Abstract
Cellular differentiation proceeds along complicated pathways, even when it is induced by extracellular signaling molecules. One of the major reasons for this complexity is the highly multidimensional internal dynamics of cells, which sometimes causes apparently stochastic responses in individual cells to extracellular stimuli. Therefore, to understand cell differentiation, it is necessary to monitor the internal dynamics of cells at single-cell resolution. Here, we used a Raman and autofluorescence spectrum analysis of single cells to detect dynamic changes in intracellular molecular components. MCF-7 cells are a human cancer-derived cell line that can be induced to differentiate into mammary-gland-like cells with the addition of heregulin (HRG) to the culture medium. We measured the spectra in the cytoplasm of MCF-7 cells during 12 days of HRG stimulation. The Raman scattering spectrum, which was the major component of the signal, changed with time. A multicomponent analysis of the Raman spectrum revealed that the dynamics of the major components of the intracellular molecules, including proteins and lipids, changed cyclically along the differentiation pathway. The background autofluorescence signals of Raman scattering also provided information about the differentiation process. Using the total information from the Raman and autofluorescence spectra, we were able to visualize the pathway of cell differentiation in the multicomponent phase space.
Introduction
Cell differentiation is a complicated process that seems to be stochastic, even in clonal cells under the same growth conditions (1). One reason for this stochasticity must be the complexity of the cellular internal dynamics. State changes in cells are directed by a complex web of intracellular metabolic reactions interacting with the complex gene-expression network. This complexity allows cells to assume multiple states even under the same environmental conditions. Therefore, it is sometimes difficult to regulate the differentiation of populations of cells effectively, although it is important to do so in many applications, including regenerative medicine (2). A method must first be established to monitor the dynamics of the intracellular components along the pathway of cell differentiation. Although various genomic, proteomic, and metabolomic technologies can be used for the precise analysis of cellular components, these omic technologies are invasive and usually are only applicable to populations of cells (3). Therefore, it is difficult to monitor the intracellular dynamics successively in individual living cells using omic technologies.
Spectroscopic technologies can be used to complement the omic technologies in monitoring cell differentiation pathways because they allow successive measurements to be made in single living cells. Spectroscopic measurements can potentially provide multidimensional information about the many chemical components of living cells with low invasiveness, especially when long light wavelengths are used to monitor the cells. Raman scattering spectroscopy is the most popular spectroscopic technology for detecting differences in the states of living cells and tissues (4). It can use long excitation wavelengths and shows good contrast in detecting longer-wavelength signals, which can be separated from the excitation light. The greatest advantage of Raman spectroscopy is probably its label-free detection of the multidimensional dynamics that reflects the differences between chemical components (5). The acquisition of multidimensional data is essential to distinguish the various cellular states without requiring prior knowledge or speculation. The problems that arise from the low signals in Raman measurements can be overridden thanks to recent improvements in the detection apparatus, including optics and photosensors.
Spectroscopic technologies can be extended straightforwardly to imaging or microscopic measurements. In fact, various cellular states or processes have been distinguished and analyzed with the use of Raman microspectroscopy in tissues, single cells, and subcellular volumes (4). For example, structure-specific signals have been detected in human skin tissue (6), and cancerous and normal cells are distinguishable in skin (7) and lung (8) tissues by Raman microscopy. In single cells, Raman spectrum analysis has successfully distinguished growth plateau conditions from exponentially proliferating conditions (9). Recent studies sought to identify differentiation markers in Raman spectra by observing undifferentiated and differentiated embryonic stem cells (10–12) and other cell types (13). However, as far as we know, most Raman spectroscopic studies to date have been directed toward static comparisons of two states of cells. Only a few studies have investigated the cellular internal dynamics during state changes in cells. Huang et al. (14) examined spatiotemporal changes in the Raman spectrum during the yeast cell cycle and detected a specific Raman band for cells growing under good conditions. Their work clearly demonstrated the power of Raman spectrum imaging measurements for detecting dynamic state transitions in cells.
In addition to Raman scattering, the autofluorescence of cellular components carries information about the cellular internal dynamics (15). Autofluorescence microspectroscopy is another label-free, multidimensional spectroscopic technology that can be naturally extended to obtain imaging measurements. It has been used to image tissues (16) and diagnose cancer (17). Autofluorescence imaging can also be used to detect the functions of tissues (18) and cells (19).
In this study, we used microspectroscopic detection of both Raman and autofluorescence signals to trace the heregulin (HRG)-induced differentiation pathway of MCF-7 cells. HRG is a small cell-signaling protein that is responsible for the development of various tissues in vertebrates (20). HRG induces differentiation of MCF-7, a cell line derived from human breast cancer, into mammary-gland-like cells, which are characterized by the formation of oil droplets and albumin secretion (21). We detected and characterized the multidimensional dynamics of the metabolic components inside cells during the HRG-induced differentiation pathway of MCF-7 cells. In this study, we concentrated on the multidimensional dynamics of the metabolites in the cell population. Therefore, we described the cell differentiation pathway in a phase space defined by the Raman and autofluorescence spectra. It is well known that cellular responses usually show wide variations, so we concentrated on the distribution of the dynamics of single cells. If we can successfully describe the population dynamics in a phase space, this description can be extended to single-cell dynamics in the future.
Materials and Methods
Cell culture and sample preparation
MCF-7 cells (obtained from the American Type Culture Collection) were maintained in Dulbecco’s minimal Eagle’s medium supplemented with 10% fetal calf serum at 37°C in 5% CO2. The cells were transferred to a 35 mm cell culture dish with a quartz coverslip on the bottom (SF-S-D27; Fine Plus International, Kyoto, Japan) for microspectroscopy. Lyophilized powder of recombinant human neuregulin1-β1/heregulin1-β1 EGF domain (HRG) was purchased from R&D Systems (Minneapolis), dissolved in PBS, and added to the culture medium at a 1/60 dilution (final concentration of 30 nM) to induce cell differentiation. The culture medium containing HRG was refreshed every 2 days. Phase-contrast micrographs of the cells were acquired with an inverted optical microscope (CK40; Nikon, Tokyo, Japan) equipped with a 20× phase-contrast objective and a digital camera.
Microspectroscopy
Two days before measurements were made, the cell culture medium was exchanged for medium without phenol red. Just before the measurements were made, the cells were washed with Hank’s balanced salt solution containing 10 mM PIPES-Na (pH 7.2; HBSS-PIPES). HBSS-PIPES shows no significant background signal in the wavelengths we observed in this study. The Raman and autofluorescence spectra of the cells were measured in HBSS-PIPES with a confocal Raman microscope (inVia; Renishaw, Gloucestershire, UK), using an upright microscope equipped with a 63× dip-type water-immersion objective (NA = 0.9, HCX APO LU-V-1; Leica, Wetzlar, Germany).
The cell cytoplasm was irradiated with a diffraction-limited spot from a 532 nm laser (7.4 mW after the objective). The spatial resolutions were 1 μm and 3–4 μm in the lateral and focal directions, respectively. The positions of the measurements were selected randomly in the cytoplasm. Evident oil droplets and other organelles were avoided, although it is possible that some small droplets were present in the measurement volumes. Seven to 24 spots in three to five cells were obtained in the cultures at each day of differentiation. Spectra from seven spots on each day for five cells without HRG treatment were also obtained as the control. The emission from the excitation spot was selected with a confocal slit, diffracted with a grating (1800 lines/mm), and accumulated for 30 s on a 576 × 384 pixel cooled CCD sensor. The spectral resolution of the system is 1.7 cm−1 according to the manufacturer. Calibration of the wavelength was done using silicon substrate. Nonuniformity of the detector response was not corrected, but was <5% over the observation wavelength.
Illumination with the excitation laser might cause some damage to cells, although the irradiation power and period used in this study were in the range of those used in the previous studies (8,22,23). To avoid possible photodamage effects, independent cell cultures were used for measurements on different days of differentiation. Cell densities were regulated to be ∼70% confluent. Cells on day 0 were in the steady state of the log-phase growth condition.
Data analysis
After subtraction of the dark current, the Raman scattering and autofluorescence signals were separated in each spectrum, as shown in Fig. S1 of the Supporting Material). The Raman spectrum was vector normalized for signals from 560 to 1790 cm–1. The signals had 616 points (2 cm–1/point). A multivariate analysis of the spectra was performed using MATLAB (The MathWorks, Natick, MA) and The Unscrambler 9.8 (CAMO Software, Tokyo, Japan).
Results
Measurement of Raman and autofluorescence spectra along the differentiation pathway of MCF-7 cells
MCF-7 cells were cultured with a continuous supply of HRG in the culture medium to induce their differentiation. Phase-contrast micrographs of the cells were acquired along the differentiation pathway (Fig. 1). The accumulation of small oil droplets in the cytoplasm was observed in a small population of cells after HRG treatment for 1–3 days (Fig. 1, B and C), and oil droplets were evident around the nucleus in most cells after 6 days of treatment (Figs. 1, D and E, and S2).
Figure 1.
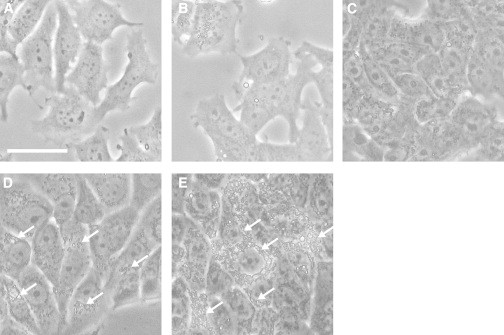
(A–E) HRG-induced differentiation of MCF-7 cells. Phase-contrast micrographs of the differentiation process in MCF-7 cells in the presence of 30 nM HRG are shown. Oil droplets in the cytoplasm are recognizable by their high refractive indices. Oil droplets are clearly present around the nuclei in cells after HRG stimulation for 6 days (arrows). Micrographs were taken before (A) and after 1 (B), 3 (C), 6 (D), and 12 days (E) of HRG stimulation. Bar: 20 μm.
To detect changes in the metabolite components of the cell cytoplasm during the differentiation process, we measured photoexcited emission spectra with a confocal microspectrometer in cells before HRG stimulation (day 0) and after HRG stimulation for 1, 3, 6, and 12 days. No measurements were made at sites that clearly contained oil droplets or other vesicles to detect changes in the cytoplasmic solution. Spectra were obtained from ∼1 μm3 of the confocal volume excited by a diffraction-limited 532 nm laser spot (Fig. 2 A). After accumulation of emission for 30 s, a spectrum with an appropriate signal/noise ratio was obtained for further analysis (Fig. 2 B). Most of the acquired signals were characteristic of the Raman scattering peaks of the metabolites. The spectra also contained a low-frequency background signal, probably caused by cellular autofluorescence. In each raw spectrum, 16 points at which the Raman signal was small were selected (the wave numbers were 560, 656, 710, 738, 772, 800, 906, 994, 1014, 1140, 1192, 1388, 1518, 1728, 1772, and 1790 cm–1). The Raman and autofluorescence signals were separated with respect to these 16 points (see Fig. S1 for details regarding separation of the Raman and autofluorescence signals). The Raman, autofluorescence, and entire spectra were vector normalized at 560–1790 cm–1 and used for further analysis (Fig. 2, C and D).
Figure 2.
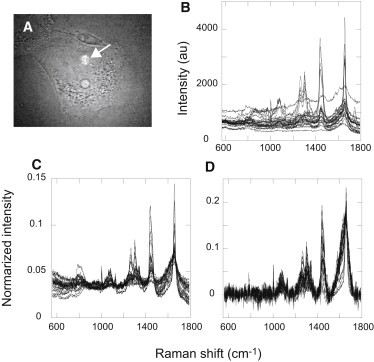
Measurement of photoexcited emission spectra in the cytoplasm of single cells. (A) Scattering of the excitation laser beam (532 nm) is shown in the cytoplasm of an MCF-7 cell (arrow). (B) Raw spectra from the cytoplasm of cells obtained with a confocal microspectrometer. Twenty-five randomly selected spectra from the 243 total spectra obtained from cells on various days of differentiation are shown. The accumulation time was 30 s. (C) Spectra after vector normalization of B. (D) Raman spectra after autofluorescence subtraction from A (see Fig. S1 for details) and vector normalization.
Changes in chemical components detected from Raman spectra
The normalized Raman spectra were averaged for each day of differentiation (Fig. 3 A) and the difference spectra relative to the average spectrum on day 0 were calculated (Fig. 3 B). The average spectrum changed with the days of differentiation, and we could assign some of the peaks to specific chemical components, including nucleic acids, proteins, lipids, carbohydrates, and cytochrome c, based on previous reports (8,10,14,24–26). The resonance Raman bands of the heme attached to the cytochrome c protein can be separated specifically from those of the other chemical components of cells by excitation at 532 nm (8). As shown in the difference spectra, the fractions of many of the chemical components of the cells changed dynamically, with individually specific patterns. The dynamics of the major Raman peaks could be categorized roughly into several groups (Fig. 3 C). Changes in the spectrum from cells without HRG treatment were small (Fig. 3, D and E).
Figure 3.
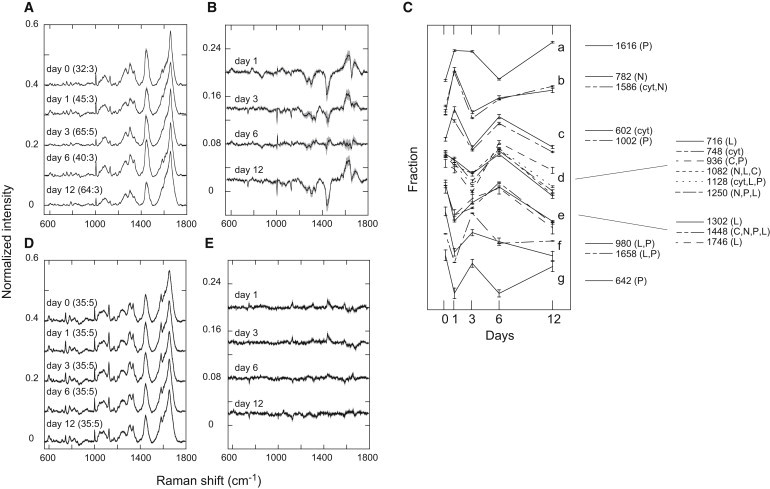
Changes in the chemical components of the cytoplasm. (A) Average Raman spectra during the period of HRG stimulation. Days of HRG treatment are indicated. (B) Differences in the average spectra on the indicated days with respect to the average spectrum on day 0. (C) Dynamics of individual chemical components. The dynamics were categorized into seven classes (a–g). These plots were made after normalization over time at individual wave numbers. Error bars indicate standard errors. C, carbohydrates; cyt, cytochrome c; L, lipids; N, nucleotides; P, proteins (see Table S1 for assignments). (D and E) Average (D) and difference (E) spectra of cells without HRG treatment. In panels A, B, D, and E, the black lines and gray regions indicate the averages and the widths of the standard errors, respectively. The baselines of individual spectra were shifted for convenience of visualization. In panels A and D, the numbers in parentheses (m:n) indicate the numbers of spectra (m) obtained on each day from (n) cells.
Although the separations were incomplete because the Raman peaks derived from different chemical species overlapped, nucleic acids, lipids, carbohydrates, and cytochrome c generally showed specific dynamics for each individual category of chemical components. Signals largely specific for nucleic acids increased on day 1, decreased on day 3, and increased again on day 12 (Fig. 3 C, b). The cytochrome c fraction showed dynamics similar to those of nucleic acids, but the fraction on day 12 was small (Fig. 3 C, c). The lipid and carbohydrate fractions were small in the early days of differentiation, but increased in the middle period of the differentiation pathway (Fig. 3 C, d and e). After the middle period, the lipid components were probably transferred to the large oil droplets excluded from the measurement. The dynamics of the protein fractions showed large variations, as expected from the different roles of each species of protein. From these oscillating patterns of change in the metabolite fractions, it can be concluded that differentiation takes a complicated pathway with time, rather than a straightforward one, from the perspective of the cytoplasmic metabolite composition.
Principal-component analysis of the Raman spectra
To characterize the differentiation pathway in a phase space, we performed a principal-component analysis (PCA) based on all of the individual Raman spectra taken at different times and in different spaces (Fig. 2 D). The loadings of the first six components and the distribution of the contribution ratios are shown (Fig. 4). Approximately 16% of the information on the cellular dynamics was contained in the first three principal components (PC1–PC3; Fig. 4 B). Although the higher-order components still contained extra information, the loadings for components higher than the third order were noisy. Therefore, for simplicity, we concentrated on PC1–PC3.
Figure 4.
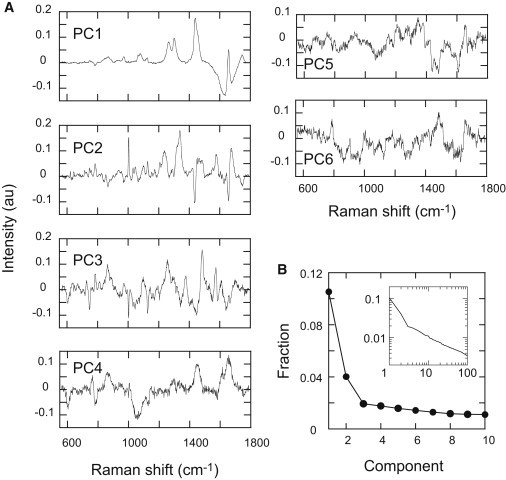
Principal components in the Raman spectra. (A) Loadings for the first six principal components (PC1–PC6) of the Raman spectra are shown. PCA was performed based on all 243 single-spot Raman spectra. (B) Distribution of the contribution ratios of the PCA loadings; 16% of the overall spectral information was expressed in PC1–PC3. Inset is the log-log plot of the fractions of the first 100 components.
Cell differentiation dynamics described in the Raman spectrum phase space
The PC1 and PC2 scores for individual spectra were plotted in a two-dimensional phase space (Fig. 5 A). Interestingly, all of the spectra were distributed in a triangular area, the bottom edge of which was parallel to the PC1 vector, and most of the spectra were aligned along the three edges of the distribution. We noted that the three vertices of this triangular distribution were characterized by spectra that were enriched with either protein, lipid, or water signals, i.e., the spectra in the middle-PC1 and high-PC2 region were rich in protein signals, those in the high-PC1 and low-PC2 region were rich in lipid signals, and those in the low-PC1 and low-PC2 region were rich in water signal (Fig. 5 B). Furthermore, the spectra for each day of differentiation showed a tendency to distribute to only one or two edges of the entire distribution (Fig. 5 A). The distribution on day 0 was on the two oblique edges, indicating protein-rich states. On day 1, the distribution moved to the oblique edge opposite the lower right vertex, indicating a reduction in the lipid fraction. On day 3, the spectra were distributed on two edges, the upper left and bottom, and on day 6, the distribution returned to the upper-right edge. The distribution on day 12 was mainly on the upper-left edge. As shown here, the changes in the fractions of chemical components circulated in this phase space in a counterclockwise direction: i.e., along the differentiation pathway, the lipid fraction decreased first, followed by reductions in protein and then a decline in the water fraction. In the last stage, another reduction in the lipid fraction occurred.
Figure 5.
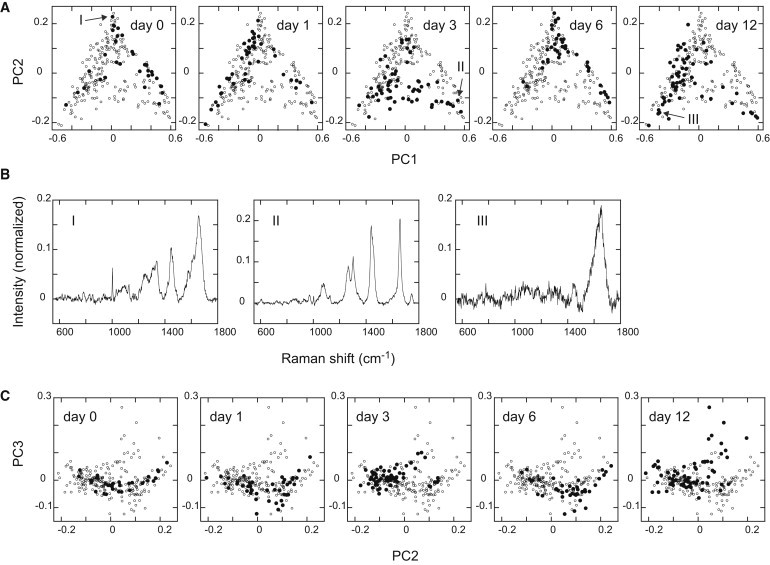
Raman dynamics of cell differentiation. (A) Plots of all Raman spectra in the PC1/PC2 plane. Each panel refers to the spectra obtained on the indicated day of differentiation (solid dots represent the indicated day, and open dots represent the other days for comparison). (B) Three characteristic spectra in the PC1/PC2 plane. The positions of the spectra in the PC1/PC2 plane are indicated by arrows in A. Spectra I–III are rich in proteins, lipids, and water, respectively. (C) Changes in the PC3 scores were plotted in the PC2/PC3 plane with the days of differentiation (see Fig. S3 for the plots in the PC1/PC3 plane).
The dynamics of PC3 were also not unimodal (Figs. 5 C and S3). Compared with the scores on day 0, a fraction of cells showed lower scores on days 1 and 6, and higher scores on days 3 and 12. Thus, the distribution of the PC3 scores oscillated along the differentiation pathway in a different phase compared with the PC1 and PC2 scores.
As shown here, the differentiation dynamics in MCF-7 cells can be characterized in a three-dimensional phase space of PC1–PC3 of the Raman spectrum. Despite the large cell-to-cell deviations on each day of differentiation, the well-differentiated population (day 12) was largely separated from the population of undifferentiated cells (day 0). However, because of the cyclic or oscillating nature of the Raman dynamics, it was hard to distinguish all of the intermediate stages in the differentiation pathway from the undifferentiated state based only on the Raman spectra. The distributions on day 6 showed especially large overlaps with those on day 0 for every PC1–PC3 score, indicating that, at this stage, it is difficult to distinguish cells in the differentiation process from those in the undifferentiated states, although once the distributions were well separated on day 3 from those on day 0. Therefore, it was difficult to distinguish undifferentiated cells from medially differentiated cells based on the cytoplasmic Raman spectra. It was not easy to separate the spectra on day 0 from those on day 6 even when higher components of the Raman spectra were considered, as shown by the residue values of a soft independent modeling of class analogy (SIMCA) analysis (Fig. S4).
Analysis using the entire information from single-cell spectra
We then analyzed the entire information of the spectra without autofluorescence subtraction. The spectra were vector normalized before the analysis (Fig. 2 C). The low-frequency changes at the baseline of the Raman signals suggest that the baseline was composed mainly of autofluorescence signals. Some biologically important metabolites, including NADH, FAD, and oxidized lipids, emit fluorescence in the visible wavelengths. Autofluorescence signals from these molecules carry information about the cellular metabolic states. To visualize all of the information in the Raman and autofluorescence spectra and thus trace the differentiation pathway, we used a SIMCA method (27) (Figs. 6 and S4). The spectra obtained on day 0 were used as the model data in the SIMCA analysis.
Figure 6.
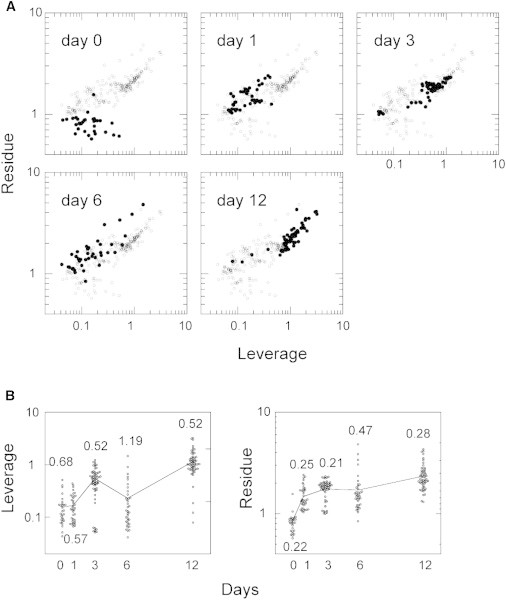
Dynamics of cell differentiation described by the SIMCA method. (A) The total spectra (including Raman and autofluorescence signals; Fig. 2C) were analyzed with the SIMCA method, using the spectra on day 0 as the model see Fig. S4A for the concept of the SIMCA method). Each panel refers to the spectra obtained on the indicated day of differentiation (solid dots represent the indicated day, and open dots represent the other days for comparison). (B) Distributions of leverage and residue values are plotted with the period of HRG treatment. Lines show the changes in the average. Numbers are coefficients of variation, showing the relative variations from the averages.
In the SIMCA plot using both Raman and autofluorescence information, the spectra on day 0 and day 6 were almost completely separated. The spectra for day 12 were distributed in an almost completely different region from those for day 0 and day 6. Therefore, the undifferentiated (day 0), medially differentiated (days 1–6), and well-differentiated (day 12) states could be distinguished in the SIMCA plot. Among the medially differentiated states, the spectra on day 1 and day 3 were well separated, but the spectra on day 6 had returned to a region similar to that occupied on day 1. This seemed to be caused by the oscillating PC3 of the Raman dynamics (Fig. 5 C). However, by observing the PC1 and PC2 components of the Raman dynamics, we could distinguish day 1 and day 6 (Fig. 5 A). SIMCA plots for the autofluorescence spectra (Fig. S5) were similar to those obtained for the entire spectra (Fig. 6). Thus, looking sterically at the phase spaces of Raman and autofluorescence spectra, intersections in the HRG-induced differentiation pathway of the MCF-7 cells at days 0 and 6 in the Raman phase space (Fig. 5 A), and at days 1 and 6 in the autofluorescence phase space (Fig. S5) were separated to show a differentiation pathway without looping. On both the leverage and residue axes in the SIMCA plot, the spectra on day 6 showed large deviations compared with those on the other days, and the distribution on day 3 was separated into two fractions, suggesting diversified pathways of cell differentiation (Fig. 6 B).
Discussion
In this study, we followed the HRG-induced differentiation pathway of MCF-7 cells by measuring single-cell photoexcited emission spectra, including the Raman and autofluorescence components. Our goal was to characterize the dynamics of the cell differentiation pathway in a multicomponent space determined from single-cell spectra. This would provide basic information for elucidating the cellular internal dynamics of differentiation. The Raman spectrum measurements of single living cells successfully detected characteristic changes in the chemical components along the differentiation pathway of a population of MCF-7 cells. The fractions of various components in the cytoplasm of cells, including nucleic acids, proteins, carbohydrates, and lipids, changed dynamically. These dynamics could be categorized into several different patterns, and no pattern showed a monotonous increase or decline. Related to this result, the pathway of differentiation was not straight in the PC1/PC2 space of the Raman spectra. Instead, the fractions of the major cytoplasmic components, including proteins and lipids, showed cyclic changes. The autofluorescence background of the Raman spectra also contained information about the differentiation dynamics. Using the entire information from the spectra, we were able to characterize the differentiation dynamics of the MCF-7 cells in multicomponent phase spaces.
Both the Raman and autofluorescence spectrum analyses revealed that in the major PC space, the distributions of cell population on each single day were wide, and the width of the distribution was sometimes larger than the distances between the averages for cells observed on different days of the differentiation pathway. These large fluctuations may be partly caused by the positional specificities of the spectra in single cells. In recent measurements, however, we observed similarly wide cell-to-cell fluctuations even after we averaged the spectra across 15 different positions in the cytoplasm of individual cells (S.T., S.M., Y.O., and Y.S., unpublished). Therefore, it is plausible that the cellular chemical components show large variations even among clonal cells under the same culture conditions. This result confirms the importance of single-cell measurements. There may be multiple tracks in the differentiation pathway in terms of the intracellular chemical components. It is at least highly likely that individual cells proceed along the differentiation pathway at various speeds. These effects might split and widen the distributions on a single day (Fig. 6 B). The rather small contribution ratios of the first three components (PC1–PC3) in the PCA reflect the large cell-to-cell and day-to-day variations in the Raman spectra.
Genome-wide studies of the changes in gene expression in MCF-7 cells in the early stage (until 72 h) of the HRG-induced differentiation pathway have been performed (28,29). Increases in the messenger RNA levels of some transcription factors, including c-FOS, EGR4, and FOSL1, began after only 1–2 h of stimulation with HRG. After 2–8 h of stimulation, other early transcription factors (FHL2 and DIPA) showed peak expression. These transcription factors induced transcription of immediate-early response genes mainly during 1–12 hr of stimulation. At 24 h, expression of the early transcription factors returned to prestimulation levels, and expression of a largely different set of genes compared with those observed during the first day was stimulated. If we assume a delay of 1–2 days from gene expression to metabolite synthesis and degradation, the switch in the gene-expression pattern observed between the first and second days of HRG stimulation seems to relate to the metabolite dynamics detected with Raman spectroscopy, which showed two peaks for the different components on day 1 and day 3 (Fig. 3 C). However, due to the highly multicomponent regulation of gene expression even with single transcription factors, along with the low resolution of Raman spectroscopy in the species of chemical compounds, it is difficult to correlate the dynamics of specific elements in the genome and spectroscopic analyses. We could not conduct genome analyses after 72 h.
We found that, like the Raman signals, the autofluorescence signals carried information about cell differentiation. The emission wavelength in our spectroscopic measurements was in the region of 550–590 nm. The major compounds of cells that show fluorescence emission in this region are FAD, NADH, and lipofuscin. FAD and NADH are involved in energy metabolism and the redox state of cells. Lipofuscin is a metabolite formed by the hyperoxidation of unsaturated lipids. Gene expression newly stimulated by HRG, which induces changes in metabolic pathway and oil drop formation, can cause dynamic changes in these fluorescent metabolites. A SIMCA analysis of the total photoexcited emission spectra showed increased diversity in the middle stages (days 3 and 6) of cell differentiation (Fig. 6 B). This diversification was not evident in the SIMCA analysis of the Raman spectra (Fig. S4), suggesting that it was mainly caused by differences in the energetic and/or redox states of individual cells (Fig. S5). Therefore, it is possible that cells can be in diverse states even when the proportions of their chemical components are similar.
In this study, we observed the dynamics of the differentiation pathway in a population of cells. In principle, the Raman and photoluminescence analyses described here can be extended to investigate the dynamics of cell differentiation and other fate-transition pathways in single cells. To that end, we must solve many problems inherent to long-term cell culture and measurements made under a microscope. Even if we could avoid photodamage during spectrum acquisitions by using low-light illumination and highly sensitive detectors, it would still be difficult to establish stable cell culture conditions and automatic cell tracking in the long term (e.g., for a week or more) under a high-resolution microscope to observe the cell differentiation pathway. If we can resolve these problems, we can then examine how single-cell fates diverge under the same environmental conditions and lead to individual and robust cell-fate decisions.
Acknowledgments
We thank Mariko Okada-Hatakeyama at RIKEN RCAI-IMS for helpful discussion.
Footnotes
Shin-ichi Morita and Sota Takanezawa contributed equally to this work.
Supporting Material
References
- 1.Johnston R.J., Jr., Desplan C. Stochastic mechanisms of cell fate specification that yield random or robust outcomes. Annu. Rev. Cell Dev. Biol. 2010;26:689–719. doi: 10.1146/annurev-cellbio-100109-104113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dean M., Fojo T., Bates S. Tumour stem cells and drug resistance. Nat. Rev. Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 3.Griffin J.L., Shockcor J.P. Metabolic profiles of cancer cells. Nat. Rev. Cancer. 2004;4:551–561. doi: 10.1038/nrc1390. [DOI] [PubMed] [Google Scholar]
- 4.Matthäus C., Bird B., Diem M. Chapter 10: Infrared and Raman microscopy in cell biology. Methods Cell Biol. 2008;89:275–308. doi: 10.1016/S0091-679X(08)00610-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Gelder J., De Gussem K., Moens L. Reference database of Raman spectra of biological molecules. J. Raman Spectrosc. 2007;38:1133–1147. [Google Scholar]
- 6.Caspers P.J., Lucassen G.W., Puppels G.J. Combined in vivo confocal Raman spectroscopy and confocal microscopy of human skin. Biophys. J. 2003;85:572–580. doi: 10.1016/S0006-3495(03)74501-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eikje N.S., Aizawa K., Ozaki Y. Vibrational spectroscopy for molecular characterisation and diagnosis of benign, premalignant and malignant skin tumours. Biotechnol. Annu. Rev. 2005;11:191–225. doi: 10.1016/S1387-2656(05)11006-0. [DOI] [PubMed] [Google Scholar]
- 8.Ohshima Y., Shinzawa H., Takenaka T., Fujita C., Sato H. Discrimination analysis of human lung cancer cells associated with histological type and malignancy using Raman spectroscopy. J. Biomed. Opt. 2010;15:017009. doi: 10.1117/1.3316296. [DOI] [PubMed] [Google Scholar]
- 9.Short K.W., Carpenter S., Mourant J.R. Raman spectroscopy detects biochemical changes due to proliferation in mammalian cell cultures. Biophys. J. 2005;88:4274–4288. doi: 10.1529/biophysj.103.038604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Notingher I., Bisson I., Hench L.L. In situ spectral monitoring of mRNA translation in embryonic stem cells during differentiation in vitro. Anal. Chem. 2004;76:3185–3193. doi: 10.1021/ac0498720. [DOI] [PubMed] [Google Scholar]
- 11.Schulze H.G., Konorov S.O., Turner R.F.B. Assessing differentiation status of human embryonic stem cells noninvasively using Raman microspectroscopy. Anal. Chem. 2010;82:5020–5027. doi: 10.1021/ac902697q. [DOI] [PubMed] [Google Scholar]
- 12.Pascut F.C., Goh H.T., Notingher I. Noninvasive detection and imaging of molecular markers in live cardiomyocytes derived from human embryonic stem cells. Biophys. J. 2011;100:251–259. doi: 10.1016/j.bpj.2010.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gentleman E., Swain R.J., Stevens M.M. Comparative materials differences revealed in engineered bone as a function of cell-specific differentiation. Nat. Mater. 2009;8:763–770. doi: 10.1038/nmat2505. [DOI] [PubMed] [Google Scholar]
- 14.Huang Y.-S., Karashima T., Hamaguchi H.O. Molecular-level investigation of the structure, transformation, and bioactivity of single living fission yeast cells by time- and space-resolved Raman spectroscopy. Biochemistry. 2005;44:10009–10019. doi: 10.1021/bi050179w. [DOI] [PubMed] [Google Scholar]
- 15.Berezin M.Y., Achilefu S. Fluorescence lifetime measurements and biological imaging. Chem. Rev. 2010;110:2641–2684. doi: 10.1021/cr900343z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mori H., Borowsky A.D., Bissell M.J. Laser scanning-based tissue autofluorescence/fluorescence imaging (LS-TAFI), a new technique for analysis of microanatomy in whole-mount tissues. Am. J. Pathol. 2012;180:2249–2256. doi: 10.1016/j.ajpath.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McWilliams A., MacAulay C., Lam S. Innovative molecular and imaging approaches for the detection of lung cancer and its precursor lesions. Oncogene. 2002;21:6949–6959. doi: 10.1038/sj.onc.1205831. [DOI] [PubMed] [Google Scholar]
- 18.Reinert K.C., Gao W., Ebner T.J. Flavoprotein autofluorescence imaging in the cerebellar cortex in vivo. J. Neurosci. Res. 2007;85:3221–3232. doi: 10.1002/jnr.21348. [DOI] [PubMed] [Google Scholar]
- 19.Skala M.C., Riching K.M., Ramanujam N. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc. Natl. Acad. Sci. USA. 2007;104:19494–19499. doi: 10.1073/pnas.0708425104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burden S., Yarden Y. Neuregulins and their receptors: a versatile signaling module in organogenesis and oncogenesis. Neuron. 1997;18:847–855. doi: 10.1016/s0896-6273(00)80324-4. [DOI] [PubMed] [Google Scholar]
- 21.Nagashima T., Shimodaira H., Hatakeyama M. Quantitative transcriptional control of ErbB receptor signaling undergoes graded to biphasic response for cell differentiation. J. Biol. Chem. 2007;282:4045–4056. doi: 10.1074/jbc.M608653200. [DOI] [PubMed] [Google Scholar]
- 22.Rösch P., Harz M., Popp J. Chemotaxonomic identification of single bacteria by micro-Raman spectroscopy: application to clean-room-relevant biological contaminations. Appl. Environ. Microbiol. 2005;71:1626–1637. doi: 10.1128/AEM.71.3.1626-1637.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogawa M., Harada Y., Takamatsu T. Label-free biochemical imaging of heart tissue with high-speed spontaneous Raman microscopy. Biochem. Biophys. Res. Commun. 2009;382:370–374. doi: 10.1016/j.bbrc.2009.03.028. [DOI] [PubMed] [Google Scholar]
- 24.Parker F.S. Plenum; New York: 1983. Applications of Infrared, Raman, and Resonance Raman Spectroscopy in Biochemistry. [Google Scholar]
- 25.Hu S., Morris I.K., Spiro T.G. Complete assignment of cytochrome c resonance Raman spectra via enzymatic reconstitution with isotopically labeled heme. J. Am. Chem. Soc. 1993;115:12446–12458. [Google Scholar]
- 26.Takai Y., Masuko T., Takeuchi H. Lipid structure of cytotoxic granules in living human killer T lymphocytes studied by Raman microspectroscopy. Biochim. Biophys. Acta. 1997;1335:199–208. doi: 10.1016/s0304-4165(96)00138-9. [DOI] [PubMed] [Google Scholar]
- 27.Wold S. Pattern recognition by means of disjoint principal components models. Pattern Recognit. 1976;8:127–130. [Google Scholar]
- 28.Saeki Y., Endo T., Ide K., Nagashima T., Yumono N., Toyoda T., Suzuki H., Hayashizaki Y., Sakaki Y., Okada-Hatakeyama M. Ligand-specific sequential regulation of transcription factors for differentiation of MCF-7 cells. BMC Genomics. 2009;10:545. doi: 10.1186/1471-2164-10-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oyama M., Nagashima T., Okada-Hatakeyama M. Integrated quantitative analysis of the phosphoproteome and transcriptome in tamoxifen-resistant breast cancer. J. Biol. Chem. 2011;286:818–829. doi: 10.1074/jbc.M110.156877. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


