Abstract
Cofilin is a ubiquitous modulator of actin cytoskeleton dynamics that can both stabilize and destabilize actin filaments depending on its concentration and/or the presence of regulatory co-factors. Three charge-reversal mutants of yeast cofilin, located in cofilin’s filament-specific secondary binding site, were characterized in order to understand why disruption of this site leads to enhanced filament disassembly. Crystal structures of the mutants showed that the mutations specifically affect the secondary actin-binding interface, leaving the primary binding site unaltered. The mutant cofilins show enhanced activity compared to wild-type cofilin in severing and disassembling actin filaments. Electron microscopy and image analysis revealed long actin filaments in the presence of wild-type cofilin, while the mutants induced many short filaments, consistent with enhanced severing. Real-time fluorescence microscopy of labeled actin filaments confirmed that the mutants, unlike wild-type cofilin, were functioning as constitutively active severing proteins. In cells, the mutant cofilins delayed endocytosis, which depends on rapid actin turnover. We conclude that mutating cofilin’s secondary actin-binding site increases cofilin’s ability to sever and depolymerize actin filaments. We hypothesize that activators of cofilin severing, like Aip1p, may act by disrupting the interface between cofilin’s secondary actin-binding site and the actin filament.
Keywords: actin cytoskeleton, S. cerevisiae, cofilin, endocytosis, severing
Introduction
The ADF/cofilin family of proteins are among the most extensively studied actin-binding proteins. In vitro, their activities have been reported to drive the actin monomer-polymer equilibrium in both directions by multiple mechanisms, depending on the conditions, the type of ADF/cofilin, and the form of actin. Cofilins bind actin filaments cooperatively (Hayden et al., 1993) (Hawkins et al., 1993) and sever filaments in a concentration-dependent manner (Maciver et al., 1991) (Andrianantoandro and Pollard, 2006) (Pavlov et al., 2007). These activities suggest a versatile protein whose ultimate purpose is to accelerate actin dynamics in vivo (Poukkula et al., 2011).
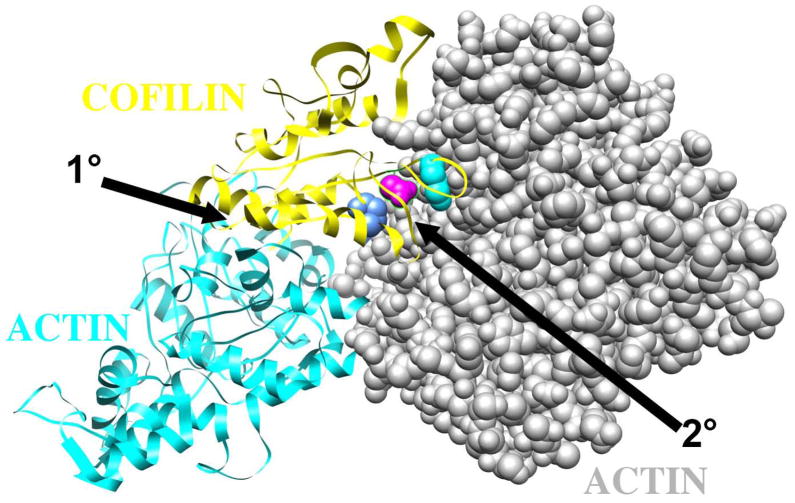
Structural and modeling approaches have characterized the interaction between cofilin and F-actin (Galkin et al., 2011) (Wong and Sept, 2011) (Fan et al., 2013) revealing that one cofilin molecule binds two consecutive actin subunits within the long-pitch helix of the filament (see Figure 3). The primary binding site, which is also the G-actin binding site (Rodal et al., 1999), is on the actin subunit closer to the pointed/minus end, between sub-domains 1 and 3 (1° on Figure 3). The secondary binding site is on sub-domain 2 of the actin subunit that is closer to the barbed/plus end (2° on Figure 3).
Figure 3. Structural modeling predicts the cofilin mutants affect the filament specific binding interface on actin.
A trimeric model for cofilin bound to two consecutive subunits in an actin filament is shown for rabbit muscle actin and human cofilin. The model (pdb file 3JOS) was derived by cryo-electron miscroscopy and image averaging (Galkin et al., 2011). The actin subunit nearer the pointed end of the filament is rendered in cyan, the actin subunit nearest the barbed end is space filled in grey, and the cofilin subunit is in yellow. K152 of human cofilin-2, equivalent to R135 of yeast cofilin, is space filled in blue. K96 of human cofilin-2, equivalent to R80 of yeast cofilin, is space filled in cyan. D98 of human cofilin-2, equivalent to K82 of yeast cofilin, is space filled in magenta. The primary (1°) and secondary (2°) binding interfaces are indicated with arrows.
Cofilin binding causes the actin subunits within a filament to assume a tilted conformation different from the G- or F- conformations (Orlova et al., 2004) (De La Cruz and Sept, 2010). These cofilin-induced structural changes mainly affect subdomain 2, where the cofilin secondary binding site lies (Orlova et al., 2004) (Galkin et al., 2011). The tilted conformation of actin alters both lateral (McGough and Chiu, 1999) (Bobkov et al., 2004) (Fan et al., 2013) and longitudinal (Bobkov et al., 2002) (Dedova et al., 2002) (Fan et al., 2013; Galkin et al., 2003) actin-actin contacts within the filament, changes that would be predicted to be destabilizing to filaments in the absence of cofilin. When cofilin binding becomes saturated, the twist of the two-start right-handed helix of the filament is increased, causing a 25% reduction in crossover length (McGough et al., 1997). When cofilin binding is sub-saturating, the conformational change that accompanies cofilin binding has been hypothesized to propagate into adjacent regions of the filament that are free of cofilin (Bobkov et al., 2006; Prochniewicz et al., 2005; Ressad et al., 1998) (Galkin et al., 2010).
The cooperative binding of cofilin to actin filaments can be described using an Ising model with a one-dimensional lattice (McGhee and von Hippel, 1974). This model predicts that the conformational changes in the filament, induced by cofilin, result in increased affinity at neighboring sites and thus cooperative binding (De La Cruz, 2009). This model is sufficient to explain both the kinetics and thermodynamics of cofilin association with actin filaments (De La Cruz, 2005) (De La Cruz and Sept, 2010) (Cao et al., 2006). Cofilin binding is largely unaffected by temperature indicating that the interaction is driven by changes in entropy not enthalpy that can be fully accounted for by cation release and increased filament flexibility (Cao et al., 2006) (McCullough et al., 2008). At sub-stoichiometric concentrations of cofilin, this cooperative behavior results in regions with cofilin decoration and regions without cofilin. At a boundary between these regions, mechanical discontinuities can then result and lead to severing at the boundary (Andrianantoandro and Pollard, 2006) (Suarez et al., 2011).
In budding yeast, cofilin is essential for viability (Iida et al., 1993) and it localizes to the actin cortical patches (Moon et al., 1993) which are the sites of endocytosis. Actin patches are highly dynamic, and actin filament assembly is believed to drive patch internalization and creation of the endocytic vesicle (Mooren et al., 2012). Endocytosis in yeast is achieved through spatiotemporal regulation of numerous proteins, associated with clathrin-mediated endocytosis and the assembly of actin (Kaksonen et al., 2005). Adapting the dendritic nucleation model (Pollard et al., 2000) to the cortical actin patch, it has been argued that branched actin assembly close to the membrane generates the force required for vesicle invagination and scission (Kaksonen et al., 2003).
Endocytosis and actin patch behavior can be divided into three phases of movement. In Phase I, the patches assemble and remain stationary. The actin filament network begins to assemble near the end of Phase I. Phase II is characterized by a slow inward movement of the actin patch, which corresponds to the invagination of plasma membrane. In phase III, the endocytic vesicle buds off from the membrane, then moves into the interior of the cell at a more rapid rate. The actin filament network disassembles during this phase of movement (Lin et al., 2010). Based on the time of its arrival, cofilin was suggested to act during Phase III (Okreglak and Drubin, 2007). However, cofilin mutants have defects that can be observed as early as Phase I (Lin et al., 2010), suggesting that cofilin could be promoting actin assembly first, during the early phases, and then disassembly during the later phases (Okreglak and Drubin, 2007).
Cortical actin patches are also enriched in the cofilin activator Aip1p, which is a ubiquitous protein, and the first one found to induce cofilin-dependent filament disassembly (Rodal et al., 1999). Aip1p functions exclusively in cooperation with cofilin to induce severing (Ono et al., 2004) likely followed by capping of the barbed end (Balcer et al., 2003) (Okada et al., 2006). Cofilin and Aip1p appear at the patch at the same time, and the localization of each one depends on the other (Rodal et al., 1999) (Okreglak and Drubin, 2010) (Lin et al., 2010). The disassembly activity of Aip1p is sensitive to cofilin stoichiometry; Aip1p is most active on filaments fully decorated with cofilin (Rodal et al., 1999). In a previous study, to investigate the molecular basis of the cofilin-Aip1p interaction, we employed molecular dynamics and molecular docking to identify possible configurations of the cofilin-Aip1p complex. One configuration agreed well with experimental data (Amberg et al., 1995) (Rodal et al., 1999) (Clark et al., 2006), and predicted a large area of contact between the molecules involving 8 stabilizing salt-bridges (Clark et al., 2006). To test this model, we constructed three charge-reversal cofilin mutants, each predicted to disrupt a salt bridge with Aip1p: cof1R80E (cof1-158p), cof1K82D (cof1-159p) and cof1R135D (cof1-157p). While one might have expected these mutants to have a high level of fitness, similar to that of an aip1Δ strain, we found that they cause a gradient of stronger in vivo defects. These mutants also showed apparent increases in actin-filament disassembly activity in vitro, which were independent of Aip1p (Clark and Amberg, 2007). Equivalent mutations in fission yeast cofilin were found to inhibit actin polymerization (Andrianantoandro and Pollard, 2006), and new models of cofilin binding to the actin filament suggest that these residues lie in the secondary binding site between cofilin and actin (Galkin et al., 2011) (Wong and Sept, 2011).
Based on these observations, we elected to use these mutants to study the contributions of the secondary binding site to cofilin activity. We employed both structural and functional biochemical approaches using exclusively proteins from S. cerevisiae. We found decreased binding and high filament severing activity for all three mutants- cof1-157p, cof1-158p, and cof1-159p. Crystal structures of the mutants, solved at high resolutions, showed only local changes in their structure. The biochemical defects of the mutant cofilins correlate well with their in vivo defects, including the assembly and dynamics of cortical actin patches. We conclude that the stabilizing and severing activities of cofilin depend on the ability of the secondary site to interact properly with the filament, and we suggest that this interaction may serve as a site for regulation by proteins that activate cofilin-induced severing, including Aip1p.
Results
Structure of mutant cofilins
To probe the molecular basis for the enhanced in vitro F-actin disassembly by the cof1-157p, cof1-158p, and cof1-159p mutant proteins (Clark and Amberg, 2007), we solved their crystal structures (Figure 1). Complete diffraction data were measured to a maximum resolution of 1.90Å, 1.45Å, and 1.10Å for cof1-157p (pdb file 4KED), cof1-158p (pdb file 4KEE), and cof1-159p (pdb file 4KEF), respectively. Atomic models for these three mutants were built and refined to an Rfactor/Rfree of 18.1%/23.3%, 13.5%/17.5%, and 12.6%/14.4%, respectively (Table 2). These atomic models have significantly higher resolutions than the structure of wild type cofilin, previously determined in two crystal forms at 2.3Å and 3Å resolution (Federov et al., 1997).
Figure 1. Crystal structures of cofilin mutants.
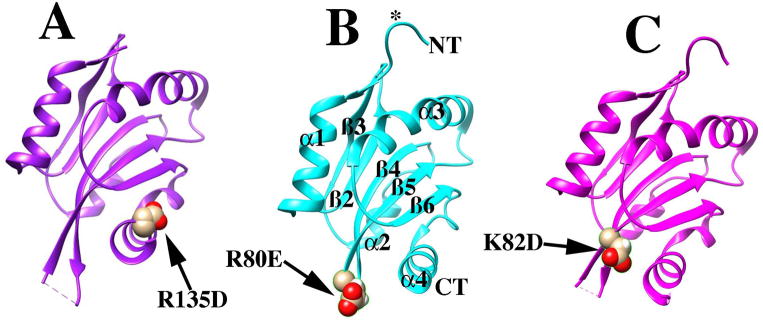
Structure of cof1-157p (A), cof1-158p (B), and cof1-159p (C) are shown in ribbon diagram. Structural elements as defined by (Federov et al., 1997) are notated in (B) for clarity. Regions mentioned in the text, residues 1-5 and the β4/5 turn are indicated with an asterisk and arrow, respectively.
Table 2.
Data collection and refinement statistics for cofilin structure determination.
| cof1-157p | cof1-158p | cof1-159p | |
|---|---|---|---|
|
| |||
| Data Collection | |||
|
| |||
| Wavelength (Å) | 0.9791 | 0.9791 | 1.0000 |
|
| |||
| Resolution (Å) | 30-1.90 (1.97-1.90) | 50-1.45 (1.50-1.45) | 30 -1.10 (1.14-1.10) |
|
| |||
| Space group | C 1 2 1 | P 1 21 1 | P 1 21 1 |
|
| |||
| Unit Cell | |||
| a, b, c (Å) | 150.0 30.3 104.3 | 28.1 69.8 32.2 | 28.1 69.5 32.0 |
| α, β, γ (°) | 90.0 133.0 90.0 | 90.0 100.4 90.0 | 90.0 100.6 90.0 |
|
| |||
| Total reflections | 162505 | 136680 | 205008 |
|
| |||
| Unique reflections | 27313 | 20869 | 49213 |
|
| |||
| Multiplicity | 5.9 (5.1) | 6.5 (3.2) | 4.2 (2.3) |
|
| |||
| Completeness (%) | 98.83 (94.61) | 95.98 (72.34) | 99.77 (98.99) |
|
| |||
| Mean I/sigma(I) | 28.7(4.2) | 14.9(2.7) | 25(2.8) |
|
| |||
| Wilson B-factor | 30.3 | 10.6 | 10.4 |
|
| |||
| R-merge | 0.05 (0.42) | 0.11 (0.54) | 0.08 (0.34) |
|
| |||
| Refinement | |||
|
| |||
| R-work | 0.181 (0.249) | 0.135 (0.159) | 0.126 (0.180) |
|
| |||
| R-free | 0.233 (0.306) | 0.175 (0.233) | 0.144 (0.203) |
|
| |||
| Number of atoms | 2279 | 2177 | 2327 |
|
| |||
| macromolecules | 2072 | 1039 | 1096 |
|
| |||
| water | 207 | 172 | 184 |
|
| |||
| RMSD (bonds) (Å) | 0.007 | 0.003 | 0.011 |
|
| |||
| RMSD (angles) (Å) | 0.97 | 0.83 | 1.41 |
|
| |||
| Ramachandran favored (%) | 100 | 100 | 100 |
|
| |||
| Ramachandran outliers (%) | 0 | 0 | 0 |
|
| |||
| Clashscore | 2.69 | 1.00 | 2.82 |
|
| |||
| Average B-factor | 39.00 | 16.60 | 17.00 |
|
| |||
| macromolecules | 38.40 | 14.40 | 14.90 |
|
| |||
| solvent | 45.20 | 29.40 | 29.30 |
Values in parentheses are for highest resolution shells. A single crystal was used for all datasets.
Highest resolution is presented here as the Bragg spacing at which I/σ(I) is at least 2.0.
Separate high and low exposure datasets were collected for Cof159p and scaled together.
The mutant cofilin structures superimpose closely with that of wild type cofilin (1COF; (Federov et al., 1997)) giving RMSDs of 0.37Å, 0.37Å, and 0.39Å for the corresponding Cα atoms of cof1-157p, cof1-158p, and cof1-159p, respectively. Using the nomenclature described in the 1COF structure, the overall topology deviates in two areas. The first is the turn linking β4 and β5 (β4/5) and second is the N-terminus (Figure 2). Unlike wild-type cofilin, the β4/5 turn of the mutant structures is disordered with the N- and C-terminal ordered residues being 74/78, 72/80, and 72/79 for cof1-157p, cof1-158p, and cof1-159p, respectively. With regard to the N-terminus, residues 1-5 are disordered in the wild type structure, as well as in cof1-158p and cof1-157p, but in cof1-159p the N-terminus is ordered. These two structural elements project away from the globular body of the structure, and the differences observed among the structures likely results from differences in crystal packing contacts.
Figure 2. Effect of mutations on cofilin structure.
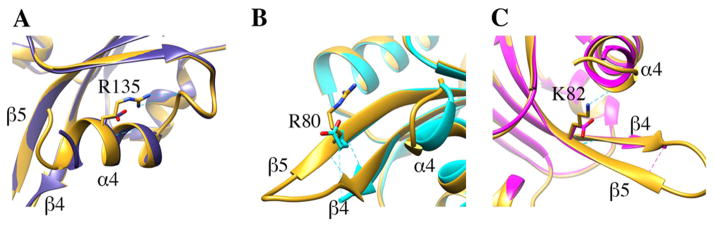
The structure of wild type cofilin (1COF; (Federov et al., 1997)) is rendered in gold and superimposed with cof1-157p in blue (A), cof1-158p in cyan (B) and cof1-159p in magenta (C). Mutations are shown in stick representation alongside corresponding wild type residues.
The structures revealed that the mutations have few effects away from the sites of the amino acid changes (Figure 2). The cof1-157p (R135D) and cof1-159p (K82D) mutations eliminate side-chain hydrogen bonding with the main-chain carbonyls of E126 and V136, respectively. However, the main-chain hydrogen bonds surrounding these mutant residues are intact and little if any deviation is observed in the backbone conformation. The cof1-158p (R80E) mutation does not affect its surrounding hydrogen-bonding network or the backbone conformation. Together, the data show that the mutations only affect residues directly involved in interactions at one of the two binding interfaces of cofilin with actin, as discussed below.
Implications for cof1-157p, cof1-158p, and cof1-159p binding to actin
We asked how the mutations affect the interaction of cofilin with the actin filament. First, we considered the structural implications of the mutations. Two high-resolution models for cofilin bound to actin filaments are available in the pdb database. Both used rabbit muscle actin and human cofilin, but our study used yeast proteins.
One model, from the Egelman lab, was derived by cryo-electron microscopy and image averaging using human cofilin-2 (Galkin et al., 2011), and the second, from the Sept lab, employed molecular dynamics and docking using human cofilin-1 (D. Sept personal communication) (Wong and Sept, 2011). An additional molecular dynamics model has been derived from the Egelman cryo-EM model (Fan et al., 2013). All the models predict that cofilin has two distinct binding interfaces that contact consecutive actin subunits within the long-pitch helix of the filament.
The Egelman model is shown in Figure 3. The human cofilin residues corresponding to our yeast mutants are shown in colored space-fill, as follows: cof1-157p (blue, R135D in yeast, is E151 in human cofilin-1 and -2); cof1-158p (cyan, R80E in yeast, is K96 in human cofilin-1 and -2); and cof1-159p (magenta, K82D in yeast, is D98 in human cofilin-1 and -2). The primary binding site (1°) contacts the actin subunit shown in cyan; this site also binds actin monomer (Rodal et al., 1999) (Lappalainen et al., 1997). The secondary binding site (2°) contacts the actin subunit shown in grey; this site is specific for the actin filament (Lappalainen et al., 1997). Our structures of cof1-157p, 158p, and 159p show that the secondary binding site is likely to be affected, but not the primary one. In the Egelman model, the mutated residue in cof1-157p (yeast R135/human E151) may form a salt bridge with actin residue E291 (Figure 3). In the Sept model, the corresponding human residue of cof1-158p (yeast R80/human K96) makes a salt bridge with actin D56. Regardless of the model-specific details, all three cofilin mutations are located at the secondary binding interface, and they have no effect on the conformation of cofilin in the crystal structures. In addition, two cofilin alanine scan mutants with altered residues at this site, cof1-16p (R80A, K82A) and cof1-22p (E134A, R135A, R138A) have defects in actin filament binding but bind actin monomer (Lappalainen et al., 1997). Therefore, we assume that the phenotypes of our three mutants result from altered interactions at the secondary binding site.
The mutations compromise filament-binding activity
In order to determine the contribution of the secondary binding interface of cofilin to filament binding, we compared mutant to wild type cofilin binding to actin filaments in a pyrene-quenching assay. Actin labeled with pyrene at cysteine 374 shows increased fluorescence on polymerization (Cooper et al., 1983), and binding of cofilin to pyrene-actin filaments quenches the fluorescence (Carlier et al., 1997) due to increased hydrophobicity of the microenvironment of the probe (Karpovich and Blanchard, 1995). We purified actin from yeast, labeled it with pyrene-iodoacetamide (Kouyama and Mihashi, 1981), and induced polymerization. We added cofilin, at a range of stoichiometries, and monitored fluorescence over time (Figure 4 and Table 3). Wild type cofilin quenched the pyrene-F-actin fluorescence within 30 s (Figure 4A), consistent with the fast binding described previously (Blanchoin and Pollard, 1999). After this decrease in fluorescence, there was no additional decrease in pyrene fluorescence suggesting that the actin filaments were not depolymerizing. Interestingly, at 1:2 cofilin:actin we observed 66% of quenching that was observed at cofilin saturation and at 2:3 cofilin:actin we observed 79% quenching (Table 3) suggestive of effects on pyrene environment beyond regions of cofilin binding.
Figure 4. Mutation of the secondary actin-binding interface in cof1-158p affects actin filament binding.
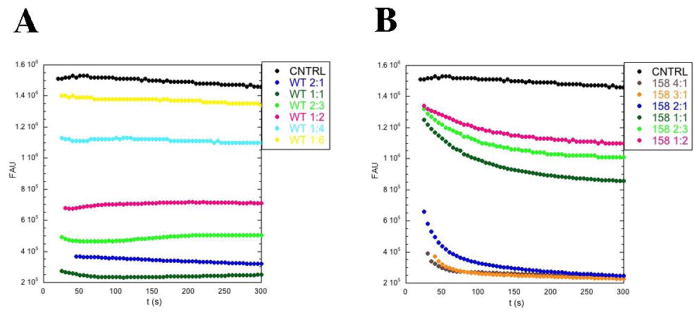
Pyrene quenching of actin filaments by wild-type cofilin (A) and cof1-158p (B) was followed at 384nm over time and plotted against fluorescence arbitrary units (FAU). At time 0, different ratios of cofilin-to-actin were added to pre-polymerized actin filaments (7.4% pyrene-iodoacetamide labeled, final concentration 2 μM). Buffer alone was added to the control reactions.
Table 3.
Cofilin-Actin Binding Data as Determined by Quenching of Pyrene Fluorescence.
| Cofilin:actin | t (s) | WT cofilin | Cof1-157p | Cof1-158p | Cof1-159p |
|---|---|---|---|---|---|
| 4:1 | 50 | ND | ND | 0.91±0.04 | ND |
| 300 | ND | ND | 0.94±0.04 | ND | |
| 3:1 | 50 | ND | ND | 0.86±0.07 | 0.92±0.06 |
| 300 | ND | ND | 0.94±0.06 | 0.94±0.06 | |
| 2:1 | 50 | 0.90±0.02 | 0.93±0.01 | 0.67±0.15 | 0.94±0.04 |
| 300 | 0.92±0.02 | 1.00±0.004 | 0.90±0.08 | 1.00±0.04 | |
| 1:1 | 50 | 0.97±0.02 | 0.87±0.02 | 0.22±0.06 | 0.44±0.04 |
| 300 | 0.96±0.02 | 0.87±0.03 | 0.41±0.05 | 0.74±0.02 | |
| 2:3 | 50 | 0.79±0.04 | 0.57±0.02 | 0.18±0.03 | 0.23±0.02 |
| 300 | 0.74±0.04 | 0.61±0.02 | 0.31±0.03 | 0.45±0.02 | |
| 1:2 | 50 | 0.66±0.03 | 0.40±0.02 | 0.12±0.07 | 0.16±0.09 |
| 300 | 0.62±0.03 | 0.43±0.02 | 0.22±0.06 | 0.33±0.07 | |
| 1:4 | 50 | 0.30±0.01 | 0.12±0.02 | 0.05±0.02 | 0.10±0.03 |
| 300 | 0.28±0.01 | 0.18±0.02 | 0.11±0.02 | 0.19±0.02 | |
| 1:6 | 50 | 0.13±0.05 | 0.04±0.03 | ND | NA |
| 300 | 0.13±0.02 | 0.07±0.04 | ND | 0.06±0.10 | |
| 1:8 | 50 | 0.13±0.04 | 0.03±0.04 | ND | ND |
| 300 | 0.13±0.02 | 0.07±0.04 | ND | ND |
Pyrene quenching of 2 μM actin filaments is expressed as a percentile fraction of the maximal quenching observed 50 and 300 seconds after cofilin addition. The values reported are the averages of three experiments.
For the mutant cofilins, we also observed rapid quenching of the pyrene-F-actin fluorescence (data for cof1-158p are shown in Figure 4B). The results for cof1-157p were very similar to those for wild-type cofilin, with slightly less quenching (Table 3), suggesting slightly compromised binding. In contrast, the results for cof1-158p and cof1-159p were very different from those for wild-type cofilin. Their concentration-dependent initial drop in quenching saturated at concentrations much higher than what was required for wild-type cofilin, indicating compromised filament binding (Figure 4B and Table 3). The results also showed a second phase with a slower drop in fluorescence consistent with filament disassembly.
Promotion of actin assembly by cofilin requires its secondary actin-binding interface
Based on observations in multiple systems, cofilin can induce actin assembly by increasing the nucleation rate (Du and Frieden, 1998) (Vartiainen et al., 2002) (Chen et al., 2004) (Yeoh et al., 2002) (Andrianantoandro and Pollard, 2006) (Kudryashov et al., 2006). In addition, the severing activity of cofilin can contribute to polymerization by increasing the number of free barbed ends of actin filaments (De La Cruz, 2009). The nucleation activity can be explained by structural models for cofilin-F-actin (Galkin et al., 2011) (Wong and Sept, 2011) (Kudryashov et al., 2006) (see Figure 3), which suggest that the two cofilin-actin interfaces position and stabilize two actin monomers in a filament-like structure. Because the three cofilin mutants displayed decreased binding to F-actin, we hypothesized that the mutations would alter filament assembly kinetics. To test this hypothesis, we assayed the activity of the mutants in actin polymerization assays.
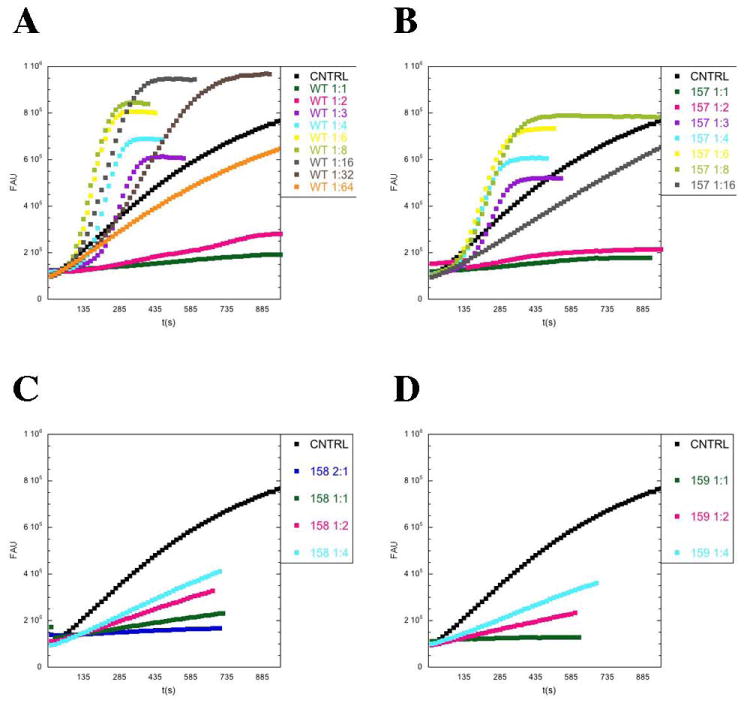
We monitored pyrene-actin assembly over time in the presence of cofilin. For wild-type cofilin, we observed that cofilin to actin ratios between 1:3 and 1:32 increased the apparent actin polymerization rate, with no effect at 1:64 (Figure 5A), similar to previous observations (Du and Frieden, 1998). At higher stoichiometries (1:1 and 1:2), pyrene-quenching masked any effects on polymerization rates. Using filament sedimentation as a complementary approach to measure filament polymerization, we confirmed that cofilin retains its polymerization properties at these high ratios (2:1, 1:1, 1:2, Supplementary Figure S1).
Figure 5. The secondary actin cofilin binding interface is required for cofilin-assisted actin polymerization/nucleation.
2±M pyrene-labeled yeast actin was polymerized in the presence of different concentrations of wild type cofilin (A), cof1-157p (B), cof1-158p (C), and cof1-159p (D). Pyrene fluorescence at 384 nm is plotted versus time.
We tested the mutant cofilins in the pyrene-actin polymerization assays. cof1-157p promoted polymerization behaving very similarly to wild-type cofilin at ratios up to 1:8 (Figure 5B). However, the activity was lost at a ratio of 1:16, compared to 1:64 for wild-type cofilin. At all ratios, cof1-157p caused less binding-induced quenching compared to wild-type (see Table 3), therefore, these data underestimate the severity of cof1-157p defects in promoting polymerization. In contrast, cof1-158p and cof1-159p did not promote polymerization and they even inhibited polymerization (Figure 5C and D) in a concentration-dependent manner. In filament sedimentation assays for polymerization, cof1-157p (Supplemental Figure S1), again resembled wild-type and did not inhibit polymerization. In contrast, cof1-158p and cof1-159p showed a concentration-dependent shift of actin to the monomer state (Supplemental Figure S1) (Clark and Amberg, 2007). Overall, while wild-type cofilin can promote actin assembly by increasing the barbed end concentration via severing and de novo nucleation, while the mutant cofilins appear to block polymerization.
Electron microscopy of actin filaments in the presence of wild type and mutant cofilins
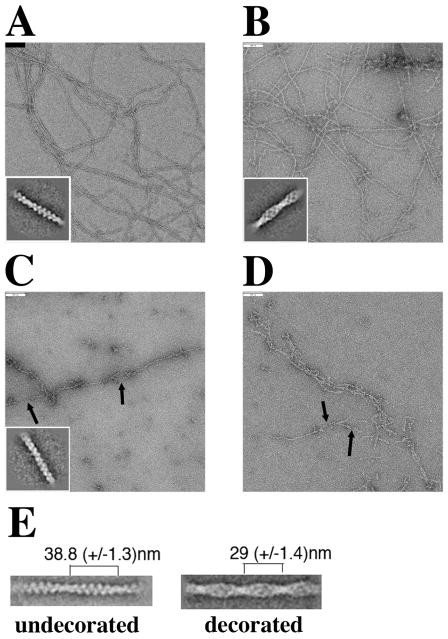
The cofilin interaction with the actin filament results in changes in filament structure that have been visualized at the macromolecular level (McGough et al., 1997) and the molecular/atomic level (Galkin et al., 2011) (Fan et al., 2013). We asked how the mutant cofilins affect the structure of the actin filament, using electron microscopy and image averaging. Figures 6A and 6B are electron micrographs and image averages for plain undecorated actin filaments for filaments decorated with wild-type yeast cofilin. In the averaged images, the cofilin bound to the filament between actin subunits, is readily resolved. We found that yeast cofilin decreases the filament crossover length by ~25%, from 38.8 nm to 29 nm (Figure 6, Panel E), similar to what was observed for human cofilin (McGough et al., 1997).
Figure 6. cof1-158p and cof1-159p affect actin filament structure leading to filament fragmentation.
Electron micrographs and image averaging (insets) of negatively stained (1% uranyl-acetate) actin filaments in the absence of cofilin (A), and presence of wild-type cofilin (B), cof1-158p (C), or cof1-159p (D). Cofilin was added to 3 ±M pre-polymerized actin shortly before it was spotted on a carbon-coated copper grid at cofilin:actin ratios of 1:1 (B and C) and 1:2 (D). Insets are of the best class sums after several rounds of multi-reference alignment/classification using the EMAN1 software package (Ludtke et al., 1999). The insets are the averages of 20 (A), 90 (B) and 77 (C) raw images. Scale bar=100 nm. Arrows indicate sites of possible filament severing. (E) Crossover lengths were measured for representative class sums of images of undecorated and wild-type cofilin-decorated filament segments. Image analysis was done using Imagic5 (van Heel et al., 1996) (Dube et al., 1993) using the same electron micrographs as in Panels A and B. The class sums have 45 (undecorated) and 43 (decorated) members.
For cof1-158p and cof1-159p, the filaments are short and their crossovers are not clearly defined (Figures 6C and 6D), in contrast to undecorated (Fig. 6A) and wild-type cofilin-decorated (Fig. 6B) filaments which long and homogeneous, with distinct crossovers. The short length of the filaments appears to be due to severing, because many short filaments are arranged in a way that suggests they were derived from a single filament that broke after adhesion to the grid (see arrows). Image averages of filaments decorated with cof1-158p (1:1) revealed a less well-resolved filamentous structure compared to undecorated or wild-type cofilin-decorated filaments, with no apparent twist (inset Figure 6C). Also the structure of the filaments appears more heterogeneous in the presence of cof1-158p, which may have complicated the image averaging obscuring effects on filament twist. These results suggest that cofilin mutants with a compromised secondary binding site are able to interact with actin filaments but that they destabilize the filaments, in contrast to wild-type cofilin.
It has been suggested that cofilin binding induces conformational changes that propagate to neighboring filament segments (Bobkov et al., 2006; Ressad et al., 1998) (Prochniewicz et al., 2005) (Galkin et al., 2010). Therefore, we would expect that image analysis on partially decorated filaments should resolve two classes that meet these criteria: twisted filaments with cofilin bound, and twisted filaments with cofilin not bound. By using a 1:2 stoichiometry of cofilin to actin, unbiased image averaging on the semi-automated EMAN1 software indeed revealed these two classes, in images from a single grid (Figure 7, Panels B and C). Panel B is a class average showing a filament that appears to have no cofilin bound (note the empty space between actin subunits) but has the same cross-over length as a class that appears to have cofilin bound (Panel C; note there is no space between actin subunits). For comparison, Panel A shows a class average of undecorated filaments from a sample lacking cofilin (same as the inset in Figure 6A) and Panel D shows a class average from fully decorated actin filaments (same as the inset in Figure 6B).
Figure 7. Yeast cofilin induces a conformational change in neighboring, undecorated filament segments.
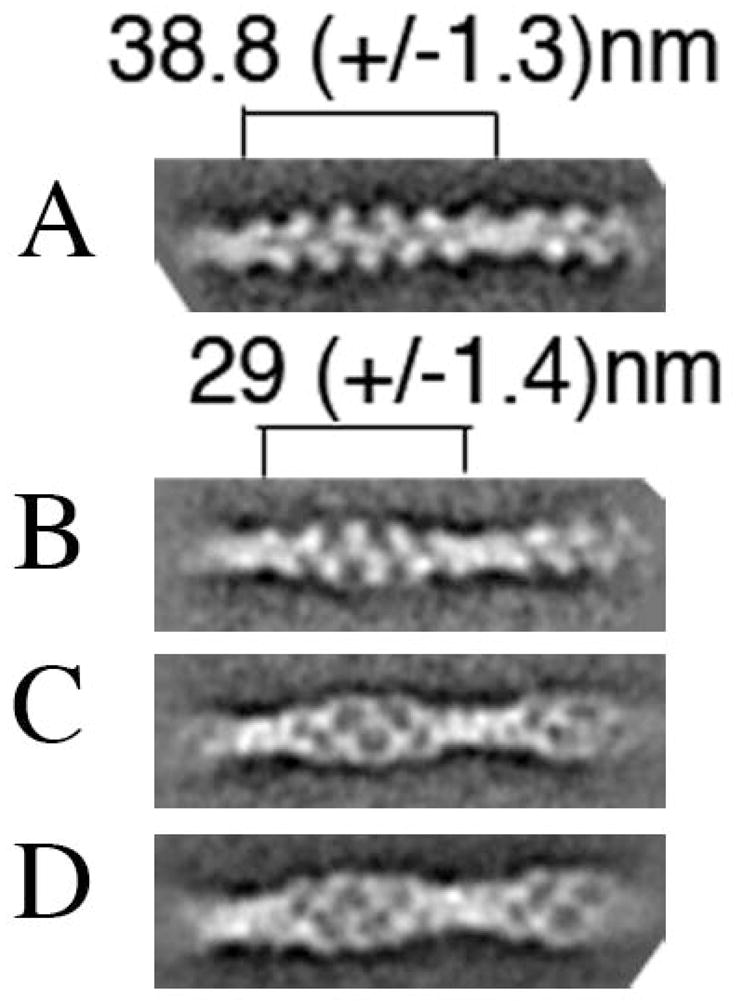
Representative class sums of incomplete wild-type cofilin-decorated filament segments after several rounds of multi-reference alignment/classification using the EMAN1 software package (Ludtke et al., 1999); averages of 241 (B) and 208 (C) raw images are shown. These class sums originated from the same set of micrographs and presumably represent cofilin-decorated (C) and undecorated (B) segments that both have the shorter crossover length. Class sums for undecorated (A) and wild type cofilin decorated (D) filament segments are shown for comparison (same as the insets Figure 6A and B).
Others have reported that this high-twist conformation is a minor one that exists naturally in undecorated actin filaments (Galkin et al., 2010). However, when we averaged undecorated filaments, using the unbiased automated software, a class that met these criteria was not prominent. Perhaps our analysis would have identified this conformer in undecorated filaments if the number of images were much larger. Regardless, our data with partially decorated filaments support the model in which cofilin stabilizes this conformer of F-actin (Galkin et al., 2011) and can propagate this less stable structure into undecorated regions of the filament.
Direct observation of severing by the cofilin mutants
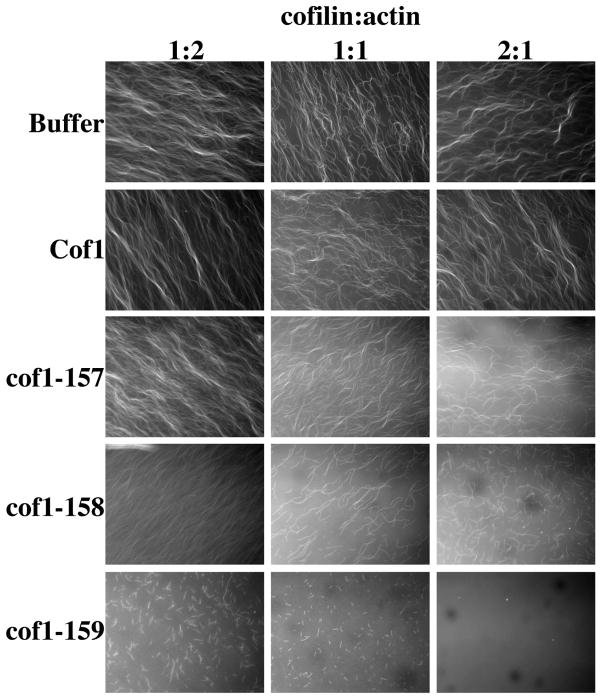
Our results suggest that the mutant cofilins, especially cof1-158p and cof1-159p, may be hyperactive for filament severing. To address this question further, we observed Oregon green-labeled actin filaments directly after they had been exposed to wild-type or the mutant cofilins. First we visualized filaments (1 μM) after they had been briefly (2 min) incubated with different ratios (2:1, 1:1, and 1:2) of wild-type versus mutant cofilin (Figure 8). Filaments incubated in the presence of wild-type cofilin showed no appreciable effects on filament density or length compared to actin filaments alone. Filaments incubated with cof1-157p at lower stoichiometries resembled those incubated with wild-type cofilin, but at a higher stoichiometry (2:1) the filaments appeared shorter and less numerous. cof1-158p and especially cof1-159p caused more dramatic effects with filaments even shorter and less numerous.
Figure 8. Microscopic visualization of filament disassembly by cof1-157p, cof1-158p, and cof1-159p.
1μM Oregon green-labeled yeast actin filaments were visualized by fluorescence microscopy after either the addition of control buffer, or after the addition of wild type cofilin, cof1-157p, cof1-158p, or cof1-159p at the indicated ratios of cofilin to actin.
In addition, we observed filaments during the time of exposure to the cofilins using epifluorescence and TIRF microscopy. For epifluorescence, 2 ±M cofilin was added to a solution of fluorescent, Oregon-green labeled F-actin, and images were collected at 200 ms intervals. With wild-type cofilin (Supplemetal Movie S1) and cof1-157p (Supplemental Movie S2), we observed very few severing events. With cof1-158p (Supplemental Movie S3) and cof1-159p (Supplemental Movie S4) many more severing events were observed, and the filament density decreased substantially over time. This was particularly pronounced for the cof1-159p mutant.
In addition, filament shortening, that appeared to occur at filament ends, was increased in the presence of cof1-158p and cof1-159p, and this contributed to the decrease in filament length and number. Figure 9A shows three consecutive frames from each of these movies. To assist the reader, we marked severing events with arrows; black before and white after the severing event. These regions are enlarged in Figure 9B. Although each sample started with the same concentration of polymerized actin, the filament density in the mutant cofilin samples was decreased prior to starting the recordings. Many more severing events can be identified in the samples with the mutant cofilins, in particular cof1-159p, particularly if one considers the density of filaments in the field of view.
Figure 9. Microscopic visualization of filament severing by cof1-157p, cof1-158p, and cof1-159p.
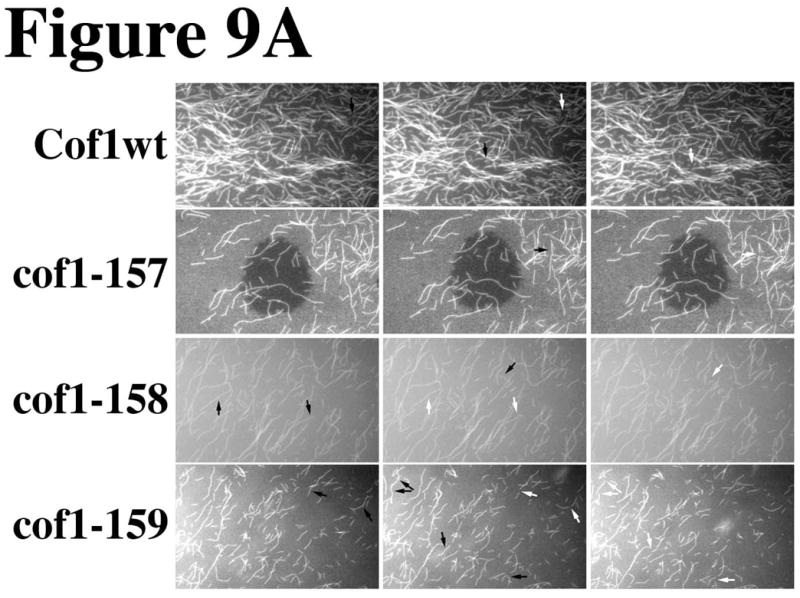
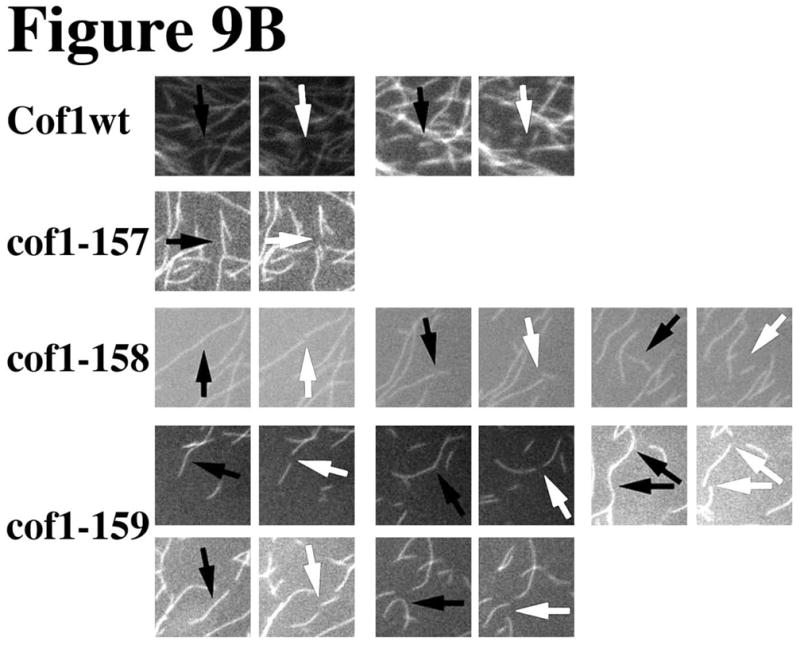
2 μM Oregon green-labeled actin filaments were placed under a coverslip and visualized by time-lapse (1 frame per 200 ms) fluorescence microscopy following the diffusion of 2μM cofilin under the coverslip. Three consecutive frames are displayed from the movies shown in the supplemental data (movies S1–S4). Arrows point at filaments before and after (white) a severing event (A). Panel B shows enlarged regions from Panel A where severing events were captured; severing sites are marked before severing with black arrows and after severing with white arrows.
For the TIRF imaging experiments, we compared the effects of wild-type cofilin and cof1-159p, our most hyperactive mutant, on actin filaments attached with myosin to glass cover-slips. Images were captured every 2.6 s and the beam was set at an angle to allow for observation of a wider volume of the specimen. We did this to allow for the visualization of filament segments not conformationally constrained by attachment to the glass coverslip, which has been shown to affect filament severing (Pavlov et al., 2007). Addition of wild type cofilin to the flow chamber did not affect filament length or density; the field was essentially unchanged during the course of the experiment (Supplemental Video S5). In contrast, in the presence of cof1-159p, the filaments were rapidly severed resulting in a decrease in the density of the filaments and the length of filament segments (Supplemental Video S6). These results were similar to those observed by epifluorescence. In conclusion, these experiments show that defects at the secondary binding site can lead to unregulated severing and filament disassembly coupled with inhibition of re-polymerization.
Actin Patch Dynamics in cof1-157 and cof1-159 mutants
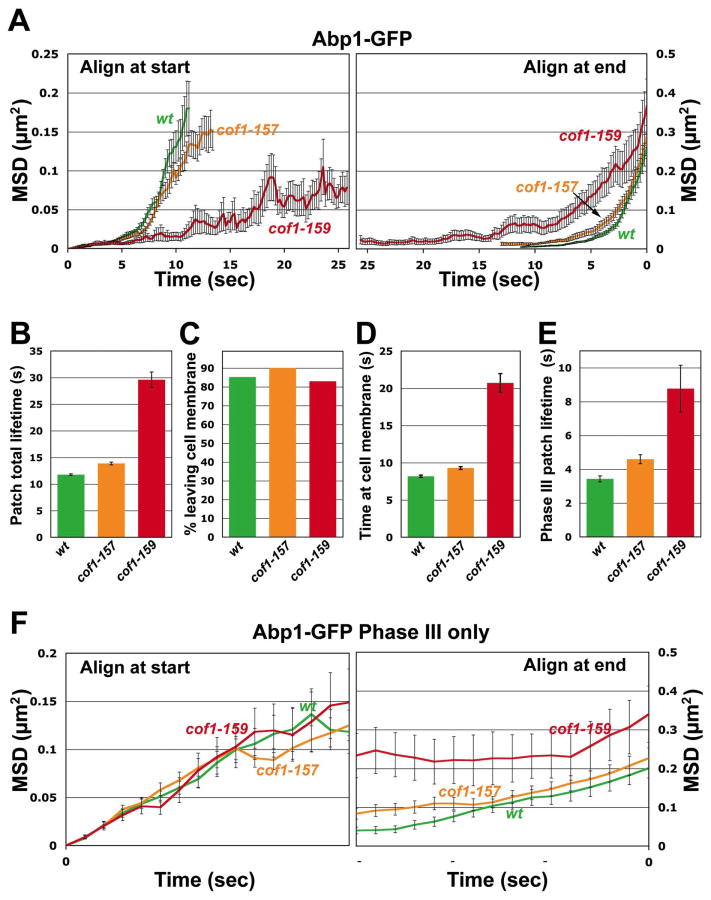
Having characterized the biochemical defects of the cofilin mutants, we next asked how these mutations affect the activity of cofilin in vivo by analyzing the dynamic assembly and movement of cortical actin patches. Using Abp1-GFP (a marker for F-actin in cortical patches), fluorescence microscopy, and particle tracking software, we analyzed actin patch dynamics in cof1-157 and cof1-159 mutant strains (Lin et al., 2010). cof1-157 cells grow as well as wild type with a relatively normal actin cytoskeleton while cof1-159 cells grow poorly, have excessive and large cortical patches and cell morphology defects (Clark and Amberg, 2007). We were unable to analyze a cof1-158 strain as this allele does not support viability. Figure 10, Panel A shows plots of mean squared displacement versus time for patches aligned either at the start or end. Patches in the cof1-159 strain showed much slower rates of movement than patches from wild type strains while patches in the cof1-157 strain were only slightly slower than in wild type. In addition, the cof1-159 patches had much longer lifetimes (Panel B), but ultimately these patches did internalize at a percentage equivalent to wild-type (Panel C). The extended lifetime of cof1-159 patches could be attributed to a slower Phase I and II (Panel D) and a slower Phase III (Panel E). The cof1-157 mutant had slightly increased patch lifetimes from wild-type.
Figure 10. Quantitative motion analysis of Abp1-GFP-labeled actin patches in wild-type, cof1-157 and cof1-159 strains.
Mean-squared displacement (MSD) plots are shown for patches aligned at the start (left) or end (right) of their lifetimes (A). For patches aligned at the start (left), the curves are truncated at the median lifetime. Average total lifetime of patches, defined as the time from the appearance of a patch until its disappearance (B). Percentage of patches that leave the membrane, defined as traveling >200 nm from their point of origin (C). Average time spent by patches within 200 nm from their point of origin (Phases I and II; D). Average lifetime of patches after they travel 200 nm from their point of origin until the time they disappear (Phase III; E). MSD plot of Phase III patch movement (F). For each patch, only data from patches that travelled more than 200 nm from the point of origin were included. Means ± s.e. (standard error of the mean) of three segregants are shown. Student’s t-tests for statistical significance were performed for B, D and E as indicated. The yeast strains used are described in Table 1. The numbers of patches analyzed were 196 for COF1wt, 212 for cof1-157, and 47 for cof1-159.
Discussion
Structural models have suggested a two-binding site mechanism for the interaction of cofilin with actin filaments (Galkin et al., 2011) (Wong and Sept, 2011). In this study we focused on three cofilin mutants that are predicted, by these models and X-ray crystallography, to specifically affect only the secondary, filament-specific binding site. The three charge-reversal mutants used in this study (cof1-157p, R135D; -158, R80E; and -159, K82D) were first described as hyper-active for filament disassembly, and they were initially designed to disrupt the interaction of cofilin with its ubiquitous activator Aip1p as discussed below (Clark and Amberg, 2007). Studies on an alanine-scan mutant, cof1-16p (R80A, K82A), which also affects this site, found that the mutant did not bind actin filaments but retained actin monomer binding, due to the primary binding site being intact (Lappalainen et al., 1997). Here, we found that the single charge-reversal mutants interact with and destabilize actin filaments. These mutants differ from wild-type cofilin and cof1-16p in that they display intermediate filament-binding activity, increased filament-severing activity, and inhibition of actin assembly. Importantly, we solved the crystal structures for all three charge-reversal mutants and these structures confirm that these mutations do not cause allosteric changes to cofilin and thus they surgically affect the mutated residues at the secondary binding site.
We chose S. cerevisiae as the model system for this study, because this allowed us to use the same organism for in vivo experiments and biochemical analyses of purified proteins. Actin was purified from yeast, and cofilin was purified from bacterial expression of yeast genes. S. cerevisiae is particularly well-suited for studying cofilin for several reasons. For both actin and cofilin, the yeast genome has only a single gene. Both genes, for actin and cofilin, are essential for viability. Neither gene displays developmental or tissue-specific regulation. For these reasons, the interactions of yeast cofilin with yeast actin should prove a sound representation of the fundamental features of how the ADF/cofilin family interacts with actin in other systems.
In order to characterize the binding of our mutant cofilins to actin filaments, a pyrene-quenching assay was employed (Dedova et al., 2002) (Figure 4). These results revealed that the mutants bind to F-actin with reduced affinity, and, in the case of cof1-158p and cof1-159p, binding is followed by actin filament disassembly, which was confirmed by pelleting assays (see Supplemental Figure S1). An unlimited, nearest-neighbor cooperativity model for ligand binding to a one-dimensional lattice (McGhee and von Hippel, 1974) (Pollard, 2010) has previously been applied to pyrene-quenching data to extrapolate binding affinity and the cooperativity factor for vertebrate cofilin binding to vertebrate F-actin (De La Cruz, 2005). However, yeast cofilin has been reported to bind filaments very strongly (Kd<50 nM) (Bobkov et al., 2002), which may limit the ability of this analysis to provide a value for cooperativity (Elam et al., 2013). In our results, wild-type cofilin occupancy, based on quenching of pyrene-actin fluorescence, appeared to be greater than the concentration of cofilin at the highest sub-saturating stoichiometries of cofilin to actin (Table 3). Given that the cofilin-induced pyrene quenching reflects cofilin-induced conformational changes in the filament that alter the environment of the probe, we attribute the greater-than-expected quenching to conformational changes in the actin filament that extend beyond regions of cofilin decoration, as suggested previously (Bobkov et al., 2006; Prochniewicz et al., 2005; Ressad et al., 1998) (Galkin et al., 2010).
To further characterize cofilin binding to F-actin we used electron microscopy with image averaging. Wild-type yeast cofilin induced a change in actin filament twist with a decrease in cross-over length similar to that reported previously (Figure 6) (McGough et al., 1997) (Galkin et al., 2002). We attempted to visualize the shorter cross-over length, resulting from propagation of this conformation beyond regions of cofilin decoration of under-decorated actin filaments, as was suggested by the pyrene quenching data. To maximize the proportion of undecorated filament regions with this conformation, we used a 1:2 ratio of cofilin to actin (Suarez et al., 2011) (Pavlov et al., 2007) (Andrianantoandro and Pollard, 2006) (Bobkov et al., 2006) (Prochniewicz et al., 2005). At this ratio, we observed quenching in 32% excess over the expected (Table 3). Indeed, we observed a class of averaged images that strongly suggests the existence of actin filament segments that lack cofilin but have the short crossover length (Figure 7B). This conformer also appears to exist in pure F-actin preparations, based on helical reconstruction studies (Egelman, 2000) (Galkin et al., 2010). This decrease in the filament crossover distance promoted by cofilin binding has been attributed to a tilted conformation of the actin subunit in the filament (Galkin et al., 2011), which differs from the flat subunit-conformation of subunits in undecorated filaments by the relative orientation of sub-domain 2 (SD2) (Galkin et al., 2011). Propagation of the tilted subunit conformation to nearest-neighbor sites has been proposed to explain the cooperative binding of cofilin (De La Cruz, 2009) (De La Cruz and Sept, 2010) and it has been proposed that this conformation can be propagated to subunits that are some distance from the interaction site (Bobkov et al., 2006) (Prochniewicz et al., 2005).
One limitation on the structural interpretation of our electron microscopy results for the effects of cofilin mutants on actin filament structure, was a high level of image heterogeneity for the two most destabilizing mutants, cof1-158p and cof1-159p (see inset in Figure 6E). The results did confirm the existence of binding as well as reveal effects on filament structure. In addition, the filaments were scarce and fragmented, suggesting increased depolymerization and severing for these mutants, compared to wild-type cofilin (Figure 6C and D).
We conclude that all three cofilin mutants have defects in promotion of polymerization, based on two complementary polymerization assays – filament sedimentation and pyrene-actin fluorescence. cof1-158p and cof1-159p, in particular, displayed no polymerization promotion activity; in contrast, they inhibited polymerization in a concentration-dependent manner, compared to wild-type cofilin. Our conclusions are supported by a three-dimensional reconstruction of a cofilin-decorated actin filament which suggested that the primary and secondary binding interfaces both participate in nucleation and filament binding (Kudryashov et al., 2006). This work implied that cofilin nucleates filaments in a tilted conformation and stabilizes pre-existing actin filaments in the tilted conformation. We needed to complement our pyrene-based polymerization assays with filament pelleting assays because at the highest cofilin to actin ratios, cofilin quenches pyrene fluorescence (Figure S1). In the pelleting assays, wild-type cofilin and cof1-157p promoted actin polymerization but even at the highest concentrations, cof1-158p and cof1-159p did not.
While cofilin is considered primarily a severing protein, its nucleation activity is likely important, considering the properties of other nucleators. We calculated the nucleation efficiency of wild-type cofilin, using yeast cofilin with yeast actin as 0.17%. On one hand, this value is low when compared to those of other nucleators: 3% for the FH1/FH2 domain of the formin Bni1p (Pring et al., 2003) and 100% for human Arp2/3 (Higgs et al., 1999). However, in the yeast cell, cofilin is highly abundant, with 20,000 molecules per cell compared to lower values for Bni1p (166) and Arp2/3 complex (6,650) (Ghaemmaghami et al., 2003) (Rodal et al., 1999). One can estimate the in vivo nucleation capacity of each nucleator as the product of its nucleation efficiency multiplied by the number of molecules in the cell. For the formin Bni1p, nucleation efficiency per molecule is about 18 times that of cofilin (3%), but its nucleation capacity in the cell corresponds to 1/7th that of cofilin.
In order to understand the mechanism of the filament destabilizing activities of cof1-158p and cof1-159p, we imaged single actin filaments in real time immediately after and during incubation with cofilin. For wild-type cofilin, we found that the actin filaments were long and numerous, prior to and immediately after the addition of wild-type cofilin. In contrast, for the mutant cofilins, the actin filaments were short and sparse, indicative of filament severing (Figure 8). Using real-time movies of actin filaments, we confirmed that cof1-158p and cof1-159p have high severing activity, as compared to wild-type cofilin (Figure 9A and B and Supplemental Movies S1–S6). For cof1-159p in particular, high concentrations caused fast filament severing and depolymerization. The severing activity may be sufficient to account for the filament depolymerization, although we cannot exclude contributions from conformation-induced filament destabilization, barbed end capping, and/or monomer sequestration.
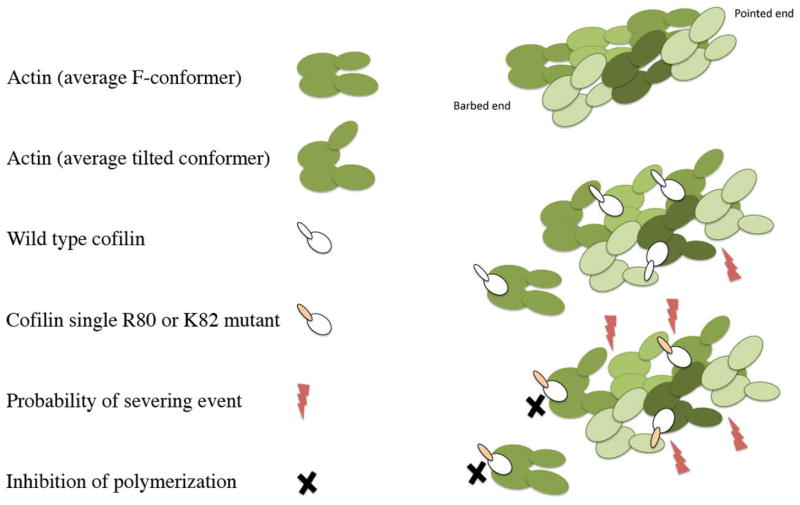
To develop a molecular model for the activities of cof1-158p and cof1-159p, we considered a proposed model for the kinetics of cofilin binding (Figure 11) in which the binding of cofilin to the filament causes a conformational isomerization of the bound actin subunit that is propagated to nearest neighbor sites (De La Cruz and Sept, 2010). This conformational isomerization in the model is analogous to and/or contributes to the formation of the tilted actin conformation seen in the structural analyses. At high concentrations of wild-type cofilin in the model, the binding of more cofilin to the nearest neighbor sites is rapid and stabilizes the filament, preventing severing. In contrast, at low cofilin:actin stoichiometries the destabilization of actin-actin contacts at boundary regions leads to severing (middle panel Figure 11). We hypothesize that the cofilin mutants studied here, due to reduced binding affinity at the secondary binding site, bind more slowly at nearest neighbor sites such that there is a loss of kinetic control leading to uncontrolled severing at all concentrations and in particular at high concentrations (see bottom panel Figure 11). If one presumes that conformation isomerization is the cause of severing, then the mutants must be capable of causing the isomerization step. In addition, since the mutants cannot stabilize new subunits in the tilted conformation at the barbed end, they inhibit nucleation and polymerization by monomer sequestration and barbed-end capping (see black X in bottom panel, Figure 11).
Figure 11. Model for filament destabilization by cof1-158p and cof1-159p.
Actin subunits are rendered in green and are shown in either the average, filament-specific conformer found in naked actin filaments or in the cofilin-stabilized tilted conformer found in cofilin-decorated filaments. Wild-type cofilin is rendered in white as are the cofilin mutants with the mutated secondary binding site shaded in pink. Severing events are indicated with lightening bolts and sites where the cofilin mutants block polymerization are marked with black X’s. The filament in the upper left has no cofilin bound and the actin subunits are shown in the normal, average F-actin conformer. The filament in the middle right is decorated with wild-type cofilin, shows that cofilin has stabilized the actin subunits in the tilted conformer, and indicates that severing is most probable at boundaries between regions decorated with cofilin and regions lacking cofilin. The model on the lower right shows a filament interacting with either cof1-158p or cof1-159p. The mutants are shown to induce the tilted conformer but are unable to compensate for destabilization of actin-actin contacts through the secondary binding site resulting very high probabilities of severing at nearest neighbor sites. In addition, the mutants cannot support subunit addition at the barbed end and sequester actin subunits in the monomer pool resulting in net filament disassembly.
In vivo, cofilin function has mainly been studied in the endocytosis-associated actin patch, with a focus on its roles in actin de-polymerization and subunit recycling. Our two viable charge reversal mutants, cof1-157 and cof1-159 displayed decreased actin patch motility and prolonged patch lifetime (Figure 10), which reflects delays in all three phases of endocytosis (Figure 10), as has been previously observed for other cofilin mutants (Lin et al., 2010). The amount of cofilin in the patch peaks in Phase III of endocytosis, the phase at which actin begins to disassemble (Okreglak and Drubin, 2007), however, considering its abundance, we assume that cofilin is always present in low concentrations as part of the cytoplasmic pool. An Arp2/3 independent actin nucleation pathway has been suggested to exist (Mooren et al., 2012), because Arp2/3 mutations that affect actin nucleation in vitro do not affect F-actin accumulation in vivo (Martin et al., 2005). Short filaments produced during patch disassembly, as a result of cofilin severing, may serve as the mother filaments in new patches (Chen and Pollard, 2013). Our results leave open the possibility that cofilin-induced actin nucleation contributes to the creation of mother-filament substrates for Arp2/3. In early stages of endocytosis (Phase I and II) the amount of cofilin in the patch is low, relative to actin, which may contribute to filament length control via clustered binding and severing of destabilized/undecorated regions (De La Cruz, 2009). In Phase III, the amount of cofilin increases to a level potentially sufficient to fully decorate filament segments, which may then become substrates for Aip1p-induced disassembly in a process that is controlled by the levels of Aip1p (Rodal et al., 1999). We suggest that the early defects observed in our mutants are attributable to decreased actin nucleation, in combination with premature and unregulated actin severing. Later, as the mutant cofilin accumulates in the patch, severing is not controlled by Aip1p levels and can occur in both ATP- and ADP-actin filament segments, thus prolonging patch lifetimes in Phase III.
Lastly, the cofilin mutants studied here suggest an elegant mechanism for the regulation of cofilin activities. The cof1-158p and cof1-159p mutants behave much like wild type cofilin in the presence of Aip1p in that they sever and block re-polymerization of the severed filaments (Rodal et al., 1999) (Ono et al., 2004) (Balcer et al., 2003) (Okada et al., 2006). Interestingly, our mutants were initially constructed to disrupt the predicted cofilin-Aip1p interface (Clark et al., 2006) (Clark and Amberg, 2007), suggesting that Aip1p may compete with actin for the secondary binding interface on cofilin. We hypothesize that Aip1p induces cofilin-dependent severing through competing for and disrupting the actin-cofilin secondary binding site, resulting in the loss of cofilin-mediated stabilization of tilted conformers. Other factors that enhance cofilin-mediated disassembly, such as Srv2p (Chaudhry et al., 2013), may employ a similar mechanism targeting the secondary actin-cofilin interface. In this manner, the filament-promoting versus filament-destabilizing activities of cofilin could be spatiotemporally regulated in cellular actin networks.
Materials and Methods
Crystallization and Structure Determination
Mutant cofilins were crystallized by mixing 1 μl purified protein at 10–20 mg/ml with 1 μl of crystallization buffer (100 mM Tris-HCl [pH 7.2 or 7.5], 24–35% PEG 3350, 100–250 mM NaCl, 1mM BME) using the hanging drop vapor diffusion method. Crystals were harvested with nylon loops, cryo-protected in crystallization buffer containing 30% glycerol, and flash cooled in liquid nitrogen. Datasets were collected from single crystals in a single pass for cof1-157p and cof1-158p. The cof1-159p dataset was collected in two passes to optimize the signal to noise of comparatively higher resolution diffraction. All datasets were processed using HKL2000 (Otwinowski and Minor, 1997). Phases were determined from molecular replacement in PHENIX (Adams et al., 2010) using 1COF (Federov et al., 1997) as a search model. Preliminary models were modified and refined iteratively using COOT (Emsley et al., 2010) and PHENIX (Adams et al., 2010). Validation was carried out using MolProbity (Chen et al., 2010). All images included in the text were prepared with UCSF Chimera (Pettersen et al., 2004).
Yeast actin purification
Yeast actin was purified by a modified DNaseI affinity purification procedure (Goode, 2002). Briefly, 100 mg DNaseI (Roche Diagnostics, Indianapolis, IN) was coupled to 3 g of swelled Sepharose 4B (Sigma-Aldrich, St. Louis, MO). The coupled beads were packed into two disposable polypropylene columns of 5 ml maximum capacity (PIERCE/ThermoScientific, Rockford, IL). Each column was washed with 25 ml 0.2 M NH4Cl in G-buffer (10 mM Tris, pH 7.5, 0.2 mM CaCl2, 0.5 mM ATP and 0.2 mM DTT) followed by 25 ml G-buffer with 0.1 mM phenylmethylsulfonyl fluoride (PMSF). ~100 g of yeast pellet (Red Star Yeast from a ~400 g brick) were thawed in ~100 ml G-buffer with 0.1 mM PMSF plus Calbiochem protease inhibitor cocktail IV (EMD Millipore Corp., Billerica, MA) diluted 1:500. The cells were passed 8 times through a micro-fluidizer (Microfluidics, Model 110 L, Newton, MA). The lysate was clarified in a Beckman JA-20 rotor at 12,000 rpm for 30 min at 4°C and then in a Beckman Ti70 rotor at 50,000 rpm for 50 min at 4°C, followed by filtration through common coffee filters. The filtered supernatant was loaded equally onto the two ~5 ml DNaseI-Sepharose affinity columns that had been pre-washed with 5 column volumes of 0.2 M NH4Cl in G-buffer and 5 column volumes of G-buffer plus 0.1mM PMSF. The columns were washed with 5 column volumes of 10% de-ionized formamide in G-buffer plus 0.1mM PMSF, 5 column volumes of 0.2 M NH4Cl in G-buffer, and 5 column volumes of G-buffer. The actin was eluted with 50% deionized formamide in G-buffer and dialyzed overnight in dialysis tubing (diameter 11.5 mm) with a molecular weight cut-off of 3,500 Da (Spectrum Laboratories, Rancho Dominiguez, CA) against 2 liters of G-buffer (25 μM ATP). After dialysis the protein was concentrated to 4–5 mls in a 3,500 Da molecular weight cut-off concentrator (Sartorius, Bohemia NY) and subjected to ultracentrifugation in a Beckman TLA100.2 rotor at 70,000 rpm for 30 min, at 4°C. The supernatant was polymerized for 20 min at room temperature by addition of 20X concentrated F-buffer (Final: 10 mM Tris, pH7.5, 25 mM KCl, 4 mM MgCl2, 1 mM EGTA, 1 mM ATP). Subsequently, 3 M KCl was added to a final concentration of 0.6 M and the incubation continued for 30 more min. The filaments were pelleted (TLA100.2, 70 krpm, 30 min., 20°C) and then washed twice with 1XG-buffer, re-suspended in the same buffer and incubated for at least 2 h on ice with frequent mixing by pipeting. The protein was subjected to a third ultracentrifugation and one more round of polymerization/depolymerization. On the second polymerization round, the KCl concentration was kept at 25 mM. Ultracentrifugation conditions were as above, with monomer actin spins performed at 4°C and filaments at 20°C. Purity was assessed by SDS-PAGE followed by Sypro-Ruby staining. Protein concentration was determined by Biorad protein determination assay. Actin was stored at 4°C as filaments.
Yeast actin labeling
Freshly depolymerized actin in modified G-buffer (10 mM HEPES, pH 8.0, 0.2 mM CaCl2, 0.5 mM ATP) was polymerized with the addition of 20X F-buffer (buffer as described previously with HEPES, pH 8.0 instead of Tris, pH 7.5) and incubated at room temperature for 30 min. 25 mM pyrene-iodoacetamide or Oregon green iodoacetimide (Invitrogen, Grand Island NY) in DMSO was added to a final concentration of 25 ±M and the reaction was incubated at 17°C in the dark overnight. After ultracentrifugation, the filaments were washed twice in G-buffer (DTT included, 10 mM Tris, pH 7.5 for pyrene labeling and 10 mM imidazole, pH 7.5 for Oregon green labeling), re-suspended in the same buffer and incubated on ice in the dark for 1 h with frequent mixing by pipetting. The pyrene-labeled actin was subjected to one more round of polymerization/depolymerization before use. Protein concentration was determined by a Biorad protein determination assay using BSA as a standard and labeling efficiency was determined by A344 using an extinction coefficient of 22,000 M−1 cm−1 (Kouyama and Mihashi, 1981) for pyrene-actin and A491 using an extinction coefficient of 77,800 M−1 cm−1 for Oregon green-actin (Kuhn and Pollard, 2005). The labeling efficiency was 5–15% for pyrene-actin and 80% for Oregon green-actin with less than 10% protein loss. Pyrene-actin was stored as F-actin at 4°C and used within 4 days of preparation. Oregon green-actin was stored in G-buffer for up to two days on ice.
Cofilin purification
Yeast cofilins (wild-type and mutant) were expressed in E. coli DH5a cells as glutathione-S-transferase (Ausubel et al., 1989) fusion proteins under the control of the Plac promoter in plasmid pGEX-2T and purified as previously described (Clark and Amberg, 2007). Briefly, cells were grown to an optical density of 0.7–0.8 in 1 liter LB supplemented with 0.1 mg/ml ampicillin and 0.2% glucose. Cofilin expression was induced with 0.5 mM IPTG for 4–5 h at 35°C. The harvested pellets were re-suspended in GST-binding buffer (4.3 mM Na2HPO4, pH 7.3, 1.5 mM KH2PO4, 140 mM NaCl, 3 mM KCl), frozen and thawed once. DNaseI and lysozyme were added to 0.05 mg/ml and 1 mg/ml respectively and the cells were incubated on ice for 30 min with frequent tube inversion. The cells were lysed by sonication and the lysate was spun in a JA-20 rotor at 12,000xg for 30 min at 4°C. The clarified supernatant was syringe filtered (0.45±M, Corning, Tewksbury MA), diluted to 50 ml total volume with GST-binding buffer and passed over an equilibrated 5 ml glutathione-agarose column (Novagen, Philadelphia PA). After washing with 4X25 ml GST-binding buffer, 20 units of thrombin (GE Healthcare, Milwaukee WI) in 6 ml GST-binding buffer supplemented with 1.75 mM CaCl2 were added to the beads, the column was capped and sealed with parafilm and the slurry was incubated at 4°C overnight on a rocker. The protein was eluted and the column was washed with 3X5 ml GST-binding buffer. The washes were combined with the eluate, concentrated to 1 ml and further purified by FPLC (GE Healthcare, Pittsburgh PA) on a superdex 75 16X50 gel filtration column. The protein was eluted in 25 mM Tris, pH 7.4, 0.5 mM EDTA, 0.1% 2-mercaptoethanol. Protein concentration was determined by Biorad protein determination assays.
Fluorimetric actin assays
For the binding assays, yeast pyrene-actin in F-buffer was mixed with cofilin or control buffer (10% volume; time zero) and placed in a 125 ±l cuvette to final concentrations of 2 ±M actin and 0.25–8 ±M cofilin. For the polymerization assays, yeast pyrene-actin in G-buffer, cofilin and 20XF-buffer were mixed (time zero) and placed in a 125 ±l cuvette to final concentrations of 2 ±M actin, 30 nM- 2 ±M cofilin and 1XF-buffer. In both cases, the pyrene fluorescence (excitation 365 nm, emission 384 nm) was monitored in a Fluoromax4 fluorimeter at 20°C beginning ~25 secs after cofilin addition.
Calculation of cofilin nucleation efficiency
The nucleation efficiency of cofilin was expressed as the percentage of actin filaments nucleated by cofilin per cofilin molecule. The critical concentration (Cc) of actin was determined by measuring the pyrene fluorescence of 0, 0.1, 0.15, 0.2, 0.25, 0.3, 0.35 and 0.4 ±M actin in G- and F-buffer at equilibrium. The fluorescence signal was plotted against the actin concentration in the two conditions. The maximum concentration at which the fluorescence signal in F-buffer is the same as in G-buffer is the Cc, and was always in the vicinity of 0.2 ±M, and the corresponding fluorescence value (FCc) is the fluorescence of G-actin that remains unpolymerized; the Cc was determined independently for each experiment. The fluorescence signals of monomer (FG) and polymer (FF) actin at our working concentration (2 ±M) were also determined by measuring fluorescence at equilibrium in F- versus G-buffer. The fluorescence attributed solely to polymerization of actin per unit of actin was calculated as FP=(FF−FG−FCc)/([A]−Cc), where [A] is 2μM. The rate of actin polymerization was calculated by dividing the slope of the linear part of the pyrene assembly assays (fluorescence units/sec) by FP expressed as μM/sec. In order to find the concentration of actin filaments, we assumed that cofilin does not affect the actin elongation rate (Andrianantoandro and Pollard, 2006). The concentration of actin filaments (or growing ends) was calculated as the rate of actin polymerization divided by the rate of actin subunit addition per filament, using the association constant at steady state (11.6 X 106 s−1 M−1) for barbed end native actin polymerization (Pollard, 1986). The nucleation efficiency of cofilin was calculated by subtracting the number of filaments nucleated in the absence of cofilin from the number nucleated in the presence of cofilin divided by the number of cofilin molecules present. These calculations assume that cofilin does not affect the actin critical concentration.
F- Actin Pelleting assays
Yeast actin in G-buffer was mixed with cofilin and 20XF-buffer to final concentrations of 2 ±M actin, 0.5–4 ±M cofilin and 1XF-buffer in a mini ultra centrifuge tube (200 μl capacity) (Beckman, Brea CA) in a final volume of 125 ±l. A 25 ±l sample was taken immediately after mixing (total). The reaction was incubated at room temperature for 30 min and then centrifuged in a Beckman TL-100 ultracentrifuge (70,000 rpm) after which the supernatants and pellets were sampled. Pellet fractions were normalized by volume to the total and supernatant fractions and all fractions were analyzed by SDS-PAGE followed by Ruby staining.
Electron Microscopy and Image Analysis
5 ±l of 3 ±M pre-polymerized actin with or without cofilin in F-buffer was applied to freshly glow-discharged carbon-coated copper grids, stained with 1% uranyl acetate for ~1 min and air dried. The grids were examined in a JEM-2100 transmission electron microscope (JEOL) operating at 200 kV. Images were recorded on a 4096 X 4096 pixels CCD camera (TVIPS F415-MP) in low dose mode at an electron optical magnification of 40,000X and at an underfocus of −1.5 ±m, placing the 1st zero of the contrast transfer function at around 20Å−1. Micrographs were displayed using the boxer program of the EMAN software package (Ludtke et al., 1999) and datasets of 721, 1539, 494, and 1688 images were collected for undecorated, wild-type cofilin-decorated, cof1-158p decorated and wild-type cofilin under-decorated (1:2 cofilin to actin) filaments using a box size of 256 X 256 pixels. Datasets of 612 and 1280 images were collected for undecorated and wild-type cofilin decorated filaments using a box size of 400 X 400 pixels. Subsequent image analysis was done with the EMAN1 (Ludtke et al., 1999) and Imagic5 (van Heel et al., 1996) packages of programs. Datasets were normalized and bandpass filtered to remove low (<0.004 Å−1) and high (>0.15 Å−1) spatial frequencies and a circular mask was applied. Datasets with 256 pixel boxes were analyzed within EMAN1 using the refine2d.py command and sorted into 20, 24, 12, and 10 classes for undecorated, wild-type cofilin, cof1-158p decorated and wild-type cofilin under-decorated filaments respectively. Datasets with 400 pixel boxes were analyzed using Imagic 5 starting with the ‘alignment by classification’ procedure (Dube et al., 1993), which led to 20 and 40 class averages for undecorated and wild type cofilin decorated filaments respectively. The best average was then used in two rounds of direct alignment followed by multi-reference alignment using the best class averages from the previous alignment step as references. The alignment was iterated until stable and the dataset was subjected to multivariate statistical analysis (MSA) and classification to extract the most characteristic views of the filaments.
Observation of actin filaments by fluorescence microscopy
For the titration reactions, 2 μM Oregon green-labeled-actin was polymerized in modified F-buffer (10 mM imidazole pH 7.5, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 100 mM DTT, 0.2 mM ATP, 15 mM glucose, 0.5% w/v methylcellulose, 20μg/ml catalase, and 100 μg/ml glucose oxidase) after which an equal volume of cofilin (in the same buffer) was added, mixed, incubated for 2 min at room temperature and visualized/photographed under the microscope. For time-lapse observation of severing events, actin was polymerized as described, spotted under a coverslip and cofilin (2 μM) was diffused between the slide and the coverslip. All cover-slips were acid-washed and nitrocellulose coated. Images and movies were obtained on a Zeiss Imager.Z1 epifluorescence microscope with a 100X Plan Apochromator objective (oil, numerical aperture of 1.46) using an Orca ER camera (Hamamatsu Photonics) and processed with Zeiss AxioVision software and Adobe Photoshop.
For the TIRF experiments, flow cells were prepared from acid washed coverslips and slides as described (Kuhn and Pollard, 2005). Instead of parafilm strips, double-sided tape was used to create chambers that could hold up to 20 μl. The flow cells were treated with 0.5 μM myosin from porcine heart (Sigma, St. Louis, MO) and inactivated with N-ethylamaleimide (Kuhn and Pollard, 2005). Freshly labeled Oregon green iodoacetamide actin in G-buffer was diluted in modified F-buffer (as described above except pH 7.0) to 1 μM, and immediately loaded into the chamber and observed using a Nikon Eclipse TE-2000E microscope equipped with a TIRF 60X Plan Apochromator objective (oil, numerical aperture of 1.45) and a Hamamatsu ORCA II CCD camera. 2 μM cofilin in the same modified F-buffer was loaded into the chamber, images were immediately captured every 2.6 s for 3 min and processed with NIS Elements, ImageJ and Adobe Photoshop software. The focal plane was maintained using Perfect focus (Nikon).
Measurement of Actin Patch Dynamics
See Table 1 for the strains used. Confocal time-lapse images were collected at room temperature with a spinning-disc microscope, as described (Galletta et al., 2008). Abp1-GFP images were collected with Piper software (Agile Automation) at 5 frames/second. Actin patches were tracked and MSD plots were created as previously described (Carlsson et al., 2002) (Galletta et al., 2008) (Kim et al., 2006). The method is based on smoothing and thresholding of the fluorescence intensity, elimination of very small putative patches, and subsequent centroid calculation to obtain patch coordinates. The total patch lifetime was the time from the emergence of a patch to its disappearance. The time on the cell membrane was the average time spent by patches within 200 nm from their point of origin. The Phase III patch lifetime and the Phase III MSD were calculated from the portion of the data collected after the time point at which a patch was greater than 200 nm from its point of origin.
Table 1.
Yeast Strains
| Name | Genotype |
|---|---|
| YJC6537 | MATα ABP1-GFP::HIS3 his3Δ leu2Δ lys2Δ0 ura3Δ |
| YJC6538 | MATa ABP1-GFP::HIS3 his3Δ leu2Δ ura3Δ |
| YJC6543 | MATa ABP1-GFP::HIS3 his3Δ leu2Δ lys2Δ ura3Δ |
| YJC7037 | MATa cof1-157::LEU2 ABP1-GFP::HIS3 his3Δ leu2Δ lys2Δ0 ura3Δ |
| YJC7038 | MATa cof1-157::LEU2 ABP1-GFP::HIS3 his3Δ leu2Δ ura3Δ |
| YJC7039 | MATa cof1-157::LEU2 ABP1-GFP::HIS3 his3Δ leu2Δ lys2Δ ura3Δ |
| YJC7040 | MATa cof1-159::LEU2 ABP1-GFP::HIS3 his3Δ leu2Δ lys2Δ ura3Δ |
| YJC7041 | MATa cof1-159::LEU2 ABP1-GFP::HIS3 his3Δ leu2Δ ura3Δ |
| YJC7042 | MATa cof1-159::LEU2 ABP1-GFP::HIS3 his3Δ leu2Δ lys2Δ ura3Δ |
Supplementary Material
Table 4.
Yeast actin nucleators, their abundance and efficiencies.
| Nucleator | Efficiency % | # of molecules per cell | Nucleation capacity |
|---|---|---|---|
| Arp2/3 | 100 (Higgs et al., 1999) | 6650 (Ghaemmaghami et al., 2003) | 6650 |
| Formin (Bni1FH1FH2) | 3 (Pring et al., 2003) | 166 (Ghaemmaghami et al., 2003) | 5 |
| Cofilin | 0.17 | 20000 (Rodal et al., 1999) | 34 |
Acknowledgments
We thank David Pruyne for assistance calculating cofilin nucleation efficiencies. We thank Mira Krendel for assistance with TIRF microscopy. We thank Nicholas Stam for assistance with electron microscopy. We thank Brian Galletta for technical assistance determining actin patch dynamics. This work was supported by NIH grant R01GM056189 to DCA, NIH grant RO1GM38542 to JAC, and NIH grant RO1GM058600 to SW.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg DC, Basart E, Botstein D. Defining protein interactions with yeast actin in vivo. Nat Struct Biol. 1995;2:28–35. doi: 10.1038/nsb0195-28. [DOI] [PubMed] [Google Scholar]
- Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: John Wiley and Sons; 1989. [Google Scholar]
- Balcer HI, Goodman AL, Rodal AA, Smith E, Kugler J, Heuser JE, Goode BL. Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin, and Aip1. Curr Biol. 2003;13:2159–2169. doi: 10.1016/j.cub.2003.11.051. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD. Mechanism of interaction of Acanthamoeba actophorin (ADF/Cofilin) with actin filaments. J Biol Chem. 1999;274:15538–15546. doi: 10.1074/jbc.274.22.15538. [DOI] [PubMed] [Google Scholar]
- Bobkov AA, Muhlrad A, Kokabi K, Vorobiev S, Almo SC, Reisler E. Structural effects of cofilin on longitudinal contacts in F-actin. Journal of molecular biology. 2002;323:739–750. doi: 10.1016/s0022-2836(02)01008-2. [DOI] [PubMed] [Google Scholar]
- Bobkov AA, Muhlrad A, Pavlov DA, Kokabi K, Yilmaz A, Reisler E. Cooperative effects of cofilin (ADF) on actin structure suggest allosteric mechanism of cofilin function. Journal of molecular biology. 2006;356:325–334. doi: 10.1016/j.jmb.2005.11.072. [DOI] [PubMed] [Google Scholar]
- Bobkov AA, Muhlrad A, Shvetsov A, Benchaar S, Scoville D, Almo SC, Reisler E. Cofilin (ADF) affects lateral contacts in F-actin. Journal of molecular biology. 2004;337:93–104. doi: 10.1016/j.jmb.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Cao W, Goodarzi JP, De La Cruz EM. Energetics and kinetics of cooperative cofilin-actin filament interactions. Journal of molecular biology. 2006;361:257–267. doi: 10.1016/j.jmb.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J Cell Biol. 1997;136:1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson AE, Shah AD, Elking D, Karpova TS, Cooper JA. Quantitative analysis of actin patch movement in yeast. Biophysical journal. 2002;82:2333–2343. doi: 10.1016/S0006-3495(02)75579-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry F, Breitsprecher D, Little K, Sharov G, Sokolova O, Goode BL. Srv2/cyclase-associated protein forms hexameric shurikens that directly catalyze actin filament severing by cofilin. Mol Biol Cell. 2013;24:31–41. doi: 10.1091/mbc.E12-08-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Bernstein BW, Sneider JM, Boyle JA, Minamide LS, Bamburg JR. In vitro activity differences between proteins of the ADF/cofilin family define two distinct subgroups. Biochemistry. 2004;43:7127–7142. doi: 10.1021/bi049797n. [DOI] [PubMed] [Google Scholar]
- Chen Q, Pollard TD. Actin filament severing by cofilin dismantles actin patches and produces mother filaments for new patches. Curr Biol. 2013;23:1154–1162. doi: 10.1016/j.cub.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MG, Amberg DC. Biochemical and genetic analyses provide insight into the structural and mechanistic properties of actin filament disassembly by the Aip1p cofilin complex in Saccharomyces cerevisiae. Genetics. 2007;176:1527–1539. doi: 10.1534/genetics.107.072066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MG, Teply J, Haarer BK, Viggiano SC, Sept D, Amberg DC. A Genetic Dissection of Aip1p’s Interactions Leads to a Model for Aip1p-Cofilin Cooperative Activities. Mol Biol Cell. 2006 doi: 10.1091/mbc.E05-10-0956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JA, Walker SB, Pollard TD. Pyrene actin: documentation of the validity of a sensitive assay for actin polymerization. J Muscle Res Cell Motil. 1983;4:253–262. doi: 10.1007/BF00712034. [DOI] [PubMed] [Google Scholar]
- De La Cruz EM. Cofilin binding to muscle and non-muscle actin filaments: isoform-dependent cooperative interactions. Journal of molecular biology. 2005;346:557–564. doi: 10.1016/j.jmb.2004.11.065. [DOI] [PubMed] [Google Scholar]
- De La Cruz EM. How cofilin severs an actin filament. Biophys Rev. 2009;1:51–59. doi: 10.1007/s12551-009-0008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cruz EM, Sept D. The kinetics of cooperative cofilin binding reveals two states of the cofilin-actin filament. Biophysical journal. 2010;98:1893–1901. doi: 10.1016/j.bpj.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedova IV, Dedov VN, Nosworthy NJ, Hambly BD, dos Remedios CG. Cofilin and DNase I affect the conformation of the small domain of actin. Biophysical journal. 2002;82:3134–3143. doi: 10.1016/S0006-3495(02)75655-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Frieden C. Kinetic studies on the effect of yeast cofilin on yeast actin polymerization. Biochemistry. 1998;37:13276–13284. doi: 10.1021/bi981117r. [DOI] [PubMed] [Google Scholar]
- Dube P, Tavares P, Lurz R, van Heel M. The portal protein of bacteriophage SPP1: a DNA pump with 13-fold symmetry. EMBO J. 1993;12:1303–1309. doi: 10.1002/j.1460-2075.1993.tb05775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- Elam WA, Kang H, De la Cruz EM. Biophysics of actin filament severing by cofilin. FEBS Lett. 2013;587:1215–1219. doi: 10.1016/j.febslet.2013.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Saunders MG, Haddadian EJ, Freed KF, De La Cruz EM, Voth GA. Molecular origins of cofilin-linked changes in actin filament mechanics. Journal of molecular biology. 2013;425:1225–1240. doi: 10.1016/j.jmb.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federov AA, Lappalainen P, Federov EV, Drubin DG, Almo SC. Structure determination of yeast cofilin. Nature Struct Biol. 1997;4:366–369. doi: 10.1038/nsb0597-366. [DOI] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Kudryashov DS, Solodukhin A, Reisler E, Schroder GF, Egelman EH. Remodeling of actin filaments by ADF/cofilin proteins. Proc Natl Acad Sci U S A. 2011;108:20568–20572. doi: 10.1073/pnas.1110109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Schroder GF, Egelman EH. Structural polymorphism in F-actin. Nature structural & molecular biology. 2010;17:1318–1323. doi: 10.1038/nsmb.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, VanLoock MS, Shvetsov A, Reisler E, Egelman EH. ADF/cofilin use an intrinsic mode of F-actin instability to disrupt actin filaments. J Cell Biol. 2003;163:1057–1066. doi: 10.1083/jcb.200308144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin VE, VanLoock MS, Orlova A, Egelman EH. A new internal mode in F-actin helps explain the remarkable evolutionary conservation of actin’s sequence and structure. Curr Biol. 2002;12:570–575. doi: 10.1016/s0960-9822(02)00742-x. [DOI] [PubMed] [Google Scholar]
- Galletta BJ, Chuang DY, Cooper JA. Distinct roles for Arp2/3 regulators in actin assembly and endocytosis. PLoS biology. 2008;6:e1. doi: 10.1371/journal.pbio.0060001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Goode BL. Purification of yeast actin and actin-associated proteins. In: Part C, Guthrie C, Fink GR, editors. Methods in Enzymology; Guide to Yeast Genetics and Molecular Biology. London: Academin Press; 2002. pp. 433–441. [DOI] [PubMed] [Google Scholar]
- Hawkins M, Pope B, Maciver SK, Weeds AG. Human actin depolymerizing factor mediates a pH-sensitive destruction of actin filaments. Biochem. 1993;32:9985–9993. doi: 10.1021/bi00089a014. [DOI] [PubMed] [Google Scholar]
- Hayden SM, Miller PS, Brauweiler A, Bamburg JR. Analysis of the interactions of actin depolymerizing factor with G- and F-actin. Biochem. 1993;32:9994–10004. doi: 10.1021/bi00089a015. [DOI] [PubMed] [Google Scholar]
- Higgs HN, Blanchoin L, Pollard TD. Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry. 1999;38:15212–15222. doi: 10.1021/bi991843+. [DOI] [PubMed] [Google Scholar]
- Iida K, Moriyama K, Matsumoto S, Kawasaki H, Nishida E, Yahara I. Isolation of a yeast essential gene, COF1, that encodes a homologue of mammalian cofilin, a low-Mr actin-binding and depolymerizing protein. Gene. 1993;124:115–120. doi: 10.1016/0378-1119(93)90770-4. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- Karpovich DS, Blanchard GJ. Relating the polarity-dependent fluorescence response of pyrene to vibronic coupling. Achieving a fundamental understanding of the py polarity scale. J Phys Chem. 1995;99:3951–3958. [Google Scholar]
- Kim K, Galletta BJ, Schmidt KO, Chang FS, Blumer KJ, Cooper JA. Actin-based motility during endocytosis in budding yeast. Mol Biol Cell. 2006;17:1354–1363. doi: 10.1091/mbc.E05-10-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouyama T, Mihashi K. Fluorimetry study of N-(1-pyrenyl)iodoacetamide-labelled F-actin. Local structural change of actin protomer both on polymerization and on binding of heavy meromyosin. Eur J Biochem. 1981;114:33–38. [PubMed] [Google Scholar]
- Kudryashov DS, Galkin VE, Orlova A, Phan M, Egelman EH, Reisler E. Cofilin cross-bridges adjacent actin protomers and replaces part of the longitudinal F-actin interface. Journal of molecular biology. 2006;358:785–797. doi: 10.1016/j.jmb.2006.02.029. [DOI] [PubMed] [Google Scholar]
- Kuhn JR, Pollard TD. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophysical journal. 2005;88:1387–1402. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen P, Federov EV, Federov AA, Almo SC, Drubin DG. Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO J. 1997;16:5520–5530. doi: 10.1093/emboj/16.18.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MC, Galletta BJ, Sept D, Cooper JA. Overlapping and distinct functions for cofilin, coronin and Aip1 in actin dynamics in vivo. J Cell Sci. 2010;123:1329–1342. doi: 10.1242/jcs.065698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Maciver SK, Zot HG, Pollard TD. Characterization of actin filament severing by actophorin from Acanthamoeba castellanii. J Cell Biol. 1991;115:1611–1620. doi: 10.1083/jcb.115.6.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AC, Xu XP, Rouiller I, Kaksonen M, Sun Y, Belmont L, Volkmann N, Hanein D, Welch M, Drubin DG. Effects of Arp2 and Arp3 nucleotide-binding pocket mutations on Arp2/3 complex function. J Cell Biol. 2005;168:315–328. doi: 10.1083/jcb.200408177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough BR, Blanchoin L, Martiel JL, De la Cruz EM. Cofilin increases the bending flexibility of actin filaments: implications for severing and cell mechanics. Journal of molecular biology. 2008;381:550–558. doi: 10.1016/j.jmb.2008.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee JD, von Hippel PH. Theoretical aspects of DNA-protein interactions: co-operative and non-cooperative binding of large ligands to a one-dimensional homogeneous lattice. Journal of molecular biology. 1974;86:469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- McGough A, Chiu W. ADF/cofilin weakens lateral contacts in the actin filament. Journal of molecular biology. 1999;291:513–519. doi: 10.1006/jmbi.1999.2968. [DOI] [PubMed] [Google Scholar]
- McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon AL, Janmey PA, Louie KA, Drubin DG. Cofilin is an essential component of the yeast cortical cytoskeleton. J Cell Biol. 1993;120:421–435. doi: 10.1083/jcb.120.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooren OL, Galletta BJ, Cooper JA. Roles for actin assembly in endocytosis. Annu Rev Biochem. 2012;81:661–686. doi: 10.1146/annurev-biochem-060910-094416. [DOI] [PubMed] [Google Scholar]
- Okada K, Ravi H, Smith EM, Goode BL. Aip1 and cofilin promote rapid turnover of yeast actin patches and cables: a coordinated mechanism for severing and capping filaments. Mol Biol Cell. 2006;17:2855–2868. doi: 10.1091/mbc.E06-02-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okreglak V, Drubin DG. Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J Cell Biol. 2007;178:1251–1264. doi: 10.1083/jcb.200703092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okreglak V, Drubin DG. Loss of Aip1 reveals a role in maintaining the actin monomer pool and an in vivo oligomer assembly pathway. J Cell Biol. 2010;188:769–777. doi: 10.1083/jcb.200909176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, Mohri K, Ono K. Microscopic evidence that actin-interacting protein 1 actively disassembles actin depolymerizing factor/cofilin-bound actin filaments. J Biol Chem. 2004;279:14207–14212. doi: 10.1074/jbc.M313418200. [DOI] [PubMed] [Google Scholar]
- Orlova A, Shvetsov A, Galkin VE, Kudryashov DS, Rubenstein PA, Egelman EH, Reisler E. Actin-destabilizing factors disrupt filaments by means of a time reversal of polymerization. Proc Natl Acad Sci U S A. 2004;101:17664–17668. doi: 10.1073/pnas.0407525102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pavlov D, Muhlrad A, Cooper J, Wear M, Reisler E. Actin filament severing by cofilin. Journal of molecular biology. 2007;365:1350–1358. doi: 10.1016/j.jmb.2006.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pollard TD. Rate Constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J Cell Biol. 1986;103:2747–2754. doi: 10.1083/jcb.103.6.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. A guide to simple and informative binding assays. Mol Biol Cell. 2010;21:4061–4067. doi: 10.1091/mbc.E10-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- Poukkula M, Kremneva E, Serlachius M, Lappalainen P. Actin-depolymerizing factor homology domain: a conserved fold performing diverse roles in cytoskeletal dynamics. Cytoskeleton (Hoboken) 2011;68:471–490. doi: 10.1002/cm.20530. [DOI] [PubMed] [Google Scholar]
- Pring M, Evangelista M, Boone C, Yang C, Zigmond SH. Mechanism of formin-induced nucleation of actin filaments. Biochemistry. 2003;42:486–496. doi: 10.1021/bi026520j. [DOI] [PubMed] [Google Scholar]
- Prochniewicz E, Janson N, Thomas DD, De la Cruz EM. Cofilin increases the torsional flexibility and dynamics of actin filaments. Journal of molecular biology. 2005;353:990–1000. doi: 10.1016/j.jmb.2005.09.021. [DOI] [PubMed] [Google Scholar]
- Ressad F, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D, Carlier MF. Kinetic analysis of the interaction of actin-depolymerizing factor (ADF)/cofilin with G- and F-actins. Comparison of plant and human ADFs and effect of phosphorylation. J Biol Chem. 1998;273:20894–20902. doi: 10.1074/jbc.273.33.20894. [DOI] [PubMed] [Google Scholar]
- Rodal AA, Tetreault JW, Lappalainen P, Drubin DG, Amberg DC. Aip1p interacts with cofilin to disassemble actin filaments. J Cell Biol. 1999;145:1251–1264. doi: 10.1083/jcb.145.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez C, Roland J, Boujemaa-Paterski R, Kang H, McCullough BR, Reymann AC, Guerin C, Martiel JL, De la Cruz EM, Blanchoin L. Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Curr Biol. 2011;21:862–868. doi: 10.1016/j.cub.2011.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- Vartiainen MK, Mustonen T, Mattila PK, Ojala PJ, Thesleff I, Partanen J, Lappalainen P. The three mouse actin-depolymerizing factor/cofilins evolved to fulfill cell-type-specific requirements for actin dynamics. Mol Biol Cell. 2002;13:183–194. doi: 10.1091/mbc.01-07-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DY, Sept D. The interaction of cofilin with the actin filament. Journal of molecular biology. 2011;413:97–105. doi: 10.1016/j.jmb.2011.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoh S, Pope B, Mannherz HG, Weeds A. Determining the differences in actin binding by human ADF and cofilin. Journal of molecular biology. 2002;315:911–925. doi: 10.1006/jmbi.2001.5280. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.