Abstract
Advanced resources for genome-assisted research in barley (Hordeum vulgare) including a whole-genome shotgun assembly and an integrated physical map have recently become available. These have made possible studies that aim to assess genetic diversity or to isolate single genes by whole-genome resequencing and in silico variant detection. However such an approach remains expensive given the 5 Gb size of the barley genome. Targeted sequencing of the mRNA-coding exome reduces barley genomic complexity more than 50-fold, thus dramatically reducing this heavy sequencing and analysis load. We have developed and employed an in-solution hybridization-based sequence capture platform to selectively enrich for a 61.6 megabase coding sequence target that includes predicted genes from the genome assembly of the cultivar Morex as well as publicly available full-length cDNAs and de novo assembled RNA-Seq consensus sequence contigs. The platform provides a highly specific capture with substantial and reproducible enrichment of targeted exons, both for cultivated barley and related species. We show that this exome capture platform provides a clear path towards a broader and deeper understanding of the natural variation residing in the mRNA-coding part of the barley genome and will thus constitute a valuable resource for applications such as mapping-by-sequencing and genetic diversity analyzes.
Keywords: barley, genomics, genetic diversity, Hordeum bulbosum, Hordeum pubiflorum, Hordeum vulgare, targeted resequencing, Triticeae
Introduction
Barley (Hordeum vulgare L.) is the fourth most important cereal crop worldwide. It is a true diploid and has been proposed as a model for genomic research in the Triticeae tribe, which includes wheat and rye (Schulte et al., 2009). The construction of a reference sequence of the barley genome has remained elusive to date, owing to its large genome size (5 Gb) and abundance of repetitive elements. The International Barley Genome Sequencing Consortium (IBSC) recently presented a gene-space assembly (The International Barley Genome Sequencing Consortium, 2012) of cultivated barley as an enabling platform for genome-assisted basic research and crop improvement. This assembly represents 86% of the entire barley gene set and, through extensive physical and genetic mapping resources, the sequence contigs have been arranged in a linear order.
The barley genome assembly is gene-focussed and enables a gene-based resequencing strategy that is relevant to both academic and applied interests. Hybridization-based exome capture is an established method for targeted resequencing of the gene space (Bamshad et al., 2011). Briefly, whole genomic DNA is hybridized to pools of oligonucleotide probes that are specific to a set of exons, capturing sequences that are homologous to the targeted regions. The probes are immobilized either by covalent attachment to a glass support (Hodges et al., 2007) or by biotin–streptavidin linkage to an insoluble matrix such as magnetic beads (Bainbridge et al., 2010). The latter approach offers the advantage that both probe and target are in solution during hybridization, decreasing hybridization time. Non-homologous sequences are removed by washing and the hybridized portion eluted and sequenced. As the region targeted for sequencing is greatly reduced, sequencing costs per genome are dramatically lower, allowing high coverage depth of targets and sensitive and accurate variant and genotype calling. Moreover, downstream computational costs per genome for data management, read mapping and variant calling are correspondingly reduced.
Restricting attention to only the mRNA-coding part of the genome can be sufficient to elucidate the molecular basis of natural or induced genetic variation. In biomedical research, exome capture has been applied successfully for the discovery of coding mutations that underlie human disease (see review of Bamshad et al., 2011) and mutant phenotypes in mice (Fairfield et al., 2011). In maize, a haplotype map (Gore et al., 2009) has been constructed by resequencing only low-copy regions of the genome in different genotypes, and sequence polymorphisms within genic regions have been estimated to contribute a large fraction of the natural variation to quantitatively inherited traits (Li et al., 2012). Furthermore, a whole exome capture assay was used to validate presence–absence variation between maize cultivars (Liu et al., 2012). In the Brassicaceae, sequence capture has been proposed as a cost-effective alternative to whole-genome resequencing for sequence-based genetic mapping (Galvao et al., 2012).
Exome capture assays designed for one species have also been successfully applied to enrich genic regions from related species. For instance, human assays have been used to capture exomes of Neanderthals and non-human primates (Burbano et al., 2010; Vallender, 2011) and an assay designed for domestic cattle has been applied to wild bison (Cosart et al., 2011). In agricultural plants, genomic tools developed for cultivated species could have great value for applicability in the less resourced wild and related species, for both phylogenetic analysis, population and evolutionary genetics research and in strategic and applied plant science (Wright et al., 2005; Ramchiary et al., 2011; Russell et al., 2011; Hufford et al., 2012; Kilian and Graner, 2012). In a breeding context, wide crosses between cultivars and wild relative species have been recognized as a means to introduce beneficial alleles into agriculture conferring, for example, resistance to biotic and abiotic stress. The development of strategies that overcome some of the practical limitations of utilizing wild germplasm in agriculture has become a major goal of crop genomic research (reviewed in Feuillet et al., 2008).
Here, we report the implementation and evaluation of a whole exome capture assay for cultivated barley, and demonstrate its applicability to genome-wide variant discovery in related species.
Results
Design of the barley exome capture platform
Target sequences were selected for probe design (Table 1), including: (i) 155 863 exons predicted by the Cufflinks pipeline from RNA-Seq reads mapped to the whole-genome shotgun (WGS) assembly of barley cv. ‘Morex’ (NCBI accession CAJW01, the ‘Morex assembly’); (ii) 35 297 full-length cDNA sequences; and (iii) 108 759 transcript contigs assembled from RNA-Seq reads. To assure that the probe design included loci previously Sanger sequenced, the design was examined for the presence of all barley genes >1000 bp available from GenBank (243 genes). Sequences were absent or partially present were added to the design (46 genes). After filtering this set of all target sequences for repetitive regions by kmer analysis, a capture space of 90.2 Mb was defined.
Table 1.
Sequence input for capture design
| Probe seta | No. of sequences | Length (bp) | Predicted coverage (%)b | Source |
|---|---|---|---|---|
| Cufflinks predictions | 155 863 | 56 418 032 | 98.5 | The International Barley Genome Sequencing Consortium (2012) |
| Full-length cDNA | 35 297 | 14 707 427 | 97.0 | Matsumoto et al. (2011) |
| RNA-Seq contigs | 108 759 | 30 663 579 | 97.9 | This study |
| Total | 300 919 | 101 789 038 | 97.8 |
Number of sequence contigs after filtering for repetitive sequences.
Predicted sequence capture coverage extending capture targets by 100 bp on each side.
Target sequences derived from full-length cDNA or RNA-Seq contigs were aligned against the Morex assembly. The Cufflinks exons are recorded as coordinates on the Morex assembly. Ninety-two percent of all transcript contigs could be positioned on the Morex assembly and overlapped with Cufflinks exons, thus their mapping collapsed the target space to 61.6 Mb of non-overlapping intervals on the Morex contigs. These target regions were used for subsequent analyzes of capture efficacy. 45.3 Mb (73.6%) of target sequence were located on sequence contigs anchored to the physical and genetic framework of barley (The International Barley Genome Sequencing Consortium, 2012). Approximately 34.6 Mb corresponded to exons of so-called high-confidence genes (The International Barley Genome Sequencing Consortium, 2012), an additional 16.4 Mb represented low confidence exons; 10.6 Mb of the capture space are not located in annotated exons. These sequences originate presumably from full-length cDNAs or from de novo RNA-Seq contigs that represent genes not included in the current annotation. Almost three-quarters (73.7%) of high-confidence exonic sequence and 40.7% of low confidence exon sequence annotated on the basis of the barley draft genome assembly (IBSC, 2012) are represented by our target regions. Note that we used a preliminary version of the RNA-Seq annotation of the Morex assembly, so that not all gene models defined in the published dataset have been included in the target space.
The design workflow for the exome capture assay is shown in Figure S1(a). A prototype design consisting of 4 021 945 oligonucleotide probes was synthesized and tested on cultivars Barke, Bowman, Morex and Steptoe. Relative performance of the probes in this design were assessed for over- or under-representation in the captured exomes. Two second-phase designs were then developed, by empirically re-balancing probe coverage of the targets to improve capture uniformity. The best of these designs was adopted as final and is available from the Roche NimbleGen by requesting SeqCap EZ Developer probe pool design 120426_Barley_BEC_D04.EZ.
Performance of the barley exome capture platform on barley cultivars
The performance of the capture design was assessed for 36 samples from 13 different barley cultivars (Table 2, Table S1). For the analysis of target coverage and single nucleotide polymorphism (SNP) calling, only properly paired non-duplicate reads were considered, i.e. those read pairs in which both single reads map to the same genomic location. A workflow chart of our analysis pipeline is given in Figure S1(c). On average, 49.6% of reads were mapped as proper pairs and passed the duplicate removal filter. Between 64 and 90% of these reads mapped within or near (±300 bp) target regions. An example of a target region with mapped reads is shown in Figure S2.
Table 2.
Barley cultivars and wild relatives included in this study
| Species | Cultivar/accession | No. of captures |
|---|---|---|
| Hordeum vulgare | Barke | 11 |
| Borwina | 1 | |
| Bonus | 1 | |
| Bowman | 3 | |
| Foma | 1 | |
| Gull | 1 | |
| Harrington | 1 | |
| Haruna Nijo | 1 | |
| Igri | 1 | |
| Kindred | 1 | |
| Morex | 11 | |
| Steptoe | 2 | |
| Vogelsanger Gold | 1 | |
| Hordeum spontaneum | B1K-04-12 | 1 |
| OUH602 | 1 | |
| B1K-03-07 | 1 | |
| Hordeum bulbosum | A42 (autotetraploid) | 1 |
| BCC 2061 (diploid) | 1 | |
| 2940/4 (diploid) | 1 | |
| Hordeum pubiflorum | BCC 2028 | 1 |
| Triticum aestivum | Chinese Spring | 1 |
For individual samples sequenced on a single HiSeq2000 lane (≥30 Gb of sequence), capture sensitivity was very high with more than 95% of all target bases covered by at least 10 reads (Figure 1). Sensitivity was slightly lower for multiplexed capture: 79–93% of targets had 10-fold coverage. Specificity was comparable between individual and pooled captures (Approximately 78% on-target reads) and capture performance was similar among different genotypes (Table S1). To test how well the sequencing output per sample is controlled by the multiplexing level, sequencing libraries from different accessions were barcoded and hybridized together in combinations with different molar ratios. The target coverage reflects the pooling ratios of the samples very well, although enrichment seems to be slightly more efficient at lower concentrations (Figure 2).
Figure 1.
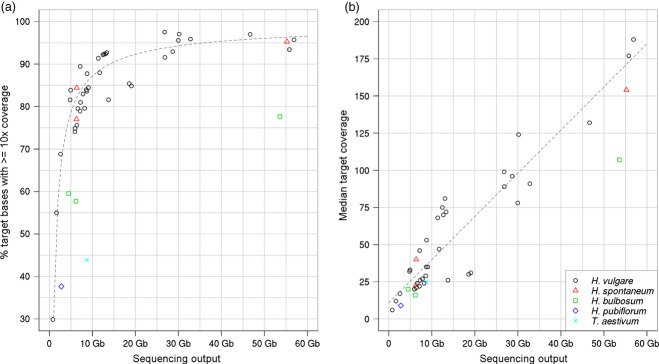
Target coverage in cultivars and related species.The percentage of target regions with at least 10-fold coverage (a), and the median coverage (b), of target regions are plotted as function of raw sequencing output. Different symbols are used for samples from different species. The legend is given in (b) for both panels. Regressions lines were obtained fitting the model log(1-y)∼log(x) (a), or a linear model (b), to the data points of Hordeum vulgare.
Figure 2.
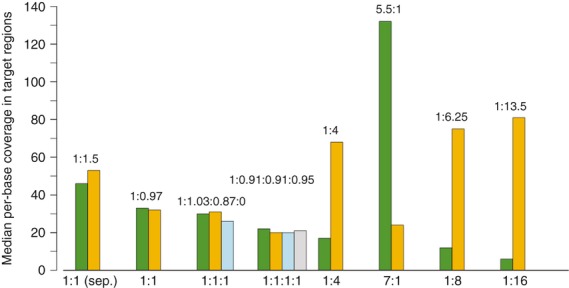
Target coverage in library combinations.The median per-base coverage in target regions is depicted cultivars Morex (green), Barke (orange), Bowman (blue) and Steptoe (gray). Libraries from these cultivars were combined at different molar ratios. The first group of bars corresponds to an experiment in which libraries from Morex and Barke were combined after hybridization. Otherwise, libraries were hybridized to the capture assay in combination. The molar ratios of the combinations are given below the bars. The observed ratios between the median target coverage are printed above the bars.
For genome-wide resequencing studies that involve a number of accessions, it is preferable that a target is covered equally across all accessions in order to maximize the amount of comparable sequence data. We determined the intervals on the Morex assembly (not necessarily within target regions) that are covered in captured samples from all 13 barley cultivars included in this study (Figure 3). The number of raw reads varied between 59 million and 133 million reads per sample. Approximately 49.4 Mb of sequence were covered at 10-fold in all 13 samples. About 92.5% of these intervals were located in or within 300 bp of target regions; 63.4% of all predefined target had at least 10-fold coverage in all 13 samples and 78.6% were covered by five or more reads. About 3.6 Mb of sequence (7.3%) were covered by 10 reads in all 13 samples, but were not within 300 bp of target regions. Seventy-six percent of this portion of sequence had a BLASTN hit with at least 50 bp alignment length and sequence identity between 80 and 98% to a predefined capture target, and indicated that reliably captured genomic regions not within the target spaces are in most cases paralogs of target sequences.
Figure 3.
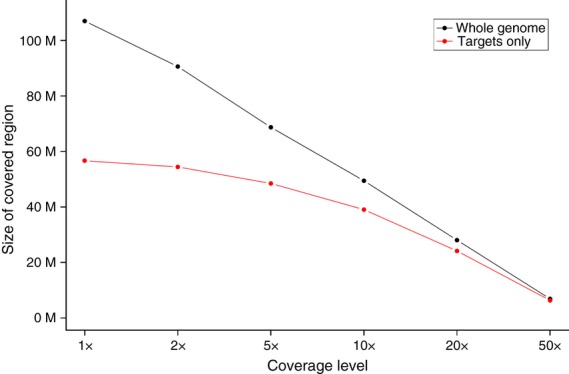
Coverage across 13 cultivars.The size of sequence intervals on the barley WGS assembly covered by at least 1, 2, 5, 10, 20 or 50 reads in all captured samples from a set of 13 cultivars. All bases positions (black dots) or only positions in target regions (red dots) were considered.
Intervals with reliable capture coverage identified in this analysis are available for download from ftp://ftp.ipk-gatersleben.de/barley_exome_capture.
Performance of the barley exome capture platform for related Hordeum species
The platform was tested for its efficacy with members of the primary, secondary and tertiary gene pools of the genus Hordeum (van Hintum and Menting, 2003). Exome capture and sequencing was performed for H. vulgare L. subsp. vulgare and subsp. spontaneum (K. Koch) Thell., H. bulbosum L. and H. pubiflorum Hook f. and Triticum aestivum. Sequencing reads from all samples were mapped against the Morex assembly. The number of reads that could be mapped to the barley assembly decreased with phylogenetic distance (Blattner, 2009). Nevertheless, a substantial proportion of the target space obtained sufficient read coverage even in a capture of the more distantly related T. aestivum (Figure 1 and Table S1). To decide whether targets not covered in related species were not captured during hybridization or whether captured reads could not be mapped to the Morex assembly because of excessive sequence divergence, we sequenced an accession of H. pubiflorum to approximately 19-fold haploid genome coverage with short Illumina reads (2 × 100 bp) and constructed a de novo gene-space assembly. Although the resulting assembly was incomplete and highly fragmented (assembly length 1.4 Gb, 1.8 million contigs with an N50 of 1662 bp), it can be expected to contain near full-length sequences of most genes as was demonstrated in the case of the Morex assembly (The International Barley Genome Sequencing Consortium, 2012). About 90% of all reads captured from H. pubiflorum could be mapped to its assembly. The number of reads that mapped as correct pairs increased by 47% from 14.8 million to 21.9 million when the H. pubiflorum assembly was used as a mapping reference. To assess capture sensitivity and specificity, we projected the target regions from the Morex assembly onto the H. pubiflorum assembly. About 82% of all target regions could be mapped to the H. pubiflorum assembly. Using the target intervals now defined on the assembly, we found that 85.5% of all reads lay within 300 bp of targets. The median depth of coverage in target regions was 20, and 66% of all targets were covered by at least 10 reads. Overall, 22.6 Mb of sequence were covered at a depth of at least 10-fold, 90% of which was comprised of on-target reads (±300 bp).
Whole-genome shotgun sequencing was performed for one accession of H. bulbosum. H. bulbosum can be either diploid or autotetraploid (Linde-Laursen et al., 1990; Brassac et al., 2012). The tetraploid accession A42 was sequenced to nine-fold polyhaploid genome coverage (assuming a diploid genome size of approximately 4.5 Gb (Jakob et al., 2004)). Assembling 82 Gb of sequencing reads resulted in a set of 2.8 million contigs with a cumulative length of 1.3 Gb and an N50 of 511 bp. The assembly statistics are substantially worse when compared with the assembly of diploid H. pubiflorum. Presumably, sequencing depth was too low or the assembler was unable to disentangle homoeologous sequences, resulting in a large number of small contigs. Mapping exome capture reads of H. bulbosum accession A42 to the A42 assembly did not substantially improve the mapping rate: 27.9 million reads could be mapped as proper pairs to this assembly, an increase of only 3% compared with 27.0 million read pairs mapped to the Morex assembly.
We conclude that, although the size of the regions efficiently captured is reduced, the specificity of our assay when applied to Hordeum species is comparable with barley cultivars. For the analysis of sequencing data from more diverse diploid species, we recommend producing a de novo WGS assembly. For polyploid Hordeum species, it is advantageous to rely on the barley assembly as de novo assemblies from next generation sequencing (NGS) data of polyploids can be too fragmented to serve as a useful reference.
Performance of the barley exome capture for hexaploid wheat
One sample of hexaploid wheat (Triticum aestivum cv. Chinese Spring) was captured with our platform and reads were mapped against WGS assemblies of barley and wheat. For wheat, there are two assemblies available (Brenchley et al., 2012) that are based on the same set of 454 reads. An orthologous group assembly (OGA) had been constructed from reads with sequence similarity to known grass genes, and a low-copy-number genome assembly (LCG) obtained from non-repetitive reads only (Brenchley et al., 2012).
About half of our exome capture reads could be mapped against the barley assembly with 96.2% of them being on target (Table S1). Mapping rates increased for the wheat assemblies: 82.8 and 63.0% of raw reads could be mapped to the low-copy-number and orthologous group assemblies, respectively. The LCG assembly incorporated more sequence information (3.6 Gb versus 437 Mb) and its contigs were, on average, about twice as long as OGA contigs (N50 of 884 bp versus 481 bp) (Brenchley et al., 2012), explaining the substantially higher amount of reads mapped to LCG contigs.
The intervals on the LCG and OGA assemblies with ≥5 × coverage had a cumulative length of 115.4 Mb and 132.6 Mb, respectively. About 46.2% of OGA contigs had been assigned to one subgenome (Brenchley et al., 2012). The ratio between the amount of sequence with five-fold coverage in the three subgenomes was 1:0.87:1.18 (A:B:D). The ratio was 1:1.20:1.30 for all OGA contigs, a finding that indicated that the barley assay is able to capture sequences from all three subgenomes, probably with slight imbalances between the three subgenomes. A whole exome capture assay based on EST and cDNA sequences has been specifically designed for hexaploid wheat (Winfield et al., 2012) and is probably more suitable for targeted resequencing in wheat. However, for diploid Aegilops, Triticum or Secale species, performance of the barley assay could be similar or better than the wheat assay, as it has been designed on the basis of a gene-space assembly and avoided probes across exon–exon junctions.
SNP discovery using the barley exome capture platform
Sequence capture has been recognized as a cost-effective means for genome-wide variant discovery (Davey et al., 2011). In a plant science context, exome sequencing could be applied, for instance, to the parents of a bi-parental mapping population and detected sequence variants converted into marker assays. To assess the usefulness of the whole exome capture for variant discovery, SNP calling was performed in the captured sample from cultivars and wild relatives with a computational pipeline consisting of BWA and SAMtools (Li and Durbin, 2009; Li, 2011) to assess the usefulness of whole exome capture for variant discovery in barley. As a reference sequence, the Morex assembly was always used. As it has been shown that variant calls in off-target regions are of high quality if coverage is sufficient (Guo et al., 2012), we did not restrict our attention to target regions, but counted any variant position on the Morex assembly. When sequencing depth was higher, so was the overall number of called variants. SNP counts in target regions, however, did not increase substantially when the sequencing output increased from approximately 10 Gb to approximately 30 Gb (Figure 4 and Table S1). Although at lower frequency, SNPs were detected relative to the Morex reference in the Morex exome capture reads (Table S1). Similar results had been reported for SNP calling from whole-genome resequencing data (The International Barley Genome Sequencing Consortium, 2012). Approximately, 68% of Morex–Morex SNPs called in the high coverage exome capture sample had also been identified from genomic data. The occurrence of these SNPs had been attributed partially to low sequence accuracy particularly at the ends of WGS contigs. Those spurious variant positions, if detected in other samples, should be treated with caution. A table of all variant positions and genotype calls is available for download from ftp://ftp.ipk-gatersleben.de/barley_exome_capture.
Figure 4.
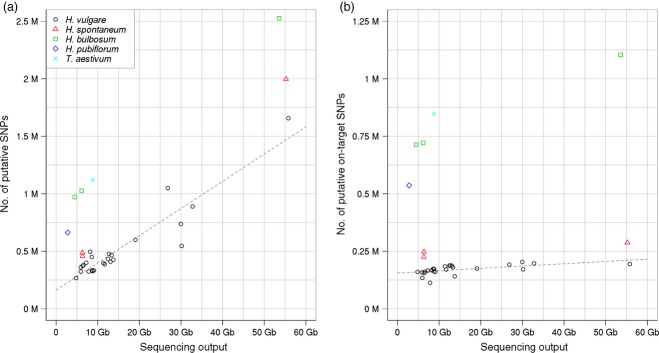
Number of detected single-nucleotide polymorphisms (SNPs).The number of SNPs detected between Morex and samples from other barley cultivars and related species across the genome (a) or only in target regions (b) is plotted as a function of sequencing depth. The legend for both panels is shown in (a). Regression lines were obtained by fitting a linear model to Hordeum vulgare data points.
We visualized the SNP frequency between Morex and other cultivars in 10 Mb windows along the physical and genetic framework of barley (Figure 5). In all samples, variant frequency decreases by about one to two orders of magnitude in the centromeric and peri-centromeric regions compared with variant frequency in the telomeric regions. We attribute this finding to a decreased frequency both of genes (and consequently targeted exons) and sequence variation in the genetic centromere of barley (The International Barley Genome Sequencing Consortium, 2012).
Figure 5.
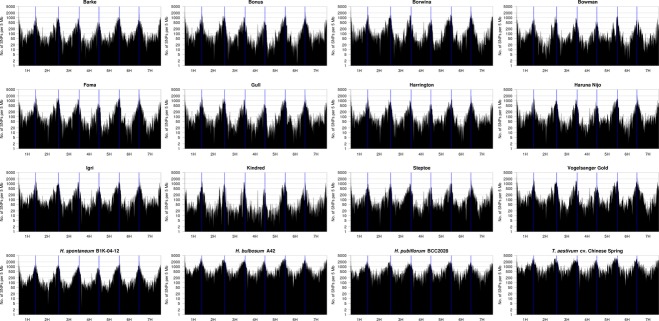
Frequency of single-nucleotide polymorphisms (SNPs) in exons along the barley chromosomes.The SNP frequency in 5 Mb windows along the physical length of the seven barley chromosomes is plotted for 12 cultivars, three accessions from species in the genus Hordeum and bread wheat cv. Chinese Spring. SNP calling was performed against a reference sequence from barley cultivar ‘Morex’. The positions of contigs were taken from the barley physical framework (IBSC, 2012).
To demonstrate the usefulness of exome sequencing for characterization of barley germplasm, we constructed a phylogenetic tree for barley cultivars and three subsp. spontaneum accessions from 122 940 SNPs with no missing data (Figure 6). Barley cultivars with a shared pedigree (e.g. Kindred and Morex or Bonus, Gull and Foma) cluster together, as expected. Wild accessions are clearly separated from each other as well as from cultivated barley.
Figure 6.
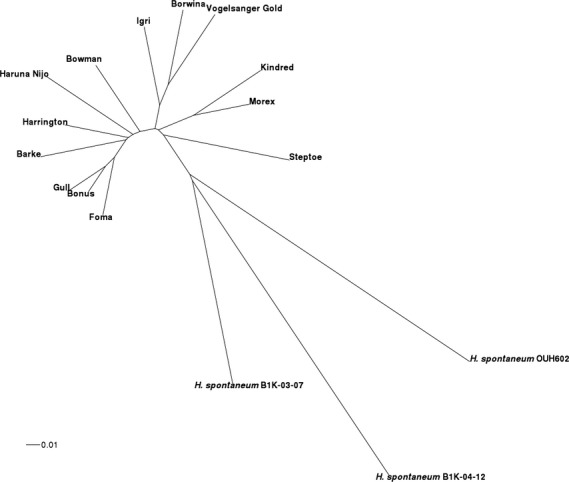
Neighbor-joining tree.The tree depicts the single-nucleotide polymorphism (SNP) distance between 13 barley cultivars and three Hordeum spontaneum accessions as inferred from genotype calls at 122 940 bi-allelic SNPs with no missing data. A tree including H. bulbosum and H. pubiflorum accessions is shown in Figure S3.
Including captured samples from wild relatives in the analysis, doubled the number of polymorphic markers and the resulting neighbor-joining tree correctly put these samples distant from both cultivated and wild barley (Figure S3). Exome sequencing of diverse panels of barley cultivars, landraces and wild accessions should overcome the ascertainment bias of array-based genotyping assays tailored for elite cultivars (Moragues et al., 2010) and enable the correct estimation of levels of genetic diversity in various sets of barley germplasm.
Discussion
We developed an assay to sequence selectively a large portion of the mRNA-coding part of the barley genome, validated the performance of our assay and outlined the path for the computational analysis of barley exome sequencing data. Read mapping and variant calling were performed against the WGS assembly of the barley cultivar ‘Morex’. The subsequent visualization of results made use of the partial order of WGS contigs in the barley physical and genetic framework (The International Barley Genome Sequencing Consortium, 2012).
To assess the performance of our assay, we mapped the portion of our capture space, that was derived from previous established transcriptome resources, onto the Morex assembly. Moreover, by comparing the coverage across different samples, we identified regions (not necessarily within target regions) expected to be captured reliably in a wide spectrum of cultivars. Regions represented in the sequencing data, but not originally targeted, are most likely to be fragmented copies or nearly identical paralogs of our targeted genes. As the capture experiments were carried out independently in four different laboratories, these results on capture sensitivity and specificity provide a robust guideline of the capture performance.
The barley genotypes we sequenced for assessing capture performance are interesting in their own right. The cultivars Morex, Barke, Igri, Harrington, Steptoe and the subsp. spontaneum accession OUH602 are among the parents of widely used mapping populations (Graner et al., 1991; Hayes et al., 1993; Yun et al., 2005; Comadran et al., 2012). The cultivar ‘Bowman’ is the recurrent parent in an extensive collection of nearly isogenic lines (Druka et al., 2011) that was developed by recurrently backcrossing independent mutant alleles to Bowman. Many of these mutants had been induced in the cultivars Bonus, Foma or Gull. Thus, the sequence variants detected in our sequencing data may contribute to further molecular characterization of this mutant collection as well as to gene isolation.
Our probe design is not only applicable to cultivated barley, but also to other species in the genus Hordeum. As the effort of designing another whole exome capture assay specific to other Hordeum species is not likely be taken in the near future, our assay may function as an effective surrogate. While a substantial proportion of the captured reads can be mapped to the Morex assembly, read mapping is more efficient if an appropriate mapping reference is used, as we demonstrated by mapping captured H. pubiflorum reads against a newly constructed H. pubiflorum shotgun assembly. Following the same approach for tetraploid H. bulbosum was less efficient due to issues of assembly quality most likely because of low read depth and the presence of two homoeologous copies for each genomic region. We estimate the necessary minimal genome coverage at 15- to 20-fold to produce a suitable de novo assembly for a diploid species. Target positions can then be redefined in terms of the new assembly. As an alternative to a de novo assembly, more lenient criteria may be used for mapping reads against the Morex assembly. Although probably resulting in an increase in the proportion of mapped reads, this approach can give rise to false-positive SNP calls due to paralogous mapping in downstream analysis.
Capture of the exome of even more distantly related bread wheat with our assay provided reasonable results. Half of the captured sequence could be mapped against the barley assembly. However, capture or read mapping may favor one homeologous copy of a gene and introduce a (locus-specific) skew towards one subgenome. Although there is a capture assay specifically designed for bread wheat (Winfield et al., 2012), our assay may be preferable in some special cases. While disentangling homoeologues from short sequencing reads remains challenging in polyploid wheat species, genotype calling and allele frequency estimation should be fairly straightforward in diploids. Exome sequencing may thus enable mapping-by-sequencing in diploid Triticeae such as diploid Einkorn wheat (T. monococcum), Aegilops tauschii or rye. The barley physical and genetic framework may serve as a reference for read mapping as long as a comparable resource is lacking for wheat and rye.
Our analysis highlights the advantages of having an established reference assembly, even if it is highly fragmented. About 80–95% of all captured reads could be mapped against the barley WGS assembly, corroborating the previous estimate that this assembly represents 86% of all barley genes. Winfield et al. (2012) could only map around 35% of their reads back onto a target space defined from ESTs. Using a more sensitive (and complicated) mapping procedure, Saintenac et al. (2011) could map 60% of reads captured in tetraploid wheat against a full-length cDNA reference. In both studies, complex intron–exon structures probably prevented the mapping of genomic reads onto a transcribed reference.
In conclusion, we expect that whole exome capture will become the most widely used approach for resequencing studies in barley and its relatives in the near future. We anticipate wide applications of this capture platform in population genetics/genomics, evolutionary studies, gene isolation and comparative genomics.
Experimental Procedures
Probe design
Input sequence to the array design was 300 919 sequences, totaling 101.8 Mb, derived from the gene models predicted from the mapping of RNA-Seq reads (The International Barley Genome Sequencing Consortium, 2012), barley full-length cDNA (Matsumoto et al., 2011), and de novo assembled RNA-Seq contigs. RNA-Seq contigs were assembled with CLC assembly cell (http://www.clcbio.com/) from RNA-Seq reads of different cultivars (The International Barley Genome Sequencing Consortium, 2012). Variable length probes (50- to 100-mers) were generated at a 5-bp step across the entire sequence space. Individual probes were repeat-masked by removing probes that had an average 15-mer frequency >200, using a 15-mer frequency table generated from the Morex genome assembly. The repeat-masked probe set was compared back to the target sequence using SSAHA (Ning et al., 2001), using a minimum match size of 30 and allowing up to five indels or gaps. Probe sequences with more than 25 matches in the target set were eliminated from consideration. A probe set, containing 2 040 943 unique probes, was generated by selecting them at an average spacing of 40 bp (measured from 5′ oligo starting position to the next 5′ oligo starting position). An array design was made using two replicates of this probe set, plus two replicates of a set of 77 192 random probes. Two slides of this array design were hybridized for 4 days with four barley cultivars (Steptoe (Cy3) versus Barke (Cy5), Morex (Cy3) versus Bowman (Cy5)) using standard CGH protocols (http://www.nimblegen.com/products/lit/NG_CGHCNV_Guide_v8p1.pdf). Based on the CGH results, 0.42% of the probes were removed as missed repetitive elements based on high signal intensity. An additional 2.16% of the probes were excluded as contaminants or non-performing, based on low signal for all four cultivars. The remaining probes were used to construct a solution-phase capture pool. Four cultivars (Morex, Steptoe, Barke and Bowman) were captured and sequenced using standard protocols with an Illumina HiSeq (2 × 76 bp). The sequencing data were used to optimize empirically the capture design, using a proprietary Roche NimbleGen method. No sequence content was changed in the optimization of the design.
Capture library preparation and pre-hybridization amplification
A schematic overview of the experimental procedures is shown Figure S1(b). Illumina TruSeq Paired End libraries (Illumina Part # 15026486) were prepared essentially as described by the manufacturer (Illumina, Inc., San Diego, CA, USA) using DNA Adapter Indexes 2, 4, 5, 6, 7 or 12, respectively. Briefly, 1 μg genomic DNA was fragmented into a 200–300 bp size range using a sample volume of 53 μl, Covaris microTUBES and a Covaris S220 Instrument (175 W ultrasonic power, 10% duty factor, 200 cycles per burst, 100 sec treatment time). Libraries were utilized either with or without size fractionation. If size fractionation was used the DNA adapter ligated products were recovered with a size between 320 and 420 bp by excision from an SYBR-Gold stained agarose gel. The TruSeq DNA libraries were purified using AmPure XP Beads (Beckman Coulter GmbH) and eluted in 30 μl Resuspension Buffer according to the manufacturer (Illumina, Inc., San Diego, CA, USA).
To enrich for correctly ligated DNA fragments, 20 μl of the TruSeq DNA library were used as template in the pre-capture LM-PCR reaction (ligation-mediated PCR; 100 μl volume) containing Illumina sequencing adapters (2 μm TS-PCR Oligo 1: AATGATACGGCGACCACCGAGA and 2 μm TS-PCR Oligo 2: CAAGCAGAAGACGGCATACGAG) and 50 μl Phusion High-Fidelity PCR Master Mix (2×, New England BioLabs GmbH, Part# F-531L). LM-PCR cycling, clean-up and elution of the sample were as described previously (Haun et al., 2011). The DNA was quantified using a Qubit 2.0 fluorometer (Invitrogen) and analyzed electrophoretically with an Agilent 2100 Bioanalyzer (Santa Clara, CA, USA) using a DNA 7500 chip (Part# 5067-1506). The library fragments of the amplified sample library were between 250 and 500 bp.
Capture library and liquid array processing
Prior to hybridization 10 μl of Roche NimbleGen's (Madison, WI, USA) proprietary Plant Capture Enhancer (PCE) were added to a 1.5-ml tube containing 1 μg of the amplified sample library. This reagent has been subsequently renamed Sequence Capture Developer Reagent (Material # 06684335001, Roche, Indianapolis, IN, USA). Next, 1 μl of 1 mm TS-HE Universal Oligo 1 (AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT) and 1 μl of the appropriate TS-INV-HE Index Oligo (1 mm) were added. The mixture was dried down in a SpeedVac at 60°C. The TS-HE Universal Oligo 1 was designed to block the universal segment of TruSeq DNA library adapters during the sequence capture hybridization. The TS-INV-HE Index Oligos were designed accordingly, to block the corresponding indexed (underlined) segment of the TruSeq DNA library adapters (TS-INV-HE Index Oligo 2: CAAGCAGAAGACGGCATACGAGATACATCGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT/term/; TS-INV-HE Index Oligo 4: CAAGCAGAAGACGGCATACGAGATTGGTCAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT/term/; TS-INV-HE Index Oligo 5: CAA GCAGAAGACGGCATACGAGATCACTGTGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT/term/; TS-INV-HE Index Oligo 6: CAAGCAGAAGACGGCATACGAGATATTGGCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT/term/; TS-INV-HE Index Oligo 7: CAAGCAGAAGACGGCATACGAGATGATCTGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT/term/; TS-INV-HE Index Oligo 12: CAAGCAGAAGACGGCATACGAGATTACAAGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT/term/; ‘/term/’indicates the addition of a dideoxy-C modification). Up to six samples provided with different indexes were pooled for hybridization. The amount of the corresponding TS-INV-HE Index Oligos was proportional to the concentration of the sample in the pool. To each dried-down sample 7.5 μl of 2 × Sequence Capture (SC) Hybridization Buffer (tube 5) and 3 μl of Hybridization Component A (tube 6) were added (Roche Part# 05634261001, Roche, Indianapolis IN, USA). The hybridization cocktail was vortexed for 10 sec and collected by centrifugation. Following denaturation in a heat block (95°C, 10 min) the sample was transferred to a 0.2 ml PCR tube containing 4.5 μl Exome Library (liquid array). The hybridization sample (15 μl) was incubated in a thermocycler (lid heated to 57°C) at 47°C for 64–72 h.
The proprietary NimbleGen SC Wash Buffers (tubes 1, 2 and 3), Stringent Wash Buffer (tube 4) and the Bead Wash Buffer (tube 7) were diluted to 1 × working solutions (Roche Part# 05634261001, Roche, Indianapolis, IN, USA). Streptavidin Dynabeads (M-270, Invitrogen) were thoroughly vortexed, aliquoted (50 μl per hybridization) into 1.5-ml tubes and prepared for the affinity purification of captured DNA. The tubes were placed in a DynaMag-2 magnet (Invitrogen, Part# 123-21D) for 2 min. The clear liquid was discarded and 100 μl of Bead Wash Buffer were added. Tubes were vortexed, placed back in the magnet, the clear liquid was removed, and the washing was repeated once. Dynabeads were resuspended in 50 μl Bead Wash Buffer, transferred into PCR plates and collected using a DynaMag-96 Side Skirted Magnetic Particle Concentrator (MPC, Invitrogen, Part# 120.27). The clear supernatant was discarded. The hybridization sample was added to the wet Dynabeads and mixed thoroughly by pipetting up and down. Using a thermocycler (lid heated to 57°C) at 47°C for 45 min the captured sample was bound to the Dynabeads. The sample was vortexed for 3 sec in 15-min intervals to ensure that the Dynabeads remain in suspension. Dynabeads plus bound DNA (15 μl) were washed by adding 100 μl SC Wash Buffer I (pre-heated to 47°C for 2 h) and vortexing for 10 sec. The suspension was transferred to a 1.5-ml tube and placed in a DynaMag-2 device, and the supernatant was discarded once clear. Washing was continued by adding 200 μl Stringent Wash Buffer (pre-heated to 47°C for 2 h). The sample was mixed by pipetting avoiding a major temperature drop and incubated for 5 min at 47°C. Following bead purification using the DynaMag-2 magnet, the liquid was discarded and the washing at 47°C with Stringent Wash Buffer was repeated once. Dynabeads plus bound DNA were purified in a DynaMag-2 device and 200 μl Wash Buffer I (room temperature) was added. The sample was vortexed for 2 min and the liquid was collected to the tube's bottom. Following magnetic concentration the liquid was discarded, and the sample was washed with 200 μl Wash Buffer II (vortexing for 1 min) followed by 200 μl Wash Buffer III (vortexing for 30 sec) as described for washing with Wash Buffer I. The tube was removed from the magnet and the bead-bound captured library was resuspended in 50 μl PCR-grade water (storage at −20°C possible).
The bead-bound captured library was amplified using LM-PCR. LM-PCR master mix (200 μl volume) contained 50 μl of bead-bound captured library, Illumina sequencing adapters (2 μm TS-PCR Oligo 1 and 2 μm TS-PCR Oligo 2) and 100 μl Phusion High-Fidelity PCR Master Mix (2 × , New England BioLabs GmbH, Part# F-531L). Next, 100 μl of the master mix were distributed into two 0.2-ml PCR tubes for amplification. LM-PCR cycling conditions were: 98°C for 30 sec, followed by 16 cycles of 98°C for 10 sec, 60°C for 30 sec and 72°C for 30 sec. The DNA was extended for 5 min at 72°C and kept at 8°C until further processing. Both LM-PCR reactions were combined and cleaned-up using the Qiaquick PCR purification kit (Qiagen, Hilden) according to the manufacturer instructions with slight modifications. To each reaction (200 μl), 1 ml of Qiagen PBI buffer was added; 600 μl of the sample were applied in two consecutive steps to the same Qiaquick column (centrifugation 16000 g for 1 min). Following washing with 750 μl Qiagen PE buffer, the flow-through was discarded and the column was spun for 1 min (16 000 g). In order to remove PE completely, the column was rotated 180° and spun again for 30 sec. DNA was eluted after addition of 50 μl EB (pre-heated to 50°C), incubation for 1 min and centrifugation (16000 g, 1 min). The A260/A280 ratio (1.7–2.0) was recorded using a NanoDrop spectrophotometer. Libraries were analyzed electrophoretically using the Agilent 2100 Bioanalyzer (Santa Clara, CA, USA) and a DNA 7500 chip (Agilent Part# 5067-1506). The library fragments of the post capture enriched sequencing libraries were between 250 and 500 bp. Quantification of libraries and sequencing of capture libraries is described in Methods S1. Library preparation for whole-genome sequencing is described in Methods S2.
Analysis of exome sequencing data
Target regions derived from transcript contigs (full-length cDNA or RNA-Seq contigs) were mapped against the WGS assembly of barley cultivar ‘Morex’ (The International Barley Genome Sequencing Consortium, 2012) with BWA (command bwasw) (Li and Durbin, 2009) and Mega BLAST (Camacho et al., 2009) and mapping positions on the assembly were collapsed into the single bed file with BEDTools (Quinlan and Hall, 2010). Sequencing reads were mapped to the Morex assembly with BWA 0.6.2 (commands aln and sampe). Coverage was computed with SAMtools depth (Li, 2011) considering only properly paired reads. Coverage statistics were calculated with AWK scripts and visualized in the R statistical environment (R Core Team, 2012). Regression analysis of sequencing output, read coverage and the number of detected SNPs was performed using standard R functions. Regions with high coverage but not located within target regions were compared with targets with Mega BLAST (Camacho et al., 2009) and BLAST output was filtered with an AWK script. SNP calling was performed with SAMtools mpileup/BCFtools (Li, 2011) (version 0.1.18) using default parameters. If an accession was captured twice or more, the sample with the highest sequencing output was chosen. Resulting SNP calls were filtered with a custom AWK script discarding positions with quality score below 40 or coverage below 10-fold. SNP frequency was visualized along the integrated physical and genetic map of barley (The International Barley Genome Sequencing Consortium, 2012) with an R script.
Assembly of WGS data
Sequencing reads from H. pubiflorum and H. bulbosum were quality trimmed and assembled with CLC assembly cell 3.2.2 (http://www.clcbio.com/) using the programs quality_trim (default parameters) and clc_novo_assemble (parameters: fb ss 100 400). Capture targets were mapped against the H. pubiflorum contigs with BWA-SW and target intervals were defined with BEDTools.
Phylogenetic tree construction
Variant positions and genotype calls were filtered according to the following criteria: (i) only bi-allelic SNPs were considered; (ii) the read depth was at least 20 in all samples (i.e. no missing calls); and (iii) the genotype score as reported by BCFtools was at least 10 in all samples. The Hamming distance (i.e. the number of different genotype calls) was calculated between any two samples. The resulting distance matrix was used to construct a neighbor-joining tree with the function nj() in the package ‘ape’ (Paradis et al., 2004).
Accession numbers
PRJEB1810 (exome capture reads), PRJEB1811 (WGS reads for H. bulbosum), PRJEB1812 (WGS reads for H. pubiflorum), PRJEB3403 (WGS assembly of H. bulbosum) and PRJEB3404 (WGS assembly of H. pubiflorum).
Acknowledgments
We thank all members of the Wheat Exome Capture consortium for helpful discussions. We are grateful to Manuela Knauft, Ines Walde, Sandra Driesslein and Katie Baker for skillful technical assistance and to Doreen Stengel for sequence data submission. We thank the laboratory of Lothar Altschmied (IPK Gatersleben) for Illumina sequencing. We greatly acknowledge the financial support provided by the following grants: GABI-BARLEX (FKZ 0314000) and TRITEX (FKZ 0315954) by German Ministry of Education and Research (BMBF) to NS, US, KFXM; SPP1530 (KI 1465/6-1) by German Research Foundation (DFG) to BK; BB/I00663X/1 and BB/I008357/1, by Biotechnology and Biological Science Research Council (BBSRC) to RW, SA, DS and PH; the project ‘The diversity and evolution of the gene component of barley peri-centromeric heterochromatin’ (BB/I022899/1) to AJF by BBSRC; the Triticeae Coordinated Agricultural Project (#2011-68002-30029) to GJM, PLM and EDA by USDA-NIFA. The authors TAR, DJG, MD and JAJ declare a conflict of interest as employees of Roche NimbleGen, Inc.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Flowcharts for assay design, experimental procedures and sequence analysis.
Example of a capture target on a WGS contig
Neighbor-joining tree with H. bulbosum and H. pubiflorum
Overview of captured samples and detailed mapping and SNP calling statistics.
Quantification of libraries and sequencing on the Illumina HiSeq2000 instrument.
Methods S2. TruSeq WGS library preparation.
References
- Bainbridge MN, Wang M, Burgess DL, et al. Whole exome capture in solution with 3 Gbp of data. Genome Biol. 2010;11:R62. doi: 10.1186/gb-2010-11-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- Blattner FR. Progress in phylogenetic analysis and a new infrageneric classification of the barley genus Hordeum (Poaceae: Triticeae) Breeding Science. 2009;59:471–480. [Google Scholar]
- Brassac J, Jakob SS, Blattner FR. Progenitor-derivative relationships of Hordeum polyploids (Poaceae, Triticeae) inferred from sequences of TOPO6, a nuclear low-copy gene region. PLoS One. 2012;7(3):e33808. doi: 10.1371/journal.pone.0033808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley R, Spannagl M, Pfeifer M, et al. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature. 2012;491:705–710. doi: 10.1038/nature11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbano HA, Hodges E, Green RE, et al. Targeted investigation of the Neandertal genome by array-based sequence capture. Science. 2010;328:723–725. doi: 10.1126/science.1188046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comadran J, Kilian B, Russell J, et al. Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat. Genet. 2012;44:1388–1392. doi: 10.1038/ng.2447. [DOI] [PubMed] [Google Scholar]
- Cosart T, Beja-Pereira A, Chen S, Ng SB, Shendure J, Luikart G. Exome-wide DNA capture and next generation sequencing in domestic and wild species. BMC Genomics. 2011;12:347. doi: 10.1186/1471-2164-12-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 2011;12:499–510. doi: 10.1038/nrg3012. [DOI] [PubMed] [Google Scholar]
- Druka A, Franckowiak J, Lundqvist U, et al. Genetic dissection of barley morphology and development. Plant Physiol. 2011;155:617–627. doi: 10.1104/pp.110.166249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairfield H, Gilbert GJ, Barter M, et al. Mutation discovery in mice by whole exome sequencing. Genome Biol. 2011;12:R86. doi: 10.1186/gb-2011-12-9-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuillet C, Langridge P, Waugh R. Cereal breeding takes a walk on the wild side. Trends Genet. 2008;24:24–32. doi: 10.1016/j.tig.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Galvao VC, Nordstrom KJ, Lanz C, Sulz P, Mathieu J, Pose D, Schmid M, Weigel D, Schneeberger K. Synteny-based mapping-by-sequencing enabled by targeted enrichment. Plant J. 2012;71:517–526. doi: 10.1111/j.1365-313X.2012.04993.x. [DOI] [PubMed] [Google Scholar]
- Gore MA, Chia JM, Elshire RJ, et al. A first-generation haplotype map of maize. Science. 2009;326:1115–1117. doi: 10.1126/science.1177837. [DOI] [PubMed] [Google Scholar]
- Graner A, Jahoor A, Schondelmaier J, Siedler H, Pillen K, Fischbeck G, Wenzel G, Herrmann RG. Construction of an RFLP map of barley. Theor. Appl. Genet. 1991;83:250–256. doi: 10.1007/BF00226259. [DOI] [PubMed] [Google Scholar]
- Guo Y, Long J, He J, Li CI, Cai Q, Shu XO, Zheng W, Li C. Exome sequencing generates high quality data in non-target regions. BMC Genomics. 2012;13:194. doi: 10.1186/1471-2164-13-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haun WJ, Hyten DL, Xu WW, et al. The composition and origins of genomic variation among individuals of the soybean reference cultivar Williams 82. Plant Physiol. 2011;155:645–655. doi: 10.1104/pp.110.166736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes P, Liu B, Knapp S, et al. Quantitative trait locus effects and environmental interaction in a sample of North American barley germ plasm. Theor. Appl. Genet. 1993;87:392–401. doi: 10.1007/BF01184929. [DOI] [PubMed] [Google Scholar]
- van Hintum T, Menting F. Diversity in ex situ genebank collections of barley. In: von Bother R, van Hintum T, Knupffer H, Sato K, editors. Diversity in Barley (Hordeum vulgare) Amsterdam: Elsevier Science B.V; 2003. pp. 247–257. [Google Scholar]
- Hodges E, Xuan Z, Balija V, et al. Genome-wide in situ exon capture for selective resequencing. Nat. Genet. 2007;39:1522–1527. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- Hufford MB, Xu X, van Heerwaarden J, et al. Comparative population genomics of maize domestication and improvement. Nat. Genet. 2012;44:808–811. doi: 10.1038/ng.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob SS, Meister A, Blattner FR. The considerable genome size variation of Hordeum species (poaceae) is linked to phylogeny, life form, ecology, and speciation rates. Mol. Biol. Evol. 2004;21:860–869. doi: 10.1093/molbev/msh092. [DOI] [PubMed] [Google Scholar]
- Kilian B, Graner A. NGS technologies for analyzing germplasm diversity in genebanks. Brief. Funct. Genomics. 2012;11:38–50. doi: 10.1093/bfgp/elr046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhu C, Yeh CT, et al. Genic and nongenic contributions to natural variation of quantitative traits in maize. Genome Res. 2012;22:2436–2444. doi: 10.1101/gr.140277.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linde-Laursen I, von Bothmer R, Jacobsen N. Giemsa C-banded karyotypes of diploid and tetraploid Hordeum bulbosum (Poaceae) Plant Syst. Evol. 1990;172:141–150. [Google Scholar]
- Liu S, Ying K, Yeh CT, et al. Changes in genome content generated via segregation of non-allelic homologs. Plant J. 2012;72:390–9. doi: 10.1111/j.1365-313X.2012.05087.x. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Tanaka T, Sakai H, et al. Comprehensive sequence analysis of 24,783 barley full-length cDNAs derived from 12 clone libraries. Plant Physiol. 2011;156:20–28. doi: 10.1104/pp.110.171579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moragues M, Comadran J, Waugh R, Milne I, Flavell AJ, Russell JR. Effects of ascertainment bias and marker number on estimations of barley diversity from high-throughput SNP genotype data. Theor. Appl. Genet. 2010;120:1525–1534. doi: 10.1007/s00122-010-1273-1. [DOI] [PubMed] [Google Scholar]
- Ning Z, Cox AJ, Mullikin JC. SSAHA: a fast search method for large DNA databases. Genome Res. 2001;11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2012. ISBN 3-900051-07-0. [Google Scholar]
- Ramchiary N, Nguyen VD, Li X, Hong CP, Dhandapani V, Choi SR, Yu G, Piao ZY, Lim YP. Genic microsatellite markers in Brassica rapa: development, characterization, mapping, and their utility in other cultivated and wild Brassica relatives. DNA Res. 2011;18:305–320. doi: 10.1093/dnares/dsr017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J, Dawson IK, Flavell AJ, Steffenson B, Weltzien E, Booth A, Ceccarelli S, Grando S, Waugh R. Analysis of >1000 single nucleotide polymorphisms in geographically matched samples of landrace and wild barley indicates secondary contact and chromosome-level differences in diversity around domestication genes. New Phytol. 2011;191:564–578. doi: 10.1111/j.1469-8137.2011.03704.x. [DOI] [PubMed] [Google Scholar]
- Saintenac C, Jiang D, Akhunov ED. Targeted analysis of nucleotide and copy number variation by exon capture in allotetraploid wheat genome. Genome Biol. 2011;12:R88. doi: 10.1186/gb-2011-12-9-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte D, Close TJ, Graner A, et al. The International Barley Sequencing Consortium–at the threshold of efficient access to the barley genome. Plant Physiol. 2009;149:142–147. doi: 10.1104/pp.108.128967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Barley Genome Sequencing Consortium. A physical, genetic and functional sequence assembly of the barley genome. Nature. 2012;491:711–716. doi: 10.1038/nature11543. [DOI] [PubMed] [Google Scholar]
- Vallender EJ. Expanding whole exome resequencing into non-human primates. Genome Biol. 2011;12:R87. doi: 10.1186/gb-2011-12-9-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winfield MO, Wilkinson PA, Allen AM, et al. Targeted re-sequencing of the allohexaploid wheat exome. Plant Biotechnol. J. 2012;10:733–742. doi: 10.1111/j.1467-7652.2012.00713.x. [DOI] [PubMed] [Google Scholar]
- Wright SI, Bi IV, Schroeder SG, Yamasaki M, Doebley JF, McMullen MD, Gaut BS. The effects of artificial selection on the maize genome. Science. 2005;308:1310–1314. doi: 10.1126/science.1107891. [DOI] [PubMed] [Google Scholar]
- Yun SJ, Gyenis L, Hayes PM, Matus I, Smith KP, Steffenson BJ, Muehlbauer GJ. Quantitative trait loci for multiple disease resistance in wild barley. Crop Sci. 2005;45:2563–2572. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flowcharts for assay design, experimental procedures and sequence analysis.
Example of a capture target on a WGS contig
Neighbor-joining tree with H. bulbosum and H. pubiflorum
Overview of captured samples and detailed mapping and SNP calling statistics.
Quantification of libraries and sequencing on the Illumina HiSeq2000 instrument.
Methods S2. TruSeq WGS library preparation.


