Abstract
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) catalyzes the oxidative phosphorylation of d-glyceraldehyde 3-phosphate (G3P) into 1,3-diphosphoglycerate (BGP) in the presence of the NAD cofactor. GAPDH is an important drug target because of its central role in glycolysis, and nonglycolytic processes such as nuclear RNA transport, DNA replication/repair, membrane fusion and cellular apoptosis. Recent studies found that GAPDH participates in the development of diabetic retinopathy and its progression after the cessation of hyperglycemia. Here, we report two structures for native bovine photoreceptor GAPDH as a homotetramer with differing occupancy by NAD, bGAPDH(NAD)4, and bGAPDH(NAD)3. The bGAPDH(NAD)4 was solved at 1.52 Å, the highest resolution for GAPDH. Structural comparison of the bGAPDH(NAD)4 and bGAPDH(NAD)3 models revealed novel details of conformational changes induced by cofactor binding, including a loop region (residues 54–56). Structure analysis of bGAPDH confirmed the importance of Phe34 in NAD binding, and demonstrated that Phe34 was stabilized in the presence of NAD but displayed greater mobility in its absence. The oxidative state of the active site Cys149 residue is regulated by NAD binding, because this residue was found oxidized in the absence of dinucleotide. The distance between Cys149 and His176 decreased upon NAD binding and Cys149 remained in a reduced state when NAD was bound. These findings provide an important structural step for understanding the mechanism of GAPDH activity in vision and its pathological role in retinopathies.
Keywords: glyceraldehyde 3-phosphate dehydrogenase, crystal structure, structural comparison, photoreceptor, NAD, oxidization, S-nitrosylation, diabetic retinopathy
Introduction
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a classic glycolytic enzyme, catalyzes the conversion of glyceraldehyde-3-phosphate (G3P) to 1,3-bisphosphoglycerate (BGP), and requires the co-factor NAD.1 In addition to its well-known glycolytic functions, new studies have shown that mammalian GAPDH is a multifunctional protein with diverse activities2 that play fundamental roles in a variety of pathologies, including diabetes, age-related neurodegenerative disorders, and malaria.3 GAPDH is physiologically nitrosylated at its active Cys149 residues.4,5 Upon S-nitrosylation, GAPDH serves as a NO-transferase, for example, it can bind to nuclear proteins and co-translocate to the nucleus and facilitate the degradation of nuclear proteins.3,6,7 In the eye, the NO level is upregulated by light and decreases in the dark.8 In cone photoreceptors, S-nitrosylation regulates the activity of L-type voltage-gated Ca2+ channels during light adaptation.9 GAPDH is also required for heme transfer into inducible nitric oxide synthase.10 GAPDH binding to heme is reversible and sensitive to cellular redox levels.11 This property expands the role of GAPDH into a potential heme-sensitive chaperone in cells.
In rod outer segments (ROS) of the eye’s photoreceptors, GAPDH is one of most abundant proteins, existing at levels higher than PDE6 subunits, but slightly lower than arrestin, but much higher than any other glycolytic enzyme by an order of magnitude.12 GAPDH also participates in the development and progression of diabetic retinopathy, one of the most severe ocular complications of diabetes. The pathogenesis of diabetic retinopathy involves multiple effector pathways13 with GAPDH considered a potential link between hyperglycemia-induced oxidative stress and major pathways implicated in the pathogenesis of diabetic complications.14 High levels of glucose in retinas can inactivate GAPDH.15 MS analysis indicated that the active site Cys149 was covalently modified in GAPDH from the muscle of diabetic animals.16 Inactivation of GAPDH could result from increased mitochondrial superoxide production that in turn nitrosylates the enzyme.17 Inactive GAPDH would divert the upstream glycolytic intermediates to alternative pathways, potentially leading to formation of advanced glycation end products that participate in the development of diabetic microvascular complications.1,14,18
Structures of GAPDH have been extensively studied in bacteria, archaea, and eukarya, providing detailed information on the folding, catalysis and regulation of this enzyme. The overall structure of GAPDH consists of identical subunits conserved among different species. GAPDH enzymatic activity requires the binding of NAD but the enzyme shows different binding characteristics depending on the tissue source.19–23 Here, we present two high resolution crystal structures of GAPDH from ROS with different occupancies of NAD. These GAPDH structures provide an updated view of their NAD-binding sites, and their conformational changes induced by NAD binding.
Results
Preparation of bovine retinal GAPDH
Bovine GAPDH is loosely associated with ROS membranes.24 In a typical preparation, we obtained about 0.2 mg of purified bGAPDH from isotonic extraction of ROS from 100 bovine retinas using NAD-agarose affinity chromatography. The purity of bGAPDH was assessed by SDS-PAGE wherein the protein exhibited a molecular weight of ∼36 kDa under denaturing conditions [Fig. 1(A)]. But size exclusion chromatography analysis demonstrated that bGAPDH had a molecular weight of about 150 kDa [Fig. 1(B)]. Like other GAPDHs from various sources, bGAPDH appeared to be a tetrameric protein in solution. The catalytic activity of bGAPDH as determined for protein isolated from ROS by NAD-affinity chromatography evidenced a Km value of 153 ± 19 µM for its G3P substrate, and a Km value of 255 ± 23 µM for the NAD cofactor [Fig. 1(C)].
Figure 1.
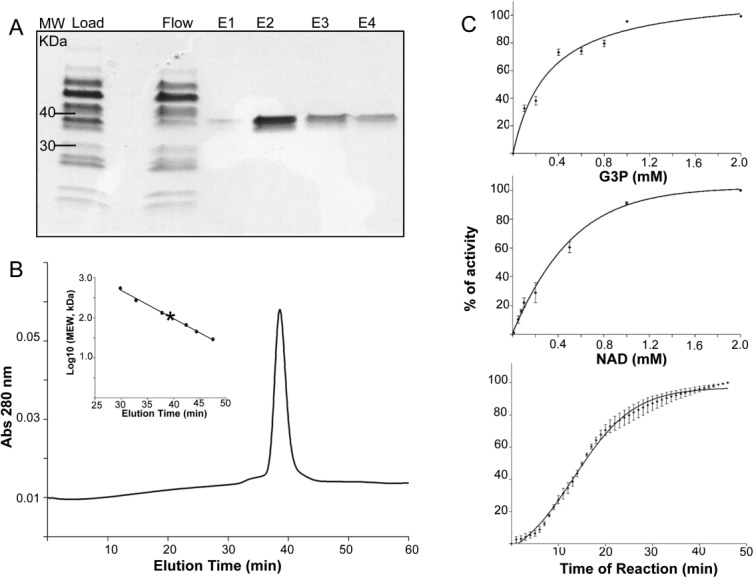
Characterization of bovine GAPDH. A. SDS-PAGE analysis of bGAPDH purified from bovine ROS. B. Size exclusion profile of purified bGAPDH. The column (Superdex 200, 10/300 GL, GE Healthcare) was calibrated with protein standards (Insert). C. Enzyme activity. Purified bGAPDH (0.5 µg/100 µL) was incubated with 0.2 mM NAD and increasing concentrations of G3P (top panel), or with 1 mM G3P and increasing concentrations of NAD (middle panel). After 30 min of incubation, GAPDH activity was monitored by measuring NADH production at 340 nm. Kinetic measurements were then done over a 45 min time course in the presence of 1 mM NAD and 1 mM G3P (bottom panel).
The overall structure of bGAPDH
bGAPDH readily formed crystals under several conditions (Supporting Information Fig. S1). Crystals of purified bGAPDH had NAD bound in all four subunits (bGAPDH(NAD)4). Alternatively, bGAPDH could be crystallized directly from isotonic extracts of ROS (Supporting Information Fig. S1B). This extraction of native bGAPDH yielded crystals diffracting to 1.93 Å with a structure containing three subunits with NAD-bound and one subunit free of NAD (bGAPDH(NAD)3). The presence of NAD further tightened the crystal packing and improved the resolution to 1.52 Å in the bGAPDH(NAD)4 structure, the highest resolution for a GAPDH structure to date. Final models of bGAPDH contained 1328 protein residues (both), 304 water molecules in bGAPDH(NAD)3, and 582 water molecules in bGAPDH(NAD)4 (Table 1). These structural models of bGAPDH showed good stereochemistry, with only Val237 appearing in disallowed regions of all four subunits.25 However, this residue had a clear electron density in all four subunits of both bGAPDH structures. The abnormal dihedral angles of this residue were also observed in other reported GAPDH structures, for example, PDB entries, 1J0X, 1U8F, and 1ZNQ. The overall folding of bGAPDH was similar to that of other previously determined eukaryotic GAPDH structures. The refined model was composed of one tetramer with four subunits shown in the typical nomenclature of GAPDH subunits’ colors of green (Q), red (P), blue (R), and yellow (O) (Fig. 2).26 The molecular axes were defined as P, Q, and R, and O subunits were related by 222 symmetry. The choice of the first monomer O in bGAPDH(NAD)3 was the subunit free of NAD. Each subunit was composed of a catalytic domain (residues 148–311) and a NAD-binding domain (residues 1–147 and 312–332). In the bGAPDH(NAD)4 model, Ser22-Gly23-Lys24 of the Q subunit, and residues Gly190-Lys191 of the O subunit were found disordered. In the bGAPDH(NAD)3 model, Gly23 (Q, R, O), Lys24 (Q, O) and Lys191 (P, Q, O) were disordered. Dual side-chain conformations were observed in Ser207 in subunits of O, Q, and R of the bGAPDH(NAD)3 model and in Ser207 of all four subunits in the bGAPDH(NAD)4 model.
Table 1.
Data Collection, Processing and Refinement Statistics for bGAPDH with Three or Four Bound Molecules of NAD
| Data collection | Average/outer shell bGAPDH(NAD)3 (Å) | Average/outer shell bGAPDH(NAD)4 (Å) |
|---|---|---|
| Wavelength (Å) | 0.9792 | 0.9792 |
| Space group | P21 | P21 |
| Unit-cell parameters (Å) | a = 79.8, b = 126.5, c = 83.8, α = 90°, β = 118o, γ = 90° | a = 79.7, b = 125.3, c = 83.6, α = 90°, β = 118°, γ = 90° |
| Diffraction resolutions (Å) | 50–2.0 (4.16–1.93) | 50–1.52 (3.27–1.52) |
| No. of observations | 439,689 | 740,361 |
| No. of unique reflections | 108,672 | 220,468 |
| Redundancy | 4.1 (3.9) | 3.1 (3.4) |
| Completeness (%) | 99.3 (98.4) | 98.5 (96.8) |
| Average I/σ (I) | 20.3 (2.8) | 10.6 (0.9) |
| Rmerge (I) | 0.088 (0.56) | 0.094 (0.97) |
| Mosaicity | 0.20–0.90 | 0.19–0.34 |
| Refinement | ||
| No. of protein residues | 1328 | 1328 |
| No. of NAD molecules | 3 | 4 |
| No. of water molecules | 304 | 582 |
| Rfact | 0.19 | 0.18 |
| Rfree | 0.24 | 0.21 |
| RMSD | 0.018 | 0.02 |
| Bond lengths (Å) | 1.92 | 2.06 |
| Bond angles (°) | 0.12 | 0.12 |
| Improper dihedrals (°) | ||
| Average B factors (Å2) | 26.1 | 17.7 |
| Protein | 26.3 | 13.0 |
| NAD+ | 24.5 | 22.5 |
| Water | ||
| Ramachandran plot | 95.9 | 96.3 |
| (Non-Pro and Gly residues) | 99.6 | 99.5 |
| Favored (%) | 0.36 | 0.45 |
| Allowed (%) | ||
| Disallowed (%) (Val237/O,P,Q, and R) |
Figure 2.
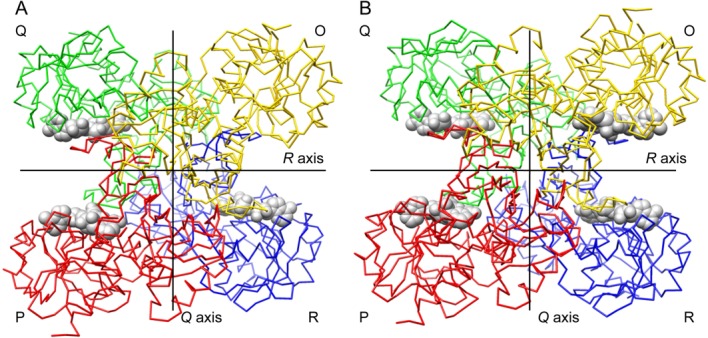
Carbon trace representations of bovine GAPDH tetramer structures. A. Structural model of bGAPDH(NAD)3. The NAD-free subunit named as “O” is shown in yellow. B. Structural model of bGAPDH(NAD)4. This view is down the P axis. Lines show the location of the Q and R twofold molecular axes. NAD cofactors are displayed in spheres. Subunit nomenclature and coloring is as described in Ref. (26). An interactive view is available in the electronic version of the article.
bGAPDH:NAD binding
NAD was bound to bGAPDH inside a typical Rossmann-fold27 composed of six parallel β-strands linked to two pairs of α-helices [Fig. 3(A)]. In bGAPDH(NAD)4, the presence of NAD in each subunit was clearly evidenced by a strong electron density map in the binding pocket. The averaged B-factors (Å)2 for NAD were 12.8 (O), 12.7 (P), 12.1 (R), and 14.4 (Q). In bGAPDH(NAD)3, the electron density map indicated the presence of NAD at high occupancy in subunits P, Q, and R. But no electron density was observed for NAD in subunit of O. Therefore, NAD was not modeled in the O subunit of bGAPDH(NAD)3. The averaged B-factors (Å)2 for NAD in bGAPDH(NAD)3 were 30.2 (P), 22.4 (Q), and 26.5 (R). Differential NAD binding to subunits of GAPDH was previously reported. For example, two NAD molecules are bound to the rabbit muscle GAPDH tetramer (PDB code: 1J0X22), whereas three NAD molecules are bound to human placental GAPDH tetramer (PDB code: 1U8F21). Differential binding of NAD in GAPDH could be the basis of the cooperativity shown by this enzyme.28–30 A diagram of the interactions between NAD and bGAPDH is shown in Figure 3(B). Details of interactions between NAD and the protein are summarized in Supporting Information Table SI.
Figure 3.
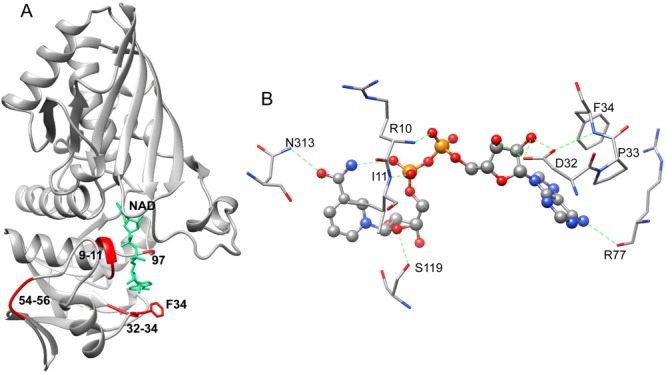
NAD conformation and interactions in bGAPDH. A. Ribbon presentation of NAD binding to bGAPDH(NAD)4 (P subunit) in a typical Rossmann fold.27 The fluctuation regions, residues 9–11 (the Gly-rich loop), 32–34 (gate for NAD entry), 54–56 (a remote loop region), and 96 are shown in red ribbons. The view of NAD binding (shown in green) and Phe34 (shown in red) in the P subunit of bGAPDH(NAD)4 is also portrayed. B. Diagram of NAD–protein interactions in the P subunit of bGAPDH(NAD). Green dotted lines indicate hydrogen bonds formed between NAD and residues from the protein. An interactive view is available in the electronic version of the article.
The high quality of electron density map of bGAPDH allowed detailed modeling of solvent in its structure. Nearly a dozen water molecules were identified that interacted with NAD with a 3.2 Å cutoff inside the binding pocket (Supporting Information Fig. S2). As summarized in Supporting Information Table SII, some of these waters formed direct hydrogen bonds to NAD whereas others mediated hydrogen bonds between NAD and the protein. For example, the backbone oxygen of Glu94 formed a water-mediated (W106) interaction with the O3D of NAD. Through water-4, the O2N of NAD interacted with the backbone nitrogen groups of Gly9, Gly12 and the oxygen group of Ser95. NH1 from the side chain of Arg13 formed a water-mediated (W62) interaction with the O3B of NAD. And the ND2 side chain of Asn6 formed a water-mediated (W182) interaction with N1A of NAD.
Structural comparison of NAD-free and NAD-bound bGAPDH
The overall structures of fully NAD-bound (bGAPDH(NAD)4) and partially NAD-bound (bGAPDH(NAD)3) were similar. The RMSD of Cα between the two tetramers was 0.24 Å, indicating that NAD binding did not cause conformational changes in the overall folding of bGAPDH.
To study the conformational changes induced by NAD binding in bGAPDH, we performed a detailed structural comparison of the NAD-free and NAD-bound subunits. Thus the NAD-free subunit (O) from bGAPDH(NAD)3, and the corresponding subunit O in bGAPDH(NAD)4 (with NAD-bound) were superimposed for comparison. In addition, the NAD-free subunit (O) was superimposed with NAD-bound subunit (P) from the same bGAPDH(NAD)3 tetramer structure. As controls, single NAD-binding subunits such as subunits P from bGAPDH(NAD)3 and bGAPDH(NAD)4, and subunits O and P from bGAPDH(NAD)4, were superimposed and compared. Atomic distances were measured for each of the paired residues. The RMSD of the Cα atoms for each residue was plotted along with the residue number [Fig. 4(A)].
Figure 4.
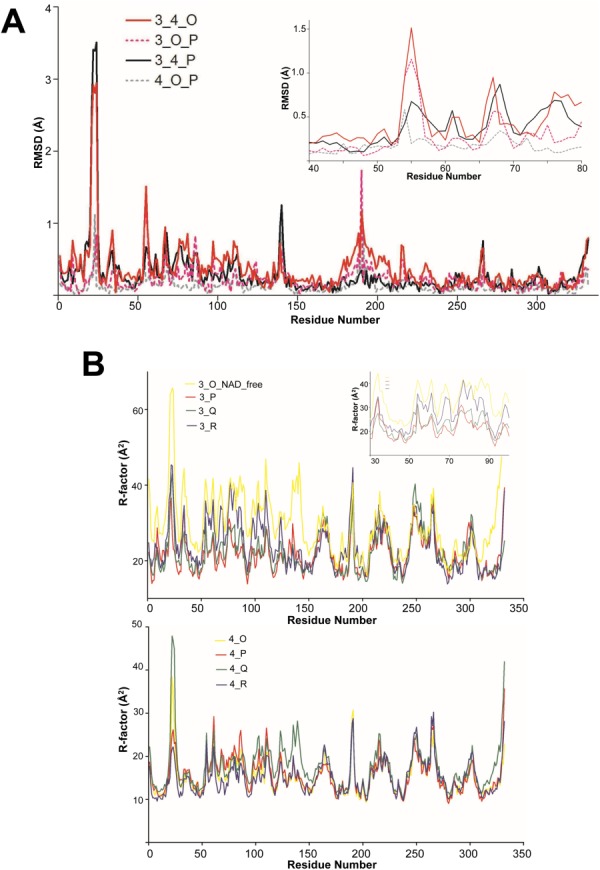
Structural comparison of the NAD-free and NAD-bound subunits. (A). The RMSD of Cα atoms in paired residues from apo- and holo-subunits of bGAPDH. The subunit of O (NAD-free) from bGAPDH(NAD)3 and subunit O (NAD-bound) from bGAPDH(NAD)4 were superimposed (solid red line, labeled as 3_4_O). Subunits of O (NAD-free) and P (NAD-bound) from bGAPDH(NAD)3 were superimposed (dotted red line, labeled as 3_O_P). Subunits of P from bGAPDH(NAD)3 and bGAPDH(NAD)4 were superimposed as one control (solid black line, labeled as 3_4_P) and subunits of O and P from fully bGAPDH(NAD)4 were superimposed as another control (dotted gray line, labeled as 4_O_P). Insert shows the plotted region comprised residues 40–80. B. Comparison of B-factors of residues in the main chains of apo- and holo-subunits of bGAPDH. The averaged B-factor values of the residue from main chain atoms from the four subunits of bGAPDH(NAD)3 were plotted along with the residue number (upper panel, solid lines, labeled as 3_O (NAD_free), 3_P, 3_Q, and 3_R). Insert shows the plot region from residues 30 to 100. The averaged B-factor values of the residue from main chain atoms from the four subunits of bGAPDH(NAD)4 were plotted along with the residue number (lower panel, solid lines, labeled as 4_O, 4_P, 4_Q, and 4_R). Each subunit was presented as a solid line colored as in Figure 2.
As illustrated in the plot, the catalytic domain (residues 148–311) with the exception of Gly190 showed only minor fluctuations, indicating that only minor conformational changes occurred in this region. In contrast, significant peaks appeared in the NAD-binding domain. These peaks were located around residues involved in direct hydrogen bond interactions with NAD, such as Arg10, Ile11, and Asp32. Together with their neighboring residues, these regions showed substantial conformational changes upon NAD binding.
The overall RMSD value was 0.397 Å for the O subunits between bGAPDH(NAD)3 and bGAPDH(NAD)4. Some residues had much higher RMSD values than the averaged overall values. Based on the plot, the regions showing the greatest fluctuation upon NAD binding were located in the overall structure [Fig. 3(A)]. The RMSD values at Cα of the residues from these regions are presented in Supporting Information Table SIII.
Residues 54–56 evidenced large peaks in the plot [Fig. 4(A), insert]. These residues were found in the loop region shown in red in Figure 3(A). This loop is located at the surface of the tetramer structure, and is not adjacent to the NAD-binding cavity. Flexibility of this loop may be required for adapting conformational changes upon NAD binding/release.
Furthermore, we compared the B-factor values of residues in each subunit of bGAPDH(NAD)3 and bGAPDH(NAD)4. The B-factor values reflect the relative flexibility of parts of a structure. Residues with low B-factors indicate a well-ordered structure, whereas residues with large B-factors indicate a flexible structure. The averaged B-factor values of Cα atoms for residues in each subunit were plotted against the residues numbers [Fig. 4(B)]. In bGAPDH(NAD)4, the four subunits showed only minor fluctuations in the NAD binding domain (residues 1–147), and almost identical values at the catalytic domain (residues 148–311) [Fig. 4(B), lower panel].
In contrast, the NAD-free subunit (3_O_NAD_free) in bGAPDH(NAD)3 had the greatest flexibility increase in the NAD binding domain, including residues 9, 34, 54, and 97. One NAD-bound subunit (3_R) had a mild flexibility increase and two NAD-bound subunits (3_P and 3_Q) had similar levels of flexibility [Fig. 4(B), upper panel and insert]. Nevertheless, the flexibility of the catalytic domain (residues 148–311) in bGAPDH(NAD)3 remained at similar levels for all four subunits.
Residues Pro33 and Phe34 formed a bottleneck for NAD binding. Both residues showed extremely large peaks in the RMSD plot. Phe34 was located at the entry of NAD-binding pocket [Fig. 3(A)] close to the adenine group of NAD. Substitution of Phe34 with smaller side chain (e.g., Gly or Leu), or polar residue (e.g., Thr) could abolish the NAD binding affinity, or reduce the protein’s catalytic efficiency.31 This residue was suggested to be important in stabilizing NAD binding.31 In the full NAD-bound bGAPDH structure, an electron density for the side chain of Phe34 was observed in all four subunits [Fig. 5(A), shown with P subunit]. In the partially NAD-bound bGAPDH structure, the electron density for the side chain of Phe34 was observed from those subunits with NAD-bound [Fig. 5(B), shown with P subunit]. In the absence of NAD, only a minor electron density was observed for the side chain of Phe34 [Fig. 5(C), shown with O subunit], indicating that this residue adopted a flexible conformation in the absence of NAD. Furthermore, Phe34 evidenced greater B-factor values in the NAD-free bGAPDH structure, as compared to much smaller B-factor values in the NAD-bound bGAPDH structure (Table 2). In the NAD-bound state, the Phe34 conformation was stabilized by an interaction with NAD. In the absence of NAD, the side chain of Phe34 exhibited much greater flexibility. The dynamic character of Phe34 explains its incomplete electron density in the partial NAD bound structure, and suggests an important role for Phe34 in stabilizing NAD binding in bovine photoreceptor GAPDH.
Figure 5.
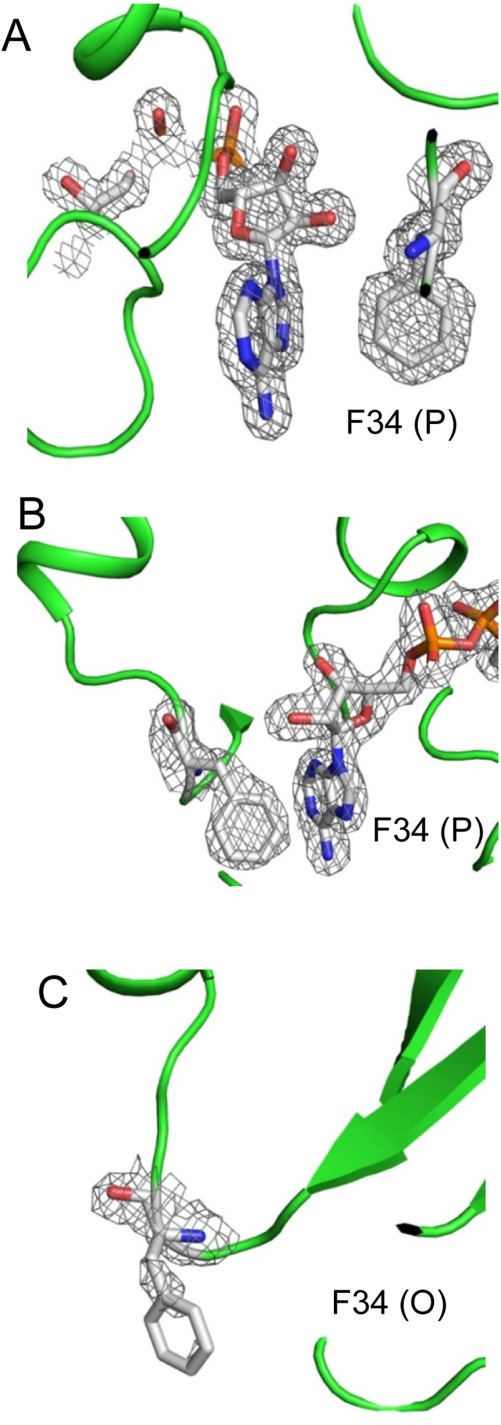
2Fo–Fc maps of Phe34 in bGAPDH NAD-binding sites. The electron density was sufficient to cover the Phe34 residue and NAD molecule in the P subunit of both bGAPDH(NAD)4 (A) and bGAPDH(NAD)3 (B). But the electron density only weakly covered the main chain of Phe34 in the NAD-free structure, namely the O subunit of bGAPDH(NAD)3 (C). 2Fo–Fc maps of Phe34 and NAD were contoured at 1.0σ and presented as gray meshes. Figures were prepared with PyMol software.
Table 2.
B-Factor Values (Å2)a of Phe34 in Two bGAPDH Structures
| Subunits | bGAPDH(NAD)3 (Å2) | bGAPDH(NAD)4 (Å2) | Difference (Å2) |
|---|---|---|---|
| O | 45.1 (NAD free) | 13.3 (+8.4) | −23.4 |
| P | 36.5 | 16.3 (+8.4) | −11.8 |
| Q | 28.6 | 16.6 (+8.4) | −3.6 |
| R | 33.0 | 14.6 (+8.4) | −10 |
| Average for whole protein | 26.1 | 17.7 | −8.4 |
The difference in overall average B-factor value was 8.4 between the two structures. To compare the B-factor values of Phe34 from the two structures, a constant value of 8.4 was added to the B-factor value for Phe34 in bGAPDH(NAD)3 that had the lower B-factor value.
bGAPDH:NAD binding and active site
In GAPDH, the active site Cys residue can be oxidized in response to changing environmental redox states. In the partially NAD-bound structure, the active Cys149 exhibited an oxidative state in all four subunits. The electron density clearly reinforced the positive peaks from Fo–Fc map around the thiol group of active Cys149 (R subunit) [Fig. 6(A)]. The active site Cys149 residues in the partially NAD-bound structure could be modeled as sulfenic acid. In contrast, the active Cys149 was in the reduced state in all four subunits of the fully NAD-bound structure [Fig. 6(B)]. No positive peak was observed from their Fo–Fc electron density maps.
Figure 6.
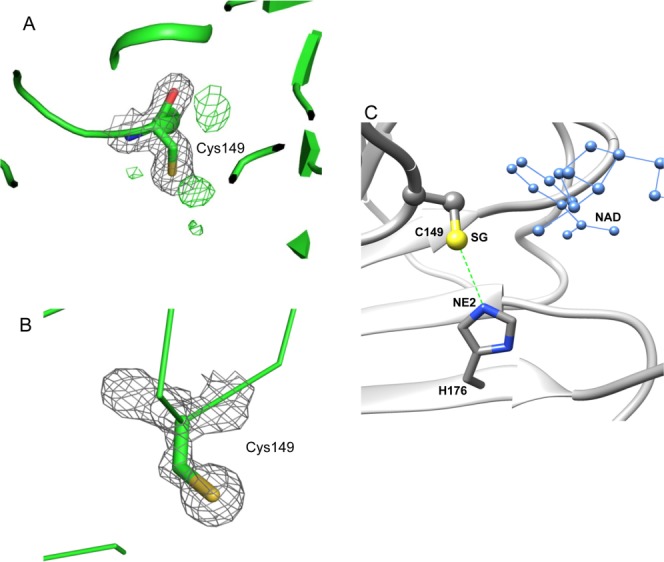
The active site Cys149 in bGAPDH and its interaction with His176. A. Electron density of Cys149 in subunit R of bGAPDH(NAD)3. The 2Fo–Fc electron density map is presented as grey meshes contoured at 1.0σ. The green meshes represent a Fo–Fc electron density map contoured at 3.0σ. The presence of a positive density next to the SG atom of Cys149 strongly suggests an oxidative state for this residue. B. Electron density of Cys149 in subunit R of bGAPDH (NAD)4. None of the four subunits exhibited a positive density next to the SG atom of Cys149 in the Fo–Fc map. Figures were prepared with PyMol software. C. Interaction between Cys149 and His176 in subunits of bGAPDH. The distance between the SG group (shown in yellow) of Cys149 and the NE2 group (shown in blue) of His176 is indicated by a dotted green line. The adjacent NAD molecule is shown in blue ball-and-sticks.
Cys149 and His176 had been identified as two important residues for the mechanism of GAPDH action. The adjacent His residue can lower the pKa value of the active site Cys residue and make it more reactive.32 To estimate the effect of NAD binding on the interaction between the active Cys149 and His176 [Fig. 6(C)], we measured the distance between the thiol group of active-site Cys149 and the NE2 atom of His176. The resulting values then were compared between NAD-free and NAD-bound subunits. As shown in Table 3, the distance was reduced in NAD-bound subunits of two bGAPDH structures by 0.04–0.16 Å (not significant), and by 0.33 Å compared within O subunits which were NAD-free in bGAPDH(NAD)3 and NAD-bound in bGAPDH(NAD)4. Thus the residue pairs of Cys149 and His176 were brought closer together upon NAD binding. This shortened distance between Cys149 and His176 could facilitate hydride transfer from activated Cys149 towards NAD through His176.
Table 3.
Distance (in Å) Between SG/Cys149 and NE2/His176
| Subunits | bGAPDH(NAD)3 (Å) | bGAPDH(NAD)4 (Å) | Difference (Å) |
|---|---|---|---|
| O | 3.75 (NAD-free) | 3.42 | −0.33 |
| P | 3.38 | 3.33 | −0.05 |
| Q | 3.5 | 3.34 | −0.16 |
| R | 3.5 | 3.36 | −0.14 |
Discussion
The overall folding of GAPDH consisting of four identical subunits that form a homotetramer is conserved among different species. GAPDH enzymatic activity requires the binding of NAD but the enzyme shows different binding characteristics depending on the tissue source. Mammalian GAPDH structures display a variety of cofactor binding. In human liver (PDB entry: 1ZNQ19) and rat sperm (PDB entry: 2VYN20), the structures were fully occupied by NAD (one in each subunit). In human placenta (PDB entry: 1U8F21), the structure had three NADs with one subunit free of cofactor. In rabbit muscle (PDB entry: 1J0X22), the structure had two NADs with a single NAD molecule per dimer of the tetrameric enzyme. Whether this diversity of NAD binding resulted from the crystallization conditions is unknown. Studies of E. coli GAPDH23 indicated that NAD bound to the subunits of GAPDH tetramer with different affinities, with two NAD molecules binding with a higher affinity than the remaining two.
Structures of bovine photoreceptor GAPDH retinas were determined as homotetramers with NAD partially and fully bound. The structure of bGAPDH(NAD)4 in this report represents the highest resolution (1.52 Å) structure for any known GAPDH. The high quality these structural models allowed a detailed view of NAD binding in bGAPDH. Key residues and waters involving NAD binding were identified. Structural comparisons of NAD-free and NAD-bound bGAPDH models revealed regions with conformational changes upon NAD binding, including a remote loop region (residues 54–56). Structure analyses confirmed the importance of Phe34 in NAD binding in bGAPDH. Furthermore, binding of NAD in bGAPDH affected the oxidative state of the active site Cys149. With partial NAD binding, Cys149 was environmentally sensitive and found to be oxidized. With full NAD binding, the Cys149 residues were more stable and remained in a reduced state. These results indicate that NAD binding in bGAPDH plays not only a cooperative role, but also a regulatory role in catalytic activity. Together, these findings provide a structural basis for understanding the mechanism of GAPDH activity in vision and its pathological role in retinal disorders.
Materials and Methods
Data collection and processing
All crystals were soaked in their reservoir solutions with added 20% glycerol for cryoprotection. Crystals were frozen in liquid nitrogen before data collection. High resolution data sets were collected at the NSLS X29 and APS NE-CAT 24-ID-C beam lines. A 1.93 Å resolution data set was collected for crystals grown from a crude bovine ROS isotonic extract. The crystal to detector distance was 400 mm with 12% transmission. A 1.52 Å resolution data set was collected for crystals grown from purified bGAPDH in the presence of NAD. The crystal to detector distance here was 290 mm with 10% transmission. Both data sets were collected at a wavelength of 0.979 Å with an exposure time of 1 s and an oscillation range set at 1.0°.
Data were processed and scaled with HKL2000.33 The space group for both bGAPDH crystals was P21, with a unit-cell dimension of a = 79.8, b = 126.5, and c = 83.8. Both data sets were virtually complete (∼99%). Statistics for data collection and processing are summarized in Table 1.
Structural determination and refinement
The structures of bGAPDH were solved by molecular replacement using the program Phaser34 in the CCP4 suite.35 The coordinates of one monomer of rabbit-muscle GAPDH (PDB code: 1J0X22) without ligand and water were used to search the initial model. Sequence identity between bovine and rabbit GAPDH is 98%. Model building calculations employed the diffraction data space group of P21 with components in the crystal asymmetric unit. The structural model of native bGAPDH indicated the presence of three NAD moieties with one subunit free of the cofactor. This model form was named bGAPDH(NAD)3. In contrast, the structural model of purified bGAPDH displayed one NAD in each subunit. This model was designated as bGAPDH(NAD)4. Both models were refined with Refmac536 at 10 cycles using automatic weighting with experimental sigma to obtain the X-ray terms. Protein sequence fitting and manual model adjustments were performed with Coot.37 Based on electron density maps, the geometry of backbone and rotamers of residue side chains were justified, followed by refinement with Refmac5. The final R-factor and R-free values for bGAPDH(NAD)3 were 0.19 and 0.23 at 1.93 Å resolution, and 0.18 and 0.21 for bGAPDH(NAD)4 at the same resolution, respectively.
These structures were validated with the MolProbity server.38 Refinement statistics (Table 1) reflected a well refined, high quality geometric structure with 96.3% of residues located in the most favored regions of a Ramachandran plot, 99.6% in generally allowed regions and only 0.4% in disallowed regions. Residues located in disallowed regions in our structures were found within similar regions in many other GAPDH structures.19–21
Water molecules were added to the present models with Coot. The peak contoured at 1.8σ in 2Fo–Fc maps was identified as a water molecule. These were positioned only when well-defined positive peaks were evident in both 2Fo–Fc and Fo–Fc electron density maps and when they could form a hydrogen bond(s) with either protein atoms or other water molecules.
Structural analyses
The programs Coot, Chimera,39,40 and PyMol (vision 1.5.0, The PyMol Molecular Graphics System, Schrodinger, LLC.) were used for structural analyses. Structure refinement was performed with Coot. Superimposition of structures and RMSD calculations were performed with Chimera. Structural visualization and analyses were performed with Chimera and PyMol. Unless otherwise specified, structure figures were prepared using Chimera.
Acknowledgments
The authors thank Mrs. Susan Farr and Satsumi Roos for assistance with ROS preparation; Drs. Philip Kiser and David Lodowski for helping with data collecting and processing; Dr. Prem Kaushal for helping with structure refinement; Dr. Leslie T. Webster, Jr. for valuable comments on the manuscript. Synchrotron data collections were conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, as well as at the National Synchrotron Light Source on the X29 beamlines, which is operated by the Case Center for Synchrotron Biosciences. K.P. is John H. Hord Professor of Pharmacology.
Glossary
- APS
advanced photon source
- BGP
1,3-diphosphoglycerate
- bGAPDH
bovine GAPDH
- BNL
Brookhaven National Laboratory
- G3P
glyceraldehyde-3-phosphate
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- bGAPDH(NAD)3 and bGAPDH(NAD)4
bovine GAPDH with NAD bound in three or four subunits
- HEPES
2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid
- MS
mass spectroscopy
- NAD
nicotinamide adenine dinucleotide
- NO
nitric oxide
- NSLS
National Synchrotron Light Source
- PDE6
phosphodiesterase 6
- PEG
polyethylene glycol
- RMSD
root-mean-square-deviation(s)
- ROS
rod outer segment(s)
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SEC
size-exclusion chromatography.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
References
- Seidler NW. Basic biology of GAPDH. Adv Exp Med Biol. 2013;985:1–36. doi: 10.1007/978-94-007-4716-6_1. [DOI] [PubMed] [Google Scholar]
- Sirover MA. New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim Biophys Acta. 1999;1432:159–184. doi: 10.1016/s0167-4838(99)00119-3. [DOI] [PubMed] [Google Scholar]
- Sirover MA. On the functional diversity of glyceraldehyde-3-phosphate dehydrogenase: biochemical mechanisms and regulatory control. Biochim Biophys Acta. 2011;1810:741–751. doi: 10.1016/j.bbagen.2011.05.010. [DOI] [PubMed] [Google Scholar]
- Padgett CM, Whorton AR. S-nitrosoglutathione reversibly inhibits GAPDH by S-nitrosylation. Am J Physiol. 1995;269:C739–C749. doi: 10.1152/ajpcell.1995.269.3.C739. [DOI] [PubMed] [Google Scholar]
- Mohr S, Stamler JS, Brune B. Posttranslational modification of glyceraldehyde-3-phosphate dehydrogenase by S-nitrosylation and subsequent NADH attachment. J Biol Chem. 1996;271:4209–4214. doi: 10.1074/jbc.271.8.4209. [DOI] [PubMed] [Google Scholar]
- Hara MR, Cascio MB, Sawa A. GAPDH as a sensor of NO stress. Biochim Biophys Acta. 2006;1762:502–509. doi: 10.1016/j.bbadis.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Hara MR, Snyder SH. Nitric oxide-GAPDH-Siah: a novel cell death cascade. Cell Mol Neurobiol. 2006;26:527–538. doi: 10.1007/s10571-006-9011-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Ohtsuka T, Stell WK. Endogenous nitric oxide enhances the light-response of cones during light-adaptation in the rat retina. Vision Res. 2011;51:131–137. doi: 10.1016/j.visres.2010.10.011. [DOI] [PubMed] [Google Scholar]
- Ko ML, Shi L, Huang CC, Grushin K, Park SY, Ko GY. Circadian phase-dependent effect of nitric oxide on L-type voltage-gated calcium channels in avian cone photoreceptors. J Neurochem. 2013;127:314–328. doi: 10.1111/jnc.12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti R, Aulak KS, Fox PL, Stuehr DJ. GAPDH regulates cellular heme insertion into inducible nitric oxide synthase. Proc Natl Acad Sci USA. 2010;107:18004–18009. doi: 10.1073/pnas.1008133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannibal L, Collins D, Brassard J, Chakravarti R, Vempati R, Dorlet P, Santolini J, Dawson JH, Stuehr DJ. Heme binding properties of glyceraldehyde-3-phosphate dehydrogenase. Biochemistry. 2012;51:8514–8529. doi: 10.1021/bi300863a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok MC, Holopainen JM, Molday LL, Foster LJ, Molday RS. Proteomics of photoreceptor outer segments identifies a subset of SNARE and Rab proteins implicated in membrane vesicle trafficking and fusion. Mol Cell Proteomics. 2008;7:1053–1066. doi: 10.1074/mcp.M700571-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee M. The pathobiology of diabetic complications: a unifying mechanism. Diabetes. 2005;54:1615–1625. doi: 10.2337/diabetes.54.6.1615. [DOI] [PubMed] [Google Scholar]
- Du Y, Miller CM, Kern TS. Hyperglycemia increases mitochondrial superoxide in retina and retinal cells. Free Radic Biol Med. 2003;35:1491–1499. doi: 10.1016/j.freeradbiomed.2003.08.018. [DOI] [PubMed] [Google Scholar]
- Madsen-Bouterse S, Mohammad G, Kowluru RA. Glyceraldehyde-3-phosphate dehydrogenase in retinal microvasculature: implications for the development and progression of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2010;51:1765–1772. doi: 10.1167/iovs.09-4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatnik M, Frizzell N, Thorpe SR, Baynes JW. Inactivation of glyceraldehyde-3-phosphate dehydrogenase by fumarate in diabetes: formation of S-(2-succinyl)cysteine, a novel chemical modification of protein and possible biomarker of mitochondrial stress. Diabetes. 2008;57:41–49. doi: 10.2337/db07-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowluru RA, Atasi L, Ho YS. Role of mitochondrial superoxide dismutase in the development of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2006;47:1594–1599. doi: 10.1167/iovs.05-1276. [DOI] [PubMed] [Google Scholar]
- Chiu CJ, Taylor A. Dietary hyperglycemia, glycemic index and metabolic retinal diseases. Prog Retin Eye Res. 2011;30:18–53. doi: 10.1016/j.preteyeres.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail SA, Park HW. Structural analysis of human liver glyceraldehyde-3-phosphate dehydrogenase. Acta Crystallogr D. 2005;61:1508–1513. doi: 10.1107/S0907444905026740. [DOI] [PubMed] [Google Scholar]
- Frayne J, Taylor A, Cameron G, Hadfield AT. Structure of insoluble rat sperm glyceraldehyde-3-phosphate dehydrogenase (GAPDH) via heterotetramer formation with Escherichia coli GAPDH reveals target for contraceptive design. J Biol Chem. 2009;284:22703–22712. doi: 10.1074/jbc.M109.004648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins JL, Tanner JJ. High-resolution structure of human d-glyceraldehyde-3-phosphate dehydrogenase. Acta Crystallogr D. 2006;62:290–301. doi: 10.1107/S0907444905042289. [DOI] [PubMed] [Google Scholar]
- Cowan-Jacob SW, Kaufmann M, Anselmo AN, Stark W, Grutter MG. Structure of rabbit-muscle glyceraldehyde-3-phosphate dehydrogenase. Acta Crystallogr D. 2003;59:2218–2227. doi: 10.1107/s0907444903020493. [DOI] [PubMed] [Google Scholar]
- Kuzminskaya EV, Asryants RA, Nagradova NK. Rabbit muscle d-glyceraldehyde-3-phosphate dehydrogenase: half-of-the-sites reactivity of the enzyme modified at arginine residues. Biochem Biophys Res Commun. 1992;187:577–583. doi: 10.1016/0006-291x(92)91233-g. [DOI] [PubMed] [Google Scholar]
- Hsu SC, Molday RS. Glyceraldehyde-3-phosphate dehydrogenase is a major protein associated with the plasma membrane of retinal photoreceptor outer segments. J Biol Chem. 1990;265:13308–13313. [PubMed] [Google Scholar]
- Ramachandran GN, Ramakrishnan C, Sasisekharan V. Stereochemistry of polypeptide chain configurations. J Mol Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- Buehner M, Ford GC, Moras D, Olsen KW, Rossmann MG. Structure determination of crystalline lobster d-glyceraldehyde-3-phosphate dehydrogenase. J Mol Biol. 1974;82:563–585. doi: 10.1016/0022-2836(74)90249-6. [DOI] [PubMed] [Google Scholar]
- Rao ST, Rossmann MG. Comparison of super-secondary structures in proteins. J Mol Biol. 1973;76:241–256. doi: 10.1016/0022-2836(73)90388-4. [DOI] [PubMed] [Google Scholar]
- Talfournier F, Colloc’h N, Mornon JP, Branlant G. Functional characterization of the phosphorylating d-glyceraldehyde 3-phosphate dehydrogenase from the archaeon Methanothermus fervidus by comparative molecular modelling and site-directed mutagenesis. Eur J Biochem. 1999;265:93–104. doi: 10.1046/j.1432-1327.1999.00681.x. [DOI] [PubMed] [Google Scholar]
- Duee E, Olivier-Deyris L, Fanchon E, Corbier C, Branlant G, Dideberg O. Comparison of the structures of wild-type and a N313T mutant of Escherichia coli glyceraldehyde 3-phosphate dehydrogenases: implication for NAD binding and cooperativity. J Mol Biol. 1996;257:814–838. doi: 10.1006/jmbi.1996.0204. [DOI] [PubMed] [Google Scholar]
- Roitel O, Vachette P, Azza S, Branlant G. P but not R-axis interface is involved in cooperative binding of NAD on tetrameric phosphorylating glyceraldehyde-3-phosphate dehydrogenase from Bacillus stearothermophilus. J Mol Biol. 2003;326:1513–1522. doi: 10.1016/s0022-2836(03)00049-4. [DOI] [PubMed] [Google Scholar]
- Tien YC, Chuankhayan P, Huang YC, Chen CD, Alikhajeh J, Chang SL, Chen CJ. Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and sulfate suggest involvement of Phe37 in NAD binding for catalysis. Plant Mol Biol. 2012;80:389–403. doi: 10.1007/s11103-012-9953-7. [DOI] [PubMed] [Google Scholar]
- Talfournier F, Colloc’h N, Mornon JP, Branlant G. Comparative study of the catalytic domain of phosphorylating glyceraldehyde-3-phosphate dehydrogenases from bacteria and archaea via essential cysteine probes and site-directed mutagenesis. Eur J Biochem. 1998;252:447–457. doi: 10.1046/j.1432-1327.1998.2520447.x. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromol Cryst A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB. Headd JJ, Keedy DA, Immormino RM, Kapral GF, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF chimera. J Struct Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Yang Z, Lasker K, Schneidman-Duhovny D, Webb B, Huang CC, Pettersen EF, Goddard TD, Meng EC, Sali A, Ferrin TE. UCSF chimera, MODELLER, and IMP: an integrated modeling system. J Struct Biol. 2012;179:269–278. doi: 10.1016/j.jsb.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


