Abstract
Filoviruses are the causative agents of a severe and often fatal hemorrhagic fever with repeated outbreaks in Africa. They are negative sense single stranded enveloped viruses that can cross species barriers from its natural host bats to primates including humans. The small size of the genome poses limits to viral adaption, which may be partially overcome by conformational plasticity. Here we review the different conformational states of the Ebola virus (EBOV) matrix protein VP40 that range from monomers, to dimers, hexamers, and RNA-bound octamers. This conformational plasticity that is required for the viral life cycle poses a unique opportunity for development of VP40 specific drugs. Furthermore, we compare the structure to homologous matrix protein structures from Paramyxoviruses and Bornaviruses and we predict that they do not only share the fold but also the conformational flexibility of EBOV VP40.
Keywords: Ebola virus, VP40, budding, assembly
Introduction
Ebola virus (EBOV) and Marburg virus constitute the two genera of the family Filoviridae. Marburg virus is represented by a single species and EBOV by five species.1 The natural reservoirs of filoviruses are a number of different bat species and human transmission is thought to occur via animal hosts including primates.1 In humans EBOV infections cause a severe hemorrhagic fever, which is mostly lethal.2 Filoviruses are membrane enveloped filamentous viruses that contain a negative sense single stranded RNA genome grouping them together with the Paramyxoviridae, Rhabdoviridae, and Bornaviridae to the order Mononegavirales. Their genome encodes five nucleocapsid-associated proteins, the nucleoprotein NP, VP35, VP30, VP24, and the RNA-dependent RNA polymerase L, in addition to the matrix protein VP40 and the glycoprotein GP. Cryo electron microscopy reconstruction of the nucleocapsid core composed of NP, VP35, and VP24 revealed helical structures containing the viral genome,3,4 which are structurally similar to other negative strand RNA virus nucleocapsids.5–7 The helical nucleocapsid complex associates with VP30,8 which is structurally similar to M2-1 of paramyxoviruses and may thus have a similar function as transcription antiterminator.9–11 Finally the polymerase L completes the nucleocapsid complex and transcription and replication are likely to resemble those of other negative strand RNA viruses.12 VP35 physically links the nucleocapsid complex to the matrix protein VP4013,14 which is the most abundant viral protein that polymerizes underneath the viral membrane.15 VP40 plays an essential role in virus budding and expression of VP40 is sufficient to form filamentous virus-like particles (VLP).16,17 VP40 also recruits the nucleocapsid and the glycoprotein GP to the plasma membrane for assembly and budding.18–20 The glycoprotein GP mediates entry into target cells via the endo-lysosomal pathway,21,22 which is mediated by extensive conformational rearrangements of GP.23,24
Transport of Viral Proteins
In order to orchestrate assembly all components need to concentrate at the budding site. Nucleocapsids are assembled within perinuclear inclusions, which are sites of virus replication.25 Marburg virus nucleocapsids are transported along actin filaments and once they reach the plasma membrane the nucleocapsid associates with VP40 and is transferred to filopodia,26 which constitute the preferred site of Marburg virus budding.27 GP reaches the plasma membrane via the secretory pathway and concentrates at sites of VP40 accumulation.28 Marburg virus VP40 is recruited to the plasma membrane via the retrograde endosomal transport29 and EBOV VP40 was shown to move along actin filaments.30 Consistent with the above data, imaging of EBOV replication demonstrated that NP, VP35, VP30, VP24, and VP40 accumulate in cytoplasmic inclusions, which enlarge near the nucleus and are subsequently transferred to the plasma membrane.31 Budding of Marburg virus occurs via a lateral interaction of the nucleocapsid with the plasma membrane in concert with VP40 and GP. Membrane wrapping then starts at one end of the nucleocapsid and proceeds to a final envelopment and virus release via membrane scission.32
VP40 Recruits the ESCRT Machinery
Final steps in budding depend on endosomal sorting complexes required for transport (ESCRTs), which are recruited to viral budding sites via late domains present in viral structural proteins.33–36 EBOV VP40 contains two overlapping late domains and Marburg virus has the same motifs present in VP40 and NP.37–39 EBOV VP40 interacts with Tsg101 via its PTAP motif in vivo40 consistent with the presence of Tsg101 in VP40 containing VLPs.41 The second late domain was first shown to bind to a WW domain from the yeast E3 ligase Rsp5 (Nedd4 homologue)42 and later to the WW domain 3 from human Nedd4 in vitro.43 Notably dominant negative mutants of Nedd4 inhibit budding of VP40 containing VLPs44 and dominant negative VPS4 inhibits Marburg virus release.45 This is consistent with the current model that stipulates that late domains present in viral structural proteins serve to provide access to ESCRT-III and the VPS4 complex, which constitute the membrane fission machinery. ESCRT-III is thought to assemble into filamentous helical structures within the neck of a budding virion, constrict the membrane neck via the formation of dome-like structures46,47 and induce scission in concert with the VPS4 ATPase.36,48 However, other factors may be able to substitute for ESCRTs during filovirus budding, because late domain mutations do not completely inhibit budding.45,49
Different Conformations of the Matrix Protein VP40
The matrix protein VP40 is the major structural protein15 and its expression in eukaryotic cells suffices to release virus-like particles (VLPs) with filamentous morphology. However, VP40 VLPs have variable diameters from 40 to 80 nm, compared to the more or less consistent 80 nm diameter of infectious virus.16–18,42,50–53 The crystal structure of VP40 revealed two structurally related N- and C-terminal beta sandwich domains (NTD and CTD), which pack against each other in a closed conformation54 [Fig. 1(A)]. The structure lacks the first 40 amino acids (30 residues had been removed for crystallization purposes). However this N-terminal region contributes to the stability of VP40; it might reach over to the CTD because its presence increases the protease resistance of the CTD and protects it from trypsin cleavage at position 212 in vitro.55 Removal of the complete CTD, the last seven residues of CTD and artificial destabilization of the NTD–CTD interface by urea leads to spontaneous oligomerization via the NTD demonstrating the metastable interaction between NTD and CTD.55,56 The oligomeric states observed in vitro are hexameric and octameric ring-like structures with dimeric NTD as the building block.55–57 The CTD, which must be linked flexibly to the NTD in these oligomers, has been implicated in membrane association.55,56 There are two regions on the CTD which have been suggested to be important for membrane interaction. One is part of a basic patch present on the disordered CTD loop connecting α helix 7 and β strand 11 [Fig. 1(B), patch 2].Deletion of this loop or mutations within this region (K274E, K275E) inhibited VLP release from cells.58 In contrast, another study implicated a CTD hydrophobic surface composed of residues Leu213, Ile293, Leu295, and Val298 at the opposite site of the CTD [Fig. 1(B), patch 1] as being critical for plasma membrane localization, VP40 oligomerization, and viral particle egress.59 The same region was shown to penetrate into the plasma membrane and was shown to be important for liposome tubulation and for VP40-induced vesicle budding from giant unilamelar vesicles.60 Furthermore mutagenesis within this region affected the intracellular localization of VP40 and caused a reduction or a complete block of VLP release.61 Due to the distant locations of patches 1 and 2 it is unlikely that both regions are directly implicated in membrane interaction. Therefore the exact orientation of VP40 CTD on membranes needs further investigation.
Figure 1.
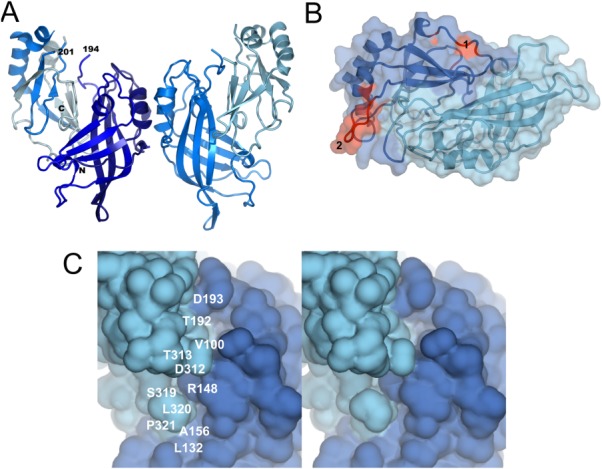
Crystal structure of VP40. (A) Structure of dimeric VP40 (pdb code 1ES6); The chain direction from N- to C-terminus is colored dark to light blue. Dimerization in solution has been confirmed.58 (B) Close-up of two proposed membrane interaction motifs located at two opposing sites of the CTD. The hydrophobic surface comprising residues Leu213, Ile293, Leu295, and Val298 is indicated as patch 1 and the loop region (disordered in the crystal structure and modeled as a coil) comprising residues K274 and K275 as patch 2. Both regions shown in red have been implicated in membrane interaction. (C) The metastable association of the NTD and CTD generates two potential drug binding pockets. Stereo image of the NTD–CTD interface. Two potential drug binding pockets (residues Leu132, Ala156, Arg148, Ser319, L320, Pro321; and residues Val100, Thr192, Asp193, Asp312, and Thr313) are indicated. Binding of small molecules within these pockets could stabilize the closed conformation and thus prevent the formation of oligomeric VP40 that is required for the Ebola virus life cycle. An interactive view is available in the electronic version of the article.
VP40 has been initially shown to be monomeric in solution based on chemical cross linking experiments performed at pH 8.8 and protein concentrations of < 2 mg/mL,55 while size exclusion chromatography and multiangle light scattering revealed dimers in solution at protein concentrations of 2 mg/mL58 and mutagenesis of the dimer interface demonstrated its relevance for virus assembly.58 The same authors showed that VP40 dimers assemble via CTD to CTD interactions into linear hexamers and they suggest that such hexamers form the building block for assembly. These hexamers would expose alternating CTDs towards the viral membrane and towards the nucleocapsid thus potentially establishing a firm link between the nucleocapsid and the membrane.58 A similar arrangement exposing the CTDs alternating above and below a ring structure has been determined for hexameric VP40 ring structures assembled in vitro.56 Until now no biological function has been assigned to these VP40 hexameric ring structures. Electron tomography analysis of EBOVs suggested that VP40 is arranged as a regular lattice with a 5 nm spacing, which was suggested to correspond to the hexameric assembly3,62 [Fig. 2(A)]. In contrast other electron microscopy studies on Marburg virus revealed only local regularity within an inner and outer layer of a VP40 lattice.14
Figure 2.
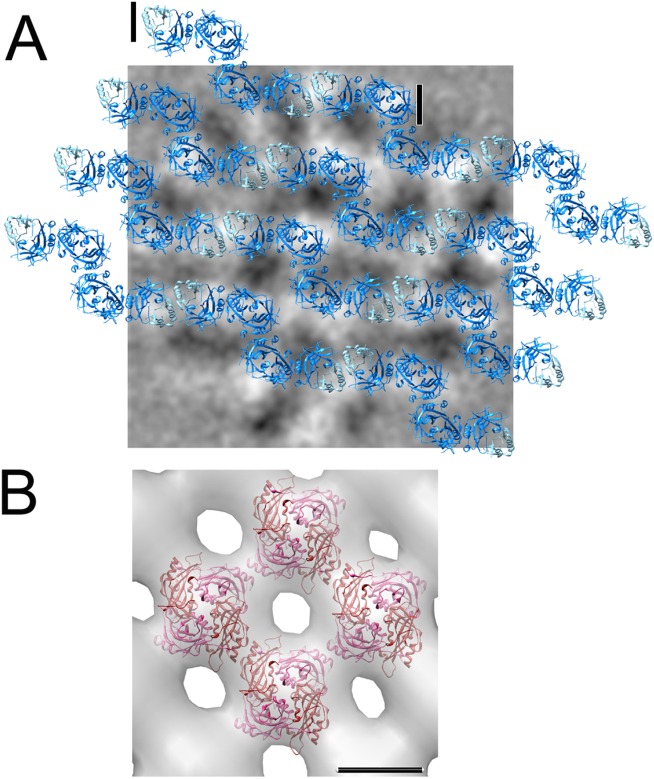
Structural organization of filovirus and paramyxovirus matrix proteins in virions. (A) VP40 filaments derived from the assembly of VP40 hexamers are modeled to scale onto 2D averages of virion tomograms. Black/white has been inverted for clarity in these images; white now corresponds to VP40 density. The hexamer unit as defined by the crystal structure is indicated by two black lines.3,58 (B) Cryo electron microscopy map of the matrix protein layer of Newcastle disease virus with the matrix protein dimer docked into the density,69 the scale bar, 5 nm.
Dissociation of the CTD leads to an octameric ring structure that specifically binds the short RNA sequence 5′-UGA-3′. The RNA stabilizes the ring structures and confers SDS resistance to the octamer57,63 [Fig. 3(A,B)]. Structural comparison of the NTD in complex with RNA and of the NTD associated with the CTD in the closed conformation revealed that the N-terminal 69 residues need to move out of the way to make the dimerization interface within the octamer accessible [Fig. 3(C)] Accordingly this region was disordered in the crystal structure. Furthermore the tip of β1 and β2 change conformation and interact with a neighboring molecule within the octamer [Fig. 3(A)]. No VP40 octamer formation has been observed in vitro in the absence of RNA.57 This is in contrast to an RNA-free octamer crystal structure.58 However, these crystals formed after proteolysis and therefore additional RNAse contamination could have removed the RNA; although Bornholdt et al.58 report oligomer formation of a mutant that can no longer bind RNA, such oligomerization is likely dependent on the relatively high protein concentration used in the analysis. In contrast VP40 octamers have been detected upon VP40 expression in mammalian cells and in infected cells as SDS-resistant oligomers detected by western blot analysis.53,63 Because we did not detect any octamer formation at low protein concentrations in the absence of RNA,57 we suggest that specific RNA binding drives octamer formation under physiological conditions. In addition to the N-terminus of NTD, the CTD also has to dissociate in order to facilitate octamer formation. A flexible linker can position the CTDs at the side, above or below the ring structure. Because the CTD binds to membranes, VP40 octamers may also function at membranes. EBOV replication was shown to occur in inclusions at the perinuclear region25,31 and the activity of VP40 octamers might be immobilized there via CTD membrane interactions. Alternatively, the CTD might associate with other viral or cellular factors required to exert its regulatory function during viral transcription.58 The function of VP40 octamers is likely limited to transcription control because octamers are not present in virus particles63 and VP40 mutants that no longer bind RNA can still form VLPs but do not support EBOV replication. The VP40 RNA binding activity is essential for the EBOV life cycle53 and we hypothesize that the sequence-specific recognition of RNA is linked to the recognition of viral RNA.
Figure 3.
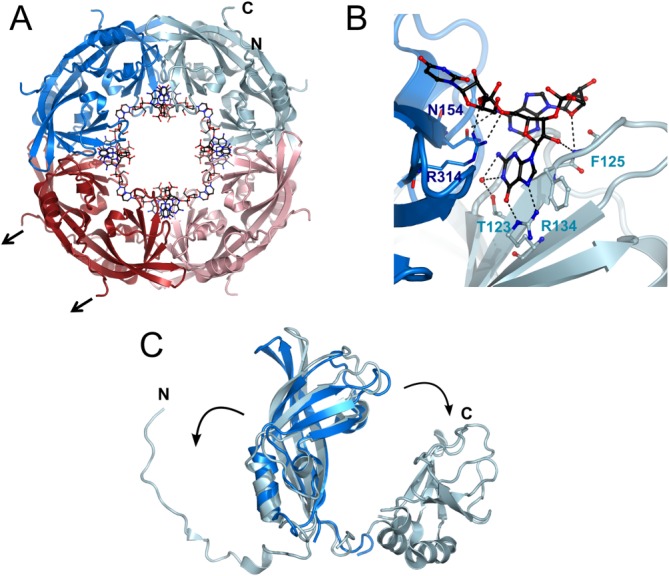
Octamer structure of the VP40 NTD bound the short RNA 5′-UGA-3′. (A) Full length VP40 can be destabilized to bind RNA in an octameric conformation mediated by the NTD (pdb code1H2C). An interactive view is available in the electronic version of the article.(B) Close-up of the sequence-specific RNA recognition (C) The N-terminal region of the VP40 NTD as well as the CTD need to dissociate from the core to facilitate octamer formation. The CTDs are connected by a long flexible linker allowing a more or less random positioning in solution (black arrows in A indicate their position).
Analyzing the NTD–CTD interface within the monomer reveals two pockets that could accommodate small molecules and thus stabilize the closed NTD–CTD conformation. Leu132, Ala156, and Arg148 form together with Ser319, L320, and Pro321 an ∼14 × 7 × 6.5 Å deep pocket and Val100 and Thr192 are at the bottom of ∼8 × 8 × 10 Å deep pocket that is surrounded by Asp193, Asp312, and Thr313 [Fig. 1(C)]. Notably removal of residues 320–326 destabilizes the NTD–CTD interaction such that the CTD dissociates, which induces spontaneous NTD oligomerization in vitro leading to VP40 hexamers and octamers in the presence of RNA (56). This suggests that the C-terminal end is critical for the NTD–CTD interaction. Therefore stabilizing the NTD–CTD interaction with small molecules should interfere with virus propagation notably because the generation of octameric ring-structures is essential for virus replication.53 Hence VP40 might be a suitable drug target in order to interfere with EBOV infection.
Conservation of the VP40 Fold in Negative Strand RNA Virus Matrix Proteins
The evolutionary relationship of negative single strand RNA viruses is confirmed by the structural conservation of the matrix proteins from Filoviridae, Paramyxoviridae, and Bornaviridae. The exception is the matrix protein structure from Vesicular stomatitis virus (VSV) a member of the Rabdoviridae. The structure of the VSV matrix protein folds into an N-terminal part composed of an anti-parallel β-sheet packed against two α-helices and a small C-terminal region64,65 [Fig. 4(A)] with no significant structural homology to EBOV VP40. In contrast the Borna disease virus matrix protein is smaller and contains only one domain with structural homology to the NTD of VP40.66 Borna virus matrix forms tetramers in solution and the crystal structure revealed that the tetramer binds single stranded RNA at its center which was modeled as cytidine-5′-monophosphate [Fig. 4(B)]. The functional role of the RNA bound conformation of the matrix protein is unclear. It has been detected in Borna virus infected brain tissue67 and could thus play a role in assembly or any other step of the viral life cycle.
Figure 4.
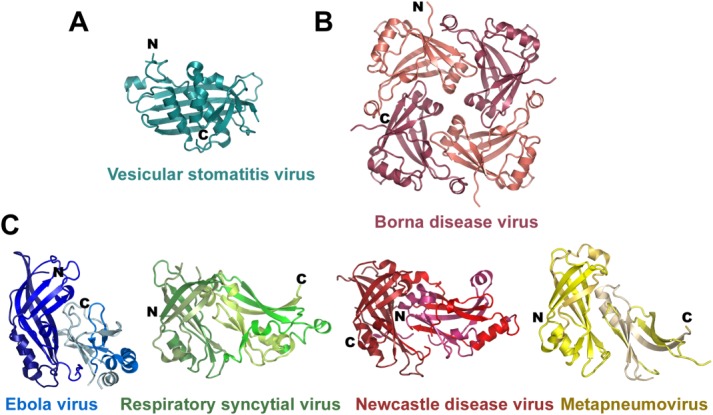
Structural conservation of matrix proteins from the Mononegavirales families. (A) Crystal structure of the vesicular stomatitis virus (VSV) matrix protein core (pdb code 2W2R). (B) Structure of the Borna disease virus tetramer (pdb code 3F1J). (C) Structure of the Ebola virus VP40 with the same NTD orientation as shown for RSV (pdb code 2VQP), Newcastle disease virus (pdb code 4G1L) and human Metapneumovirus (pdb code 4LP7) matrix protein monomers. (The chain direction is indicated with dark to light colors from the N- to the C-terminus.)
High structural conservation is found between EBOV VP40 and matrix proteins from Paramyxoviridae such as Respiratory syncytial virus matrix,68 Newcastle disease virus matrix,69 and human Metapneumovirus matrix.70 They are all composed of two beta sandwich domains homologous to VP40 NTD and CTD that associate with each other. However the mode of interaction and the orientations of the CTDs with respect to the NTDs is different in all four viral matrix proteins [Fig. 4(C)]. In addition, the matrix proteins from Metapneumovirus and Newcastle disease virus were reported to form dimers in solution and in the crystal. Dimerization positions the NTD and CTD at the same side, and produces a pseudo-tetrameric arrangement, which is different from the VP40 dimers (Fig. 5), but resembles the quaternary structure of Borna disease virus matrix protein tetramers [Fig. 4(B)].70 Its physiological relevance is indirectly supported by the fact that three different viral matrix proteins assemble into a similar quaternary structure. Human Metapneumovirus matrix also binds calcium, which is coordinated by two solvent exposed loops of the NTD. The presence of calcium was shown to substantially increase the thermostability of M.70
Figure 5.
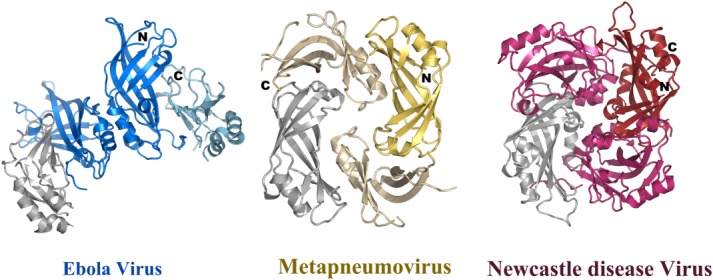
Comparison of dimeric structures of the matrix proteins. Dimeric conformation of the human Metapneumovirus (pdb code 4LP7) matrix protein and of Newcastle disease virus matrix (pdb code 4G1L) in comparison to the Ebola virus VP40 dimer. All three structures have one NTD (grey) in the same orientation. This demonstrates that not only the orientations of the NTD with respect to the CTD are different as shown in Figure 4, but also their mode of dimerization. It should be noted however, that the dimerization mode is quite similar for Metapneumovirus and Newcastle disease virus matrix proteins.
Incubation of human Metapneumovirus matrix with 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) produced tubular helical structures with variable diameters of ∼40 nm. Helical three-dimensional reconstruction from electron microscopy images revealed a subunit arrangement with a rise of 5.16 Å. The repeating unit fits the dimeric crystal structure, when positioned with its dimeric symmetry axis orthogonal to the long axis of the filament. This would position the concave surface of the dimer toward the inside producing a non-physiological positive curvature. Similar tubular structures of M have been observed for RSV M71 and are found to form irregular helical assemblies in RSV virions.72 In contrast the crystal packing of M from Metapneumovirus revealed the opposite negative curvature that is more relevant for M assembly in vivo.70 In fact electron microscopy analysis of Newcastle disease virus showed an M protein layer that fits the dimeric M structure with the concave membrane binding surface positioned toward the viral membrane implicating the CTD in membrane interaction [Fig. 2(B)]. Contacts between dimers and between filaments are made by NTDs and CTDs of symmetry related units.69 This is different from the EBOV assembly model, where NTD–NTD interactions form the filament (Fig. 2A) and the CTD contacts both the membrane and the nucleocapsid.58
An open question is whether the matrix proteins from Paramyxoviruses can also adopt different quaternary structures as it is the case for EBOV VP40. Money and colleagues noted that the NTD and CTD of RSV M are only kept together by weak interactions and cleavage within the linker region leads to the dissociation of NTD and CTD.68 Furthermore Leyrat and colleagues noted that NTD–CTD interface residues are conserved within EBOV and Metapneumovirus matrix proteins despite their overall low sequence similarity.70 Comparison of the NTD–CTD interfaces of the known paramyxovirus matrix protein structures indicates that their interaction is relatively weak in agreement with their calculated total buried surface area that range from ∼700 to 1600 Å2 and corresponding dissociation energies (ΔG) ranging from −21.7 to −9.9 kcal/mol, with Ebola VP40 having the highest ΔG (Table 1). This suggests that these matrix protein structures can also change their NTD–CTD arrangements to produce new conformations that might play additional role(s) during the life cycle of these viruses.
Table 1.
Calculated Total Buried Area by the NTD–CTD Interface and Binding Energies (ΔG Indicates the Solvation Free Energy Gain upon Formation of the Assembly) for NTD–CTD Interactions of Ebola Virus VP40, Respiratory Syncytial Virus Matrix, Newcastle Disease Virus Matrix and Human Metapneumovirus Matrix using the PISA Server73
| Buried area (Å2) | ΔG (kcal/mol) | |
|---|---|---|
| Ebola virus VP40 | 1365 | −21.7 |
| Respiratory syncytila virus matrix | 977 | −10.6 |
| Newcastle disease virus matrix | 1679 | −18.4 |
| Human Metapneumovirus matrix | 760 | −9.9 |
| Gp41-HK20 Fab complex | 5590 | −35 |
For comparison the total buried surface and calculated ΔG are indicated for a high affinity antigen-antibody interaction (gp41-HK20).74
Conclusions
The different conformations and quaternary structures of VP40, monomer/dimer, octamers in complex with RNA, ring-like hexamers observed in vitro and linear hexamers observed in the crystal are prime examples of how the small size of a viral genome can enlarge its coding capacity by packing different functions into different conformations of the same protein. Comparison of the matrix protein structures from three different families of negative single stranded RNA viruses has unraveled their evolutionary relationship that was not evident from the primary sequences highlighting once more the contribution of structural biology to the understanding of evolutionary relationships. Although the current structures have brought much insight into how these matrix proteins assemble into higher order protein networks that drive and coordinate the budding process, further work is required to determine whether the conformational plasticity observed for EBOV VP40 is also conserved in case of the matrix proteins from paramyxoviruses and Borna virus.
Acknowledgments
Work in the author’s lab is supported by the CNRS, the University Joseph Fourier and the Grenoble Instruct center (ISBG; UMS 3518 CNRS-CEA-UJF-EMBL) with support from FRISBI (ANR-10-INSB-05-02) and GRAL (ANR-10-LABX-49-01). G.E. is supported by a postdoctoral fellowship from the ANRS (Agence nationale de recherché sur le sida et les hépatites virales) and J.R. by the Labex GRAL (ANR-10-LABX-49-01).
References
- Olival KJ, Hayman DT. Filoviruses in bats: current knowledge and future directions. Viruses. 2014;6:1759–1788. doi: 10.3390/v6041759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JH. Filoviruses. A compendium of 40 years of epidemiological, clinical, and laboratory studies. Arch Virol Suppl. 2008;20:13–360. [PubMed] [Google Scholar]
- Beniac DR, Melito PL, Devarennes SL, Hiebert SL, Rabb MJ, Lamboo LL, Jones SM, Booth TF. The organisation of Ebola virus reveals a capacity for extensive, modular polyploidy. PloS One. 2012;7:e29608. doi: 10.1371/journal.pone.0029608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat TA, Noda T, Riches JD, Kraehling V, Kolesnikova L, Becker S, Kawaoka Y, Briggs JA. Structural dissection of Ebola virus and its assembly determinants using cryo-electron tomography. Proc Natl Acad Sci USA. 2012;109:4275–4280. doi: 10.1073/pnas.1120453109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertini AA, Schoehn G, Weissenhorn W, Ruigrok RW. Structural aspects of rabies virus replication. Cell Mol Life Sci. 2008;65:282–294. doi: 10.1007/s00018-007-7298-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruigrok RW, Crepin T, Kolakofsky D. Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Curr Opin Microbiol. 2011;14:504–510. doi: 10.1016/j.mib.2011.07.011. [DOI] [PubMed] [Google Scholar]
- Bakker SE, Duquerroy S, Galloux M, Loney C, Conner E, Eleouet JF, Rey FA, Bhella D. The respiratory syncytial virus nucleoprotein-RNA complex forms a left-handed helical nucleocapsid. J Gen Virol. 2013;94:1734–1738. doi: 10.1099/vir.0.053025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartlieb B, Muziol T, Weissenhorn W, Becker S. Crystal structure of the C-terminal domain of Ebola virus VP30 reveals a role in transcription and nucleocapsid association. Proc Natl Acad Sci USA. 2007;104:624–629. doi: 10.1073/pnas.0606730104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondot ML, Dubosclard V, Fix J, Lassoued S, Aumont-Nicaise M, Bontems F, Eleouet JF, Sizun C. Structure and functional analysis of the RNA- and viral phosphoprotein-binding domain of respiratory syncytial virus M2-1 protein. PLoS Pathog. 2012;8:e1002734. doi: 10.1371/journal.ppat.1002734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner SJ, Ariza A, Richard CA, Kyle HF, Dods RL, Blondot ML, Wu W, Trincao J, Trinh CH, Hiscox JA, Carroll MW, Silman NJ, Eleouet JF, Edwards TA, Barr JN. Crystal structure of the essential transcription antiterminator M2-1 protein of human respiratory syncytial virus and implications of its phosphorylation. Proc Natl Acad Sci USA. 2014;111:1580–1585. doi: 10.1073/pnas.1317262111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyrat C, Renner M, Harlos K, Huiskonen JT, Grimes JM. Drastic changes in conformational dynamics of the antiterminator M2-1 regulate transcription efficiency in Pneumovirinae. eLife. 2014;3:e02674. doi: 10.7554/eLife.02674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I, Yabukarski F, Ruigrok RW, Jamin M. Structural insights into the rhabdovirus transcription/replication complex. Virus Res. 2011;162:126–137. doi: 10.1016/j.virusres.2011.09.025. [DOI] [PubMed] [Google Scholar]
- Johnson RF, McCarthy SE, Godlewski PJ, Harty RN. Ebola virus VP35-VP40 interaction is sufficient for packaging 3E-5E minigenome RNA into virus-like particles. J Virol. 2006;80:5135–5144. doi: 10.1128/JVI.01857-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat TA, Riches JD, Kolesnikova L, Welsch S, Krahling V, Davey N, Parsy ML, Becker S, Briggs JA. Cryo-electron tomography of Marburg virus particles and their morphogenesis within infected cells. PLoS Biol. 2011;9:e1001196. doi: 10.1371/journal.pbio.1001196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert TW, Jahrling PB. Differentiation of filoviruses by electron microscopy. Virus Res. 1995;39:129–150. doi: 10.1016/0168-1702(95)00080-1. [DOI] [PubMed] [Google Scholar]
- Jasenosky LD, Neumann G, Lukashevich I, Kawaoka Y. Ebola virus VP40-induced particle formation and association with the lipid bilayer. J Virol. 2001;75:5205–5214. doi: 10.1128/JVI.75.11.5205-5214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmins J, Scianimanico S, Schoehn G, Weissenhorn W. Vesicular release of ebola virus matrix protein VP40. Virology. 2001;283:1–6. doi: 10.1006/viro.2001.0860. [DOI] [PubMed] [Google Scholar]
- Kolesnikova L, Berghofer B, Bamberg S, Becker S. Multivesicular bodies as a platform for formation of the Marburg virus envelope. J Virol. 2004;78:12277–12287. doi: 10.1128/JVI.78.22.12277-12287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikova L, Ryabchikova E, Shestopalov A, Becker S. Basolateral budding of Marburg virus: VP40 retargets viral glycoprotein GP to the basolateral surface. J Infect Dis. 2007;196:S232–S236. doi: 10.1086/520584. [DOI] [PubMed] [Google Scholar]
- Mittler E, Kolesnikova L, Strecker T, Garten W, Becker S. Role of the transmembrane domain of marburg virus surface protein GP in assembly of the viral envelope. J Virol. 2007;81:3942–3948. doi: 10.1128/JVI.02263-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, Dal Cin P, Dye JM, Whelan SP, Chandran K, Brummelkamp TR. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote M, Misasi J, Ren T, Bruchez A, Lee K, Filone CM, Hensley L, Li Q, Ory D, Chandran K, Cunningham J. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 2011;477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenhorn W, Carfi A, Lee KH, Skehel JJ, Wiley DC. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenen T, Shabman RS, Groseth A, Herwig A, Weber M, Schudt G, Dolnik O, Basler CF, Becker S, Feldmann H. Inclusion bodies are a site of ebolavirus replication. J Virol. 2012;86:11779–11788. doi: 10.1128/JVI.01525-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schudt G, Kolesnikova L, Dolnik O, Sodeik B, Becker S. Live-cell imaging of Marburg virus-infected cells uncovers actin-dependent transport of nucleocapsids over long distances. Proc Natl Acad Sci USA. 2013;110:14402–14407. doi: 10.1073/pnas.1307681110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikova L, Bohil AB, Cheney RE, Becker S. Budding of Marburgvirus is associated with filopodia. Cell Microbiol. 2007;9:939–951. doi: 10.1111/j.1462-5822.2006.00842.x. [DOI] [PubMed] [Google Scholar]
- Mittler E, Kolesnikova L, Herwig A, Dolnik O, Becker S. Assembly of the Marburg virus envelope. Cell Microbiol. 2013;15:270–284. doi: 10.1111/cmi.12076. [DOI] [PubMed] [Google Scholar]
- Kolesnikova L, Bamberg S, Berghofer B, Becker S. The matrix protein of Marburg virus is transported to the plasma membrane along cellular membranes: exploiting the retrograde late endosomal pathway. J Virol. 2004;78:2382–2393. doi: 10.1128/JVI.78.5.2382-2393.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adu-Gyamfi E, Digman MA, Gratton E, Stahelin RV. Single-particle tracking demonstrates that actin coordinates the movement of the Ebola virus matrix protein. Biophys J. 2012;103:L41–L43. doi: 10.1016/j.bpj.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanbo A, Watanabe S, Halfmann P, Kawaoka Y. The spatio-temporal distribution dynamics of Ebola virus proteins and RNA in infected cells. Scientific Rep. 2013;3:1206. doi: 10.1038/srep01206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch S, Kolesnikova L, Krahling V, Riches JD, Becker S, Briggs JA. Electron tomography reveals the steps in filovirus budding. PLoS Pathog. 2010;6:e1000875. doi: 10.1371/journal.ppat.1000875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartlieb B, Weissenhorn W. Filovirus assembly and budding. Virology. 2006;344:64–70. doi: 10.1016/j.virol.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Usami Y, Popov S, Popova E, Inoue M, Weissenhorn W, Gottlinger HG. The ESCRT pathway and HIV-1 budding. Biochem Soc Trans. 2009;37:181–184. doi: 10.1042/BST0370181. [DOI] [PubMed] [Google Scholar]
- Peel S, Macheboeuf P, Martinelli N, Weissenhorn W. Divergent pathways lead to ESCRT-III-catalyzed membrane fission. Trends Biochem Sci. 2011;36:199–210. doi: 10.1016/j.tibs.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Sundquist WI, Krausslich HG. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med. 2012;2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sundquist WI. Retrovirus budding. Ann Rev Cell Devel Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- Schmitt AP, Lamb RA. Escaping from the cell: assembly and budding of negative-strand RNA viruses. Curr Top Microbiol Immun. 2004;283:145–196. doi: 10.1007/978-3-662-06099-5_5. [DOI] [PubMed] [Google Scholar]
- Dolnik O, Kolesnikova L, Stevermann L, Becker S. Tsg101 is recruited by a late domain of the nucleocapsid protein to support budding of Marburg virus-like particles. J Virol. 2010;84:7847–7856. doi: 10.1128/JVI.00476-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J, Zang T, Bieniasz PD. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nature Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- Licata JM, Simpson-Holley M, Wright NT, Han Z, Paragas J, Harty RN. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J Virol. 2003;77:1812–1819. doi: 10.1128/JVI.77.3.1812-1819.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty RN, Brown ME, Wang G, Huibregtse J, Hayes FP. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc Natl Acad Sci USA. 2000;97:13871–13876. doi: 10.1073/pnas.250277297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmins J, Schoehn G, Ricard-Blum S, Scianimanico S, Vernet T, Ruigrok RW, Weissenhorn W. Ebola virus matrix protein VP40 interaction with human cellular factors Tsg101 and Nedd4. J Mol Biol. 2003;326:493–502. doi: 10.1016/s0022-2836(02)01406-7. [DOI] [PubMed] [Google Scholar]
- Yasuda J, Nakao M, Kawaoka Y, Shida H. Nedd4 regulates egress of Ebola virus-like particles from host cells. J Virol. 2003;77:9987–9992. doi: 10.1128/JVI.77.18.9987-9992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikova L, Strecker T, Morita E, Zielecki F, Mittler E, Crump C, Becker S. Vacuolar protein sorting pathway contributes to the release of Marburg virus. J Virol. 2009;83:2327–2337. doi: 10.1128/JVI.02184-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weissenhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrikant G, Lata S, Riches JD, Briggs JA, Weissenhorn W, Kozlov MM. Computational model of membrane fission catalyzed by ESCRT-III. PLoS Comput Biol. 2009;5:e1000575. doi: 10.1371/journal.pcbi.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenhorn W, Poudevigne E, Effantin G, Bassereau P. How to get out: ssRNA enveloped viruses and membrane fission. Current Opin Virol. 2013;3:159–167. doi: 10.1016/j.coviro.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Ebihara H, Takada A, Noda T, Kobasa D, Jasenosky LD, Watanabe S, Kim JH, Feldmann H, Kawaoka Y. Ebola virus VP40 late domains are not essential for viral replication in cell culture. J Virol. 2005;79:10300–10307. doi: 10.1128/JVI.79.16.10300-10307.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavari S, Bosio CM, Wiegand E, Ruthel G, Will AB, Geisbert TW, Hevey M, Schmaljohn C, Schmaljohn A, Aman MJ. Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J Exper Med. 2002;195:593–602. doi: 10.1084/jem.20011500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Sagara H, Suzuki E, Takada A, Kida H, Kawaoka Y. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J Virol. 2002;76:4855–4865. doi: 10.1128/JVI.76.10.4855-4865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson DL, Warfield KL, Kuehl K, Larsen T, Hevey MC, Schmaljohn A, Bavari S, Aman MJ. Generation of Marburg virus-like particles by co-expression of glycoprotein and matrix protein. FEMS Immun Med Microbiol. 2004;40:27–31. doi: 10.1016/S0928-8244(03)00273-6. [DOI] [PubMed] [Google Scholar]
- Hoenen T, Volchkov V, Kolesnikova L, Mittler E, Timmins J, Ottmann M, Reynard O, Becker S, Weissenhorn W. VP40 octamers are essential for Ebola virus replication. J Virol. 2005;79:1898–1905. doi: 10.1128/JVI.79.3.1898-1905.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessen A, Volchkov V, Dolnik O, Klenk HD, Weissenhorn W. Crystal structure of the matrix protein VP40 from Ebola virus. EMBO J. 2000;19:4228–4236. doi: 10.1093/emboj/19.16.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruigrok RW, Schoehn G, Dessen A, Forest E, Volchkov V, Dolnik O, Klenk HD, Weissenhorn W. Structural characterization and membrane binding properties of the matrix protein VP40 of Ebola virus. J Mol Biol. 2000;300:103–112. doi: 10.1006/jmbi.2000.3822. [DOI] [PubMed] [Google Scholar]
- Scianimanico S, Schoehn G, Timmins J, Ruigrok RH, Klenk HD, Weissenhorn W. Membrane association induces a conformational change in the Ebola virus matrix protein. EMBO J. 2000;19:6732–6741. doi: 10.1093/emboj/19.24.6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmins J, Schoehn G, Kohlhaas C, Klenk HD, Ruigrok RW, Weissenhorn W. Oligomerization and polymerization of the filovirus matrix protein VP40. Virology. 2003;312:359–368. doi: 10.1016/s0042-6822(03)00260-5. [DOI] [PubMed] [Google Scholar]
- Bornholdt ZA, Noda T, Abelson DM, Halfmann P, Wood MR, Kawaoka Y, Saphire EO. Structural rearrangement of ebola virus VP40 begets multiple functions in the virus life cycle. Cell. 2013;154:763–774. doi: 10.1016/j.cell.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adu-Gyamfi E, Soni SP, Xue Y, Digman MA, Gratton E, Stahelin RV. The Ebola virus matrix protein penetrates into the plasma membrane: a key step in viral protein 40 (VP40) oligomerization and viral egress. J Biol Chem. 2013;288:5779–5789. doi: 10.1074/jbc.M112.443960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni SP, Adu-Gyamfi E, Yong SS, Jee CS, Stahelin RV. The Ebola virus matrix protein deeply penetrates the plasma membrane: an important step in viral egress. Biophys J. 2013;104:1940–1949. doi: 10.1016/j.bpj.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynard O, Nemirov K, Page A, Mateo M, Raoul H, Weissenhorn W, Volchkov VE. Conserved proline-rich region of Ebola virus matrix protein VP40 is essential for plasma membrane targeting and virus-like particle release. J Infect Dis. 2011;204:S884–S891. doi: 10.1093/infdis/jir359. [DOI] [PubMed] [Google Scholar]
- Booth TF, Rabb MJ, Beniac DR. How do filovirus filaments bend without breaking? Trends Microbiol. 2013;21:583–593. doi: 10.1016/j.tim.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Gomis-Ruth FX, Dessen A, Timmins J, Bracher A, Kolesnikowa L, Becker S, Klenk HD, Weissenhorn W. The matrix protein VP40 from Ebola virus octamerizes into pore-like structures with specific RNA binding properties. Structure. 2003;11:423–433. doi: 10.1016/S0969-2126(03)00050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudier M, Gaudin Y, Knossow M. Crystal structure of vesicular stomatitis virus matrix protein. EMBO J. 2002;21:2886–2892. doi: 10.1093/emboj/cdf284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham SC, Assenberg R, Delmas O, Verma A, Gholami A, Talbi C, Owens RJ, Stuart DI, Grimes JM, Bourhy H. Rhabdovirus matrix protein structures reveal a novel mode of self-association. PLoS Pathog. 2008;4:e1000251. doi: 10.1371/journal.ppat.1000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann P, Lieber D, Meyer S, Dautel P, Kerth A, Kraus I, Garten W, Stubbs MT. Crystal structure of the Borna disease virus matrix protein (BDV-M) reveals ssRNA binding properties. Proc Natl Acad Sci USA. 2009;106:3710–3715. doi: 10.1073/pnas.0808101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoyloff R, Strecker A, Bode L, Franke P, Ludwig H, Hucho F. The glycosylated matrix protein of Borna disease virus is a tetrameric membrane-bound viral component essential for infection. Eur J Biochem/FEBS. 1997;246:252–257. doi: 10.1111/j.1432-1033.1997.t01-2-00252.x. [DOI] [PubMed] [Google Scholar]
- Money VA, McPhee HK, Mosely JA, Sanderson JM, Yeo RP. Surface features of a Mononegavirales matrix protein indicate sites of membrane interaction. Proc Natl Acad Sci USA. 2009;106:4441–4446. doi: 10.1073/pnas.0805740106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battisti AJ, Meng G, Winkler DC, McGinnes LW, Plevka P, Steven AC, Morrison TG, Rossmann MG. Structure and assembly of a paramyxovirus matrix protein. Proc Natl Acad Sci USA. 2012;109:13996–14000. doi: 10.1073/pnas.1210275109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyrat C, Renner M, Harlos K, Huiskonen JT, Grimes JM. Structure and self-assembly of the calcium binding matrix protein of human metapneumovirus. Structure. 2014;22:136–148. doi: 10.1016/j.str.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhee HK, Carlisle JL, Beeby A, Money VA, Watson SM, Yeo RP, Sanderson JM. Influence of lipids on the interfacial disposition of respiratory syncytical virus matrix protein. Langmuir. 2011;27:304–311. doi: 10.1021/la104041n. [DOI] [PubMed] [Google Scholar]
- Liljeroos L, Krzyzaniak MA, Helenius A, Butcher SJ. Architecture of respiratory syncytial virus revealed by electron cryotomography. Proc Natl Acad Sci USA. 2013;110:11133–11138. doi: 10.1073/pnas.1309070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Sabin C, Corti D, Buzon V, Seaman MS, Lutje Hulsik D, Hinz A, Vanzetta F, Agatic G, Silacci C, Mainetti L, Scarlatti G, Sallusto F, Weiss R, Lanzavecchia A, Weissenhorn W. Crystal structure and size-dependent neutralization properties of HK20, a human monoclonal antibody binding to the highly conserved heptad repeat 1 of gp41. PLoS Pathog. 2010;6:e1001195. doi: 10.1371/journal.ppat.1001195. [DOI] [PMC free article] [PubMed] [Google Scholar]


