Abstract
Maintenance and faithful transmission of genomic information depends on the efficient execution of numerous DNA replication, recombination, and repair pathways. Many of the enzymes that catalyze steps within these pathways require access to sequence information that is buried in the interior of the DNA double helix, which makes DNA unwinding an essential cellular reaction. The unwinding process is mediated by specialized molecular motors called DNA helicases that couple the chemical energy derived from nucleoside triphosphate hydrolysis to the otherwise non-spontaneous unwinding reaction. An impressive number of high-resolution helicase structures are now available that, together with equally important mechanistic studies, have begun to define the features that allow this class of enzymes to function as molecular motors. In this review, we explore the structural features within DNA helicases that are used to bind and unwind DNA. We focus in particular on “aromatic-rich loops” that allow some helicases to couple single-stranded DNA binding to ATP hydrolysis and “wedge/pin” elements that provide mechanical tools for DNA strand separation when connected to translocating motor domains.
Keywords: helicase, nucleic acid, molecular motor, aromatic-rich loops, pins, wedges
Many DNA replication, recombination, and repair reactions use single-stranded DNA (ssDNA) as templates for directing their genome copying and editing functions. However, since double-stranded DNA (dsDNA) is generally more stable than its single-stranded form under cellular conditions, a thermodynamic barrier must be overcome to allow genome maintenance enzymes access to ssDNA templates. DNA helicases provide the solution to this problem by acting as molecular motors that couple the energy derived from hydrolyzing phosphoanhydride bonds present in nucleoside triphosphates (NTPs) to DNA unwinding.1–7 Given their importance in genome biology, the mechanisms by which helicases carry out their core DNA unwinding functions have been an area of intense study since their discovery nearly 40 years ago.8 This research has produced physical models that account for the DNA unwinding functions of several helicases and explained how individual helicase families have adapted to fit the demands of diverse biological pathways. In this review, we survey what has been learned about DNA helicases using structural approaches. In particular, we examine specialized structural elements that allow a subset of DNA helicases to couple ssDNA binding to ATPase activities (aromatic-rich loops) and that mediate DNA unwinding when coupled to a translocation motor (wedges/pins).
General overview of helicases
In terms of their biological and biochemical functions, helicases can vary significantly.1–7 First, helicases typically act on either DNA or RNA, with only a few examples of enzymes acting on both. Second, helicases differ in the particular nucleic acids structures upon which they operate. For example, some DNA helicases preferentially act on blunt duplex DNA whereas others require ssDNA extensions (either 3′ or 5′) for loading and directional translocation. Third, the rates of helicase movement on nucleic acids (translocation) and the average number of base pairs unwound per helicase engagement event (processivity) can differ radically among helicases. Finally, many helicases function as components in multiprotein complexes in which the protein partners can alter helicase substrate targeting and unwinding efficiency. These differences likely reflect adaptations that have allowed the core activity of nucleic acid unwinding to evolve into diverse molecular motors with fine-tuned functions that suit a broad set of cellular needs.
Based upon sequence conservation, biochemical activities, and three-dimensional structures, helicases can be classified in a number of ways.1,4,9 An early classification system divided helicases into five different “superfamilies” based on conservation of a series of protein sequence motifs.4 Superfamilies 1 and 2 (SF1 and SF2) comprise the largest number of helicase families and members are involved in a wide array of cellular functions that require manipulation of DNA or RNA structures. SF3 helicases are found in RNA and DNA viruses. SF4 and SF5 are hexameric DNA helicases that function as replicative and transcription termination factors, respectively. A sixth family has also been proposed for helicases that belong to the AAA+ (ATPases associated with various cellular activities) class of proteins.1 Helicase superfamilies can also be subdivided into those that translocate along DNA and unwind in a 3′–5′ direction (“A” type, e.g., SF1A) or a 5′–3′ direction (“B” type, e.g., SF1B).1 Many recent comprehensive reviews of helicase superfamilies are available.1–7 Here we briefly introduce general features of SF1 and SF2 helicases as a backdrop for exploring specific structural elements found within subsets of helicases in greater detail.
SF1 and SF2 helicases can be identified based on evolutionary conservation of seven sequence motifs (I, Ia, II–VI) that are required for ATP binding/hydrolysis, nucleic acid binding, and/or translocation (Fig. 1).4 Within these, motifs I and II are the most highly conserved and are essential for ATP hydrolysis. Motif I (also known as the Walker A element or P-loop) binds to phosphate groups in ATP and contains an invariant Lys residue that helps to stabilize the transition state intermediate during ATP hydrolysis. Motif II (also known as the Walker B or DExx element) binds to a catalytic Mg2+ ion that also stabilizes the ATP hydrolysis intermediate and provides a residue side chain that acts as a general base to activate a water nucleophile for phosphoanhydride hydrolysis.3,12
Figure 1.
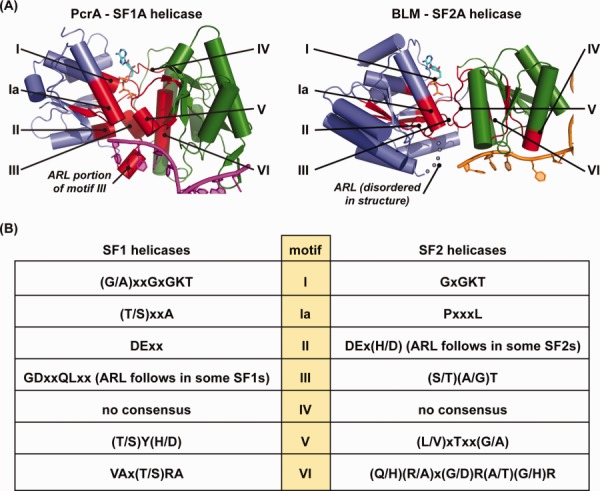
Overview of SF1 and SF2 helicase domains structures. (A) Comparison of the helicase domains from selected SF1 and SF2 helicases. (left) The helicase domain from the SF1A DNA helicase PcrA bound to partial-duplex DNA10 (PDB 3PJR) is shown with the N-terminal and C-terminal domains shown in blue and green, respectively. Helicase motifs are colored red and labeled I, Ia, II–VI. DNA is colored magenta. Domains outside of the core helicase domain have been removed for clarity. (right) The helicase domain from the SF2A DNA helicase BLM bound to partial-duplex DNA11 (PDB 4O3M) is shown with the N-terminal and C-terminal domains shown in blue and green, respectively. Helicase motifs are colored red and labeled I, Ia, II–VI. DNA is colored orange. Domains outside of the core helicase domain have been removed for clarity. (B) Helicase motifs from SF1 and SF2 helicases. Conserved helicase motifs I, Ia, and II–VI as defined by the Jankowsky lab9 are listed using the single-letter amino acid code.
Helicase motifs Ia and III–VI have greater sequence variation among the helicase superfamilies than that observed within motifs I and II. These motifs play important roles in ATP and nucleic acid binding and in coordinating ATPase function with translocation/unwinding reactions.4 Additional sequence motifs have been defined that confer important functional adaptations in specific subsets of helicases (e.g., TxGx, Q-motif, motif 4a, and TRG).13–16
Crystal structures of several SF1 and SF2 helicases have shown that these enzymes include a conserved core helicase domain that is comprised of two subdomains that share similarity with RecA ATPase/recombinase enzyme family (Fig. 1). This domain contains all of the helicase motifs, with I, Ia, II, and III found in the N-terminal RecA-like subdomain and IV–VI in the C-terminal subdomain (Fig. 1). The two subdomains abut one another to create an ATP binding/hydrolysis active site on one face of the domain and a nucleic acid binding surface along the opposite face.1 The composite nature of the ATPase active site helps to link ATPase cycle-dependent conformational changes within the helicase domain to translocation along nucleic acids. Helicases generally also encode additional domains that regulate substrate binding, provide additional enzymatic functions (e.g., nuclease domains), and/or mediate interactions with other protein partners.
A total of nine X-ray crystal structures of SF1 and SF2 helicases bound to partial-duplex DNA have been determined to date. The SF1 helicase/DNA complexes are G. stearothermophilus PcrA,10 E. coli UvrD,17 D. radiodurans UvrD,18 E. coli RecB in the RecBCD complex,19 and B. subtilis AddA in the AddAB complex,20,21 which are all SF1A enzymes. The SF2 helicase/DNA complexes are A. fulgidus Hel308,22,23 human RecQ1 (unpublished, PDB: 2WWY), and 2 human BLM structures (Ref.11 and unpublished, PDB: 4CGZ)), which are all SF2A enzymes. When these structures are superimposed using the structural similarity of their helicase domains, an intriguing pattern emerges that highlights how different groups of helicases associate with DNA. Two groups of SF1 arrangements are observed—one projects the duplex portion of the substrate orthogonally into the helicase domain for the closely related single-subunit PcrA and UvrD helicases and a second wraps the duplex “behind” the helicase domain in the multisubunit RecBCD and AddAB enzymes (Fig. 2). In contrast, the duplex projects more laterally into the helicase domain of the single-subunit SF2 enzymes RecQ1, BLM, and Hel308 (Fig. 2). From this comparison it is also apparent that both the helicase domain and nonhelicase domains of SF1 and SF2 enzymes are involved in binding to DNA and that the ssDNA along which these enzymes track is threaded across a common face of the helicase domain in both helicase superfamilies (Figs. 1 and 2). A second difference becomes apparent when the substrates from the helicase/DNA structures are compared (individual examples are shown in Fig. 3). The single-subunit SF1 helicases induce a ∼90–110° bend angle at the ss/dsDNA junction whereas the DNA bend angles of both the SF2 and multisubunit SF1 enzymes are only ∼20–60°. Larger bend angles could contribute to unwinding activity by wrenching apart the duplex as the enzyme translocates along ssDNA.17 One aspect that makes it difficult to draw functional conclusions from structural analysis of single-subunit helicases, however, is that many of these enzymes function optimally (or perhaps obligatorily) as homooligomers,24–31 but structural studies to date have captured only 1:1 helicase:DNA complexes. Thus alternative ss/dsDNA bend angles in higher order structures could be possible. It will be interesting to see whether future structures of DNA helicases bound to partial-duplex DNA conform to the general DNA binding arrangements established by the structures that are currently available.
Figure 2.
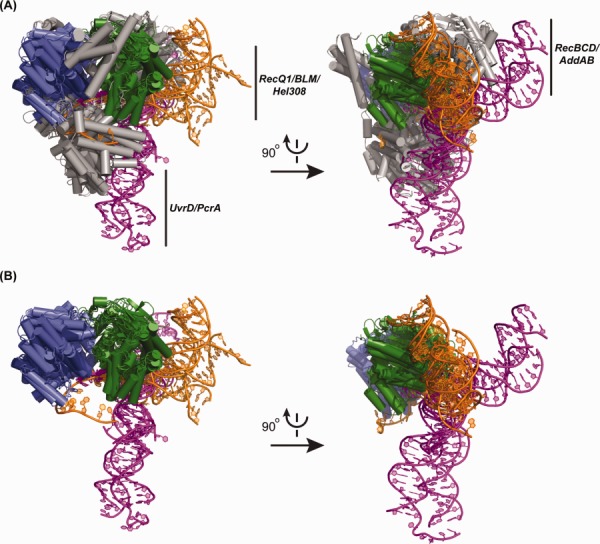
Overlay of substrate-bound SF1 and SF2 helicase structures. (A) Orthogonal views of crystal structures of SF1 enzymes [E. coli UvrD17 (PDB 2IS1), D. radiodurans UvrD18 (PDB 4C30), G. stearothermophilus PcrA10 (PDB 3PJR), E. coli RecB in the RecBCD complex19 (PDB 1W36), and B. subtilis AddA in the AddAB complex20,21 (PDB 3U44)] and SF2 enzymes [(A. fulgidus Hel30822 (PDB 2P6R), human RecQ1 (unpublished structure, PDB 2WWY), and two human BLMs11 (PDB 4O3M and unpublished structure, PDB 4CGZ)]. The helicase domains (blue = N-terminal subdomain and green = C-terminal subdomain) in each structure are aligned and non-helicase core domains are colored in grey. Only the helicase domains are shown for RecBCD and AddAB due to the large sizes of each complex. The DNA is colored in magenta for SF1/DNA complexes and in orange for SF2/DNA complexes. (B) The overlay from Figure 2(A) is shown with the non-helicase domain elements removed to better visualize the arrangement of DNA relative to the helicase domain.
Figure 3.
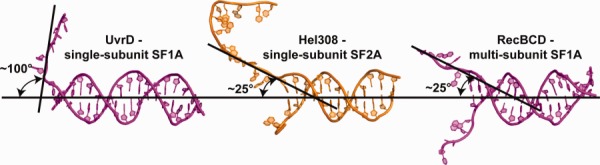
DNA bend angles in substrate-bound SF1 and SF2 helicase structures. Comparison of ss/ds bend angles of representative DNAs from single-subunit SF1A (UvrD, left), single-subunit SF2A (Hel308, middle), and multi-subunit SF1A (RecBCD, right) helicase/DNA complex crystal structures.
The remainder of this review focuses on two structural elements that have been found to be important for DNA binding and unwinding in SF1 and SF2 DNA helicases. The first is the aromatic-rich loop, or ARL, which is a sequence element embedded within the helicase domains of several SF1 and SF2 enzymes. ARLs directly contact ssDNA along which the helicases translocate and they function as coupling elements that link DNA binding/translocation and ATPase functions. The second element is the “wedge” or “pin,” which, when combined with a translocase motor domain, acts as a physical barrier to separate the two strands of duplex DNA. Wedge/pin elements have proven to be surprisingly diverse, ranging from simple β-hairpin elements projecting from the helicase domain to separate folded domains, some of which appear to be stabilized by bound metal ions.
Aromatic-rich loops as coupling motifs that link DNA binding and ATP hydrolysis
The conserved SF1 and SF2 helicase motifs mediate ATP binding and hydrolysis and convert the released chemical energy into the mechanical energy required for translocation and DNA unwinding. Precisely how this coupling is achieved appears to be different between SF1 and SF2 enzymes. In particular, the role of motif III is distinct in this regard. Motif III includes two functional elements in SF1 enzymes—an N-terminal portion that is weakly homologous between SF1 and SF2 enzymes and a C-terminal ARL that has only been found in SF1 helicases (Fig. 4). Co-crystal structures of SF1A helicases PcrA,10 UvrD,17,18 Rep,32 RecBCD,19 and AddAB20,21 with ssDNA or partial-duplex DNA demonstrate direct DNA binding by conserved aromatic (Trp or Phe) and electropositive (Arg) residues within the ARLs via stacking with ssDNA bases and gripping the phosphodiester backbone, respectively [Fig. 4(A)]. Similarly, co-crystal structures of SF1B helicases RecD233 and Dda34 with ssDNA also show direct ssDNA binding by a conserved Val residue in a manner that mimics that of the Trp/Phe from SF1A helicases [Fig. 4(B)]. The SF1 motif III sequence also has a highly conserved Gln residue N-terminal to the ARL that interacts with the γ-phosphate of ATP.10,17,18 In E. coli UvrD, a role for this Gln in positioning a water molecule that is bound by the presumed general base (Glu from motif II) has also been noted.17 This water is in a position to act as a nucleophile for in-line attack of ATP. Thus the SF1 motif III provides a direct intra-peptide link between DNA binding and ATP hydrolysis that can allow it to act as a “sensor” connecting nucleotide and DNA binding activities. Consistent with such a role, SF1 helicase variants in which selected residues within the N-terminal or ARL portions of motif III are altered display defective ATP hydrolysis, DNA binding, and/or helicase activities.38–41
Figure 4.
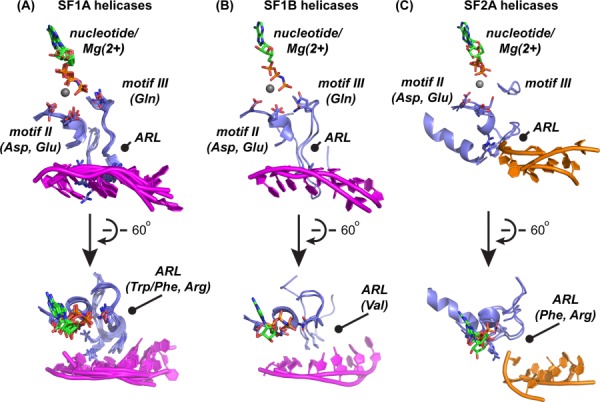
Comparison of ssDNA-binding aromatic-rich loops in DNA helicases. (A) Sixty degree rotated views of selected elements extracted from crystal structures of SF1A enzymes [E. coli UvrD17 (PDB 2IS4), D. radiodurans UvrD18 (PDB 4C30), G. stearothermophilus PcrA10 (PDB 3PJR), E. coli Rep32 (PDB 1UAA), E. coli RecB in the RecBCD complex19 (PDB 1W36), and B. subtilis AddA in the AddAB complex20,21 (PDB 3U44)]. Motifs II, III, and the ARL are labeled and key sidechains are shown in stick form. Color coding is the same as in Figure 1 (blue = segments from the N-terminal helicase subdomain; magenta = DNA from SF1 structures). (B) Sixty degree rotated views of selected elements extracted from crystal structures of SF1B enzymes [D. radiodutans RecD233 PDB 3GPL), and T4 Dda34 PDB 3UPU)]. Motifs II, III, and the ARL are labeled and key sidechains are shown in stick form. (C) Sixty degree rotated views of selected elements extracted from crystal structures of SF2A enzymes that have ARLs [human RecQ1 (unpublished structure, PDB 2WWY), and two human BLMs11 (PDB 4O3M and unpublished structure, PDB 4CGZ)]. Motifs II, III, and the ARL are labeled and key sidechains are shown in stick form. Color coding is the same as in Figure 1 (blue = segments from the N-terminal helicase subdomain; orange = DNA from SF2 structures). Hel308,22,23 which is also an SF2A enzyme, and XPD,35–37 the only SF2B enzyme of known structure, are omitted from this comparison since both lack apparent ARLs.
Motifs III also exist in SF2 helicases, however they are significantly shorter and lack the characteristic aromatic and positively charged residues that allow coordination of DNA binding and ATP hydrolysis in SF1 enzymes [Fig. 4(C)]. How then do SF2 helicases regulate ATP hydrolysis in a DNA-dependent manner? It appears that there may be multiple mechanisms that allow such coordination in SF2 helicases although, interestingly, at least two SF2A enzymes (RecQ and PriA) present ARLs on the same face of their helicase domains as the SF1 enzymes (Figs. 1 and 4).42–44 These ARLs are not part of motif III but are instead C-terminal to motif II in the primary sequence. Hel308,22,23 which is also an SF2A enzyme, and XPD,35–37 the only SF2B enzyme of known structure, both lack apparent ARLs.
Do SF2 helicase ARLs couple DNA binding to ATPase function even though they lack an apparent direct contact to ATP? This question has been examined in E. coli RecQ in which mutagenesis studies highlighted its similar coupling role to that of SF1 enzymes.43 E. coli RecQ variants with altered aromatic or basic ARL residues retain DNA-dependent ATPase activity, but require substantially higher DNA concentrations to stimulate ATPase functions and have greatly diminished helicase activity. One variant in which the conserved Phe from the ARL was altered had a strong increase in ATPase activity in the absence of DNA, indicating that the ARL in RecQ helps to suppress ATPase activity that is not coupled to DNA binding. Each of the RecQ variants retained similar ssDNA and ATP binding affinities to those observed in the wild-type enzyme, suggesting that the sequence changes selectively decouple DNA binding and ATPase activities.43 Structures of DNA-bound human RecQ proteins have not revealed the mechanism by which the ARL functions since the 3′ ssDNA extensions included in the substrates have been too short to reach the ARL [Fig. 4(C)]. However, our lab has recently determined the structure of a bacterial RecQ/DNA complex that shows direct ssDNA binding by the ARL (Manthei and Keck, submitted). In this structure, ssDNA binding stabilizes a conformation of the ARL that is distinct from that observed in the unbound structure. Complex formation also alters the ATPase active site in a manner that appears to poise the enzyme for ATP hydrolysis. Thus, similarly to the effect in SF1 helicases, DNA binding at the RecQ ARL appears to modulate the position of residues within the ATPase active site to regulate enzyme function. Whether similar effects will be observed in the recently identified ARL of PriA helicase and if additional ARLs are utilized in other DNA or RNA helicases remain open questions.
Pins and wedges: Structural aids to duplex DNA unwinding
Precisely how helicases unwind DNA remains an active area of research. However, a growing number of examples point to structural elements that, when coupled to a translocation motor, act as physical barriers that help plow apart the strands of dsDNA. These structures, commonly called “pins” or “wedges,” range from simple β-hairpins to more complicated domains (Fig. 5). The structures of SF1 DNA helicases Dda,34 RecBCD,19 RecD2,33 AddAB,20,21 PcrA,12 Rep,32 and UvrD17,18 and SF2 DNA helicases RecG,45 RecQ family,42,46,47 Hel308,22,23 NS3,22,48–51 XPD,35–37 and PriA44 all possess elements that have been proposed to act as pins or wedges. With the diversity of pin and wedge domains, protein primary structures have proven to be poor guides in predicting the positions of these unwinding elements. Such diversity seems essential given the broad range of substrates that helicases must process and the myriad biological processes in which helicases function.
Figure 5.
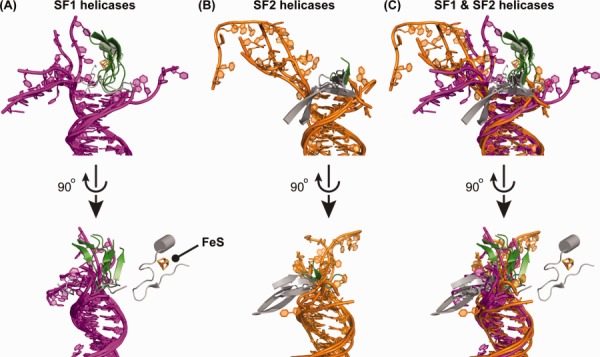
Pins/wedges in DNA helicases. (A) Orthogonal views of the SF1/DNA complexes. The alignment superimposes duplex regions of the DNA with the ss/dsDNA junction fixed among each of the substrates. Pin/wedge elements from each protein and DNA are colored as in Figure 1 (green = segments from the C-terminal helicase subdomain; grey = non-helicase domain elements; magenta = DNA from SF1 structures). An iron–sulfur cluster from AddAB is labeled. (B) Orthogonal views of the SF2/DNA complexes. Pin/wedge elements from each protein and DNA are colored as in Figure 1 (green = segments from the C-terminal helicase subdomain; grey = non-helicase domain elements; orange = DNA from SF2 structures). (C) Orthogonal views of overlayed DNA and pin/wedge elements from SF1/and SF2/DNA complexes.
Pins comprised of β-hairpin-like structures are the simplest and most commonly observed elements that are found at the ss/ds junction in known helicase/DNA complex structures. Often, pins use aromatic residues that base stack with duplex DNA at the ss/dsDNA junction but other residues can be used to cap the duplex region as well. These structures can have remarkable diversity even among closely related family members, varying greatly in terms of size, of their importance in helicase function, and in their sequence composition. For example, in E. coli RecQ the pin is very short and can be mutated without any apparent effect on DNA unwinding, indicating that it is not critical for function.42,47 In contrast, pins observed in three human RecQ proteins (RecQ1, BLM, and WRN) form elongated structures that project away from the protein to rest atop dsDNA [Fig. 5(B)] and mutations in pin residues that bind DNA eliminate unwinding activity.11,30,46,47 Interestingly, even among the pins in human RecQ proteins, variety is apparent—the dsDNA-interacting residues from pins in RecQ1, BLM, and WRN are Tyr, Asn, and a Phe-Met dipeptide, respectively. Moreover, the RecQ pins appear to be functionally diverse, with the RecQ1 pin playing a role in oligomerization as well.30
Structural studies of the closely related SF1A helicases UvrD and PcrA have provided the most detailed views of pin dynamics during DNA unwinding.10,17,18 The UvrD/PcrA pin is a 12-residue β-hairpin encoded within the helicase domain that binds at the ss/ds DNA junction point by buttressing the duplex DNA. A Tyr or Phe residue at the tip of the pin in UvrD or PcrA, respectively, binds to dsDNA through a base stacking interaction with the first base pair of the duplex.10,17,18 An outline of how UvrD couples ATPase function to unwinding at the pin has been derived from different nucleotide-bound UvrD structures.17 In this model, ATP binding leads to separation of a single base pair and the Tyr pin flips to a vertical position that is no longer base stacked with the duplex. ADP release allows the Tyr to resume its base stacking position but, due to translocation of the helicase domain, it does so with the next base pair. Mutation of the pin residue reduces, but does not abolish, DNA unwinding in UvrD17 or PcrA,39 indicating that although the pin/DNA interaction is important for helicase function, additional mechanisms can compensate for alterations within the pin. As pointed out earlier, the DNA bend angle introduced by UvrD or PcrA could be the driving force that accounts for this residual activity.
Wedges can form larger domains that act as physical unwinding elements in helicases. The locations of these domains vary based on the function of the helicase and these accessory domains often play multifunctional roles in the protein. RecG, a bacterial SF2A helicase that processes stalled replication forks to form Holliday junctions for subsequent repair and restart processes, was one of the first structures in which a wedge element was identified.45 The protein contains a canonical SF2 helicase domain as well as a separate wedge domain, which specifically binds at the junction of DNA arms. Aromatic residues within this domain are essential for this substrate recognition. Wedge domains are not necessarily essential for helicase function as a RecG wedge domain deletion still displays helicase activity, but with reduced substrate affinity.52,53 In addition to its ability to act as a physical unwinding element, the wedge domain is seen as a substrate specificity factor and a processivity factor. The domain by itself is capable of binding Holliday junctions with high affinity, but not replication forks.53,54 Strong binding of RecG to DNA junctions appears to allow for proper positioning of the helicase domains in unwinding of duplex DNA. Leading and lagging strands wrap around the wedge domain. Pulling the template strands across the wedge domain allows for unwinding of both leading and lagging strands at three way junctions and subsequent formation of the Holliday junction.45,53,55
Several recent examples of metal-containing domains have been suggested to function as wedges within XPD,56,57 AddAB,20,21 and PriA DNA helicases.44 These examples add helicases to the growing list of metalloproteins that catalyze nucleic acid metabolic reactions. Iron-sulfur (FeS) clusters form the metal cofactors in XPD and AddAB. These cluster are typically in 2Fe-2S, 3Fe-4S, 4Fe-4S, or 8Fe-7S arrangements where cysteines provide sulfur groups that coordinate the Fe atom. FeS clusters serve a variety of biological roles including electron transport, iron storage, protein stabilization, and oxidative stress sensors.58 The presence of FeS clusters in nucleic acid processing enzymes was first discovered in the DNA glycosylase endonuclease III from E. coli.59 Since then, FeS clusters have been found in human primase60–62 and several SF1 and SF2 helicases [XPD,35–37 FANCJ,63 ChlR1,64 DinG,65 and AddAB20].
FeS clusters appear to have multiple roles in DNA helicases including electrochemical functional modulation and stabilization in DNA binding and unwinding wedge domains. An example of the former is provided by S. acidocaldarius XPD, an SF2B DNA helicase with an ATP-dependent FeS redox potential that has been linked to a possible mechanism for selecting damaged DNA sites and for its cooperation with Endonuclease III.66,67 In combination with a second domain (called the “arch”), the FeS cluster in XPD is also thought to play a role in stabilizing the helicase’s unwinding wedge.35–37,56,57,68 The arch domain is a mixed αβ fold that is encoded between motifs II and III, forming an insertion in the first RecA-like subdomain of the helicase. The arch domain and FeS cluster abut one another with the FeS cluster-containing domain forming a wedge element that has been proposed to unwind DNA.56 The arch domain also folds over the first motor domain, creating a channel that is large enough to accommodate ssDNA, but not dsDNA. Mutations of the FeS cluster liganding residues lead to defects in helicase activity69 and removal of the FeS cluster causes dramatic changes in the crystal structure,37 supporting the role of the FeS cluster as a factor stabilizing the XPD enzyme.
In addition to XPD, the AddAB helicase-nuclease appears to rely on a wedge domain FeS cluster for DNA unwinding.20,21,70 Structural and functional analyses have shown that the FeS cluster is adjacent to the ss/ds junction of partially unwound DNA [Fig. 5(A)] and that it is essential for DNA unwinding. The portion of AddAB that coordinates the FeS cluster is in close proximity to an apparent pin residue from the protein, implying that the two elements may jointly function in DNA unwinding.20
The recent structure of the PriA helicase revealed the first example of a Zn2+-binding domain that appears to function as a DNA unwinding wedge.44 PriA is an SF2A DNA helicase that initiates the process of DNA replication restart in bacteria.71 In addition to the conserved helicase motifs, sequence comparisons of PriA have shown that the protein has a highly conserved series of 8 Cys residues in Cys-Xxx-Xxx-Cys motifs embedded between motifs IV and V. PriA variants with Cys sequence changes within this element retain ssDNA-dependent ATPase activity but have lost helicase/translocation activities that, in some cases, could be partially rescued with the addition of Zn2+.72 PriA Cys variants additionally abolish the ability of PriA to interact with another DNA replication restart protein (PriB), consistent with the Zn2+-binding motif playing roles both in DNA unwinding and in mediating protein-protein interactions.73 The recent PriA structure showed that the Zn2+-binding element forms a compact 40-residue fold that coordinates two Zn2+ ions via the conserved Cys residues.44 A β-hairpin within this Zn2+-binding domain has been proposed to serve as a DNA unwinding pin. Taken together, these data strongly suggest a role for the Zn2+-binding element in PriA unwinding mechanisms.
Conclusions and future perspectives
Helicases display a remarkable versatility in their DNA unwinding mechanisms. Their biochemical abilities include structure-specific nucleic acid binding and unwinding, directional translocation, and protein interactions to seed macromolecular protein complex formation. Subfamilies of helicases within each of the superfamilies have adapted the core helicase domain to produce enzymes that are capable of processing a wide range of nucleic acid structures and of functioning in diverse biological contexts. In terms of the DNA helicases reviewed here, the ssDNA binding and DNA unwinding components (ARLs and pins/wedges, respectively) show distinctions in their positions within helicases and their structures but appear to have adopted broadly similar roles across these enzymes. The features described herein represent an incomplete list of tools utilized by helicases to deal with the complicated nucleic acid structures they must face. These features are often not readily identifiable from the primary structures of helicases, making sustained structural efforts essential for determining the level of conservation of these features as well as for identifying new mechanistic features of helicases.
Acknowledgments
The authors thank Maria Spies, Kevin Raney, and Sarah Wessel for critical evaluation of the manuscript. JLK is cofounder of Replisoma, Inc.
References
- Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Ann Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- Ye J, Osborne AR, Groll M, Rapoport TA. RecA-like motor ATPases—lessons from structures. Biochim Biophys Acta. 2004;1659:1–18. doi: 10.1016/j.bbabio.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV. Helicases—amino-acid-sequence comparisons and structure-function-relationships. Curr Opin Struct Biol. 1993;3:419–429. [Google Scholar]
- Wu CG, Spies M. Overview: what are helicases? Adv Exp Med Biol. 2013;767:1–16. doi: 10.1007/978-1-4614-5037-5_1. [DOI] [PubMed] [Google Scholar]
- Beyer DC, Ghoneim MK, Spies M. Structure and mechanisms of SF2 DNA helicases. Adv Exp Med Biol. 2013;767:47–73. doi: 10.1007/978-1-4614-5037-5_3. [DOI] [PubMed] [Google Scholar]
- Raney KD, Byrd AK, Aarattuthodiyil S. Structure and mechanisms of SF1 DNA helicases. Adv Exp Med Biol. 2013;973:E1. doi: 10.1007/978-1-4614-5037-5_14. [DOI] [PubMed] [Google Scholar]
- Abdel-Monem M, Durwald H, Hoffmann-Berling H. Enzymic unwinding of DNA. 2. Chain separation by an ATP-dependent DNA unwinding enzyme. Eur J Biochem/FEBS. 1976;65:441–449. doi: 10.1111/j.1432-1033.1976.tb10359.x. [DOI] [PubMed] [Google Scholar]
- Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- Swan MK, Legris V, Tanner A, Reaper PM, Vial S, Bordas R, Pollard JR, Charlton PA, Golec JM, Bertrand JA. Structure of human Bloom’s syndrome helicase in complex with ADP and duplex DNA. Acta Cryst. 2014;D70:1465–1475. doi: 10.1107/S139900471400501X. [DOI] [PubMed] [Google Scholar]
- Subramanya HS, Bird LE, Brannigan JA, Wigley DB. Crystal structure of a DExx box DNA helicase. Nature. 1996;384:379–383. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner NK, Cordin O, Banroques J, Doere M, Linder P. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol Cell. 2003;11:127–138. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- Korolev S, Yao N, Lohman TM, Weber PC, Waksman G. Comparisons between the structures of HCV and Rep helicases reveal structural similarities between SF1 and SF2 super-families of helicases. Protein Sci. 1998;7:605–610. doi: 10.1002/pro.5560070309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdi AA, Briggs GS, Sharples GJ, Wen Q, Lloyd RG. A model for dsDNA translocation revealed by a structural motif common to RecG and Mfd proteins. EMBO J. 2003;22:724–734. doi: 10.1093/emboj/cdg043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter M, Acajjaoui S, McSweeney S, Timmins J. Structural and mechanistic insight into DNA unwinding by Deinococcus radiodurans UvrD. PloS One. 2013;8:e77364. doi: 10.1371/journal.pone.0077364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- Saikrishnan K, Yeeles JT, Gilhooly NS, Krajewski WW, Dillingham MS, Wigley DB. Insights into Chi recognition from the structure of an AddAB-type helicase-nuclease complex. EMBO J. 2012;31:1568–1578. doi: 10.1038/emboj.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajewski WW, Fu X, Wilkinson M, Cronin NB, Dillingham MS, Wigley DB. Structural basis for translocation by AddAB helicase-nuclease and its arrest at chi sites. Nature. 2014;508:416–419. doi: 10.1038/nature13037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol. 2007;14:647–652. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- Richards JD, Johnson KA, Liu H, McRobbie AM, McMahon S, Oke M, Carter L, Naismith JH, White MF. Structure of the DNA repair helicase hel308 reveals DNA binding and autoinhibitory domains. J Biol Chem. 2008;283:5118–5126. doi: 10.1074/jbc.M707548200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali JA, Maluf NK, Lohman TM. An oligomeric form of E. coli UvrD is required for optimal helicase activity. J Mol Biol. 1999;293:815–834. doi: 10.1006/jmbi.1999.3185. [DOI] [PubMed] [Google Scholar]
- Byrd AK, Raney KD. Protein displacement by an assembly of helicase molecules aligned along single-stranded DNA. Nat Struct Mol Biol. 2004;11:531–538. doi: 10.1038/nsmb774. [DOI] [PubMed] [Google Scholar]
- Byrd AK, Raney KD. Increasing the length of the single-stranded overhang enhances unwinding of duplex DNA by bacteriophage T4 Dda helicase. Biochemistry. 2005;44:12990–12997. doi: 10.1021/bi050703z. [DOI] [PubMed] [Google Scholar]
- Jia H, Korolev S, Niedziela-Majka A, Maluf NK, Gauss GH, Myong S, Ha T, Waksman G, Lohman TM. Rotations of the 2B sub-domain of E. coli UvrD helicase/translocase coupled to nucleotide and DNA binding. J Mol Biol. 2011;411:633–648. doi: 10.1016/j.jmb.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maluf NK, Fischer CJ, Lohman TM. A dimer of Escherichia coli UvrD is the active form of the helicase in vitro. J Mol Biol. 2003;325:913–935. doi: 10.1016/s0022-2836(02)01277-9. [DOI] [PubMed] [Google Scholar]
- Brendza KM, Cheng W, Fischer CJ, Chesnik MA, Niedziela-Majka A, Lohman TM. Autoinhibition of Escherichia coli Rep monomer helicase activity by its 2B subdomain. Proc Natl Acad Sci USA. 2005;102:10076–10081. doi: 10.1073/pnas.0502886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucic B, Zhang Y, King O, Mendoza-Maldonado R, Berti M, Niesen FH, Burgess-Brown NA, Pike AC, Cooper CD, Gileadi O, Vindigni A. A prominent beta-hairpin structure in the winged-helix domain of RECQ1 is required for DNA unwinding and oligomer formation. Nucleic Acids Res. 2011;39:1703–1717. doi: 10.1093/nar/gkq1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton SA, Tolun G, Kamath-Loeb AS, Loeb LA, Griffith JD. The Werner syndrome protein binds replication fork and holliday junction DNAs as an oligomer. J Biol Chem. 2008;283:24478–24483. doi: 10.1074/jbc.M803370200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- Saikrishnan K, Powell B, Cook NJ, Webb MR, Wigley DB. Mechanistic basis of 5′-3′ translocation in SF1B helicases. Cell. 2009;137:849–859. doi: 10.1016/j.cell.2009.03.036. [DOI] [PubMed] [Google Scholar]
- He X, Byrd AK, Yun MK, Pemble CWT, Harrison D, Yeruva L, Dahl C, Kreuzer KN, Raney KD, White SW. The T4 phage SF1B helicase Dda is structurally optimized to perform DNA strand separation. Structure. 2012;20:1189–1200. doi: 10.1016/j.str.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Rudolf J, Johnson KA, McMahon SA, Oke M, Carter L, McRobbie AM, Brown SE, Naismith JH, White MF. Structure of the DNA repair helicase XPD. Cell. 2008;133:801–812. doi: 10.1016/j.cell.2008.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolski SC, Kuper J, Hanzelmann P, Truglio JJ, Croteau DL, Van Houten B, Kisker C. Crystal structure of the FeS cluster-containing nucleotide excision repair helicase XPD. PLoS Biol. 2008;6:e149. doi: 10.1371/journal.pbio.0060149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Fuss JO, Cheng QJ, Arvai AS, Hammel M, Roberts VA, Cooper PK, Tainer JA. XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell. 2008;133:789–800. doi: 10.1016/j.cell.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillingham MS, Soultanas P, Wigley DB. Site-directed mutagenesis of motif III in PcrA helicase reveals a role in coupling ATP hydrolysis to strand separation. Nucleic Acids Res. 1999;27:3310–3317. doi: 10.1093/nar/27.16.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillingham MS, Soultanas P, Wiley P, Webb MR, Wigley DB. Defining the roles of individual residues in the single-stranded DNA binding site of PcrA helicase. Proc Natl Acad Sci USA. 2001;98:8381–8387. doi: 10.1073/pnas.131009598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh RM, Matson SW. A partially functional DNA helicase II mutant defective in forming stable binary complexes with ATP and DNA—a role for helicase motif III. J Biol Chem. 1996;271:25360–25368. doi: 10.1074/jbc.271.41.25360. [DOI] [PubMed] [Google Scholar]
- Brosh RM, Matson SW. A point mutation in Escherichia coli DNA helicase II renders the enzyme nonfunctional in two DNA repair pathways—evidence for initiation of unwinding from a nick in vivo. J Biol Chem. 1997;272:572–579. doi: 10.1074/jbc.272.1.572. [DOI] [PubMed] [Google Scholar]
- Bernstein DA, Zittel MC, Keck JL. High-resolution structure of the E. coli RecQ helicase catalytic core. EMBO J. 2003;22:4910–4921. doi: 10.1093/emboj/cdg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zittel MC, Keck JL. Coupling DNA-binding and ATP hydrolysis in Escherichia coli RecQ: role of a highly conserved aromatic-rich sequence. Nucleic Acids Res. 2005;33:6982–6991. doi: 10.1093/nar/gki999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya B, George NP, Thurmes TM, Zhou R, Jani N, Wessel SR, Sandler SJ, Ha T, Keck JL. Structural mechanisms of PriA-mediated DNA replication restart. Proc Natl Acad Sci USA. 2014;111:1373–1378. doi: 10.1073/pnas.1318001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR, Scaife S, Wigley DB. Structural analysis of DNA replication fork reversal by RecG. Cell. 2001;107:79–89. doi: 10.1016/s0092-8674(01)00501-3. [DOI] [PubMed] [Google Scholar]
- Kitano K, Kim SY, Hakoshima T. Structural basis for DNA strand separation by the unconventional winged-helix domain of RecQ helicase WRN. Structure. 2010;18:177–187. doi: 10.1016/j.str.2009.12.011. [DOI] [PubMed] [Google Scholar]
- Pike AC, Shrestha B, Popuri V, Burgess-Brown N, Muzzolini L, Costantini S, Vindigni A, Gileadi O. Structure of the human RECQ1 helicase reveals a putative strand-separation pin. Proc Natl Acad Sci USA. 2009;106:1039–1044. doi: 10.1073/pnas.0806908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Rice CM. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc Natl Acad Sci USA. 2010;107:521–528. doi: 10.1073/pnas.0913380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam AM, Keeney D, Frick DN. Two novel conserved motifs in the hepatitis C virus NS3 protein critical for helicase action. J Biol Chem. 2003;278:44514–44524. doi: 10.1074/jbc.M306444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JL, Morgenstern KA, Griffith JP, Dwyer MD, Thomson JA, Murcko MA, Lin C, Caron PR. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- Mackintosh SG, Lu JZ, Jordan JB, Harrison MK, Sikora B, Sharma SD, Cameron CE, Raney KD, Sakon J. Structural and biological identification of residues on the surface of NS3 helicase required for optimal replication of the hepatitis C virus. J Biol Chem. 2006;281:3528–3535. doi: 10.1074/jbc.M512100200. [DOI] [PubMed] [Google Scholar]
- Briggs GS, Mahdi AA, Wen Q, Lloyd RG. DNA binding by the substrate specificity (wedge) domain of RecG helicase suggests a role in processivity. J Biol Chem. 2005;280:13921–13927. doi: 10.1074/jbc.M412054200. [DOI] [PubMed] [Google Scholar]
- McGlynn P, Lloyd RG. Rescue of stalled replication forks by RecG: simultaneous translocation on the leading and lagging strand templates supports an active DNA unwinding model of fork reversal and Holliday junction formation. Proc Natl Acad Sci USA. 2001;98:8227–8234. doi: 10.1073/pnas.111008698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn P, Lloyd RG. RecG helicase activity at three- and four-strand DNA structures. Nucleic Acids Res. 1999;27:3049–3056. doi: 10.1093/nar/27.15.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn P, Lloyd RG, Marians KJ. Formation of Holliday junctions by regression of nascent DNA in intermediates containing stalled replication forks: RecG stimulates regression even when the DNA is negatively supercoiled. Proc Natl Acad Sci USA. 2001;98:8235–8240. doi: 10.1073/pnas.121007798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh RA, Honda M, Leesley H, Thomas A, Lin Y, Nilges MJ, Cann IK, Spies M. The iron-containing domain is essential in Rad3 helicases for coupling of ATP hydrolysis to DNA translocation and for targeting the helicase to the single-stranded DNA-double-stranded DNA junction. J Biol Chem. 2008;283:1732–1743. doi: 10.1074/jbc.M707064200. [DOI] [PubMed] [Google Scholar]
- Pugh RA, Wu CG, Spies M. Regulation of translocation polarity by helicase domain 1 in SF2B helicases. EMBO J. 2012;31:503–514. doi: 10.1038/emboj.2011.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Ann Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- Cunningham RP, Asahara H, Bank JF, Scholes CP, Salerno JC, Surerus K, Munck E, McCracken J, Peisach J, Emptage MH. Endonuclease III is an iron-sulfur protein. Biochemistry. 1989;28:4450–4455. doi: 10.1021/bi00436a049. [DOI] [PubMed] [Google Scholar]
- Klinge S, Hirst J, Maman JD, Krude T, Pellegrini L. An iron-sulfur domain of the eukaryotic primase is essential for RNA primer synthesis. Nat Struct Mol Biol. 2007;14:875–877. doi: 10.1038/nsmb1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner BE, Huang H, Dattilo BM, Nilges MJ, Fanning E, Chazin WJ. An iron-sulfur cluster in the C-terminal domain of the p58 subunit of human DNA primase. J Biol Chem. 2007;282:33444–33451. doi: 10.1074/jbc.M705826200. [DOI] [PubMed] [Google Scholar]
- Vaithiyalingam S, Warren EM, Eichman BF, Chazin WJ. Insights into eukaryotic DNA priming from the structure and functional interactions of the 4Fe-4S cluster domain of human DNA primase. Proc Natl Acad Sci USA. 2010;107:13684–13689. doi: 10.1073/pnas.1002009107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Sommers JA, Suhasini AN, Leonard T, Deakyne JS, Mazin AV, Shin-Ya K, Kitao H, Brosh RM., Jr Fanconi anemia group J mutation abolishes its DNA repair function by uncoupling DNA translocation from helicase activity or disruption of protein-DNA complexes. Blood. 2010;116:3780–3791. doi: 10.1182/blood-2009-11-256016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung I, Schertzer M, Rose A, Lansdorp PM. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat Gen. 2002;31:405–409. doi: 10.1038/ng928. [DOI] [PubMed] [Google Scholar]
- Ren B, Duan X, Ding H. Redox control of the DNA damage-inducible protein DinG helicase activity via its iron-sulfur cluster. J Biol Chem. 2009;284:4829–4835. doi: 10.1074/jbc.M807943200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mui TP, Fuss JO, Ishida JP, Tainer JA, Barton JK. ATP-stimulated, DNA-mediated redox signaling by XPD, a DNA repair and transcription helicase. J Am Chem Soc. 2011;133:16378–16381. doi: 10.1021/ja207222t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontz PA, Mui TP, Fuss JO, Tainer JA, Barton JK. DNA charge transport as a first step in coordinating the detection of lesions by repair proteins. Proc Natl Acad Sci USA. 2012;109:1856–1861. doi: 10.1073/pnas.1120063109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuper J, Wolski SC, Michels G, Kisker C. Functional and structural studies of the nucleotide excision repair helicase XPD suggest a polarity for DNA translocation. EMBO J. 2012;31:494–502. doi: 10.1038/emboj.2011.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf J, Makrantoni V, Ingledew WJ, Stark MJ, White MF. The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Mol Cell. 2006;23:801–808. doi: 10.1016/j.molcel.2006.07.019. [DOI] [PubMed] [Google Scholar]
- Yeeles JT, Cammack R, Dillingham MS. An iron-sulfur cluster is essential for the binding of broken DNA by AddAB-type helicase-nucleases. J Biol Chem. 2009;284:7746–7755. doi: 10.1074/jbc.M808526200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabbai CB, Marians KJ. Recruitment to stalled replication forks of the PriA DNA helicase and replisome-loading activities is essential for survival. DNA Repair. 2010;9:202–209. doi: 10.1016/j.dnarep.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavitz KH, Marians KJ. Helicase-deficient cysteine to glycine substitution mutants of Escherichia coli replication protein PriA retain single-stranded DNA-dependent ATPase activity. Zn2+ stimulation of mutant PriA helicase and primosome assembly activities. J Biol Chem. 1993;268:4337–4346. [PubMed] [Google Scholar]
- Liu J, Nurse P, Marians KJ. The ordered assembly of the phiX174-type primosome. III. PriB facilitates complex formation between PriA and DnaT. J Biol Chem. 1996;271:15656–15661. doi: 10.1074/jbc.271.26.15656. [DOI] [PubMed] [Google Scholar]


