Abstract
ADP-ribosylation is a post-translational protein modification, in which ADP-ribose is transferred from nicotinamide adenine dinucleotide (NAD+) to specific acceptors, thereby altering their activities. The ADP-ribose transfer reactions are divided into mono- and poly-(ADP-ribosyl)ation. Cellular ADP-ribosylation levels are tightly regulated by enzymes that transfer ADP-ribose to acceptor proteins (e.g. ADP-ribosyltransferases, poly-(ADP-ribose) polymerases (PARP)) and those that cleave the linkage between ADP-ribose and acceptor (e.g. ADP-ribosyl-acceptor hydrolases (ARH), poly-(ADP-ribose) glycohydrolases (PARG)), thereby constituting an ADP-ribosylation cycle. This review summarizes current findings related to the ARH family of proteins. This family comprises three members (ARH1-3) with similar size (39 kDa) and amino acid sequence. ARH1 catalyzes the hydrolysis of the N-glycosidic bond of mono-(ADP-ribosyl)ated arginine. ARH3 hydrolyzes poly-(ADP-ribose) (PAR) and O-acetyl-ADP-ribose. The different substrate specificities of ARH1 and ARH3 contribute to their unique roles in the cell. Based on a phenotype analysis of ARH1−/− and ARH3−/− mice, ARH1 is involved in the action by bacterial toxins as well as in tumorigenesis. ARH3 participates in the degradation of PAR that is synthesized by PARP1 in response to oxidative stress-induced DNA damage; this hydrolytic reaction suppresses PAR-mediated cell death, a pathway termed parthanatos.
Keywords: ADP-ribosylation, ADP-ribose-acceptor hydrolase, tumorigenesis, cholera toxin, Parthanatos, ARH
1. ADP-ribosylation
ADP-ribosylation is a reversible post-translational modification of proteins in which the ADP-ribose moiety of nicotinamide adenine dinucleotide (NAD+) is transferred to acceptors such as the amino acid residues of proteins, altering their activity and thus critical cellular functions [1–3]. These modifications are broadly grouped into two categories, mono- and poly-(ADP-ribosyl)ation.
1.1. Mono-(ADP-ribosyl)ation
ADP-ribosylation was initially discovered as the mechanism by which some bacterial toxins and cytotoxins e.g., Pseudomonas exoenzyme S, pertussis toxin, cholera toxin, diphtheria toxin, exert their effects [4–7]. This family of bacterial toxins comprises mono-ADP-ribosyltransferases, which transfer a single ADP-ribose moiety to specific amino acids, e.g., arginine, cysteine, diphthamide, asparagine, of acceptor proteins in host cells, thereby disrupting host cell biosynthetic, regulatory and metabolic pathways.
Mammalian cells also contain ADP-ribosyltransferases, which catalyze reactions similar to those of the bacterial toxins [8–10]. A family of ecto-ADP-ribosyltransferases (ART1-5) from avian and mammalian tissues has been cloned and characterized [11–17]. ART1-4 are anchored to the plasma membrane through a glycosylphosphatidylinositol moiety, whereas ART5 is a secreted protein. ART1, ART2, and ART5 catalyze the stereospecific transfer of an ADP-ribose from NAD+ to the guanidino moiety of arginine (protein), forming an α-anomeric N-glycosidic linkage of ADP-ribose to arginine, whereas amino acid substrates of ART3 and ART4 have not been identified. These extracellular enzymes are involved in the modification of secreted and membrane proteins, as well as cell surface receptors, including the P2X7 purinergic receptor, human neutrophil peptide 1 (HNP-1), integrin α7, platelet-derived growth factor-BB (PDGF-BB), and fibroblast growth factor-2 (FGF-2) [18–22]. By mono-(ADP-ribosyl)ation of target proteins, ARTs appear to regulate innate immunity and cell-cell and cell-matrix interactions. However, mono-(ADP-ribosyl)ation also occurs intracellularly. This reaction is catalyzed, in part, by members of the sirtuin family of NAD+-dependent deacetylases (SIRT) [23–27]. The SIRT family comprises seven members (SIRT1-7), which are widely distributed in intracellular organelles. SIRT1, 2, 4, and 6 possess intrinsic mono-(ADP-ribosyl)ation activity, transferring a single ADP-ribose to an arginine residue of specific target proteins [23–26]. Glutamate dehydrogenase (GDH) is a specific target for mono-(ADP-ribosyl)ation catalyzed by SIRT4; cysteine residue at position 119 of human GDH is ADP-ribosylated [26, 28]. The mono-(ADP-ribosyl)ation of GDH negatively regulates its activity, resulting in inhibition of insulin secretion from the pancreas. In addition, some members of poly-(ADP-ribose) polymerase (PARP) family have also been reported to possess mono-ADP-ribosyltransferase activity [29–31].
1.2. Poly-(ADP-ribosyl)ation
Poly-(ADP-ribosyl)ation in mammal cells has a crucial role in cellular functions including mitosis, DNA repair, and cell death [31–33]. It is initiated by transferring ADP-ribose primarily to carboxyl groups of glutamate and aspartate residues and ε–amino group of lysine residue(s) of target proteins to create O- and N-glycosidic bonds, respectively, of ADP-ribose to proteins; this reaction is followed by chain elongation and branching, resulting in the formation of a long, branched chain of poly-(ADP-ribose) (PAR). PAR is a polymer composed of several hundred ADP-ribose units, and thus is negatively charged; its formation is catalyzed by PARP. PARP1, the best studied protein in the PARP family, is a nuclear, chromatin-associated protein that is found in most eukaryotes except for yeast [34–36]. PARP1 is the most abundant and most active PARP and constitutes the founding member of the PARP superfamily. On the basis of sequence similarities to the catalytic domain of PARP1, seventeen PARP enzymes have been identified in the human genome [31–33]. Within this family of seventeen proteins, the enzymes capable of catalyzing poly-(ADP-ribosyl)ation are PARP1, PARP2, PARP3, PARP4, Tankyrase1 (PARP5A), and Tankyrase2 (PARP5B), whereas PARP10, PARP12, PARP14, PARP15, and PARP16 are mono-ADP-ribosyltransferases [29–31, 37–39]. PARP9 and PARP13 appear to be enzymatically inactive, because of the lack of NAD+-binding residues. Other PARP isoforms are predicted to be mono-ADP-ribosyltransferases [31].
PARP1 is critical for cell survival under conditions in which DNA damage is induced by oxidation, alkylating agents, and ionizing radiation. Basal activity of PARP1 is increased by 500-fold in response to DNA single- and double-strand breaks by binding to DNA breaks through its zinc finger, DNA-binding domains, which initiate poly-(ADP-ribosyl)ation of glutamate, aspartate, and lysine residues of acceptor proteins [40–44]. Based on results obtained with PARP1 and PARP2-deficient cells, more than 90% of PAR production results from PARP1 activity [39]. PAR with its negative charge alters the physical and biological properties of target proteins such as histones, topoisomerase I, and DNA protein kinases, resulting in DNA remodeling and repair [44–48]. PARP1 itself is also auto-modified by PAR via its auto-modification and DNA-binding domains [41, 43, 49, 50]. Its modification promotes interaction with several proteins such as XRCC1, DNA ligase III, and the Ku70 subunit of the DNA-dependent protein kinase, recruiting them to DNA-damage sites for DNA repair [34, 45, 51]. Alternatively, PAR(P) has a role in cell injury and death. A PAR-dependent death pathway has been demonstrated in several disease models including brain and myocardial ischemia–reperfusion injury, glutamate excitotoxicity, streptozotocin-induced diabetes, and 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced Parkinsonism [52–58]. PAR production by excessive activation of PARP1 causes significant consumption of cellular NAD+, followed by depletion of ATP, which results in necrotic cell death [59, 60]. In addition, non-covalent PAR itself has been reported to have a role as a death signal [54, 58, 61, 62]. PAR may induce release of apoptosis-inducing factor (AIF) from mitochondria, which is involved in caspase-independent cell death, a process termed parthanatos, which was named after Thanatos, the personification of death in Greek mythology [62]. PAR generated by PARP1 in the nucleus translocates to the cytoplasm [61]. Cytoplasmic PAR associates with apoptosis-inducing factor (AIF) anchored on mitochondrial membranes, resulting in release of AIF into the cytoplasm [58]. AIF translocates to the nucleus via its nuclear localization sequence and induces large-scale DNA fragmentation and chromatin condensation [62].
1.3. Enzymes involved in termination of ADP-ribosylation
Thus, as ADP-ribosylation participates in several important biological processes, it must be controlled both spatially and temporally. In fact, ADP-ribosylation is reversibly regulated by several enzymes including poly-(ADP-ribose) glycohydrolase (PARG), macrodomain proteins, and ADP-ribosyl-acceptor hydrolases (ARH). PARG has been thought to be the primary enzyme responsible for termination of poly-(ADP-ribosyl)ation by catalyzing the hydrolysis of the O-glycosidic bond of PAR chains [63, 64]. Alternative splicing of a single PARG gene gives rise to several PARG isoforms with different sizes, activities, and localizations; a nuclear 110-kDa protein, cytoplasmic 103-kDa, 99-kDa, and 60-kDa proteins, and a mitochondrial 55-kDa protein [65–68]. Absence of the PARG gene results in embryonic lethality, because of excessive accumulation of PAR in nuclei and subsequent induction of cell death [69].
However, as PARG is unable to hydrolyze the O-glycosidic bond of the first ADP-ribose directly attached to glutamate residues of target proteins, its activity generates mono-(ADP-ribosyl)ated proteins. Macrodomains, evolutionally conserved modules of 130–190 amino acids discovered as a domain of a core histone variant macroH2A, bind ADP-ribose monomers and polymers as well as the sirtuin product O-acetyl-ADP-ribose (OAADPr) [70, 71]. In mammalian cells, MacroD1, MacroD2, and C6orf130 which is also known as Terminal ADP-Ribose protein Glycohydrolase (TARG1), possess mono-ADP-ribosyl-acceptor hydrolase activity that cleaves the O-glycosidic linkage of ADP-ribose to glutamate, leading to regeneration of unmodified protein [72–75]. Homozygous mutation of the TARG1 gene, which generates a non-functional truncated variant, results in severe neurodegeneration in humans, indicating the importance of TARG1 function on termination of poly-(ADP-ribosyl)ation [72].
The ARH family consists of three members (ARH1-3) with substantial amino acid sequence similarity (Table 1) [16, 76–80]. ARH1 and ARH3 have different substrate specificities. ARH1 cleaves mono-ADP-ribosylated substrate with the modification on an arginine, while ARH3 hydrolyzes PAR and OAADPr [76, 78, 81, 82]. ARH2 has not been shown to have enzymatic activities related to mono- and poly-ADP-ribose [81, 82]. The ARH family is described in more detail below.
Table 1. Similarity of amino acid sequences and relative enzymatic activities of the ARH family.
Amino acid sequences of ARH2 and ARH3 were compared to ARH1 sequence [76]. Their relative enzymatic activities were quantified using a five-grade scale. “-” indicates that no activity was detected. ARH1 is the only enzyme responsible for the hydrolysis of ADP-ribosylated arginine [89]. ARH3 hydrolyzes the O-glycosidic bond of PAR chains and OAADPr as does ARH1. ARH1, however, has < 1% of the specific activity of ARH3 [76, 81, 82]. Enzymatic substrates of ARH2 have not been identified.
| aa | Similarity to ARH1 (%) | ADP-ribosyl-arginine | PAR | OAADPr | |
|---|---|---|---|---|---|
|
| |||||
| ARH1 | 357 | +++++ | + | + | |
| ARH2 | 354 | 68 | − | − | − |
| ARH3 | 363 | 41 | − | +++ | ++++ |
2. ARH family
The ARH family of proteins (ARH1-3) exhibit similar size (39 kDa) and amino acid sequence [16, 76–80]. ARH1 is a mono-ADP-ribosyl-arginine hydrolase, which catalyzes the hydrolysis of the N-glycosidic bond linking ADP-ribose to the guanidino group of arginine, leading to release of ADP-ribose, with formation of arginine [78]. ARH3 catalyzes the hydrolysis of the O-glycosidic bond in PAR, generating ADP-ribose [76, 77]. These reactions are stereospecific. ARH1 and ARH3 preferentially cleave the α-anomer at the C-1″ position of ADP-ribose attached to arginine and ADP-ribose in PAR, respectively. ARH3 hydrolyzes OAADPr in the same manner [81, 82]. Similar to ARH3, ARH1 cleaves PAR and OAADPr, but ARH1 activity is less than 1% that of ARH3, indicating that ARH1 might possess weak hydrolytic activity toward the O-glycosidic bond [81]. OAADPr itself, independent of protein deacetylation, regulates SIRT activity, TRPM2 ion-channel gating, embryo development, and cellular redox [83–86]. Through OAADPr metabolism, ARH1 and ARH3 may participate in these signal transduction pathways. On the other hand, ADP-ribosylated cysteine, asparagine, and diphthamide, synthesized by bacterial toxins, are not substrates hydrolyzed by ARH1 or ARH3 [76]. ARH1 and ARH3 activities are competitively inhibited by ADP-ribose, but not ribose 5-phosphate, AMP, ADP or NAD+ [80–82]. Thus, ARH1 and ARH3 recognize the ADP-ribose moiety of the substrate. Although ARH2 binds ADP-ribose, it has not been shown to be enzymatically active [81, 82].
Enzymatic activities of both ARH1 and ARH3 require Mg2+ [76–78]. According to the crystal structure of ARH3, the active site is defined by the position of two Mg2+ ions [77]. ARH3 contains a pair of vicinal acidic amino acids, aspartates, at positions 77 and 78, that are essential to coordinate two Mg2+ ions and required for its enzymatic activity [76, 77, 81]. ARH1 also contains two aspartates at positions 60 and 61 [87]. Replacement of the pair with asparagine or alanine in ARH1 or ARH3 results in a dramatic reduction in activity, although the mutant proteins are structurally intact and retain their ability to bind ADP-ribose. In ARH2, the vicinal amino acid residues at positions 60 and 61 are aspartate-asparagine, which may explain why ARH2 lacks enzymatic activity [76].
In some species, ARH1 enzymatic activity also requires thiol; rat and mouse ARH1 requires the presence of thiol, whereas human, calf and guinea pig ARH1 do not [78]. In human ARH1, a critical cysteine residue is responsible for thiol-dependence; in mouse, it is replaced by serine [88]. Likewise, human and mouse ARH3 do not require thiol, possibly because ARH3 contains valine at the position that determines thiol-sensitivity of human ARH1 [76].
Based on phenotype analysis of ARH1−/− and ARH3−/− mice and cells, both enzymes regulate several cellular responses through their activities. In this review, we highlight current knowledge of ARH1 and ARH3 related to cellular distributions and physiological and pathological functions.
2.1. ARH1 distribution and functions
ARH1 is a cytoplasmic protein ubiquitously expressed in mammalian tissues and cells [78]. In ARH1−/− mouse embryonic fibroblasts (MEFs) and tissues, the ability to hydrolyze mono-(ADP-ribosyl)ated arginine is lost [89], suggesting that ARH1 is the only cytoplasmic enzyme that hydrolyzes the ADP-ribose-arginine bond. It appears to be a component of a mono-(ADP-ribosyl)ation cycle (Fig. 1). A role for ARH1 in disease was first found in a mouse model of cholera toxin-induced disease [89]. Cholera toxin, secreted by Vibrio cholerae, is a multimeric protein, which transfers an ADP-ribose to arginine residues of the α-subunit of the stimulatory guanine nucleotide-binding (Gαs) protein of the adenylyl cyclase system [6]. Its ADP-ribosylation stabilizes Gs-GTP by inhibiting the GTPase activity of Gαs, resulting in prolonged activation of adenylyl cyclase. Activation of adenylyl cyclase in intestine increases intracellular cAMP, leading to abnormalities of fluid and electrolyte transport characteristic of cholera. ARH1−/− mice and MEFs exhibit enhanced sensitivity to cholera toxin compared to their wild-type (WT) counterparts [89]. Cholera toxin increased the mono-(ADP-ribosyl)ation levels of Gαs; mono-(ADP-ribosyl)ation of Gαs was further increased and prolonged in ARH1−/− MEFs and mice, compared to their WT counterparts. Moreover, intestinal loops of ARH1−/− mice showed significantly enhanced fluid accumulation following exposure to cholera toxin than did those of WT mice. Thus, these data support a role for ARH1 in the intoxication process seen in cholera.
Fig. 1. Arginine-specific mono-(ADP-ribosyl)ation.
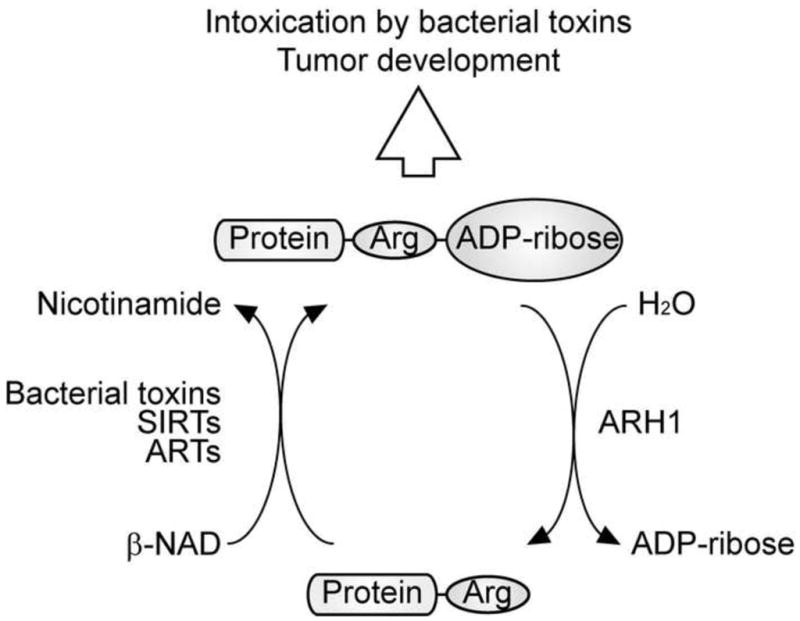
Bacterial toxins, ARTs, and some members of the SIRT family transfer the ADP-ribose moiety of NAD+ to arginine residues of acceptor proteins with release of nicotinamide [6, 7, 9, 24]. Mono-(ADP-ribosyl)ation may be involved in tumorigenesis as well as in disruption of host regulatory biosynthetic and metabolic pathways by bacterial toxins [89, 90]. ARH1 is responsible for catalyzing the hydrolysis of the N-glycosidic bond linking ADP-ribose to the guanidino group of arginine, which occurs at the C-1″ position of ADP-ribose. Hydrolysis leads to release ADP-ribose with regeneration of arginine. ARH1 deficiency results in enhanced sensitivity to cholera toxin as well as other toxins and tumor development.
ARH1-mediated mono-ADP-ribosylarginine hydrolase activity is also involved in intracellular signal transduction. ARH1−/− and ARH1+/− mice are prone to tumor development (Table 2) [90]. Further, metastasis and multi-tumor occurrences are seen more frequently in ARH1−/− and ARH1+/− mice than in WT mice. Approximately 25% of ARH1−/− mice and 13% of ARH1+/− mice developed tumors in the age range of 3–12 months when tumors were infrequent in WT mice. In ARH1−/− mice, several types of tumors (e.g., carcinoma, sarcoma, lymphoma) developed in a variety of tissues/organs (e.g., lung, liver, spleen, lymph nodes, mammary gland, uterus, skeletal muscle) in an age-dependent manner. Thus, loss of function of ARH1 was strongly correlated with tumorigenesis. Consistent with a high frequency of tumorigenesis in ARH1−/− mice, ARH1−/− MEFs have characteristics of tumor cells; ARH1−/− MEFs proliferated faster and formed more and larger colonies in soft agar than did WT MEFs, and subcutaneous injection of ARH1−/− MEFs in athymic nude mice resulted in the formation of tumor-like masses. By stable expression of ARH1, but not an inactive ARH1 mutant, all the phenotypes seen in ARH1−/− MEFs were reversed. ARH1−/− MEFs exhibited a shorter G1 phase of the cell cycle. Abnormal cell cycle progression is possibly linked to genomic instability and uncontrolled cell growth, leading to tumorigenesis. Thus, the proper control of mono-(ADP-ribosyl)ation by ARH1 has a crucial role in cell proliferation and cancer suppression.
Table 2. Effect of ARH1 genotype on incidence of tumors.
Incidence of tumors (%) in ARH1 genotype was evaluated. Metastasis and multi-tumor occurrences were seen more frequently in ARH1−/− and ARH1+/− mice than in WT mice. In ARH1−/− and ARH+/− mice, several types of tumors (e.g., carcinoma, sarcoma, lymphoma) developed at a younger age than was seen with WT mice. Lymphoma was seen most frequently in ARH1−/− and ARH+/− mice.
| Carcinoma | Lymphoma | Sarcoma | Total | ||||
|---|---|---|---|---|---|---|---|
| Age | Genotype | Adeno- | * HC- | Hemangio- | Rhabdomyo- | ||
| 3–12 months | +/+ | 0 | 0 | 0 | 0 | 0 | 0.0 |
| +/− | 4.1 | 1.1 | 7.9 | 0.2 | 0.1 | 13.4 | |
| −/− | 6.2 | 2.1 | 12.9 | 4.0 | 0 | 25.2 | |
| 13–20 months | +/+ | 0 | 0 | 2.9 | 0.6 | 0 | 3.5 |
| +/− | 3.0 | 0.6 | 7.1 | 0.6 | 0 | 11.2 | |
| −/− | 5.8 | 0.6 | 8.9 | 5.0 | 0 | 20.3 | |
Hepatocellular (HC). This table was modified from Kato et al. [90].
Interestingly, tumorigenesis seen in ARH1−/− mice and MEFs is gender-specific [91]. ARH1−/− and ARH1+/− female mice developed tumors and metastasis more frequently and at a younger age than did male mice. ARH1−/− MEFs subcutaneously injected into female nude mice formed tumor-like masses more rapidly than they did in male mice. Gender effect of ARH1 deficiency on incidence of tumor growth may result from effects of estrogen. Ovariectomized female nude mice exhibited reduced growth rate of tumors from following injection of ARH1−/− MEFs, whereas estrogen treatment of male and ovarictomized female nude mice increased the frequency and rate of tumor development. Further, estrogen promoted the survival of ARH1−/− MEFs in the circulation and thus enhanced their metastatic potential. Based on these and other data, it appeared that ARH1 is involved in estrogen-stimulated tumorigenesis.
2.2. ARH3 distribution and functions
ARH3 is ubiquitously expressed in mouse and human tissues [76]. ARH3 is mainly located in the cytoplasm (65%), followed by mitochondria (25%), and nucleus (10%) [92, 93]. ARH3 contains a mitochondrial-targeting sequence near its N-terminus, which allows ARH3 to be expressed in the mitochondrial matrix [77, 92]. Niere et al. provided evidence that ARH3 might be the primary enzyme responsible for putative PAR degradation in mitochondria [65, 92]. Overexpression of ARH3 lowered mitochondrial PAR content artificially driven by expression of the catalytic domain of PARP1 with a mitochondrial targeting sequence (mito-PARP1cd) [92]. In ARH3−/− MEFs expressing mito-PARP1cd, mitochondrial PAR content was greater than in WT MEFs expressing mito-PARP1cd [65]. In the presence of a PARP inhibitor, turnover of mito-PARP1cd-induced PAR in mitochondria was significantly slower in ARH3−/− MEFs than in WT MEFs. In human cultured cells (e.g., HEK 293, HepG2, HeLaS3, SH-SY5Y cells), small PARG isoforms, PARG60 and PARG55, resulting from alternative splicing of PARG transcripts, were expressed in cytoplasm and mitochondria, but unable to degrade PAR, because they lack exon 5, which encodes a region critical for PARG activity [65, 68]. Although some reports indicated that PAR is present in mitochondria [94], a mitochondrial PARP has not been identified and ARH3 deficiency did not increase PAR content in mitochondria under resting conditions. Therefore, the function of ARH3 in mitochondria is unclear. However, SIRT3, SIRT4, and SIRT5 are located in the mitochondrial matrix [95]. SIRT3 and SIRT5 exhibit deacetylase activity, generating OAADPr. By degradation of OAADPr, ARH3 might participate in the regulation of functions involving mitochondrial SIRTs.
ARH3 also participates in nuclear and cytoplasmic PAR degradation under oxidative stress conditions induced by hydrogen peroxide (Fig. 2) [93]. ARH3−/− MEFs were more susceptible to hydrogen peroxide-induced cytotoxicity than WT MEFs. ARH3 expression in ARH3−/− MEFs reduced their sensitivity to hydrogen peroxide. Cell death in ARH3−/− MEFs occurred in a caspase-independent manner, accompanied by nuclear shrinkage, chromatin condensation, and exposure of phosphatidylserine on the cell surface. Genetic studies using PARP1 shRNA indicated that PARP1 activation has a role as an initiation step to induce the cell death signal following oxidative stress, by synthesizing PAR in the nucleus. In ARH3−/− MEFs, nuclear PAR content in response to hydrogen peroxide was significantly increased as early as 10 min, compared to WT MEFs. After 30 min, PAR gradually translocated from nucleus to the cytoplasm in ARH3−/− MEFs, whereas PAR accumulation was less in WT MEFs, suggesting that PAR hydrolytic activity of ARH3 lowers PAR content in the nucleus and cytoplasm, preventing PAR translocation and accumulation in the cytoplasm. As suggested by the above, cytoplasmic PAR associates with AIF on the mitochondrial surface, releasing it to the cytoplasm [54, 58, 61, 62]. Although AIF is an anchored protein present in the inner mitochondrial membrane, some AIF (20–30%) is also located on the cytoplasmic side of the outer-mitochondrial membrane where it can be accessed by cytoplasmic PAR [58, 96]. Cytoplasmic PAR seen in ARH3−/− MEFs appeared to enhance AIF release from mitochondria and translocation to the nucleus, leading to large-scale DNA fragmentation and chromatin condensation. Thus, ARH3 serves as a suppressor of PARP1-dependent cell death, parthanatos, under oxidative stress.
Fig. 2. Regulation of parthanatos by PARP1, PARG, and ARH3.
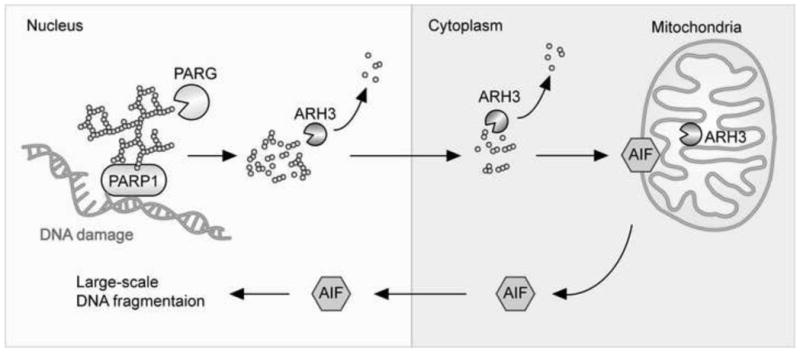
DNA damage secondary to oxidative stress activate PARP1, results in increased PAR synthesis in the nucleus. Protein-associated PAR is cleaved by the endoglycosidic activity of nuclear PARG, generating protein-free PAR, which then translocates to the cytoplasm. Cytoplasmic PAR releases AIF attached to the outer surface of mitochondrial membranes. AIF released into the cytoplasm then translocates to the nucleus and induces large-scale DNA fragmentation and chromatin condensation. This figure was modified from that in Mashimo et al. [93].
Interestingly, PAR export from the nucleus may result from the action of nuclear 110-kDa PARG, which releases protein-free PAR from acceptor proteins such as PARP1. PARG depletion by shRNA resulted in increased and prolonged poly-(ADP-ribosyl)ation of PARP1, inhibited PAR translocation from the nucleus to cytoplasm, and attenuated AIF-mediated cell death [93]. These findings are consistent with the facts that selective knockout of nuclear 110-kDa PARG has a protective effect against renal and intestinal injury induced by ischemia/reperfusion in mice [36, 97, 98]. PARG has both endo- and exo-glycosidase activities that hydrolyze the glycosidic linkage between ADP-ribose moieties of PAR chains [99–101]. Endo-glycosidase activity of PARG might account for production of protein-free PAR, which can pass through nuclear pores and then bind to AIF on mitochondrial membranes. This model is consistent with biochemical studies that show that (1) PARG preferentially cleaves long, rather than short PAR chains, (2) the Km value for long PAR chains is approximately ~1% of that for small ones, and that (3) PARG hydrolyzes covalently protein-bound PAR more rapidly than it does protein-free PAR [63, 102]. On the other hand, according to the three-dimensional structure of ARH3, the cavity of ARH3 docks only on the terminal ADP-ribose moiety of the PAR chain [77], suggesting that ARH3 has exoglycosidase activity, and thus is unable to substitute for PARG. Despite that the fact that ARH3 has lower PAR-degrading activity than PARG [76], the different substrates for PAR degradation and the cellular localization may contribute to their unique roles in the regulation of parthanatos.
3. Concluding remarks
We describe the properties of ARH1 and ARH3, focusing on their enzymatic activities, cellular distributions, and physiological and pathological functions. Based on phenotype analysis of ARH1−/− and ARH3−/− mice, we found that mono-ADP-ribosyl-acceptor hydrolase activity of ARH1 is involved in reversing the response to cholera toxin, possibly serving as a novel host defense mechanism [89]. Moreover, ARH1 appears to act as a tumor suppressor gene [90]. In contrast, ARH3 has poly-ADP-ribosyl-acceptor hydrolase activity and is involved in PAR degradation and the induction of parthanatos under oxidative stress conditions [65, 92, 93]. Thus, although ARH1 shares substantial amino acid sequence similarity with ARH3, they have different enzymatic activities.
We demonstrate that, through their substrate-specific hydrolase activities, both ARH1 and ARH3 regulate ADP-ribose levels; abnormalities in the regulation of either of these enzymes may lead to defective regulation of metabolism and potentially to disease. Both mono- and poly-(ADP-ribosyl)ation may thus have significant roles in health and disease. Understanding these functions may result in novel therapeutic targets.
Highlights.
The ADP-ribose-acceptor hydrolase (ARH) family consists of three 39-kDa proteins.
ARH1 is involved in tumorigenesis and the response to bacterial toxin transferases.
ARH3 participates in the induction of parthanatos, PARP1-mediated cell death.
Acknowledgments
This study was supported by the Intramural Research Program, National Institutes of Health, National Heart, Lung, and Blood Institute.
Abbreviations
- MPTP
1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine
- Gαs
α-subunit of the stimulatory guanine nucleotide-binding protein
- ARH
ADP-ribosyl-acceptor hydrolase
- AIF
Apoptosis-inducing factor
- mito-PARP1cd
Catalytic domain of PARP1 with a mitochondrial targeting sequence
- ART
ADP-ribosyltransferase
- FGF-2
Fibroblast growth factor-2
- GDH
Glutamate dehydrogenase
- HNP-1
Human neutrophil peptide 1
- MEFs
Mouse embryonic fibroblasts
- NAD+
Nicotinamide adenine dinucleotide
- OAADPr
O-acetyl-ADP-ribose
- PDGF-BB
Platelet-derived growth factor-BB
- PARG
Poly-(ADP-ribose) glycohydrolase
- PARP
Poly-(ADP-ribose) polymerase
- TARG1
Terminal ADP-ribose protein glycohydrolase
- WT
Wild-type
Footnotes
4. Conflict of interest
The authors confirm there is no conflict of interest, financial, or otherwise.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ueda K, Hayaishi O. ADP-ribosylation. Annu Rev Biochem. 1985;54:73–100. doi: 10.1146/annurev.bi.54.070185.000445. [DOI] [PubMed] [Google Scholar]
- 2.Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vaughan M, Moss J. Mono (ADP-ribosyl)transferases and their effects on cellular metabolism. Curr Top Cell Regul. 1981;20:205–246. doi: 10.1016/b978-0-12-152820-1.50010-9. [DOI] [PubMed] [Google Scholar]
- 4.Iglewski BH, Kabat D. NAD-dependent inhibition of protein synthesis by Pseudomonas aeruginosa toxin. Proc Natl Acad Sci U S A. 1975;72:2284–2288. doi: 10.1073/pnas.72.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pappenheimer AM., Jr Diphtheria toxin. Annu Rev Biochem. 1977;46:69–94. doi: 10.1146/annurev.bi.46.070177.000441. [DOI] [PubMed] [Google Scholar]
- 6.Cassel D, Pfeuffer T. Mechanism of cholera toxin action: covalent modification of the guanyl nucleotide-binding protein of the adenylate cyclase system. Proc Natl Acad Sci U S A. 1978;75:2669–2673. doi: 10.1073/pnas.75.6.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamura M, Nogimori K, Murai S, Yajima M, Ito K, Katada T, Ui M, Ishii S. Subunit structure of islet-activating protein, pertussis toxin, in conformity with the A–B model. Biochemistry. 1982;21:5516–5522. doi: 10.1021/bi00265a021. [DOI] [PubMed] [Google Scholar]
- 8.Moss J, Stanley SJ, Watkins PA. Isolation and properties of an NAD- and guanidine-dependent ADP-ribosyltransferase from turkey erythrocytes. J Biol Chem. 1980;255:5838–5840. [PubMed] [Google Scholar]
- 9.Moss J, Vaughan M. NAD: arginine mono-ADP-ribosyltransferases from animal cells. Methods Enzymol. 1984;106:430–437. doi: 10.1016/0076-6879(84)06046-8. [DOI] [PubMed] [Google Scholar]
- 10.Moss J, Stanley SJ, Oppenheimer NJ. Substrate specificity and partial purification of a stereospecific NAD- and guanidine-dependent ADP-ribosyltransferase from avian erythrocytes. J Biol Chem. 1979;254:8891–8894. [PubMed] [Google Scholar]
- 11.Okazaki IJ, Moss J. Characterization of glycosylphosphatidylinositiol-anchored, secreted, and intracellular vertebrate mono-ADP-ribosyltransferases. Annu Rev Nutr. 1999;19:485–509. doi: 10.1146/annurev.nutr.19.1.485. [DOI] [PubMed] [Google Scholar]
- 12.Seman M, Adriouch S, Haag F, Koch-Nolte F. Ecto-ADP-ribosyltransferases (ARTs): emerging actors in cell communication and signaling. Curr Med Chem. 2004;11:857–872. doi: 10.2174/0929867043455611. [DOI] [PubMed] [Google Scholar]
- 13.Okazaki IJ, Moss J. Glycosylphosphatidylinositol-anchored and secretory isoforms of mono-ADP-ribosyltransferases. J Biol Chem. 1998;273:23617–23620. doi: 10.1074/jbc.273.37.23617. [DOI] [PubMed] [Google Scholar]
- 14.Zolkiewska A, Nightingale MS, Moss J. Molecular characterization of NAD:arginine ADP-ribosyltransferase from rabbit skeletal muscle. Proc Natl Acad Sci U S A. 1992;89:11352–11356. doi: 10.1073/pnas.89.23.11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okazaki IJ, Kim HJ, Moss J. Cloning and characterization of a novel membrane-associated lymphocyte NAD:arginine ADP-ribosyltransferase. J Biol Chem. 1996;271:22052–22057. doi: 10.1074/jbc.271.36.22052. [DOI] [PubMed] [Google Scholar]
- 16.Glowacki G, Braren R, Firner K, Nissen M, Kuhl M, Reche P, Bazan F, Cetkovic-Cvrlje M, Leiter E, Haag F, Koch-Nolte F. The family of toxin-related ecto-ADP-ribosyltransferases in humans and the mouse. Protein Sci. 2002;11:1657–1670. doi: 10.1110/ps.0200602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moss J, Balducci E, Cavanaugh E, Kim HJ, Konczalik P, Lesma EA, Okazaki IJ, Park M, Shoemaker M, Stevens LA, Zolkiewska A. Characterization of NAD:arginine ADP-ribosyltransferases. Mol Cell Biochem. 1999;193:109–113. [PubMed] [Google Scholar]
- 18.Jones EM, Baird A. Cell-surface ADP-ribosylation of fibroblast growth factor-2 by an arginine-specific ADP-ribosyltransferase. Biochem J. 1997;323(Pt 1):173–177. doi: 10.1042/bj3230173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saxty BA, Yadollahi-Farsani M, Upton PD, Johnstone SR, MacDermot J. Inactivation of platelet-derived growth factor-BB following modification by ADP-ribosyltransferase. Br J Pharmacol. 2001;133:1219–1226. doi: 10.1038/sj.bjp.0704187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paone G, Wada A, Stevens LA, Matin A, Hirayama T, Levine RL, Moss J. ADP ribosylation of human neutrophil peptide-1 regulates its biological properties. Proc Natl Acad Sci U S A. 2002;99:8231–8235. doi: 10.1073/pnas.122238899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zolkiewska A, Moss J. Integrin alpha 7 as substrate for a glycosylphosphatidylinositol-anchored ADP-ribosyltransferase on the surface of skeletal muscle cells. J Biol Chem. 1993;268:25273–25276. [PubMed] [Google Scholar]
- 22.Seman M, Adriouch S, Scheuplein F, Krebs C, Freese D, Glowacki G, Deterre P, Haag F, Koch-Nolte F. NAD-induced T cell death: ADP-ribosylation of cell surface proteins by ART2 activates the cytolytic P2X7 purinoceptor. Immunity. 2003;19:571–582. doi: 10.1016/s1074-7613(03)00266-8. [DOI] [PubMed] [Google Scholar]
- 23.Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 24.Hawse WF, Wolberger C. Structure-based mechanism of ADP-ribosylation by sirtuins. J Biol Chem. 2009;284:33654–33661. doi: 10.1074/jbc.M109.024521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 26.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 27.Tsang AW, Escalante-Semerena JC. CobB, a new member of the SIR2 family of eucaryotic regulatory proteins, is required to compensate for the lack of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase activity in cobT mutants during cobalamin biosynthesis in Salmonella typhimurium LT2. J Biol Chem. 1998;273:31788–31794. doi: 10.1074/jbc.273.48.31788. [DOI] [PubMed] [Google Scholar]
- 28.Choi MM, Huh JW, Yang SJ, Cho EH, Choi SY, Cho SW. Identification of ADP-ribosylation site in human glutamate dehydrogenase isozymes. FEBS Lett. 2005;579:4125–4130. doi: 10.1016/j.febslet.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 29.Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW, Shilton BH, Luscher B. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 30.Di Paola S, Micaroni M, Di Tullio G, Buccione R, Di Girolamo M. PARP16/ARTD15 is a novel endoplasmic-reticulum-associated mono-ADP-ribosyltransferase that interacts with, and modifies karyopherin-ss1. PLoS One. 2012;7:e37352. doi: 10.1371/journal.pone.0037352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 32.Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 33.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 34.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- 36.Heeres JT, Hergenrother PJ. Poly(ADP-ribose) makes a date with death. Curr Opin Chem Biol. 2007;11:644–653. doi: 10.1016/j.cbpa.2007.08.038. [DOI] [PubMed] [Google Scholar]
- 37.Leung AK, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell. 2011;42:489–499. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boehler C, Gauthier LR, Mortusewicz O, Biard DS, Saliou JM, Bresson A, Sanglier-Cianferani S, Smith S, Schreiber V, Boussin F, Dantzer F. Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression. Proc Natl Acad Sci U S A. 2011;108:2783–2788. doi: 10.1073/pnas.1016574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ame JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, Muller S, Hoger T, Menissier-de Murcia J, de Murcia G. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J Biol Chem. 1999;274:17860–17868. doi: 10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- 40.Langelier MF, Planck JL, Roy S, Pascal JM. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science. 2012;336:728–732. doi: 10.1126/science.1216338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawaichi M, Ueda K, Hayaishi O. Multiple autopoly(ADP-ribosyl)ation of rat liver poly(ADP-ribose) synthetase. Mode of modification and properties of automodified synthetase. J Biol Chem. 1981;256:9483–9489. [PubMed] [Google Scholar]
- 42.Ali AA, Timinszky G, Arribas-Bosacoma R, Kozlowski M, Hassa PO, Hassler M, Ladurner AG, Pearl LH, Oliver AW. The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat Struct Mol Biol. 2012;19:685–692. doi: 10.1038/nsmb.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009;37:3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogata N, Ueda K, Kagamiyama H, Hayaishi O. ADP-ribosylation of histone H1. Identification of glutamic acid residues 2, 14, and the COOH-terminal lysine residue as modification sites. J Biol Chem. 1980;255:7616–7620. [PubMed] [Google Scholar]
- 45.D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 46.Jungmichel S, Rosenthal F, Altmeyer M, Lukas J, Hottiger MO, Nielsen ML. Proteome-wide Identification of Poly(ADP-Ribosyl)ation Targets in Different Genotoxic Stress Responses. Mol Cell. 2013;52:272–285. doi: 10.1016/j.molcel.2013.08.026. [DOI] [PubMed] [Google Scholar]
- 47.Gagne JP, Pic E, Isabelle M, Krietsch J, Ethier C, Paquet E, Kelly I, Boutin M, Moon KM, Foster LJ, Poirier GG. Quantitative proteomics profiling of the poly(ADP-ribose)-related response to genotoxic stress. Nucleic Acids Res. 2012;40:7788–7805. doi: 10.1093/nar/gks486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krupitza G, Cerutti P. ADP-ribosylation of ADPR-transferase and topoisomerase I in intact mouse epidermal cells JB6. Biochemistry. 1989;28:2034–2040. doi: 10.1021/bi00431a011. [DOI] [PubMed] [Google Scholar]
- 49.Ogata N, Ueda K, Kawaichi M, Hayaishi O. Poly(ADP-ribose) synthetase, a main acceptor of poly(ADP-ribose) in isolated nuclei. J Biol Chem. 1981;256:4135–4137. [PubMed] [Google Scholar]
- 50.Chapman JD, Gagne JP, Poirier GG, Goodlett DR. Mapping PARP-1 Auto-ADP-ribosylation Sites by Liquid Chromatography-Tandem Mass Spectrometry. J Proteome Res. 2013 doi: 10.1021/pr301219h. [DOI] [PubMed] [Google Scholar]
- 51.Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pacher P, Szabo C. Role of poly(ADP-ribose) polymerase 1 (PARP-1) in cardiovascular diseases: the therapeutic potential of PARP inhibitors. Cardiovasc Drug Rev. 2007;25:235–260. doi: 10.1111/j.1527-3466.2007.00018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Komjati K, Besson VC, Szabo C. Poly (adp-ribose) polymerase inhibitors as potential therapeutic agents in stroke and neurotrauma. Curr Drug Targets CNS Neurol Disord. 2005;4:179–194. doi: 10.2174/1568007053544138. [DOI] [PubMed] [Google Scholar]
- 54.Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M, Klaus JA, Otsuka T, Zhang Z, Koehler RC, Hurn PD, Poirier GG, Dawson VL, Dawson TM. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci U S A. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee Y, Karuppagounder SS, Shin JH, Lee YI, Ko HS, Swing D, Jiang H, Kang SU, Lee BD, Kang HC, Kim D, Tessarollo L, Dawson VL, Dawson TM. Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss. Nat Neurosci. 2013;16:1392–1400. doi: 10.1038/nn.3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mandir AS, Przedborski S, Jackson-Lewis V, Wang ZQ, Simbulan-Rosenthal CM, Smulson ME, Hoffman BE, Guastella DB, Dawson VL, Dawson TM. Poly(ADP-ribose) polymerase activation mediates 1-methyl-4-phenyl-1, 2,3,6-tetrahydropyridine (MPTP)-induced parkinsonism. Proc Natl Acad Sci U S A. 1999;96:5774–5779. doi: 10.1073/pnas.96.10.5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Masutani M, Suzuki H, Kamada N, Watanabe M, Ueda O, Nozaki T, Jishage K, Watanabe T, Sugimoto T, Nakagama H, Ochiya T, Sugimura T. Poly(ADP-ribose) polymerase gene disruption conferred mice resistant to streptozotocin-induced diabetes. Proc Natl Acad Sci U S A. 1999;96:2301–2304. doi: 10.1073/pnas.96.5.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y, Kim NS, Haince JF, Kang HC, David KK, Andrabi SA, Poirier GG, Dawson VL, Dawson TM. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos) Sci Signal. 2011;4:ra20. doi: 10.1126/scisignal.2000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Szabo C, Zingarelli B, O’Connor M, Salzman AL. DNA strand breakage, activation of poly (ADP-ribose) synthetase, and cellular energy depletion are involved in the cytotoxicity of macrophages and smooth muscle cells exposed to peroxynitrite. Proc Natl Acad Sci U S A. 1996;93:1753–1758. doi: 10.1073/pnas.93.5.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ha HC, Snyder SH. Poly(ADP-ribose) polymerase is a mediator of necrotic cell death by ATP depletion. Proc Natl Acad Sci U S A. 1999;96:13978–13982. doi: 10.1073/pnas.96.24.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu SW, Andrabi SA, Wang H, Kim NS, Poirier GG, Dawson TM, Dawson VL. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc Natl Acad Sci U S A. 2006;103:18314–18319. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 63.Hatakeyama K, Nemoto Y, Ueda K, Hayaishi O. Purification and characterization of poly(ADP-ribose) glycohydrolase. Different modes of action on large and small poly(ADP-ribose) J Biol Chem. 1986;261:14902–14911. [PubMed] [Google Scholar]
- 64.Miwa M, Sugimura T. Splitting of the ribose-ribose linkage of poly(adenosine diphosphate-robose) by a calf thymus extract. J Biol Chem. 1971;246:6362–6364. [PubMed] [Google Scholar]
- 65.Niere M, Mashimo M, Agledal L, Dolle C, Kasamatsu A, Kato J, Moss J, Ziegler M. ADP-ribosylhydrolase 3 (ARH3), not poly(ADP-ribose) glycohydrolase (PARG) isoforms, is responsible for degradation of mitochondrial matrix-associated poly(ADP-ribose) J Biol Chem. 2012;287:16088–16102. doi: 10.1074/jbc.M112.349183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meyer-Ficca ML, Meyer RG, Coyle DL, Jacobson EL, Jacobson MK. Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp Cell Res. 2004;297:521–532. doi: 10.1016/j.yexcr.2004.03.050. [DOI] [PubMed] [Google Scholar]
- 67.Winstall E, Affar EB, Shah R, Bourassa S, Scovassi AI, Poirier GG. Poly(ADP-ribose) glycohydrolase is present and active in mammalian cells as a 110-kDa protein. Exp Cell Res. 1999;246:395–398. doi: 10.1006/excr.1998.4321. [DOI] [PubMed] [Google Scholar]
- 68.Meyer RG, Meyer-Ficca ML, Whatcott CJ, Jacobson EL, Jacobson MK. Two small enzyme isoforms mediate mammalian mitochondrial poly(ADP-ribose) glycohydrolase (PARG) activity. Exp Cell Res. 2007;313:2920–2936. doi: 10.1016/j.yexcr.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koh DW, Lawler AM, Poitras MF, Sasaki M, Wattler S, Nehls MC, Stoger T, Poirier GG, Dawson VL, Dawson TM. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc Natl Acad Sci U S A. 2004;101:17699–17704. doi: 10.1073/pnas.0406182101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, Ladurner AG. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kustatscher G, Hothorn M, Pugieux C, Scheffzek K, Ladurner AG. Splicing regulates NAD metabolite binding to histone macroH2A. Nat Struct Mol Biol. 2005;12:624–625. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]
- 72.Sharifi R, Morra R, Appel CD, Tallis M, Chioza B, Jankevicius G, Simpson MA, Matic I, Ozkan E, Golia B, Schellenberg MJ, Weston R, Williams JG, Rossi MN, Galehdari H, Krahn J, Wan A, Trembath RC, Crosby AH, Ahel D, Hay R, Ladurner AG, Timinszky G, Williams RS, Ahel I. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 2013;32:1225–1237. doi: 10.1038/emboj.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Feijs KL, Forst AH, Verheugd P, Luscher B. Macrodomain-containing proteins: regulating new intracellular functions of mono(ADP-ribosyl)ation. Nat Rev Mol Cell Biol. 2013;14:443–451. doi: 10.1038/nrm3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rosenthal F, Feijs KL, Frugier E, Bonalli M, Forst AH, Imhof R, Winkler HC, Fischer D, Caflisch A, Hassa PO, Luscher B, Hottiger MO. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat Struct Mol Biol. 2013;20:502–507. doi: 10.1038/nsmb.2521. [DOI] [PubMed] [Google Scholar]
- 75.Jankevicius G, Hassler M, Golia B, Rybin V, Zacharias M, Timinszky G, Ladurner AG. A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat Struct Mol Biol. 2013;20:508–514. doi: 10.1038/nsmb.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oka S, Kato J, Moss J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J Biol Chem. 2006;281:705–713. doi: 10.1074/jbc.M510290200. [DOI] [PubMed] [Google Scholar]
- 77.Mueller-Dieckmann C, Kernstock S, Lisurek M, von Kries JP, Haag F, Weiss MS, Koch-Nolte F. The structure of human ADP-ribosylhydrolase 3 (ARH3) provides insights into the reversibility of protein ADP-ribosylation. Proc Natl Acad Sci U S A. 2006;103:15026–15031. doi: 10.1073/pnas.0606762103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moss J, Stanley SJ, Nightingale MS, Murtagh JJ, Jr, Monaco L, Mishima K, Chen HC, Williamson KC, Tsai SC. Molecular and immunological characterization of ADP-ribosylarginine hydrolases. J Biol Chem. 1992;267:10481–10488. [PubMed] [Google Scholar]
- 79.Moss J, Jacobson MK, Stanley SJ. Reversibility of arginine-specific mono(ADP-ribosyl)ation: identification in erythrocytes of an ADP-ribose-L-arginine cleavage enzyme. Proc Natl Acad Sci U S A. 1985;82:5603–5607. doi: 10.1073/pnas.82.17.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moss J, Oppenheimer NJ, West RE, Jr, Stanley SJ. Amino acid specific ADP-ribosylation: substrate specificity of an ADP-ribosylarginine hydrolase from turkey erythrocytes. Biochemistry. 1986;25:5408–5414. doi: 10.1021/bi00367a010. [DOI] [PubMed] [Google Scholar]
- 81.Ono T, Kasamatsu A, Oka S, Moss J. The 39-kDa poly(ADP-ribose) glycohydrolase ARH3 hydrolyzes O-acetyl-ADP-ribose, a product of the Sir2 family of acetyl-histone deacetylases. Proc Natl Acad Sci U S A. 2006;103:16687–16691. doi: 10.1073/pnas.0607911103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kasamatsu A, Nakao M, Smith BC, Comstock LR, Ono T, Kato J, Denu JM, Moss J. Hydrolysis of O-acetyl-ADP-ribose isomers by ADP-ribosylhydrolase 3. J Biol Chem. 2011;286:21110–21117. doi: 10.1074/jbc.M111.237636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121:515–527. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 84.Borra MT, O’Neill FJ, Jackson MD, Marshall B, Verdin E, Foltz KR, Denu JM. Conserved enzymatic production and biological effect of O-acetyl-ADP-ribose by silent information regulator 2-like NAD+-dependent deacetylases. J Biol Chem. 2002;277:12632–12641. doi: 10.1074/jbc.M111830200. [DOI] [PubMed] [Google Scholar]
- 85.Grubisha O, Rafty LA, Takanishi CL, Xu X, Tong L, Perraud AL, Scharenberg AM, Denu JM. Metabolite of SIR2 reaction modulates TRPM2 ion channel. J Biol Chem. 2006;281:14057–14065. doi: 10.1074/jbc.M513741200. [DOI] [PubMed] [Google Scholar]
- 86.Tong L, Lee S, Denu JM. Hydrolase regulates NAD+ metabolites and modulates cellular redox. J Biol Chem. 2009;284:11256–11266. doi: 10.1074/jbc.M809790200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Konczalik P, Moss J. Identification of critical, conserved vicinal aspartate residues in mammalian and bacterial ADP-ribosylarginine hydrolases. J Biol Chem. 1999;274:16736–16740. doi: 10.1074/jbc.274.24.16736. [DOI] [PubMed] [Google Scholar]
- 88.Takada T, Iida K, Moss J. Cloning and site-directed mutagenesis of human ADP-ribosylarginine hydrolase. J Biol Chem. 1993;268:17837–17843. [PubMed] [Google Scholar]
- 89.Kato J, Zhu J, Liu C, Moss J. Enhanced sensitivity to cholera toxin in ADP-ribosylarginine hydrolase-deficient mice. Mol Cell Biol. 2007;27:5534–5543. doi: 10.1128/MCB.00302-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kato J, Zhu J, Liu C, Stylianou M, Hoffmann V, Lizak MJ, Glasgow CG, Moss J. ADP-ribosylarginine hydrolase regulates cell proliferation and tumorigenesis. Cancer Res. 2011;71:5327–5335. doi: 10.1158/0008-5472.CAN-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shim B, Pacheco-Rodriguez G, Kato J, Darling TN, Vaughan M, Moss J. Sex-specific lung diseases: effect of oestrogen on cultured cells and in animal models. Eur Respir Rev. 2013;22:302–311. doi: 10.1183/09059180.00002813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Niere M, Kernstock S, Koch-Nolte F, Ziegler M. Functional localization of two poly(ADP-ribose)-degrading enzymes to the mitochondrial matrix. Mol Cell Biol. 2008;28:814–824. doi: 10.1128/MCB.01766-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mashimo M, Kato J, Moss J. ADP-ribosyl-acceptor hydrolase 3 regulates poly (ADP-ribose) degradation and cell death during oxidative stress. Proc Natl Acad Sci U S A. 2013;110:18964–18969. doi: 10.1073/pnas.1312783110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Scovassi AI. Mitochondrial poly(ADP-ribosylation): from old data to new perspectives. FASEB J. 2004;18:1487–1488. doi: 10.1096/fj.04-1841rev. [DOI] [PubMed] [Google Scholar]
- 95.Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 96.Yu SW, Wang Y, Frydenlund DS, Ottersen OP, Dawson VL, Dawson TM. Outer mitochondrial membrane localization of apoptosis-inducing factor: mechanistic implications for release. ASN Neuro. 2009;1 doi: 10.1042/AN20090046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Patel NS, Cortes U, Di Poala R, Mazzon E, Mota-Filipe H, Cuzzocrea S, Wang ZQ, Thiemermann C. Mice lacking the 110-kD isoform of poly(ADP-ribose) glycohydrolase are protected against renal ischemia/reperfusion injury. J Am Soc Nephrol. 2005;16:712–719. doi: 10.1681/ASN.2004080677. [DOI] [PubMed] [Google Scholar]
- 98.Cuzzocrea S, Di Paola R, Mazzon E, Cortes U, Genovese T, Muia C, Li W, Xu W, Li JH, Zhang J, Wang ZQ. PARG activity mediates intestinal injury induced by splanchnic artery occlusion and reperfusion. FASEB J. 2005;19:558–566. doi: 10.1096/fj.04-3117com. [DOI] [PubMed] [Google Scholar]
- 99.Barkauskaite E, Brassington A, Tan ES, Warwicker J, Dunstan MS, Banos B, Lafite P, Ahel M, Mitchison TJ, Ahel I, Leys D. Visualization of poly(ADP-ribose) bound to PARG reveals inherent balance between exo- and endo-glycohydrolase activities. Nat Commun. 2013;4:2164. doi: 10.1038/ncomms3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Braun SA, Panzeter PL, Collinge MA, Althaus FR. Endoglycosidic cleavage of branched polymers by poly(ADP-ribose) glycohydrolase. Eur J Biochem. 1994;220:369–375. doi: 10.1111/j.1432-1033.1994.tb18633.x. [DOI] [PubMed] [Google Scholar]
- 101.Brochu G, Duchaine C, Thibeault L, Lagueux J, Shah GM, Poirier GG. Mode of action of poly(ADP-ribose) glycohydrolase. Biochim Biophys Acta. 1994;1219:342–350. doi: 10.1016/0167-4781(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 102.Uchida K, Suzuki H, Maruta H, Abe H, Aoki K, Miwa M, Tanuma S. Preferential degradation of protein-bound (ADP-ribose)n by nuclear poly(ADP-ribose) glycohydrolase from human placenta. J Biol Chem. 1993;268:3194–3200. [PubMed] [Google Scholar]


