicates that substitution of 395th amino acid had no effect on CYP1B1 enzyme activity. Maegok T395P was also generated to confirm the effect of the amino acid change. Each bar represents the mean ± s.d. from multiple independent samples (n = 3). Significant differences against Tanabe allele were identified using Games–Howell tests; *p < 0.05. (Online version in colour.)
3. Results and discussion
(a). The anal fin of medaka fish is a sexually selected trait
We initially quantified the differences in degrees of sexual dimorphism in anal fin morphology within- and between-medaka populations of Tanabe and Maegok, and their influence on reproductive success. To statistically compare anal fin characteristics, 92 individuals were sampled from each population. Adult fish were selected on the basis of body length (more than 15 mm), as secondary sexual characters of anal fin lengths were easily observable [35]. A-AFL and P-AFL (figure 1e) were measured, and AFL ratios (A-AFL/P-AFL) were calculated to compare sexual differences in anal fin shape between individuals from Tanabe and Maegok (figure 1f–h and electronic supplementary material, table S1). Statistically significant differences in AFL ratios were observed between males and females from Tanabe (p = 5.5 × 10−6; figure 1i), indicating pronounced sexual dimorphism. By contrast, no significant differences in AFL ratios were observed between males and females from Maegok, indicating less dimorphism.
The morphological differences in the anal fin can be characterized by genetic polymorphisms, and the anal fin might have evolved through strong intra-sexual competition, as indicated by the sexual function of the anal fin. Assuming that morphological variations in the anal fin affect mating behaviours, they may be positively correlated with reproductive success and may reflect strong sexual selection. We examined whether males with a larger AFL ratio had greater reproductive success by observing competitive mating behaviours of two males (from Tanabe and Maegok) and one female (from Tanabe or Maegok) medaka fish in a tank (electronic supplementary material, figure S3). We collected fertile eggs after each mating event of eight groups (total of 198 eggs from 55 events) (electronic supplementary material, tables S2 and S3), and determined which males contributed to the fertile eggs by genotyping in each event. As a result, the male carrying the larger AFL ratio mated more frequently with females (figure 1j; Spearman's rank correlation, ρ = 0.470, p = 0.004; electronic supplementary material, table S4), whereas there were no significant correlations with their genetic background (kinship between parents) and the difference in males' body size (electronic supplementary material, table S4). This indicated that males with greater AFL ratios were reproductively more successful than those with a ratio closer to zero. Thus, anal fin shape is significantly associated with reproductive success and represents a feature of sexual selection in medaka fish.
(b). CYP1B1 alleles show high and low enzyme activities
Assuming that associations exist between CYP1A and CYP1B1 variants and medaka anal fin shape, functional allelic differences may exist between the two populations. To assess variations in CYP enzyme activities, we cloned the first and second most frequent haplotypes of CYP1A and CYP1B1 from S.JPN and W.KOR, respectively, into pUAST expression vectors and expressed these in Drosophila S2 cells. Because medaka from Tanabe carried major CYP haplotypes (H02 in oCYP1A and H03 in oCYP1B1; electronic supplementary material, figure S2a,b), enzyme activities were expressed relative to those of CYP1A and CYP1B1 in fish from Tanabe. Kasumi's CYP1A haplotype (H03 oCYP1A) exhibited the highest relative activity (1.24-fold that from Tanabe medaka, p = 0.019), but large differences were not observed between the four haplotypes examined (range, 0.70–1.24; figure 2a). By contrast, relative enzyme activities of CYP1B1 haplotypes were significantly lower than those of the Tanabe haplotypes (figure 2b). Maegok haplotypes (H08 oCYP1B1) had substantially lower activities (26%; p = 0.017) than the major Tanabe haplotype (H03 oCYP1B1). Subsequent western blotting experiments confirmed that the observed inter-haplotype differences in CYP1B1 activity were not due to the relative amount of protein expression, but due to functional differences causing enzyme activity variation based on amino acid sequences (electronic supplementary material, figure S2f). Thus, the two CYP1B1 haplotypes H03 (Tanabe-type) and H08 (Maegok-type) had significantly higher and lower enzyme activities, respectively.
Figure 2.
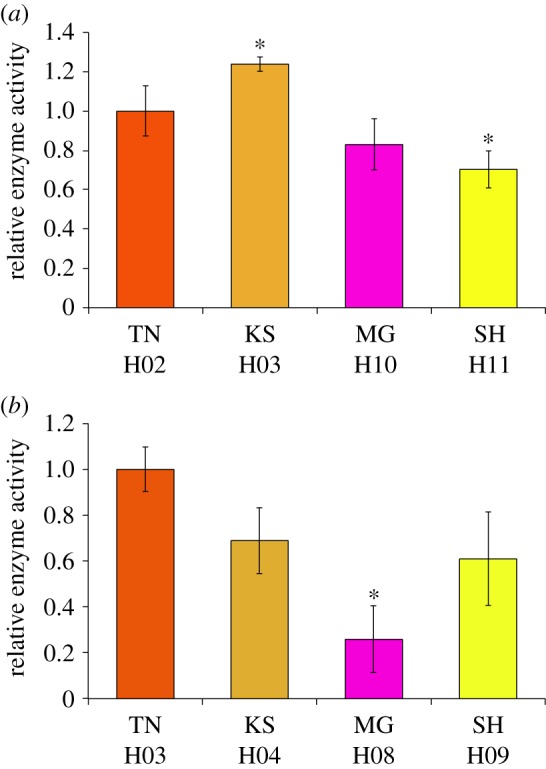
Relative enzyme activities of medaka CYP1A and CYP1B1. The x-axis displays the names of local wild medaka populations and corresponding CYP1A (a) and CYP1B1 (b) haplotypes. Each bar represents the mean ± s.d. from multiple independent samples. Each colour corresponds with that in the electronic supplementary material, figure S2a,b. Significant differences against Tanabe alleles were identified using Games–Howell tests; *p < 0.05.
(c). CYP1B1 alleles affect the difference in the degree of sexual dimorphism
Given that the CYP1B1 enzyme was involved in oestrogen metabolism in fish [36], the presence of the higher activity Tanabe-type alleles in a hybrid would increase anal fin dimorphism. Firstly, we used genetic and phenotypic data from 102 F2 fish obtained by genetic crossing Tanabe and Maegok medaka fish to examine the relationship between CYP1B1 genotype and anal fin morphology (electronic supplementary material, table S5). AFL ratios (A-AFL/P-AFL) significantly differed between the sexes in CYP1B1 Tanabe-type homozygotes (TT; p = 0.027, effect size (rTT) from Mann–Whitney's U = 0.946) and in heterozygotes of Tanabe and Maegok types (TM; p = 2.7 × 10−4, rTM = 0.815), but did not in Maegok-type homozygotes (MM; p = 0.22, rMM = 0.528; figure 3a). This pattern, rTT > rTM > rMM, is consistent with the number of high-activity alleles in the diploid. Namely, a greater male–female difference in AFL ratio for TT homozygotes and a smaller difference for heterozygotes were observed in F2 generation.
Figure 3.
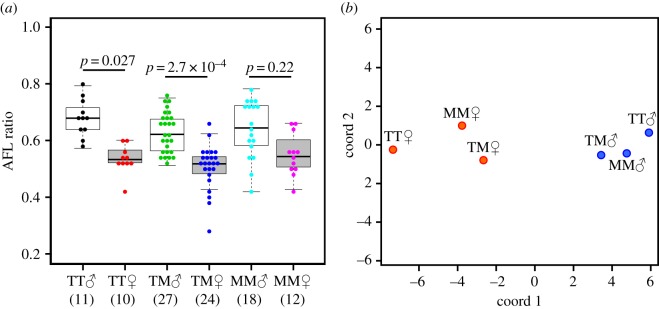
Comparisons of anal fin shape among F2 individuals. (a) Boxplots show differences in AFL ratios between combinations of CYP1B1 genotypes and sexes. Coloured dot plots show distributions of data within each group: TT, homozygote from TN; TM, heterozygote from TN and MG; MM, homozygotes from MG. Significant differences were identified using Kruskal–Wallis' test and Scheffe's type multiple tests. Numbers in parentheses represent the numbers of individual fish examined. (b) Multidimensional scaling plots of C-score-based Mahalanobis' generalized distances (D2) show marked morphological sex differences between CYP1B1 genotypes (see also the electronic supplementary material, table S7). Blue and red dots represent males and females, respectively.
Secondly, we analysed our F2 data using a generalized linear model (GLM) framework and obtained the best-fit model including CYP genotypes, AFL ratio and an interaction of the CYP genotypes and AFL ratio. The best-fit model was selected from a global model with all investigated explanatory variables based on the AIC values; i.e. we evaluated all the models that have every conceivable combination of variables and could explain our results of the genetic cross experiment (electronic supplementary material, table S6). Furthermore, a likelihood ratio test also showed model ‘L’, including the CYP genotype × AFL ratio interaction, was significantly more fitting to our F2 data than another model ‘H’ in which each variable was independent (p = 0.0078; table 1). In this model, ‘L’ including the interaction, significant effects of the CYP genotype, of the AFL ratio, and of their interaction were observed (table 1; p = 0.015, <0.001 and 0.017, respectively), whereas no significant effect of body size (p = 0.060) was detected. The plot of expectancies from the best-fit model ‘L’ showed a more gradual slope of the sex prediction by the AFL ratio with the Maegok-type homozygote than that with the Tanabe-type homo- and heterozygote, indicating that the Maegok-type allele reduces the sexual dimorphism (electronic supplementary material, figure S4). The best-fit model strongly supports the observation that CYP alleles interact significantly with anal fin shape, i.e. our GLM analysis thus showed a significant effect of genotype on the dimorphism.
Table 1.
Coefficients and standard errors (s.e.) of the two models by GLM framework. AIC, Akaike's information criteria; P, number of parameters; LLV, log likelihood value.
| model | variables | coefficient | s.e. | p-value |
|---|---|---|---|---|
| L | (intercept) | 9.421 | 4.427 | 0.033 |
| AIC = 90.9 | CYP genotypes | −7.831 | 3.23 | 0.015 |
| P = 5 | AFL ratio | −28.087 | 6.308 | <0.001 |
| LLV = −40.45 | CYP genotypes × AFL ratio | 13.296 | 5.574 | 0.017 |
| standard length | 0.378 | 0.201 | 0.06 | |
| H | (intercept) | 6.107 | 3.753 | 0.104 |
| AIC = 96 | CYP genotypes | −0.120 | 0.277 | 0.666 |
| P = 4 | AFL ratio | −21.540 | 4.309 | <0.001 |
| LLV = −43.99 | standard length | 0.354 | 0.187 | 0.058 |
Additionally, this GLM analysis gives us an insight into the role of CYP polymorphisms. The mean of the model including the CYP genotype × AFL ratio interaction was selected and indicates that the CYP1B1 gene is not responsible for an elongation/atrophy of anal fin length, but enhancement/decline of the sexual dimorphism. Indeed, although the AFL ratio between genotypes in males or females showed no significant difference, the between-sex difference in mean AFL ratio is different for each genotype (as above effect size). Recently, several genes related to fin formation in medaka were reported [22,37]. This GLM result suggests the possibility that the different enzyme activities between CYP1B1 polymorphisms contribute to the geographical variation in the development of fin ray formation in which those genes are involved.
Finally, we calculated C-score-based Mahalanobis' generalized distances (D2), which were corrected for the influence of differences in the sizes of analysed traits between groups of F2 individuals that were categorized according to CYP1B1 genotypes. D2 values indicated pronounced sex differences, and between-sex D2 values among Tanabe-type homozygotes (TT♂–TT♀) were approximately 1.6 times higher than those among Maegok-type homozygotes (MM♂–MM♀; electronic supplementary material, table S7). These were presented in two-dimensional deployments of D2 values using classical multidimensional scaling (figure 3b). TT males and females were more distant from each other than MM and TM males and females, indicating that homozygote males and females from Tanabe were more sexually dimorphic than those from Maegok. After excluding the size factor from each variable, this analysis still showed a strong association between CYP genotype and the degree of sexual dimorphism.
A candidate gene approach cannot, in general, eliminate a possibility that another gene(s) adjacent to the candidate locus is responsible for the trait under focus. However, we found that the possibility could be ruled out in this case based on database information and published data. First, in a recent medaka population genomic study by Spivakov et al. [38], the authors reported that linkage disequilibrium (LD) for medaka estimated from 16 trio genome sequences was approximately 12.5 kb on average, which is one-third that of human LD. We looked for the gene(s) in 12.5 kb upstream and downstream of CYP1B1 and found no genes in the total 25 kb region. Second, expanding the genomic region four times (50 kb, total 100 kb), we found only the NANP (N-acetyl neuraminic acid phosphatase) gene, which seems to have no relationship with fin morphogenesis and/or androgens. Thus, these results show that CYP1B1 is a predominant predictor gene for the difference in the degree of sexually selected traits.
(d). Detecting a signal of selective pressures acting on CYP1B1 alleles
Because males with larger anal fins mated more successfully (figure 1j), we expected that anal fin-derived sexual selection would be accompanied by alleles that give rise to high enzyme activity. To evaluate whether there have been selective pressures on CYP1B1, we estimated likelihood ratios [31], tested constancy of non-synonymous SNPs over synonymous SNPs ratios (dN/dS = ω) among evolutionary lineages (electronic supplementary material, table S8) and showed significant non-constancy of the ω value for each lineage (p < 0.05; electronic supplementary material, table S9). Interestingly, only a single amino acid substitution was found in the lineage from the common ancestral allele compared with the Tanabe-type allele, as indicated by a ω value (0.001) of significantly less than 1 (figure 4a and electronic supplementary material, table S9). In the S.JPN, nucleotide diversity of the CYP1B1 coding region was lower than that of other CYP genes (electronic supplementary material, figure S5), indicating evolutionary conservation of CYP1B1. These observations suggest that purifying selection has operated on the Tanabe-type allele. Moreover, reconstructed ancestral CYP1B1 showed high enzyme activity equivalent to that of the Tanabe-type allele (figure 4b), suggesting functional conservation of this allele for the 5 Myr since divergence from the common ancestral population (electronic supplementary material, table S10). These results support the association between the high enzyme activity (Tanabe-type) allele and the preponderance of sexual dimorphism, and strongly suggest that this allele has been maintained by sexual selection.
Because Maegok females mate more readily with males carrying larger AFL ratio (figure 1j), the ‘indicator model’ was ruled out, and an additional process to sexual selection may have contributed to reduced sexual dimorphism. In the phylogenetic tree, the ω value (3.1/0.0) significantly increased only on the branch of the ancestral allele that was common to both Maegok and Shanghai alleles (p < 0.02; figure 4a and electronic supplementary material, table S9), implying that positive selection operated on this allele. Given that previous studies have shown toxicity of CYP1B1 metabolites of 17β-oestradiol [39], and survival advantages of CYP inactivation in vertebrates under certain conditions with several environmental toxicants [25,40], selective pressures on the common ancestral population may have been environmental (e.g. water pollution). Hence, positive selection on the ancestral branch indicates that the low-activity allele may have been advantageous for survival of the ancestral population. These results indicate that adaptive evolution of this allele may have reduced sexual dimorphism as a by-product. This agrees with a theoretical explanation that the effects of positive selection must have predominated over those of sexual selection [6].
(e). A potential model animal of population genetics in vertebrates
In this study, we have used medaka to unravel evolutionary and molecular mechanisms of the paradox that genetic diversity is retained under sexual selection. Our data have shown that (i) medaka anal fin has geographical difference in the degree of sexual dimorphism, (ii) medaka CYP polymorphisms have variations of enzyme activity, (iii) corresponding to the difference of geographical populations, and (iv) the high-enzyme-activity allele has been plausibly maintained by sexual selection, while the low-enzyme-activity allele seems to be retained probably by environmental adaptation beyond sexual selection. The considerable criticism is that these might have been generated by genetic fixation during maintaining small populations bred in a laboratory for many generations. In order to see genetic diversity of medaka populations, we have preliminary conducted population genetic analyses for the medaka populations maintained in the closed colony. We have compared the diversity of CYP1B1 sequences to that of mtDNA D-loop, which is a potentially neutral genetic marker, for 12 and eight individuals from 12 S.JPN and eight W.KOR geographical populations, respectively. The eight W.KOR individuals are given by NBRP medaka, which are bred independently from our closed colonies. It is clear that Tanabe-type and Maegok-type alleles are not found in a particular local population, but very common in each S.JPN and W.KOR group (electronic supplementary material, figure S6). Furthermore, gene diversities calculated based on nucleotide sequences from the mtDNA D-loop region show substantial values, 0.955 and 0.964, in each group, suggesting that the medaka individuals used in the experiments of present study are not genetically identical between populations. This indicates that minor alleles have been fixed in neither TN nor MG populations: a possibility that unknowingly artificially selection due to the fixation of rare mutations has occurred by chance in our laboratory is quite low. Rather than that possibility, the distribution of the high- and low-activity alleles suggest that balancing selection between the sexual selection of anal fin morphology and the deleterious effects from an environmental toxicant may be the major driving force for variation of sexual dimorphism in the medaka population. Thus, even closed colonies have maintained their genetic diversity obtained in nature which has been formed under their evolutionary history, probably because medaka have originally high between-population genetic diversity which is not lost immediately by laboratory circumstances [14,16]. The characteristics of medaka as a model animal must be useful and powerful for addressing the various issues in the fields of evolutionary biology, morphology and behaviour, using genetics and population genetic theory.
We have interpreted that polymorphisms in genes encoding xenobiotic metabolizing enzymes can decrease sexual dimorphism under past adaptive evolution, despite the strong phenotypic influence of sexual selection. This mechanism can be distinct from the widely accepted mechanism that all sexually dimorphic traits are fundamental products of sex-limited gene expression [21]. However, we have not examined a gene expression of CYP1B1 in various tissues and stages of development, though a large difference in CYP1B1 gene expression was not observed between sexes in each medaka adult (electronic supplementary material, figure S7), which would be required in the near future. Although whole genome analyses are also required to identify genes related to the differences between geographical populations because variance of the sexual dimorphism cannot be explained solely by CYP genotypes, our candidate gene approach considerably examined differences in traits between geographically isolated populations of vertebrates and managed to find the gene related to sexual dimorphism. In addition, this study suggests analogous relationships between polymorphisms and phenotypic traits in wild medaka populations to those in humans (electronic supplementary material, text S1 and figure S8) as likely seen in other model fish [41]. Comparisons of distinct vertebrate populations with common genetic polymorphisms may provide insights into the evolutionary significance of functional differences between alleles.
Supplementary Material
Acknowledgements
We thank Mrs Shizuko Chiba, Mrs Sumiko Tomizuka, Dr Atsuko Shimada and Prof. Emeritus Akihiro Shima (University of Tokyo) for keeping medaka stocks from wild populations. We also thank Dr Naoki Osada (National Institute of Genetics) for his useful comments, Takeshi Matsuya and Tetsuya Yamada (University of Tokyo) for their support in western blotting experiments and measurements of CYP enzyme activity, and Dr Shuhei Mano (Institute of Statistical Mathematics) for his helpful advice in statistical analysis. We are also thankful to the National BioResource Project (NBRP)-medaka for providing wild medaka.
Data accessibility
Nucleotide sequence data were deposited into the international DNA database DDBJ/EMBL/GeneBank (accession nos.: AB829613–AB829721 and AB830922–AB830964). Phenotype–genotype and enzyme activities data were archived in the Dryad repository (doi:10.5061/dryad.vm060).
Funding statement
This study was supported in part by grants-in-aid for Challenging Exploratory Research from the Japan Society for the Promotion of Science, nos. 21657065 and 23657164 to H.O. and S.K., respectively. T.K. was supported by a grant-in-aid from JSPS Research Fellow (22-6207).
References
- 1.Darwin C. 1871. The descent of man, and selection in relation to sex. London, UK: J. Murray. [Google Scholar]
- 2.Kingsolver JG, et al. 2001. The strength of phenotypic selection in natural populations. Am. Nat. 157, 245–261. ( 10.1086/319193) [DOI] [PubMed] [Google Scholar]
- 3.Hartl D, Clark A. 2007. Principles of population genetics. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 4.Pomiankowski A, Moller AP. 1995. A resolution of the Lek paradox. Proc. R. Soc. Lond. B 260, 21–29. ( 10.1098/rspb.1995.0054) [DOI] [Google Scholar]
- 5.Promislow DEL. 1992. Costs of sexual selection in natural populations of mammals. Proc. R. Soc. Lond. B 247, 203–210. ( 10.1098/rspb.1992.0030) [DOI] [Google Scholar]
- 6.Andersson MB. 1994. Sexual selection. Princeton, NJ: Princeton University Press. [Google Scholar]
- 7.Tomkins JL, Radwan J, Kotiaho JS, Tregenza T. 2004. Genic capture and resolving the lek paradox. Trends Ecol. Evol. 19, 323–328. ( 10.1016/j.tree.2004.03.029) [DOI] [PubMed] [Google Scholar]
- 8.Butler MA, Sawyer SA, Losos JB. 2007. Sexual dimorphism and adaptive radiation in Anolis lizards. Nature 447, 202–205. ( 10.1038/nature05774) [DOI] [PubMed] [Google Scholar]
- 9.Reimchen TE, Nosil P. 2004. Variable predation regimes predict the evolution of sexual dimorphism in a population of threespine stickleback. Evolution 58, 1274–1281. ( 10.1111/j.0014-3820.2004.tb01706.x) [DOI] [PubMed] [Google Scholar]
- 10.Candolin U, Heuschele J. 2008. Is sexual selection beneficial during adaptation to environmental change? Trends Ecol. Evol. 23, 446–452. ( 10.1016/j.tree.2008.04.008) [DOI] [PubMed] [Google Scholar]
- 11.Johnston SE, Gratten J, Berenos C, Pilkington JG, Clutton-Brock TH, Pemberton JM, Slate J. 2013. Life history trade-offs at a single locus maintain sexually selected genetic variation. Nature 502, 93–95. ( 10.1038/nature12489) [DOI] [PubMed] [Google Scholar]
- 12.Hughes KA, Houde AE, Price AC, Rodd FH. 2013. Mating advantage for rare males in wild guppy populations. Nature 503, 108–110. ( 10.1038/nature12717) [DOI] [PubMed] [Google Scholar]
- 13.Katsumura T, et al. 2012. A population genetic study on the relationship between medaka fish and the spread of wet-rice cultivation across the Japanese archipelago. Anthropol. Sci. 120, 81–89. ( 10.1537/ase.110525) [DOI] [Google Scholar]
- 14.Katsumura T, Oda S, Mano S, Suguro N, Watanabe K, Mitani H, Oota H, Kawamura S. 2009. Genetic differentiation among local populations of medaka fish (Oryzias latipes) evaluated through grid- and deme-based sampling. Gene 443, 170–177. ( 10.1016/j.gene.2009.04.017) [DOI] [PubMed] [Google Scholar]
- 15.Omran H, et al. 2008. Ktu/PF13 is required for cytoplasmic pre-assembly of axonemal dyneins. Nature 456, 611–616. ( 10.1038/nature07471) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kasahara M, et al. 2007. The medaka draft genome and insights into vertebrate genome evolution. Nature 447, 714–719. ( 10.1038/nature05846) [DOI] [PubMed] [Google Scholar]
- 17.Wittbrodt J, Shima A, Schartl M. 2002. Medaka: a model organism from the far East. Nat. Rev. Genet. 3, 53–64. ( 10.1038/nrg704) [DOI] [PubMed] [Google Scholar]
- 18.Kawajiri M, Yamahira K. 2011. Ontogenetic variation in fin ray segmentation between latitudinal populations of the medaka, Oryzias latipes. Environ. Biol. Fish. 92, 285–293. ( 10.1007/s10641-011-9839-6) [DOI] [Google Scholar]
- 19.Oka TB. 1931. On the processes on the fin-rays of the male of Oryzias latipes and other sex characters of this fish. J. Fac. Sci. Univ. Tokyo Sect. IV Zool. 2, 209–219. [Google Scholar]
- 20.Koseki Y, Takata K, Maekawa K. 2000. The role of the anal fin in fertilization success in male medaka, Oryzias latipes. Fish. Sci. 66, 633–635. ( 10.1046/j.1444-2906.2000.00103.x) [DOI] [Google Scholar]
- 21.Williams TM, Carroll SB. 2009. Genetic and molecular insights into the development and evolution of sexual dimorphism. Nat. Rev. Genet. 10, 797–804. ( 10.1038/nrg2687) [DOI] [PubMed] [Google Scholar]
- 22.Ogino Y, Hirakawa I, Inohaya K, Sumiya E, Miyagawa S, Denslow N, Yamada G, Tatarazako N, Iguchi T. 2014. Bmp7 and Lef1 are the downstream effectors of androgen signaling in androgen-induced sex characteristics development in medaka. Endocrinology 155, 449–462. ( 10.1210/en.2013-1507) [DOI] [PubMed] [Google Scholar]
- 23.Ngamniyom A, Magtoon W, Nagahama Y, Sasayama Y. 2009. Expression levels of hormone receptors and bone morphogenic protein in fins of Medaka. Zool. Sci. 26, 74–79. ( 10.2108/zsj.26.74) [DOI] [PubMed] [Google Scholar]
- 24.Mizuta T, Kubokawa K. 2007. Presence of sex steroids and cytochrome P450 genes in amphioxus. Endocrinology 148, 3554–3565. ( 10.1210/en.2007-0109) [DOI] [PubMed] [Google Scholar]
- 25.Nebert DW, Dalton TP. 2006. The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nat. Rev. Cancer 6, 947–960. ( 10.1038/nrc2015) [DOI] [PubMed] [Google Scholar]
- 26.Omura T, Sato R. 1962. A new cytochrome in liver microsomes J. Biol. Chem. 237, PC1375–PC1376. [PubMed] [Google Scholar]
- 27.Devlin RH, Nagahama Y. 2002. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture 208, 191–364. ( 10.1016/S0044-8486(02)00057-1) [DOI] [Google Scholar]
- 28.Andersson T, Förlin L. 1992. Regulation of the cytochrome P450 enzyme system in fish. Aquat. Toxicol. 24, 1–19. ( 10.1016/0166-445X(92)90014-E) [DOI] [Google Scholar]
- 29.Yoshiyama-Yanagawa T, et al. 2011. The conserved Rieske oxygenase DAF-36/Neverland is a novel cholesterol-metabolizing enzyme. J. Biol. Chem. 286, 25 756–25 762. ( 10.1074/jbc.M111.244384) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cali JJ, Ma D, Sobol M, Simpson DJ, Frackman S, Good TD, Daily WJ, Liu D. 2006. Luminogenic cytochrome P450 assays. Expert. Opin. Drug Metab. Toxicol. 2, 629–645. ( 10.1517/17425255.2.4.629) [DOI] [PubMed] [Google Scholar]
- 31.Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. ( 10.1093/molbev/msm088) [DOI] [PubMed] [Google Scholar]
- 32.Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. New York, NY: Oxford University Press. [Google Scholar]
- 33.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 8, 275–282. ( 10.1093/bioinformatics/8.3.275) [DOI] [PubMed] [Google Scholar]
- 34.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. ( 10.1093/molbev/msr121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawajiri M, Kokita T, Yamahira K. 2009. Heterochronic differences in fin development between latitudinal populations of the medaka Oryzias latipes (Actinopterygii: Adrianichthyidae). Biol. J. Linn. Soc. 97, 571–580. ( 10.1111/j.1095-8312.2009.01233.x) [DOI] [Google Scholar]
- 36.Scornaienchi ML, Thornton C, Willett KL, Wilson JY. 2010. Cytochrome P450-mediated 17beta-estradiol metabolism in zebrafish (Danio rerio). J. Endocrinol. 206, 317–325. ( 10.1677/JOE-10-0075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iida Y, Hibiya K, Inohaya K, Kudo A. 2014. Eda/Edar signaling guides fin ray formation with preceding osteoblast differentiation, as revealed by analyses of the medaka all-fin less mutant afl. Dev. Dyn. 243, 765–777. ( 10.1002/dvdy.24120) [DOI] [PubMed] [Google Scholar]
- 38.Spivakov M, et al. 2014. Genomic and phenotypic characterization of a wild medaka population: towards the establishment of an isogenic population genetic resource in fish. Genes Genomes Genet. 4, 433–445. ( 10.1534/g3.113.008722) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gajjar K, Martin-Hirsch PL, Martin FL. 2012. CYP1B1 and hormone-induced cancer. Cancer Lett. 324, 13–30. ( 10.1016/j.canlet.2012.04.021) [DOI] [PubMed] [Google Scholar]
- 40.Wirgin I, Roy NK, Loftus M, Chambers RC, Franks DG, Hahn ME. 2011. Mechanistic basis of resistance to PCBs in Atlantic tomcod from the Hudson River. Science 331, 1322–1325. ( 10.1126/science.1197296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller CT, Beleza S, Pollen AA, Schluter D, Kittles RA, Shriver MD, Kingsley DM. 2007. cis-Regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell 131, 1179–1189. ( 10.1016/j.cell.2007.10.055) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Nucleotide sequence data were deposited into the international DNA database DDBJ/EMBL/GeneBank (accession nos.: AB829613–AB829721 and AB830922–AB830964). Phenotype–genotype and enzyme activities data were archived in the Dryad repository (doi:10.5061/dryad.vm060).