Abstract
We aimed to validate genetic variants as instruments for insulin resistance and secretion, to characterise their association with intermediate phenotypes, and to investigate their role in T2D risk among normal-weight, overweight and obese individuals.We investigated the association of genetic scores with euglycaemic-hyperinsulinaemic clamp- and OGTT-based measures of insulin resistance and secretion, and a range of metabolic measures in up to 18,565 individuals. We also studied their association with T2D risk among normal-weight, overweight and obese individuals in up to 8,124 incident T2D cases. The insulin resistance score was associated with lower insulin sensitivity measured by M/I value (β in SDs-per-allele [95%CI]:−0.03[−0.04,−0.01];p=0.004). This score was associated with lower BMI (−0.01[−0.01,−0.0;p=0.02) and gluteofemoral fat-mass (−0.03[−0.05,−0.02;p=1.4×10−6), and with higher ALT (0.02[0.01,0.03];p=0.002) and gamma-GT (0.02[0.01,0.03];p=0.001). While the secretion score had a stronger association with T2D in leaner individuals (pinteraction=0.001), we saw no difference in the association of the insulin resistance score with T2D among BMI- or waist-strata(pinteraction>0.31). While insulin resistance is often considered secondary to obesity, the association of the insulin resistance score with lower BMI and adiposity and with incident T2D even among individuals of normal weight highlights the role of insulin resistance and ectopic fat distribution in T2D, independently of body size.
Keywords: Genetics, type 2 diabetes, insulin resistance, insulin secretion, adipose expandability
Introduction
Type 2 diabetes (T2D) develops when insulin secretion is insufficient to maintain normoglycaemia, often in the context of an obesity-induced increase in insulin demand i.e. insulin resistance (1). Despite the importance of obesity as a risk factor for T2D, clinical heterogeneity exists in pathways leading to T2D. A recent report from the EPIC-InterAct study showed that over 10% of incident cases of T2D occurred among individuals of normal weight, and over 50% occurred in individuals who were non-obese at baseline (2). It was also shown that waist circumference was associated with risk of T2D within BMI strata, suggesting that for a given BMI the pattern of fat storage is an important determinant of T2D risk. Indeed, being overweight, but with a high waist circumference made future risk of T2D comparable to that of obese individuals (2).
Most genetic variants associated with T2D are implicated in beta-cell function (3). Recent studies revealed a stronger effect of these variants on T2D in lean individuals (4), highlighting the role of impaired insulin secretion in individuals who develop T2D in the absence of obesity. The relative role of insulin resistance in T2D, independent of obesity, has been more difficult to disentangle. This is partly attributable to the strong correlation between obesity and insulin resistance and also because gold standard measures are seldom feasible in large-scale prospective studies. Rare monogenic examples of severe insulin resistance in lean patients have been described (5). Amongst these syndromes, patients with lipodystrophy exhibit severe insulin resistance, metabolic dyslipidaemia and diabetes resulting from impaired adipose tissue function. However, the role of common genetic variants associated with insulin resistance in the aetiology of T2D, particularly among non-obese individuals remains poorly documented.
Initial evidence that genetic approaches can highlight specific aetiological pathways comes from recent investigations showing that individuals carrying body-fat-lowering alleles at the IRS1 locus are insulin resistant and have a higher risk of dyslipidemia, T2D and CHD (6). As part of the Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC), we have recently identified 19 SNPs associated with fasting insulin (including IRS1), 10 of which were also associated with a dyslipidaemic profile suggestive of a role in insulin resistance (7,8). Such genetic variants allow the opportunity to investigate the correlates and consequences of lifelong genetic susceptibility to insulin resistance and/or insulin secretion independently of obesity (7-9).
This study therefore aimed to a) validate the use of recently identified common genetic variants as specific markers for insulin resistance or secretion; b) characterise associations between these variants and detailed metabolic measures including measures of body size, fat mass and distribution; and c) use these instruments to investigate the contributions of insulin resistance and secretion to the risk of T2D in normal-weight, overweight and obese individuals.
Research Design and Methods
Cohort characteristics
Up to 1,374 (range by phenotype (Nrange): 1136-1374) individuals without diabetes who attended phase 3 of the MRC Ely study (10) had relevant phenotypic measurements and genotyping from the Illumina CardioMetabochip (Metabochip). Up to 4,322 individuals from the Fenland study (11) without diabetes and with Metabochip genotyping (Nrange=2,618-2973) or imputed into 1000 Genomes (12) using Impute from the Affymetrix 5.0 genotyping chip (Nrange: 1223-1357) were included. The definition of regional compartments in DXA data is shown in Supplementary Figure 1. We performed sensitivity analyses subtracting the gynoid component from leg estimates to avoid double-counting of gynoid mass. Up to 1,031 (Nrange: 923-1031) individuals from the RISC study (13) who underwent a euglycemic-hyperinsulinemic clamp were included. Genotyping was performed at KBioscience and imputed into 1000 Genomes using MACH and minimac. Up to 909 (Nrange: 884-909) non-diabetic participants from the ULSAM study (14) were included and had genotyping available from Metabochip. Participants were of European ancestry. Participant characteristics and measurement availability for each study are shown in Table 1, while the number of participants included in each analysis is also shown in Figures 1-3.
Table 1.
Study descriptives of each of the five participating studies, along with details of the genetic risk scores. M/I in RISC is reported in (micromol/kgFFM/min/nmol/l)/1000, while M/I in ULSAM is reported as mg/kg bw/min/mU/l*100.
| Study |
||||||
|---|---|---|---|---|---|---|
| MRC-Ely | RISC | ULSAM | Fenland (Metabochip) | Fenland (Genomewide) | EPIC-InterAct subcohort | |
| N (M/F) | 1374 (629/745) | 1031(453/578) | 907 (907/0) | 2978 (1403/1575) | 1357 (597/760) | 16,154 (6,111/10043) |
| Age (years) | 60.9 (9.2) | 43.96(8.37) | 70.98 (0.59) | 47.0 (7.1) | 45.0 (7.3) | 52.4 (9.2) |
| Height (cm) | 167.5 (8.9) | 170.91 (9.32) | 175.12 (5.95) | 170.6 (9.5) | 169.8 (9.3) | 166.2 (9.3) |
| BMI (kg/m2) | 27.03 (4.68) | 25.47 (4.01) | 25.97 (3.22) | 26.7 (4.9) | 27.1 (4.9) | 26.0 (4.2) |
| Waist circumference (cm) | 92.57 (13.2) | 86.46 (12.70) | 93.85 (9.12) | 90.5 (13.4) | 92.2 (13.6) | 86.4 (12.7) |
| M/I | 0.14(0.07) | 5.43(2.42) | ||||
| Matsuda Index | 7.09 (4.60) | 6.42(1.86) | 4.30(2.66) | |||
| Insulinogenic index (pmolinsulin/mmolglucose) |
115.70 (108.0) |
30.42 (19.86) |
||||
| Fasting glucose (mmol/L) | 4.97 (0.54) | 5.05(0.56) | 5.34(0.53) | 4.8 (0.71) | 4.89 (0.61) | |
| Fasting insulin (pmol/L) | 55.84 (34.64) | 34.4(18.68) | 72.41(40.70) | 45.1 (11.2) | 46.4 (32.9) | |
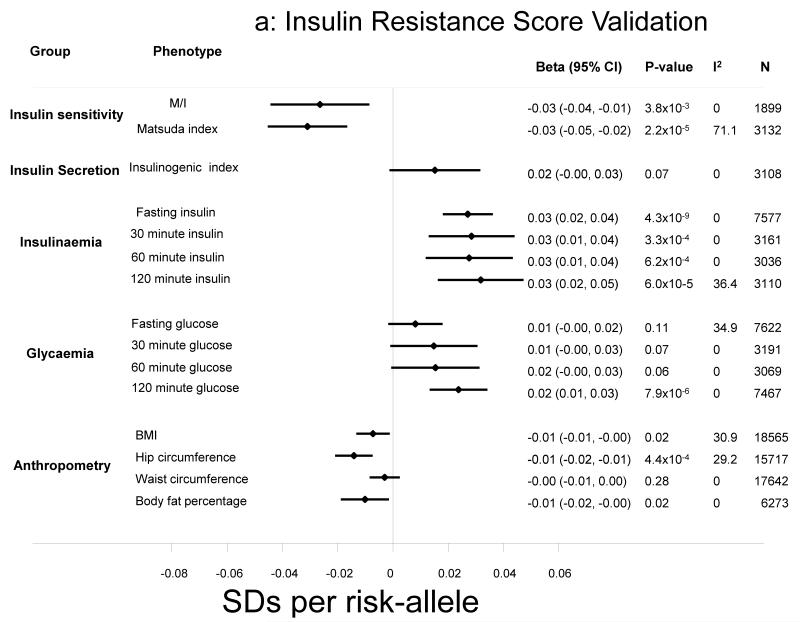
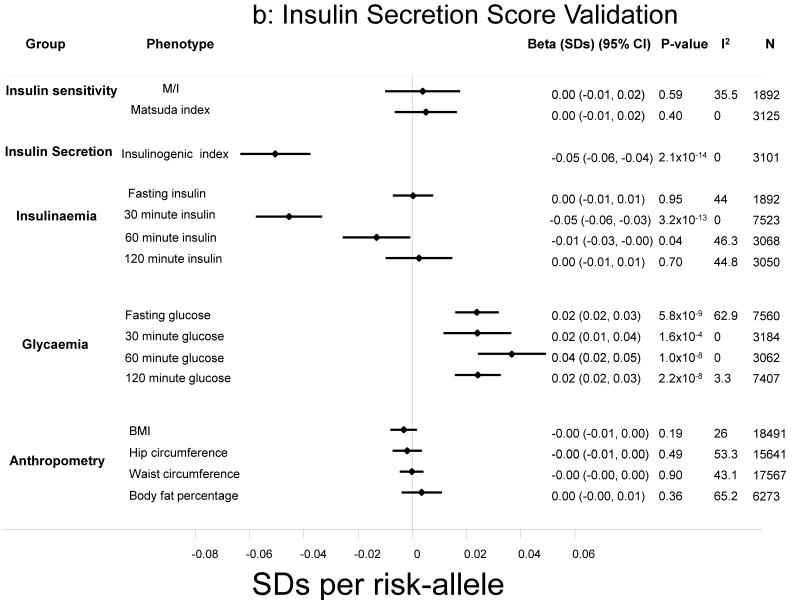
Figure 1.
a-b: Association of the insulin resistance and secretion risk scores with a range of standardised outcomes. Effect sizes are expressed per-risk allele. All models were adjusted for age, sex and BMI, other than anthropometric traits, which were adjusted only for age and sex.
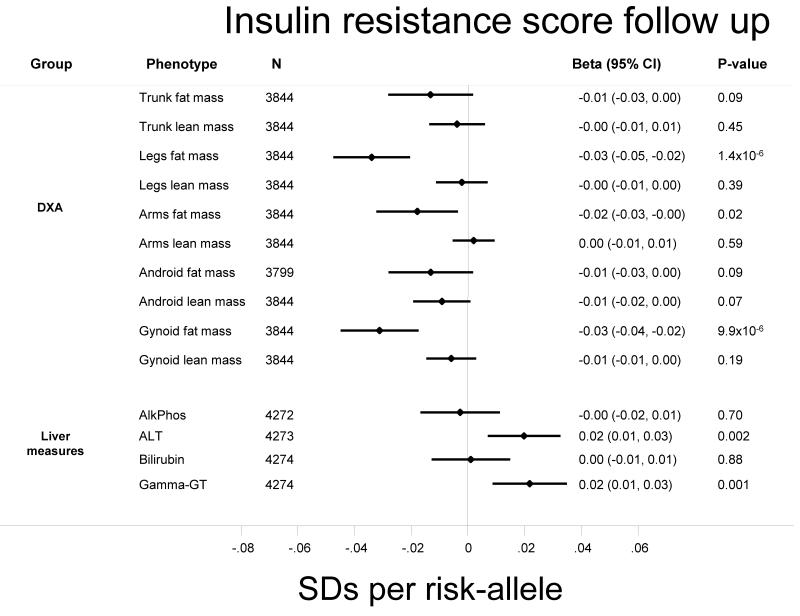
Figure 2.
Association of the insulin resistance score on standardised anthropometric traits in the Fenland study. Effect sizes are expressed per-risk allele. All models were adjusted for age and sex.
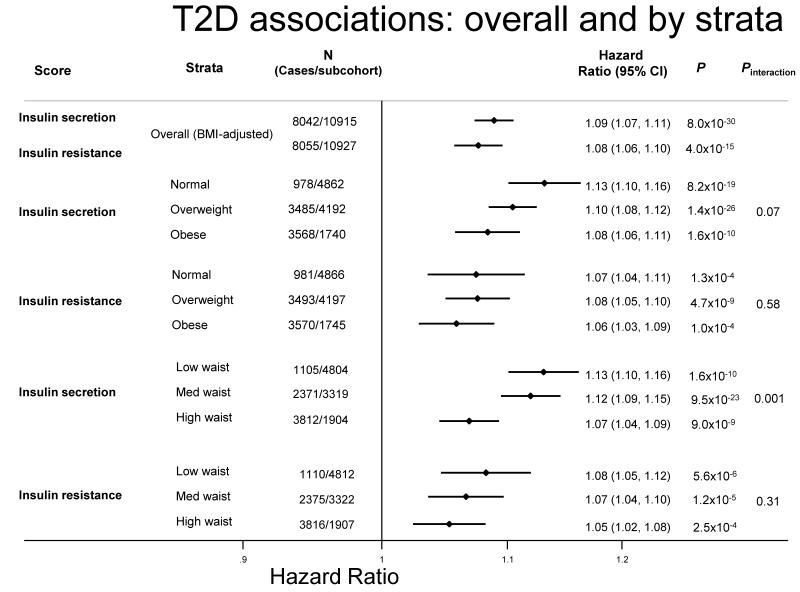
Figure 3.
Association of the risk scores with type 2 diabetes in the EPIC-InterAct study. Associations are shown overall and by strata of BMI and waist circumference at baseline. BMI strata were defined by WHO BMI cutoffs and waist circumference strata were defined by sex-specific tertiles (low: M<94cm, F<78.5cm; med: M>94-103cm, F>78.5-90cm; high: M>103cm, F>90cm).
We tested associations between genetic risk scores and incident diabetes in the EPIC-InterAct study (15), a case-cohort study nested within European Prospective Investigation into Cancer and Nutrition (EPIC) cohorts which includes 12,403 incident cases of T2D and a subcohort of 16,154 individuals (including 778 randomly selected incident T2D cases). A maximum of 18,676 participants (8,136 incident cases, 10,540 non-cases) had genotypes available from the Metabochip (N=9,361) or Illumina 660W-Quad Chip (N=9,290) imputed into 1000 Genomes and were included in the current study. Up to 10,923 participants (Nrange: 10,029-10,923) from the EPIC-InterAct subcohort were also included in quantitative trait analyses (Table 1).
All participants gave written informed consent, and studies were approved by local ethics committees and the Internal Review Board of the International Agency for Research on Cancer.
Genetic risk scores
We created unweighted (i.e. per-allele) genetic risk scores for insulin resistance and impaired insulin secretion using effect alleles defined from the literature as shown in Supplementary Table 1. The insulin resistance genetic score comprised variants associated with fasting insulin in recent meta-analyses (8). In order to improve specificity we restricted the insulin resistance score to the 10 variants showing association (p<0.05) with lower HDL and higher triglycerides (8,16): a hallmark of common insulin resistance. This excluded TCF7L2, associated principally with insulin secretion (17), and FTO, whose effect on insulin levels was entirely mediated by BMI (8). Variants included were those in or near the IRS1, GRB14, ARL15, PPARG, PEPD, ANKRD55/MAP3K1, PDGFC, LYPLAL1, RSPO3, and FAM13A1 genes (Supplementary Table 1). For the insulin secretion score, from loci associated with T2D and related traits (18-21) we undertook literature searches to identify SNPs showing an association with impaired early insulin secretion. In addition, we investigated the literature to identify additional candidate genes associated with early insulin secretion. Up to 21 variants associated with decreased early insulin secretion were included in the insulin secretion score. Where SNPs were missing, we included a proxy where available (Supplementary Table 1), and where no proxy was available, we did not impute missing SNPs. Each SNP, the reason for inclusion in the score and its availability in each study is shown in Supplementary Table 1. The genetic score distributions in each study are shown in for the insulin secretion and insulin resistance scores in Supplementary Figure 2a and b, respectively.
Statistical analysis
In order to meta-analyse data from multiple studies centrally, each study first natural-log transformed and standardised the phenotype, such that for each variable the mean was equal to zero and SD equal to one. Each study then fit linear regression models on each of these outcomes using the genetic risk scores as exposures, adjusted for age and sex (with and without adjustment for BMI). Genetic risk scores were unweighted and effect sizes expressed per fasting insulin-raising or insulin secretion-lowering allele, respectively. We investigated the association of these risk scores with euglycaemic-hyperinsulinaemic clamp (22) and OGTT-based measures of insulin sensitivity and secretion (23,24). We also investigated the associations of scores with glycaemia and insulinaemia during the OGTT, lipids, BMI, waist and hip-circumferences, and body fat percentage (assessed by bioimpedance in RISC, and by DXA in Fenland). We performed fixed-effect, inverse-variance weighted meta-analyses using Stata SE-12.1 software (StataCorp LP, College Station, TX). Associations with T2D in the EPIC-InterAct study were investigated using Prentice-weighted Cox regression with age as the underlying time variable, adjusted for age at entry (to account for potential cohort effects), sex and centre of recruitment. BMI strata were defined by WHO cutoffs and waist circumference strata were defined by sex-specific tertiles. Interactions of risk scores with BMI and waist circumference were tested by including the product term of risk scores and BMI categories or waist circumference tertiles. Effect sizes were expressed as hazard ratios (HR) per-risk allele.
Results
Validation of genetic risk scores: associations with insulin sensitivity and secretion
The insulin resistance score was associated with lower whole-body insulin sensitivity based on the M/I value from euglycaemic-hyperinsulinaemic clamps (β in standard deviations per-allele [95% CI]: −0.03 [−0.04, −0.01]; p=0.004) (Figure 1a) and with lower Matsuda index (β = −0.03 [−0.05, −0.02], p=2.2×10−5) calculated from frequently-sampled OGTTs (Figure 1a). The score was not associated with insulinogenic index, but was associated with higher insulin levels throughout the OGTT (p<0.001) and higher levels of glycaemia, albeit only statistically significant for 2-h glucose (Figure 1a).
In contrast, the insulin secretion score was associated with lower insulinogenic index (−0.05 [−0.06, −0.04], p=2.1×10−14) and lower 30-minute insulin levels (−0.05 [−0.06, −0.03], p=3.2×10−13), but showed no associations with any of the measures of insulin resistance including M/I (0.00 [−0.01, 0.02], p=0.59), Matsuda index (0.00 [−0.01, 0.02], p=0.40) or fasting insulin (0.00 [−0.01, 0.01], p=0.95) (Figure 1b). Unlike the insulin resistance score, associations of the insulin secretion score with lower post-challenge insulin were accompanied by higher glucose levels at all time-points (p-values<1.7×10−4).
Associations with detailed anthropometric and metabolic traits
The insulin resistance score was strongly associated with both higher triglycerides (0.03 [0.02, 0.03], p=3.5×10−20) and lower HDL-cholesterol (−0.02 [−0.03, −0.02], p=1.6×10−14). It was also associated with lower BMI (−0.01 [−0.01, −0.00], p=0.02), smaller hip circumference (−0.01 [−0.02, −0.01], p=4.4×10−4) and lower body fat percentage (−0.01 [−0.02, −0.00], p=0.02) (Figure 1a). Also, the insulin resistance score was also associated with lower BMI when we restricted analyses to incident cases of T2D in the EPIC-InterAct study (N=7577; −0.02 [−0.03, −0.01], p=0.001). Further investigation of detailed anthropometric measures obtained by DXA in Fenland participants highlighted inverse associations of the score with fat mass in different body compartments (Supplementary Figure 1). The strongest associations were observed for leg (−0.03 [−0.05, −0.02], p=1.4×10−6) and gynoid fat mass (−0.03 [−0.04, −0.02], p=9.9×10−6). These associations remained after excluding PPARG and IRS1 variants from the genetic score (leg (−0.03 [−0.04, −0.01], p=1.2×10−4) and gynoid fat mass (−0.03 [−0.04, −0.01], p=3.4×10−4). Sensitivity analysis on leg fat mass removing the gynoid region, showed that the association remained highly significant (−0.03 [−0.05, −0.02], p=9.2×10−7). For these associations, we saw similar magnitudes of association in men and women, which were statistically significant (p<0.05) in both genders. We also saw an association with arm fat mass (−0.02 [−0.03, −0.00], p=0.02), but not with lean mass measurements (Figure 2).
In order to investigate the possibility that lower levels of gluteofemoral fat mass might limit subcutaneous fat storage and hence increase ectopic fat deposition, we also investigated the association of the insulin resistance score with estimates of liver damage. The score was associated with both higher ALT (0.02 [0.01, 0.03], p=0.002) and gamma-GT (0.02 [0.01, 0.03], p=0.001). The insulin resistance score was not associated with self-reported alcohol intake (p=0.66) and associations with liver enzymes were unchanged after adjustment alcohol intake (ALT: 0.02 [0.01, 0.04], p=0.002. gamma-GT: 0.02 [95% CI 0.01, 0.04], p=0.003).
The insulin secretion score was not associated with triglycerides or HDL-cholesterol (p>0.1), nor with any of the anthropometric traits (p>0.18) (Figure 1b). The secretion score was nominally associated with higher android fat mass (p=0.04), but with no other parameters in the DXA data. We saw a weak association of the insulin secretion score with higher levels of ALT (0.01 [0.00, 0.02], p=0.02), but not gamma-GT (0.00 [−0.00, 0.02], p=0.24).
Associations with T2D
Both the insulin secretion (Hazard ratio (HR) [95%CI]: 1.09 [1.07, 1.11]; p=8.0×10−30) and resistance scores (1.08 (1.06, 1.10); p=4.0×10−15) were associated with incident T2D (Figure 3). To investigate the relative importance of genetically predicted insulin resistance and secretion on T2D incidence at different levels of BMI, we examined the effect of the score on incident T2D in normal-weight, overweight and obese individuals and found no difference in associations between strata (normal-weight: HR=1.07 [1.04, 1.11], p=1.3×10−4; overweight: HR=1.08 [1.05, 1.10], p=4.7×10−9; obese: HR=1.06 [1.03, 1.09], p=1.0×10−4; pinteraction=0.58) (Figure 3). Nor was there any difference between strata of waist circumference (pinteraction=0.31) (Figure 3). In contrast, the insulin secretion score showed an interaction with waist circumference on the risk of T2D (pinteraction=0.001). The association was stronger in individuals with smaller waist circumference than in those with large waist circumference (lowest third: HR=1.13 [1.10, 1.16], p= 1.6×10−10; middle third:HR=1.12 [1.09, 1.15], p= 9.5×10−23; highest third HR=1.07 [1.04, 1.09], p= 9.0×10−9). There was a similar but non-statistically significant trend for BMI (pinteraction=0.07), with a tendency toward stronger associations in leaner compared to obese individuals (Figure 3).
Discussion
While rare monogenic examples of insulin resistance highlight the causal role of inadequate subcutaneous adipose tissue in the aetiology of cardiometabolic disease, the causal role of impaired adipose expandability and ectopic lipid accumulation in “common” cardiometabolic disease remains largely unproven. We observe that a genetic score for insulin resistance displays a pattern of association (lower subcutaneous adipose tissue and T2D) similar to that observed in monogenic forms of lipodystrophy, implicating a role for inadequate capacity to store surplus lipids in the aetiology of T2D. Furthermore, we show that these genetic scores are associated with incident T2D even in individuals of normal weight, highlighting the role of impaired adipose expandability in T2D independently of BMI.
Validation of the genetic risk scores
We found that the genetic risk score comprising variants previously associated with fasting insulin was associated with euglycaemic-hyperinsulinaemic clamp-based insulin sensitivity. Furthermore, a genetic score comprising variants previously associated with early insulin secretion was strongly associated with 30-minute insulin and insulinogenic index, but not with insulin sensitivity. These associations validated the utility of these risk scores as specific and sensitive genetic instruments to understand the role of both insulin resistance and beta-cell dysfunction in the aetiology of diabetes and other disease outcomes.
Association of genetic risk scores with other metabolic traits
The insulin resistance score was associated with lower BMI, hip circumference, and body fat percentage, and particularly with lower gynoid and leg fat mass (Figure 2): adipose tissue depots considered protective against the complications of ectopic fat deposition (25). A prevailing hypothesis for the pathogenesis of insulin resistance proposes that the capacity of adipose tissue to expand in the face of sustained positive energy balance is finite and that exceeding this limit results in lipid storage in tissues less well adapted to this need (26). This phenomenon of ectopic lipid accumulation has been strongly associated with insulin resistance in multiple studies and plausible, albeit still largely unproven, explanations exist linking this lipid accumulation to impaired insulin action (lipotoxicity) (27). Lipodystrophic disorders are characterised by a primary lack of adipose tissue and present an extreme example of a mismatch between the need and capacity to store surplus lipids. These extremely rare disorders are associated with particularly severe ectopic fat accumulation, dyslipidaemia, insulin resistance and diabetes. Recent reports suggests novel forms of lipodystrophy which may be more common (28). Here, we report evidence that common genetic variants show associations similar to those observed in rare, monogenic lipodystrophies (Figure 2), and highlight a potential role a mismatch between the need and capacity to store surplus lipids in “common” metabolic disease. Associations with elevated ALT and gamma-GT are indicative of hepatic fat deposition, consistent with ectopic lipid accumulation. While elevated ALT and gamma-GT are associated with fatty liver (29), they can be elevated in response to other forms of liver injury or disease (30) including alcohol consumption, medication or hepatitis. While we could not exclude the possibility that the insulin resistance score was associated with higher ALT and gamma-GT via mechanisms other than fatty liver, associations with ALT and gamma-GT were unchanged after adjustment for self-reported alcohol intake, and other forms of disease sufficiently rare that in this healthy middle-aged population we consider them unlikely to explain our findings.
Associations with incident disease
While obesity is a major risk factor for insulin resistance and T2D, there is considerable inter-individual variation in the metabolic response to obesity, with some individuals apparently protected from the typical consequences of obesity (31). It has previously been shown that insulin sensitive obese individuals have lower visceral and hepatic fat content than insulin resistant obese individuals as well as a lower intima-media thickness (32), further implicating ectopic fat deposition as a determinant of the metabolic consequences of obesity. Despite negative confounding by BMI, we saw an association of the insulin resistance score with incident T2D. As the adipose tissue of obese individuals is placed under greater demand for fat storage, we hypothesized that the insulin resistance score would have a larger effect on T2D risk in these individuals than in normal-weight individuals. However, the insulin resistance score was associated with incident T2D even in normal-weight individuals and those with the lowest waist circumference (Figure 3), with similar effect sizes to obese individuals. This suggests that the relationship between adipose expandability and positive energy balance is not subject to a threshold effect, but may result in degrees of ectopic fat accumulation even in people with a “normal” BMI. This is reminiscent of what is observed in South Asian individuals who have been reported to have higher visceral fat and exacerbated metabolic consequences for a given BMI (33). While the role of beta-cell function has been highlighted in the aetiology of T2D among lean individuals (4), our findings also highlight the role of impaired adipose expandability and insulin resistance in T2D in lean individuals.
Recent analyses have highlighted the causal role of increased adiposity in impaired cardiometabolic health (34), and we now highlight a causal link between insulin resistance and incident disease, completely independent of BMI. The obesity epidemic is heavily implicated in driving the increased incidence of metabolic disease (35,36). However, while there is some suggestion that hyperinsulinaemia can be a cause and consequence of obesity (37), we observe that genetically predicted insulin resistance and hyperinsulinaemia are associated with lower adiposity (Figure 2). However, as we restricted our genetic score to those variants associated with dyslipidaemia (to improve specificity), we cannot exclude the possibility that primary insulin resistance of another form could cause obesity, or display different associations with metabolic traits or disease. While insulin resistance is strongly associated with obesity and the secular trends in obesity raise concerns about the growing consequences of insulin resistance (35), of the 19 loci recently found to be associated with fasting insulin levels, only one (FTO) was mediated entirely by higher BMI, highlighting the role of other pathways in the aetiology of insulin resistance. Furthermore, the association of 10 of the 19 SNPs with dyslipidaemia (indicative of postreceptor-mediated insulin resistance (38)), implicate this as a prevalent form of common insulin resistance.
As our cross-sectional analyses were restricted to individuals without diagnosed diabetes, we considered the possibility that the insulin resistance score association with lower BMI and adiposity were are as a result of a truncation effect. I.e. participants with both higher BMI and higher genetic predisposition to insulin resistance had a higher risk of T2D and were preferentially excluded from the sample (3). However, we observed the same association of the score with lower BMI in the incident cases of T2D in the EPIC-InterAct study, suggesting that this association is not wholly attributable to truncation effects.
A limitation of our approach is that we cannot ascribe a specific direction to the associations of the score with insulin resistance and adiposity. For example, while these loci were among the top signals in a genome-wide association study of fasting insulin, it is unclear whether they have a primary association with insulin resistance or with adipocyte function. Indeed, a variant near IRS1 is included in this list, and while absence of IRS1 is known to result in insulin resistance through impaired insulin signal transduction (39), IRS1 also influences adipocyte differentiation (40). Indeed, IRS1 was previously associated with body fat percentage (6), where the allele associated with higher body fat percentage was associated with a favourable metabolic profile, including lower risk of T2D and cardiovascular disease. Here, we see the same pattern of association for our genetic risk score and thereby highlight a number of genetic variants that may be influential in the ability to store surplus lipids optimally. The association with insulin resistance was apparently paradoxically accompanied by lower body fat percentage and lower BMI. However, these results are paradoxical only when considered in the context of the observational epidemiological association between higher adiposity and insulin resistance. Our results, in combination with observations from individuals with monogenic lipodystrophy, suggest that these loci may have primary effects on subcutaneous adipocyte function, which then results in insulin resistance via ectopic lipid deposition. While we perform analyses using a combined genetic score, our findings implicate each of these loci in the aetiology of insulin resistance and body fat distribution. This conclusion is supported by findings in an accompanying article (Yaghootkar et al, Diabetes, submitted), which independently identify the same variants in our score as being associated with a “monogenic lipodystrophy-like” phenotype using a hypothesis-free clustering approach. The inclusions of PPARG and IRS1 in the insulin resistance score further highlight the likely role of adipocyte function in their associations with insulin resistance. However, even after removing PPARG and IRS1 variants from the insuiln resistance score, we observed consistent associations with body fat distribution. Furthermore, LYPLAL1, GRB14 and RSPO3 have been associated with waist-hip ratio at genome-wide levels of significance (41).
Conclusions
Genetic scores for insulin resistance and secretion based on common variants are valid tools to study the role of these features in a range of disease processes. In particular, while insulin resistance as a cause of T2D is largely considered to be a consequence of obesity, we highlight the role of polygenic insulin resistance in the development of T2D independent of body size. Furthermore, the association of these variants with lower subcutaneous fat mass and suggestion of ectopic fat deposition highlight the role of impaired adipose expandability and body fat distribution in T2D even among lean individuals.
Supplementary Material
Supplementary Table 1: SNPs included in the genetic risk scores, with the study in which they were identified or implicated in insulin secretion or resistance. The risk allele for each of these SNPs is also shown. Where the lead SNP was not available, the proxy used is listed for each study. Where a suitable proxy was not available, the SNP is marked “x”.
Supplementary Figure 1: Regional compartment definition in the Fenland DXA data. The trunk region extends from the chin to the top of the pelvis. The leg regions are defined by a cut across the femoral neck, not touching the pelvis. The arm regions are defined by a cut placed as close to the body as possible. The android region is a quadrilateral box, where the lower boundary is the pelvis, the lateral boundaries are the arm cuts and sthe upper boundary is above the pelvis cut by 20% of the distance between the pelvis and neck cuts. The gynoid region is another quadrilateral box where the lateral boundaries are the arm cuts. The upper boundary is below the pelvis cut by 1.5 x the height of the android region. The Lower boundary is below the upper boundary by 2 x the height of the android region
Supplementary Figure 2: Histograms of genetic risk scores from each study for a) insulin secretion and b) insulin resistance scores.
Acknowledgements
The MRC-Ely Study was funded by the Medical Research Council (MC_U106179471) and Diabetes UK. We are grateful to all the volunteers, and to the staff of St. Mary’s Street Surgery, Ely and the study team. The Fenland Study is funded by the Medical Research Council (MC_U106179471) and Wellcome Trust. We are grateful to all the volunteers for their time and help, and to the General Practitioners and practice staff for assistance with recruitment. We thank the Fenland Study Investigators, Fenland Study Co-ordination team and the Epidemiology Field, Data and Laboratory teams. DBS and RKS are funded by the Wellcome Trust, the U.K. NIHR Cambridge Biomedical Research Centre and the MRC Centre for Obesity and Related Metabolic Disease. Genotyping in ULSAM was performed by the SNP&SEQ Technology Platform in Uppsala (www.genotyping.se), which is supported by Uppsala University, Uppsala University Hospital, Science for Life Laboratory - Uppsala and the Swedish Research Council (Contracts 80576801 and 70374401). The RISC Study was supported by European Union grant QLG1-CT-2001-01252 and AstraZeneca. The RISC Study Project Management Board: B Balkau, F Bonnet, SW Coppack, JM Dekker, E Ferrannini, A Golay, A Mari, A Natali, J Petrie, M Walker. We thank all EPIC participants and staff for their contribution to the study. We thank the lab team at the MRC Epidemiology Unit for sample management and Nicola Kerrison of the MRC Epidemiology Unit for data management. Funding for the EPIC-InterAct project was provided by the EU FP6 programme (grant number LSHM_CT_2006_037197). In addition, EPIC-InterAct investigators acknowledge funding from the following agencies: PWF: Swedish Research Council, Novo Nordisk, Swedish Diabetes Association, Swedish Heart-Lung Foundation; LCG: Swedish Research Council; NS: Health Research Fund (FIS) of the Spanish Ministry of Health; Murcia Regional Government (N° 6236); LA: We thank the participants of the Spanish EPIC cohort for their contribution to the study as well as to the team of trained nurses who participated in the recruitment; RK: German Cancer Aid, German Ministry of Research (BMBF); TJK: Cancer Research UK; PMN: Swedish Research Council; KO: Danish Cancer Society; SP: Compagnia di San Paolo; JRQ: Asturias Regional Government; OR: The Västerboten County Council; AMWS and DLvdA: Dutch Ministry of Public Health, Welfare and Sports (VWS), Netherlands Cancer Registry (NKR), LK Research Funds, Dutch Prevention Funds, Dutch ZON (Zorg Onderzoek Nederland), World Cancer Research Fund (WCRF), Statistics Netherlands; RT: AIRE-ONLUS Ragusa, AVIS-Ragusa, Sicilian Regional Government; IS: Verification of diabetes cases was additionally funded by NL Agency grant IGE05012 and an Incentive Grant from the Board of the UMC Utrecht; IB: Wellcome Trust grant 098051 and United Kingdom NIHR Cambridge Biomedical Research Centre; MIM: InterAct, Wellcome Trust (083270/Z/07/Z), MRC (G0601261); ER: Imperial College Biomedical Research.
Footnotes
None of the authors declared a conflict of interest. Inês Barroso and her spouse own stock in the companies GlaxoSmithKline (GSK) and Incyte (INCY).
References
- 1.Kahn SE. The relative contributions of insulin resistance and beta-cell dysfunction to the pathophysiology of Type 2 diabetes. Diabetologia. 2003 Jan;46(1):3–19. doi: 10.1007/s00125-002-1009-0. [DOI] [PubMed] [Google Scholar]
- 2.Langenberg C, Sharp SJ, Schulze MB, Rolandsson O, Overvad K, Forouhi NG, et al. PLoS medicine. 6. Vol. 9. Public Library of Science; Jan 5, 2012. Long-term risk of incident type 2 diabetes and measures of overall and regional obesity: the EPIC-InterAct case-cohort study; p. e1001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dimas AS, Lagou V, Barker A, Knowles JW, Mägi R, Hivert M-F, et al. Impact of type 2 diabetes susceptibility variants on quantitative glycemic traits reveals mechanistic heterogeneity. Diabetes. 2013 Dec 2;:3–75. doi: 10.2337/db13-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perry JRB, Voight BF, Yengo L, Amin N, Dupuis J, Ganser M, et al. Stratifying type 2 diabetes cases by BMI identifies genetic risk variants in LAMA1 and enrichment for risk variants in lean compared to obese cases. PLoS genetics. 2012 May;8(5):e1002741. doi: 10.1371/journal.pgen.1002741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Semple RK, Savage DB, Cochran EK, Gorden P, O’Rahilly S. Genetic syndromes of severe insulin resistance. Endocrine reviews. 2011 Aug;32(4):498–514. doi: 10.1210/er.2010-0020. [DOI] [PubMed] [Google Scholar]
- 6.Kilpeläinen TO, Zillikens MC, Stančákova A, Finucane FM, Ried JS, Langenberg C, et al. Nature genetics. 8. Vol. 43. Nature Publishing Group; Aug, 2011. Genetic variation near IRS1 associates with reduced adiposity and an impaired metabolic profile; pp. 753–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manning AK, Hivert M-F, Scott Ra, Grimsby JL, Bouatia-Naji N, Chen H, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nature genetics. 2012 Jun;44(6):659–69. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scott R a, Lagou V, Welch RP, Wheeler E, Montasser ME, Luan J, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nature genetics. 2012 Sep;44(9):991–1005. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ingelsson E, Langenberg C, Hivert M, Prokopenko I, Ma R, Sharp S, et al. Insulin Metabolism in Humans. 2010 May;59 [Google Scholar]
- 10.Forouhi NG, Luan J, Hennings S, Wareham NJ. Incidence of Type 2 diabetes in England and its association with baseline impaired fasting glucose: the Ely study 1990-2000. Diabetic medicine: a journal of the British Diabetic Association. 2007 Feb;24(2):200–7. doi: 10.1111/j.1464-5491.2007.02068.x. [DOI] [PubMed] [Google Scholar]
- 11.Rolfe EDL, Loos RJF, Druet C, Stolk RP, Ekelund U, Griffin SJ, et al. Association between birth weight and visceral fat in adults. The American journal of clinical nutrition. 2010 Aug;92(2):347–52. doi: 10.3945/ajcn.2010.29247. [DOI] [PubMed] [Google Scholar]
- 12.Abecasis GR, Auton A, Brooks LD, DePristo M a, Durbin RM, Handsaker RE, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012 Nov 1;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hills SA, Balkau B, Coppack SW, Dekker JM, Mari A, Natali A, et al. The EGIR-RISC STUDY (The European group for the study of insulin resistance: relationship between insulin sensitivity and cardiovascular disease risk): I. Methodology and objectives. Diabetologia. 2004 Mar;47(3):566–70. doi: 10.1007/s00125-004-1335-5. [DOI] [PubMed] [Google Scholar]
- 14.Zethelius B, Byberg L, Hales CN, Lithell H, Berne C. Proinsulin and acute insulin response independently predict Type 2 diabetes mellitus in men--report from 27 years of follow-up study. Diabetologia. 2003 Jan;46(1):20–6. doi: 10.1007/s00125-002-0995-2. [DOI] [PubMed] [Google Scholar]
- 15.Langenberg C, Sharp S, Forouhi NG, Franks PW, Schulze MB, Kerrison N, et al. Design and cohort description of the InterAct Project: an examination of the interaction of genetic and lifestyle factors on the incidence of type 2 diabetes in the EPIC Study. Diabetologia. 2011 Sep;54(9):2272–82. doi: 10.1007/s00125-011-2182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, et al. Nature. 7307. Vol. 466. Nature Publishing Group; Aug 5, 2010. Biological, clinical and population relevance of 95 loci for blood lipids; pp. 707–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyssenko V, Lupi R, Marchetti P, Guerra S, Del, Orho-melander M, Almgren P, et al. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. 2007;117(8):2155–63. doi: 10.1172/JCI30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nature genetics. 2010 Jul;42(7):579–89. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strawbridge RJ, Dupuis J, Prokopenko I, Barker A, Ahlqvist E, Rybin D, et al. Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes. 2011 Oct;60(10):2624–34. doi: 10.2337/db11-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saxena R, Hivert M-F, Langenberg C, Tanaka T, Pankow JS, Vollenweider P, et al. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nature genetics. 2010 Feb;42(2):142–8. doi: 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nature genetics. 2010 Feb;42(2):105–16. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeFronzo RA, Tobin JD, Andres R. Glucose clamp technique: a method for quantifying insulin secretion and resistance. The American journal of physiology. 1979 Sep;237(3):E214–23. doi: 10.1152/ajpendo.1979.237.3.E214. [DOI] [PubMed] [Google Scholar]
- 23.Matsuda M, DeFronzo RA. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes care. 1999 Sep;22(9):1462–70. doi: 10.2337/diacare.22.9.1462. [DOI] [PubMed] [Google Scholar]
- 24.Seltzer HS, Allen EW, Herron a L, Brennan MT. Insulin secretion in response to glycemic stimulus: relation of delayed initial release to carbohydrate intolerance in mild diabetes mellitus. The Journal of clinical investigation. 1967 Mar;46(3):323–35. doi: 10.1172/JCI105534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manolopoulos KN, Karpe F, Frayn KN. Gluteofemoral body fat as a determinant of metabolic health. International journal of obesity (2005) 2010 Jun;34(6):949–59. doi: 10.1038/ijo.2009.286. Nature Publishing Group. [DOI] [PubMed] [Google Scholar]
- 26.Virtue S, Vidal-Puig A. Biochimica et biophysica acta. 3. Vol. 1801. Elsevier B.V.; Mar, 2010. Adipose tissue expandability, lipotoxicity and the Metabolic Syndrome--an allostatic perspective; pp. 338–49. [DOI] [PubMed] [Google Scholar]
- 27.Savage DB, Petersen KF, Shulman GI. Disordered Lipid Metabolism and the Pathogenesis of Insulin Resistance. 2007;(32):507–20. doi: 10.1152/physrev.00024.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strickland LR, Guo F, Lok K, Garvey WT. Type 2 diabetes with partial lipodystrophy of the limbs: a new lipodystrophy phenotype. Diabetes care. 2013 Aug;36(8):2247–53. doi: 10.2337/dc12-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Angulo P. Nonalcoholic fatty liver disease. The New England journal of medicine. 2002 Apr 18;346(16):1221–31. doi: 10.1056/NEJMra011775. [DOI] [PubMed] [Google Scholar]
- 30.Pratt DS, Kaplan MM. Evaluation of abnormal liver-enzyme results in asymptomatic patients. The New England journal of medicine. 2000 Apr 27;342(17):1266–71. doi: 10.1056/NEJM200004273421707. [DOI] [PubMed] [Google Scholar]
- 31.Sims E a. Are there persons who are obese, but metabolically healthy? Metabolism: clinical and experimental. 2001 Dec;50(12):1499–504. doi: 10.1053/meta.2001.27213. [DOI] [PubMed] [Google Scholar]
- 32.Stefan N, Kantartzis K, Machann J, Schick F, Thamer C, Rittig K, et al. Identification and characterization of metabolically benign obesity in humans. Archives of internal medicine. 2008 Aug 11;168(15):1609–16. doi: 10.1001/archinte.168.15.1609. [DOI] [PubMed] [Google Scholar]
- 33.Sniderman AD, Bhopal R, Prabhakaran D, Sarrafzadegan N, Tchernof A. Why might South Asians be so susceptible to central obesity and its atherogenic consequences? The adipose tissue overflow hypothesis. International journal of epidemiology. 2007 Feb;36(1):220–5. doi: 10.1093/ije/dyl245. [DOI] [PubMed] [Google Scholar]
- 34.Fall T, Hägg S, Mägi R, Ploner A, Fischer K, Horikoshi M, et al. The role of adiposity in cardiometabolic traits: a mendelian randomization analysis. PLoS medicine. 2013 Jun;10(6):e1001474. doi: 10.1371/journal.pmed.1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 2006 Dec 14;444(7121):840–6. doi: 10.1038/nature05482. [DOI] [PubMed] [Google Scholar]
- 36.Li S, Zhao JH, Luan J, Langenberg C, Luben RN, Khaw KT, et al. Genetic predisposition to obesity leads to increased risk of type 2 diabetes. Diabetologia. 2011 Apr;54(4):776–82. doi: 10.1007/s00125-011-2044-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehran AE, Templeman NM, Brigidi GS, Lim GE, Chu K-Y, Hu X, et al. Cell metabolism. 6. Vol. 16. Elsevier Inc; Dec 5, 2012. Hyperinsulinemia drives diet-induced obesity independently of brain insulin production; pp. 723–37. [DOI] [PubMed] [Google Scholar]
- 38.Semple RK, Sleigh A, Murgatroyd PR, Adams CA, Bluck L, Jackson S, et al. Postreceptor insulin resistance contributes to human dyslipidemia and hepatic steatosis. 2009;119(2) doi: 10.1172/JCI37432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamemoto H, Kadowaki T, Tobe K, Yagi T, Sakura H, Hayakawa T, et al. Insulin resistance and growth retardation in mice lacking insulin receptor substrate-1. Nature. 1994 Nov 10;372(6502):182–6. doi: 10.1038/372182a0. [DOI] [PubMed] [Google Scholar]
- 40.Fasshauer M, Klein J, Kristina M, Ueki K, Benito M, Ronald C, et al. Essential Role of Insulin Receptor Substrate 1 in Differentiation of Brown Adipocytes Essential Role of Insulin Receptor Substrate 1 in Differentiation of Brown Adipocytes. 2001 [Google Scholar]
- 41.Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, Steinthorsdottir V, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nature genetics. 2010 Nov;42(11):949–60. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: SNPs included in the genetic risk scores, with the study in which they were identified or implicated in insulin secretion or resistance. The risk allele for each of these SNPs is also shown. Where the lead SNP was not available, the proxy used is listed for each study. Where a suitable proxy was not available, the SNP is marked “x”.
Supplementary Figure 1: Regional compartment definition in the Fenland DXA data. The trunk region extends from the chin to the top of the pelvis. The leg regions are defined by a cut across the femoral neck, not touching the pelvis. The arm regions are defined by a cut placed as close to the body as possible. The android region is a quadrilateral box, where the lower boundary is the pelvis, the lateral boundaries are the arm cuts and sthe upper boundary is above the pelvis cut by 20% of the distance between the pelvis and neck cuts. The gynoid region is another quadrilateral box where the lateral boundaries are the arm cuts. The upper boundary is below the pelvis cut by 1.5 x the height of the android region. The Lower boundary is below the upper boundary by 2 x the height of the android region
Supplementary Figure 2: Histograms of genetic risk scores from each study for a) insulin secretion and b) insulin resistance scores.