Abstract
• Background and Aims Aglaonema is an important ornamental foliage plant genus, but genetic relationships among its species and cultivars have not been reported. This study analysed genetic relatedness of 54 cultivars derived from nine species using amplified fragment length polymorphism (AFLP) markers.
• Methods Initially, 48 EcoRI + 2/MseI + 3 primer set combinations were screened, from which six primer sets that showed clear scoreable and highly polymorphic fragments were selected and used for AFLP reactions. AFLP fragments were scored and entered into a binary data matrix as discrete variables. Jaccard’s coefficient of similarity was calculated for all pair‐wise comparisons among the 54 cultivars, and a dendrogram was constructed by the unweighted pair‐group method using the arithmetic average (UPGMA).
• Key Results The number of AFLP fragments generated per primer set ranged from 59 to 112 with fragment sizes varying from 50 to 565 bp. A total of 449 AFLP fragments was detected, of which 314 were polymorphic (70 %). All cultivars were clearly differentiated by their AFLP fingerprints. The 54 cultivars were divided into seven clusters; cultivars within each cluster generally share similar morphological characteristics. Cluster I contains 35 cultivars, most of them are interspecific hybrids developed mainly from A. commutatum, A. crispum or A. nitidum. However, Jaccard’s similarity coefficients among these hybrids are 0·84 or higher, suggesting that these popular hybrid cultivars are genetically much closer than previously thought. This genetic similarity may imply that A. nitidum and A. crispum are likely progenitors of A. commutatum.
• Conclusions Results of this study demonstrate the efficiency and ease of using AFLP markers for investigating genetic relationships of ornamental foliage plants, a group usually propagated vegetatively. The AFLP markers developed will help future Aglaonema cultivar identification, germplasm conservation and new cultivar development.
Key words: AFLP markers, Araceae, Aglaonema, Chinese evergreens, genetic variation, ornamental tropical foliage plants
INTRODUCTION
The genus Aglaonema Schott belongs to the family Araceae Juss. and comprises 21 species (Nicolson, 1969). All species are herbaceous evergreens native to south‐east Asia, north‐eastern India, across southern China, and into Indonesia and New Guinea where they inhabit humid and heavily shaded tropical forests (Nicolson, 1969; Mayo et al., 1997). Flowers are unisexual, dichogamous in nature and, thus, most species are open‐pollinated. The chromosome number varies from 2n = 42 to 60 or 120 (Nicolson, 1969) depending on species. Due to their attractive foliar variegation and tolerance to low light, Aglaonema species have been cultivated in China and other Asian countries for centuries as indoor ornamental foliage plants or houseplants. As a result, Aglaonema are commonly referred to as Chinese evergreens. Chinese evergreens were introduced into the Royal Botanic Gardens, Kew in 1885 (Brown, 1885).
Aglaonema cultivation in the USA started in the 1930s (Smith and Scarborough, 1981) and soon became a great success in Florida, where most foliage plants are now produced. The success can be partly attributed to an increasing release of attractive interspecific hybrids (Chen et al., 2002) as the number of commercial cultivars grown in Florida increased from ten in 1975 to more than 50 in 2002. Many of the cultivars have gained wide recognition and contribute significantly to the ornamental foliage plant industry. For example, ‘Silver Queen’, a hybrid selected from a cross between A. commutatum Schott ‘Treubii’ and A. nitidum (Jack) Kunth ‘Curtisii’, brings about $4 million to the Florida state economy annually (Elliott et al., 1998). With the discovery of new Aglaonema species in the Philippines and Thailand in the 1970s (Brown, 2001), wild collections as well as new hybrids have been introduced to Florida. A total of 29 Aglaonema hybrids received USA patents during the 1990s (Henny and Chen, 2003). Genetic information on some of these hybrids, however, is unclear.
Although Aglaonema has become an important ornamental foliage genus, neither the US National Plant Germplasm System (NPGS) nor the International Board for Plant Genetic Resources (IBPGR) had been actively involved in conservation of Aglaonema or other foliage plant germplasm until recently. The Ornamental Plant Germplasm Center (OPGC) established in 1999 at Ohio State University is considering conservation of five genera from the family Araceae including Aglaonema. Currently, most Aglaonema resources have been collected and maintained by private plant collectors or public institutions such as botanical gardens or conservatories (Henny and Chen, 2003). Because of the expense of maintaining tender tropical plants in glasshouses, it is conceivable that changes in funding or in individual research interests could result in a loss of plant diversity in these invaluable collections.
Scientific conservation and utilization of genetic resources require accurate information on genetic relatedness among species and cultivars. However, attempts to classify Aglaonema species based on morphology have been difficult and controversial. In his monograph A Revision of the Genus Aglaonema (Araceae), Nicolson (1969) identified 21 species. The monumental Hortus III (Bailey and Bailey, 1976) described ten species, Jervis (1980) identified 17 species, and the New Royal Horticultural Society Dictionary of Gardening documented 11 species (Huxley, 1994). Among the species, the origin of A. commutatum has been at the centre of controversy due to its morphological variability. Nicolson (1969) hypothesized that A. commutatum might be of hybrid origin since it shares some morphological characters with A. simplex Blume, A. nitidum and A. marantifolium Blume. Later, Jervis (1980) proposed that A. commutatum might originate from introgressive hybridization of A. crispum (Pitcher & Manda) Nicols., A. simplex and A. philippense Engl. However, neither of these two hypotheses has been tested.
Amplified fragment length polymorphism (AFLP) is a novel PCR‐based assay for plant DNA fingerprinting that reveals significant levels of DNA polymorphism (Vos et al., 1995). Molecular marker profiles based on AFLPs can be used to detect variation in the DNA level and have proved to be extremely effective in distinguishing closely related genotypes. AFLP has been used in the assessment of genetic relationships of a wide range of species, including some ornamentals such as Caladium Venten. spp. (Loh et al., 1999, 2000), Hemerocallis L. spp. (Tomkins et al., 2001), Musa acuminata Colla (Wong et al., 2001), Impatiens hawkeri W. Bull (Carr et al., 2003), Orchis simia Vill. (Qamaruz‐Zaman et al., 1998) and Pelargonium peltatum (L.) L’Her. ex Ait. (Barcaccia et al., 1999). However, genetic relationships of Aglaonema species and cultivars at DNA levels have not been determined thus far. The objectives of this study were to assess the usefulness of AFLP for cultivar identification and to determine genetic relationships of common Aglaonema species and culitvars.
MATERIALS AND METHODS
Plant material
Fifty‐four Aglaonema cultivars, along with one cultivar each of five other ornamental aroid genera (Alocasia (Schott) G. Don., Anthurium Schott, Dieffenbachia Schott, Philodendron Schott and Spathiphyllum Schott), were collected from either the research glasshouses at the Mid‐Florida Research and Education Center (MREC), University of Florida or ornamental foliage plant nurseries in central Florida (Table 1).
Table 1.
List of 54 Aglaonema cultivars and five other ariod genera used in this study
| Cultivars | Background | Sources |
| Treubii | A. commutatum | UF/MREC* |
| Curtissi | A. nitidum | UF/MREC |
| Ernesto’s Favorite | A. nitidum | UF/MREC |
| Silver Queen | Interspecific hybrid | UF/MREC |
| 277028 | Interspecific hybrid | UF/MREC |
| Emerald Bay | Somaclonal variant | UF/MREC |
| Golden Bay | Interspecific hybrid | UF/MREC |
| Moonlight Bay | Somaclonal variant | UF/MREC |
| Diamond Bay | Somaclonal variant | UF/MREC |
| 257‐12 | Interspecific hybrid | UF/MREC |
| Echo | A. commutatum | UF/MREC |
| Cory | Interspecific hybrid | UF/MREC |
| Costatum Hybrid | Unknown | Twyford International, Inc.† |
| Queen of Siam | Interspecific hybrid | Stewart’s Greenhouse, Inc.‡ |
| White Rain | Hybrid | Stewart’s Greenhouse, Inc. |
| Stripes | Hybrid | UF/MREC |
| White Lance | Hybrid | Stewart’s Greenhouse, Inc. |
| Green Lady | Interspecific hybrid | Stewart’s Greenhouse, Inc. |
| Jubilee | Hybrid | Green Star Foliage, Inc.§ |
| Jubilee Petite | Hybrid | Stewart’s Greenhouse |
| Maria Christina | Unknown | Stewart’s Greenhouse |
| Painted Princess | Hybrid | Stewart’s Greenhouse |
| J.G. | Hybrid | UF/MREC |
| 1502 | Hybrid | UF/MREC |
| Amelia | Hybrid | Stewart’s Greenhouse |
| B.J. Freeman | A. crispum | Stewart’s Greenhouse |
| Camouflage | Hybrid | Stewart’s Greenhouse |
| Jewel of India | Interspecific hybrid | Green Star Foliage |
| Maria | Unknown | Green Star Foliage |
| Mary Ann | Hybrid | Stewart’s Greenhouse |
| Patricia | Hybrid | Stewart’s Greenhouse |
| Royal Ripple | Hybrid | Stewart’s Greenhouse |
| Silver Ribbon | Hybrid | Stewart’s Greenhouse |
| Silver Bay | Interspecific hybrid | UF/MREC |
| Commatum | Unknown | UF/MREC |
| Deborah | Sport | Green Star Foliage |
| Emerald Star | Interspecific hybrid | Green Star Foliage |
| Stars | Interspecific hybrid | Stewart’s Greenhouse |
| Donna Carmen Mutant | Sport | Twyford International, Inc |
| Peacock | Interspecific hybrid | Stewart’s Greenhouse |
| III | Unknown | Twyford International, Inc. |
| JT2000 | Unknown | Stewart’s Greenhouse |
| Pride of Sumatra | Interspecific hybrid | Twyford Inc. |
| Red Gold | Interspecific hybrid | Twyford international, Inc. |
| Donna Carmen | Interspecific hybrid | Stewart’s Greenhouse |
| Modestum | A. modestum | UF/MREC |
| 714‐12 | Unknown | UF/MREC |
| Mod × Rot Clone 1 | Interspecific hybrid | Twyford Inc. |
| Mod × Rot Clone 2 | Interspecific hybrid | Twyford Inc. |
| Mod × Rot Clone 3 | Interspecific hybrid | Twyford Inc. |
| Mod × Rot Clone 4 | Interspecific hybrid | Twyford Inc. |
| Pictum Tricolor | A. pictum | UF/MREC |
| XII | Unknown | Twyford Inc. |
| XIV | Unknown | Twyford Inc. |
| Red Hot | Anthurium × | UF/MREC |
| 64706 | Spathiphyllum × | UF/MREC |
| Moonlight | Philodendron × | Agri‐Starts, Inc.¶ |
| Compacta | Dieffenbachia maculata ‘Compacta’ | UF/MREC |
| Polly | Alocasia × | Agri‐Starts, Inc. |
* University of Florida, Mid‐Florida Research and Education Center, Apopka, FL, USA.
† Twyford International, Inc. Apopka, FL, USA.
‡ Stewart’s Greenhouse, Inc. Mt Dora, FL, USA.
§ Green Star Foliage, Inc. Apopka, FL, USA.
¶ Agri‐Starts, Inc. Apopka, FL, USA
Fluorescent‐AFLP analysis
Total DNA was extracted from young leaves of 60 samples (54 Aglaonema cultivars, one duplicated A. commutatum ‘Treubii’, and five other aroid genera) using the DNeasy system (Qiagen, Valencia, CA, USA). After DNA quantification using a Hoefer DyNA Quant 200 (Pharmacia Biotech, Piscataway, NJ, USA), AFLP analysis was conducted using the GIBCO BRL AFLP System II (Life Tecnologies, Grand Island, NY, USA) and visualized with the LI‐COR IR automated sequencer 4000‐L (Li‐COR Inc., Lincoln, NE, USA). Total DNA (125 ng) from all 60 samples was digested with 1 µl of a mixture of EcoRI/MseI (1·25 units µl–1) at 37 °C overnight and ligated to EcoRI/MseI adapters with 1·5 µl (1 unit µl–1) of T4 DNA ligase at 25 °C for at least 6 h. The adaptor‐ligated DNA was amplified using a mixture of 2·5 µl of DNA from the ligation reaction, 20 µl of pre‐amplification mix II, 2·5 µl of 10 × PCR buffer, and 0·2 µl of Taq DNA polymerase (5 units µl–1). The pre‐amplification reactions were performed on a MJR Cycle LR™ (MJ Research, Inc., Watertown, MA, USA) using the following cycling parameters: 30 cycles at 94 °C for 15 s, 56 °C for 30 s plus 1 extra s per cycle, and 72 °C for 1 min plus 1 extra s per cycle, then 1 cycle at 72 °C for 3 min. The pre‐amplified PCR product was quantified in the fluorometer, and the amount of template for the subsequent PCR was diluted to 125 µg µl–1. Selective amplification was performed using a reaction mix composed of 2 µl of DNA from pre‐amplification, 2 µl of MseI primer (8·3 µm), 0·5 µl of IRD700‐labeled EcoRI primer (1 µm), 0·5 µl of IRD800‐labeled EcoRI primer (1 µm), 1 µl of 10 × PCR buffer, 4 µl of H2O, and 0·16 µl of Taq DNA polymerase (5 units µl–1).
The selective amplification PCRs were performed as follows: 13 cycles at 94 °C for 15 s, 65 °C for 30 s minus 0·7 s per cycle, and 72 °C for 1 min, then 30 cycles at 94 °C for 15 s, 56 °C for 30 s plus 1 extra s per cycle, 72 °C for 1 min plus 1 extra s per cycle, then 72 °C for 3 min. Both pre‐ and selective‐amplification conditions were modified according to Myburg et al. (2000). The products from the selective amplification were electrophoresed on 25 cm × 0·25 mm 8 % denaturing polyacrylamide Long Ranger® Gel Solution (BMA, Rockland, ME, USA) in 0·8 × TBE buffer using a LI‐COR automated sequencer 4000‐L. The gel was pre‐run for 10–20 min at 1500 V, 40 mA, and 40 W until the gel temperature reached 50 °C. The samples were denatured at 95 °C for 3 min and immediately placed on ice. Electrophoresis was performed at 1500 V, 50 °C for 3·5 h after 1·15 µl samples and 1 µl of a mixture of IRD700 and IRD800 size markers were loaded (Li‐COR Inc., Lincoln, NE, USA).
A total of 48 AFLP primers were initially screened, from which six primer sets (IRD700 E + TC/M + CAA; IRD800 E + AA/M + CAA; IRD700 E + TC/M + CAT; IRD800 E + AA/M + CAT; IRD700 E + GG/M + CAA; and IRD800 E + CC/M + CAA) that showed clear scoreable and also highly polymorphic fragments were selected for fluorescent‐AFLP reactions with the samples of the Aglaonema cultivars and five other aroid genera.
Data analysis
For the genetic similarity analysis, AFLP fragments were visually scored as present (1) or absent (0) to create a binary data set. The data was entered into a binary data matrix as discrete variables. Jaccard’s coefficient of similarity (Sneath and Sokal, 1973) was calculated for all pair‐wise comparisons among the Aglaonema cultivars and five other aroid genera as follows: Jaccard = NAB/ (NAB + NA + NB), where NAB is the number of fragments shared by two cultivars (A and B), NA represents amplified fragments in cultivar A and NB represents fragments in cultivar B. A dendrogram was constructed using NTSYS version 2·1 (Exeter Software, Setauket, NY, USA) (Rohlf, 2000) based on the unweighted pair group method of the arithmetic average (UPGMA).
RESULTS AND DISCUSSION
AFLP profiles and analysis
The six primer sets generated clear‐cut AFLP profiles for the 54 Aglaonema cultivars and five other aroid genera. The duplicate cultivar ‘Treubii’ showed identical AFLP patterns with each primer set. An example of fluorescent‐AFLP profiles of 30 Aglaonema cultivars with five other aroid genera using primer IRD700 E + GG/M + CAA is shown in Fig. 1. In a theoretical consideration of the number of primers needed to produce accurate estimates of genetic relatedness, Ellis et al. (1997) believed that by choosing the six best combinations of primers, it is possible to explain more than 80 % of the expected relatedness. Using six primer combinations selected from our initial screening of 48, it was found that the AFLP fragments ranged from 50 to 565 bp, with a majority of the polymorphism distributed between 150 and 350 bp. A total of 449 fragments were scored, of which 314 (70 %) were polymorphic (Table 2), suggesting that each primer generated an average of 74·8 fragments and 52·3 of them were polymorphic. The level of polymorphism was similar to that detected in other plants when the AFLP technique was used for measuring genetic variability. Aggarwal et al. (2002) identified 501 AFLP markers from Basmati rice (Oryza sativa L.) with 65 % of them being polymorphic. Singh et al. (1999) reported a total of 422 fragments were obtained from Azadiracht indica A. Juss, of which 297 (70 %) were polymorphic. Tomkins et al. (2001) determined that the average percentage of polymorphism was 79 % per primer combination in daylily (Hemerocallis spp).
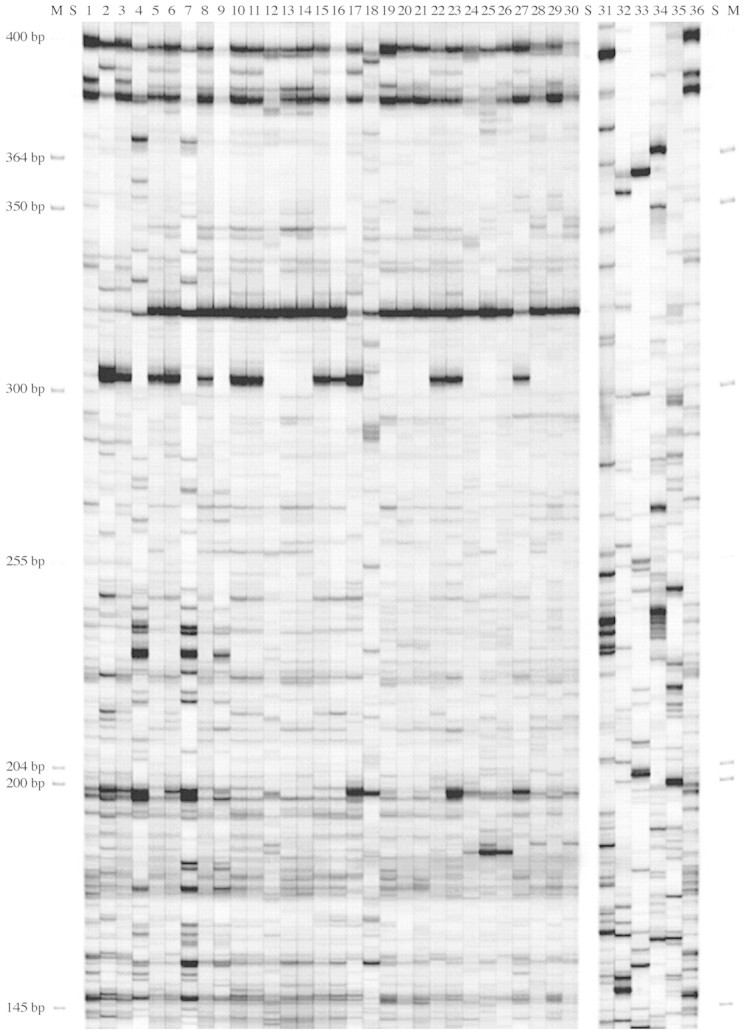
Fig. 1. Fluorescent‐AFLP profiles of 30 Aglaonema cultivars, five cultivars from other aroid genera, and a duplicate A. commutatum ‘Treubii’ using primer IRD700 E + GG/M + CAA. The samples are arranged from left to the right in the order of (M) size markers, (S) space, (1) ‘Treubii’, (2) ‘Curtisii’, (3) ‘Silver Queen’, (4) ‘Modestum’, (5) ‘277028’, (6) ‘257‐12’, (7) ‘714‐12’, (8) ‘Echo’, (9) ‘Peacock’, (10) ‘Emerald Bay’, (11) ‘Golden Bay’, (12) ‘Donna Carmen’, (13) ‘J.G.’, (14) ‘1502’, (15) ‘Moonlight Bay’, (16) ‘Diamond Bay’, (17) ‘Ernesto’s Favorite’, (18) ‘Pictum Tricolor’, (19) ‘Amelia’, (20) ‘B.J. Freeman’, (21) ‘Camouflage’, (22) ‘Cory’, (23) ‘Costatum Hybrid’, (24) ‘Deborah’, (25) ‘Donna Carmen Mutant’, (26) ‘Emerald Star’, (27) ‘Green Lady’, (28) ‘III’, (29) ‘Jewel of India’, (30) ‘JT2000’, (S) space, (31) Anthurium ‘Red Hot’, (32) Spathiphyllum ‘64760’, (33) Philodendron ‘Moonlight’, (34) Dieffenbachia ‘Compacta’, (35) Alocasia ‘Polly’, (36) Aglaonema ‘Treubii’, (S) space, and (M) size markers.
Table 2.
AFLP primer combinations, primer sequences, total number of bands generated by each primer set, number of polymorphic bands detected, and percentages of polymorphic bands used in the study of Aglaonema cultivars
| Primer | Total no. of bands | No. of polymorphic bands | % of polymorphic bands |
| IRD700 E + TC/M + CAA | 69 | 43 | 62 |
| IRD800 E + AA/M + CAA | 66 | 45 | 68 |
| IRD700 E + TC/M + CAT | 74 | 54 | 73 |
| IRD800 E + AA/M + CAT | 69 | 46 | 67 |
| IRD700 E + GG/M + CAA | 112 | 86 | 77 |
| IRD800 E + CC/M + CAA | 59 | 40 | 68 |
| Total | 449 | 314 | 70 |
| Average | 74·8 | 52·3 | 70 |
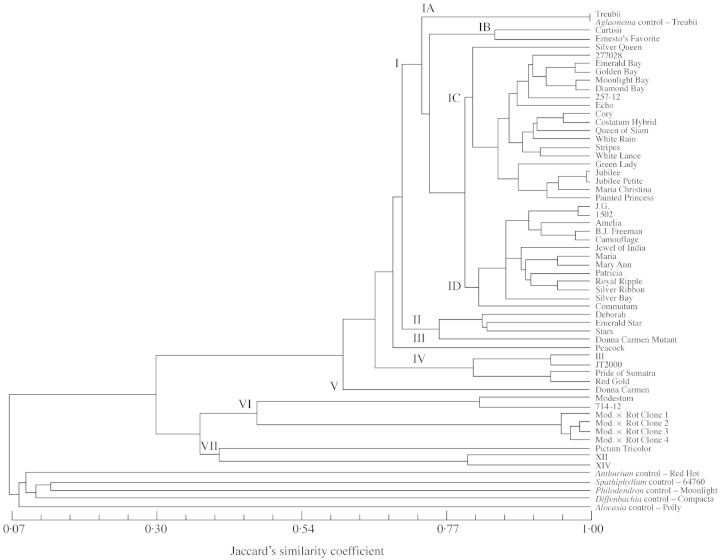
Genetic relationships among cultivars
Based on the UPGMA analysis, a dendrogram for the Aglaonema cultivars and also other five aroid genera was constructed (Fig. 2). Fifty‐four Aglaonema cultivars were grouped into seven clusters. Cluster I is the largest, comprising 35 cultivars which can be divided into four subclusters, IA, IB, IC and ID. Subcluster IA includes A. commutatum ‘Treubii’ only, and IB includes A. nitidum ‘Curtissi’ and A. nitidum ‘Ernesto’s Favorite’. Aglaonema commutatum generally displays foliar variegation confluent in bars along the primary lateral veins of the leaf, rarely in irregularly scattered spots (Nicolson, 1969). Foliar variegations of A. nitidum include narrow, silvery stripes along lateral veins or silvery blotches around the midrib.
Fig. 2. Dendrogram of 54 Aglaonema cultivars, a duplicate A. commutatum ‘Treubii’, and five cultivars from other aroid genera resulting from a UPGMA cluster analysis based on Jaccard estimates of similarity obtained from 449 polymorphic AFLP bands.
Subcluster IC contains 19 cultivars, which can be further broken into four groups. The cultivar in the first group is ‘Silver Queen’, an interspecific hybrid selected from a cross of A. commutatum ‘Treubii’ and A. nitidum ‘Curtisii’. The second group includes an interspecific hybrid ‘277028’, developed from a cross between A. commutatum ‘Treubii’ and A. nitidum ‘Ernesto’s Favorite’. ‘Emerald Bay’ is a somaclonal variant selected from ‘Golden Bay’ (Henny et al., 2003). ‘Golden Bay’ was derived from a cross of ‘277028’ and ‘MREC 1501’ (Henny and Chen, 2001). The ‘MREC 1501’ was a hybrid selected from a cross of A. commutatum ‘Tricolor’ and A. × ‘Manila’. Whereas Aglaonema × ‘Manila’ was collected from the Philippines in 1962 and believed to be a hybrid resulting from a cross of A. commutatum ‘Tricolor’ with an unknown species (Brown, 2001). ‘MREC 1501’ and A. × ‘Manila’ were not included in this study as they were not available. Both ‘Moonlight Bay’ and ‘Diamond Bay’ are somaclonal variants selected from ‘Silver Bay’, which was a hybrid of A. × ‘Manila Whirl’ and A. nitidum ‘Ernesto’s Favorite’ (Henny et al., 1992). Cultivar ‘257‐12’ was developed from a cross of ‘1502’ and A. nitidum ‘Curtisii’. Another cultivar, ‘Echo’, also belongs to the second group. ‘Echo’ was found in Manila, the Philippines (Jervis, 1980) and is a cultivar of A. commutatum.
There are six cultivars in the third group of IC. ‘Cory’ is a hybrid selected from a cross of A. nitidum ‘Curtisii’ with an unknown plant. ‘Costatum hybrid’, which has a Jaccard’s coefficient of 0·96 with ‘Cory’, is most likely not a cultivar from A. costatum Schott since cultivars related to A. costatum are positioned in cluster II. ‘Queen of Siam’ is an interspecific hybrid selected from A. commutatum ‘Tricolor’ and A. nitidum ‘Ernesto’s Favorite’. ‘White Rain’ is a hybrid of A. nitidum ‘Ernesto’s Favorite’ crossed with A. cv. ‘Panayensis’. There is no species information available about A. cv. ‘Panayensis’, and this cultivar was not used in this study. ‘Stripes’ is a hybrid of A. ‘Manila’ and A. nitidum ‘Curtisii’. ‘White Lance’ is a hybrid of A. cv. ‘Bayonet’ and A. nitidum ‘Ernesto’s Favorite’. Again, A. cv. ‘Bayonet’ has an unknown origin and was not included in this study. There are five cultivars in the fourth group of IC. ‘Green Lady’, according to the USA plant patent no. 10271 (or PP10271), was selected from a cross between A. commutatum and A. pictum (Roxb.) Kunth. This claim is probably untrue because A. pictum has ten unique fragments, of which none are present in ‘Green Lady’. ‘Jubilee’ is a hybrid of A. nitidum ‘Curtisii’ and A. × ‘Manila’, ‘Jubilee Petite’ a hybrid of A. nitidum ‘Curtisii’ and A. commutatum ‘Tricolor’. The origin of ‘Maria Christina’ is not completely clear, but it could be a hybrid of A. nitidum and A. commutatum as it morphologically resembles ‘Silver Queen’. ‘Painted Princess’ (USA PP9081) was claimed as a hybrid from unidentified parents. However, its parents could be related to A. nitidum and A. commutatum as well since it is genetically similar to the cultivars in the fourth group of IC (Jaccard’s similarity coefficient = 0·90).
Cluster ID includes 13 cultivars, which fall into two groups. There are three subgroups in the first group. Both ‘J.G.’ and ‘1502’ are hybrids selected from the same cross between A. commutatum ‘Tricolor’ and A. × ‘Manila’. ‘Amelia’ (USA PP8977) is a hybrid without identified parents. ‘B.J. Freeman’ is a cultivar of A. crispum. ‘Camouflage’ is a hybrid developed from a cross of A. × ‘Manila’ and ‘FB‐23’. The origin of ‘FB‐23’ is unknown; but it may be related to A. crispum. Due to its unavailability, ‘FB‐23’ was not used in this study. A distinct feature of A. crispum is its ‘ashy’ variegation that completely covers the leaves except for the midvein and margin. The second subgroup includes ‘Jewel of India’, a hybrid of A. commutatum ‘Malay Lady’ and A. crispum ‘Dow Hybrid’. ‘Maria’ has been a common cultivar in the foliage plant industry for decades, and it may have a relationship with A. commutatum. ‘Mary Ann’ (USA PP8976), ‘Patricia’ (USA PP10269) and ‘Royal Ripple’ (USA PP9076) all have no identified parents even though the USA patents were issued. Due to its genetic (Jaccard’s similarity coefficient = 0·95) and morphological (bi‐coloured leaves marked with silver‐green on dark green background) similarity to ‘Maria’, ‘Mary Ann’ may be related to A. commutatum. The solid‐silver colour between the midrib and leaf margin led us to believe that ‘Patricia’ may have genetic makeup similar to A. crispum. ‘Silver Ripple’ is both genetically and morphologically close to ‘Silver Ribbon’. ‘Silver Ribbon’ was developed in India as a hybrid of A. commutatum var. ‘Picturatum’ and A. commutatum var. ‘Silver III’. ‘Silver Bay’, a hybrid of A. × ‘Manila Whirl’ and A. nitidum ‘Ernesto’s Favorite’ (Henny et al., 1992) as mentioned previously, is the only cultivar in the third subgroup of ID subcluster. There is only one cultivar, ‘Commatum’, in the second group of ID, which may be a selection within A. commutatum.
Cluster II includes four cultivars. ‘Deborah’ is assumed to be a sport of ‘Queen of Siam’. ‘Emerald Star’ is another hybrid introduced from India, it was selected from of a cross of A. commutatum ‘Elegans’ and A. costatum. Leaf characteristics of A. costatum include scattered white or yellow spots on both sides of leaves and a white midrib. ‘Stars’ is a hybrid of a cross of A. commutatum and A. brevispathum (Engl.) Engl. Foliar variegation of A. brevispathum is also characterized by small white and yellow spots. Since A. brevispathum and A. costatum are closely related (Nicolson, 1969), ‘Emerald Star’ and ‘Stars’ are genetically close and morphologically similar. ‘Donna Carmen Mutant’ is presumed to be a sport of ‘Donna Carmen’. However, its similarity to cultivars from either A. brevispathum or A. costatum raises a doubt about this presumption.
There is only one cultivar in cluster III – ‘Peacock’, which is a hybrid selected from a cross of A. commutatum ‘Echo’ and A. marantifolium Blume ‘Tricolor’. Aglaonema marantifolium ‘Tricolor’ has narrow areas of mottled yellow and green extended along the midrib and lateral veins (Jervis, 1980). ‘Peacock’ leaves have a mottled green colour with random dark green spots and yellow green midribs.
Cluster IV contains four cultivars. There is no information available as to the origin and pedigree of ‘III’ and ‘JT2000’. ‘Pride of Sumatra’ was from Indonesia and most likely relates to A. rotundum and A. marantifolium since its morphology resembles ‘Red Gold’. ‘Red Gold’ is a hybrid developed by Sithiporn Donavanik of Thailand who crossed A. rotundum with A. marantifolium ‘Tricolor’. Aglaonema rotundum has pink to red midribs and veins on a dark, glossy green upper surface. ‘Red Gold’ has red veins strongly resembling a Codiaeum variegatum and was later named A. sithiporn. Twyford Plant Laboratories, Inc., Sebring, Florida renamed it as ‘Red Gold’ (Brown, 2001).
Only one cultivar, ‘Donna Carmen’ is positioned in cluster V. This cultivar was developed by botanist Gregory Hambali of Bogar, Indonesia. It also has red leaves and resulted from crosses of A. rotundum with another, unknown, variety.
Cluster VI has A. modestum and hybrids of A. modestum crossed by other species. Based on the AFLP polymorphism, A. modestum is clearly separated from other Aglaonema species. This species has solid green leaves and has been cultivated in the East for centuries as a symbol of good fortune. The origin of ‘714‐12’ is unclear. The ‘Mod. × Rot. Clone’ series were believed to be crosses between A. modestum and A. rotundum.
Three cultivars are situated in cluster VII; subcluster VIIA has A. pictum ‘Tricolor’, and subcluster VIIB has cultivars ‘XII’ and ‘XIV’. The Jaccard’s similarity coefficient of Aglaonema pictum ‘Tricolor’ is the lowest (0·31) with all other Aglaonema species based on the AFLP polymorphism. Cultivars ‘XII’ and ‘XIV’ are of unknown origins. Their AFLP polymorphisms are different from other Aglaonema cultivars tested and their origins remain unclear.
Cultivars clustered based on the similarity of AFLP polymorphisms are also morphologically related. Variegation patterns of cultivars in cluster I include silvery stripes along the principle lateral veins, ash blotches on the centre of the leaf or between the midrib and margin, or irregular grey blotches adjacent to primary lateral veins. These variegation patterns are characteristics of A. commutatum, A. crispum, A. nitidum or hybrids derived from two of the three species (Henny, 1983, 1986). Foliar variegation patterns of cluster II cultivars most resemble A. costatum or A. brevispathum with scattered white or yellowish spots on both sides of the leaves. ‘Peacock’ in cluster III has a unique pattern of leaf variegation, characterized by a mottled green colour with random dark green spots. This pattern is probably a combination of A. commutatum ‘Echo’ and A. marantifolium ‘Tricolor’. Cultivars in cluster IV and V are distinguished by red‐veined leaves, which is the characteristic of A. rotundum. Cluster VI cultivars, like A. modestum, are either solid green or slightly variegated. Foliar variegation in cluster VII cultivars generally has rather ashy to slivery irregular blotches that are close to the pattern of A. pictum.
Five other genera of Araceae (Alocasia, Anthurium, Dieffenbachia, Philodendron and Spathiphyllum) were included in this study. The five genera were properly positioned outside the Aglaonema cluster. They share genetic similarity no greater than 0·10 based on the Jaccard’s coefficient with any Aglaonema samples and the genetic similarities among these five genera are < 0·14.
Genetic relationships among species
Seven of the 54 cultivars are not of hybrid origin: A. commutatum ‘Echo’ and ‘Treubii’, A. crispum ‘B.J. Freeman’, A. nitidum ‘Curtisii’ and ‘Ernesto’s Favorite’, A. modestum and A. pictum ‘Tricolor’. Other species that were indirectly involved in this study include A. brevispathum, A. costatum, A. marantifolium and A. rotundum; they were parents of the hybrids used in the study. Aglaonema pictum and A. modestum are rather dissimilar since their Jaccard’s similarity coefficient is only 0·31, and both are distant from the other species (Table 3). The distant relationships between these two species and from the others may be attributed to their originating geographical locations. Aglaonema pictum is native to Sumatra and inhabits the slopes of major volcanoes between 1000 and 2000 m a.s.l (Nicolson, 1969), which may isolate it from the other species. The origin of A. modestum in southern China, northern Laos and northern Thailand is certainly separate from the others, which are generally native to Malaysia, Indonesia and the Philippines. Hybrid cultivars related to A. marantifolium and A. rotundum are situated in clusters III, IV and V; Jaccard’s similarity coefficients across the three clusters range from 0·61 to 0·68. Aglaonema marantifolium originates from Moluccas through New Guinea, whereas A. rotundum is native to Sumatra (Jervis, 1980). However, genetic similarity manifested by their hybrids may imply that the two species are relatively close. Aglaonema costatum and A. brevispathum are native to continental south‐east Asia and were considered to be morphologically similar by Nicolson (1969). When both were crossed with A. commutatum, their hybrids, ‘Emerald Star’ and ‘Stars’, resemble each other and share a Jaccard’s similarity coefficient of 0·82. Cultivars related to A. crispum, A. nititum and A. commutatum are mainly gathered in cluster I, and most of them have a Jaccard’s similarity coefficient above 0·80. Aglaonema commutatum originates from the Philippines and north‐eastern Celebes (Nicolson, 1969). Both Nicolson (1969) and Jervis (1980) believed that A. commutatum could actually be hybrid in origin because chromosome counts of A. commutatum cultivars are polyploid (2n = 120) (Jones, 1957; Nicolson, 1969). Aglaonema crispum is native to Mount Bulusan, Sorsogon Province, and Luzon, Philippines with a chromosome number of 2n = 60 (Nicolson, 1969). Aglaonema nitidum is more widely distributed from Southern Burma, Malaysia and Sumatra to north Borneo and possibly north‐eastern Celebes. However, there is no documented information about the chromosome number of A. nitidum. The locations where A. crispum, A. nitidum and A. commutatum are all in a close proximity, and the close genetic similarity among the three species (Table 3) and their related cultivars in cluster 1, lead us to believe that A. crispum and A. nitidum are probably progenitors of A. commutatum. Further studies with more species combined with cytological analyses are warranted to test the origin of A. commutatum.
Table 3.
Jaccard’s similarity coefficients among seven cultivars of A. commutatum, A. crispum, A. modestum, A. nitidum or A. pictum using six primer sets
| A. commutatum ‘Echo’ | A. commutatum ‘Trebuii’ | A. crispum ‘B.J. Freeman’ | A. modestum | A. nitidum ‘Curtisii’ | A. nitidum ‘Ernesto’s Favorite’ | A. pictum ‘Tricolor’ | |
| A. commutatum ‘Echo’ | – | 0·69 | 0·79 | 0·40 | 0·72 | 0·75 | 0·32 |
| A. commutatum ‘Trebuii’ | – | – | 0·74 | 0·38 | 0·67 | 0·63 | 0·33 |
| A. crispum ‘B.J. Freeman’ | – | – | – | 0·42 | 0·66 | 0·70 | 0·33 |
| A. modestum | – | – | – | – | 0·37 | 0·38 | 0·31 |
| A. nitidum ‘Curtisii’ | – | – | – | – | – | 0·85 | 0·31 |
| A. nitidum ‘Ernesto’s Favorite’ | – | – | – | – | – | – | 0·32 |
| A. pictum ‘Tricolor’ | – | – | – | – | – | – | – |
Implications for Aglaonema breeding
Clear separation of all 54 Aglaonema cultivars proves the sensitivity of AFLP markers for cultivar identification. This sensitivity is further illustrated by the fact that two cultivars selected from the same cross can be distinguished. For example, ‘J.G.’ and ‘1502’ were F1 progenies of the same cross between A. commutatum ‘Tricolor’ and A. × ‘Manila’. Due to their parental heterozygosities, F1 is actually a segregating generation. Selection can thus be initiated in the F1 generation, and desirable individuals can be reproduced by vegetative propagation. The AFLP analysis can also detect differences between somaclonal variants. Both ‘Moonlight Bay’ and ‘Diamond Bay’ are somaclonal variants selected from ‘Silver Bay’; Jaccard’s similarity coefficient between ‘Silver Bay’ and the two variants was 0·79. In addition to proving the discriminatory power of AFLP analysis, this separation may suggest that some somaclonal variants are actually significantly different genetically. Genetic differences between somaclonal variants and their parents have also been detected in Arabidopsis thaliana (L.) Heynh. (Polanco and Ruiz, 2002), Carya illinoinensis (Wangenh.) C. Koch (Vendrame et al., 2000), Dieffenbachia (Chen et al., 2003) and Quercus suber L. (Hornero et al., 2001) by AFLP analysis. Additionally, it was possible to identify unique fragments for four cultivars: one fragment for ‘Treubii’, two fragments for ‘714‐12’, ten for A. pictum ‘Tricolor’ and four for ‘XIV’.
Furthermore, cultivars with some similar physiological traits are also clustered. For example, one of the most important problems in Aglaonema production and interior use is chilling injury. ‘Emerald Star’ and ‘Stars’, two of the most chilling‐resistant cultivars (Chen et al., 2001), are clustered together in the dendrogram. Basal shoot formation is also an important trait for Aglaonema because a cultivar with more basal shoots will need fewer cuttings per container to produce a full appearance in the final product. Basal shoots can also be removed and used as propagules since Aglaonema is propagated mainly by shoot cuttings or division. Cultivars ‘277028’, ‘Emerald Bay’, ‘Golden Bay’, ‘Moonlight Bay’ and ‘257‐12’, all clustered together in the dendrogram, produce multiple shoots, requiring fewer cuttings per container in production. The correlation between AFLP markers and these desirable traits will help future selection of cultivars that are resistant to chilling or that produce multiple shoots.
However, with most hybrid cultivars gathered in cluster I with Jaccard’s similarity coefficients of more than 0·84, there must be a serious concern over the potential genetic vulnerability of Aglaonema cultivars to either diseases or pests because these hybrids are among the most popular and most commonly grown cultivars in production. This finding is unexpected because many of them are interspecific hybrids, and presumed to be genetically diverse. The AFLP analysis suggests that due to the natural hybridization among some species, parental materials used in interspecific hybridization are actually genetically rather similar. To avoid genetic vulnerability, parental materials with wide genetic diversity should be considered for future hybrid development.
As far as is known, this is the first molecular investigation of the genetic relationships of Aglaonema species and cultivars. It was possible to identify 54 Aglaonema cultivars based on polymorphism generated by six primer sets. The AFLP profiles and the clusters identified can be used as a basis for comparison of other Aglaonema species or cultivars. The genetic similarity among cultivars established can help future Aglaonema germplasm identification, conservation and new cultivar development.
ACKNOWLEDGEMENTS
We thank Agri‐Starts, Inc., Apopka, FL, Green Star Foliage, Inc., Apopka, FL, Stewart’s Greenhouse, Inc., Mt Dora, FL, and Twyford International, Inc., Apopka, FL for providing plant materials used in this study and Kelly Everitt for critically reading this manuscript. This research was supported in part by the Florida Agricultural Experiment Station, California Agricultural Experiment Station, and University of California Cooperative Extension.
Supplementary Material
Received: 24 March 2003;; Returned for revision: 11 September 2003; Accepted: 24 October 2003
References
- AggarwalRK, Shenoy VV, Ramadevi J, Rajkumar R, Singh L.2002. Molecular characterization of some Indian Basmati and other elite rice genotypes using fluorescent–AFLP. Theoretical and Applied Genetics 105: 680–690. [DOI] [PubMed] [Google Scholar]
- BaileyLH, Bailey EZ.1976.Hortus III. New York: Macmillon Publishing Co. [Google Scholar]
- BarcacciaG, Albertini E, Falcinelli M 1999. AFLP fingerprinting in Pelargonium peltatum: its development and potential in cultivar identification. Journal of Horticultural Science and Biotechnology 74: 243–250. [Google Scholar]
- BrownFB.2001.The amazing Aglaonema houseplant to the world. Valkaria: Valkaria Tropical Gardens. [Google Scholar]
- BrownNE.1885.Aglaonema acutispathum.Gardener’s Chronicle 24: 39. [Google Scholar]
- CarrJ, Xu M, Dudley JW, Korban SS.2003. AFLP analysis of genetic variability in New Guinea impatiens. Theoretical and Applied Genetics 106: 1509–1516. [DOI] [PubMed] [Google Scholar]
- ChenJ, Henny RJ, Norman DJ, Devanand PS, Chao CT.2004. Analysis of genetic relatedness of Dieffenbachia cultivars using AFLP markers. Journal of American Society for Horticultural Science 129: 81–87. [Google Scholar]
- ChenJ, Henny RJ, Caldwell RD, Robinson CA.2001.Aglaonema cultivar differences in resistance to chilling temperatures. Journal of Environmental Horticulture 19: 198–202. [Google Scholar]
- ChenJ, Henny RJ, McConnell DB.2002. Development of new foliage plant cultivars. In: Janick J, Whipkey A, eds. Trends in new crops and new uses Alexandra: American Society of Horticultural Science Press, 466–472. [Google Scholar]
- ElliottMS, Griffis JL, McConnell DB, Zettler FW.1998. A light and scanning electron microscope study of bent‐tip in Aglaonema ‘Silver Queen’. Proceeding of Florida State Horticultural Society 110: 107–110. [Google Scholar]
- EllisRP, McNicol JW, Baird E, Booth A, Lawrence P.1997. The use of AFLPs to examine genetic relatedness in barley. Molecular Breeding 3: 359–369. [Google Scholar]
- HennyRJ 1983. Inheritance of foliar variegation in three Aglaonema species. Journal of Heredity 74: 475–476. [Google Scholar]
- HennyRJ.1986. Single locus, multiallelic inheritance of foliar variegation in Aglaonema Journal of Heredity 77: 214–215. [Google Scholar]
- HennyRJ, Chen J.2001. ‘Golden Bay’ Aglaonema HortScience 36: 1142–1143. [Google Scholar]
- HennyRJ, Chen J.2003. Foliage plant cultivar development. Plant Breeding Review 21: 245–290. [Google Scholar]
- HennyRJ, Chen J, Norman DJ.2003. ‘Diamond Bay’ and ‘Emerald Bay’ Aglaonema HortScience 38: 1446–1447. [Google Scholar]
- HennyRJ, Poole RT, Conover CA.1992. ‘Silver Bay’ Aglaonema HortScience 27: 1238. [Google Scholar]
- HorneroJ, Martinez I, Celestino C, Gallego FJ, Torres V, Toribio M.2001 Early checking of genetic stability of cork oat somatic embryos by AFLP analysis. International Journal of Plant Science 162: 827–833. [Google Scholar]
- HuxleyA.1994.The New Royal Horticultural Society Dictionary of Gardening. London: Macmillon Press. [Google Scholar]
- JervisRN.1980.Chinese evergreens: Aglaonema grower’s notebook. Clearwater: R. Jervis. [Google Scholar]
- JonesGE.1957.Chromosome numbers and phylogenetic relationship in the Araceae. Ph.D. dissertation. University of Virginia, Charlottesville, VA. [Google Scholar]
- LohJP, Kiew R, Hay A, Kee A, Gan LH, Gan YY.2000. Intergeneric and interspecific relationships in Araceae tribe Caladieae and development of molecular markers using amplified fragment length polymorphism (AFLP). Annals of Botany 85: 371–378. [Google Scholar]
- LohJP, Kiew R, Kee A, Gan LH, Gan YY.1999. Amplified fragment length polymorphism (AFLP) provides molecular markers for the identification of Caladium bicolor cultivars. Annals of Botany 84: 155–161. [Google Scholar]
- MayoSJ, Bogner J, Boyce PC.1997.The genera of Araceae. London: Royal Botanic Gardens, Kew. [Google Scholar]
- MyburgAA, O’Malley D, Sederoff RR, Whetten R.2000. High‐throughput multiplexed AFLP analysis of interspecific hybrids of Eucalyptus trees species. In: Plant & Animal Genome VIII Conference Abstract Book, San Diego, USA, 544. [Google Scholar]
- NicolsonDH.1969. A revision of the genus Aglaonema (Araceae). Smithsonian Contributions to Botany 1: 1–69. [Google Scholar]
- PolancoC, Ruiz ML.2002. AFLP analysis of somaclonal variation in Arabidopsis thaliana regenerated plants. Plant Science 162: 817–824. [DOI] [PubMed] [Google Scholar]
- Qamaruz‐ZamanF, Fay MF, Parker JS, Chase MW.1998. The use of AFLP fingerprinting in conversation genetics: a case study of Orchis simia (Orchidaceae). Lindleyana 13: 125–133. [Google Scholar]
- RohlfFJ.2000.NTSYSpc numerical taxonomy and multivariate analysis system, version 2·1 user guide. New York: Exeter Software. [Google Scholar]
- SinghA, Negi MS, Rajagopal J, Bhatia S, Tomar UK, Srivastava PS, Lakshmikumaran M 1999. Assessment of genetic diversity in Azadirachta indica using AFLP markers. Theory and Applied Genetics 99: 272–279. [Google Scholar]
- SmithCN, Scarborough EF.1981. Status and development of foliage plant industries. In: Joiner J, ed. Foliage plant production Englewood Cliffs: Prentice‐Hall, 1–39. [Google Scholar]
- SneathPHA, Sokal RR.1973.Numerical taxonomy: the principles and practice of numerical classification. San Francisco: W.H. Freeman. [Google Scholar]
- TomkinsJP, Wood TC, Barnes LS, Westman A, Wing RA.2001. Evaluation of genetic variation in the daylily (Hemerocallis spp.) using AFLP markers. Theoretical and Applied Genetics 102: 489–496. [Google Scholar]
- VendrameWA, Kochert GD, Sparks D, Wetzstein.2000. Field performance and molecular evaluations of pecan trees regenerated from somatic embryogenic cultures. Journal of the American Society for Horticultural Science 125: 542–546. [Google Scholar]
- VosP, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kupier M, Zabeau M.1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research 23: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WongC, Kiew R, Loh JP, GanLH, Set O, Lee SK, Lum S, Gan YY.2001. Genetic diversity of the wild banana Musa acuminata Colla in Malaysia as evidenced by AFLP. Annals of Botany 88: 1017–1025. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.