Abstract
Chronic exposure to drugs of abuse or stress regulates transcription factors, chromatin modifying enzymes, and histone posttranslational modifications in discrete brain regions. Due to the promiscuity of the enzymes involved, it has not yet been possible to obtain direct causal evidence to implicate the regulation of transcription and consequent behavioral plasticity by chromatin remodeling that occurs at a single gene. Here, we investigate the mechanism linking chromatin dynamics to neurobiological phenomena by applying engineered transcription factors to selectively modify chromatin at a specific gene in vivo. We found that histone methylation or acetylation at the FosB locus in nucleus accumbens—a brain reward region—is sufficient to control drug- and stress-evoked transcriptional and behavioral responses via interactions with the endogenous transcriptional machinery. This approach allows us to relate the epigenetic landscape at a given gene directly to regulation of its expression and to its subsequent effects on reward behavior.
Introduction
Drug addiction and depression are associated with altered expression or activity of particular transcription factors and histone-modifying enzymes, in both rodent models and human patients; experimental manipulation of such molecules in discrete brain regions alters the cellular and behavioral responses to drug or stress exposures in animals1-4. Studies of the mechanisms underlying altered gene expression in response to drugs or stress have involved analyses of transcription factor binding and enrichment of modified histones throughout the genome5,6. Interestingly, despite global enrichment in histone acetylation and depletion in histone methylation within a given brain region, many genes show alterations opposite to these, posing a key question as to the mechanism and relevance of gene-specific chromatin remodeling in the context of drug- and stress-evoked epigenetic changes. While work to date has laid the foundation for investigating the role of such histone modifications in regulating drug or stress responses, previous approaches have lacked the specificity needed to demonstrate the causal mechanisms by which a given chromatin modification at a specific gene is necessary and sufficient to regulate drug- or stress-evoked transcriptional and behavioral plasticity.
To overcome this limitation, we focused on the transcription factor, ΔFosB, which plays important roles, acting in the nucleus accumbens (NAc)—a key brain reward region, in drug and stress action7,8. ΔFosB is induced in the NAc by chronic cocaine exposure in rodents and in human cocaine addicts4,9, but is reduced by certain forms of chronic stress in rodents and in depressed humans10,11. The histone methyltransferase, G9a (EHMT2), which catalyzes dimethylation of histone H3 lysine 9 (H3K9me2), a repressive histone modification12, appears to be crucial in mediating this regulation: H3K9me2 enrichment at FosB is suppressed in the NAc by repeated cocaine12 and we show here that it is increased at FosB in the NAc of depressed humans. Moreover, we have demonstrated that local overexpression or knockdown of G9a in NAc potently controls drug and stress responses in rodent11,12. Nevertheless, this quality of proof is inherently limited because drugs and stress, as well as G9a manipulations, affect H3K9me2 enrichment at hundreds of loci in this brain region1,12,13. These previous studies underscore the fact that all current evidence for the epigenetic basis of reward pathology has relied on correlating the effects of global manipulations of histone posttranslational modifications (HPTMs) with affected behavior and physiology, resulting in a limited analysis of the transcriptional mechanisms linking epigenetic modifications at a single gene to the transcriptional activity of that gene and subsequent reward behavior.
Here, we sought to achieve a stronger level of proof by use of engineered transcription factors, zinc-finger proteins (ZFPs)14 or transcription activator-like effectors (TALEs)15, that can be designed in silico to recognize and bind to a specific 18 to 20-bp locus in the genome. Fusion of the DNA-binding domain to a given catalytic domain allows gene-targeted transcriptional regulation at a site of interest. ZFPs and TALEs have been applied in vitro and shown to efficiently activate or repress gene expression when fused to any of several functional domains. More recently, ZFPs and TALEs have been used in vitro to promote epigenetic remodeling when fused to chromatin modifying enzymes14,16,17. Although very few studies to date have utilized this approach in vivo, one study showed reversal of a Parkinsonian phenotype in a mouse model by overexpression of a ZFP-transcriptional activator directed against the promoter of glial cell line-derived neurotrophic factor (Gdnf), which conferred protection from a striatal chemical lesion18. We thus took advantage of this novel approach to investigate the causal role of cocaine- and stress-evoked epigenetic regulation of the FosB locus in the NAc in vivo.
Results
Generation of ZFPs that target the FosB gene in vivo
To direct locus-specific epigenetic changes to a specific gene we screened a suite of cys2-his2 six-finger ZFPs19 for regulation of FosB transcription, designed in silico to span the murine FosB promoter at various 18-20 bp motifs within –1000 to +200 bp relative to the transcription start site (TSS) (Fig. 1a, c). As an initial proof of principle, each ZFP was fused to the p65 domain of the mammalian transcription factor NFkB, which activates transcription by promoting histone acetylation via recruitment of p300/CBP (CREB binding protein)20. Histone acetylation has been shown to modulate drug- and stress-evoked behavior21-23 and gene expression1,24, in part by regulating drug-associated memory formation25,26. A subset of the FosB-ZFP-p65 constructs were transfected into Neuro2a (N2a) cells and their relative efficacy was determined by qRTPCR analysis of FosB/ΔFosB mRNA expression (Supplementary Fig. 1). ZFP25, -35, and -61 were selected as potent activators of FosB transcription and subjected to further analysis. To study regulation of the FosB gene in vivo, we injected mouse NAc (Fig. 1b and Supplementary Fig. 2a) with herpes simplex virus (HSV) vectors expressing various FosB-ZFP-p65 or -NFD (no functional domain) constructs and quantified FosB/ΔFosB mRNA using qRT-PCR.
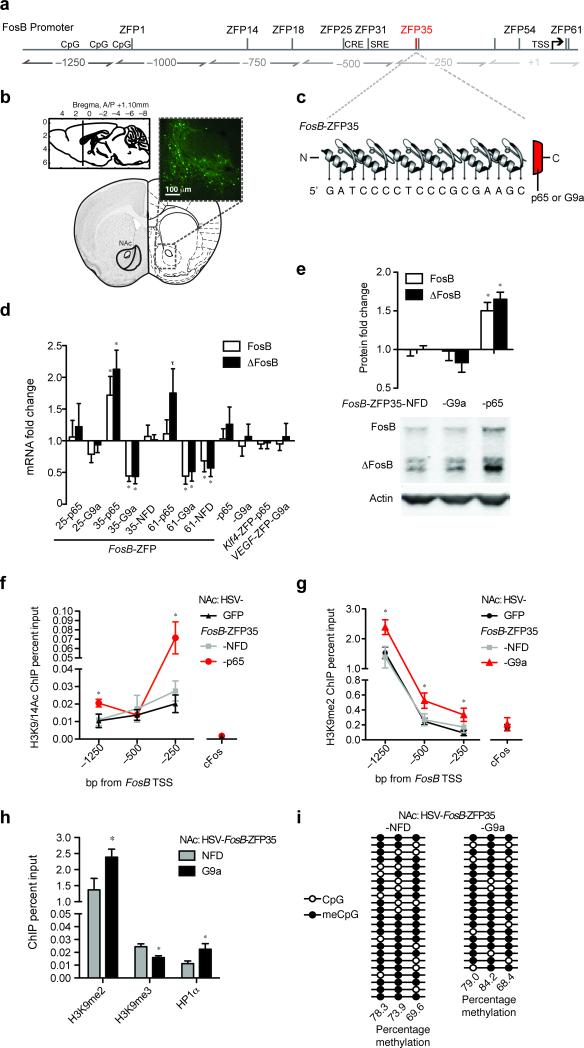
Figure 1.
Engineered transcription factors targeting the FosB promoter bidirectionally regulate gene expression in NAc neurons via epigenetic manipulation. (a) Locations of FosB-ZFP binding relative to the FosB TSS. The location of functional SRF and CREB binding sites are shown. CpG: methylation sites analyzed in Fig. 1i. (b) HSV injection into the mouse NAc43 drives robust transgene expression in neurons. (c) The 6-finger ZFP35 recognizes the FosB promoter at approximately 250 bp upstream from the FosB TSS. (d) At 72 hours post-injection, FosB/ΔFosB mRNA expression in the NAc was significantly induced by HSV-FosB-ZFP35-p65 compared to control virus [FosB: t7=2.37, *P=0.049; ΔFosB: t7=3.83, *P=0.007; n=4, 5], , with a trend for ΔFosB induction by FosB-ZFP61-p65 [t7=2.03, τP=0.076; n=4, 5]. Compared to controls, FosB/ΔFosB mRNA in the NAc were significantly repressed by HSV-FosB-ZFP35-G9a [FosB: t6=4.84, *P=0.003; ΔFosB: t6=3.40, *P=0.015; n=5, 3], -FosB-ZFP61-NFD [FosB t7=2.39, *P=0.048; ΔFosB: t7=2.18, τP=0.066; n=4, 5], and FosB-ZFP61-G9a [FosB: t8=2.98, *P=0.017; ΔFosB: t8=2.37, *P=0.047; n=5, 5]. HSV-G9a, -p65, -Klf4-ZFP-p65, VEGF-ZFP-G9a and -FosB-ZFP35-NFD had no effect on FosB/ΔFosB mRNA expression. Student's unpaired t-test P>0.05 for all other comparisons (see Supplementary Table 3). (e) Compared to HSV-FosB-ZFP35-NFD [n=9], NAc injection of HSV-FosB-ZFP35-p65 caused increased FosB/ΔFosB protein level [FosB: t13=3.73, *P=0.003; ΔFosB: t13=6.65, *P=0.000; n=6], while HSV-FosB-ZFP35-G9a had no effect at 72 hours post-injection [FosB: t14=0.137, P=0.8929; ΔFosB: t14=1.25, P=0.230; n=8. Representative western blot shown. Complete western blot shown in Supplementary Figure 5. (f) NAc injection of HSV-FosB-ZFP35-p65 causes enrichment of the activating mark H3K9/14ac at the FosB promoter at –1250 [t8=2.65, *P=0.029; n=5, 5] and –250 bp [t8=2.9, *P=0.021; n=5, 5] relative to the TSS compared to control HSV, with no change in tissue treated with HSV-FosB-ZFP35-NFD [t10=0.00, P=1.00; n=6, 6] or at the cFos promoter [t7=0.217, P=0.834; n=5, 4]. IgG control IP was undetectable in >80% of the sample wells by qRT-PCR. (g) NAc injection of HSV-FosB-ZFP35-G9a caused enrichment of the repressive mark H3K9me2 at the FosB promoter at –1250 [t26=2.7, *P=0.011; n=14, 14], –500 [t14=2.3, *P=0.041; n=7, 9], and –250 bp [t13=2.9, *P=0.031; n=7, 8] relative to the TSS compared to control HSV, with no change in tissue treated with HSV-FosB-ZFP35-NFD [–1250: t19=0.45, P=0.658, n=14, 7; –500: t8=0.03, P=0.977, n=7, 3; –250: t9=1.51, P=0.167, n=8, 3] or at the cFos promoter [t16=0.42 P=0.680; n=7, 11]. IgG control IP was undetectable in >80% of the sample wells by qPCR. (h) NAc injection of HSV-FosB-ZFP35-G9a caused depletion of the repressive mark H3K9me3 [t8=3.3, *P=0.011; n=5, 5] and enrichment of HP1α [t7=2.5, *P=0.039; n=4, 4] at the FosB promoter –1250 bp upstream from the TSS compared to control HSV-FosB-ZFP35-NFD. (i) Bisulfite sequencing analysis performed on three CpG sites located –1141, –1101 and –1036 bp upstream of the FosB TSS, in a region that corresponds to the observed H3K9me2 and HP1α enrichment (See Fig. 1a). There was no difference in the percentage of methylated CpGs in NAc infected with HSV-FosB-ZFP35-G9a [n=19] compared to HSV-FosB-ZFP35-NFD [n=23]. Each row represents analysis performed on one clone. Data are presented as mean ± s.e.m.
We found that HSV-FosB-ZFP35 and –ZFP65-p65 constructs activated transcription of FosB/ΔFosB in the NAc relative to HSV-GFP or -NFDs (Fig. 1d). To address the mechanism of the repressive histone mark H3K9me2 in regulation of FosB expression, we generated FosB-ZFP-G9a constructs, in which each of three distinct ZFPs was fused to the minimal catalytic domain of G9a14,27. HSV delivery of FosB-ZFP35-G9a and -ZFP61-G9a to the NAc efficiently repressed FosB/ΔFosB mRNA expression (Fig. 1d). The changes in the amount of FosB/ΔFosB mRNA we observed in the NAc are physiologically relevant, and similar in magnitude to drug- and stress-induced changes observed previously1,24. In addition to regulation of basal mRNA expression, we found by immunoblotting that FosB/ΔFosB protein is elevated in NAc expressing HSV-FosB-ZFP35-p65 (Fig. 1e), while there is no effect in NAc infected with HSV-FosB-ZFP35-G9a, relative to the -NFD at 72 hours post infection. To more carefully examine FosB repression by HSV-FosB-ZFP35-G9a, we performed immunohistochemistry and found that the percentage of FosB/ΔFosB immunopositive cells decreased dramatically as a percentage of HSV infected (GFP+) cells in NAc infected with HSV-FosB-ZFP35-G9a compared to a control vector (Supplementary Fig. 2b). These findings confirm that the ZFPs exert their function within the HSV-infected neurons that express FosB/ΔFosB.
The ability to site-specifically regulate chromatin architecture enables direct examination of the function of HPTMs in regulating gene transcription in the brain. We used semi-quantitative chromatin immunoprecipitation (qChIP) and found enrichment of the activating histone modification, H3K9/K14 acetylation, at the FosB promoter (Fig. 1f) in NAc infected with HSV-FosB-ZFP35-p65 - an effect that was not observed in NAc infected with HSV-FosB-ZFP35-G9a (Fig. 2a). Conversely, NAc infection with HSV-FosB-ZFP35-G9a led to enrichment of the repressive histone modification, H3K9me2, at the FosB promoter (Fig. 1g). Importantly, neither HPTM was enriched at the promoter of the related immediate early gene, cFos (Fig. 1f, 1g), nor was H3K9me2 enriched globally in total NAc extracts [H3K9me2: t(1, 15)=1.35, p=0.20, n=8 NFD, n=9 G9a]. Taken together, these results demonstrate that viral expression of engineered ZFPs efficiently enrich their respective HPTMs at the FosB promoter and bi-directionally control FosB gene expression in vivo.
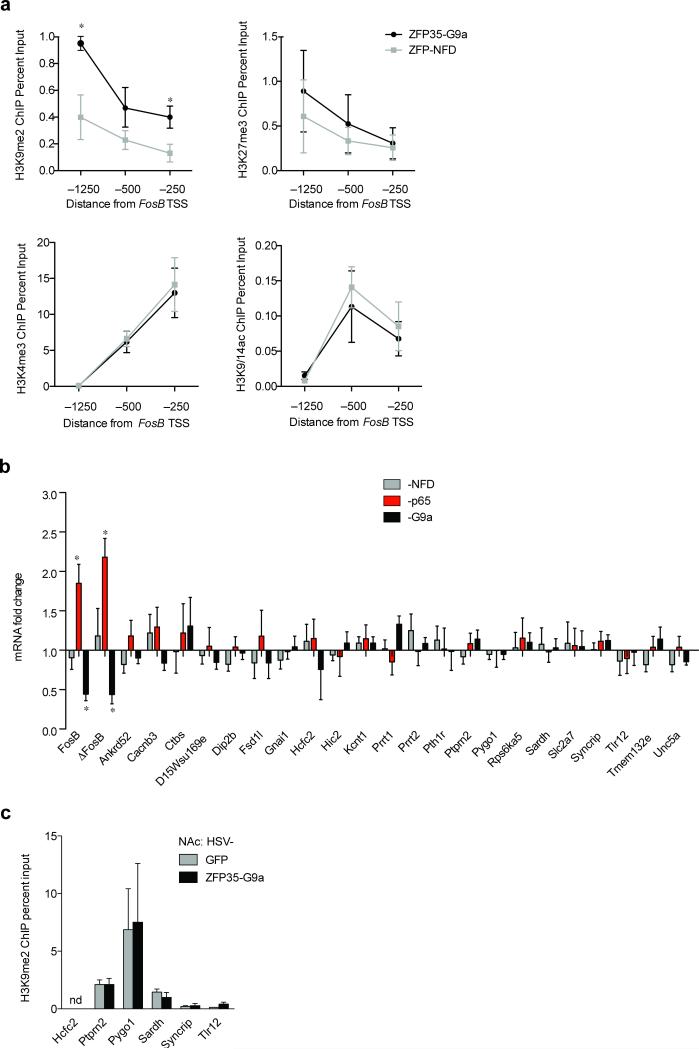
Figure 2.
HSV-FosB-ZFP35-p65 and -G9a in the NAc specifically regulate FosB/ΔFosB expression. (a) Induction of H3K9me2 at the FosB promoter by HSV-FosB-ZFP-G9a in NAc occurs relative to the TSS at –1250 bp [t5=2.75, *P=0.040; n=3, 4] and –250 bp [t5=2.69, *P=0.043; n=3, 4] without changes in the activating marks H3K9/14ac [–1250: t5=1.49, P=0.197; –500: t5=0.50, P=0.636; –250: t5=0.39, P=0.715; n=3, 4] or H3K4me3 [–1250: t5=0.37, P=0.728; –500: t5=0.24, P=0.820; –250: t5=0.22, P=0.835; n=3, 4] or the repressive mark H3K27me3 [–1250: t5=0.46, P=0.666; –500: t5=0.59, P=0.583; –250: t5=0.23, P=0.831; n=3, 4] as compared to the control (HSV-FosB-ZFP-NFD). (b) cDNA was generated from NAc injected with HSV-FosB-ZFP35-p65, -G9a or NFD. qRT-PCR was used to measure expression of potential off-target genes (See Supplementary Table 2) in samples that show regulation of FosB/ΔFosB by HSV-FosB-ZFP35-p65 [FosB: t7=4.73, *P=0.002; ΔFosB: t7=4.83, *P=0.002; n=4, 5] and HSV-FosB-ZFP35-G9a [FosB: t8=2.40, *P=0.043; ΔFosB: t8=2.88, *P=0.021; n=5, 5] but not HSV-FosB-ZFP35-G9a [FosB: t8=0.38, P=0.715; ΔFosB: t8=0.52, P=0.619; n=5, 5]. Data are normalized to HSV-GFP. Student's unpaired t-test P>0.05 for all other comparisons (see Supplementary Table 3). (c) HSV-FosB-ZFP35-G9a in the NAc does not cause H3K9me2 enrichment at off-target loci as measured by qChIP using primers that flank the off-target binding site [Ptprn2: t4=0.03, P=0.97; Pygo1: t5=0.10, P=0.922; Sardh: t4=0.93, P=0.407; Syncrip: t4=0.34, P=0.775; Tlr12: t4=1.80, P=0.152; n=3, 3].
The analysis of control constructs was critical to our investigation, as both a ZFP with no functional domain (NFD)28 or the functional domain alone29 may have nonspecific effects on transcription. We found that neither the p65 nor G9a domains alone regulated expression of FosB when virally expressed in the NAc (Fig. 1d). Furthermore, ZFPs targeted to unrelated genes, Klf4-ZFP-p65 and VEGF-ZFP-G9a, did not affect FosB/ΔFosB expression in the NAc (Fig. 1d). Finally, NAc expression of HSV-FosB-ZFP35-NFD had no effect on FosB/ΔFosB mRNA levels (Fig. 1d), or on enrichment of HPTMs at the FosB promoter (Fig. 1f, g). We did find that FosB-ZFP61-NFD repressed FosB expression (Fig. 1d), perhaps because its binding site is just downstream from the TSS (see Fig. 1a), and thus excluded this ZFP from further study.
To determine specificity of FosB gene regulation by FosB-ZFP35, we examined the mouse genome for 19 bp motifs that differed from the ZFP35 binding motif by 1-, 2- or 3-nucleotides and were located within 3 kb of a transcription start site (no site shared the identical 19 bp sequence; Supplementary Table 2). We found by qRT-PCR that the expression of these 28 off-target genes was unaffected by NAc expression of HSV-FosB-ZFP35-p65, -G9a or -NFD (22 genes) or was undetectable (6 genes) (Fig. 2b). Finally, we selected those loci located within 1000 bp upstream of their respective TSSs and found that HSV-FosB-ZFP35-G9a in NAc did not alter H3K9me2 binding at any of these 6 off-target sites (Fig. 2c). This detailed analysis of specificity goes further than previous publications to assess the off-target regulation by ZFPs in vivo. Nonetheless, we cannot rule out the possibility of undetected off-target regulation of additional genes that may contribute to the transcriptional and behavioral changes induced by FosB-ZFP35-G9a or -p65. Based on this detailed characterization, we focused on FosB-ZFP35 for the remainder of the experiments in this study.
Mechanisms of action of FosB-ZFP-G9a in NAc
To investigate the potential mechanism by which FosB promoter enrichment of H3K9me2 regulates expression of FosB/ΔFosB, we examined additional histone modifications and histone binding proteins, which may contribute to gene repression. We found that expression of HSV-FosB-ZFP-G9a in NAc did not alter enrichment of tri-methylated histone H3 lysine 27 (H3K27me3) or histone H3 lysine 4 (H3K4me3), known repressive and activating marks, respectively, at the FosB gene (Fig. 2). We next examined two marks that have been strongly implicated in repression, acting in concert with H3K9me2,, tri-methylated histone H3 lysine 9 (H3K9me3) and heterochromatin protein 1α (HP1α)30,31. We found that HSV-FosB-ZFP-G9a in NAc decreased levels of H3K9me3, but induced a ~2-fold enrichment of HP1α, at the FosB promoter compared to HSV-FosB-ZFP-NFD (Fig. 1h). However, the level of H3K9me3 and HP1α binding to FosB is nearly an order of magnitude below that of H3K9me2, raising the possibility that other factors are primarily involved in mediating FosB repression associated with increased H3K9me2 at euchromatic regions in adult brain. Interestingly, both H3K9me3 depletion and HP1α enrichment were detected only at a site -1200 bp upstream from the FosB TSS, which we found to be the major site of enrichment of H3K9me2 in response to HSV-FosB-ZFP35-G9a (Fig. 1g) and under physiological conditions in both mouse12,13 and human (Fig. 3g) NAc. This site is ~1000 bp upstream of the ZFP35 binding site (see Fig. 1a), suggesting that the binding of G9a to a proximal site on the FosB promoter induces a spreading repressive mechanism that affects a relatively large span of the FosB gene. Based on these findings, we further investigated an additional epigenetic signature, DNA methylation. While we observed methylation of the 3 predicted CpG sites within this -1200 bp region of FosB by bisulfite conversion, there was no effect of HSV-FosB-ZFP35-G9a (Fig. 1i). Importantly, the site targeted by ZFP35 coincides with a region where we have demonstrated regulation of endogenous G9a binding in response to cocaine treatments12, supporting the physiological relevance of FosB-ZFP35-G9a action in regulating FosB expression, and the potential use of this tool for understanding mechanisms controlling FosB expression in vivo.
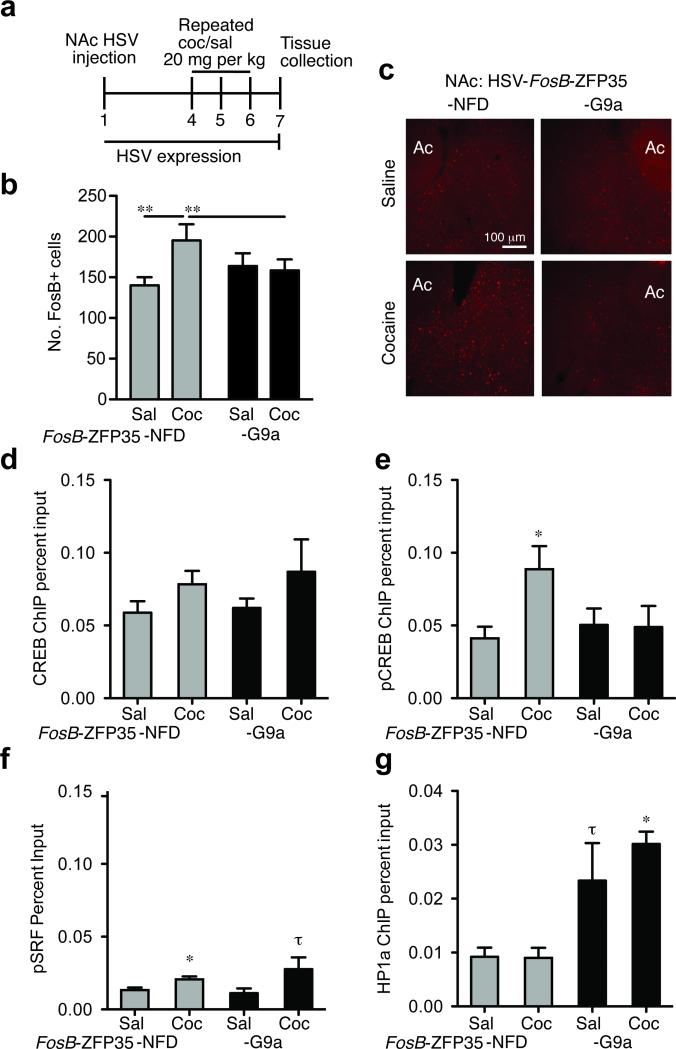
Figure 3.
Cocaine induction of FosB/ΔFosB protein expression and endogenous transcription factor binding is blocked by HSV-FosB-ZFP35-G9a in the NAc. (a) Mice were injected intra-NAc with HSV-FosB-ZFP35-NFD or -G9a and treated with repeated cocaine (20 mg/kg) or saline. (b) Cocaine induction of FosB/ΔFosB+ cells in the NAc is suppressed by FosB-ZFP35-G9a [two-way ANOVA: interaction between virus (NFD, G9a) and drug (saline, cocaine) (F(1,28)=6.6, *P=0.016, n=8); no main effects of virus (F(1,28)=0.98, P=0.330, n=8) or drug (F(1,28)=1.18, P=0.286, n=8) alone]. Cocaine enhances FosB+ cell levels within NFD-infected tissue [t14=2.48, *P=0.027; n=8] but not G9a-infected tissue [t14=1.20, P=0.291; n=8]. Among animals receiving cocaine, G9a repressed FosB/ΔFosB+ cells [t14=2.25, *P=0.041; n=8]. (c) Representative images from mice infected with HSV-FosB-ZFP35-NFD in the left hemisphere and HSV-FosB-ZFP35-G9a into the right hemisphere. Ac, anterior commissure. (d) Compared to saline, repeated cocaine treatment caused enrichment of phospho-CREB(S133) at the FosB promoter in NAc injected with HSV-FosB-ZFP35-NFD [t17=2.38, *P=0.029; n=9, 11], but not with HSV-FosB-ZFP-G9a [t15=0.09, P=0.932; n=8, 9]. (e) No change was observed in total CREB at the FosB promoter after cocaine treatment in NAc injected with either virus [NFD: t18=1.55, P=0.138; n=9, 11; G9a: t15=1.13, P=0.274; n=8, 9]. (f) Compared to saline, repeated cocaine treatment causes enrichment of phospho-SRF(S103) at the FosB promoter in NAc injected with HSV-FosB-ZFP35-NFD [t(1, 6)= 2.7, p=0.034; n=4, 4] compared to repeated saline with only a modest effect of HSV-FosB-ZFP-G9a in blocking this enrichment [t(1, 8)= 1.9, τP=0.097; n=5, 5]. (g) NAc injection of HSV-FosB-ZFP-G9a causes enrichment of HP1α at the FosB promoter compared to HSV-FosB-ZFP-NFD under cocaine conditions [t(1, 7)= 1.24, *P=0.0002, n=4, 5] and a trend under saline treatment conditions [t(1, 8)= 1.97, τP =0.085; n=4, 5].
Given that cocaine-induced ΔFosB protein accumulation critically contributes to the addiction phenotype, we sought to interfere with this physiological response by inducing H3K9me2 specifically at the FosB locus. Mice were injected with HSV-FosB-ZFP35-G9a and - NFD into the right and left NAc, respectively, and then treated with repeated saline or cocaine (20 mg/kg) over a time course designed to coincide with HSV-expression (72 hours) while still sufficiently long enough to lead to FosB/ΔFosB protein accumulation in NAc (Fig. 3a). Immunohistochemical analysis using an anti-FosB/ΔFosB antibody demonstrated that, while cocaine increased the number of FosB immunopositive NAc neurons by 39% in NAc infected with control virus, expression of HSV-FosB-ZFP35-G9a blocked this induction (Fig. 3b, c). At this time point (7 days after HSV injection) HSV-mediated transgene expression has fully dissipated, thus, it is not possible to selectively examine the infected cells, which likely accounts for the fact that expression of FosB/ΔFosB in saline-treated animals was not affected by HSV-FosB-ZFP35-G9a (Fig. 3b, c).
To further our understanding of cocaine-regulated FosB expression, we sought to discover a molecular mechanism by which an increase in H3K9me2 solely at the FosB promoter blocks cocaine induction of FosB/ΔFosB protein in vivo. We reasoned that this repressive histone modification might interfere with activation (i.e., phosphorylation) of cAMP response element binding protein (phospho-CREB[S133]) and/or serum response factor (phospho-SRF[S103]), both of which are required for FosB induction in the NAc by repeated cocaine32. We injected mouse NAc with either control HSV or HSV-FosB-ZFP35-G9a, and then treated the animals with either repeated saline or cocaine (20 mg/kg). qChIP revealed that cocaine treatment results in a 3.9-fold enrichment of phospho-CREB at the FosB promoter compared to saline treated mice (Fig. 3d). This enrichment was blocked by NAc injection of HSV-FosB-ZFP35-G9a (Fig. 3d). As previously demonstrated32, cocaine had no significant effect on the enrichment of total CREB protein at the FosB promoter, nor did HSV-FosB-ZFP35-G9a alter levels of total CREB binding (Fig. 3e). Interestingly, cocaine treatment also caused enrichment of phospho-SRF binding at the FosB promoter, but there was only a modest effect of HSV-FosB-ZFP35-G9a (Fig. 3f), despite the close proximity of the CREB and SRF binding sites (see Fig. 1a). We also examined the enrichment of HP1α at the FosB promoter and found that while cocaine does not regulate binding of this protein to FosB in NAc, HSV-FosB-ZFP35-G9a expression leads to enrichment of HP1α in either the cocaine or saline context (Fig. 3g), as shown earlier (Fig. 1h). These data demonstrate that, in fully differentiated adult neurons in vivo, a primary increase in H3K9me2 at a given gene controls enrichment of an activated transcription factor at that gene, and the gene's expression, in response to an environmental stimulus.
Regulation of FosB by transcription activator-like effectors
To gain a broader understanding of transcriptional regulation by engineered transcription factors, we targeted the FosB gene with transcription activator-like effectors (TALEs)17,33, a distinct type of designer DNA binding domain, as well as with a variety of transcriptional activators, including VP64, VP16, and 2xp65 (Supplementary Fig. 3). FosB-TALEs fused to the transactivator, VP64, efficiently upregulated expression of FosB/ΔFosB when virally expressed in the NAc, while Klf4-TALE-VP64 had no effect (Supplementary Fig. 3b). Furthermore, VP16, VP64, or 2xp65 activated FosB/ΔFosB expression when fused to FosB-ZFP35 and expressed in N2a cells (Supplementary Fig. 3d).
Such a comprehensive analysis demonstrated that engineered transcription factors are potent, bidirectional regulators of FosB/ΔFosB expression, yet we also found that their effect in cell culture was not always an accurate predictor of their function in brain. For example, FosB-ZFP25-p65 robustly activated FosB/ΔFosB expression in N2a cells (Supplementary Fig. 1), but failed to activate transcription when expressed in NAc neurons in vivo (Fig. 1d). We also observed that several FosB-ZFP-G9a constructs activated FosB expression in N2a cells (Supplementary Fig. 1) but repressed expression in NAc neurons in vivo (Fig. 1d). To investigate a potential explanation for these discrepancies, we compared the expression level of ZFP constructs in vitro vs. in vivo and found several orders of magnitude greater expression in N2a cells than in NAc (Supplementary Fig. 4a). However, titration of the amount of FosB-ZFP35-G9a expressed in N2a cells affected the expression of ΔFosB, but not FosB, mRNA, indicating that ZFP expression levels account only in part for its distinct function in vitro and in vivo (Supplementary Fig. 4b). An alternative explanation is provided by the fact that chromatin state is a major determinant of transcription binding and function and that such states differ between cell types34,35. These data underscore the importance of our approach in using an in vivo system to characterize engineered transcription factors as tools to study epigenetic mechanisms of gene regulation in reward regions of mammalian brain.
Regulation of reward behavior by FosB-ZFPs
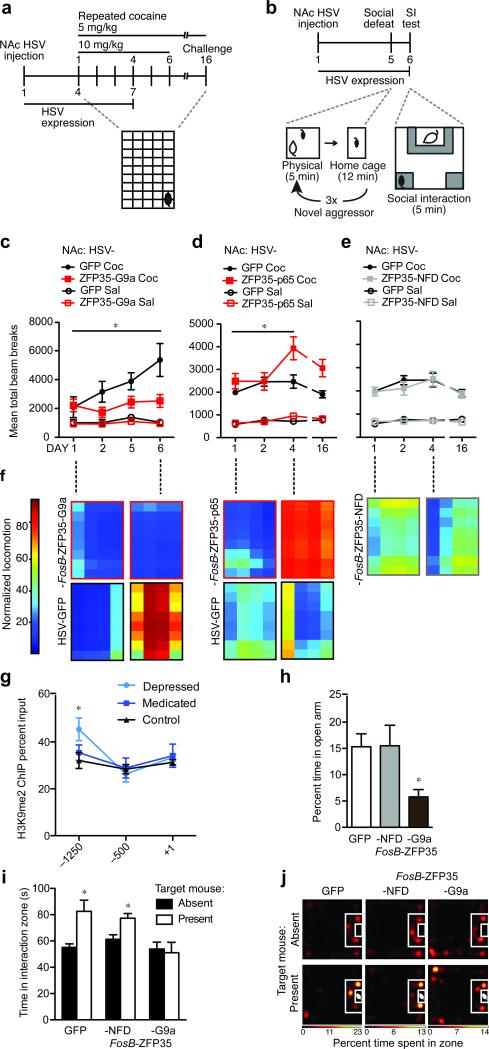
Having established a mechanism whereby engineered transcription factors can specifically regulate ΔFosB expression in vivo, we next investigated their ability to modulate reward behaviors, studying first the regulation of cocaine action. Animals were injected intra-NAc with HSV-FosB-ZFP35, -TALE1, or control vectors and their locomotor activity was recorded over several days of cocaine treatment at either a sensitizing dose (10 mg/kg) or a subthreshold dose (5 mg/kg) that does not lead to locomotor sensitization in control animals (Fig. 4a, Supplementary Fig. 3e). We found that HSV-FosB-ZFP35-G9a expression in NAc efficiently blocked cocaine locomotor sensitization (10 mg/kg) (Fig. 3c), while both HSV-FosB-ZFP35-p65 and HSV-TALE1-VP64 augmented the sensitizing effects of the drug, with no effect of HSV-FosB-ZFP35-NFD (5 mg/kg) (Fig. 4d, e and Supplementary Fig. 3e). None of the constructs affected baseline locomotor activity seen in the absence of cocaine compared to control virus (Fig. 4c-e and Supplementary Fig. 3e).
Figure 4.
Engineered transcription factors bidirectionally modulate cocaine- and stress-evoked behaviors. (a) Locomotor activity was assessed during repeated cocaine exposure in mice injected intra-NAc with HSV-FosB-ZFP35-G9a [n=7], -p65 [n=9], -NFD [n=9] or control virus [n=5 (10mg/kg) or n=10 (5mg/kg)]. (b) Social behavior was measured 24 hours after subthreshold social defeat in mice injected with HSV-FosB-ZFP35-G9a or control virus into the NAc. (c) At high doses of cocaine (10 mg/kg), locomotor behavior sensitizes over time and this effect was blocked by HSV-FosB-ZFP35-G9a in NAc. Repeated measures ANOVA revealed an interaction between day, cocaine, and virus [F(3,18)=4.00, *P=0.024] on locomotor behavior. Among cocaine treated mice, there is a main effect of virus [F(1,10)=8.81, *P=0.026] and a trend for an interaction between virus and day [F(2,8)=3.33, τP=0.077] such that GFP locomotor behavior is enhanced above FosB-ZFP35-G9a levels by treatment day 6 [t(1,10)=2.61, *P =0.026]. (d) At low doses (5 mg/kg), cocaine-induced locomotor behavior sensitizes over time with FosB-ZFP35-p65 in NAc [repeated measures: interaction between day, treatment, and virus [F(3,32)=4.71, *P=0.008]. Among cocaine treated mice, there is an interaction between virus and day [F(2,15)=7.08, *P=0.003] and a trend for a main effect of day among FosB-ZFP35-p65 [F(1,32)=2.85, τP=0.053] but not GFP cocaine treated animals. Among cocaine-treated animals, FosB-ZFP35-p65 enhances locomotor behavior above GFP levels by treatment day 4 [t17=2.58, *P=0.020] through day 16 [t17=2.92, *P=0.009]. (e) NAc injection of HSV-FosB-ZFP35-NFD, like controls, did not display cocaine locomotor sensitization to a low dose of cocaine. Repeated measures failed to find an interaction among day, treatment, and virus [F(1,28)=0.13, P=0.944]. There is no effect of day among cocaine-treated mice [F(1,28)=0.86, P=0.471]. HSV-GFP data are the same as in (Fig. 4d). (f) Heat maps show representative locomotor data within the chamber for mice over the course of repeated cocaine exposure. (g) H3K9me2 is significantly enriched at –1250 bp from the FosB TSS in depressed humans [t22=2.19, p=0.040, n=8, 17] compared to levels in control subjects. (h) FosB-ZFP35-G9a in the NAc reduced exploration of the open arm in the elevated plus maze, compared to control virus [t12=2.36, *P=0.036; n=7]. (i) HSV-FosB-ZFP35-G9a in the NAc blocked increased exploration of a novel aggressor mouse after exposure to subthreshold social defeat, compared to control virus [t123.1, *P=0.009; n=7] with no effect of HSV-FosB-ZFP35-NFD [t12=3.2, *P=0.008; n=7]. (j) Representative heatmaps of social interaction after a subthreshold defeat stress show a preference for the interaction zone when a target mouse is present for mice injected with control virus and HSV-FosB-ZFP35-NFD, but not HSV-FosB-ZFP35-G9a.
Recent evidence has also implicated a reduction of ΔFosB levels in the NAc in the etiology of depression in humans and in chronic stress models in rodents8. Indeed, we found that depressed human NAc exhibited a 37% enrichment of H3K9me2 at the FosB promoter (Fig. 3g), specifically at a site 1200 bp upstream from the TSS, coincident with the site of greatest H3K9me2 enrichment at the FosB promoter in mouse NAc (Fig. 1g). Based on these results, we examined the effect of artificially inducing H3K9me2 selectively at the FosB gene in mouse NAc, thus mimicking this single pathological histone modification in depressed humans. We relied on a variant of the chronic social defeat stress (CSDS) paradigm, in which animals are subjected to a subthreshold social stress that has no effect on depression-related behaviors, such as social avoidance, in normal animals36. Mice were injected with HSV-FosB-ZFP-G9a into NAc, allowed to recover for five days, and then exposed to an aggressor for three bouts of five minutes each (Fig. 4b). In contrast to control virus, mice that were infected with HSV-FosB-ZFP35-G9a showed reduced social interaction when tested 24 hours after the aggressive encounter, a pro-depression-like behavioral response (Fig. 4i). Further, we found that these mice spent significantly less time exploring the open arms in the elevated plus maze, indicative of increased anxiety-related behavior (Figure 3h). Thus, mimicking a single HPTM abnormality seen at a single gene in the NAc of depressed humans is sufficient to promote behavioral susceptibility to stress.
Discussion
In the current study, we used engineered transcription factors to site-selectively remodel chromatin in a discrete brain region to control both the molecular and behavioral effects of drug- and stress-exposure. We targeted the FosB gene because of its known role in the pathophysiology of drug addiction and depression in both rodents and humans, which has been linked to epigenetic remodeling at this locus. Specifically, drug-induced activation of FosB is correlated with increased histone acetylation and decreased histone methylation at its promoter, yet it has not previously been possible to elucidate the functional relevance of such changes due to the genome-wide context within which they occur. To overcome this limitation, we designed a suite of engineered transcription factors that target the FosB locus and applied them to the expression of FosB/ΔFosB under both basal and cocaine-induced conditions. We found that FosB-ZFPs and -TALEs fused to the transcriptional activators, p65 or VP64, respectively, activate FosB/ΔFosB expression both in cultured cells and when virally expressed in NAc. NAc expression of FosB-ZFP35-p65 induces histone acetylation across the FosB promoter, a modification previously implicated in cocaine-induced activation of this locus1. We show here that acetylation of FosB by FosB-ZFP35-p65 is sufficient to enhance the sensitizing effects of cocaine, indicating that ZFP-induced acetylation may act synergistically with cocaine-activation of this gene to produce an enhanced behavioral response. In a complementary paradigm, we found that histone methylation by FosB-ZFP35-G9a is sufficient to repress basal expression of FosB/ΔFosB in vivo and block cocaine locomotor sensitization. These results represent the first demonstration of ZFP-induced chromatin modifications in vivo, and the first use of engineered transcription factors to elucidate transcriptional dysregulation underlying neuropsychiatric disease.
Importantly, we included numerous control constructs in our study to rule out the effects of stoichiometric interference by ZFPs in the transcription of FosB. We found that FosB-ZFP35 without a functional domain did not affect FosB/ΔFosB expression or chromatin modification, in either the basal or cocaine contexts. To further establish the specificity of our engineered transcription factors in targeting FosB selectively, we analyzed the regulation of 28 potential off-target binding motifs in vivo, each of which differs from the ZFP35 binding site by 1 to 3 nucleotides, and found no transcriptional regulation of their associated genes by FosB-ZFP35-p65 or -G9a. In addition, we found no enrichment of H3K9me2 at 6 of these sites examined after expression of FosB-ZFP35-G9a. Our results are in accordance with a still small number of in vitro studies that have examined ZFP or TALE specificity, and concluded that there is little to no genome-wide transcription regulation by gene-targeted engineered transcription factors17,37,38. Our data are the first to demonstrate the functional specificity of engineered transcription factors in neurons in vivo, thus greatly enhancing their application to the study of the epigenetic underpinnings of neuropsychiatric disease.
Previous studies have demonstrated the dynamic regulation of histone methylation by both chronic cocaine and stress11,12. In particular, repeated cocaine reduces G9a expression and the enrichment of H3K9me2 in the NAc, resulting in derepression at hundreds of genes, including FosB; viral overexpression of G9a in NAc increases H3K9me2 at FosB, represses ΔFosB expression, and inhibits cocaine reward behavior12. Our results improve upon this previous approach by targeting G9a specifically to the FosB gene, thereby arguing against effects of G9a across the genome, and directly implicating G9a-induced H3K9me2 at FosB in the specific mechanism of FosB gene repression.
Histone methylation is thought to repress gene expression via the recruitment of additional methyltransferases and HP1, which can lead to the formation of silent heterochromatin30,39. We found that FosB-ZFP35-G9a induces H3K9me2 enrichment across the FosB promoter, up to 1000 bp upstream from the ZFP recognition motif. This site, 1250 bp upstream from the FosB TSS, shows the greatest basal enrichment in H3K9me2 in both mouse and human NAc, and we found that following ZFP-induced H3K9me2, binding of HP1α is increased only at this location. In non-nervous tissues, repression by H3K9me2 often involves not only the recruitment of HP1 but also generation of H3K9me3 and can spread to adjacent genomic regions40,41. Given the widespread methylation induced by FosB-ZFP35-G9a, we investigated H3K9me3 binding across the FosB promoter and found, surprisingly, that this modification was depleted at the site of HP1α and H3K9me2 co-enrichment, with no regulation elsewhere in the FosB promoter. We also measured DNA methylation at three CpGs located adjacent to this region, as DNA methyltransferase is also often recruited by HP1 and H3K9me2/342, but did not find regulated DNA methylation regulation coincident with induced H3K9me2. Taken together, our results demonstrate a causal mechanism of gene repression at a euchromatic locus that is initiated by G9a recruitment, leading to enrichment of H3K9me2 and binding of HP1α, a mechanism of G9a/H3K9me2 action within fully differentiated adult neurons in vivo that differs from models based on other systems.
We next investigated a potential mechanism by which ZFP-induced H3K9me2 enrichment blocked cocaine-evoked FosB expression in NAc. We found that this manipulation was sufficient to prevent the increase in phospho-CREB at the FosB promoter, which is required for cocaine-evoked FosB/ΔFosB expression32 and known to be important for cellular and behavioral plasticity relevant to drug exposure8. Our results show no effect of H3K9me2 on the binding of total CREB or of phospho-SRF, indicating a specific action of H3K9me2 in preventing the phosphorylation of CREB already bound to FosB. These results thus provide the first direct evidence by which a drug-evoked histone modification regulates gene expression by controlling transcription factor activity. Further studies will be needed to shed light on the mechanism by which H3K9me2 enrichment prevents CREB phosphorylation, especially given the ~1000 bp distance between their binding sites.
FosB has also been implicated in the cellular and behavioral correlates of stress and depression. In particular, previous studies have demonstrated that FosB/ΔFosB expression in the NAc is deficient in the social isolation model of depression in mice and in this region of postmortem human depressed patients, and that increased FosB/ΔFosB expression in NAc protects animals from the deleterious effects of chronic stress8. To complement these data, we found that H3K9me2 is enriched at the FosB promoter in NAc of human depressed patients relative to controls, implicating this repressive epigenetic modification in the repression of FosB in this population. Indeed, we found that ZFP-induced enrichment of H3K9me2 at FosB in mouse NAc was not only sufficient to reduce FosB/ΔFosB expression, it also induced a prodepressive state, as manifested by depression- and anxiety-like behaviors after social stress. These data thus suggest a causal epigenetic mechanism for FosB repression in the pathophysiology of depression.
In summary, we have demonstrated that histone methylation or acetylation per se at the FosB locus in the NAc is sufficient to dynamically regulate gene expression at this locus and thereby control the behavioral effects of cocaine or stress exposure. The ability to site-specifically modify chromatin enabled us to model a pathological epigenetic marker of human addiction and depression and observe the effects on drug and stress susceptibility in mice. Our findings also suggest that a primary change in H3K9me2 is sufficient to drive downstream changes in transcription factor binding and gene expression, which is in contrast to current models of HPTMs occurring mostly secondarily to alterations in transcription factors. By expanding this novel approach to target additional genes and HPTMs regulated by emotional stimuli, it will be possible to gradually build a detailed understanding of the epigenetic basis of addiction and depression.
Methods
Animals and treatments
Male 7-8 week old C57BL/6J mice and 6-month old CD1 retired breeders (CD1 aggressor) were housed at 22-25°C in a 12-hr light/dark cycle and provided food and water ad libitum. Animals were housed five per cage and habituated in our facility for at least 1 week before experimentation and all tests were conducted during the light cycle. Members of the same cage were randomly assigned to different experimental groups for behavioral studies and the order of testing was distributed across groups. Wherever possible, the experimenter conducting the data analysis was blind to treatment conditions of the animals. All experiments were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee at Mount Sinai. Group size determination was based on published protocols for the behavioral paradigm in use; no statistical test was used to predetermine sample size. For repeated cocaine experiments (immunohistochemistry, chromatin immunoprecipitation [ChIP]), animals received daily injections of either saline (5 treatments saline, i.p.), or saline followed by cocaine (1 treatment saline, 4 treatments cocaine, 20 mg/kg, i.p.) and sacrificed 24 hours after the last injection. Sample size was determined based on empirical data in preliminary experiments and published protocols.
Transcription factor engineering
All zinc finger proteins (ZFPs) were manufactured by the CompoZr ZFN Operations Group at Sigma-Aldrich Biotechnology and cloned in frame N-terminal to the activation or repression domain. The ZFPs recognize the following unique 18-20 bp motifs in the FosB promoter: ZFP1: TCCGTGACAAAGCTAGTGG; ZFP14: ACCTTCTCCAACCCGGGT; ZFP25: CAACAGCGGGCGCGCGCAG; ZFP29: CCAAACAAACACTGGGCCGC; ZFP31:GGCCGCGCTGGCCGAGCTCC; ZFP35: GATCCCCTCCCGCGAAGCC; ZFP54: TGCGGGTGACGCAAGCGCG; ZFP61: GCCGGGGACACGCGGAGCC and Klf4 promoter: GTGACCCGCGCCCATGGCC. All transactivator-like effector (TALE) constructs were generated as described previously15. Briefly, TALE binding sites, 18-nt targets with T as the first base, were selected from the promoter region of FosB. All TALE repeat arrays were constructed using modular assembly followed by insertion into cloning backbones containing the non-repetitive TALE capping regions and the VP64 transcription activator. The identities of repeat variable diresidues (RVDs) in each TALE repeat array are as follows (Name, Target Site (5’ to 3’), Repeat Array RVDs) or published33: FosB-TALE1, TCCCCTCCCGCGAAGCCC, HD-HD-HD-HD-NG-HD-HD-HD-NN-HD-NN-HI-HINN-HD-HD-HD; FosB-TALE2, TCACCCGCAACTCCATGC, HD-NI-HD-HD-HD-NN-HD-NI-NI-HD-NG-HD-HD-NI-NG-NN-HD; FosB-TALE3, TACGCCGGGGACACGCGG, NI-HD-NN-HD-HD-NN-NN-NN-NN-NI-HD-NI-HD-NN-HD-NN-NN.
Bioinformatics
Bowtie was used to align FosB-ZFP35 motif to mouse genome (mm9)44, and three mismatches at maximum was allowed (parameter: -v 3). Then region_analysis.pl, a genome
N2a transfection and RNA isolation
Neuro2a cells (ATCC CCL-131; manufacturer's authentication available online) were cultured and transfected with 400 ng plasmid DNA using Effectene reagent (Qiagen) and RNA was isolated using the RNeasy Mini Kit (Qiagen) according to the manufacturers instructions. qPCR and data analysis were performed as described11, using the primers listed in Table S1. Data were analyzed by comparing C(t) values of the experimental construct with mock-transfected cells using the ΔΔC(t) method.
Viral-mediated gene transfer
Mice were anesthetized with a mixture of ketamine (100 mg/kg) and xylazine (10 mg/kg) and prepared for stereotaxic surgery. Thirty-three gauge syringe needles (Hamilton) were used to bilaterally infuse 0.5-1.0 ml of virus into NAc at a rate of 0.1 ml/min at 1.6 mm A/P, +1.5 mm lateral, and 4.4 mm D/V from Bregma. We used bicistronic p1005 HSV vectors expressing GFP alone or GFP plus FosB-ZFP or FosB-TALE. In this system, GFP expression is driven by a cytomegalovirus (CMV) promoter, whereas the select gene of interest is driven by the IE4/5 promoter12. As with other HSV vectors, we detected no change in neuronal viability with any of the vectors utilized in this study.
NAc RNA isolation and qRT-PCR
72 hours after viral infection, when transgene expression is maximal, mice were rapidly decapitated, and brains were removed and placed on ice. A fluorescence microscope (Leica) was used to visualize accurate NAc targeting of GFP-expressing virus and to accurately dissect only infected tissue. Dissections of NAc were taken with a 1 mm aluminum micro-punch (Harris) and quickly frozen on dry ice until RNA was extracted. RNA isolation, qPCR, and data analysis were performed as described11, using the expressing virus and to accurately dissect only infected tissue. Dissections of NAc were taken with a 1 mm aluminum micro-punch (Harris) and quickly frozen on dry ice until RNA was extracted. RNA isolation, qPCR, and data analysis were performed as described11, using the primers listed in Table S2. Data were analyzed by comparing C(t) values of the experimental virus with control virus using the ΔΔC(t) method.
Western blotting
Frozen NAc tissue was homogenized in 30 μl of RIPA buffer containing 10 mM Tris, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100, 1% Sodium Deoxycholate, and protease inhibitors (Roche, Basel, Switzerland) using an ultrasonic processor (Cole Parmer, Vernon Hills, IL, USA). Protein concentrations were determined using a DC protein assay (Bio-Rad, Hercules, CA, USA), and 50 μg of protein were loaded onto 4-15% gradient Tris-HCl polyacrylamide gels for electrophoresis fractionation (Bio-Rad). Proteins were transferred to nitrocellulose membranes, blocked with Odyssey® blocking buffer (Li-Cor, NE, USA), and incubated overnight at 4°C with primary antibodies (FosB/ΔFosB: Cell signalling 255112 H3K9me2: Abcam ab122012) in Odyssey® blocking buffer. After thorough washing with 1x Tris-Buffered Saline plus 0.1% Tween-20, membranes were incubated with IRDye® secondary antibodies (1/5000 to 1/10000; Li-Cor, Lincoln, NE, USA) dissolved in Odyssey® blocking buffer for 1 hr at room temperature. For analysis, the blots were imaged with the Odyssey® Infrared Imaging system (Li-Cor, Lincoln, NE, USA) and quantified by densitometry using ImageJ (NIH, Bethesda, Maryland, USA). The amount of protein blotted onto each lane was normalized to levels of actin (MP BioMedicals Clone C4). Representative image and data shown are from one experiment of two biological replicate experiments using two animal cohorts.
Immunohistochemistry, imaging and cell counting
Adult males were terminally anesthetized (chloral hydrate) and transcardially perfused with PBS followed by 4% paraformaldehyde. Brains were then post-fixed overnight in 4% paraformaldehyde at 4°C, cryoprotected in 30% sucrose at 4°C until isotonic, and stored at -80°C. 35 μm coronal sections were sliced on a freezing microtome. Striatal sections were washed in PBS and blocked with 3% normal donkey serum (NDS) before incubating in primary antibody (rabbit-anti-FosB, 1:1000, Santa Cruz Biotechnology sc-4846; rabbit-anti-DARPP32, 1:1000, AbCam ab4080147; mouse-anti-NeuN, 1:10000, Millipore MAB37746,48; chicken-anti-GFP, 1/8000, Aves Labs GFP 10-1012) in 0.1% Triton-X100/3%NDS/PBS at 4°C overnight. Sections were then washed in PBS, incubated in secondary antibody (Cy3 conjugated donkey-anti-rabbit, or -mouse, Cy2 conjugated goat anti-chicken, Jackson Immuno) in 0.1% Tween-20/PBS, washed, and mounted on poly-L-lysine coated slides. Slides were washed briefly in 75% ethanol, dried, and coverslipped with ProLong Gold anti-fade with DAPI (Life Technologies). Slides were stored in a light-proof box at 4°C before and after imaging. Slides were imaged on an Axio Observer Inverted Microscope (Zeiss) fitted with fluorescent filters. For each animal, 4-8 sections along the A-P axis of the NAc were imaged at 20x magnification (AxioVision imaging software, Zeiss) by an observer blind to conditions. For anatomical consistency, images included a portion of the anterior commissure as a marker of the NAc. Images were taken simultaneously for DAPI, GFP, and FosB (Cy3). Adjustments were made as necessary to accurately capture fluorescent intensity. Analysis of FosB+ cells was performed using ImageJ. Within the red channel, threshold was adjusted to accurately count moderate- to highly-expressing FosB+ cells. An elliptical of a constant size selected a region of interest through the core and shell of the NAc, and particles were analyzed (size: 15-175; circularity: 0.01-1) for each image. Representative image and data shown are from one experiment of two technical replicate experiments, in which sections from the same cohort of animals were subject to immunohistochemical analysis.
Chromatin immunoprecipitation
ChIP was performed on bilateral NAc 15-gauge punches pooled from 4-5 mice 72 hours after viral infection or 24 hours after the last drug treatment, and dissected as described above, or (2) 5 × 106 N2a cells 48 hours after transfection. Chromatin was prepared as described previously1, and sheared using a diogenode bioruptor XL at high sonication intensity for 30 sec on/ 30 sec off for either 2x25 minutes. Fragment size was verified with an Agilent 2100 bioanalyzer at 150-300 bp or 1000-3000 bp respectively. Sheared chromatin was incubated overnight with the following antibodies previously bound to magnetic beads (Dynabeads M-280, Life Technologies): anti-H3K9me2 antibody (AbCam ab122012), anti-H3K27me3 (AbCam ab60025), anti-H3K4me3 (EMD Millipore 17-6145), H3K9/14ac (EMD Millipore 17-6151), anti-H3K9me3 (EMD Millipore 07-44249), anti-HP1α (EMD Millipore 05-68950), anti-CREB (EMD Millipore 17-60032), anti-phospho-CREB (EMD Millipore 17-1013132), anti-phospho-SRF (AbCam ab53130). After reverse cross-linking and DNA purification (Qiagen Spin Column), binding to the FosB promoter was determined by qRT-PCR using the primers listed in Table S2.
DNA methylation
NAc tissue from three each of HSV-FosB-ZFP35-NFD and HSV-FosB-ZFP35-G9a viral injected animals was collected at Day 4 after surgery. Genomic DNA was isolated following standard protocol and treated with sodium bisulfite using Zymo EZ DNA Methylation kit. PCR was run with a primer pair (See Supplementary Table 1) flanking a region (873-1192 bp) upstream from the FosB TSS. TOPO cloning was then performed following the manufacturer's instructions (Invitrogen). Minipreps were prepared and sequenced by Sanger sequencing (Macrogen).
Locomotor sensitization
Locomotor sensitization was measured per published protocols7 with minor modifications. Activity was assessed in the x and y planes for horizontal ambulations in a 75 cm2 chambers using Ethovision XT (Noldus). On each day of sensitization, mice were injected with saline and analyzed for 10-15 minutes, then immediately injected with cocaine (5 or 10 mg/kg) and analyzed for 45-60 min. In some cases, locomotor activity was measured seven days after the last cocaine injection after mice were given a challenge injection of cocaine (5 mg/kg). Heatmap color-scales were generated by pooling individual pixel values across animals and treatments. Each animal's pixel values were computed from raw x- and y-beam break counts normalized to that animals total count. The pairwise summation of 4 x and 8 y-beam break counts resulted in 32 pixel values for each animal; each value was divisively normalized by the total sum of the 12 beam break counts and rounded to the nearest integer. Pixel values from all animals in a given plot were then pooled, and the number of unique integers in the pool determined the number of colors comprising a scale starting with dark blue, ranging through blue, cyan, green, yellow, and red, and ending with dark red. Calculations and image generation were performed using MATLAB R2012b (The Mathworks, Natick, MA). Representative images show the locomotor activity of one animal in each group, selected as that which displayed total locomotor activity closest to the mean on day 1 of the experiment.
Subthreshold social defeat and social interaction test
This paradigm is used to detect the mechanisms that promote a depression-like phenotype36. Male C57BL/6J mice were exposed to a novel CD1 male aggressor for 5 min, 3 times with 15 min intervals between each exposure. A social interaction test was performed 24 hours later. For this test, the subject is placed in an open field arena divided into an interaction zone and two opposing corner zones. A social target (novel CD1 mouse) is placed in a metal mesh-plastic box in the interaction zone, which allows sensory but not physical interaction. Ethovision XT (Noldus) tracking software was used to measure the duration (s) the subject mouse spent in the interaction zone with and without the target CD1 present (2.5 mi). Heat maps and post-acquisition processing were conducted in MATLAB (Mathworks) according to published protocols51. Representative images show the locomotor activity of one animal in each group from a single experiment. This experiment was repeated twice with separate animal cohorts.
Human postmortem brain tissue
Human postmortem brain tissue was obtained from the Dallas Brain Collection (DBC), where tissue is collected from the Dallas Medical Examiner's Office and the UT Southwestern Tissue Transplant Program after consent from the next of kin. Tissue was analyzed from 9 depressed patients (5 male, 4 female; age 44±7, PMI 15±2) and 18 controls (15 male, 3 female; age 51±3; PMI 16±1). The UT Southwestern IRB reviewed and approved the collection of this tissue for research use. A direct informant interview was carried out for each depression sample at a later date, where information regarding the individual's illness was documented; a consensus diagnosis of major depressive disorder was made by psychiatrists using DSM-IV criteria. None of the samples included in the study had blood toxicology screens positive for drugs of abuse, alcohol, or prescription drugs, including antidepressants. NAc dissection, tissue processing, and chromatin preparation were performed as reported previously52. qChIP was performed using anti-H3K9me2 antibody (ab1220, AbCam) previously bound to magnetic beads (Dynabeads M-280, Life Technologies). Analysis of relative enrichment was performed using qRT-PCR as described.
Statistics
The appropriate statistical test was determined based on the number of comparisons being done. Student's t-tests were used for comparison of two groups, in the analysis of qRTPCR, qChIP, and social interaction data. One-way ANOVA was used for analysis of three or more experimental groups, with a Bonferroni correction test post-hoc, when appropriate, in the analysis of mouse locomotor data. Main and interaction effects were considered significant at p < 0.05. Data are expressed as mean ± SEM. The Grubbs test was used when appropriate to identify outliers. F tests of variance were conducted on all datasets to ensure that the data followed a normal distribution. All experiments were carried out one to three times, and data replication was observed in instances of repeated experiments. Experimental sample sizes as indicated in the figure legends were determined gave the reported s.e.m. values that were sufficiently low to allow meaningful interpretation of the data.
Supplementary Material
Acknowledgements
This work was supported by grants from NIDA (EAH, EJN) and NIMH (EJN). The authors wish to thank Drs. Garret Stuber and Randall Ung (UNC) for their help in generating social interaction heatmaps.
Footnotes
Contributions
E.A.H. designed and executed biochemical, molecular and behavioral experiments (viral packaging constructs, qRT-PCR expression analysis, chromatin immunoprecipitation (mouse and human), immunohistochemical preparation, behavioral assays). H.C. and H.S. generated viral packaging constructs, conducted expression analysis and prepared chromatin. C.J.P. and S.A.G. analyzed immunohistochemical data. N.S. and L.S. conducted genome-wide sequence analysis. J.F. and E.A.H. performed DNA methylation sequencing. S.A.G. and S.J.R. generated human chromatin. E.A.H., H.C., H.S., J.J.W., M.M-R., D.F. and M.H. performed stereotaxic mouse surgery. J.P.H. generated mouse locomotor heat maps. C.S.T. prepared human postmortem brain tissue. R.L.N. generated HSV viral vectors. H.S.Z. generated the G9a catalytic domain construct. F.Z. generated the TALE constructs. C.J.P., E.A.H. and H.M.C. analyzed the data conducted statistical analyses. E.A.H. and E.J.N. discussed the data and wrote the manuscript.
Competing financial interests
The authors wish to declare no competing financial interests.
Contributor Information
Elizabeth A. Heller, Fishberg Department of Neuroscience and Friedman Brain Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Hannah M. Cates, Fishberg Department of Neuroscience and Friedman Brain Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Catherine J. Peña, Fishberg Department of Neuroscience and Friedman Brain Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Haosheng Sun, Fishberg Department of Neuroscience and Friedman Brain Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA..
Ningyi Shao, Fishberg Department of Neuroscience and Friedman Brain Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA..
Jian Feng, Fishberg Department of Neuroscience and Friedman Brain Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA..
Sam A. Golden, Fishberg Department of Neuroscience and Friedman Brain Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
James P. Herman, National Eye Institute, National Institutes of Health, Bethesda, MD, USA.
Jessica J. Walsh, Department of Pharmacology and System Therapeutics, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Michelle Mazei-Robison, Fishberg Department of Neuroscience and Friedman Brain Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA..
Deveroux Ferguson, Fishberg Department of Neuroscience and Friedman Brain Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA..
Scott Knight, Sigma Aldrich, Saint Louis, MO, USA..
Mark A. Gerber, Sigma Aldrich, Saint Louis, MO, USA.
Christian Nievera, Sigma Aldrich, Saint Louis, MO, USA..
Ming-Hu Han, Department of Pharmacology and System Therapeutics, Icahn School of Medicine at Mount Sinai, New York, NY, USA..
Scott J. Russo, Fishberg Department of Neuroscience and Friedman Brain Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Carol S. Tamminga, Department of Psychiatry, University of Texas Southwestern Medical Center, Dallas, TX 75390, USA.
Rachael L. Neve, Department of Brain and Cognitive Sciences, Massachusetts Institute of Technology, Cambridge, MA, USA.
Li Shen, Fishberg Department of Neuroscience and Friedman Brain Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA..
H. Steve Zhang, Sangamo Biosciences Inc., Richmond, CA..
Feng Zhang, McGovern Institute for Brain Research, Massachusetts Institute of Technology, Cambridge, USA..
Eric J. Nestler, Fishberg Department of Neuroscience and Friedman Brain Institute, Icahn School of Medicine at Mount Sinai, New York, NY, USA..
References
- 1.Renthal W, et al. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron. 2009;62:335–348. doi: 10.1016/j.neuron.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kennedy PJ, et al. Class I HDAC inhibition blocks cocaine-induced plasticity by targeted changes in histone methylation. Nature Neuroscience. 2013;16:434–440. doi: 10.1038/nn.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malvaez M, Mhillaj E, Matheos DP, Palmery M, Wood MA. CBP in the nucleus accumbens regulates cocaine-induced histone acetylation and is critical for cocaine-associated behaviors. J Neurosci. 2011;31:16941–16948. doi: 10.1523/JNEUROSCI.2747-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nature Publishing Group. 2011;12:623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng J, et al. Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome Biol. 2014;15:R65. doi: 10.1186/gb-2014-15-4-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar A, et al. SOM Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 7.Kelz MB, et al. Expression of the transcription factor deltaFosB in the brain controls sensitivity to cocaine. Nature. 1999;401:272–276. doi: 10.1038/45790. [DOI] [PubMed] [Google Scholar]
- 8.Vialou V, et al. DeltaFosB in brain reward circuits mediates resilience to stress and antidepressant responses. Nature Neuroscience. 2010;13:745–752. doi: 10.1038/nn.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robison AJ, et al. Behavioral and structural responses to chronic cocaine require a feedforward loop involving ΔFosB and calcium/calmodulin-dependent protein kinase II in the nucleus accumbens shell. J Neurosci. 2013;33:4295–4307. doi: 10.1523/JNEUROSCI.5192-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maze I, et al. Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. Proceedings of the National Academy of Sciences. 2011;108:3035–3040. doi: 10.1073/pnas.1015483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Covington HE, et al. A role for repressive histone methylation in cocaine-induced vulnerability to stress. Neuron. 2011;71:656–670. doi: 10.1016/j.neuron.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maze I, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun H, et al. Morphine epigenomically regulates behavior through alterations in histone H3 lysine 9 dimethylation in the nucleus accumbens. J Neurosci. 2012;32:17454–17464. doi: 10.1523/JNEUROSCI.1357-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snowden AW, Gregory PD, Case CC, Pabo CO. Gene-specific targeting of H3K9 methylation is sufficient for initiating repression in vivo. Current biology : CB. 2002;12:2159–2166. doi: 10.1016/s0960-9822(02)01391-x. [DOI] [PubMed] [Google Scholar]
- 15.Sanjana NE, et al. A transcription activator-like effector toolbox for genome engineering. Nat Protoc. 2012;7:171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konermann S, et al. NOT FINAL PROOF. Nature. 2013:1–17. doi:10.1038/nature 12466. [Google Scholar]
- 17.Mendenhall EM, et al. Locus-specific editing of histone modifications at endogenous enhancers. Nat Biotechnol. 2013 doi: 10.1038/nbt.2701. doi:10.1038/nbt.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laganiere J, et al. An engineered zinc finger protein activator of the endogenous glial cell line-derived neurotrophic factor gene provides functional neuroprotection in a rat model of Parkinson's disease. J Neurosci. 2010;30:16469–16474. doi: 10.1523/JNEUROSCI.2440-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beerli RR, Barbas CF. Engineering polydactyl zinc-finger transcription factors. Nat Biotechnol. 2002;20:135–141. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- 20.Gerritsen ME, et al. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc Natl Acad Sci USA. 1997;94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Covington HE, et al. Antidepressant actions of histone deacetylase inhibitors. J Neurosci. 2009;29:11451–11460. doi: 10.1523/JNEUROSCI.1758-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malvaez M, Mhillaj E, Matheos DP, Palmery M, Wood MA. CBP in the nucleus accumbens regulates cocaine-induced histone acetylation and is critical for cocaine-associated behaviors. J Neurosci. 2011;31:16941–16948. doi: 10.1523/JNEUROSCI.2747-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kennedy PJ, et al. Class I HDAC inhibition blocks cocaine-induced plasticity by targeted changes in histone methylation. Nature Neuroscience. 2013;16:434–440. doi: 10.1038/nn.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar A, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 25.Peixoto L, Abel T. The role of histone acetylation in memory formation and cognitive impairments. Neuropsychopharmacology. 2013;38:62–76. doi: 10.1038/npp.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogge GA, Singh H, Dang R, Wood MA. HDAC3 is a negative regulator of cocaine-context-associated memory formation. J Neurosci. 2013;33:6623–6632. doi: 10.1523/JNEUROSCI.4472-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rea S, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 28.Garriga-Canut M, et al. Synthetic zinc finger repressors reduce mutant huntingtin expression in the brain of R6/2 mice. Proceedings of the National Academy of Sciences. 2012;109:E3136–45. doi: 10.1073/pnas.1206506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith AE, Hurd PJ, Bannister AJ, Kouzarides T, Ford KG. Heritable gene repression through the action of a directed DNA methyltransferase at a chromosomal locus. J Biol Chem. 2008;283:9878–9885. doi: 10.1074/jbc.M710393200. [DOI] [PubMed] [Google Scholar]
- 30.Fritsch L, et al. A Subset of the Histone H3 Lysine 9 Methyltransferases Suv39h1, G9a, GLP, and SETDB1 Participate in a Multimeric Complex. Molecular Cell. 2010;37:46–56. doi: 10.1016/j.molcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 31.Filion GJ, van Steensel B. Reassessing the abundance of H3K9me2 chromatin domains in embryonic stem cells. Nature Publishing Group. 2010;42:5–6. doi: 10.1038/ng0110-4. 4–author reply. [DOI] [PubMed] [Google Scholar]
- 32.Vialou V, et al. Serum Response Factor and cAMP Response Element Binding Protein Are Both Required for Cocaine Induction of FosB. J Neurosci. 2012;32:7577–7584. doi: 10.1523/JNEUROSCI.1381-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang F, et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maniatis T, Goodbourn S, Fischer JA. Regulation of inducible and tissue-specific gene expression. Science. 1987;236:1237–1245. doi: 10.1126/science.3296191. [DOI] [PubMed] [Google Scholar]
- 35.Ernst J, Kellis M. Interplay between chromatin state, regulator binding, and regulatory motifs in six human cell types. Genome Res. 2013;23:1142–1154. doi: 10.1101/gr.144840.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaudhury D, et al. Rapid regulation of depression-related behaviours by control of midbrain dopamine neurons. Nature. 2013;493:532–536. doi: 10.1038/nature11713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith DJ, Konarska MM. A critical assessment of the utility of protein-free splicing systems. RNA. 2008;15:1–3. doi: 10.1261/rna.1322709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grimmer MR, et al. Analysis of an artificial zinc finger epigenetic modulator: widespread binding but limited regulation. Nucleic Acids Res. 2014;42:10856–10868. doi: 10.1093/nar/gku708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 40.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 41.Hathaway NA, et al. Dynamics and Memory of Heterochromatin in Living Cells. Cell. 2012:1–14. doi: 10.1016/j.cell.2012.03.052. doi:10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 43.Paxinos G, Franklin KB. J. Paxinos and Franklin's the Mouse Brain in Stereotaxic Coordinates. 2012 [Google Scholar]
- 44.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen L, et al. diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS ONE. 2013;8:e65598. doi: 10.1371/journal.pone.0065598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lobo MK, et al. ΔFosB induction in striatal medium spiny neuron subtypes in response to chronic pharmacological, emotional, and optogenetic stimuli. J Neurosci. 2013;33:18381–18395. doi: 10.1523/JNEUROSCI.1875-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Becker A, et al. Glutaminyl cyclase-mediated toxicity of pyroglutamate-beta amyloid induces striatal neurodegeneration. BMC Neurosci. 2013;14:108. doi: 10.1186/1471-2202-14-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guidelines for the Care and Use of Mammals in Neuroscience and Behavioral Research. National Academies Press (US) 2003. National Research Council (US) Committee on Guidelines for the Use of Animals in Neuroscience and Behavioral Research. [PubMed] [Google Scholar]
- 49.Maze I, et al. Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. Proceedings of the National Academy of Sciences. 2011;108:3035–3040. doi: 10.1073/pnas.1015483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allan RS, et al. An epigenetic silencing pathway controlling T helper 2 cell lineage commitment. Nature. 2012;487:249–253. doi: 10.1038/nature11173. [DOI] [PubMed] [Google Scholar]
- 51.Jennings JH, et al. Distinct extended amygdala circuits for divergent motivational states. Nature. 2013;496:224–228. doi: 10.1038/nature12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Golden SA, et al. Epigenetic regulation of RAC1 induces synaptic remodeling in stress disorders and depression. Nat Med. 2013;19:337–344. doi: 10.1038/nm.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.