Abstract
Association studies implicate the multiple PDZ domain protein (MUPP1/MPDZ) gene in risk for alcoholism in humans and alcohol withdrawal in mice. Although manipulation of the Mpdz gene by homologous recombination and bacterial artificial chromosome transgenesis has suggested that its expression affects alcohol withdrawal risk, the potential confounding effects of linked genes and developmental compensation currently limit interpretation. Here, using RNA interference, we directly test the impact of Mpdz expression on alcohol withdrawal severity and provide brain regional mechanistic information. Lentiviral-mediated delivery of Mpdz short hairpin RNA (shRNA) to the caudolateral substantia nigra pars reticulata significantly reduces Mpdz expression and exacerbates alcohol withdrawal convulsions compared to control mice delivered a scrambled shRNA. Neither baseline nor pentylenetetrazol enhanced convulsions differed between Mpdz shRNA and control animals, indicating that Mpdz expression in the caudolateral substantia nigra pars reticulata does not generally affect seizure susceptibility. To our knowledge, these represent the first in vivo Mpdz RNA interference analyses, and provide the first direct evidence that Mpdz expression impacts behavior. Our results confirm that Mpdz is a quantitative trait gene for alcohol withdrawal and demonstrate that its expression in the caudolateral substantia nigra pars reticulata is crucially involved in risk for alcohol withdrawal.
Keywords: Mpdz, MUPP1, alcohol, substantia nigra, RNA interference, withdrawal, mouse, HICs, lentivirus, genetics
Introduction
Alcohol is widely abused for its euphoric and sedative effects, and dependence on alcohol (alcoholism) has tremendous cost to afflicted individuals and society. A multitude of twin and adoption studies demonstrate that risk for alcoholism is 50-65% genetically determined (Kendler, 2009, Palmer, 2012, Reich, 1999). Unfortunately, genetic determinants of risk remain largely unknown, hindering effective prevention and treatment of dependent individuals.
Although no animal model exactly duplicates alcoholism, models for specific factors, such as risk for physiological dependence and associated alcohol withdrawal (AWD) episodes, which constitute a motivational force that perpetuates alcohol use/abuse and contribute to relapse (Little et al., 2005), are useful to identify potential genetic determinants of liability in humans. Using a robust behavioral model of physiological dependence, positional cloning, and molecular analyses, we identified Mpdz as a putative quantitative trait gene (QTG) for AWD in mice, demonstrating allelic variation in sequence (structure) and expression, either one or both of which could contribute to the quantitative trait locus (QTL) phenotypic effect on predisposition to AWD (Fehr, 2002, Shirley, 2004). Mpdz and its human homolog (MPDZ) encode the multiple PDZ domain protein (MUPP1/MPDZ) (Simpson et al., 1999, Ullmer et al., 1998). As a member of the PDZ protein family, MUPP1 may impact the trafficking and function of the protein signaling complexes it scaffolds (Romero et al., 2011).
Recent studies using conventional Mpdz knockout heterozygotes and bacterial artificial chromosome (BAC) transgenic mouse models further support an inverse relationship between Mpdz expression and AWD severity (Milner et al., 2013), but cannot disentangle Mpdz actions from potential effects of developmental compensation and/or linked genes. Fortunately, RNA interference (RNAi) has emerged as a powerful approach to disentangle target gene effects from these potential confounds (Bahi & Dreyer, 2012). Further, RNAi spread beyond the microinjection site is minimal, thus limiting target gene knockdown to a discrete region to provide additional mechanistic information (Bahi & Dreyer, 2012). Success of the RNAi approach is therefore dependent upon targeting the appropriate brain region(s). Here, we target the caudolateral substantia nigra pars reticulata (clSNr). cFos induction analyses have implicated the SNr as involved in genetically determined differences in AWD in a manner dependent upon QTL status, and bilateral lesions of the clSNr, but not rostromedial SNr, significantly mitigate AWD using acute and repeated alcohol exposure models (Chen et al., 2011b, Chen et al., 2008), confirming that the clSNr itself (rather than fibers of passage) is intrinsically involved in AWD.
For these reasons, the present studies use an RNAi approach to directly and rigorously test the hypothesis that reduced Mpdz expression in the clSNr results in more severe AWD, without generally affecting seizure susceptibility. In both humans and mice, spontaneous convulsions are rare, but seizure threshold for some stimuli is reduced during AWD. The present studies assess AWD using the handling-induced convulsion (HIC), a quantitative index of AWD, which is highly correlated with other signs of AWD and substantially genetically determined (Crabbe et al., 1991a, Crabbe et al., 1983).
Materials and methods
Animals
A total of 80 male DBA/2J (D2) strain mice (Jackson Laboratories) were acclimated to our vivarium for 1-2 weeks. Mice were 50-57 days old at the time of surgery and behaviorally assessed starting at 70-77 days of age. Colony and procedure rooms were maintained on a 12 hr light/dark cycle with lights on from 06:00 to 18:00, and the temperature maintained at 21±1°C. All mice received water and food (LabDiet 5001Rodent Diet) ad libitum. All procedures were approved by the Oregon Health & Science University and VA Medical Center Care and Use Committees in accordance with United States Department of Agriculture and United States Public Health Service guidelines.
Mpdz RNAi in mouse NS20Y cells
Five Sigma MISSION shRNA clones for Mpdz, designed and developed by the RNAi Consortium at the Broad Institute of MIT and Harvard, were additionally analyzed using BLAST to ensure specificity to reduce endogenous Mpdz mRNA expression in mouse neuroblastoma (NS20Y) cells compared to a control (scrambled) shRNA clone. Plasmid DNA (1 μg) was transfected into 60-70% confluent NS20Y cells using lipofectamine. Following selection for shRNA transfected cells (in 2 μg/ml puromyocin for two weeks), the cells were harvested and total RNA isolated. Relative Mpdz mRNA expression was assessed using a quantitative polymerase chain reaction (QPCR) assay as in previous work (Shirley, 2004). Knockdown of Mpdz mRNA expression was calculated relative to a scrambled control, which does not target any gene sequence in the mouse genome.
Intra-clSNr RNAi
Mice were anesthetized using isoflurane (induced at a flow rate of 1.5 liter/min oxygen, 4% isoflurane, and maintained at 0.9 liter/min oxygen, 2.5% isoflurane), injected with meloxicam (10 mg/kg ip, in 0.9% saline), and placed in a mouse stereotaxic instrument (Kopf Instruments). The skull surface was exposed and burr holes drilled at the appropriate coordinates. The clSNr coordinates were based on the Mouse Brain Atlas (Paxinos & Franklin, 2001) and empirically determined for D2 mice as follows: anterior-posterior (AP) = −3.1, medial-lateral (ML) = ±1.7, and dorsal-ventral (DV) = −4.5.
In order to use green fluorescent protein (GFP) to assess targeting and spread, the Mpdz shRNA and scrambled control were subcloned into the pLKO.1-hPGK-puro-CMV-TurboGFP vector, then packaged into lentivirus to a high titer (108 titer units [TU]/ml; Sigma). Microinjectors were created as in previous work (Lasek & Azouaou, 2010) by affixing a 33 gauge stainless steel hypodermic tube within a shorter 26 gauge stainless steel hypodermic tube, and were attached to polyethylene-20 tubing affixed to a 10 μl Hamilton syringe. Microinjectors were lowered bilaterally to the designated coordinates and 1 μl (up to 105 TU) per side microinjected over 5 min using an infusion pump, and left in place for an additional 10 min. During the two days post-surgery the mice received additional meloxicam and were monitored for potential weight loss, pain, and distress. Behavioral testing began 3 weeks later.
Baseline, and alcohol withdrawal enhanced HICs
Physiological dependence is characterized by the appearance of physical disturbances (withdrawal) after alcohol administration is suspended. Alcohol withdrawal induced seizures are a distinct measure of central nervous system (CNS) physiological dependence occurring in both humans, as well as rodents. McQuarrie & Fingl (1958), first reported that AWD is apparent in mice following a single hypnotic dose, and was later shown to be genetically determined (Crabbe et al., 1991b). The detection and fine-mapping of Mpdz utilized this acute model (Buck, 1997, Fehr, 2002, Shirley, 2004), which is therefore used in the present studies. Importantly, this acute AWD model assesses CNS sensitivity to ethanol, while chronic models can be confounded by tolerance (Crabbe et al., 1991a).
AWD was assessed in two experiments (experiments 1 and 2). D2 mice were used because they demonstrate robust AWD using both acute and chronic models, allowing detection of increased or decreased AWD severity with experimental manipulation. Details of the AWD procedure using the 7-point HIC scale have been published (Chen et al., 2011b, Metten & Crabbe, 2005). Individual mice and different genetic models can differ in baseline HIC scores. Therefore, three weeks after surgery, mice were scored twice (30 min apart) for baseline (pre-ethanol) HICs prior to AWD testing. Immediately thereafter, mice received a single hypnotic dose of ethanol (4 g/kg, i.p., 20% v/v in 0.9% physiological saline) and were scored hourly between 2 and 12 hr, and at 24 and 25 hr post-ethanol. In order to create an index of AWD that is independent of individual differences in baseline HIC scores and that reflects differences in withdrawal convulsion severity, post-ethanol HIC scores were corrected for the individual's average baseline HIC score as in our previous work (Chen et al., 2008). Individual AWD severity scores were calculated as the area under the curve (AUC), which is calculated as a sum of the corrected HIC scores 2-12 hr post-ethanol as in our previous work (Chen et al., 2011b).
Pentylenetetrazol (PTZ) enhanced HICs
One week later, the animals in experiment 2 were assessed for HICs in response to PTZ (30 mg/kg i.p.), which blocks GABAA receptor-mediated transmission. Previous studies show that the severity of PTZ-enhanced HICs is not influenced by prior testing for AWD or surgery (Chen et al., 2008). This dose was used because it enhances HIC intensity without inducing other convulsions (e.g., tonic hindlimb extensor) associated with higher doses. Mice were scored for HICs at 10 time points from 1 to 65 min post-PTZ injection. PTZ enhanced HIC scores were corrected for individual differences in baseline HIC scores to create an index of response to PTZ independent of variability in individual baseline (pre-PTZ) scores. The PTZ enhanced HIC severity scores were calculated as AUC scores and in our previous work (Chen et al., 2011b).
Histology and laser capture microdissection (LCMD)
Within 2 hr of completing the behavioral testing, Mpdz shRNA and control animals were sacrificed by cervical dislocation and brains immediately removed and frozen (−80°C) until sectioning (coronal, 30 μm). Alternating slices containing the SNr were mounted on glass slides (for histology and GFP analyses), and polyethylene napthalate (PEN) membrane glass slides (for LCMD), and stored (−80°C). Slides were dried for 5 min, then placed in 0.1% thionin for 5 sec, washed twice in diethylpyrocarbonate treated water for 15 sec, washed in ethanol (75%, 95%, and 100%, 30 sec each), and dried for 5 min. For histology, slides were examined using a fluorescent microscope (Leica Microsystems, 10x) to assess injection placement and the extent of GFP expression; this produced a template for confident LCMD of the region of interest (ROI, i.e., the clSNr) which was carefully determined from an adjacent section. Using LCMD (Leica Microsystems, 6.3x) four clSNr tissue samples per side were isolated, combined, and stored in 50 μl RNAlater until processing for expression analyses.
Mpdz RNA expression
Total RNA was isolated from NS20Y cells using Qiagen RNeasy mini kit and from LCMD clSNr samples using the Ambion RNAqueous-Micro Kit. cDNA synthesis used a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems), with preamplification (in vivo experiment only) using a TaqMan PreAmp Master Mix kit and probes for Mpdz and a control gene (Reep5). Reep5 was used because it is a highly stable reference gene expressed throughout the mouse brain (www.brain-map.org) (Kozell et al., 2009). Quantitative polymerase chain reaction (QPCR) utilized Mpdz and Reep5 TaqMan probes (Mm00447849_m1 and Mm00492230_m1, respectively), and was performed on an ABI Prism7500 thermal cycler using TaqMan Universal PCR Master Mix and standard reaction conditions. A standard relative quantification method (ΔΔCt) was used which corrects for run to run variability by normalization of target and reference gene expression to calibrator sample expression (preamplified cDNA pooled from all samples) (Livak & Schmittgen, 2001, Shen & Johnson, 1997). The calibrator was set to “1” and gene expression in clSNr samples was evaluated in reference to the calibrator.
Statistical analyses
The RNAi behavioral and Mpdz expression data were normally distributed (Shapiro-Wilks test, p>0.05), so two-sample t-tests were used for comparisons of the Mpdz shRNA and control groups for AWD severity, baseline and PTZ enhanced HIC severities, and Mpdz mRNA expression. Data were analyzed using Systat 13 statistical software (Systat Software). Throughout, the significance level was set at p<0.05 (two-tailed).
Results
Mpdz RNAi efficacy in mouse neuroblastoma cells
Table 1 summarizes the results for five Mpdz shRNAs tested for their efficacies to affect endogenous Mpdz expression in mouse NS20Y cells compared to a scrambled control. This validated negative control engages the RNAi pathway but does not target any sequence in the mouse genome. The TRCN0000103482 plasmid demonstrated the highest in vitro knockdown of Mpdz expression (i.e., a 77% reduction relative to the scrambled control), and was therefore utilized for the subsequent in vivo RNAi experiments.
Table 1.
Mpdz knockdown in NS20Y cells. Five Mpdz shRNA plasmids were assessed for relative knockdown of Mpdz expression in NS20Y cells compared to scrambled control using QPCR. Each hairpin sequence is comprised of a 21 base stem (with the 21 underlined bases indicating the bases that hybridize to the Mpdz sequence in Exon 10) and a 6 base loop.
| Plasmid | Targeted Sequence | Mpdz expression |
|---|---|---|
| TRCN0000103480 | CCGGCGCACCAGAATGAGTGTGTTTCTCGAGAAACACACTCATTCTGGTGCGTTTTTG | 1.05 |
| TRCN0000103481 | CCGGGCTCTGATAGATACACCTGATCTCGAGATCAGGTGTATCTATCAGAGCTTTTTG | 0.24 |
| TRCN0000103482 | CCGGGCCTTCAGGAATCTTTGTAAACTCGAGTTTACAAAGATTCCTGAAGGCTTTTTG | 0.23 |
| TRCN0000103483 | CCGGGCATCTGAAATTCAGGGACTACTCGAGTAGTCCCTGAATTTCAGATGCTTTTTG | 0.62 |
| TRCN0000103484 | CCGGCATCTGAAATTCAGGGACTAACTCGAGTTAGTCCCTGAATTTCAGATGTTTTTG | 0.50 |
| SCH002 | CCGGCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTT | 1.00 |
Intra-clSNr Mpdz RNAi
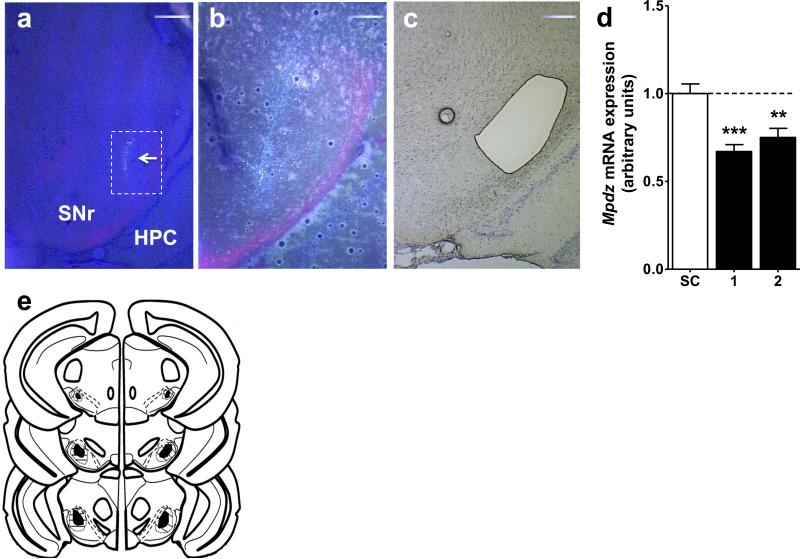
Mpdz shRNA (TRCN0000103482) and a scrambled control shRNA were subcloned into pLKO.1-hPGK-puro-CMV and packaged in lentivirus to a high titer (108 TU/ml). Figures 1a and 1b show representative photomicrographs of the clSNr microinjection site at 4x and 10x magnification, respectively, evident by GFP expression. Figure 1c shows a representative LCMD tissue section of the clSNr, which were used for Mpdz mRNA quantification by QPCR (Fig 1d). Figure 1e is a schematic illustration of the extent of the bilateral clSNr microinjection sites and GFP expression spread, which extended from −3.1 to −3.8 mm (anterior-posterior) from Bregma, with occasional spread into the SN lateralis (Paxinos & Franklin, 2001).
Figure 1. Bilateral Mpdz RNAi targeting of the clSNr.
Representative photomicrograph showing fluorescence at the microinjection site (arrow) overlain a light photomicrograph to visualize structure and GFP expression at (a) 4X and (b) 10X (of the area outlined in white within panel a). (c) 4X Thionin stained light photomicrograph image with the clSNr dissected by LCMD. (d) Data presented are for animals with confirmed bilateral targeting of the clSNr. Mpdz expression in LCMD clSNr from Mpdz shRNA animals in two experiments conducted 4 months apart (black bars) compared to scrambled control (SC) animals (open bar). (e) Schematic diagram of a coronal brain section depicting microinjection sites and extent of GFP expression (indicating viral infection) in the clSNr. The darkened regions indicate the sites of clSNr microinjections from all confirmed bilateral hits, identified by GFP. The striped region indicates the extent of GFP spread (including injection sites). Based on coordinates from the mouse brain atlas (Paxinos & Franklin, 2001), the confirmed microinjection sites and lentiviral spread ranged from −3.1 to −3.8 mm AP from Bregma in the clSNr. Additional abbreviations: HPC, hippocampus. Scale bars are located in the upper right hand corner of each panel. **p<0.01, ***p<0.001.
Baseline and AWD enhanced HICs in Mpdz shRNA and control animals
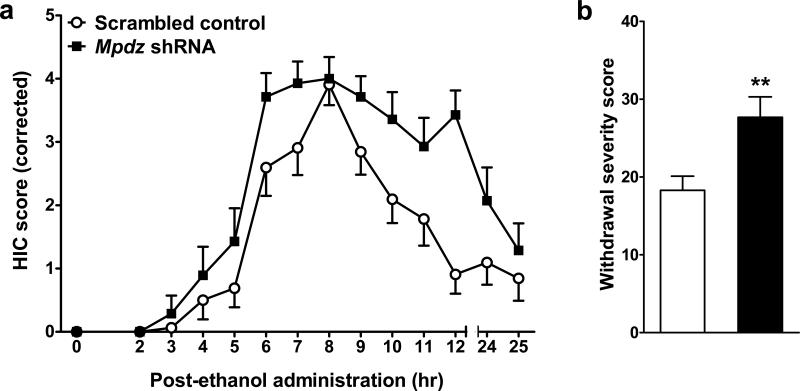
Baseline (pre-ethanol) and AWD enhanced HICs were assessed in two experiments which were combined for increased statistical power. For those animals with confirmed bilateral targeting of the clSNr, Mpdz shRNA and control animals did not differ in baseline (pre-ethanol) HIC severity (mean score ± SEM = 0.5±1.1 and 0.2±0.6, n=14 and 16, respectively, t1,28=0.99, p=0.33, NS). However, Mpdz shRNA mice demonstrated more severe AWD than controls (t1,28=3.0, p=0.006, Fig 2). To our knowledge, these represent the first in vivo Mpdz RNAi analyses, and provide the first direct evidence that Mpdz expression impacts behavior. Taken together, our results conclusively prove that Mpdz is a QTG for AWD risk.
Figure 2. AWD severity is increased by intra-clSNr Mpdz RNAi.
(a) Data represent the mean HIC score values ± SEM (corrected for individual baseline scores) for mice with confirmed bilateral targeting of the clSNr. Around 4 hr post-ethanol, HIC scores increase above baseline, indicating a state of withdrawal hyperexcitability. (b) Individual withdrawal severity scores were calculated as the sum of the corrected HIC scores 2-12 hr post-ethanol as in previous work (Chen et al., 2011b). AWD is significantly more severe in Mpdz shRNA (N=14) compared to control animals (N=16). **p<0.01, ***p<0.001.
In animals in which clSNr targeting was unilateral only, neither baseline HICs (mean score ± SEM = 0.4±0.4 and 0.4±0.7, n=14 and 20, respectively; t1,32=0.01, p=0.9) nor AWD severity (mean score ± SEM = 22.3±7.4 and 24.3±10.0, n=14 and 20, respectively, t1,32=0.7, p=0.5, NS) differed between Mpdz shRNA and control animals. These results are consistent with our previous data showing that bilateral but not unilateral clSNr lesions mitigate AWD (Chen et al., 2011b, Chen et al., 2008)
PTZ enhanced HICs in Mpdz shRNA and control animals
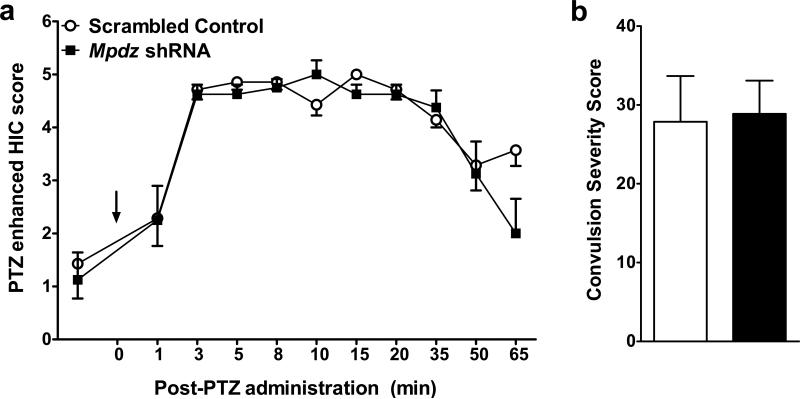
In order to assess whether Mpdz RNAi might have more general effects on enhanced HICs beyond AWD, the animals in experiment 2 were assessed for PTZ enhanced HICs. Mpdz shRNA and control animals with confirmed bilateral targeting of the clSNr did not differ in baseline (pre-PTZ) HIC scores (mean score ± SEM = 1.1±1.5 and 1.4±1.7, n=8 and 7, respectively, t1,13=0.36, p=0.7, NS; Fig 3a) or PTZ enhanced HIC severity (t1,13=0.14, p=0.9, NS, Fig 3).
Figure 3. PTZ enhanced convulsion severity is not altered by intra-clSNr Mpdz RNAi.
(a) Data represent the mean HIC score values ± SEM for mice with confirmed bilateral targeting of the clSNr. HICs were assessed at 10 time points from 1 to 65 min post-PTZ administration. (b) Individual HIC severity scores were calculated as the sum of the corrected HIC scores 2-12 hr post-ethanol as in previous work (Chen et al., 2011b). PTZ enhanced convulsions did not differ between Mpdz shRNA (N=8) compared to control (N=7) animals.
RNAi significantly reduces intra-clSNr Mpdz expression
In order to assess the impact of RNAi on target gene expression it is important to specifically examine the ROI using a robust, quantitative method. We therefore used LCMD followed by QPCR to determine expression of Mpdz in the clSNr. As indicated in Figure 2d,Mpdz shRNA significantly reduced Mpdz mRNA expression in LCMD-clSNr compared to control animals. Mpdz mRNA was reduced by 33% in experiment 1 (mean score ± SEM = 0.88±0.05 and 0.59±0.04, respectively, t1,28 = 4.2, p = 0.0002). A significant reduction in Mpdz mRNA was also apparent in experiment 2 (mean score ± SEM = 1.5±0.07 and 1.2±0.08, respectively, t1,28= 3.6, p = 0.001), and, not surprisingly, was somewhat smaller in magnitude (25% decrease) than in experiment 1 owing to the expected reduction in titer over time.
Discussion
Naturally-occurring genetic (allelic) variation affects Mpdz/MPDZ sequence and expression in mice and humans, but the functional and behavioral consequences remain largely unknown. Association studies implicate the mouse and human homologs in risk for alcohol physiological dependence and alcoholism (Fehr, 2002, Karpyak et al., 2009, Shirley, 2004, Tabakoff, 2009). However, despite the recent creation of Mpdz transgenic and knockout heterozygote models (Milner et al., 2013), direct evidence that Mpdz impacts these (or any) behavior has been lacking. Here, we report the creation of the first in vivo Mpdz RNAi animal model, and provide the first direct evidence that Mpdz expression impacts behavior. Specifically, we demonstrate that Mpdz shRNA reduces Mpdz expression in the clSNr, resulting in more severe AWD severity compared to control animals, but no change in baseline and PTZ enhanced convulsions. These data provide conclusive proof that Mpdz is a QTG affecting AWD risk.
The SNr is a main output of the basal ganglia circuit and has been implicated in CNS hyperexcitability including AWD (Chen et al., 2008) and limbic seizures (Shehab et al., 1996, Veliskova et al., 2005). A growing body of literature indicates that the caudal and rostral subregions are functionally distinct in their regulation of AWD as well as related limbic seizure phenotypes (Shehab et al., 1996, Veliskova et al., 2005). The caudal SNr, active during pre-clonic seizures, is thought to be an early gateway for seizure initiation/propagation, and is a sensitive target for the suppression of tonic seizures (Shehab et al., 1996, Veliskova et al., 2005). In contrast, the rostral subregion becomes activated after a motor seizures occurs (Collins et al., 1986, Shehab et al., 1996, Veliskova et al., 2005). Interestingly, the present studies find that intra-clSNr Mpdz RNAi impacts AWD, but not baseline or PTZ enhanced HICs, demonstrating that the clSNr, and particularly Mpdz expression within the clSNr, has an important role in AWD but does not generally affect seizure susceptibility. This is consistent with and builds upon our data showing that clSNr (but not rostral SNr) lesions mitigate AWD but do not affect baseline or PTZ enhanced HICs (Chen et al., 2011b, Chen et al., 2008). Taken together, these studies consistently demonstrate that the clSNr subregion plays a crucial role in mediating AWD convulsions, and that Mpdz expression is inversely related to AWD severity.
We recently demonstrated that Mpdz knockout heterozygotes voluntarily consume less alcohol than wildtype littermates, providing the first evidence that Mpdz may also influence alcohol consumption and reward (Milner et al., 2013). Recent association studies using human clinical populations also implicate MPDZ in alcohol consumption and dependence (Gizer, 2011, Karpyak et al., 2009, Tabakoff, 2009) in a manner dependent upon AWD risk (Karpyak et al., 2009), apparently similar to our results in mouse Mpdz genetic models (Milner et al., 2013). Although, to our knowledge, the clSNr has not been implicated in alcohol consumption phenotypes, it receives robust projections from the rostroventral striatum (Chen & Buck, 2010), which in turn has reciprocal connections to the dorsal striatum, a region known to have an important role in alcohol seeking behavior, its habitual use, and alcoholism risk (Chen et al., 2011a, Jeanblanc et al., 2009). The lateral SNr also receives projections from the central nucleus of the amygdala and the intercalated cells of the amygdala (Shinonaga et al., 1992), another region with a proven role in alcohol consumption and preference in mice (Dhaher et al., 2008, Funk et al., 2006). Future studies will be needed to rigorously test the role of MUPP1 in one or more of these brain regions, as well as the role of its expression in the clSNr beyond AWD to alcohol seeking, consumption, and preference behaviors.
MUPP1 and other PDZ domain proteins impact signaling by regulating supramolecular complex assembly and synaptic protein localization, as well as the rate and fidelity of the signal transduction pathways of the receptors and proteins which they scaffold (Romero et al., 2011, Sheng & Sala, 2001). MUPP1 is not thought to have an intrinsic catalytic domain, suggesting that its impact on AWD and alcohol consumption is mediated by the protein complexes that it scaffolds. MUPP1 contains thirteen PDZ domains allowing for a large diversity of interactions, so multiple MUPP1 partners may be involved in MUPP1 actions on AWD, as well as alcohol consumption/preference. Yeast two hybrid analyses have begun to identify MUPP1 association partners, including G-protein coupled GABAB and 5-HT2 receptors (Balasubramanian et al., 2007, Ullmer et al., 1998) and the NMDA2B-SynGAP (synaptic GTPase-activating protein)-CaMKII (Ca2+/calmodulin-dependent kinase) complex (Krapivinsky et al., 2004), all three of which have been implicated in AWD and/or alcohol consumption/reward (Chen et al., 2011a, Humeniuk et al., 1994, Lal et al., 1993, Panocka & Massi, 1992, Vlachou & Markou, 2010). In addition, we have previously demonstrated that the degree to which a selective GABAB receptor agonist and a selective 5-HT2C receptor antagonist enhance HICs may be dependent upon QTL/Mpdz status (Reilly et al., 2008). Furthermore, ongoing neurophysiological in clSNr neurons and behavioral studies indicate that GABAB and 5-HT2 receptor function are altered in MUPP1 knockdown mice (K Buck, L Kruse, and D Rossi, unpublished data) which also show enhanced AWD severity compared to wildtype (Milner et al., 2013). In addition, the role of GABAB receptors in alcohol dependence can be extended beyond AWD to alcohol consumption, since baclofen, a selective GABAB receptor agonist, is currently being evaluated in clinical trials as a therapeutic treatment for alcoholism (Addolorato et al., 2011, Leggio et al., 2013). Taken together with the knowledge that MUPP1 plays a role in mediating GABAB and 5-HT2 receptor function, both of which have been implicated in AWD and alcohol consumption, the MUPP1 interaction with one or both of these receptors could be a potential mechanism contributing to Mpdz's role in alcohol physiological dependence and associated AWD. Future studies will be important to begin to dissociate the functional importance of MUPP1 interactions with 5-HT2 receptors (at PDZ10) from its interactions with GABAB and SynGAP/NMDA2B receptors (at PDZ13) on AWD, as well as on alcohol consumption (e.g., using the CRISPR/Cas9 system) to further elucidate the role of Mpdz in alcohol dependence.
To our knowledge, the present studies are the first to employ state-of-the-art RNAi techniques for targeted gene expression to advance the understanding of the important interplay between brain region specific gene expression and its influence on the severity of AWD. These studies present clear evidence that Mpdz is a QTG for physiological dependence and associated AWD, and demonstrate that its expression within the clSNr is crucially involved. Further, taken together with previous work from our lab (Chen et al., 2011b, Chen et al., 2008), these studies suggest that the clSNr plays an important role as a critical modulator of AWD severity. Though our work contributes significantly to the understanding of the genetic determinants of alcohol physiological dependence and associated withdrawal, there are some limitations. First, in order to confidently assess Mpdz expression in the clSNr we employed LCMD followed by QPCR to quantify Mpdz mRNA. Although this technique did not allow us to quantify MUPP1 protein in the clSNr, our previous work demonstrates that reduced Mpdz mRNA results in a comparable reduction in MUPP1 protein in the brain (Shirley, 2004). Importantly, while the present RNAi studies were performed in D2 mice, evidence supporting the role of Mpdz expression in AWD is not limited to this strain. Recently, we have demonstrated that Mpdz knockout heterozygote mice (on a C57BL/6J genetic background) with an approximate 53% reduction of Mpdz expression, also demonstrate increased AWD severity (Milner et al., 2013). Finally, our results are based on a single measure of AWD, and future studies will be needed to assess the broader role of Mpdz/MUPP1 in additional measures of AWD (e.g., anxiety-like and depression-like behaviors), alcohol drinking and preference, and beyond to further understand Mpdz's contributing role in alcohol use and abuse. Mpdz's influence on risk for alcohol dependence is of particular interest since drugs with known MUPP1 partners as an initial target are currently being used in clinical trials evaluating their efficacies to reduce craving and relapse (Addolorato et al., 2011, Leggio et al., 2013). As more information becomes available on precise MUPP1 partner interactions and the relevant neural circuit, the mechanism by which MUPP1 regulates AWD and its contribution to alcohol consumption, preference, and reward will become apparent and could identify “drugable” targets for new pharmacotherapies for alcohol dependence.
Acknowledgements
This data was supported by T32 AA007468, RO1 DA005228, RO1 AA011114, P60 AA010760, and the VA.
References
- Addolorato G, Leggio L, Ferrulli A, Cardone S, Bedogni G, Caputo F, Gasbarrini G, Landolfi R. Dose-response effect of baclofen in reducing daily alcohol intake in alcohol dependence: secondary analyses of a randomized, double-blind, placebo-controlled trial. Alcohol Alcohol. 2011;46:312–317. doi: 10.1093/alcalc/agr017. [DOI] [PubMed] [Google Scholar]
- Bahi A, Dreyer J. Involvement of nucleus accumbens dopamine D1 receptors in ethanol drinking, ethanol-induced conditioned place preference, and ethanol-induced psychomotor sensitization in mice. Psychopharm. 2012;222 doi: 10.1007/s00213-011-2630-8. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Fam S, Hall R. GABAB receptor association with the PDZ scaffold Mupp1 alters receptor stability and function. J Biol Chem. 2007;282:4162–4171. doi: 10.1074/jbc.M607695200. [DOI] [PubMed] [Google Scholar]
- Buck KJ, Metten P, Belknap JK, Crabbe JC. Quantitative trait loci involved in genetic predisposition to acute alcohol withdrawal in mice. J Neurosci. 1997;17:3946–3955. doi: 10.1523/JNEUROSCI.17-10-03946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Kozell LB, Hitzemann R, Buck KJ. Involvement of the limbic basal ganglia in ethanol withdrawal convulsivity in mice is influenced by a chromosome 4 locus. J Neurosci. 2008;28:9840–9849. doi: 10.1523/JNEUROSCI.1713-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Buck KJ. Rostroventral caudate putamen involvement in ethanol withdrawal is influenced by a chromosome 4 locus. Genes Brain Behav. 2010;9:768–776. doi: 10.1111/j.1601-183X.2010.00611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Cuzon Carlson VC, Wang J, Beck A, Heinz A, Ron D, Lovinger DM, Buck KJ. Striatal involvement in human alcoholism and alcohol consumption, and withdrawal in animal models. Alcohol Clin Exp Res. 2011a;35:1739–1748. doi: 10.1111/j.1530-0277.2011.01520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Kozell LB, Buck KJ. Substantia nigra pars reticulata is crucially involved in barbiturate and ethanol withdrawal in mice. Behav Brain Res. 2011b;218:152–157. doi: 10.1016/j.bbr.2010.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins R, Santori E, Der T, Toga A, Lothman E. Functional metabolic mapping during forelimb movement in rat. I. Stimulation of motor cortex. J Neuro. 1986;6:448–462. doi: 10.1523/JNEUROSCI.06-02-00448.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC, Young E, Kosobud A. Genetic correlations with ethanol withdrawal severity. Pharm Biochem Behav. 1983;18:541–547. doi: 10.1016/0091-3057(83)90233-2. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Merrill C, Belknap JK. Acute dependence on depressant drugs is determined by common genes in mice. J Pharmacol Exp Ther. 1991a;257:663–667. [PubMed] [Google Scholar]
- Crabbe JC, Merrill C, Belknap JK. Effects of convulsants on handling-induced convulsions in mice selected for ethanol withdrawal severity. Brain Res. 1991b;550:1–6. doi: 10.1016/0006-8993(91)90397-e. [DOI] [PubMed] [Google Scholar]
- Dhaher R, Finn DA, Snelling C, Hitzemann R. Lesions of the extended amygdala in C57BL/6J mice do not block the intermittent ethanol vapor-induced increase in ethanol consumption. Alcohol Clin Exp Res. 2008;32:197–208. doi: 10.1111/j.1530-0277.2007.00566.x. [DOI] [PubMed] [Google Scholar]
- Fehr C, Shirley RL, Belknap JK, Crabbe JC, Buck KJ. Congenic mapping of alcohol and pentobarbital withdrawal liability loci to a <1 centimorgan interval of murine chromosome 4: Identification of Mpdz as a candidate gene. J Neurosci. 2002;22:3730–3738. doi: 10.1523/JNEUROSCI.22-09-03730.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk C, O'Dell L, Crawford E, Koob G. Corticotropin-releasing factor within the central nucleus of the amygdala mediates enhanced ethanol self-administration in withdrawn, ethanol-dependent rats. J Neurosci. 2006;26:11324–11332. doi: 10.1523/JNEUROSCI.3096-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gizer IR, Elhers CL, Vieten C, Seaton-Smith KL, Feiler HS, Lee JV, Segall SK, Gilder DA, Wilhelmsen KC. Linkage scan of alcohol dependence in the UCSF Family Alcoholism Study. Drug Alcohol Depend. 2011;133:125–132. doi: 10.1016/j.drugalcdep.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humeniuk R, White J, Ong J. The effects of GABAB ligands on alcohol withdrawal in mice. Pharm Biochem Behav. 1994;49:561–566. doi: 10.1016/0091-3057(94)90069-8. [DOI] [PubMed] [Google Scholar]
- Jeanblanc J, He D-Y, Carnicella S, Kharazia V, Janak PH, Ron D. Endogenous BNDF in the dorsolateral striatum gates alcohol drinking. J Neuro. 2009;29:13494–13502. doi: 10.1523/JNEUROSCI.2243-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpyak V, Kim J, Biernacka J, Wieben E, Mrazek D, Black J, Choi D. Sequence variations of the human MPDZ gene and association with alcoholism in subjects with european ancestry. Alcohol Clin Exp Res. 2009;33:712–721. doi: 10.1111/j.1530-0277.2008.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Prescott CA, Neale MC, Pedersen NL. Temperance board registration for alcohol abuse in a national sample of Swedish male twins, born 1902 to 1949. Arch Gen Psychiatry. 2009;54:178–184. doi: 10.1001/archpsyc.1997.01830140090015. [DOI] [PubMed] [Google Scholar]
- Kozell L, Walter N, Milner L, Wickman K, Buck KJ. Mapping a barbiturate withdrawal locus to a 0.44 Mb interval and analysis of a novel null mutant identify a role for Kcnj9 (GIRK3) in withdrawal from pentobarbital, zolpidem, and ethanol. J Neurosci. 2009;29:11662–11673. doi: 10.1523/JNEUROSCI.1413-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapivinsky G, Medina I, Krapivinsky L, Gapon S, Clapham D. SynGAP-MUPP1-CaMKII synaptic complexes regulate p38 MAP kinase activity and NMDA receptor-dependent synaptic AMPA receptor potentiation. Neuron. 2004;43:563–574. doi: 10.1016/j.neuron.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Lal H, Prather P, Rezazadeh S. Potential role of 5HT1C and/or 5HT2 receptors in the mianserin-induced prevention of anxiogenic behaviors occurring during ethanol withdrawal. Alcohol Clin Exp Res. 1993;17:411–417. doi: 10.1111/j.1530-0277.1993.tb00785.x. [DOI] [PubMed] [Google Scholar]
- Lasek A, Azouaou N. Virus-delivered RNA interference in mouse brain to study addiction-related behaviors. Methods Mol Biol. 2010;602:283–298. doi: 10.1007/978-1-60761-058-8_17. [DOI] [PubMed] [Google Scholar]
- Leggio L, Zywiak W, McGeary J, Edwards S, Fricchione S, Shoaff J, Addolorato G, Swift R, Kenna G. A human laboratory pilot study with baclofen in alcoholic individuals. Pharmacol Biochem Behav. 2013;103:784–791. doi: 10.1016/j.pbb.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little H, Stephens D, Ripley T, Borlikova G, Duka T, Schubert M, Albrecht D, Becker H, Lopez MF, Weiss F, Drummond C, Peoples M, Cunningham C. Alcohol withdrawal and conditioning. Alcohol Clin Exp Res. 2005;29:453–464. doi: 10.1097/01.alc.0000156737.56425.e3. [DOI] [PubMed] [Google Scholar]
- Livak K, Schmittgen T. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Metten P, Crabbe JC. Alcohol withdrawal severity in inbred mouse (Mus Musculus) strains. Behav Neuro. 2005;119:911–925. doi: 10.1037/0735-7044.119.4.911. [DOI] [PubMed] [Google Scholar]
- Milner LC, Shirley RL, Kozell LB, Walter NA, Kruse LC, Komiyama NH, Grant SG, Buck KJ. Novel MPDZ/Mupp1 transgenic and knockdown models confirm Mpdz's role in ethanol withdrawal and support its role in voluntary ethanol consumption. Addict Biol. 2013 doi: 10.1111/adb.12087. Under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer RHC, Button TM, Rhee SH, Corley RP, Young SE, Stallings MC, Hopfer CJ, Hewitt JK. Genetic etiology of the common liability to drug dependence: Evidence of common and specific mechanisms for DSM-IV dependence symptoms. Drug Alcohol Depend. 2012;123S:S24–S32. doi: 10.1016/j.drugalcdep.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panocka I, Massi M. Long-lasting suppression of alcohol preference in rats following serotonin receptor blockade by ritanserin. Brain Res Bull. 1992;28:493–496. doi: 10.1016/0361-9230(92)90052-y. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Franklin K. The mouse brain in stereotaxic coordinates. Academic Press; Orlando, FL: 2001. [Google Scholar]
- Reich T, Hinrichs A, Culverhouse R, Bierut L. Genetic studies of alcoholism and substance dependence. Am J Hum Genet. 1999;65:599–605. doi: 10.1086/302561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly M, Milner L, Shirley R, Crabbe JC, Buck KJ. 5-HT2C and GABAB receptors influence handling-induced convulsion severity in chromosome 4 congenic and DBA/2J background strain mice. Brain Res. 2008;1198:124–131. doi: 10.1016/j.brainres.2008.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero G, Von Zastrow M, Friedman P. Role of PDZ proteins in regulating trafficking, signaling, and function of GPCRs: Means, motif, and opportunity. Adv Pharmacol. 2011;62:279–314. doi: 10.1016/B978-0-12-385952-5.00003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehab S, Simkins M, Dean P, Redgrave P. Regional distribution of the anticonvulsant and behavioural effects of muscimol injected into the substantia nigra of rats. Eur J Neurosci. 1996;4:749–757. doi: 10.1111/j.1460-9568.1996.tb01260.x. [DOI] [PubMed] [Google Scholar]
- Shen K, Johnson S. Presynaptic GABAB and adenosine A1 receptors regulate synaptic transmission to rat substantia nigra pars reticulata neurones. J Physiol. 1997;505:153–163. doi: 10.1111/j.1469-7793.1997.153bc.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Sala C. PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- Shinonaga Y, Takada M, Mizuno N. Direct projections from the central amygdaloid nucleus to the globus pallidus and substantia nigra in the cat. Neuroscience. 1992;51:691–703. doi: 10.1016/0306-4522(92)90308-o. [DOI] [PubMed] [Google Scholar]
- Shirley RL, Walter NAR, Reilly MT, Fehr C, Buck KJ. Mpdz is a quantitative trait gene for drug withdrawal seizures. Nat Neurosci. 2004;7:669–670. doi: 10.1038/nn1271. [DOI] [PubMed] [Google Scholar]
- Simpson E, Suffolk R, Jackson I. Identification, sequence, and mapping of the mouse multiple PDZ domain protein gene. Mpdz. Genomics. 1999;59:102–104. doi: 10.1006/geno.1999.5853. [DOI] [PubMed] [Google Scholar]
- Tabakoff B, Saba L, Printz M, Flodman R, Hodgkinson C, Goldman D, Koob G, Richardson HN, Kechric K, Bell RL, Hubner N, Heinig M, Pravenec M, Mangion J, Legault L, Dongier M, Conigrave KM, Whitfield JB, Saunders J, Grant B, Hoffman PL, WHO/IBSRA Study on State and Trait Markers of Alcoholism Genetical genomic determinants of alcohol consumption in rats and humans. BMC Biol. 2009;7 doi: 10.1186/1741-7007-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullmer C, Schmuck K, Figge A, Lubbert H. Cloning and characterization of MUPP1, a novel PDZ domain protein. FEBS Letters. 1998;424:63–68. doi: 10.1016/s0014-5793(98)00141-0. [DOI] [PubMed] [Google Scholar]
- Veliskova J, Miller A, Nunes M, Brown L. Regional neural activity within the substantia nigra during peri-ictal flurothyl generalized seizure stages. Neurobiol Disease. 2005;20:752–759. doi: 10.1016/j.nbd.2005.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachou S, Markou A. GABAB receptors in reward processes. Adv Pharmacol. 2010;58:315–371. doi: 10.1016/S1054-3589(10)58013-X. [DOI] [PubMed] [Google Scholar]