Abstract
Numerous selective breeding experiments have been performed with rodents, in an attempt to understand the genetic basis for innate differences in preference for alcohol consumption. QTL analysis has been used to determine regions of the genome that are associated with the behavioral difference in alcohol preference/consumption. Recent work suggests that differences in gene expression represent a major genetic basis for complex traits. Therefore, the QTLs are likely to harbor regulatory regions (eQTLs) for the differentially expressed genes that are associated with the trait. In the present study, we examined brain gene expression differences over generations of selection of the third replicate lines of High and Low Alcohol Preferring (HAP3 and LAP3) mice, and determined regions of the genome that control the expression of these differentially expressed genes (deeQTLs). We also determined eQTL regions (rveQTLs) for genes that showed a decrease in variance of expression levels over the course of selection. We postulated that deeQTLs that overlap with rveQTLs, and also with phenotypic QTLs, represent genomic regions that are affected by the process of selection. These overlapping regions controlled the expression of candidate genes (that displayed differential expression and reduced variance of expression) for the predisposition to differences in alcohol consumption by the HAP3/LAP3 mice.
Keywords: selective breeding, genetics of gene expression, alcohol preference, QTL analysis, HAP and LAP mice
Introduction
Selective breeding of rodents has been widely used in alcohol research as a means to understand the genetic basis for complex alcohol-related traits (Erwin & Deitrich, 1996, Erwin et al., 1976, Grahame et al., 1999, Li et al., 1993, Lumeng et al., 1995, Oberlin et al., 2011, Phillips & Crabbe, 1991). The effect of selective breeding is to change the phenotype of the selected trait in the offspring, as a result of a change in the frequency of genes (alleles) that are associated with the phenotype.
Quantitative trait locus (QTL) mapping is a means to identify the regions of the genome that are associated with the selected trait (Bice et al., 2011, Lander & Botstein, 1989). The assumption is that these regions of the genome harbor “candidate genes” that influence the selected trait. These genes may have coding polymorphisms that affect the function of the gene product. However, recent evidence indicates that differences in gene expression levels often represent the genetic basis for complex traits (e.g., Emilsson et al., 2008, Li & Burmeister, 2005, Schadt et al., 2003), and in this case, the regions of the genome associated with the trait may be regions that regulate the expression levels of the candidate genes (expression (e)QTLs). Therefore, genes whose expression levels diverge as selection for the behavioral phenotype proceeds may be considered as endophenotypes, and determination of the QTLs for expression of these genes provides a method to identify regions of the genome affected by selection. This approach has the advantage of also identifying potential candidate (differentially expressed) genes that contribute to the phenotype.
Furthermore, selective breeding not only affects the population mean of the selected phenotypic trait (and of endophenotypes such as gene expression levels), but also reduces the variance of the selected trait (Falconer & Mackay, 1996). Identification of eQTLs for genes that show a significant decrease in expression variance during selection is therefore another means to identify regions of the genome affected by the selective breeding process.
In the current study, we have used these approaches to identify genomic regions that may be affected during the third replicate selective breeding experiment for high alcohol-preferring (HAP3) and low alcohol-preferring (LAP3) mice (Oberlin et al., 2011). These selectively bred lines (Grahame et al., 1999, Oberlin et al., 2011) are derived from heterogeneous stock (HS) mice, which were originally generated from an eight-way cross of inbred mouse strains (Mcclearn et al., 1970). In the current study, we obtained brain tissue from several generations of naïve (no alcohol exposure) HAP3 and LAP3 mice as selection proceeded, as well as from the progenitor HS mice. We used microarray analysis to track changes in brain gene expression levels and changes in the amount of variation of gene expression levels at each generation of selection. These data allowed us to take a novel approach to assessing the effect of selective breeding, where eQTL analyses identified candidate regions of the genome, and potential candidate genes associated with the predisposition to drink differing amounts of alcohol.
Materials and Methods
Animals
HS/Ibg mice generated by an 8-way cross of inbred stains (Mcclearn et al., 1970) were selectively bred for alcohol preference based on 24-hr two-bottle choice consumption of a 10% alcohol solution (Grahame et al., 1999, Oberlin et al., 2011). All experiments were performed under a protocol approved by the IUPUI Institutional Animal Care and Use Committee.
Brain Gene Expression Analysis
Alcohol-naïve high alcohol-preferring (HAP3) and low alcohol-preferring (LAP3) male mice (10–12 weeks old) from generations 6, 8, 9, 10, 12 and 13 of selection, in addition to male HS mice (generation 0), were used for whole brain gene expression analysis, as previously described (Hu et al., 2008, Saba et al., 2006, Tabakoff et al., 2008). Mice were sacrificed and brains were removed and stored in RNALater at −80°C until RNA extraction. RNA from each individual mouse brain was hybridized to a separate Affymetrix Mouse Genome 430 v.2.0 Array. To control for batch effects, samples were processed in batches that included four samples per line from a single generation (e.g., generation 8) and one sample per line from the previously analyzed generation (e.g., generation 6). Brains were obtained from 5 HAP3 and 5 LAP3 mice from generations 6, 8, 9, 10 and 12; 4 HAP3 and 4 LAP3 mice from generation 13; and 4 HS mice, for a total of 62 samples. RNA was extracted, processed, and hybridized to arrays as previously described (Hu et al., 2008, Saba et al., 2006, Tabakoff et al., 2008). All experiments were performed under a protocol approved by the University of Colorado Denver Institutional Animal Care and Use Committee. All microarray data are available at http://phenogen.ucdenver.edu.
Microarray Data Analysis
Quality control measures were assessed using the PhenoGen informatics website (http://phenogen.ucdenver.edu, Bhave et al., 2007). A detailed description of the methods for quality control are available in Saba et al. (2006). No arrays that were used for these studies deviated substantially in any of the quality control measures.
Prior to normalization and quality control assessment, probes were removed if their sequence did not uniquely map to the mouse genome (NCBIM37) or if their target region included a known SNP (see Hoffman et al., 2011). The probe mask was based on SNPs between any of the 8 founding strains of the heterogeneous stock used for phenotypic selection. SNPs were identified from the whole genome sequence data generated by the Sanger Institute for 6 of the 8 founding strains (Keane et al., 2011) and from the Imputed Genotype Resource from the Jackson Laboratory, http://csbio.unc.edu/imputation/, for the remaining two strains. Entire probesets were eliminated if less than 4 associated probes remained after filtering (124,731 probes/4,002 probesets removed). Data were normalized using robust multi-array average (rma package, Irizarry et al., 2003) and adjusted for batch effects using the ComBat method (Johnson et al., 2007) in R. Present/absent calls for each probeset were extracted using the MAS 5.0 algorithm (Affymetrix, 2001), and the rma expression data were filtered to include only those probesets “present” in all 62 samples.
Determination of Maintained Differential Expression
We assumed that transcripts that are differentially expressed due to selection in a particular generation would remain differentially expressed in the same direction (e.g., always higher in HAP3) in the subsequent generations. For each probeset, we assessed differential expression between HAP3 and LAP3 mice within each generation using a moderated t-test (Smyth, 2004). We then evaluated each transcript for differential expression in six different scenarios based on the generation at which differential expression began (Table S1). For example, in scenario 1, we declared a transcript to be differentially expressed due to selection at generation 6 if it was differentially expressed at generation 6 and in ALL subsequent generations. Because transcripts that are differentially expressed early in the selection process have to surpass a threshold of significance in more generations (comparisons) than transcripts that become differentially expressed at a later generation, we developed separate significance thresholds for differential expression for each scenario. These thresholds were applied to all generations relevant to that scenario and were dependent on the number of generations that the transcript had to maintain the statistically significant difference. We set a more conservative significance threshold when a transcript had to maintain differential expression in only a few generations and we allowed for a more liberal significance threshold when the transcript had to surpass the threshold in many generations. The goal was to maintain a p-value of 3.28×10−6 (0.05/15,259 probesets; Bonferroni adjustment due to testing of multiple probesets) for each transcript regardless of the number of generations examined. Assuming the null distribution is that the transcript is not differentially expressed at ANY generation, the combined p-value is equal to the product of the p-values for each individual generation. For example, a transcript is declared differentially expressed beginning at generation 6, and differential expression is maintained in subsequent generations, if the individual p-values for each generation are below 0.122, i.e., 3.28×10−6 ≅ (0.122)6. If a transcript did not show differential expression until generation 12, the transcript had to reach a more stringent p-value of 0.0018 for generation 12 and generation 13, i.e., 3.28×10−6 = (0.0018)2. Because the tests for differential expression of a transcript across generations may not be independent, we also calculated meta-analysis p-values post hoc for each transcript and each scenario that did account for dependence (Stouffer-Liptak Test; (Kechris et al., 2010)) and applied a false discovery rate correction for multiple testing (Benjamini & Hochberg, 1995) to verify the significance of differential expression of “priority genes” as defined below.
Determination of Maintained Differential Variance
Each probeset was assessed for a significant reduction in expression variance at each generation, compared to the HS mice (generation 0), using a one-sided F-test. Probesets were interrogated for reduced variance between the HAP3 mice and HS mice and between the LAP3 and HS mice. Similar to maintained differential expression described above, probesets with reduced expression variance were categorized by the initial generation where reduced variance was detected and then maintained. Significance thresholds were also determined in the same manner as for differential expression.
Quantitative Trait Locus (QTL) Analyses
Phenotype QTLs (QTLs)
Quantitative trait loci (QTLs) for alcohol consumption/preference were obtained from studies of Bice et al. (2011), based on an F2 generation derived from HAP1 and LAP1 mice; Belknap and Atkins (2001), based on a meta-analysis of studies using mapping populations derived from C57BL/6 and DBA/2 strains; and Rodriguez et al. (1995), based on BXD recombinant inbred (RI) strains. Bice et al (2006) also reported QTLs for HAP2 and LAP2 mice, but in their study, only chromosomal regions that had already been demonstrated to harbor significant QTLs for alcohol preference, as reported by Belknap and Atkins (2001), were investigated. Since we also used the data reported by Belknap and Atkins (2001) to identify QTLs, there were no QTLs found for HAP2 and LAP2 mice that differed from those used in the present study.
The QTL data in the analysis by Bice et al. (2011) were converted from cM to Mb using the Mouse Map Converter from the Center for Genome Dynamics at the Jackson Laboratory (http://cgd.jax.org/mousemapconverter) with sex-averaged cM. The QTL data in the meta-analysis by Belknap and Atkins (2001) were also reported as cM values, and the QTL range in the paper represents the range of peak LOD scores. Assuming a symmetric confidence interval, the results from each study in the meta-analysis were translated into a cM region, which was converted to Mb as described. For the current analysis, the intervals from the Belknap and Atkins (2001) analysis are defined as the minimum lower confidence limit, and the maximum upper confidence limit, across studies within the meta-analysis. We chose to use these broad ranges in order to be inclusive, given that QTL location is only one of the filters used for our identification of priority QTLs and genes. When multiple peaks were reported, the average of the cM locations was used as the center of the confidence region. QTLs were recalculated from the data of Rodriguez et al. (1995) using the Wellcome Trust-CTC Mouse Strain SNP Genotype Set (http://mus.well.ox.ac.uk/mouse/INBREDS/) with mm37 locations. Markers with LOD>2.0 were identified as QTLs, and confidence intervals were defined as 10 Mb upstream and downstream from the peak LOD marker. All QTLs used for this study are reported in Table S2.
Expression QTLs (eQTLs)
We used gene expression data generated by our laboratory (Colorado) from whole brain tissue of the BXD recombinant inbred (RI) panel (Tabakoff et al., 2008, http://phenogen.ucdenver.edu) for eQTL calculations. For these expression data, we generated a separate probe mask that took advantage of the full genome sequence of the panel’s parental strains (Keane et al., 2011). This mask eliminated 82,292 probes and 3,456 probesets. Expression data were normalized and summarized into probesets using rma and present/absent calls were determined using the MAS 5.0 algorithm (Affymetrix, 2001). A new method for removing batch effects, while retaining confounded strain effects, was used (personal communication, Evan Johnson, Boston University). This method is similar to the empirical Bayes method, ComBat (Johnson et al., 2007) that was used for the HAP3/LAP3 microarray analysis. eQTLs were calculated as described previously (Tabakoff et al., 2008), using genotype information from the Wellcome Trust-CTC Mouse Strain SNP Genotype Set (http://mus.well.ox.ac.uk/mouse/INBREDS/) and a weighted marker regression.
Expression QTLs for Differentially Expressed Genes and Genes that show Reduced Variance (deeQTLs and rveQTLs)
We identified shared eQTLs for a group of probesets that displayed differential expression or reduced expression variance using a meta-analysis approach, with eQTLs derived from the BXD recombinant inbred strains. First, a locus-specific empirical p-value (10,000 permutations) was calculated for each SNP (representing a unique strain distribution pattern) and probeset combination. To identify shared eQTLs, we calculated a meta-analysis p-value at each SNP across the group of probesets in which we were interested by combining p-values using a Z-score method (Hedges & Olkin, 1985), with equal weight given to each transcript. To ensure that the shared eQTLs are specific to the group of transcripts, we calculated a permutation p-value by randomly selecting the same number of transcripts from the pool of expressed transcripts and calculating the meta-analysis p-value for each SNP. We repeated this random permutation 10,000 times to generate a locus-specific null distribution. Empirical p-values based on the null distribution were corrected for multiple testing across loci using a false discovery rate (FDR, Benjamini & Hochberg, 1995). For differential expression QTLs (deeQTLs), shared eQTLs were identified separately for each generation to determine which areas of the genome were being fixed due to selection and when these areas became fixed. Likewise for the reduced variance QTLs (rveQTLs), shared eQTLs were identified by generation, separately for decreased variance in LAP3 mice and decreased variance in HAP3 mice. Loci with an FDR less than 0.05, and the genomic region 10 Mb on either side, are reported as eQTLs.
To determine if multiple eQTLs were contributing independently or even synergistically to the differential expression of a transcript, we identified the most parsimonious multi-locus model for each differentially expressed transcript using the eQTLs identified in the same generation as initial differential expression. This model was determined using a backward selection process that began with a model including all eQTLs and eliminated one locus at a time until all remaining loci were significant (p-value <0.05).
Identification of High Priority Genes
Initially we identified “high priority” regions of the mouse genome by focusing on eQTLs for differentially expressed transcripts that overlapped a QTL for alcohol preference and also overlapped an eQTL for transcripts with reduced variance (either HAP3 or LAP3). We then identified genes that were differentially expressed between HAP3 and LAP3 mice, and had a significant reduction in variance of expression levels in either the HAP3 or LAP3 line. Genes with these expression properties, and whose expression was significantly controlled by at least one of the “high priority” genomic regions, were considered to be “high priority” candidate genes.
Validation of Endogenous Gnb1 Differential Expression: Quantitative Reverse Transcriptase Real-Time PCR (qRT-PCR)
RNA was isolated from brains of HAP3 and LAP3 mice (generation 12 of selection) using the RNAeasy Midi Kit (Qiagen, Gathersburg, MD) and RNAeasy Mini kit (Qiagen) for cleanup. RNA quality was assessed using a Bioanalyzer (Agilent Technologies, Santa Clara, CA). Quantitative RT-PCR was performed with the Roche LightCycler 480II (Roche Applied Science, Indianapolis, IN). Sequence-specific TaqMan probes and primer sets (Mm 00515002 for Gnb1) were obtained from ABI (Applied Biosystems, Foster City, CA) and qRT-PCR was performed according to the manufacturer’s protocol (ABI Master Mix, 15 μl/sample, 20 ng cDNA/sample). Samples from three mice per line were assayed in triplicate on a single plate. Following correction for two endogenous controls (Gapdh and Pkg1), relative quantities of the transcripts were calculated using the LightCycler 480 Software release 1.5.0 SP4.
Functional Validation of Gnb1 Effect on Alcohol Consumption
All experiments were performed under a protocol approved by the University of Colorado Denver Institutional Animal Care and Use Committee.
Production of adenovirus expressing Gnb1 or scrambled shRNA
Small interfering RNAs targeting Gnb1 were designed by Dr. Amy Lasek (University of Illinois, Chicago) using the siDesign Center at Thermo Fisher Scientific (http://www.dharmacon.com/DesignCenter/DesignCenterPage.aspx). These 19 nt sequences were incorporated into DNA oligonucleotides encoding short hairpin RNAs (shRNAs) and cloned into the lentiviral-vector pLL3.7 (Lasek et al., 2007), which includes a U6 promoter for shRNA expression and a CMV promoter for eGFP expression. The targeting sequence used for the Gnb1 shRNA is: GGAAGAATCCAAATGCGGA. The viral plasmids containing the shRNAs were tested in Neuro2A cells for ability to reduce expression (mRNA) levels of Gnb1 (Lasek et al., 2007, Lasek et al., 2010). The shRNA constructs (Gnb1 and scrambled [Scr]) provided by Dr. Lasek were used to produce adenoviral vectors. The pLL3.7 vector was digested with EcoRI, blunted and digested with XbaI for cloning into the pSHUTTLE vector (He et al., 1998), which was modified by insertion of a chromosomal insulator (P2) to reduce inflammation caused by the vector, and a polyadenylation site (pA). The constructs were verified by sequencing to contain the cloned shRNAs and eGFP. The plasmids were then linearized with PmeI and contransformed by electroporation into E. Coli BJ5183 cells with an adenoviral backbone vector, pAdEasy (He et al., 1998). Recombinants were selected for kanamycin resistance. The recombinant plasmids were digested with PacI to release the viral chromosome, and the DNA was used to transfect an adenovirus packaging cell line, HEK293A (Life Technologies), using Lipofectamine 2000, to produce virus. Cells were harvested after 21 days, when most showed a cytopathic effect, freeze-thawed to release virus, and centrifuged to remove cell debris. The supernatant was stored at −80°C. HEK293A cells were infected with the crude lysate to increase the virus titer, and virus was collected as described. Virus was titered by measuring absorbance at 260 nm (virus particles/ml) or by using the Adeno-X Rapid Titer Kit (Clontech, Mountain View, CA), which provides a measure of infectious units (ifu). To assay the Gnb1 shRNA effect, Neuro2A cells were infected in suspension with the adenoviral stock (MOI 50) and plated on 12-well plates. Protein lysates were prepared at various times after plating and assayed for Gβ1 by immunoblotting (Figure S1).
Stereotactic Injections
A. Effect of Gnb1 shRNA on G protein β1 subunit (Gβ1) protein levels
Male DBA/2 mice (~25g) were anesthetized with pentobarbital and placed in a stereotaxic frame. Holes were drilled in the skull above the target area (nucleus accumbens). Infusions of the adenoviral vectors (1.75 μl/side) were made with a Hamilton syringe, connected to an infusion pump. Virus (Gnb1 shRNA, 1.46 × 106 virus particles/μl or Scr shRNA, 1.46 × 106 virus particles/μl) was injected bilaterally into the nucleus accumbens (coordinates are in reference to bregma: AP, +1.25 mm; ML, ± 0.9 mm; DV, −3.75 mm). Virus was allowed to diffuse for 5 min before the needle was removed. Mice were singly housed and levels of Gβ1 protein were assessed by immunohistochemistry at 26 days after virus injection.
B. Effect of Gnb1 shRNA on alcohol consumption
A different set of male DBA/2 mice (~25 g) were anesthetized with pentobarbital and placed in a stereotaxic frame, and injected as described above. Virus (Gnb1 shRNA, 1.2410e11 ifu or Scr shRNA, 1.410e11 ifu) was injected bilaterally into the nucleus accumbens (AP, +1.25 mm; ML, ±0.9 mm; DV, −3.75 mm, relative to bregma). Mice were allowed to recover for one week prior to initiating alcohol consumption experiments.
Alcohol Consumption
Mice were singly-housed and given 24-hr free choice access to water or alcohol solutions (3% w/v for 5 days, 6% w/v for seven days), as previously described (Blednov et al., 2001, Yoneyama et al., 2008). The experiments were carried out in standard cages. Drinking tubes were placed through holes in the cage top and two tubes were continuously available to each mouse. Tubes were weighed daily, and food was available ad libitum. Mice were sacrificed after the last day of access to 6% ethanol, approximately 3 weeks after virus injection.
Immunohistochemistry
Mice were anesthetized with pentobarbital, and subjected to intracardiac perfusion with 0.9% cold heparinized saline and 4% paraformaldehyde in phosphate buffer. The brain was removed and allowed to remain in 4% paraformaldehyde for one day before being cryoprotected in 30% sucrose for an additional day. Brains were sectioned in the coronal plane on a freezing sliding microtome at 30 um thickness and sections were stored in a cryoprotectant solution. To determine the number of Gβ1-stained cells in hippocampus, sections containing nucleus accumbens were processed immunohistochemically using antigen retrieval and double fluorescence labeling for GFP (mouse monoclonal antibody, 1:1000, Millipore Corp.) and for Gβ1 (rabbit polyclonal antibody, 1:1000, Santa Cruz Biotechnology). Secondary antibodies were Dylight 594-conjugated AffiniPure Donkey anti-rabbit IgG (1:500, Jackson ImmunoResearch Laboratories) for Gβ1, and for GFP, AlexaFluor 488-conjugated AffiniPure Donkey anti-mouse IgG (1:200, Jackson ImmunoResearch Laboratories). Sections were mounted on slides and coverslipped with an antifade fluorescent mounting medium. Examination of the stained sections was done with a spinning disc confocal system (Intelligent Imaging Innovations (3i) Marianas SDC) mounted on a Zeiss LSM510 microscope. The system is equipped with lasers having 488 nm (for excitation of green-emitting fluorophores) and 561 nm (for excitation of red-emitting fluorophores) excitation wavelengths. Images were obtained with Slidebook 5.5 software (3i), and images from the sections containing the nucleus accumbens (range, Bregma +0.86 to Bregma +1.54) were imported into ImageJ to separate the channels, and then imported into Adobe Photoshop where equally sized areas from the nucleus accumbens on each section were outlined, and all Gβ1-stained cells and GFP-stained cells within this outline were counted.
Brains from DBA2 mice that had been treated with virus expressing Gnb1 shRNA or Scr shRNA and had consumed alcohol were obtained and sectioned as described above, and, to evaluate targeting of the injected vectors, GFP fluorescence was directly visualized in sections containing nucleus accumbens (range, Bregma +0.26 to Bregma +2.96) on an Olympus AX70 microscope.
Results
Selective Breeding of HAP3 and LAP3 Mice for Alcohol Preference
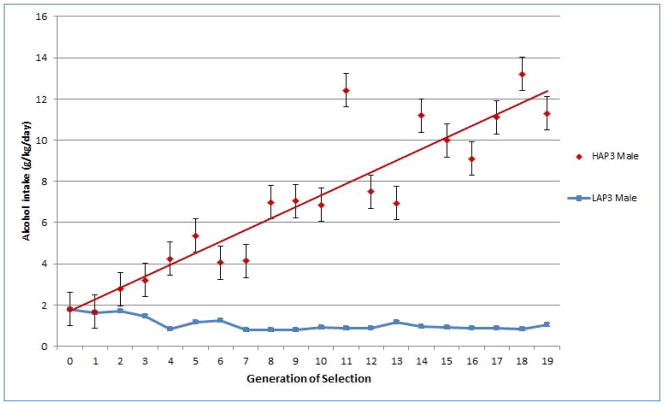
Figure 1 shows the alcohol consumption of male HAP3 and LAP3 mice over generations of selection. The relatively slow response to selection reflects the heritability of the selected trait of alcohol preference/consumption, which was estimated at 0.3 for the first 10 generations of HAP3/LAP3 mice (Falconer & Mackay, 1996, Oberlin et al., 2011). Alcohol consumption by the LAP3 mice appears to reach a “floor” by generation 4. The level of alcohol intake reached by the HAP3 mice at generation 13 is less than that reached by male HAP1 or HAP2 mice at generation 13 (Oberlin et al., 2011). However, there is a substantial difference in alcohol intake between HAP3 and LAP3 mice by this generation, which continues to increase through generation 19 (Figure 1).
Figure 1. Alcohol Consumption during Selective Breeding of HAP3 and LAP3 Mice.
Selective breeding was based on consumption of a 10% alcohol solution in a 24 hr access, 2-bottle choice paradigm, as reported previously (Grahame et al., 1999, Oberlin et al., 2011). Values represent mean ± SEM (n=32–43 mice per line in each of generations 6–18). Mice used for gene expression analysis did not consume alcohol, but were from second litters in each generation. Consumption was significantly different between HAP3 and LAP3 mice in generation 6–18 (p<6.8 × 10−5 at each generation, t-test).
It has been shown that HAP3 mice achieve blood alcohol levels high enough to have pharmacological effects when measured at the highest point of alcohol intake (Matson & Grahame, 2011). It must be emphasized, however, that the gene expression measurements in this study were performed on HAP3 and LAP3 mice that never consumed alcohol. Our experiments are designed to determine genomic regions (deeQTLs, rveQTLs) that affect gene expression levels during the course of a selective breeding experiment, and to identify potential candidate genes that may predispose to differences in alcohol consumption, not to identify changes in gene expression levels that occur as a result of alcohol consumption.
Transcript Expression Differences Measured During Selection of HAP3 and LAP3 Mice
After filtering for sequences that contained known SNPs, normalization and filtering for present calls (i.e., present in all 62 samples), 15,259 probesets (37% of original probesets on the arrays) remained for analysis. The batch adjustment was applied to this set of 15,259 probesets. We examined the effect of batch on the expression data before and after adjustment using hierarchial clustering (Figure S3) and a principal component analysis. Prior to adjustment, the first principal component accounted for 98% of the variation in expression. This principal component had a strong association with batch (p<1.0 × 10−7) even after accounting for the line (HAP vs. LAP) and for generation. After adjustment, the first principal component (99% of variation) was no longer associated with batch (p=0.56).
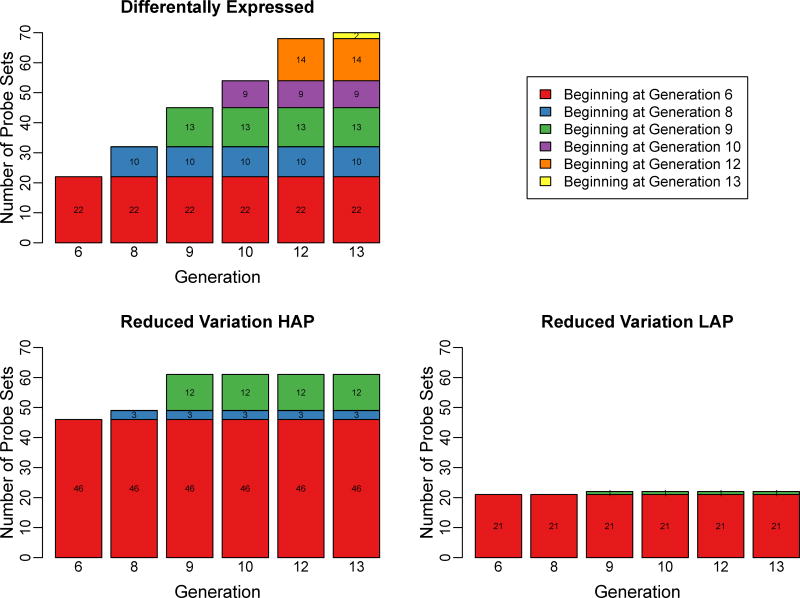
As shown in Figure 2, using the significance threshold described in Methods, at generation 6, 22 probesets (19 genes) were differentially expressed and maintained that difference in expression thereafter (up to generation 13). The number of differentially expressed probesets at each measured generation then remained relatively constant as selection proceeded (Figure 2). All of these differentially expressed transcripts are listed in Table S3.
Figure 2. Differentially Expressed Probesets, and Probesets Displaying Reduced Variance of Expression During Selection for HAP3 and LAP3 Mice.
Brain gene expression was determined using Affymetrix MOE430 v2 arrays from 4–5 mice at each generation. Following normalization and filtering for expression above background, differences in expression at each generation, that were maintained throughout selection (until generation 13), were determined using a moderated t-test (Panel A). Similarly, probesets that displayed reduced variance of expression, compared to generation 0 (HS mice), and that maintained that reduced variance throughout selection, were determined (Panels B and C). The cumulative number of probesets that displayed these characteristics at each generation of selection is indicated. Colors indicate the generation of which differential expression or reduced variance began.
Reduction in Variance of Transcript Expression Levels Measured During Selection of HAP3 and LAP3 Mice
Using the significance threshold described in Methods, there were 46 probesets (40 annotated genes) that showed reduced variance in expression levels in HAP3 mice, and 21 probesets (18 annotated genes) in LAP3 mice, between generation 0 (HS mice) and generation 6 of selection, and that maintained that reduced variance until generation 13 (Figure 2). Three transcripts showed reduced variance of expression levels at generation 8, and 12 at generation 9, in HAP3 mice. In LAP3 mice, the largest number of probesets with reduced variance of expression was observed between generation 0 and 6, with only one probeset showing reduced variance of expression at the later generations of selection (Figure 2). All of these probesets with reduced expression variance are listed in Table S4.
eQTLs for Differentially Expressed Transcripts (deeQTLs) and Transcripts with Reduced Variance of Expression (rveQTLs)
At each generation of selection where transcript expression levels were measured, common eQTLs were identified for transcripts that displayed significant differential expression between the lines, or reduced variance of expression in each line, and maintained the differential expression or reduced variance of expression through generation 13. As shown in Table S5, there were five significant (FDR<0.05) deeQTLs at generation 6, one significant deeQTL each at generations 8, 9, and 10, and two at generation 13. rveQTLs were identified separately for the HAP3 and LAP3 lines. For LAP3 mice, all 6 significant (FDR<0.05) rveQTLs were identified in generation 6, while in the HAP3 mice, 1 rveQTL was identified in generation 6, 11 rveQTLs were identified in generation 8, and 13 rveQTLs were identified in generation 9 (Table S5). If the deeQTLs and rveQTLs are important for the selected trait of alcohol consumption/preference, then one might expect that the deeQTLs and rveQTLs would occur in common regions of the genome, and would also occur in regions of the genome that have previously been associated with alcohol consumption (QTLs) (Table S2). Table 1 shows the deeQTLs that overlap with rveQTLs and QTLs. It is of particular interest that all but two (5/7) of the deeQTLs that overlap with a rveQTL also overlap with a QTL previously identified for alcohol consumption/preference (Table 1 and Table S5).
Table 1.
Differential Expression QTLs (deeQTLs) with Overlapping Reduced Variation QTLs (rveQTLs) and Alcohol Preference/Consumption QTLs (QTLs)
| deeQTLa | Overlapping rveQTLa | Overlapping Alcohol Preference QTL |
|---|---|---|
| 3:99.4 - 151.7 (generation 6) | 3:118.0 - 138.0 (HAP3 - generation 9) 3:135.1 - 155.1 (LAP3 - generation 6) |
3:0 - 224.84 (Belknap and Atkins 2001) 3:128.52 - 129.96 (Bice et al 2011) 3:137.82 - 138 (Bice et al 2011) 3:145.36 - 145.56 (Bice et al 2011) |
| 4:144.37 - 165.38 (generation 6) | 4:139.68 - 159.68 (HAP3 - generation 8) 4:134.56 – 165.38 (LAP3 – generation 6) |
4:52.01 - 202.78 (Belknap and Atkins 2001) |
| 7:116.33 – 136.33 (generation 6) | 7:71.49 – 155.53 (HAP3 - generation 8) | 7:114.4 - 136.3 (Rodriguez et al 1995) |
| 1:3.98 - 30.77 (generation 8) | 1:3.37 - 35.63 (LAP3 - generation 6) | 1:1.8 - 13.57 (Bice et al 2011) |
| 7:85.16 - 131.84 (generation 13) | 7:71.49 - 155.33 (HAP3 - generation 8) | 7:114.4 - 136.3 (Rodriguez et al 1995) |
QTLs are reported as Chromosome: Mb range
The region reported represents adjacent SNPs with an FDR less than 0.05. The number of SNPs per deeQTL ranges from 3 to 10. Calculation of Mb range for alcohol consumption/preference QTLs is described in the text.
Candidate Gene Identification
The genomic regions defined by the deeQTLs and rveQTLs control the expression levels of a higher proportion of differentially expressed transcripts, or of transcripts with reduced expression variance, than would be expected by chance. However, these genomic regions do not control the expression of all of the differentially expressed transcripts or transcripts with reduced variance of expression. In order to prioritize particular transcripts as potential candidates that contribute to differences in the selected trait of alcohol consumption, we used multi-locus models to identify transcripts that were differentially expressed and that were significantly associated with (strongly controlled by) a deeQTL that overlapped an rveQTL and a phenotypic QTL (the deeQTL was identified in the generation where the transcript first displayed differential expression). The “candidate genes” identified in this manner are shown in Table 2, and their patterns of expression over generations are shown in Figure 3. We verified the significance of differential expression of these “candidate genes” by accounting post hoc for dependence across generations and multiple testing (FDR<1%). These genes were further prioritized based on whether they also displayed reduced variance of expression levels. Five transcripts (Gnb1, Peg3, Fam168a, Usp53 and Acer3) met these criteria, and represent the highest priority candidate genes. The other genes shown in Table 2 were differentially expressed and associated with the deeQTL/rveQTL/QTL regions, but did not show a significant reduction in variance of expression. Some of the highest priority genes had a major (strongly associated, usually cis) eQTL that was in the same location as the deeQTL (e.g., Gnb1, Usp53, Acer3), while for the others, the major eQTL was located outside of the deeQTL. When the major eQTL was in the same location as the deeQTL, the multivariate model explained a relatively large proportion of the variance in transcript expression (Table 2).
Table 2.
Priority Genes With Expression Differences from Selective Breeding For Alcohol Preference
| Gene Symbol | Gene Name | QTL (SNP) associated in multivariate modela (chromosome: Mb range) | Proportion of variance in the gene’s expression explained by multivariate model | Generation Where Significant Differential Expression Began | Generation Where Significant Reduced Variation Began (Line of RV) | Physical Location of Probe Set | Location of Major eQTL [chromosome: Mb (p-value)] | Difference in Expression at Generation 13 [log2 HAP3/LAP3 (p-value)] |
|---|---|---|---|---|---|---|---|---|
| Gnb1 | guanine nucleotide binding protein (G protein), beta 1 | rs13478067 (4:144.37 - 165.38) | 0.74 | 6 | 6 (LAP) | 4:154.93 Mb | 4:155.24 Mb (<0.0001) | −1.98 (<0.0001) |
| Usp53 | ubiquitin specific peptidase 53 | rs13477323 (3:99.4 - 151.7) rs13478067 (4:144.37 - 165.38) |
0.73 | 6 | 6 (LAP) | 3:122.69 Mb | 3:118.29 Mb (<0.0001) | −0.47 (<0.0001) |
| Peg3 | paternally expressed 3 | rs3669485 (1:3.98 - 30.77) | 0.14 | 8 | 6 (HAP) | 7:6.66 Mb | 3:23.72 Mb (0.0920) | 1.33 (0.0009) |
| Fam168a | familiy sequence similarity 168, member A | rs3669485 (1:3.98 - 30.77) | 0.20 | 8 | 8 (HAP) | 7:107.99 Mb | 7:103.13 Mb (<0.0001) | 1.2 (<0.0001) |
| Acer3 | alkaline ceramidase 3 | rs13479416 (7:85.16 - 131.84) | 0.98 | 13 | 8 (HAP) | 7:105.36 Mb | 7:103.13 Mb (<0.0001) | 1.46 (<0.0001) |
| Ccnh | cyclin H | rs6241342 (7:116.33 - 136.33) | 0.15 | 6 | 13:85.35 Mb | 8:14.76 Mb (0.2070) | 0.13 (0.0749) | |
| Map3k7 | mitogen-activated protein kinase kinase kinase 7 | rs3701432 (4:19.19 - 44.04) rs13478067 (4:144.37 - 165.38) |
0.50 | 6 | 4:32.11 Mb | 4:31.65 Mb (0.0260) | −0.39 (0.0004) | |
| Mphosph9 | M-phase phosphoprotein 9 | rs13477323 (3:99.4 - 151.7) | 0.25 | 6 | 5:124.7 Mb | 5:119.76 Mb (0.0370) | −1.13 (<0.0001) | |
| Vps41 | vacuolar protein sorting 41 (yeast) | rs6241342 (7:116.33 - 136.33) | 0.15 | 6 | 13:18.92 Mb | 12:82.69 Mb (0.6730) | −0.35 (0.0001) | |
| Xaf1 | XIAP associated factor 1 | rs13478067 (4:144.37 - 165.38) rs6241342 (7:116.33 - 136.33) |
0.28 | 6 | 11:72.13 Mb | 11:69.42 Mb (<0.0001) | −0.42 (0.0002) | |
| Odz4 | odd Oz/ten-m homolog 4 (Drosophila) | rs13479416 (7:85.16 - 131.84) | 0.20 | 13 | 7:104.06 Mb | 7:117.12 Mb (0.3440) | −0.92 (<0.0001) |
priority QTLs (deeQTLs shown in Table 1) are represented in red
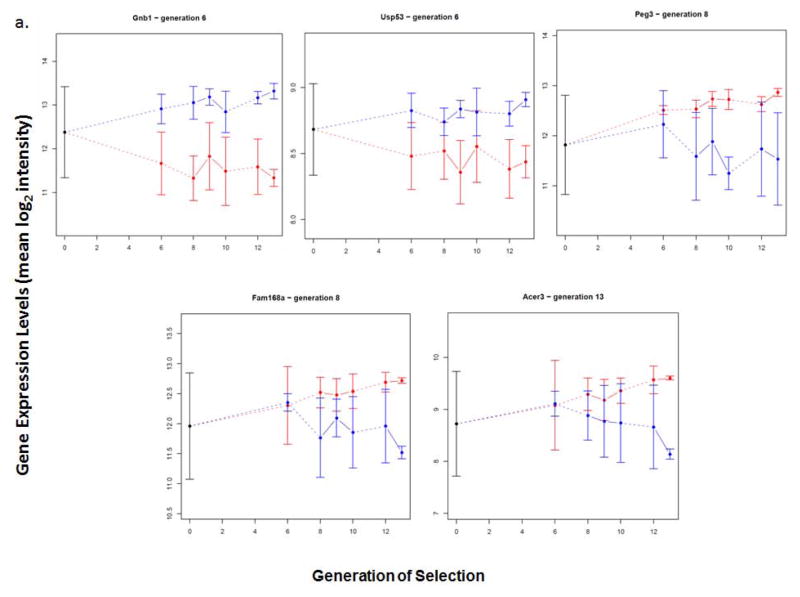
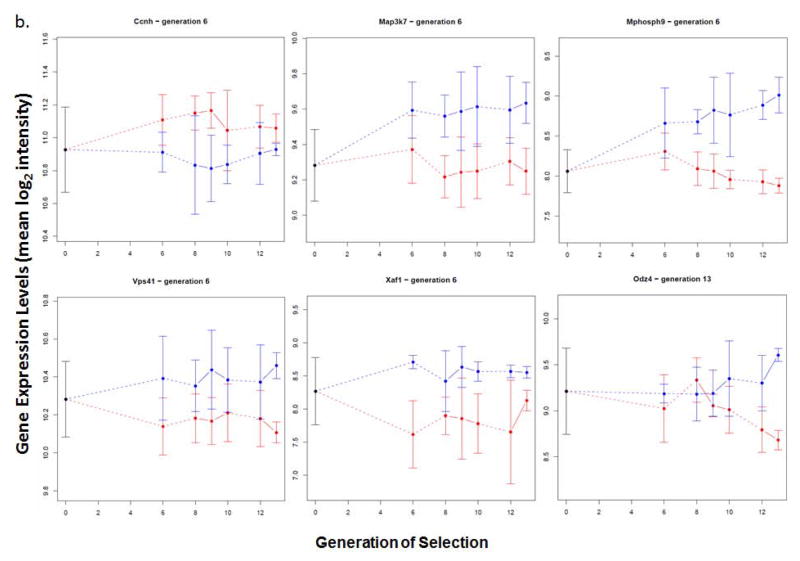
Figure 3. Candidate Transcripts for Alcohol Consumption in HAP3 and LAP3 Mice.
Brain expression patterns over generations of selection for the transcripts listed in Table 2 are shown (log base 2 expression level, mean ± SEM). Each of these transcripts, indicated by the gene symbol, displayed significantly different expression levels between HAP3 (red) and LAP3 (blue) mice beginning at the indicated generation (Table 2), and maintained differential expression until the last measurement, at generation 13. A. Gnb1, Usp53, Peg3, Fam168a and Acer3 also showed reduced variance of expression, compared to HS mice (generation 0) in at least one of the lines. B. The other differentially expressed transcripts did not show significantly reduced variance of expression.
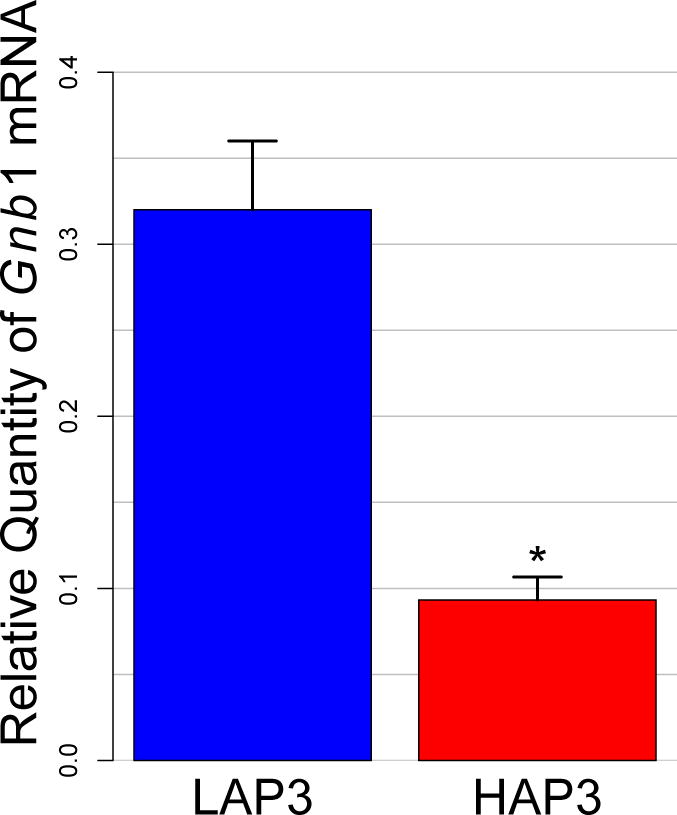
Gnb1, the G protein beta 1 subunit (Gβ1), and Usp53, ubiquitin specific peptidase 53, are expressed at higher levels in brains of LAP mice. Gnb1 was identified and validated in our previous analyses as a candidate gene associated with differences in predisposition to consume alcohol (Mulligan et al., 2006, Saba et al., 2006, 2011, Tabakoff et al., 2008), and its differential expression between HAP3 and LAP3 mice was verified by qRT-PCR analysis (Figure 4). Acer3, alkaline ceramidase 3, Peg3, paternally expressed 3 (also known as Pw1, Zfp102 and Gcap4), and Fam168a, family with sequence similarity 168, member A (also known as TCRP1), are expressed at higher levels in brains of HAP3 mice.
Figure 4. Verification of Differential Expression of Gnb1 in HAP3 vs LAP3 Mice by qRT-PCR.
RNA was isolated from brains of three individual HAP3 and LAP3 mice (generation 12). qRT-PCR was performed on the Roche LightCycler 480 using primers and probes from ABI. Samples from all mice were assayed in triplicate in the same assay. Data were corrected for two endogenous controls (Gapdh and Pkg1), and relative quantities (ratio of target to standard, mean ± SEM) were calculated using the LightCycler 480 software. *P<0.006 (Student’s t-test).
Functional Validation: Effect of Gβ1 on Alcohol Consumption
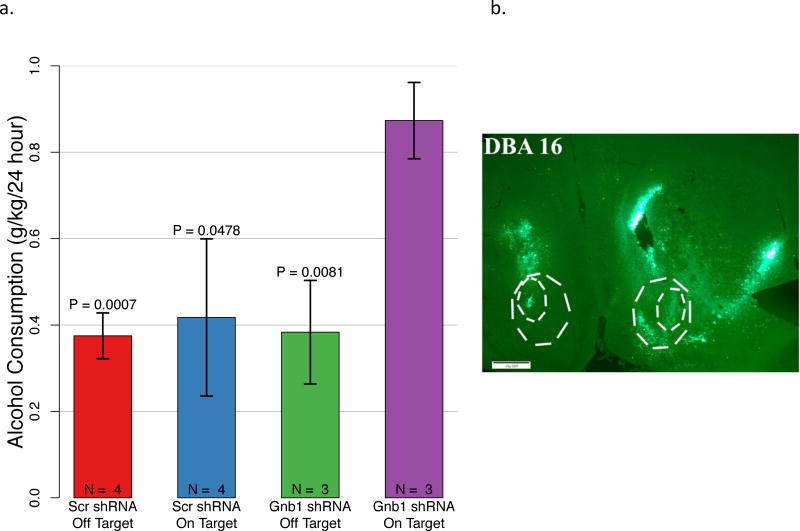
To evaluate the effect of Gβ1 on alcohol consumption, we targeted the viral vector to nucleus accumbens, a brain area that is crucial for the rewarding effects of ethanol, as well as other drugs (Koob & Volkow, 2010). We previously reported that whole brain expression levels of Gnb1 significantly negatively correlate with alcohol consumption levels in BXD recombinant inbred (RI) mice (Tabakoff et al., 2008). Using gene expression data available from Gene Network (www.genenetwork.org), we also found that Gnb1 expression in the nucleus accumbens (GN accession # GN156) significantly negatively correlated with alcohol consumption in the BXD RI panel (p=0.015). Because Gnb1 levels are higher in brains of mice that drink low amounts of alcohol (Saba et al., 2011), we elected to lower Gβ1 levels in a strain of low-drinking mice (DBA/2), hypothesizing that this manipulation would increase alcohol consumption. In one group of DBA/2 mice, we determined that treatment with adenoviral vector expressing Gnb1 shRNA lowered the number of cells expressing Gβ1 in nucleus accumbens by about 40% (Figure S2). In a separate group of DBA/2 mice, which were used to assess alcohol consumption, we verified the targeting of the adenoviral vector injection by visualization of GFP fluorescence (Figure 5B). When GFP was observed in the nucleus accumbens on both sides of brain, injections were classified as “on target”. Otherwise, injections were classified as “off target” (regardless of whether GFP was observed outside of the nucleus accumbens). Groups of DBA/2 mice injected with adenovirus expressing scrambled (Scr) shRNA, whether the injection was “on target” or “off target”, as well as mice injected “off target” with the adenovirus expressing Gnb1 shRNA, consumed amounts of 6% alcohol, averaged over a period of 7 days, that were not significantly different (Figure 5A), and these amounts were comparable to previously reported levels of alcohol consumption by DBA/2 mice in a 24 hour, 2-bottle choice paradigm (Belknap et al., 1993, Wahlsten et al., 2006). However, mice that were injected “on target” into the nucleus accumbens with the adenovirus expressing Gnb1 shRNA (Figure 5B) showed a significant increase in alcohol intake, compared to all other groups (Figure 5A) (ANOVA corrected for heterogeneous variance indicated a significant effect of treatment on alcohol consumption, F(3,10) = 8.16, P = 0.0048; Gnb1 shRNA “on target” group significantly different from all other groups, see Figure 5A). Similar results were found for preference for 6% alcohol (ml alcohol/ml total fluid × 100): Gnb1 shRNA “on target”, 10.2 ± 1.5% preference (mean ± SD); Scr shRNA “on target”, 5.0 ± 4.5% preference; Scr shRNA “off target”, 5.0 ± 1.6% preference; Gnb1 shRNA “off target”, 4.5 ± 2.4% preference (ANOVA corrected for heterogeneous variance, F(3,19) = 8.45, P = 0.0043; Gnb1 shRNA “off target” group, P = 0.005, Scr shRNA “off target” group, P = 0.001, Scr shRNA “on target” group, P = 0.054, compared to Gnb1 shRNA “on target” group, Fisher’s LSD test).
Figure 5. Functional Validation of the Role of Gnb1 in alcohol Consumption.
DBA/2 mice were injected with an adenoviral vector expressing Gnb1 shRNA or scrambled (Scr) shRNA targeted bilaterally into the nucleus accumbens. One week later, mice were given access to alcohol in a 24-hr, 2-bottle choice paradigm. A. Mice treated with the Gnb1 shRNA (n=3) that was targeted correctly (bilaterally to the nucleus accumbens;” on target”) showed increased consumption of 6% ethanol (g/kg/24 hr) compared to mice treated with scrambled (Scr) shRNA (“on target”, n=4, “off target”, n=4) or to mice treated with Gnb1 shRNA that was not correctly targeted (“off target”, n = 3). Results are the average consumption of alcohol over a 7-day period of access to 6% alcohol. ANOVA corrected for heterogeneous variance showed a significant effect of treatment (F (3, 10) = 8.16, P = 0.0048). Post-hoc analysis (Fisher’s LSD test) showed that the “ on target” Gnb1 shRNA group was significantly different from all other groups (P values shown in Figure). B. GFP fluorescence, indicating infection by the adenovirus, is visible in the nucleus accumbens (indicated by white dotted lines outlining shell and core regions) of a mouse treated with Gnb1 shRNA, which showed increased alcohol consumption. This representative section is located at approximately Bregma +1.78 mm. Similar results were found in various sections throughout the nucleus accumbens, and for other mice that showed an increase in alcohol consumption.
Discussion
Previous work by us and others has used the “genetical genomic/phenomic” approach for identification of candidate genes that affect complex traits through variation in their expression levels. This method involves determining the correlation of gene expression levels with the complex trait across a panel of inbred or recombinant inbred animals, or another segregating population, and using the overlap of phenotypic and expression QTLs (QTLs and eQTLs) as a filter to identify candidate genes among the correlated genes. In our studies, we chose to examine whole brain gene expression data because the predisposition to alcohol use involves numerous brain regions (see Crabbe et al., 2006, Koob & Volkow, 2010). Furthermore, we have reported that gene expression and eQTL data from whole brain captures the majority of transcripts that are associated with alcohol consumption in relevant brain areas (Vanderlinden et al., 2013). Using this method with mice and rats, we have previously identified candidate genes and signaling pathways in brain that are proposed to influence the predisposition of the animals to consume varying amounts of alcohol (Saba et al., 2006, Tabakoff et al., 2008, 2009).
During the independent selective breeding experiment to generate the HAP3 and LAP3 lines from HS mice, we had the unique opportunity to track the development not only of differential gene expression levels, but also changes in the variance of gene expression levels, as selection proceeded. To our knowledge, this is one of the only studies of this type to be performed using a rodent model for a complex behavioral trait, and these data allowed us to identify common eQTL regions (deeQTLs and rveQTLs) that are postulated to reflect regions of the genome affected by selective breeding. We considered that genomic regions associated with both differential gene expression and reduced variance of gene expression (overlapping deeQTL and rveQTL) would be the most likely to be associated with the process of selection for the phenotypic trait. The utility of the filter is supported by the finding that 5/7 of the overlapping deeQTL/rveQTL genomic regions also overlap with previously identified QTL regions associated with alcohol preference, based on 2-bottle choice procedures (Belknap & Atkins, 2001, Bice et al., 2011, Rodriguez et al., 1995).
A limitation of this analysis is that the alcohol consumption QTLs are derived from studies of HAP1/LAP1 mice and from mapping populations derived from C57BL/6 and DBA/2 mice, not from the progeny of the selected lines of HAP3 and LAP3 mice. The eQTLs are also derived from BXD recombinant inbred mice. However, C57BL/6 and DBA/2 mouse strains were included in the cross that produced the HS mice from which the HAP and LAP mice are derived (Mcclearn et al., 1970) and, of the eight inbred strains in the cross, C57BL/6 show the highest alcohol preference, and DBA/2 are among the strains showing the lowest alcohol preference. Therefore, it is likely that alleles from these strains are major determinants of genetically-mediated differences in alcohol consumption/preference, and it is notable that phenotypic QTLs derived from progeny of HAP1/LAP1 and HAP2/LAP2 mice are in the same genomic regions as those found for mice derived from C57BL/6 and DBA/2 strains (Bice et al., 2009, 2006, 2011). The finding that most of the overlapping deeQTL/rveQTL regions also overlap with QTLs for alcohol consumption/preference serves as a proof of concept for the approach used in this study, but it must be acknowledged that QTLs derived from other strains included in the HS cross cannot be detected in this analysis.
The differentially expressed genes listed in Table 2 highlight the fact that the expression levels of many transcripts in brain are controlled by multiple genetic loci (also see Table S3). Three of the transcripts in Table 2 (Usp53, Map3k7, Xaf1) had more than one deeQTL contributing independently to their expression level. The influence of multiple QTL regions on gene expression levels may also be evident from the expression patterns of the transcripts as selection proceeds (Figure 3). Most of the differentially expressed transcripts, including the candidate transcripts that show differential expression beginning at generation 6 (Gnb1, Usp53), maintain the same magnitude of differential expression through generation 13. This pattern suggests that transcript expression levels are regulated from a small number of eQTLs that have strong effects. However, some transcripts, notably Mphosph9 (Figure 3b), show continual increases in the magnitude of differential expression that parallel the changes in the phenotype of alcohol consumption over generations of selection. Transcripts such as this are likely to be regulated by numerous eQTLs, each of which has a relatively small effect on expression levels, i.e., at each generation, more eQTLs with small effects on expression become evident. By determining deeQTLs for transcripts such as this only at the first generation where differential expression occurs, we may miss other loci that become fixed at later generations.
Numerous studies have identified candidate genes for alcohol consumption as genes with expression levels that correlate with phenotypic values (Edenberg et al., 2005, Hu et al., 2008, Kimpel et al., 2007, Mcbride et al., 2013, Mulligan et al., 2008, Mulligan et al., 2006, Saba et al., 2006, Tabakoff et al., 2008, Tabakoff et al., 2009), including differences in expression levels between lines of animals selected for high and low alcohol preference. Fewer studies have investigated reduced variance of gene expression as an indicator of a genomic locus affected by selection. Hughes and Buitenhuis (2010) found reduced variance of transcript expression in a line of chickens selected for high feather picking, compared to controls, but did not find differences in mean expression values between these groups. Ho et al. (2008), studying variability of gene expression in populations with and without disease, also noted that a number of genes that appeared to be relevant to a particular disease displayed differential variability of expression in the absence of mean differences in expression levels. As suggested by Hughes and Buitenhuis (2010), genes with reduced variance of expression in a selected line may reflect the influence of selective breeding, but it is not clear that such genes (if they are not differentially expressed) contribute to the selected behavior. We propose that our “high priority” transcripts, which display both differential expression between the selected lines and reduced expression variance, are the most likely to represent genes that are influenced by selection, and are valid candidate genes for predisposition to differences in alcohol preference.
The highest priority candidate genes that we identified have been implicated in cell signaling and cell survival/proliferation/apoptosis. The Gβ1 protein (product of Gnb1) plays a key role in cell signaling, including modulation of activity of adenylyl cyclases, potassium channels, phospholipase C and N-type calcium channels, and has also been implicated in epigenetic regulation of gene expression, synaptic function (e.g., receptor trafficking, neurotransmitter release), and regulation of Map kinase activity (Dupre et al., 2009). The protein product of Usp53 is a member of a protease family (ubiquitin-specific proteases) of deubquitylating enzymes (Quesada et al., 2004). In general, these proteases remove ubiquitin and ubiquitin-like domains from ubiquitin-conjugated proteins. Ubiquitylation of proteins plays a role in many cellular processes, including cell cycle progression, DNA repair, vesicular trafficking, signal transduction, etc. (Glickman & Ciechanover, 2002, Hochstrasser, 2000, Weissman, 2001). However, the human USP53 showed no catalytic activity in a deubiquitylating assay (Quesada et al., 2004). It was suggested that this non-protease member of the USP family could function as a regulatory or inhibitory (“dominant negative”) factor (Quesada et al., 2004). On the other hand, a study of the proteins that interact with deubquitylating enzymes suggested that Usp53 may be involved in mitotic processes (Sowa et al., 2009). The protein product of Acer3 is an alkaline ceramidase. Ceramidases hydrolyze ceramides to form sphingosine. Ceramide, sphingosine and sphingosine phosphates are bioactive lipids that mediate cell proliferation, differentiation, and apoptosis, as well as cell migration and cell adhesion (Hu et al., 2010, Mao & Obeid, 2008). Ceramidase has been reported to regulate synaptic vesicle exocytosis and trafficking (Rohrbough et al., 2004). The protein product of Peg3 is a maternally imprinted transcription factor, which is expressed only from the paternally inherited allele (Thiaville et al., 2013). Knocking out this gene results in increased apoptosis in numerous brain regions important for reproductive behavior, olfactory processing and reward (Broad et al., 2009). Adult knockout mice show a complex olfactory deficit that affects estrous odor preferences (Swaney et al., 2008). Little is known about the function of the protein product of Fam168a in rodents. In humans this protein is called “tongue cancer resistance-associated protein”, TCRP, and it has been found to promote resistance to radiation and the chemotherapeutic agent cisplatin by activating the PI-3 kinase/Akt/NF- B cell survival pathway (Gu et al., 2011, Peng et al., 2012).
The identification of a gene product that plays an important role in olfactory processing (Peg3), in combination with gene products involved in cell proliferation and survival (e.g., Gnb1, Acer3, Usp53, Fam168a), suggests the possibility of differences between HAP3 and LAP3 mice in the olfactory system, including neurogenesis and neuroplasticity in this system that occurs in the adult (Whitman & Greer, 2009). This conclusion is compatible with our previous work, in which both the function and brain regional localization of candidate genes led us to propose that olfactory cues, as well as learning processes, may regulate voluntary alcohol intake by mice in the two-bottle choice paradigm (Tabakoff et al., 2008). The role of flavor perception, which involves taste, olfactory and chemosensory information, has often been discussed with regard to its influence on initial alcohol consumption (Bachmanov et al., 2003, Kampov-Polevoy et al., 1990). Using a “lickometer” apparatus, we have found that HAP mice prefer the taste of alcohol, while LAP mice do not (Hoffman et al., 2010), and recent studies have shown similar results with an alcohol-preferring rat line (Brasser et al., 2012). The increase in alcohol consumption/preference that resulted from lowering levels of Gβ1 in brains of DBA/2 mice is also consistent with a role for orosensory perception in alcohol consumption by these mice. The amount of alcohol consumed by the DBA/2 mice, even after lowering Gβ1 levels, is unlikely to result in pharmacological (“rewarding”) effects. The observed increase in consumption is more likely to reflect a decrease in taste or smell aversion, given the intimate connections between central taste and olfactory pathways and the reward system, including the nucleus accumbens (Wesson & Wilson, 2011, Yamamoto, 2006). In future studies, it would be of interest to determine whether changes in Gβ1 levels would alter preference or avoidance of tastants such as sucrose or quinine. Since only one differentially expressed candidate transcript (Gnb1) was found in common in all three of the HAP and LAP selection experiments, as well as between C57BL/6 vs DBA/2 mice (Saba et al., 2011), we conclude that the same behavioral phenotype can be achieved through varying combinations of differentially expressed genes that arise during the processes of selective breeding or inbreeding.
The current study suggests an approach to the identification of genomic regions that are affected during a selective breeding experiment. This approach, which is based on the genetics of gene expression (eQTL analysis), has the added advantage of identifying specific candidate genes associated with the selected phenotype, in this case, the predisposition to variation in alcohol consumption. It can be envisioned that analysis of the genetics of gene expression in relevant tissues could be used to design and/or track the progress of selective breeding for other phenotypes.
Supplementary Material
Acknowledgments
This work was supported in part by NIAAA (U01AA016649, U01AA016663, INIA West Consortium; R24AA013162; K01AA016922; R24AA015512; P60AA007611) and the Banbury Fund. We thank Dr. Amy Lasek, University of Illinois, Chicago, for providing the Gnb1 and Scr shRNA constructs (INIA West Consortium, U01AA016654). Imaging experiments were performed in the University of Colorado Anschutz Medical Campus Advanced Light Microscopy Core, supported in part by NIH/NCATS Colorado CTSI Grant No. UL1 TR001802. Contents are the authors’ responsibility and do not necessarily represent official NIH views. We thank Adam Chapman, Yinni Yu, Seija Tillanen and Barbara Blanchard for expert technical assistance.
Footnotes
References
- Affymetrix. Statistical algorithms reference guide. Affymetrix, Inc; Santa Clara, CA: 2001. [Google Scholar]
- Bachmanov AA, Kiefer SW, Molina JC, Tordoff MG, Duffy VB, Bartoshuk LM, Mennella JA. Chemosensory factors influencing alcohol perception, preferences, and consumption. Alcohol Clin Exp Res. 2003;27:220–231. doi: 10.1097/01.ALC.0000051021.99641.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belknap JK, Atkins AL. The replicability of QTLs for murine alcohol preference drinking behavior across eight independent studies. Mammalian Genome. 2001;12:893–899. doi: 10.1007/s00335-001-2074-2. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Crabbe JC, Young ER. Voluntary consumption of ethanol in 15 inbred mouse strains. Psychopharmacology (Berl) 1993;112:503–510. doi: 10.1007/BF02244901. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc. 1995;B.57:289–300. [Google Scholar]
- Bhave SV, Hornbaker C, Phang TL, Saba L, Lapadat R, Kechris K, Gaydos J, McGoldrick D, Dolbey A, Leach S, Soriano B, Ellington A, Ellington E, Jones K, Mangion J, Belknap JK, Williams RW, Hunter LE, Hoffman PL, Tabakoff B. The PhenoGen informatics website: tools for analyses of complex traits. BMC Genet. 2007;8:59. doi: 10.1186/1471-2156-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bice P, Valdar W, Zhang L, Liu L, Lai D, Grahame N, Flint J, Li TK, Lumeng L, Foroud T. Genomewide SNP screen to detect quantitative trait loci for alcohol preference in the high alcohol preferring and low alcohol preferring mice. Alcoholism: Clinical and Experimental Research. 2009;33:531–537. doi: 10.1111/j.1530-0277.2008.00866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bice PJ, Foroud T, Carr LG, Zhang L, Liu L, Grahame NJ, Lumeng L, Li TK, Belknap JK. Identification of QTLs influencing alcohol preference in the High Alcohol Preferring (HAP) and Low Alcohol Preferring (LAP) mouse lines. Behav Genet. 2006;36:248–260. doi: 10.1007/s10519-005-9019-6. [DOI] [PubMed] [Google Scholar]
- Bice PJ, Lai D, Zhang L, Foroud T. Fine mapping quantitative trait loci that influence alcohol preference behavior in the high and low alcohol preferring (HAP and LAP) mice. Behav Genet. 2011;41:565–570. doi: 10.1007/s10519-010-9414-5. [DOI] [PubMed] [Google Scholar]
- Blednov YA, Stoffel M, Chang SR, Harris RA. Potassium channels as targets for ethanol: studies of G-protein-coupled inwardly rectifying potassium channel 2 (GIRK2) null mutant mice. J Pharmacol Exp Ther. 2001;298:521–530. [PubMed] [Google Scholar]
- Brasser SM, Silbaugh BC, Ketchum MJ, Olney JJ, Lemon CH. Chemosensory responsiveness to ethanol and its individual sensory components in alcohol-preferring, alcohol-nonpreferring and genetically heterogeneous rats. Addict Biol. 2012;17:423–436. doi: 10.1111/j.1369-1600.2011.00415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broad KD, Curley JP, Keverne EB. Increased apoptosis during neonatal brain development underlies the adult behavioral deficits seen in mice lacking a functional paternally expressed gene 3 (Peg3) Dev Neurobiol. 2009;69:314–325. doi: 10.1002/dneu.20702. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Harris RA, Arends MA, Koob GF. Alcohol-related genes: contributions from studies with genetically engineered mice. Addict Biol. 2006;11:195–269. doi: 10.1111/j.1369-1600.2006.00038.x. [DOI] [PubMed] [Google Scholar]
- Dupre DJ, Robitaille M, Rebois RV, Hebert TE. The role of Gbetagamma subunits in the organization, assembly, and function of GPCR signaling complexes. Annu Rev Pharmacol Toxicol. 2009;49:31–56. doi: 10.1146/annurev-pharmtox-061008-103038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Strother WN, McClintick JN, Tian H, Stephens M, Jerome RE, Lumeng L, Li TK, McBride WJ. Gene expression in the hippocampus of inbred alcohol-preferring and -nonpreferring rats. Genes Brain Behav. 2005;4:20–30. doi: 10.1111/j.1601-183X.2004.00091.x. [DOI] [PubMed] [Google Scholar]
- Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, Mouy M, Steinthorsdottir V, Eiriksdottir GH, Bjornsdottir G, Reynisdottir I, Gudbjartsson D, Helgadottir A, Jonasdottir A, Styrkarsdottir U, Gretarsdottir S, Magnusson KP, Stefansson H, Fossdal R, Kristjansson K, Gislason HG, Stefansson T, Leifsson BG, Thorsteinsdottir U, Lamb JR, Gulcher JR, Reitman ML, Kong A, Schadt EE, Stefansson K. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- Erwin VG, Deitrich RA. Genetic selection and characterization of mouse lines for acute functional tolerance to ethanol. J Pharmacol Exp Ther. 1996;279:1310–1317. [PubMed] [Google Scholar]
- Erwin VG, Heston WD, McClearn GE, Deitrich RA. Effect of hypnotics on mice genetically selected for sensitivity to ethanol. Pharmacol Biochem Behav. 1976;4:679–683. doi: 10.1016/0091-3057(76)90219-7. [DOI] [PubMed] [Google Scholar]
- Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. Longman Group Ltd; Essox, England: 1996. [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Grahame NJ, Li TK, Lumeng L. Selective breeding for high and low alcohol preference in mice. Behav Genet. 1999;29:47–57. doi: 10.1023/a:1021489922751. [DOI] [PubMed] [Google Scholar]
- Gu Y, Fan S, Liu B, Zheng G, Yu Y, Ouyang Y, He Z. TCRP1 promotes radioresistance of oral squamous cell carcinoma cells via Akt signal pathway. Mol Cell Biochem. 2011;357:107–113. doi: 10.1007/s11010-011-0880-8. [DOI] [PubMed] [Google Scholar]
- He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges LV, Olkin I. Statistical methods for meta-analysis. Academic Press; Orlando: 1985. [Google Scholar]
- Ho JW, Stefani M, dos Remedios CG, Charleston MA. Differential variability analysis of gene expression and its application to human diseases. Bioinformatics. 2008;24:i390–398. doi: 10.1093/bioinformatics/btn142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M. Evolution and function of ubiquitin-like protein-conjugation systems. Nat Cell Biol. 2000;2:E153–157. doi: 10.1038/35019643. [DOI] [PubMed] [Google Scholar]
- Hoffman PL, Bennett B, Saba LM, Bhave SV, Carosone-Link PJ, Hornbaker CK, Kechris KJ, Williams RW, Tabakoff B. Using the Phenogen website for ‘in silico’ analysis of morphine-induced analgesia: identifying candidate genes. Addict Biol. 2011;16:393–404. doi: 10.1111/j.1369-1600.2010.00254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman PL, Jones J, Bennett B, Carosone-Link PJ, Grahame N, Finger TE, Tabakoff B. Does taste influence alcohol preference? Alcohol Clin Exp Res. 2010;34:84A. [Google Scholar]
- Hu W, Saba L, Kechris K, Bhave SV, Hoffman PL, Tabakoff B. Genomic insights into acute alcohol tolerance. J Pharmacol Exp Ther. 2008;326:792–800. doi: 10.1124/jpet.108.137521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Xu R, Sun W, Szulc ZM, Bielawski J, Obeid LM, Mao C. Alkaline ceramidase 3 (ACER3) hydrolyzes unsaturated long-chain ceramides, and its down-regulation inhibits both cell proliferation and apoptosis. J Biol Chem. 2010;285:7964–7976. doi: 10.1074/jbc.M109.063586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Buitenhuis AJ. Reduced variance of gene expression at numerous loci in a population of chickens selected for high feather pecking. Poult Sci. 2010;89:1858–1869. doi: 10.3382/ps.2010-00827. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Kampov-Polevoy AB, Kasheffskaya OP, Sinclair JD. Initial acceptance of ethanol: gustatory factors and patterns of alcohol drinking. Alcohol. 1990;7:83–85. doi: 10.1016/0741-8329(90)90065-k. [DOI] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, Furlotte NA, Eskin E, Nellaker C, Whitley H, Cleak J, Janowitz D, Hernandez-Pliego P, Edwards A, Belgard TG, Oliver PL, McIntyre RE, Bhomra A, Nicod J, Gan X, Yuan W, van der Weyden L, Steward CA, Bala S, Stalker J, Mott R, Durbin R, Jackson IJ, Czechanski A, Guerra-Assuncao JA, Donahue LR, Reinholdt LG, Payseur BA, Ponting CP, Birney E, Flint J, Adams DJ. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kechris KJ, Biehs B, Kornberg TB. Generalizing moving averages for tiling arrays using combined p-value statistics. Stat Appl Genet Mol Biol. 2010;9:Article 29. doi: 10.2202/1544-6115.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimpel MW, Strother WN, McClintick JN, Carr LG, Liang T, Edenberg HJ, McBride WJ. Functional gene expression differences between inbred alcohol-preferring and -non-preferring rats in five brain regions. Alcohol. 2007;41:95–132. doi: 10.1016/j.alcohol.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Volkow ND. Neurocircuitry of addiction. Neuropsychopharmacology. 2010;35:217–238. doi: 10.1038/npp.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Botstein D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasek AW, Janak PH, He L, Whistler JL, Heberlein U. Downregulation of mu opioid receptor by RNA interference in the ventral tegmental area reduces ethanol consumption in mice. Genes Brain Behav. 2007;6:728–735. doi: 10.1111/j.1601-183X.2007.00303.x. [DOI] [PubMed] [Google Scholar]
- Lasek AW, Kapfhamer D, Kharazia V, Gesch J, Giorgetti F, Heberlein U. Lmo4 in the nucleus accumbens regulates cocaine sensitivity. Genes Brain Behav. 2010;9:817–824. doi: 10.1111/j.1601-183X.2010.00620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Burmeister M. Genetical genomics: combining genetics with gene expression analysis. Hum Mol Genet. 2005;14(Spec No 2):R163–169. doi: 10.1093/hmg/ddi267. [DOI] [PubMed] [Google Scholar]
- Li TK, Lumeng L, Doolittle DP. Selective breeding for alcohol preference and associated responses. Behav Genet. 1993;23:163–170. doi: 10.1007/BF01067421. [DOI] [PubMed] [Google Scholar]
- Lumeng L, Waller MB, McBride WJ, Li T-K. Genetic influences on alcohol preference in animals. In: Begleiter H, Kissin B, editors. The Genetics of Alcoholism. Oxford University Press; New York: 1995. pp. 165–220. [Google Scholar]
- Mao C, Obeid LM. Ceramidases: regulators of cellular responses mediated by ceramide, sphingosine, and sphingosine-1-phosphate. Biochim Biophys Acta. 2008;1781:424–434. doi: 10.1016/j.bbalip.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson LM, Grahame NJ. Pharmacologically relevant intake during chronic, free-choice drinking rhythms in selectively bred high alcohol-preferring mice. Addict Biol. 2011 doi: 10.1111/j.1369-1600.2011.00412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride WJ, Kimpel MW, McClintick JN, Ding ZM, Hyytia P, Colombo G, Liang T, Edenberg HJ, Lumeng L, Bell RL. Gene expression within the extended amygdala of 5 pairs of rat lines selectively bred for high or low ethanol consumption. Alcohol. 2013 doi: 10.1016/j.alcohol.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClearn GE, Wilson JR, Meredith W. In: Contributions to Behavior - Genetic Analysis The Mouse as a Prototype. Lindzey G, Lindsey GG, Thiesson DD, editors. Appleton-Century Crofts; New York: 1970. pp. 3–22. [Google Scholar]
- Mulligan MK, Ponomarev I, Boehm SL, 2nd, Owen JA, Levin PS, Berman AE, Blednov YA, Crabbe JC, Williams RW, Miles MF, Bergeson SE. Alcohol trait and transcriptional genomic analysis of C57BL/6 substrains. Genes Brain Behav. 2008;7:677–689. doi: 10.1111/j.1601-183X.2008.00405.x. [DOI] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, Crabbe JC, Blednov YA, Grahame NJ, Phillips TJ, Finn DA, Hoffman PL, Iyer VR, Koob GF, Bergeson SE. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci U S A. 2006;103:6368–6373. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberlin B, Best C, Matson L, Henderson A, Grahame N. Derivation and characterization of replicate high- and low-alcohol preferring lines of mice and a high-drinking crossed HAP line. Behav Genet. 2011;41:288–302. doi: 10.1007/s10519-010-9394-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng B, Gu Y, Xiong Y, Zheng G, He Z. Microarray-assisted pathway analysis identifies MT1X & NFkappaB as mediators of TCRP1-associated resistance to cisplatin in oral squamous cell carcinoma. PLoS One. 2012;7:e51413. doi: 10.1371/journal.pone.0051413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips TJ, Crabbe JC. Behavioral studies of genetic differences in alcohol action. In: Crabbe JC, Harris RA, editors. The Genetic Basis of Alcohol and Drug Actions. Plenum Press; New York: 1991. pp. 25–104. [Google Scholar]
- Quesada V, Diaz-Perales A, Gutierrez-Fernandez A, Garabaya C, Cal S, Lopez-Otin C. Cloning and enzymatic analysis of 22 novel human ubiquitin-specific proteases. Biochem Biophys Res Commun. 2004;314:54–62. doi: 10.1016/j.bbrc.2003.12.050. [DOI] [PubMed] [Google Scholar]
- Rodriguez LA, Plomin R, Blizard DA, Jones BC, McClearn GE. Alcohol acceptance, preference, and sensitivity in mice. II. Quantitative trait loci mapping analysis using BXD recombinant inbred strains. Alcoholism: Clinical and Experimental Research. 1995;19:367–373. doi: 10.1111/j.1530-0277.1995.tb01517.x. [DOI] [PubMed] [Google Scholar]
- Rohrbough J, Rushton E, Palanker L, Woodruff E, Matthies HJ, Acharya U, Acharya JK, Broadie K. Ceramidase regulates synaptic vesicle exocytosis and trafficking. The Journal of Neuroscience. 2004;24:7789–7803. doi: 10.1523/JNEUROSCI.1146-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba L, Bhave SV, Grahame N, Bice P, Lapadat R, Belknap J, Hoffman PL, Tabakoff B. Candidate genes and their regulatory elements: alcohol preference and tolerance. Mammalian Genome. 2006;17:669–688. doi: 10.1007/s00335-005-0190-0. [DOI] [PubMed] [Google Scholar]
- Saba LM, Bennett B, Hoffman PL, Barcomb K, Ishii T, Kechris K, Tabakoff B. A systems genetic analysis of alcohol drinking by mice, rats and men: influence of brain GABAergic transmission. Neuropharmacology. 2011;60:1269–1280. doi: 10.1016/j.neuropharm.2010.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, Linsley PS, Mao M, Stoughton RB, Friend SH. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney WT, Curley JP, Champagne FA, Keverne EB. The paternally expressed gene Peg3 regulates sexual experience-dependent preferences for estrous odors. Behav Neurosci. 2008;122:963–973. doi: 10.1037/a0012706. [DOI] [PubMed] [Google Scholar]
- Tabakoff B, Saba L, Kechris K, Hu W, Bhave SV, Finn DA, Grahame NJ, Hoffman PL. The genomic determinants of alcohol preference in mice. Mammalian Genome. 2008;19:352–365. doi: 10.1007/s00335-008-9115-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabakoff B, Saba L, Printz M, Flodman P, Hodgkinson C, Goldman D, Koob G, Richardson HN, Kechris K, Bell RL, Hubner N, Heinig M, Pravenec M, Mangion J, Legault L, Dongier M, Conigrave KM, Whitfield JB, Saunders J, Grant B, Hoffman PL. Genetical genomic determinants of alcohol consumption in rats and humans. BMC Biol. 2009;7:70. doi: 10.1186/1741-7007-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiaville MM, Huang JM, Kim H, Ekram MB, Roh TY, Kim J. DNA-binding motif and target genes of the imprinted transcription factor PEG3. Gene. 2013;512:314–320. doi: 10.1016/j.gene.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderlinden LA, Saba LM, Kechris K, Miles MF, Hoffman PL, Tabakoff B. Whole brain and brain regional coexpression network interactions associated with predisposition to alcohol consumption. PLoS One. 2013;8:e68878. doi: 10.1371/journal.pone.0068878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlsten D, Bachmanov A, Finn DA, Crabbe JC. Stability of inbred mouse strain differences in behavior and brain size between laboratories and across decades. Proc Natl Acad Sci U S A. 2006;103:16364–16369. doi: 10.1073/pnas.0605342103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- Wesson DW, Wilson DA. Sniffing out the contributions of the olfactory tubercle to the sense of smell: hedonics, sensory integration, and more? Neurosci Biobehav Rev. 2011;35:655–668. doi: 10.1016/j.neubiorev.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman MC, Greer CA. Adult neurogenesis and the olfactory system. Prog Neurobiol. 2009;89:162–175. doi: 10.1016/j.pneurobio.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T. Neural substrates for the processing of cognitive and affective aspects of taste in the brain. Arch Histol Cytol. 2006;69:243–255. doi: 10.1679/aohc.69.243. [DOI] [PubMed] [Google Scholar]
- Yoneyama N, Crabbe JC, Ford MM, Murillo A, Finn DA. Voluntary ethanol consumption in 22 inbred mouse strains. Alcohol. 2008;42:149–160. doi: 10.1016/j.alcohol.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.