Abstract
Finegoldia magna is a Gram-positive anaerobic commensal of the human skin microbiota, but also known to act as an opportunistic pathogen. Two primary virulence factors of F. magna are the subtilisin-like extracellular serine protease SufA and the adhesive protein FAF. This study examines the molecular mechanisms F. magna uses when colonizing or establishing an infection in the skin. FAF was found to be essential in the initial adherence of F. magna to human skin biopsies. In the upper layers of the epidermis FAF mediates adhesion through binding to galectin-7 – a keratinocyte cell marker. Once the bacteria moved deeper into the skin to the basement membrane layer, SufA was found to degrade collagen IV which forms the backbone structure of the basement membrane. It also degraded collagen V, whereby F. magna could reach deeper dermal tissue sites. In the dermis, FAF interacts with collagen V and fibrillin, which presumably helps the bacteria to establish infection in this area. The findings of this study paint a clear picture of how F. magna interacts with human skin and explain how it is such a successful opportunistic pathogen in chronic wounds and ulcers.
Introduction
Skin is the largest organ in the human body and constitutes an important physical barrier to external stresses such as microorganisms, ultraviolet radiation, toxins, allergens and mechanical insults. It is composed of four structural layers: the epidermis, basement membrane, dermis and the subcutaneous layer (Koziel and Potempa, 2013). The skin is inhabited by a diverse array of bacteria, fungi and viruses, which vary between individuals and different sites on the skin (Schommer and Gallo, 2013). Colonization of the skin by commensal bacteria such as Staphylococcus, Finegoldia, Micrococcus and Corynebacterium sp., helps protect the host against colonization by more pathogenic microbes by depleting available nutrients and preventing their adherence and translocation across skin layers. However, when the host immune defence is compromised or the microbiota balance is disrupted, some of these commensals can act as opportunistic pathogens and cause infection (Nagy et al., 2011).
The human skin inhabitant Finegoldia magna is a Gram positive anaerobic coccus (GPAC), whose first complete genome sequence of strain ATCC 29328 was published in 2008 (Goto et al., 2008). Genomic analysis revealed that it can utilize fructose as an energy source, but also amino acids, due to its high number of amino peptidases and oligo-peptide transporters. It was found to harbour more aminopeptidase activities than other GPAC species, indicating a higher pathogenicity (Murphy and Frick, 2013). This is clearly seen in clinical infection, as F. magna is the most frequently isolated GPAC species in pure culture from various infection sites (Bourgault et al., 1980; Murphy and Frick, 2013). It is typically isolated from infections such as wound infections, soft tissue abscesses and bone and prosthetic joint infections (Fitzgerald et al., 1982; Davies et al., 1988; Brook and Frazier, 2000; Brazier et al., 2008; Brook, 2008; Levy et al., 2009). Due to being a member of the skin microbiota, it was found to be one of the most common anaerobes isolated from skin specimens and also highly prevalent in chronic wounds, diabetic ulcers and pressure ulcers (Hansson et al., 1995; Higaki and Morohashi, 2003; Stephens et al., 2003; Dowd et al., 2008a,b,; Murphy and Frick, 2013).
The prominence of F. magna in GPAC infections could potentially be explained due to its expression of proteins that enhance virulence. The superantigen, protein L, is a surface protein with high affinity for immunoglobulin light chains and can induce the release of pro-inflammatory mediators (Björck, 1988; Genovese et al., 2003). Protein L is expressed by approximately 10% of F. magna isolates and is known to be associated with bacterial vaginoses (Kastern et al., 1990). Another surface protein that could promote virulence, is the albumin binding protein PAB, which was found to give the expressing strain a significant increase in growth rate (de Château and Björck, 1994; de Château et al., 1996). Protein FAF (F. magna adhesion factor) is expressed by more than 90% of F. magna isolates. Its surface associated form causes bacterial clumping and helps bacterial adhesion to the basement membrane in skin by binding to BM-40 (Frick et al., 2008). This form and a released extracellular form help neutralize the activity of the human antibacterial peptide LL-37, and human histones H2B and H4 which exhibit strong antibacterial activities (Frick et al., 2008; Murphy et al., 2014). Another important virulence factor of F. magna is the subtilase-like enzyme SufA, which protects the bacterium from antibacterial activities of LL-37, histones and MIG/CXCL9 by proteolytic degradation (Karlsson et al., 2007; Murphy and Frick, 2013). F. magna also has the capability to produce a capsule and the enzymes collagenase and gelatinase, which could be other important pathogenicity factors (Brook, 1986; Krepel et al., 1992).
In this study, the path of F. magna from commensal to opportunistic pathogen was studied by analysing binding and interaction of FAF and SufA, expressed by the majority of F. magna strains, to proteins in the epidermis and dermis of human skin. The epidermis is mainly composed of keratinocytes, which make up 90% of this layer (Tortora and Derrickson, 2009). Keratinocytes produce the structural protein keratin, which assembles into a web-like pattern of intracellular filaments bound together by the matrix protein filaggrin into tight bundles (Proksch et al., 2008). It is in the epidermal layer that the skin microbiota reside, although a recent report suggests that some bacteria or their products exist below this, in the dermis and dermal adipose layers (Nakatsuji et al., 2013). The basement membrane separates the epidermal and dermal layers and is made up of independent networks of collagen IV and laminin, which are linked by molecules such as nidogen and BM-40 (for a review, see Timpl, 1996). It has previously been shown that FAF binds the non-collagenous glycoprotein BM-40, allowing F. magna to reside at the basement membrane (Frick et al., 2008). The dermis provides nutrients and physical support to the epidermis. It is composed of strong connective tissue containing collagen and elastic fibres that form a network providing strength and support (Pringle and Penzer, 2002).
In the present work, we discovered binding of protein FAF to galectin-7, collagen V and fibrillin, all of which are found in different layers of human skin. Furthermore, SufA was found to cleave collagen IV and V, which would help F. magna reach deeper tissue sites during infection.
Results
F. magna binds to galectin-7 in the epidermis of human skin
In a recent publication where the F. magna cell wall adhesion protein FAF was tested for an interaction with epidermal skin proteins, galectin-7 was identified as one of the ligands (Murphy et al., 2014). Galectin-7 is a member of the galectin family of animal lectins that have an affinity for β-galactose containing oligosaccharides (Saussez and Kiss, 2006). In this current study, the interaction between FAF and Galectin-7 was further investigated. Binding of galectin-7 to protein FAF was examined in a slot binding assay together with proteins PAB and SufA, other potential virulence factors in F. magna, see Fig. 1A. This blot shows a concentration dependent binding of galectin-7 to FAF, but no interaction with SufA and protein PAB. Binding of FAF to galectin-7 was further confirmed through surface plasmon resonance analysis which showed a significant binding with a KD of 0.53 nM. In order to map the binding of galectin-7 to a particular region of protein FAF, various recombinantly expressed FAF fragments were applied onto a PVDF membrane and probed with galectin-7, see Fig. 1B. The fragments, which were constructed in another study, correspond to the N-terminal FAF I (amino acids 28–115, 10 kDa) and FAF II (amino acids 28–317, 32 kDa) and the C-terminal FAF III (amino acids 239–616, 42 kDa) (Frick et al., 2008). The blot shows that FAF III, covering the C-terminal region of the molecule, is responsible for binding to galectin-7. There is no binding to FAF fragments I or II (Fig. 1B).
Figure 1.
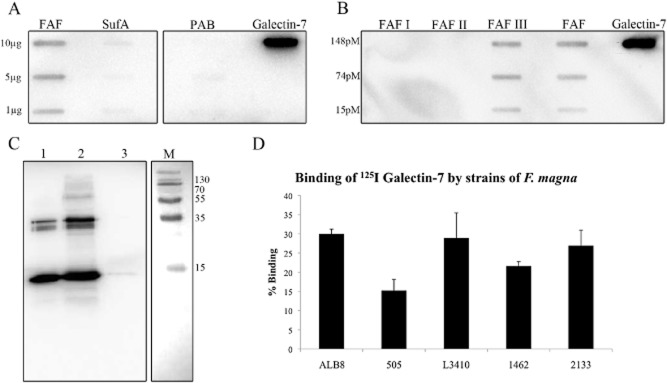
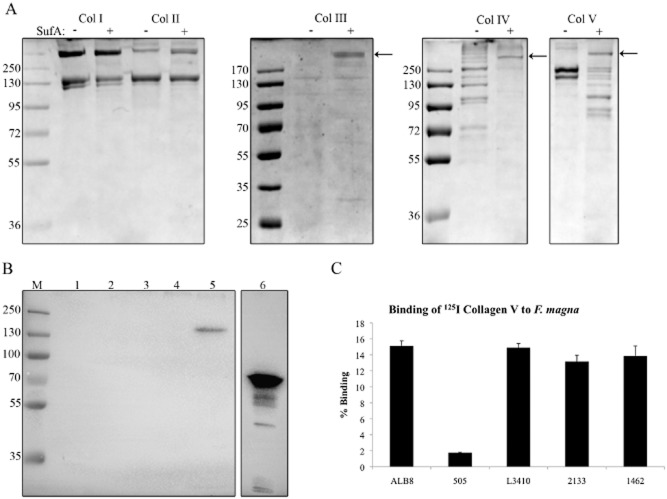
Protein FAF binds to galectin-7.A. Proteins FAF, SufA and PAB were applied in various amounts to a PVDF membrane. The membrane was incubated with a 1.33 μM solution of galectin-7 followed by probing with a rabbit anti-galectin-7 antibody. Galectin-7 was included as a positive control.B. Different recombinant length fragments of protein FAF (see Experimental procedures) were applied to a PVDF membrane and binding to galectin-7 was investigated by immunoblotting as in (A).C. A solution of F. magna strain ALB8 (2 × 109 cfu ml−1) in PBS was incubated with galectin-7 for 1 h at 37°C. Protein bound to the bacterial surface was eluted with low pH buffer and analysed by SDS-PAGE and immunoblotting using a galectin-7 antibody. M: Biorad Broad Range Molecular Weight Marker; Lane 1: Galectin-7 positive control 1 μg; Lane 2: F. magna incubated with galectin-7; Lane 3: F. magna incubated with PBS.D. Percentage binding of 125I-labelled galectin-7 to different strains of F. magna. Bars represent mean ± S.E. of at least three experiments.
Next, FAF-expressing bacteria (strain ALB8) were incubated with galectin-7 and bound ligand was eluted by low pH buffer followed by Western blot analysis using a rabbit anti-galectin-7 antibody. Bacteria incubated with buffer only served as a negative control to ensure that any positive bands seen were not due to bacterial surface proteins cross reacting with the antibodies. As shown in Fig. 1C, lane 2, there is a strong binding by F. magna to galectin-7 in solution as judged by the immuno-reactive band corresponding to the molecular weight of 14 kDa for galectin-7. Galectin-7 is known to form dimers (Leonidas et al., 1998), and this probably corresponds to the band close to the 35 kDa marker (Fig. 1C). There is no cleavage of galectin-7 by SufA present on the bacterial surface (Fig. 1C, lane 2) or when galectin-7 was incubated with purified SufA (data not shown).
Finally, binding of radiolabelled galectin-7 to different strains of F. magna was investigated, see Fig. 1D. The binding to FAF-expressing strains (ALB8, L3410, 1462 and 2133) varied between 20–30% of added radiolabelled galectin-7, while the non-FAF expressing strain 505 showed a significantly lower level of binding at 15% ± 3%. Taken together, the results of the binding experiments demonstrate that FAF interacts with galectin-7.
SufA of F. magna degrades collagens found in the skin and its surface protein FAF binds to collagen V
Collagens form a major part of the structural component of skin and thus the ability of F. magna to bind and degrade various collagens was investigated. Initially, the protease SufA was incubated with collagens I–V and cleavage was examined by SDS-PAGE, see Fig. 2A. The > 250 kDa band represents SufA and is indicated by a black arrow. This band is not visible in the analysis of collagen I and II cleavage due to the size of the collagen protein bands. The results clearly show that there is no breakdown of collagen I, II or III by SufA. However, collagen IV appears to be completely cleaved and collagen V is degraded into smaller fragments by SufA. Type IV collagen is the most important structural component of basement membranes separating the epidermis and dermis of skin (Gelse et al., 2003), while type V collagen is located on or adjacent to the basement membrane (Birk et al., 1988).
Figure 2.
SufA and FAF interact with collagens.A. SufA cleavage of collagens type I–V was determined by co-incubation for 3 h at 37°C followed by analysis on 8% SDS-PAGE. Lanes containing collagen co-incubated with SufA are indicated with a + sign. The position of SufA is indicated by a black arrow (not visible in cleavage of collagen I and II due to the position of the collagen bands). As a control, collagens were also incubated without SufA for the same length of time, indicated by a – sign.B. Binding of FAF to collagens I, II, III, IV and V was determined by electrophoresing 3.5 μg of each collagen on an 8% SDS-PAGE gel followed by incubation with FAF and immunoblotting with an anti-FAF antibody. M: Biorad Broad Range Molecular Weight Marker; Lane 1: Collagen I; Lane 2: Collagen II; Lane 3: Collagen III; Lane 4: Collagen IV; Lane 5: Collagen V; Lane 6: FAF 0.8 μg.C. Percentage binding of 125I-labelled collagen V to different strains of F. magna. Bars represent mean ± S.E. of at least three experiments.
To investigate a possible interaction between collagens and protein FAF, collagen type I-V were separated by SDS-PAGE followed by Western blotting. The membrane was then incubated with protein FAF and bound protein was detected by antibodies against FAF. Only collagen V interacts with FAF as judged by the prominent band just above 130 kDa representing collagen V (Fig. 2B, lane 5). In order to confirm this interaction using another method, recombinant collagen V was radiolabelled with iodine and its interaction with different strains of F. magna was examined, see Fig. 2C. Again, F. magna strains expressing FAF bound collagen V, while strain 505, which does not express FAF, bound almost no collagen V.
These results signify that F. magna can interact with collagens of the skin and the basement membrane. By breaking down collagens of the basement membrane, it could assist the bacteria to reach deeper tissue sites during infection.
F. magna binds to fibrillin through its surface adhesion protein FAF
Moving beyond the basement membrane into the dermis, fibrillin microfibrils are found which play vital roles in maintaining structural integrity and in regulating extracellular growth factors (Jensen et al., 2012). Fibrillin makes up 10–12 nm microfibrils, which form a scaffold for elastin deposition during elastogenesis allowing for elasticity and extensibility of connective tissues. As part of the path of F. magna into deeper dermal sites during infection, we investigated if it could bind and interact with fibrillin as a means of establishing infection in the dermis. Binding of 125I-labelled FAF to various fibrillin recombinant fragments was investigated in a slot binding assay, see Fig. 3A. This blot shows a concentration dependent binding of FAF to the N-terminal fragment of fibrillin-2 (rFBN2-N) and a slightly lower binding intensity to the N-terminal fragment of fibrillin-1 (rFBN1-N). There appears to be low/background binding of FAF to the C-terminal fragment of fibrillin-1 (rFBN1-C) and it does not appear to be concentration dependent. There is no binding at all to the C-terminal fragment of fibrillin-2 (rFBN2-C). Due to the strong interaction of FAF with the N-terminal of fibrillin-2, the interaction of FAF with the N-terminal of fibrillin-1 was also further investigated by radiolabelling both rFBN1-N and rFBN2-N with 125I and examining binding to different strains of F. magna, see Fig. 3B. Highest binding for both fragments was seen with F. magna strain ALB8 which displayed 24% binding to rFBN1-N and 17% binding to rFBN2-N. The pattern of binding for all strains was similar for both fragments, with a generally higher binding to rFBN1-N, than to rFBN2-N. Again, strain 505 showed the lowest level of binding, due to its lack of expression of protein FAF. This experiment confirms the results seen in the slot blot in Fig. 3A.
Figure 3.
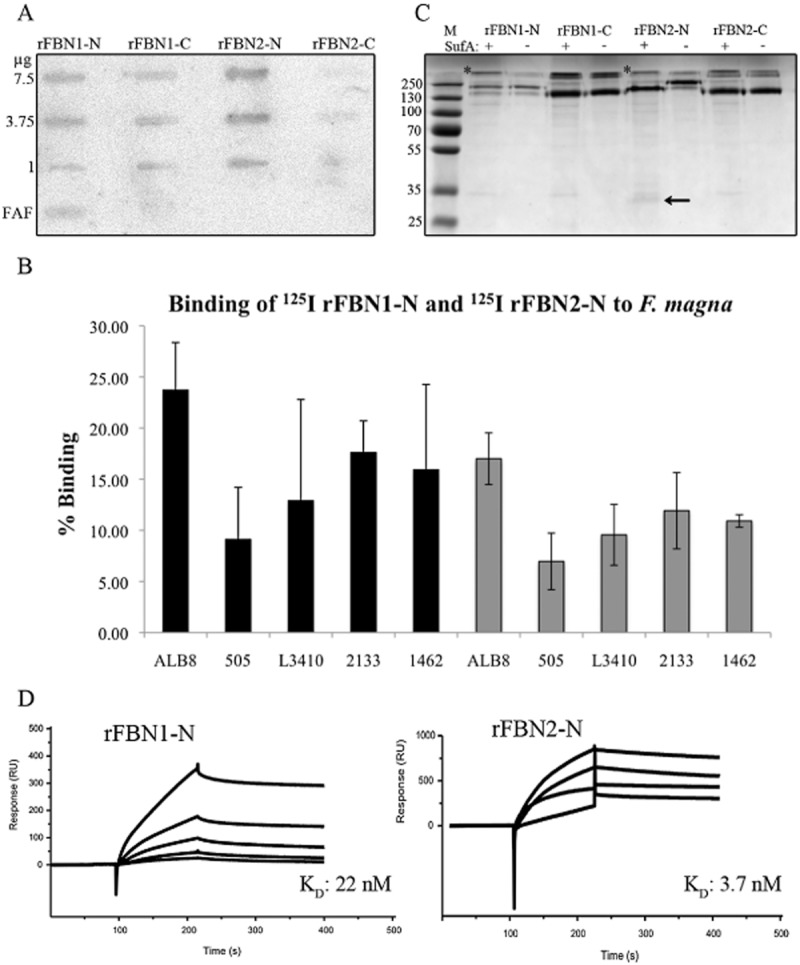
Examination of the interaction of FAF and SufA with fibrillin.A. Indicated amounts of various recombinant fibrillin fragments and FAF (1 mg) were applied in slots to a PVDF membrane. The membrane was incubated with 125I-labelled FAF and bound FAF was determined using the Fuji Imaging system.B. Percentage binding of 125I-labelled fibrillin fragments rFBN1-N and rFBN2-N to different strains of F. magna. The black bars represent binding to rFBN1-N and the grey bars represent binding to rFBN2-N. Bars represent mean ± S.E. of at least three experiments.C. SufA cleavage of fibrillin was determined by co-incubation with the various fragments for 3 h at 37°C followed by analysis of cleavage on an 8% SDS-PAGE gel. Lanes containing fibrillin co-incubated with SufA are indicated with a + sign. As a control, fibrillin fragments were also incubated without SufA for the same length of time, indicated by a – sign. The black arrow indicates the 35 kDa fragment released by SufA from rFBN2-N. A protein band representing SufA is clearly seen above the N-terminal fibrillin fragments and is indicated by a star. The SufA band is hidden by the C-terminal fibrillin fragments due to their size.D. FAF binds rFBN1-N and rFBN2-N in surface plasmon resonance experiments. FAF was immobilized on a sensor chip, and increasing concentrations of rFBN1-N/rFBN2-N were injected over the surface. RU, response units.
As SufA had previously been seen to degrade some collagen proteins, degradation of fibrillin components by SufA was also investigated, see Fig. 3C. No degradation of rFBN1-N, rFBN1-C or rFBN2-C was observed. However, SufA appears to release a 35 kDa fragment from rFBN2-N, as indicated by the black arrow. This 35 kDa fragment was cut out from the SDS-PAGE gel and analysed by tandem mass spectrometry (MS/MS), see Fig. S1. Results from this analysis show that the fragment is cleaved from the NH2-terminus of rFBN2-N, see Fig. S1. However, it is unknown if this cleavage is enough to allow F. magna to reach deeper tissue sites or if it interferes with tissue homeostasis.
Further investigations into the binding of FAF to fibrillin with surface plasmon resonance showed binding of FAF to both N-terminal fragments of fibrillin-1 and -2, with a slightly higher binding to rFBN2-N (KD: 3.7 nM) than rFBN1-N (KD: 22 nM), see Fig. 3D.
Protein FAF binds fibrillin both at the bacterial surface and in solution
Visual confirmation of F. magna binding to fibrillin via protein FAF was carried out using transmission electron microscopy. F. magna strains ALB8 and 505 were incubated with antibodies against FAF and recombinant fibrillin fragments labelled with colloidal gold, 5 and 20 nm respectively. Strain 505 was used as a negative control as it does not express FAF and therefore, no binding of fibrillin fragments to the surface should be seen. As shown in Fig. 4, immuno-staining identifies FAF on the surface of ALB8 bacteria and also shows binding of the N-terminal fibrillin fragments rFBN1-N and rFBN2-N to the surface. Furthermore, colocalization between FAF and bound fibrillin can be clearly seen on the bacterial surface. No binding of fibrillin fragments rFBN1-C and rFBN2-C to ALB8 bacteria could be observed (Fig. 4). As expected, no binding of FAF antibodies or gold labelled fibrillin fragments to the bacterial surface of strain 505 could be seen. Complexes formed, in solution, between FAF and fibrillin fragments were then analysed by electron microscopy following rotary shadowing, see Fig. 5. Fibrillin fragments are seen as long rod like structures (black arrows) and FAF molecules are labelled with colloidal gold (black arrowheads). Fig. 5A and C show complexes formed between FAF and the N-terminal fibrillin fragments rFBN1-N and rFBN2-N, while no complex formation could be observed between FAF and the C-terminal fragments of fibrillin (Fig 5B and D). These results confirm that protein FAF mediates binding of fibrillin to F. magna, both at the bacterial surface and in solution.
Figure 4.
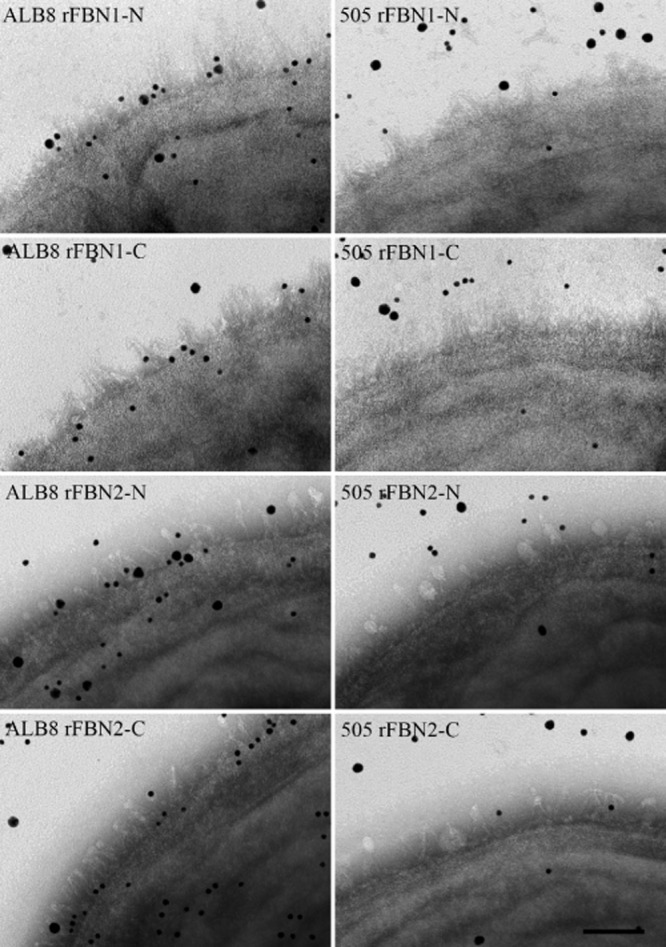
Transmission electron microscopy demonstrating binding of protein FAF on the ALB8 surface to the N-terminal region of fibrillin. F. magna strains ALB8 and 505 were incubated with anti-FAF antibodies and the various fibrillin fragments labelled with colloidal gold (anti-FAF antibody, 5 nm gold and fibrillin fragments, 20 nm gold). Samples were then prepared for transmission electron microscopy. Left panels: F. magna strain ALB8. Right panels: F. magna strain 505. The scale bar represents 50 nm.
Figure 5.
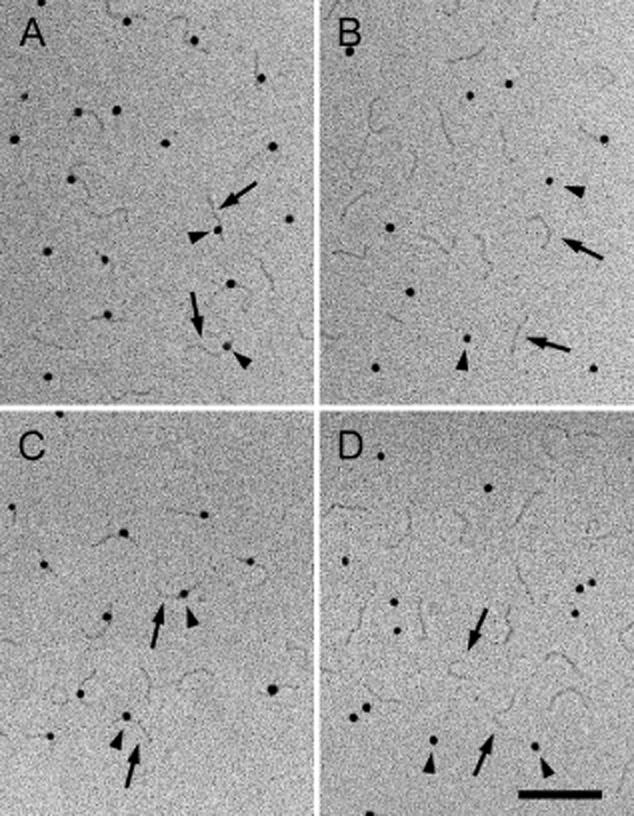
Electron micrographs after rotary shadowing showing complex formation between FAF and N-terminal fragments of fibrillin. The arrowheads point to FAF labelled with 10 nm colloidal gold and the arrows point to extended fibrillin molecules. The FAF molecules exhibit a globular appearance due to decoration with gold particles. A: Interaction of FAF with rFBN1-N; B: Interaction of FAF with rFBN1-C; C: Interaction of FAF with rFBN2-N; D: Interaction of FAF with rFBN2-C. The scale bar represents 100 nm.
SufA is essential in the invasion of F. magna into deeper dermal skin layers
In order to investigate F. magna interaction with human skin, and the effects of FAF and SufA expression, different strains of F. magna were incubated with human skin biopsies for 1 h. Following extensive washing to remove unbound bacteria the biopsies were prepared for scanning electron microscopy or incubated anaerobically for another 72 h, see Fig. 6. F. magna strain ALB8 expresses both FAF and SufA, strain 505 was used as it naturally does not express FAF and F. magna strain ΔSufA represents the ALB8 strain with SufA knocked out. Fig. 6, panel A, shows F. magna ALB8 bacteria bound to the skin epidermal surface at time point 0 h. This is in stark contrast to Panel B, which shows no binding of bacteria to the skin surface. This result underlines the importance of FAF in initial binding and adherence to the skin surface during colonization. Panel C shows that the absence of SufA from ALB8 has no effect on the adhesion of F. magna to host tissue as similar amounts of bound bacteria can be seen in Panel A and C. Here, the basement membrane is indicated by the letter B. Panels D to F represent the dermal layer, located below the epidermis, at time point 0 h. These micrographs demonstrate that the connective tissue, composed of collagen, fibrillin and elastic fibres, is intact. Also, no bacteria are present. Panels G to I represent the dermal layer after 72 h of incubation with the different strains of F. magna. Panel G shows that the ALB8 strain has caused significant destruction to the microfibrillar structure of the dermal layer. Also, strain 505, which expresses SufA, caused significant breakdown of the dermal fibres (Fig. 6H). Although no adherent 505 bacteria could initially be observed on the epidermal surface (Fig. 6B), some bacteria were present in the dermis after 72h. Obviously low affinity adhesion of strain 505 is mediated by a mechanism different from FAF. Fig. 6I shows an entirely different picture, where the dermal structure is completely undamaged and intact. This image represents ALB8 ΔSufA and displays how SufA could be important in the infection process for F. magna. It can also be seen that there are far lower numbers of F. magna in panel I, when compared with panel G. Apparently, the absence of SufA has hindered the ability of F. magna to reach the deeper dermal layers. However, low amounts of F. magna can still reach the deeper skin layers through binding with FAF, but it cannot cause any tissue destruction or damage. This data has been evaluated in further detail by examing the bacterial cell density of 30 tissue profiles in electron microscopy, see Table 1. This data agrees with Fig. 6 and, significantly, shows that there is a 1000-fold lower density of F. magna strain 505 in the skin biopsies, compared to ALB8. Clearly, protein FAF is an important factor in the adhesion of F. magna to host tissue. Furthermore, the data in the table indicate, that after 72 h, most of the ALB8 strain have transitioned from the epidermis to the dermis. Whereas, both strain 505 and ΔSufA remain mostly at the epidermis and are hindered in delving into deeper tissue layers due to the absence of FAF and SufA. Taken together, the results demonstrate that FAF is needed for optimal adhesion to human skin and SufA contributes to the breakdown of connective tissue to reach deeper tissue layers.
Figure 6.
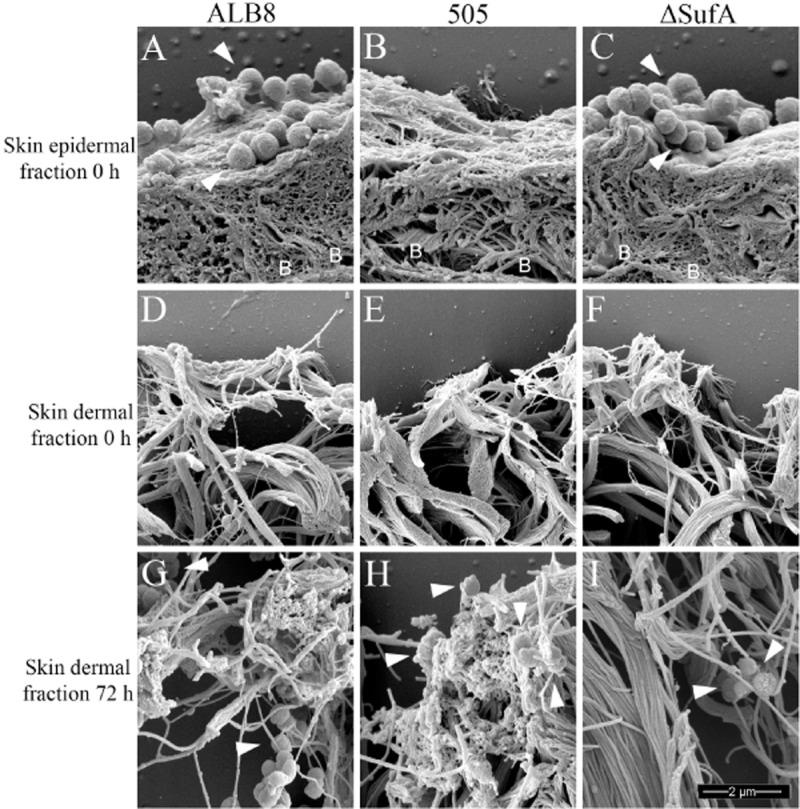
Scanning electron microscopy demonstrating the effect of FAF and SufA on binding and interaction of F. magna with human skin biopsies. Different strains of F. magna were incubated with human skin biopsies anaerobically for 1 h at 37°C, followed by washing to remove unbound bacteria. Skin harbouring bound F. magna was then anaerobically incubated for 72 h at 37°C to examine the effects of FAF and SufA. The biopsies were then prepared for scanning electron microscopy. Bacteria are indicated by white arrowheads and the location of the basement membrane is shown by letters B. Images represent: Skin epidermal fraction at 0 h incubated with (A) F. magna ALB8 (B) F. magna 505 (C) F. magna ALB8 ΔSufA; Skin dermal fraction at 0 h incubated with (D) F. magna ALB8 (E) F. magna 505 (F) F. magna ALB8 ΔSufA; Skin dermal fraction at 72 h co-incubated with (G) F. magna ALB8 (H) F. magna 505 (I) F. magna ALB8 ΔSufA. The scale bar represents 2 μm. Evaluation of the data is seen in Table 1.
Table 1.
Evaluation in electron microscopy of percentage cell density of different F. magna strains after 0, 24 and 72 h from 30 skin biopsy tissue profiles
| Strain | Cell density | Location | T0 | T24 | T72 |
|---|---|---|---|---|---|
| ALB8 | 2.3 × 106 mm−2 | Epidermis | 98% | 67% | 38% |
| Dermis | 2% | 32% | 62% | ||
| 505 | 1.8 × 103 mm−2 | Epidermis | 99% | 88% | 81% |
| Dermis | 1% | 12% | 19% | ||
| ΔSufA | 2.2 × 106 mm−2 | Epidermis | 100% | 97% | 94% |
| Dermis | 0% | 3% | 6% |
For a representative image, see Fig. 6.
Electron microscopy analysis shows colocalization of F. magna to galectin-7, BM-40 and fibrillin in different skin compartments
F. magna ALB8 bacteria were allowed to adhere to human skin biopsies for 1 h. The biopsies were then anaerobically incubated for another 24, 48 or 72 h and binding of the bacteria to proteins in the skin was examined by transmission electron microscopy. Fig. 7A, C and E show the path of F. magna into the deeper layers of the skin over time and that ALB8 bacteria are present in the epidermis after 24 h (Panel A), at the basement membrane after 48 h (Panel C) and in the dermis after 72 h (Panel E). Taking a closer look using immuno electron microscopy (Fig. 7B, D and F) colocalization of FAF with galectin-7, BM-40 and fibrillin present in the various skin layers could be demonstrated. Thus, the biopsies were incubated with antibodies against FAF, labelled with 5 nm colloidal gold, and antibodies against galectin-7, BM-40 and fibrillin, each labelled with 20 nm colloidal gold. In these images, colocalization between FAF and these proteins can clearly be seen. Following binding to the epidermal surface initially at 0 h, see Fig. 6A, F. magna enters the epidermal layer after 24 h facilitated by binding of FAF to galectin-7 on the keratinocyte cell surface (Fig. 7B). After 48 h, F. magna reaches the basement membrane, where it attaches through a FAF-BM-40 interaction (Fig. 7D). After 72 h, F. magna has already reached the dermal layer where colonization is promoted by attachment and binding to fibrillin microfibrils through FAF interaction (Fig. 7F). These images summarize our earlier experiments and visualize the path of F. magna through the skin in the establishment of infection.
Figure 7.
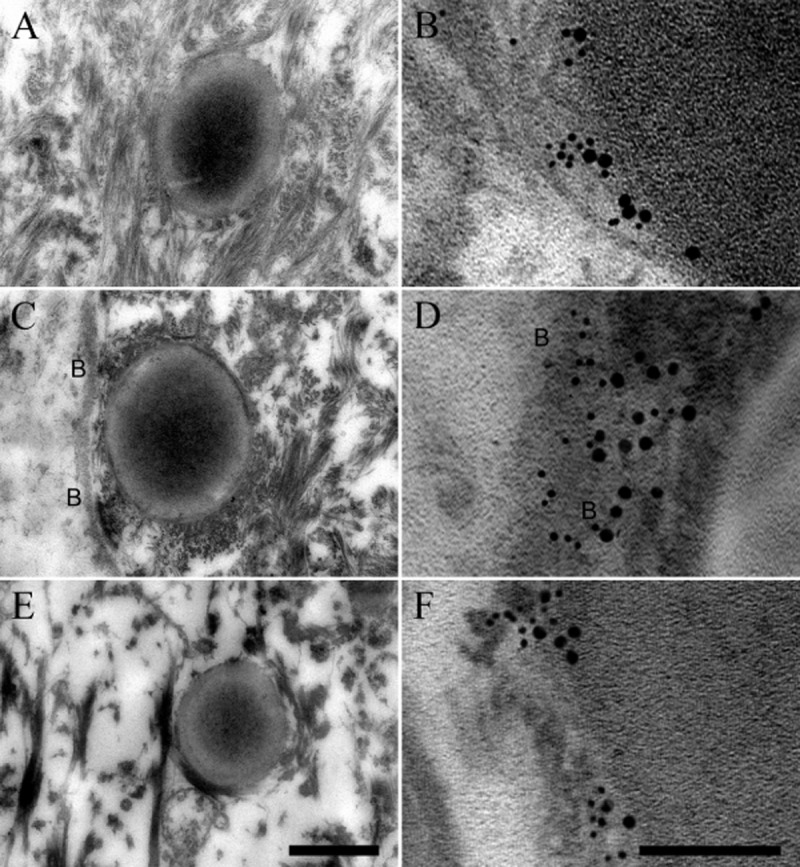
Transmission electron micrographs visualizing F. magna ALB8 interacting with galectin-7, BM-40 and fibrillin in human skin. Human skin biopsies were anaerobically incubated with F. magna strain ALB8 for 1 h at 37°C. Non-adherent bacteria were removed and the biopsies were anaerobically incubated for 24–72 h at 37°C and prepared for ultrathin sectioning and transmission electron microscopy. Sections were also subjected to immunolabelling with anti-FAF antibody (labelled with 5 nm colloidal gold) and anti-galectin-7, anti-BM-40 and anti-rFBN2-N antibodies (labelled with 20 nm colloidal gold) respectively. The location of the basement membrane is indicated by letters B. Trans section of F. magna ALB8 at the (A) skin epidermis after 24 h; (C) basement membrane after 48 h; (E) dermis after 72 h. The scale bar represents 0.5 μm. Gold labelled FAF on the surface of F. magna ALB8 interacting with (B) Gold labelled galectin-7 in the epidermis after 24 h; (D) Gold labelled BM-40 in the basement membrane after 48 h; (F) Gold labelled rFBN2-N in the dermis after 72 h. The scale bar represents 100 nm.
Discussion
F. magna is a commensal commonly found on the skin, but under some circumstances, has been known to act as an opportunistic pathogen. It is probably one of the most pathogenic species of the GPAC and has been isolated in pure culture from a variety of clinical infection sites (Bourgault et al., 1980; Murphy and Frick, 2013). In recent years, two proteins with importance for virulence and colonization, were identified in F. magna – the surface adhesion protein FAF and the subtilisin-like serine protease SufA (Karlsson et al., 2007; Frick et al., 2008). In this study, we investigated the role these two proteins play in helping F. magna adhere to the skin and invade deeper tissue layers during the establishment of infection. Binding and interaction of FAF and SufA with proteins in the different layers of skin was studied, in order to identify the mechanisms by which F. magna reaches deeper tissue layers during infection.
Ligands for FAF in the epidermis was investigated using human skin epidermal extracts (Murphy et al., 2014). Galectin-7 was identified as one of the major binding partners for FAF in the epidermis. This lectin is thought to be a keratinocyte cell type marker and its expression is maintained in all living layers of the epidermis (Magnaldo et al., 1995; Saussez and Kiss, 2006). Galectin-7 can be found in the cytoplasm, in the nucleus or in the extracellular space and its function may vary, according to its cellular localization (Saussez and Kiss, 2006). It is especially found in the upper layers of the human epidermis in areas of cell-to-cell contact (Madsen et al., 1995). It is also thought to play a key role in the re-epithelialization process of epidermal wounds and is involved in epithelial cell migration (Saussez and Kiss, 2006). It modulates cell proliferation and cell interactions and plays an important role in the formation of stratified epithelia (Saussez and Kiss, 2006). As galectin-7 expression is highly concentrated in the upper levels of the epithelium, it may serve as an important first step in the binding of F. magna to the skin and binding interactions may help it pass to deeper levels of the epithelium. SufA releases a 53 kDa FAF fragment from the bacterial surface (Frick et al., 2008). Binding of extracellular FAF to galectin-7 could possibly help affect and prevent re-epithelialization during wounding, thus maintaining an open chronic wound. However, this could not be conclusively proven (data not shown) and requires future studies.
Looking deeper into the basement membrane and dermis compartments of the skin, F. magna was found to have strong interactions with both collagen IV and V that have important structural roles in this area (Madri and Furthmayr, 1979; Gelse et al., 2003). SufA was found to completely degrade collagen IV while it was capable of breaking down collagen V into smaller fragments. Collagen IV is the backbone for the basement membrane layer of skin and it is thought that collagen V acts as an intermediary collagen, linking collagen IV in the basement membrane to collagen III in the dermal layer (Madri and Furthmayr, 1979; Roll et al., 1980; Konomi et al., 1984). The ability of F. magna to degrade collagen IV and break through the basement membrane allows it to overcome a significant hurdle in its passage to deeper tissue layers. Furthermore, F. magna was found to bind to collagen V via its surface adhesion protein FAF. Once F. magna breaks through the basement membrane, binding to collagen V via FAF would help the bacteria to adhere to the dermal layer.
Another important structural component of the dermis is fibrillin. Fibrillin microfibrils provide important structural integrity to the connective tissue and dermal layer of the skin and enable properties such as elasticity and extensibility by acting as a scaffold for the deposition of elastin (Kewley et al., 1978; Jones et al., 1980). Recombinant protein fragments of both fibrillin-1 and -2 were used to investigate F. magna interaction. Fibrillin-1 is the main component of structural microfibrils, while fibrillin-2 is integrated during tissue development and remodelling (Cain et al., 2006; Massam-Wu et al., 2010; Ramirez and Sakai, 2010). Binding experiments demonstrated that FAF binds to the N-terminal half of fibrillin-1 and -2, which would allow F. magna to adhere to fibrillin microfibrils and to establish infection in the dermis. It was also discovered that SufA can cleave off a 35 kDa fragment from the NH2-terminus of fibrillin-2. It is however unknown if this fragment cleavage is significant enough to allow F. magna to disrupt fibrillin microfibrils or whether it could negatively affect interactions of the NH2-terminus, which is rich in epidermal-growth-factor (EGF)-like domains, to microfibril associated proteins.
Rotary shadowing enabled the visualization of complexes formed in solution between protein FAF and fibrillin recombinant fragments. These results confirmed the binding studies, showing that FAF interacts with the N-terminal region of fibrillin-1 and -2.
The majority of F. magna clinical isolates express FAF and SufA (Karlsson et al., 2007; Frick et al., 2008), suggesting that these proteins play important roles in the F. magna colonization and infection process. This was also visually illustrated, see Fig. 6, where expression of FAF seems to aid the bacteria in the initial binding and colonization step. However, F. magna strain 505, despite lacking FAF, appeared to adhere in low amounts to human skin as some bacteria were present in the dermis layer after 72 h. Obviously, other mechanisms mediate adhesion of this strain. However, as can be seen in Table 1, strain 505 is present in a 1000-fold lower cell density than ALB8 and is hindered in the transmigration over the basement membrane to the dermal layer. The F. magna ΔSufA strain was also seen to be still mostly present at the epidermis, even after 72 h, suggesting that expression of SufA is of importance for the bacteria to reach the dermal layer. Once in the dermal layer, SufA causes significant breakdown and destruction of the collagen and fibrillin microfibrillar structures of the dermal matrix. These findings may explain why F. magna is so often found in ulcers and chronic wounds (Fitzgerald et al., 1982; Davies et al., 1988; Brook, 2008; Martin et al., 2009; Murphy and Frick, 2013). The ability of F. magna to invade deep dermal layers and to cause ongoing tissue destruction would severely delay the wound healing process, in particular if the bacterium was not identified and appropriate antibiotic treatment undertaken.
The presence of extracellular and surface located FAF and SufA have proven to play an important role in assisting F. magna to colonize skin epidermal sites. As F. magna is, for the most part, a commensal bacterium, SufA would probably not interfere with the host and cause extracellular matrix destruction under normal conditions. SufA probably comes more into play after an initial external wounding event, allowing F. magna greater access to dermal tissue sites. Here, it could transform from a commensal to an opportunistic pathogen and use SufA to further destruct tissue, enabling it to initiate an infection deep in the anaerobic tissue.
Host–microbe relationships are based on a multitude of molecular interactions, and the present investigation revealed that F. magna uses a combination of proteolytic and adhesive mechanisms to penetrate and colonize human skin. F. magna is also part of the indigenous microbiota in the oral cavity and the gastrointestinal and genitourinary tracts, and future studies will show if F. magna has a similar strategy to colonize these sites.
Experimental procedures
Bacteria and growth conditions
F. magna strains ALB8, 505, L3410, 2133 and 1462 were isolated at the Department of Clinical Microbiology, Skåne University Hospital, Sweden and have been described earlier (de Château and Björck, 1994; Frick et al., 2008). The strains were isolated from various clinical infection sites: scrotal abscess (ALB8), urethra (505), intra-abdominal abscess (L3410), foot abscess (2133) and foot wound (1462). For cultivation of F. magna ALB8 mutant ΔSufA which was produced in a previous study (Karlsson et al., 2009), 200 μg ml−1 kanamycin was added to the culture medium. The bacteria were grown under strict anaerobic conditions in Todd-Hewitt broth (TH) (Difco) supplemented with 0.5% Tween-80 at 37°C.
Proteins, plasma, antibodies and reagents
Galectin-7 was purchased from Nordic BioSite. Horseradish peroxidase (HRP) conjugated goat anti-rabbit IgG was purchased from Pierce. Collagen I, II and IV were purchased from Sigma and collagen III and V were purchased from Millipore. SufA was purified natively from F. magna ALB8 as previously described (Karlsson et al., 2007) and protein PAB was produced recombinantly as described (de Château and Björck, 1994). The purification of fibrillin recombinant fragments has been described previously (Jensen et al., 2001; Lin et al., 2002). Recombinant fragments included rF16 (N-terminal half of fibrillin-1; rFBN1-N), rF6H (C-terminal half of fibrillin-1, rFBN1-C), rFBN2-2 (N-terminal half of fibrillin-2, rFBN2-N) and rFBN2-1 (C-terminal half of fibrillin-2, rFBN2-C). Recombinantly expressed protein FAF [amino acid (AA) 28–616], and FAF fragments I (AA 28–115), II (AA 28–317) and III (AA 239–616), were obtained as fusion proteins with glutathione S-transferase (GST) as described (Frick et al., 2008). Prior to binding experiments, the GST tag was removed using PreScission Protease as described by the manufacturer (Amersham Biosciences). Anti-FAF antibodies were raised in rabbits as described (Frick et al., 2008) and rabbit anti-galectin-7 antibodies were purchased from AbD Serotec. The preparation of FAF-Au conjugates and gold-labelled anti-FAF antibodies was carried out as reviewed in (Baschong and Wrigley, 1990). Galectin-7, collagen V, rFBN1-N, rFBN2-N and FAF were radiolabelled with 125I using iodobeads (Pierce) as described by the manufacturer. Binding of radiolabelled proteins to bacteria was carried out as previously described (Björck and Kronvall, 1984).
Slot-binding, SDS-PAGE and Western blot analysis
SDS-PAGE was performed as described by Neville (1971). Samples were prepared for boiling in sample buffer containing 2% SDS and 5% β-mercaptoethanol for 5 min and 8% or 15% SDS-PAGE gels were used for sample analysis. Separated proteins were visualized by Coomassie Blue staining. For Western Blot analysis, proteins were electrophoretically transferred to a polyvinylidene difluoride (PVDF) membrane (Amersham Biosciences). For slot-binding analysis, proteins were directly applied to PVDF membranes using a Milliblot-D system (Millipore). Membranes were blocked in a phosphate buffered saline-Tween [PBS containing 0.1% Tween 20 (PBS-T)] solution containing 5% (w/v) skim milk powder at 37°C for 30 min. The membranes were then incubated with galectin-7 (0.71 μM) or FAF (0.23 μM) for 1 h at 37°C in blocking buffer. Membranes were washed three times with PBS-T for 5 min followed by incubation with primary antibodies (rabbit anti-FAF 1:1000 dilution, rabbit anti-galectin-7 1:10 000 dilution) in blocking buffer at 37°C for 30 min. Membranes were washed three times with PBS-T for 5 min followed by incubation with HRP-conjugated goat anti-rabbit secondary antibody (1:3000 dilution) in blocking buffer at 37°C for 30 min. Following a repeat of the wash steps, bound antibodies were detected by chemiluminescence as described by Nesbitt and Horton (1992).
Binding of 125I-labelled FAF to fibrillin protein fragments
The fibrillin fragments were applied to a PVDF membrane using a Milliblot-D system (Millipore). The membrane was washed in PBS for 30 min at room temperature and then blocked with PBS + 3% BSA (Bovine serum albumin) for 1 h at room temperature. The membrane was incubated with radiolabelled FAF (0.2 × 105 cpm ml−1) in 10 ml PBS + 3% BSA for 3 h at room temperature, followed by washing twice for 30 min with PBS + 0.5% Tween-20. The membrane was dried, exposed with film and developed for radioactivity using a Bas 2000 radio imaging system (Fuji, Tokyo, Japan).
Cleavage of proteins by SufA
SufA (0.11 μM) was incubated with collagen I, II, III, IV and V and fibrillin fragments rFBN1-N, rFBN1-C, rFBN2-N and rFBN2-C (3.5 μg of each protein) in 50 mM Tris-HCl pH 7.5. As a control, SufA was substituted with buffer. Following incubation for 3 h at 37°C, the samples were subjected to analysis by SDS-PAGE using a polyacrylamide concentration of 8%.
Binding of galectin-7 to whole bacteria in solution
A stationary culture of F. magna ALB8 was washed twice in PBS and bacteria were resuspended in a 2 × 109 bacteria ml−1 solution. One hundred microlitres of this bacterial solution was added to 7.2 μM galectin-7 in 100 μl PBS. As a control, the bacteria were added to PBS only. The solutions were incubated for 1 h at 37°C, end over end rotation. The bacteria were then washed four times in PBS to remove unbound protein, followed by incubation in 0.1 M glycine-HCl buffer, pH 2, for 30 min at room temperature to elute bound protein. The bacterial cells were removed by spinning at 6000 g for 5 min and the supernatant was transferred to a fresh tube where the pH was neutralized using 1 M Tris. Proteins were precipitated using 5% TCA and analysis of galectin-7 binding was carried out by SDS-PAGE and western blotting as described above.
Surface plasmon resonance assay (BIAcore)
FAF protein was immobilized on a CM5 sensor chip (GE Healthcare). Affinity measurements were monitored in a BIAcore 2000 instrument. Different concentrations of Galectin-7/rFBN1-N/rFBN2-N were injected over the coated surfaces. The association (ka) and dissociation (kd) rate constants were determined simultaneously using the equation for 1:1 Langmuir binding in the BIA Evaluation 4.1 software (GE Healthcare). The binding curves were fitted locally and the equilibration dissociation constants (KD) were calculated from mean values of the obtained rate constants.
Rotary shadowing of FAF-fibrillin complexes
Fibrillin samples in complex with FAF-Au (10 nm Au) were dissolved in 0.2 M ammonium hydrogen carbonate at final concentrations of 10–50 μg ml−1, mixed with an equal volume of 80% glycerol and then sprayed on to freshly cleaved mica pieces, followed by rotary shadowing with platinum/carbon at a 9° angle. Details of the electron microscopic procedures and methods of data evaluation have been described previously (Engel and Furthmayr, 1987).
Negative staining and transmission electron microscopy of fibrillin fragments and FAF on the F. magna ALB8 surface
F. magna ALB8 grown to stationary phase were washed with TBS (50 mM Tris-HCl, 150 mM NaCl, pH 7.5) and adjusted to 2 × 109 bacteria ml−1. One microlitre of 2 × 109 bacteria ml−1 was incubated with 5 μl of 5 nm Au-labelled anti-FAF antibody and 5 μl of 20 nm Au-labelled fibrillin fragments for 30 min at 37°C. Following incubation, the bacteria were recovered by centrifugation at 3000 g and samples were prepared for negative staining by adsorption onto 400 mesh carbon-coated copper grids and staining with 0.75% (w/v) uranyl formate as described (Bober et al., 2010). Samples were observed in an FEI Tecnai Spirit BioTWIN transmission electron microscope (North America NanoPort, Hillsboro, OR) operating at an accelerating voltage of 60 kV. Images were recorded using an Eagle™ CCD camera.
Transmission and scanning electron microscopy of skin sections
For transmission electron microscopy of skin sections, a stationary phase culture of F. magna ALB8 was washed twice in PBS and the concentration adjusted to 2 × 109 bacteria ml−1 in PBS. Skin specimens were obtained as excess healthy tissue from skin surgery, under protocols approved by the Ethics Committee at Lund University (permit No. LU 762-02). A 4 cm2 section of a skin specimen was washed 3 times in PBS and 4 mm circular punches of epidermis were extracted. A measure of 2 × 109 bacteria was incubated with each skin biopsy for 1 h at 37°C under anaerobic conditions. Following incubation, skin sections were removed to a 24-well plate, washed 3 times with PBS to remove unbound bacteria, followed by addition of 500 μl MEM (Life Technologies). Skin sections with bound bacteria were incubated for 24 h, 48 h or 72 h at 37°C under anaerobic conditions. The skin specimen was then added to 1 ml of 2.5% glutaraldehyde in cacodylate buffer overnight at 4°C.
For transmission electron microscopy, samples were washed with cacodylate buffer and post-fixed for 1 h at room temperature in 1% osmium tetroxide in cacodylate buffer, dehydrated in a graded series of ethanol, and then embedded in Epon 812 (SPI Supplies) using acetone as an intermediate solvent. Specimens were sectioned with a diamond knife into 50–70 nm thick ultrathin sections on an LKB ultramicrotome. Immunolabelling was carried out with rabbit anti FAF antibody, titre 1:10, labelled with 5 nm colloidal gold and rabbit anti galectin-7 antibody (titre 1:100), rabbit anti BM-40 (titre 1:100) or rabbit anti-rFBN2-N (titre 1:10) all labelled with 20 nm colloidal gold. Analysis was carried out using a JEOL JEM 1230 transmission electron microscope (JEOL, Peabody, MA) as previously described (Svensson et al., 2011).
For scanning electron microscopy, specimens were fixed in 2.5% glutaraldehyde in 0.15 M sodium cacodylate, pH 7.4 for 2 h at room temperature, washed and stored in 0.15 M cacodylate buffer, pH 7.4. Fixed specimens were dehydrated for 10 min at each step of an ascending ethanol series and inserted into a Balzers critical point dryer using 100% ethanol as the intermediate solvent. The pressure chamber was then extensively flushed three times with carbon dioxide to remove all traces of residual ethanol. The samples were critical point dried, mounted on aluminium holders, palladium/gold sputtered and examined in a Jeol ST300 SEM.
Acknowledgments
This work was supported by the Swedish Research Council (project 7480), the Foundations of Knut and Alice Wallenberg, Crafoord, Bergvall, Anders Österlund, O. E. and Edla Johansson, Sigurd and Elsa Goljes, Greta and Johan Kock, the Royal Physiographic Society of Lund, Hansa Medical AB and the Swedish Government Funds for Clinical Research (ALF). We acknowledge the help with protein analysis from the SCIBLU Proteomics Resource Centre at Lund University.
Supporting Information
Additional supporting information may be found in the online version of this article at the publisher's web-site.
References
- Baschong W. Wrigley NG. Small colloidal gold conjugated to Fab fragments or to immunoglobulin G as high-resolution labels for electron microscopy: a technical overview. J Electron Microsc Tech. 1990;14:313–323. doi: 10.1002/jemt.1060140405. [DOI] [PubMed] [Google Scholar]
- Birk DE, Fitch JM, Babiarz JP. Linsenmayer TF. Collagen type I and type V are present in the same fibril in the avian corneal stroma. J Cell Biol. 1988;106:999–1008. doi: 10.1083/jcb.106.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björck L. Protein L. A novel bacterial cell wall protein with affinity for Ig L chains. J Immunol. 1988;140:1194–1197. [PubMed] [Google Scholar]
- Björck L. Kronvall G. Purification and some properties of streptococcal protein G, a novel IgG-binding reagent. J Immunol. 1984;133:969–974. [PubMed] [Google Scholar]
- Bober M, Enochsson C, Collin M. Mörgelin M. Collagen VI is a subepithelial adhesive target for human respiratory tract pathogens. J Innate Immun. 2010;2:160–166. doi: 10.1159/000232587. [DOI] [PubMed] [Google Scholar]
- Bourgault AM, Rosenblatt JE. Fitzgerald RH. Peptococcus magnus: a significant human pathogen. Ann Intern Med. 1980;93:244–248. doi: 10.7326/0003-4819-93-2-244. [DOI] [PubMed] [Google Scholar]
- Brazier J, Chmelar D, Dubreuil L, Feierl G, Hedberg M, Kalenic S, et al. European surveillance study on antimicrobial susceptibility of Gram-positive anaerobic cocci. Int J Antimicrob Agents. 2008;31:316–320. doi: 10.1016/j.ijantimicag.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Brook I. Encapsulated anaerobic bacteria in synergistic infections. Microbiol Rev. 1986;50:452–457. doi: 10.1128/mr.50.4.452-457.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook I. Microbiology and management of joint and bone infections due to anaerobic bacteria. J Orthop Sci. 2008;13:160–169. doi: 10.1007/s00776-007-1207-1. [DOI] [PubMed] [Google Scholar]
- Brook I. Frazier EH. Aerobic and anaerobic microbiology in intra-abdominal infections associated with diverticulitis. J Med Microbiol. 2000;49:827–830. doi: 10.1099/0022-1317-49-9-827. [DOI] [PubMed] [Google Scholar]
- Cain SA, Morgan A, Sherratt MJ, Ball SG, Shuttleworth CA. Kielty CM. Proteomic analysis of fibrillin-rich microfibrils. Proteomics. 2006;6:111–122. doi: 10.1002/pmic.200401340. [DOI] [PubMed] [Google Scholar]
- de Château M. Björck L. Protein PAB, a mosaic albumin-binding bacterial protein representing the first contemporary example of module shuffling. J Biol Chem. 1994;269:12147–12151. [PubMed] [Google Scholar]
- de Château M, Holst E. Björck L. Protein PAB, an albumin-binding bacterial surface protein promoting growth and virulence. J Biol Chem. 1996;271:26609–26615. doi: 10.1074/jbc.271.43.26609. [DOI] [PubMed] [Google Scholar]
- Davies UM, Leak AM. Dave J. Infection of a prosthetic knee joint with Peptostreptococcus magnus. Ann Rheum Dis. 1988;47:866–868. doi: 10.1136/ard.47.10.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM, James GA. Wolcott RD. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008a;8:43. doi: 10.1186/1471-2180-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd SE, Wolcott RD, Sun Y, McKeehan T, Smith E. Rhoads D. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP) PLoS ONE. 2008b;3:e3326. doi: 10.1371/journal.pone.0003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel J. Furthmayr H. Electron microscopy and other physical methods for the characterization of extracellular matrix components: laminin, fibronectin, collagen IV, collagen VI, and proteoglycans. Methods Enzymol. 1987;145:3–78. doi: 10.1016/0076-6879(87)45003-9. [DOI] [PubMed] [Google Scholar]
- Fitzgerald RH, Jr, Rosenblatt JE, Tenney JH. Bourgault AM. Anaerobic septic arthritis. Clin Orthop Relat Res. 1982;164:141–148. [PubMed] [Google Scholar]
- Frick IM, Karlsson C, Mörgelin M, Olin AI, Janjusevic R, Hammarstrom C, et al. Identification of a novel protein promoting the colonization and survival of Finegoldia magna, a bacterial commensal and opportunistic pathogen. Mol Microbiol. 2008;70:695–708. doi: 10.1111/j.1365-2958.2008.06439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelse K, Poschl E. Aigner T. Collagens – structure, function, and biosynthesis. Adv Drug Deliv Rev. 2003;55:1531–1546. doi: 10.1016/j.addr.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Genovese A, Borgia G, Björck L, Petraroli A, de Paulis A, Piazza M. Marone G. Immunoglobulin superantigen protein L induces IL-4 and IL-13 secretion from human Fc epsilon RI+ cells through interaction with the kappa light chains of IgE. J Immunol. 2003;170:1854–1861. doi: 10.4049/jimmunol.170.4.1854. [DOI] [PubMed] [Google Scholar]
- Goto T, Yamashita A, Hirakawa H, Matsutani M, Todo K, Ohshima K, et al. Complete genome sequence of Finegoldia magna, an anaerobic opportunistic pathogen. DNA Res. 2008;15:39–47. doi: 10.1093/dnares/dsm030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson C, Hoborn J, Moller A. Swanbeck G. The microbial flora in venous leg ulcers without clinical signs of infection. Repeated culture using a validated standardised microbiological technique. Acta Derm Venereol. 1995;75:24–30. doi: 10.2340/00015555752430. [DOI] [PubMed] [Google Scholar]
- Higaki S. Morohashi M. Characteristics of anaerobes from skin specimens. Drugs Exp Clin Res. 2003;29:153–155. [PubMed] [Google Scholar]
- Jensen SA, Reinhardt DP, Gibson MA. Weiss AS. Protein interaction studies of MAGP-1 with tropoelastin and fibrillin-1. J Biol Chem. 2001;276:39661–39666. doi: 10.1074/jbc.M104533200. [DOI] [PubMed] [Google Scholar]
- Jensen SA, Robertson IB. Handford PA. Dissecting the fibrillin microfibril: structural insights into organization and function. Structure. 2012;20:215–225. doi: 10.1016/j.str.2011.12.008. [DOI] [PubMed] [Google Scholar]
- Jones CJ, Sear CH. Grant ME. An ultrastructural study of fibroblasts derived from bovine ligamentum nuchae and their capacity for elastogenesis in culture. J Pathol. 1980;131:35–53. doi: 10.1002/path.1711310104. [DOI] [PubMed] [Google Scholar]
- Karlsson C, Andersson ML, Collin M, Schmidtchen A, Björck L. Frick IM. SufA – a novel subtilisin-like serine proteinase of Finegoldia magna. Microbiology. 2007;153:4208–4218. doi: 10.1099/mic.0.2007/010322-0. [DOI] [PubMed] [Google Scholar]
- Karlsson C, Mörgelin M, Collin M, Lood R, Andersson ML, Schmidtchen A, et al. SufA – a bacterial enzyme that cleaves fibrinogen and blocks fibrin network formation. Microbiology. 2009;155:238–248. doi: 10.1099/mic.0.021311-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastern W, Holst E, Nielsen E, Sjöbring U. Björck L. Protein L, a Bacterial Immunoglobulin-Binding Protein and Possible Virulence Determinant. Infect Immun. 1990;58:1217–1222. [Google Scholar]
- Kewley MA, Williams G. Steven FS. Studies of elastic tissue formation in the developing bovine ligamentum nuchae. J Pathol. 1978;124:95–101. doi: 10.1002/path.1711240205. [DOI] [PubMed] [Google Scholar]
- Konomi H, Hayashi T, Nakayasu K. Arima M. Localization of type V collagen and type IV collagen in human cornea, lung, and skin. Immunohistochemical evidence by anti-collagen antibodies characterized by immunoelectroblotting. Am J Pathol. 1984;116:417–426. [PMC free article] [PubMed] [Google Scholar]
- Koziel J. Potempa J. Protease-armed bacteria in the skin. Cell Tissue Res. 2013;351:325–337. doi: 10.1007/s00441-012-1355-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krepel CJ, Gohr CM, Walker AP, Farmer SG. Edmiston CE. Enzymatically active Peptostreptococcus magnus: association with site of infection. J Clin Microbiol. 1992;30:2330–2334. doi: 10.1128/jcm.30.9.2330-2334.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonidas DD, Vatzaki EH, Vorum H, Celis JE, Madsen P. Acharya KR. Structural basis for the recognition of carbohydrates by human galectin-7. Biochemistry. 1998;37:13930–13940. doi: 10.1021/bi981056x. [DOI] [PubMed] [Google Scholar]
- Levy PY, Fenollar F, Stein A, Borrione F. Raoult D. Finegoldia magna: a forgotten pathogen in prosthetic joint infection rediscovered by molecular biology. Clin Infect Dis. 2009;49:1244–1247. doi: 10.1086/605672. [DOI] [PubMed] [Google Scholar]
- Lin G, Tiedemann K, Vollbrandt T, Peters H, Batge B, Brinckmann J. Reinhardt DP. Homo- and heterotypic fibrillin-1 and -2 interactions constitute the basis for the assembly of microfibrils. J Biol Chem. 2002;277:50795–50804. doi: 10.1074/jbc.M210611200. [DOI] [PubMed] [Google Scholar]
- Madri JA. Furthmayr H. Isolation and tissue localization of type AB2 collagen from normal lung parenchyma. Am J Pathol. 1979;94:323–331. [PMC free article] [PubMed] [Google Scholar]
- Madsen P, Rasmussen HH, Flint T, Gromov P, Kruse TA, Honore B, et al. Cloning, expression, and chromosome mapping of human galectin-7. J Biol Chem. 1995;270:5823–5829. doi: 10.1074/jbc.270.11.5823. [DOI] [PubMed] [Google Scholar]
- Magnaldo T, Bernerd F. Darmon M. Galectin-7, a human 14-kDa S-lectin, specifically expressed in keratinocytes and sensitive to retinoic acid. Dev Biol. 1995;168:259–271. doi: 10.1006/dbio.1995.1078. [DOI] [PubMed] [Google Scholar]
- Martin J, Bemer P, Touchais S, Asseray N. Corvec S. Recurrent abscesses due to Finegoldia magnaDermabacter hominis and Staphylococcus aureus in an immunocompetent patient. Anaerobe. 2009;15:201–203. doi: 10.1016/j.anaerobe.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Massam-Wu T, Chiu M, Choudhury R, Chaudhry SS, Baldwin AK, McGovern A, et al. Assembly of fibrillin microfibrils governs extracellular deposition of latent TGF beta. J Cell Sci. 2010;123:3006–3018. doi: 10.1242/jcs.073437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy EC. Frick IM. Gram-positive anaerobic cocci – commensals and opportunistic pathogens. FEMS Microbiol Rev. 2013;37:520–553. doi: 10.1111/1574-6976.12005. [DOI] [PubMed] [Google Scholar]
- Murphy EC, Mohanty T. Frick IM. FAF and SufA: proteins of Finegoldia magna that modulate the antibacterial activity of histones. J Innate Immun. 2014;6:394–404. doi: 10.1159/000356432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy I, Filkor K, Nemeth T, Hamari Z, Vagvolgyi C. Gacser A. In vitro interactions of Candida parapsilosis wild type and lipase deficient mutants with human monocyte derived dendritic cells. BMC Microbiol. 2011;11:122. doi: 10.1186/1471-2180-11-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji T, Chiang HI, Jiang SB, Nagarajan H, Zengler K. Gallo RL. The microbiome extends to subepidermal compartments of normal skin. Nat Commun. 2013;4:1431. doi: 10.1038/ncomms2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbitt SA. Horton MA. A nonradioactive biochemical characterization of membrane proteins using enhanced chemiluminescence. Anal Biochem. 1992;206:267–272. doi: 10.1016/0003-2697(92)90365-e. [DOI] [PubMed] [Google Scholar]
- Neville DM., Jr Molecular weight determination of protein-dodecyl sulfate complexes by gel electrophoresis in a discontinuous buffer system. J Biol Chem. 1971;246:6328–6334. [PubMed] [Google Scholar]
- Pringle F. Penzer R. Normal skin: its function and care. In: Penzer R, editor; Nursing Care of the Skin. Oxford: Butterworth Heinimann; 2002. pp. 20–45. [Google Scholar]
- Proksch E, Brandner JM. Jensen JM. The skin: an indispensable barrier. Exp Dermatol. 2008;17:1063–1072. doi: 10.1111/j.1600-0625.2008.00786.x. [DOI] [PubMed] [Google Scholar]
- Ramirez F. Sakai LY. Biogenesis and function of fibrillin assemblies. Cell Tissue Res. 2010;339:71–82. doi: 10.1007/s00441-009-0822-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll FJ, Madri JA, Albert J. Furthmayr H. Codistribution of collagen types IV and AB2 in basement membranes and mesangium of the kidney. an immunoferritin study of ultrathin frozen sections. J Cell Biol. 1980;85:597–616. doi: 10.1083/jcb.85.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saussez S. Kiss R. Galectin-7. Cell Mol Life Sci. 2006;63:686–697. doi: 10.1007/s00018-005-5458-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer NN. Gallo RL. Structure and function of the human skin microbiome. Trends Microbiol. 2013;21:660–668. doi: 10.1016/j.tim.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens P, Wall IB, Wilson MJ, Hill KE, Davies CE, Hill CM, et al. Anaerobic cocci populating the deep tissues of chronic wounds impair cellular wound healing responses in vitro. Br J Dermatol. 2003;148:456–466. doi: 10.1046/j.1365-2133.2003.05232.x. [DOI] [PubMed] [Google Scholar]
- Svensson KJ, Kucharzewska P, Christianson HC, Skold S, Lofstedt T, Johansson MC, et al. Hypoxia triggers a proangiogenic pathway involving cancer cell microvesicles and PAR-2-mediated heparin-binding EGF signaling in endothelial cells. Proc Natl Acad Sci USA. 2011;108:13147–13152. doi: 10.1073/pnas.1104261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpl R. Macromolecular organization of basement membranes. Curr Opin Cell Biol. 1996;8:618–624. doi: 10.1016/s0955-0674(96)80102-5. [DOI] [PubMed] [Google Scholar]
- Tortora GJ. Derrickson BH. Principles of Anatomy and Pysiology: Organisation, Support and Movement and Control Systems of the Human Body. Hoboken, NJ: John Wiley and Sons; 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.