Abstract
Wnt4 gene plays a role in developmental processes in mammals. However, little is known regarding its function in teleosts. We cloned and characterized the full-length half-smooth tongue sole (Cynoglossus semilaevis) wnt4a gene (CS-wnt4a). CS-wnt4a cDNA was 1746 bp in length encoding 353aa. CS-wnt4a expression level was highest in the testis, and gradually increased in the developing gonads until 1 year of age. In situ hybridization revealed that CS-wnt4a expression level was highest in stage II oocytes and sperm in the adult ovary and testis, respectively. CS-wnt4a expression level was significantly up-regulated in the gonads after exposure to high temperature. The level of methylation of the CS-wnt4a first exon was negatively correlated with the expression of CS-wnt4a. The branch-site model suggested that vertebrate wnt4a differed significantly from that of wnt4b, and that the selective pressures differed between ancestral aquatic and terrestrial organisms. Two positively selected sites were found in the ancestral lineages of teleost fish, but none in the ancestral lineages of mammals. One positively selected site was located on the α-helices of the 3D structure, the other on the random coil. Our results are of value for further study of the function of wnt4 and the mechanism of selection.
Wnt4 (wingless-type MMTV integration site family, member 4) plays a crucial role in ovarian differentiation in mammals. In mice, wnt4 is expressed in the mesonephors, the gonad, and the Müllerian duct, suggesting that wnt4 plays an important role in development of the reproductive system1. The absence of wnt4 in female embryos (XX) leads to masculinization2. However, over-expression of wnt4 does not result in sex reversal in males3. The phenotype of human female that have a wnt4 mutation is similar to that of female wnt4 knockout mice4. Individuals who have a duplication of the chromosome containing the wnt4 gene exhibit sex reversal5,6. In addition to humans and mice, ovarian wnt4 expression has been reported in a range of mammals, including the bonnet macaque (Macaca radiata)7, tammar wallaby (Macropus eugenii)8, and goats (Capraaegagrus hircus)9. These studies have focused primarily on the role of wnt4 in ovarian differentiation. In contrast, little is known about the potential role in testis development. In mice, Sertoli cell differentiation is compromised in wnt4 mutant testes. In this instance, the expression of the testis-determining gene Sry was down-regulation, whereas expression of the Sertoli cell marker genes, Sox9 and Dhh, was up-regulated10. These observations suggest that wnt4 is involved in mammalian testis determination. In mice that have transgenic mis-expression of wnt4, Leydig cell differentiation is not affected but migration of the steroidogenic adrenal precursors into the gonad is repressed3. Similarly, transgenic mice that have a human wnt4 gene exhibit a dramatic reduction in steroidogenic acute regulatory protein expression and testicular androgen levels11. These observations suggest that wnt4 acts as an anti-male factor12. Taken together, the studies described above provide evidence that wnt4 plays an important role in gonad development. Wnt4 function has also been evaluated in other organisms, including the millipedes Glomeris marginata (Myriapoda: Diplopoda)13, frogs (Rana rugosa)14, garden lizard (Calotes versicolor)15 and chickens (Gallus gallus)16. In teleosts, the role of wnt4 in gonad development has only been studied in rainbow trout (Oncorhynchus mykiss)17, and the protandrous black porgy (Acanthopagrus schlegeli)18. Interestingly, in most teleosts, the wnt4 gene was duplicated due to the teleost-specific whole-genome duplication17. This second duplication resulted in two paralogs, wnt4a and wnt4b. Additionally, a third wnt4 gene, wnt4a2, has been documented in some teleost species17. The two wnt4a genes (wnt4a1 and wnt4a2) exhibit dimorphic expression in the gonad. This can be explained in two ways. Gene duplication may have lead to new function for the gene (neo-functionalization) which may be different from the paralogs's function19,20,21. Alternatively, genome duplication can lead to a high rate of molecular evolution in fish, after which the duplicated gene causes functional division of the tasks22.
Half-smooth tongue sole (Cynoglossus semilaevis) is an economically important farmed marine fish that are widely distributed along the Chinese coast. Because of their desirable taste and nutritional and economic value, it was selected for aquaculture. Half-smooth tongue sole exhibit significant sexual dimorphism in growth whereby the female grows faster than the male23. Thus, breeding all female or a high proportion female stock has been proposed as a way to increase production. To achieve this, there is a need to better understand the mechanism of sex determination and sex differentiation. In the previous research, we demonstrated that exposure of genetic female half-smooth tongue sole to high temperature resulted in a phenotypic male with the female genotype (neomale: ZW)24. Interestingly, methylation appears to play an important role in sex determination and sex reversal25. Additionally, we have measured the expression of a number of sex related genes, including Foxl2, Cyp19a1, Sox9, Ubc9, and Dmrt1 during gonadal development26,27,28,29,30. Furthermore, the whole genomic sequence of the half-smooth tongue sole was sequenced30. Comparison of the tongue sole sex chromosomes with those of mammals and birds during the early phase of sex chromosome evolution has revealed the loss of large numbers of genes in tongue sole during sex chromosome evolution30. Despite the availability of the whole genomic sequence, the mechanism of its sex determination remains unclear.
The role of the wnt4 gene in half-smooth tongue sole, particularly in sex determination and gonad differentiation, remains uncharacterized. Our objective was to identify and characterize the role of the CS-wnt4a in sex determination and gonadal differentiation. We measured the pattern of tissue expression and the temporal pattern of expression in the gonads. Additionally, we evaluated the correlation between promoter methylation and CS-wnt4a expression. We found evidence for positive selection of wnt4 genes in the various groups: teleosts, mammals and invertebrates.
Results
cDNA characterization of half-smooth tongue sole CS-wnt4a
We obtained the full length CS-wnt4a gene following RACE primer amplification (Table S1). The complete cDNA sequence of CS-wnt4a was 1746 nucleotides in length (GenBank accession No: KJ825677), consisting of a 5′-untranslated region (UTR) of 435 bp and a 3′UTR of 252 bp. The open reading frame (ORF) was 1059 bp in length and encoded a protein of 353 amino acids with a predicted molecular weight of 39.38 kDa. The position of the signal peptide was predicted and the signal cleavage site was identified between Ala22 and Ser23 (Fig. S1). A typical WNT domain (containing 308 amino acids) was identified in CS-wnt4a gene. Additionally, twenty glycosylation sites (Thr-2, Asn-21, Ser-23, Asn-24, Ser-60, Asn-88, Thr-103, Ser-116, Ser-146, Ser-168, Ser-181, Ser-182, Asn-187, Asn-191, Ser-200, Thr-219, Asn-232, Thr-252, Thr-286, Asn-298, and Thr-350) were predicted using bio-informatics (Fig. S1).
Tissue distribution and temporal expression in the gonads
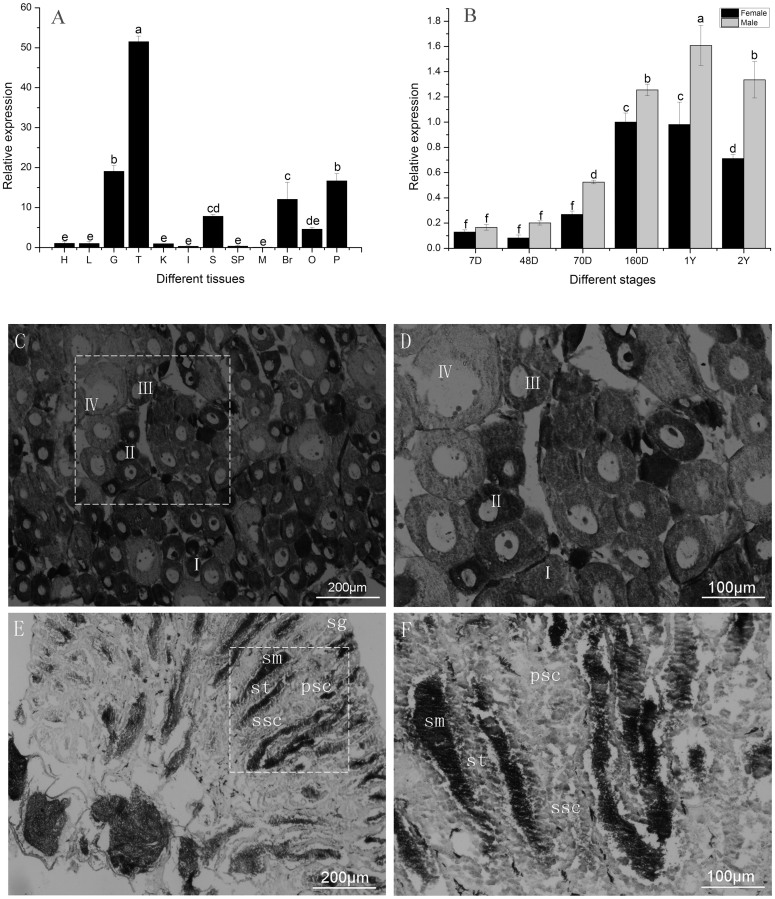
We measured mRNA levels in 13 tissues and throughout development in the gonads of the half-smooth tongue sole. The level of expression differed significantly among the tissues (p < 0.05). The level of expression was high in the testis (T), moderate in the gill (G), skin (S), brain (B), pituitary (P), and ovary (O), and low in the remaining tissues (Fig. 1A).
Figure 1. The expression of CS-wnt4a in C.semilaevis.
(A). The expression of CS-wnt4a in different tissues H: heart, L: liver, G: gill, S: skin, K: kidney, I: intestine, Br: brain, Sp: spleen, M: muscle, P: pituitary, O: ovary, T: testis. (B). The expression profile of CS-wnt4a at different developmental stages in the gonads was revealed by real-time quantitative PCR. 7D: 7 day age, 48D: 48 day age, 70D: 70 day age, 160D: 160 day age, 1Y: 1 year age, 2Y: 2 year age. (C) Low magnification showing the adult ovary architecture including four stages of oocytes by in situ hybridization: stage I–IV. (D) Large magnification of frame area in (C). (E) Low magnification showing the adult testis architecture including spermatogonaia (sg), primary spermatocytes (psc), secondary spermatocytes (ssc), spermatids (st), and sperm (sm) by in situ hybridization. (F) Large magnification of frame area in (E) highlighting the distribution of CS-wnt4a RNA.
To measure the expression of CS-wnt4a genes during the period of gonad differentiation, CS-wnt4a expression was detected in female and male gonads at different stages by qRT-PCR. The level of expression was weak from 7 d (7D) to 48 d (48D) after hatch, then increased rapidly at 70D and continued to increase gradually up to 1 year (1Y). The level of expression was highest at 1Y, after which there was a rapid decrease at 2 years (2Y) (Fig 1B). There was no difference in CS-wnt4a expression between female gonads and male gonads from 7D to 160D. However, expression was significantly higher in male gonads than in female gonads from 1–2Y (p < 0.05).
The location of CS-wnt4a expression in the gonad was determined using in situ hybridization (ISH) in adult tongue sole. The adult tongue sole ovary contains somatic cell and several different developmental stages of oocytes from small (stage I) to mature oocytes (stage IV). ISH revealed that CS-want4a expression was strong in stage I–III oocytes, but weak in stage IV oocytes (Fig. 1C). The strongest signal was observed at stage II (Fig. 1D). The distribution of CS-wnt4a RNA expression was uniform in stages I–III oocytes, and perinuclear in stage IV oocytes (Fig. 1D). The adult tongue sole testis contain spermatogonaia (sg), primary spermatocytes (psc), secondary spermatocytes (ssc), spermatids (st), and sperm (sm). The transcription of CS-wnt4a was detected in sg, psc, ssc, st, and sm. The signal was strong in sm, moderate in ssc and st, and weak in sg and psc (Fig. 1E and F). We did not detect any signal from the sense probe (Fig. S2).
Expression and methylation during sex reversal
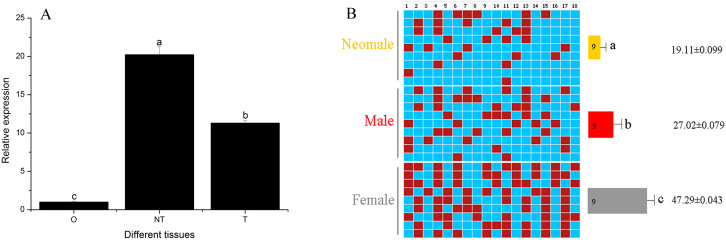
We measured CS-wnt4a expression in the testis and ovary of normal males and females and the testis of neomales. CS-wnt4a expression was high in the testis of neomales, moderate in the testis of normal males, and low in the ovary of normal females (Fig. 2A).
Figure 2.
(A). Expression analysis of CS-wnt4a by relative quantitative real-time PCR in gonads of females, males, and neomales in 1-year-old individuals. O: ovary, T: testis, NT: neomale testis. Bars with different letters are significantly different (p < 0.05). (B). Differences in CS-wnt4a first exon methylation in the gonads of females, males, and neomales at 1 year of age. Numbers in the first row indicate CpG positions. The red box and blue box denote methylated and unmethylated positions, respectively. The number inside the bar indicates sample size. Results represent the mean ± SE. Groups with different letters are significantly different (P < 0.05).
To test the relationship between methylation and expression, the first exon of CS-wnt4a was examined and the 298 bp CpG dinucleotide was selected (Fig. S1). The level of gonad methylation in the CS-wnt4a first exon was determined using bisulfite sequencing in one-year-old half-smooth tongue sole males, females, and neomales. There was a significant difference in average DNA methylation levels in the half-smooth tongue sole CS-wnt4a first exon among these three groups. The level of CS-wnt4a methylation was twice as high in females as in sex reversed tongue sole (mean ± SEM: 47.29 ± 0.043% versus 19.11 ± 0.099%, Fig. S3). In addition, the levels were significantly higher in females than in males (27.02 ± 0.079%) (Fig. 2B). Interestingly, the level of RNA transcription was negatively correlated with the level of methylation suggesting that methylation of the CS-wnt4a first exon inhibits RNA transcription.
Multiple alignment and phylogenetic analysis
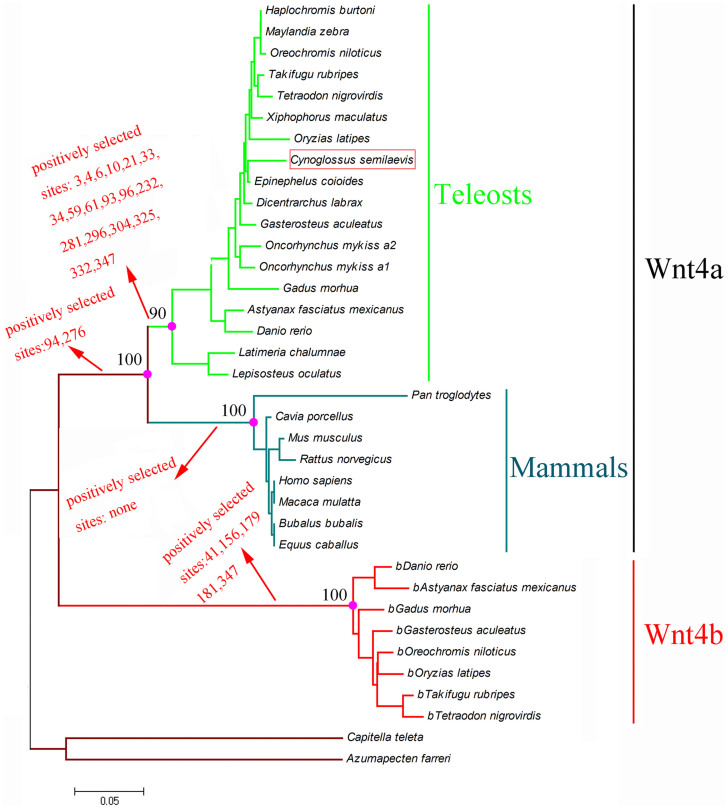
After a comprehensive search in GenBank and Ensemble, we downloaded 36 sequences of wnt4 in 28 species, including Takifugu rubripes, Gasterosteus aculeatus, Oreochromis niloticus, Tetraodon nigrovirdis, Haplochromis burtoni, Maylandia zebra, Dicentrarchus labrax, Oncorhynchus mykiss, Xiphophorus maculatus, Oryzias latipes, Gadus morhua, Danio rerio, Rattus norvegicus, Homo sapiens, and Macaca mulatta (Table S2). The protein sequences were aligned (Fig. S4) in Clustalx to produce the phylogenetic tree of the wnt4 genes using the Bayesian method. To evaluate clade support, we used the Bayesian posterior probability (PP) method. The sequences clustered in two main groups: a wnt4a cluster containing teleosts (pp = 0.90) and mammals (pp = 1.00), and a wnt4b cluster that contained only teleosts (Fig. 3).
Figure 3. Phylogenetic tree of the nucleotide sequence of wnt4 reconstructed by the Bayesian approach.
Molecular evolution analysis
To analyze whether there were positively selected sites in the ancestral lineages of the wnt4 gene tree, all vertebrate wnt4 sequences were utilized to test whether different environments had affected the selective pressure on wnt4 genes in ancestral lineages of aquatic or terrestrial organisms. Firstly, the ω for all branches was 0.036 (p = 0, Table 1), suggesting that all wnt4 genes were under purifying selection. Secondly, comparison of the free-ratio model with the one ratio model revealed that each branch has independent ω values (p = 5.93715E-07). Lastly, the branch-site model was applied to determine whether a positive selection site was existed in the lineages of vertebrate, wnt4a, wnt4b, mammalian and teleost. We found evidence for positive selection in wnt4a, wnt4b and teleosts (Table 1). A site model was then used to identify the positive selection site in the wnt4 gene of mammals and teleosts. In mammals, the likelihood ratio test (LRT) value was obtained from comparison of the two models. The M3-M0 comparison was 107.74 (p < 0.01 Table 2). The M2a-M1a and M7–M8 comparison was 0 (p = 1), and M8 and M2a were rejected. A positive selection site was not found in mammals. In teleosts, 4 and 2 positively selected sites were found in M3-M0 and M8-M7, respectively, with LRT values of 212.94 (p < 0.01) and 20.48 (p < 0.01). Conversely, no positively selected site was found in the M2a-M1a comparison and the LRT value was 0 (p = 1) (Table 2).
Table 1. Likelihood ratio test (LRT) of branch-model and branch-site-models for wnt4a genes.
| Model | Npa | Ln likelihood | Parameter estimates | Model compare | Positive Selection site | 2ΔlnL(p-value)b |
|---|---|---|---|---|---|---|
| Branch model | ||||||
| 1.One-ratio | 71 | −14470.28 | ω = 0.036 | |||
| 2.Omega = 1 | 70 | −17334.08 | ω = 1 | 2vs1 | 5727.6(p = 0) | |
| 3.Free-ratio | 74 | −14486.15 | variable v by branch | 3vs1 | 31.74(p = 5.93715E-07) | |
| Branch-site model | ||||||
| 4:Null b | 73 | −14417.88 | ||||
| 5:b | 74 | −14415.45 | 3vs4 | 41,156**,179,181,347 | 4.86(p = 0.027) | |
| 6:Null-a | 73 | −14412.85 | ||||
| 7:a | 74 | −14406.24 | 5vs6 | 94,276 | 13.22(p = 0.000277) | |
| 8:Null-Mamc | 73 | −14419.26 | ||||
| 9:Mam | 74 | −14417.72 | 7vs8 | Not allowed | 3.08(p = 0.079) | |
| 10:Null-Teld | 73 | −14419.26 | ||||
| 11:Tel | 74 | −14406.53 | 10vs9 | 3,4*,6,10,21*,33,34,59,61,93,96,232,281,296,304,325,332,347 | 25.46(p = 4.51651E-07) |
aNumber of parameters.
bTwice the difference of ln [likelihood] between the two models compared.
cMam = Ancestor branch of the mammals examined in present study.
dTel = Ancestor branch of the Teleosts examined in present study.
The posterior probabilities p > 0.95 and p > 0.99 are indicated by * and **, respectively.
Table 2. Site model tests on wnt4a genes in mammalian and fish.
| Model | Npa | Ln likelihood | Parameter estimates | Model compare | Positive Selection site | 2ΔlnL(p-value)b |
|---|---|---|---|---|---|---|
| Mammalian | ||||||
| M0:one-ratio | 15 | −3063.91 | ω = 0.056 | |||
| M3:discrete | 19 | −3010.04 | ω0 = 0.000, p0 = 0.213 | M3vsM0 | Not found | 107.74(p = 2.2075E-22) |
| ω1 = 0.000, p1 = 0.586 | ||||||
| ω2 = 0.339, p2 = 0.201 | ||||||
| M1a:nearly neutral | 16 | −3026.45 | ω0 = 0.020, p0 = 0.909 | |||
| ω1 = 1.000, p1 = 0.090 | ||||||
| M2a:positive selection | 18 | −3026.44 | ω0 = 0.020, p0 = 0.909 | M2vsM1 | Not allowed | 0 (p = 1) |
| ω1 = 1.000, p1 = 0.070 | ||||||
| ω2 = 1.000, p2 = 0.020 | ||||||
| M7:β | 16 | −3011.21 | p = 0.091, q = 1.168 | |||
| M8:β and ω | 18 | −3011.14 | p0 = 0.994, p1 = 0.006 | M8vsM7 | Not allowed | 0 (p = 1) |
| ω = 1.000, p = 0.094, | ||||||
| q = 1.306 | ||||||
| Teleosts | ||||||
| M0:one-ratio | 31 | −5834.91 | ω = 0.033 | |||
| M3:discrete | 35 | −5728.44 | ω0 = 0.005, p0 = 0.771 | M3vsM0 | 5,7,39**,94** | 212.94(p = 6.193E-45) |
| ω1 = 0.123, p1 = 0.22 | ||||||
| ω2 = 1.075, p2 = 0.009 | ||||||
| M1a:nearly neutral | 32 | −5779.92 | ω0 = 0.026,p0 = 0.978 | |||
| ω1 = 1.000,p1 = 0.022 | ||||||
| M2a:positive selection | 34 | −5779.92 | ω0 = 0.026, p0 = 0.978 | M2vsM1 | Not allowed | 0 (p = 1) |
| ω1 = 1.000, p1 = 0.022 | ||||||
| ω2 = 40.005, p2 = 0.00 | ||||||
| M7:β | 32 | −5739.53 | p = 0.18, q = 3.84 | |||
| M8:β and ω | 34 | −5729.29 | p0 = 0.993, p1 = 0.007 | M8vsM7 | 39,94 | 20.48(p = 3.5712E-5) |
| ω = 1.291, p = 0.214 | ||||||
| q = 5.962 |
aNumber of parameters.
bTwice the difference of ln [likelihood] between the two models compared.
The posterior probabilities p > 0.95 and p > 0.99 are indicated by * and **, respectively.
Structure analysis and positive selection sites on the three dimensional structure
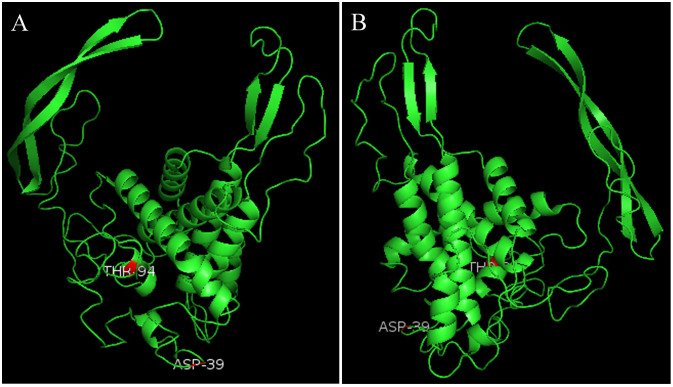
To map positively selected sites onto CS-wnt4a 3D model and present the results of our study more visualized, two positively selected sites were identified and mapped onto the surface of the 3D structure (Fig. 4). The 3D structure contained 8 α-helices and 4β-strands, and one positively selected site (94T) on the α-helices, the other one (39D) on the random coil.
Figure 4. Three dimensional models of CS-wnt4a gene based on the tongue sole sequences.
Images depicted represent two perspectives (the angles of view are 0°and 180°).Shown is the positive selection sites (red). 94T positive selection sites located in the α-helices; 39D located in the random coils.
Discussion
Wnt4 genes are involved in many important developmental processes in vertebrates, including reproduction, sex determination, and sex inversion13,31,32. New functions for wnt4 genes have been found in mammals, fishes, and other vertebrates33,34,35. Wnt4 genes have been described in some teleosts, though few species have been comprehensively and systematically studied. We cloned CS-wnt4a, and then evaluated the pattern of expression and methylation. The open reading frame shared many characteristic features with wnt4a from other vertebrates. The putative conservative domain of CS-wnt4a possessed high identity with wnt4a from other species (Fig. S4). CS-wnt4a was divided into two regions, a signal peptide and a mature peptide. Additionally, twenty glycosylation sites were found in the CS-wnt4a putative amino acid peptide. The number of glycosylation sites is positively associated with the diversity of protein function36,37, suggesting that the wnt4a gene has a variety of biological functions.
Wnt4 gene is found in both invertebrates14,38,39 and vertebrates11,40,41. Our results demonstrated that most teleost fish have two wnt4 genes, wnt4a and wnt4b, whereas invertebrates and other vertebrates possess only a single wnt4 gene. This suggested that teleost fish underwent a whole-genome duplication that yielded two wnt4 paralogs42. The relationship of a duplicated wnt4 gene (wnt4b), which formed a cluster on one branch with mammals (Fig. 4), was not supported by the phylogenetic analysis. This inconsistent phenomenon could be explained by a long-branch attraction effect due to the divergence of wnt4b sequences43. A high rate of molecular evolution has occurred during genome duplication in fish such that duplicate genes are often associated with a functional shift24. An additional duplication of wnt4 occurred in some teleost species resulting in two copies of the wnt4a gene, wnt4a1 and wnt4a217.
In half-smooth tongue sole, wnt4a was primarily expressed in the gonads, gill, and brain. This is consistent with observations in other teleost fish, including zebrafish44 and rainbow trout17. Similarly, wnt4 is expressed at a high level in the testis and moderate level in the ovary of the tammar wallaby45. The level of CS-wnt4a expression increased significantly at day 70 then increased gradually up to 1 year. The pattern of expression was consistent with the developmental process in the gonads. For example, proliferation is most rapid in the ovary at 62 days and spermatogonial cells begin mitosis at 80 days. Additionally, the level of CS-wnt4a expression was significantly higher in the testis than in the ovary at both 1 and 2 years of age (p < 0.05). In mice, steroidogenesis occurs earlier in male embryonic gonads than in female embryonic gonads. Steroidogenic cell recruitment and steroidogenic gene expression appears to be inhibited by elevated expression of wnt4a in the future ovary3,46. However, in half-smooth tongue sole, steroidogenesis begins earlier in female embryonic gonads than in male embryonic gonads47. So, we speculated that the over-expression of CS-wnt4a in male half-smooth tongue sole gonads might cause a delay of steroidogenesis in males. A similar phenomenon was reported in rainbow trout11. Therefore, we speculated that CS-wnt4a expression was associated with the gonad differentiation in the half-smooth tongue sole. Indeed, the role of wnt4 in gonad differentiation has been demonstrated in the frog (Rana rugosa)16. black porgy (Acanthopagrus schlegeli)18, and rainbow trout (Oncorhynchus mykiss)17.
In situ hybridization of the adult ovary revealed that CS-wnt4a was primarily expressed in the oocytes. The strongest signal was observed in stage II oocytes. CS-wnt4a expression increased gradually from stage I and then decreased after expression peaked during stage II (Fig. 1). In the testis, CS-wnt4a expression was identified during all stages of spermatogenesis. The signal strength gradually increased during spermatogenesis and peaked in the mature sperm. These observations were consistent with the pattern of CS-wnt4a expression determined by qRT-PCR.
To evaluate whether CS-wnt4a played a role in sex reversal, we measured the level of CS-wnt4a transcripts in the gonads of females, males, and neomales. The expression was significantly (p < 0.05) higher in neomales than in males and females, the latter of which had low levels of expression. In mice, loss of wnt4 is sufficient to up-regulate Fgf9 and results in partial testis development in XX gonads33. In humans, wnt4 over-expression leads to up-regulated of DAX1, which results in an XY female phenotype48. Gonadal expression of wnt4a is higher in males than in females in rainbow trout17, marsupials45, and tongue sole, which is in contrast to the pattern during gonadal development in mice and humans. Thus, wnt4a over-expression is associated with sex reversal.
To evaluate the mechanism of differential expression in gonads of males, females, and neomales, we measured the level of methylation in the first exon of CS-wnt4a in tongue sole. The level was highest in the gonads of females, moderate in males, and lowest in neomales. The level of methylation was negatively correlated with CS-wnt4a expression suggesting that methylation of the CS-wnt4a first exon was associated with the CS-wnt4a expression. In many species, DNA methylation inhibits expression of the target gene. For example, DNA methylation of GATAD1 in humans decreases expression of GATAD148. In European sea bass, DNA methylation of the Cyp19a1 promoter decreases expression and induces masculinization49. In Japanese flounder, DNA methylation of Dmrt1 and the Cyp19a1 promoter results in sexual dimorphism50.
We constructed a phylogenetic tree based on the nucleotide sequences from 36 taxa to analyze the evolutionary context of CS-wnt4a. CS-wnt4a was genetically related to those of Epinephelus coioides (Fig. 3). The results of the phylogeny analysis were consistent with the highest amino acid sequence identity in the homolog comparison. Mammalian and teleost wnt4a genes formed one branch and the teleost wnt4b gene formed another branch. We assumed that genome duplication in teleosts resulted in two wnt4 genes and the wnt4 subgroup likely originated very early during metazoan evolution17. Additionally, the high ratio of molecular evolution for many genes following the genome duplications in fish has resulted in functional shifts22. Therefore, wnt4b and wnt4a did not cluster together on a single branch. Our phylogenetic tree was similar to that constructed for rainbow trout17.
To study the molecular evolution of the wnt4 sequences, we constructed a series of models. The phylogenetic tree was also used to detect positive selection of wnt4 sequences within the ancestral lineage of vertebrates, mammals, and teleosts. If the ratio of the nonsynonymous nucleotide difference (dN) to the synonymous nucleotide difference (dS) between sequences was significantly greater than one, then positive selection was involved51. If the ratio of dN/dS(ω) was significantly less than one, then purity selection is inferred. We compared the one-ratio model with the one-ratio model (ω = 1). The wnt4 gene was strongly conserved and underwent purity selection (p < 0.01, Table 1). In general, most genes undergo purity selection whereas rare genes undergo position selection during evolution52. Subsequently, the free-ratio model was used to analyze the ratios among the different branches, revealing that each branch had independent dN/dS (ω). Additionally, a number of positive selection sites were detected in the different groups using the branch-site model. For example, we found 2 and 5 positively selected sites in wnt4a and wnt4b, respectively, in the ancestor vertebrate. Genome duplication occurred 320 million years ago in teleost fish, and the high rate of molecular evolution for many genes following genome duplication drove the functional shift24,43. We detected 18 positively selected sites in teleosts for the wnt4a gene. Rensch noted that positive selection is associated with a functional and environmental shift53. Thus, the positively selected sites in mammals and teleosts may be associated with occupation of different habitats, changes in oxygen consumption, or sex differentiation.
Additionally, we used a site model to detect positive selection. We detected 2 positively selected sites in teleosts, but none in mammals. Water represents a relative stable environment, thus selection pressure is thought to be weaker than in terrestrial environments and the rate of molecular evolution should be correspondingly lower53. However, our observations did not support this, likely because genome duplication resulted in two wnt4 genes in teleosts (wnt4a and wnt4b), and the duplicate gene had a high rate of molecular evolution. Additionally, the wnt4 gene is primarily expressed in the gonads, with expression being higher in the ovary than the testis in mammalian. In teleosts, wnt4 gene RNA transcription is also higher in the ovary than in the testis in normal fish. However, the pattern is reversed in partial fish. As exampled, Rainbow trout17, Zebrafish44 and Tongue sole. Thus, the functional diversity in fish may be a result of wnt4 gene evolution. As we known, positive selection plays a role in the diversification of protein function. Thus, we mapped the positively selected sites on the 3D structure to assess their biological significance. We mapped one positively selected site on the α-helices and the other positively selected sites on the random coils. The two positively selected sites were on the outside region of the 3D structure. This indicated that outside region may be necessary for protein function.
Teleosts occupy water of a wide range of salinities, which can have an effect on reproduction54,55,56. We speculated that as part of the adaptation to freshwater, the ancestral wnt4 gene may have shifted its function by molecular evolution. Our observations of the positively selected wnt4a gene sites may be used to analyze genome duplication and the reproduction system in tongues sole.
Methods
Fish sampling and exposure to high temperature
Healthy tongue sole were collected from the Haiyang 863 High-Tech Experiment Base (Haiyang, China). The tongue sole were acclimated to laboratory culture for at least one week. The fish were fed twice daily with commercial pellets. To determine the role of the CS-wnt4a gene in sex reversal, the tongue sole were treated at 28°C on day 23–30 after hatching until day 100, then reared to adulthood at a natural water temperature (22°C)29.To determine the pattern of CS-wnt4a tissue expression, samples of one year old age were collected from the heart, liver, gill, skin, blood, kidney, intestine, brain, spleen, muscle, pituitary, ovary, and testis. Additionally, gonad samples were collected from healthy tongue sole throughout the rearing period (70D, 160D, 1Y, 2Y). The samples were immediately frozen in liquid nitrogen and then stored at −80°C until RNA extraction. To visualize the location of RNA transcription in gonad cells, the gonad was fixed in 4% parafarmaldehyde (PFA) and subject to in situ hybridization (ISH).
Genetic sex and phenotypic sex
To determine the genetic sex, genomic DNA was extracted from the fin samples of tongue sole using a standard phenol-chloroform method. The gonads were collected and fixed in Bouin's solution. The genetic sex and phenotype sex was identified followed Deng's description27.
PCR amplification and cloning
To obtain the full length cDNA sequence, one pair of primers (Wnt4a-5′GSP and Wnt4a-3′GSP) was designed according to the partial sequence which was obtained from our laboratory transcriptome data30. The PCR reaction mixture (50 μl) was prepared according to the manufacturer's instructions using a BD SMART™ RACE cDNA Amplification kit. The PCR reactions were performed under the following conditions: 5 cycles of 94°C for 30 s and 72°C for 3 min, 5 cycles of 94°C for 30 s, 70°C for 30 s, and 72°C for 3 min, then 27 cycles of 94°C for 30 s, 68°C for 30 s, and 72°C for 3 min. All of the amplification products were run on a 1% agarose gel using the D2000 marker (Takara, Dalian, China). The purified fragment was cloned into a PMD-18-T vector, inserted into E. coli TOP10, and then sequenced.
CS-wnt4a expression in tongue sole and localization in gonadal cells
Samples from three individuals were mixed for RNA preparation. RNA was extracted from the various tissues, from the gonads collected at different stages, and from sex reversed gonads using Trizol (Qiagen, Düsseldorf, Germany) following the manufacturer's instruction. cDNA synthesis was performed using a QuantScript RT kit (Takara, Dalian, China) according to the manufacturer's protocol. The real-time RT-PCR primers were designed based on the CS-wnt4a gene sequence (Table S1). Real-time RT-PCR was performed on a 7500 real-time PCR system (Applied Biosystems, California, USA) using 10 ml SYBR Premix Ex Taq (Takara, Dalian, China), 0.4 ml of Wnt4a-S1 and Wnt4a-A1, 1 ml cDNA, and 0.4 ml ROX reference dye following the manufacturer's protocol. PCR amplification was performed in triplicate wells and the conditions were as follows: 10 s at 95°C, followed 40 cycles at 95°C for 5 s and 60°C for 34 s. A disassociation curve analysis was performed to determine target specificity. β-Actin was used as the internal control. The standard curve was performed at three dilutions of the template (10, 100, and 1000 times). The concentration of cDNA in each sample was reflected by the crossing point (Ct) values, which were compared using the relative quantification method and 7500 system SDS software (Applied Biosystems, California, USA). The data were log-transformed. The transformed data were analyzed by one-way ANOVA followed by Duncan multiple comparison tests using SPSS 17.0 (IBM, New York, NY, USA). Significance was set at p < 0.05. To locate CS-wnt4a expression in the gonad cell, one pair of primers (Table S1) was designed for in situ hybridization (ISH) following the method described in Bhat57.
Analysis of CS-wnt4a first exon methylation
The majority of gonadal DNA methylation in half-smooth tongue sole occurred at the first exon of the gene25. The CS-wnt4a gene structure was determined using the complete cDNA sequence and the whole genome sequence30. A methylation primer was designed using online software http://www.urogene.org/methprimer/index1.html. To account for individual differences, we collected the gonads of eight females, eight males, and eight neomales for methylation analysis. The DNA was extracted and the concentration was quantified. Then, equimolar gonadal DNA from each individual was mixed in the same group. The mixed DNA was treated using a DNA methylation kit (Zymo, California, USA) and subject to PCR amplification following the manufacturer's protocol with the treated DNA as the reaction template. The purified fragment was cloned into a PMD-18-T vector and conveyed into E. coli TOP10. We then sequenced nine positive clones from each group.
Sequences analysis, alignment, and phylogenetic analysis
The wnt4 cDNA sequences were obtained from the GenBank (http://www.ncbi.nlm.nih.gov/) and Ensemble (http://www.ensembl.org/index.html) databases, including 34 vertebrates and 2 invertebrates as the outgroup (Table S2). The signal was predicted by the online software http://www.cbs.dtu.dk/services/SignalP/, the protein conserved domain was identified by http://www.ncbi.nlm.nih.gov/cdd/, glycosylation sites were determined using http://www.cbs.dtu.dk/services/NetOGlyc/, and the methylation island was determined using http://www.urogene.org/methprimer/. The online software (http://www.ebi.ac.uk/Tools/msa/muscle/) was used to align the putative amino acid sequences. The jmodeltest 2 software was used to assess the optimal model for the nucleotide sequences under the Bayesian Information Criterion (BIC). The GTR+I+G model was selected as the best model to structure the phylogenetic tree using the Bayesian inference in MrBayes 3.1 with 25% tree burned under 5,000,000 generations. TreeView software was used to edit the phylogenetic tree.
Evolutionary analysis
The phylogenetic tree reconstructed using a Bayesian approach was analyzed to determine whether the environment had influenced the selective pressure on the ancestors of aquatic or terrestrial species. We performed a rigorous statistical analysis with wnt4 gene sequences, including branch-specific dN/dS ratio tests, branch-site dN/dS ratio tests, and site-specific dN/dS ratio tests. All these analyses used the maximum likelihood (ML) method in the Codeml program of PAML version 452. The maximum likelihood estimates the selective pressure via the nonsynonymous (dN) and synonymous (dS) nucleotide substitution rate ratio (dN/dS) with the dN/dS(ω) = 1 representing neutral evolution, ω > 1 representing positive selection, and ω < 1 representing purifying selection. First, the one-ratio model was used to assess the selective pressure among all wnt4 genes. Then, the free model was used to determine whether every branch allows the varied ω ratio via the likelihood ratio test by comparing with the one ratio model. Additionally, the branch-site model was used to detect the interested foreground lineage. A total of six site models were used to test for positive selection in individual codons of wnt4 sequences. A two-fold difference in the log-likelihood values (2ΔinL) between the two nested models was calculated using a chi-square test with the degrees of freedom equal to the difference in the number of parameters between the nested models. The Bayes empirical Bayes (BEB) was used to assess the Bayesian posterior probability (BPP) of the site model under positive selection. To present the result of our study more visualized, three dimensional models of CS-wnt4a which can map the positively selected sites was constructed by the iterative threading assembly refinement (I-TASSER) server58. The PYMOL software v1.5 was used to analyze the structure.
Author Contributions
Q.M.H. and S.L.C. conceived and designed the experiments. Q.M.H., Y.Z. and Y.L. performed the experiments. Q.M.H. and N.W. analyzed the data. Q.M.H. and S.L.C. wrote the paper. All authors reviewed the manuscript.
Supplementary Material
supplementary data
Acknowledgments
This work was supported by State 863 High-Technology R&D Project of China (2012AA092203;2012AA10A403-2), National Nature Science Foundation (31130057), and Taishan Scholar Project Fund of Shandong of China.
References
- Bernard P. & Harley V. R. Wnt4 action in gonadal development and sex determination. Int J Biochem Cell Biol 39, 31–43 (2007). [DOI] [PubMed] [Google Scholar]
- Vainio S., Heikkilā M., Kispert A., Chin N. & McMahon A. P. Female development in mammals is regulated by Wnt-4 signalling. Nature 397, 405–409 (1999). [DOI] [PubMed] [Google Scholar]
- Jeays-Ward et al. Endothelial and steroidogenic cell migration are regulated by WNT4 in the developing mammalian gonad. Development 130, 3663–3670 (2003). [DOI] [PubMed] [Google Scholar]
- Biason-Lauber A., Konrad D., Navratil F. & Schoenle E. J. A WNT4 mutation associated with Mullerian-duct regression and virilization in a 46 XX woman. N. Engl. J. Med 351, 792–798 (2004). [DOI] [PubMed] [Google Scholar]
- Elejalde B. R. et al. Tandem dup (1p) within the short arm of chromosome 1 in a child with ambiguous genitalia and multiple congenital anomalies. Am J Med Genet 17, 723–730 (1984). [DOI] [PubMed] [Google Scholar]
- Mohammed F. M. et al. Directduplication of chromosome 1, dir dup(1) (p21.2–p32) in a Bedouin boy with multiple congenital anomalies. Am. J. Med. Genet 32, 353–355 (1989). [DOI] [PubMed] [Google Scholar]
- Deshpandea S. N., Vijayakumarb G. & Rao A. J. Oestrogenic regulation and differential expression of WNT4 in the bonnet monkey and rodent epididymis. Reprod Biomed Online 18, 555–561 (2009). [DOI] [PubMed] [Google Scholar]
- Pask A. J., Calatayud N. E., Shaw G., Wood W. M. & Renfree M. B. Oestrogen blocks the nuclear entry of SOX9 in the developing gonad of a marsupial mammal. BMC Biol 8, 113 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pailhoux E. et al. Ontogenesis of female-to-male sex-reversal in XX polled goats. Dev Dyn 224, 39–50 (2002). [DOI] [PubMed] [Google Scholar]
- Jeays-Ward K., Dandonneau M. & Swain A. Wnt4 is required for proper male as well as female sexual development. Dev Biol. 276, 431–440 (2004). [DOI] [PubMed] [Google Scholar]
- Jordan B. K., Shen J. H., Olaso R., Ingraham H. A. & Vilain E. Wnt4 over expression disrupts normal testicular vasculature and inhibits testosterone synthesis by repressing steroidogenic factor 1/beta-catenin synergy. Proc Natl Acad Sci U S A 100, 10866–10871 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson S. A., Lin Y. T. & Capel B. Testis development requires the repression of Wnt4 by Fgf signaling. Dev Biol 370, 24–32 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen R. & Posnien N. Identification and embryonic expression of Wnt2, Wnt4, Wnt5 and Wnt9 in the millipede Glomeris marginata (Myriapoda: Diplopoda). Gene Expr Patterns 14, 55–61 (2014). [DOI] [PubMed] [Google Scholar]
- Oshima Y., Hayashi T., Tokunaga S. & Nakamura M. Wnt4 expression in the differentiating gonad of the frog Rana rugosa. Zoolog Sci 22, 689–693 (2005). [DOI] [PubMed] [Google Scholar]
- Tripathi V. & Raman R. Identification of Wnt4 as the ovary pathway gene and temporal disparity of its expression vis-a-vis testis genes in the garden lizard, Calotes versicolor. Gene 449, 77–84 (2010). [DOI] [PubMed] [Google Scholar]
- Smith C. A. et al. Cloning and expression of R-Spondin1 in different vertebrates suggests a conserved role in ovarian development. BMC Dev Biol 8, 72 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol B., Guerin A., Fostier A. & Guiguen Y. Ovary-predominant wnt4 expression during gonadal differentiation is not conserved in the rainbow trout (Oncorhynchus mykiss). Mol Reprod Dev 79, 51–63 (2012). [DOI] [PubMed] [Google Scholar]
- Wu G. C. & Chang C. F. Wnt4 is associated with the development of ovarian tissue in the protandrous black Porgy. Acanthopagrus schlegeli.Biol Reprod 81, 1073–1082 (2009). [DOI] [PubMed] [Google Scholar]
- Lynch M. & Conery J. S. The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155 (2000). [DOI] [PubMed] [Google Scholar]
- Hughes A. L. The evolution of functionally novel proteins after gene duplication. Proc Biol Sci 256, 119–124 (1994). [DOI] [PubMed] [Google Scholar]
- Assis R. & Bachtrog D. Neofunctionalization of young duplicate genes in Drosophila. Proc Natl Acad Sci U S A 110, 17409–17414 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinke D., Salzburger W., Braasch I. & Meyer A. Many genes in fish have species-specific asymmetric rates of molecular evolution. BMC Genomics 7, 20 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. L. et al. Isolation of female-specific AFLP markers and molecular identification of genotypic sex in half-smooth tongue sole (Cynoglossus semilaevis). Mar Biotechnol 9, 273–280 (2007). [DOI] [PubMed] [Google Scholar]
- Hu Q. M. et al. Differences in sex reversion and growth between normal and neomale stock in half-smooth tongue sole. Cynoglossus semilaevis.Aquacult Int 22, 1437–1449 (2014). [Google Scholar]
- Shao C. W. et al. Epigenetic modification and inheritance in sexual reversal of fish. Genome Res 24, 604–615 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X. L., Chen S. L., JI X. S. & Shao C. W. Molecular cloning, characterization and expression analysis of Sox9a and Foxl2 genes in half-smooth tongue sole (Cynoglossus semilaevis). Acta Oceanol. Sin 30, 68–77 (2011). [Google Scholar]
- Deng S. P., Chen S. L., Xu J. Y. & Liu B. W. Molecular cloning, characterization and expression analysis of gonadal P450 aromatase in the half-smooth tongue-sole, Cynoglossus semilaevis. Aquaculture 287, 211–218 (2009). [Google Scholar]
- Deng S. P., Chen S. L. & Liu B. W. Molecular cloning and expression analysis of FTZ-F1 in the half-smooth tongue sole. Cynoglossus semilaevis.Zoological Research 29, 592−598 (2008). [Google Scholar]
- Hu Q. M. & Chen S. L. Cloning, genomic structure and expression analysis of ubc9 in the course of development in the half-smooth tongue sole (Cynoglossus semilaevis). Comp Biochem Physiol B Biochem Mol Biol 165, 181–188 (2013). [DOI] [PubMed] [Google Scholar]
- Chen S. L. et al. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat Genet 46, 253–260 (2014). [DOI] [PubMed] [Google Scholar]
- Inohaya K., Takano Y. & Kudo A. Production of Wnt4b by floor plate cells is essential for the segmental patterning of the vertebral column in medaka. Development 137, 1807–1813 (2010). [DOI] [PubMed] [Google Scholar]
- Kim Y. et al. Fgf9 and wnt4 act as antagonistic signals to regulate mammalian sex determination. PLoS Biol 4, e187 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Castro B. et al. Restoration of WNT4 inhibits cell growth in leukemia-derived cell lines. BMC Cancer 13, 557 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiRocco D. P., Kobayashi A., Taketo M. M., McMahon A. P. & Humphreys B. D. Wnt4/β-catenin signaling in medullary kidney myofibroblasts. J Am Soc Nephrol 24, 1399–412 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunskaite-Hyyryläinen R. et al. Wnt4, a pleiotropic signal for controlling cell polarity, basement membrane integrity, and antimüllerian hormone expression during oocyte maturation in the female follicle. FASEB J 28, 1568–81 (2013). [DOI] [PubMed] [Google Scholar]
- B.Parekh R. Effects of glycosylation on protein function. Curr Opin Struct Biol 1, 750–754 (1991). [Google Scholar]
- Moremen K. W., Tiemeyer M. & Nairn A. V. Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol 13, 448–462 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graba Y. et al. DWnt4, a novel Drosophila Wnt gene acts downstream of homeotic complex genes in the visceral mesoderm. Development 121, 209–718 (1995). [DOI] [PubMed] [Google Scholar]
- Ferkowicz M. J., Stander M. C. & Raff R. A Phylogenetic relationships and developmental expression of three sea urchin Wnt genes. Mol Biol Evol 15, 809–819 (1998). [DOI] [PubMed] [Google Scholar]
- Shan J., Jokela T., Peltoketo H. & Vainio S. Generation of an allele to inactivate Wnt4 gene function conditionally in the mouse. Genesis 47, 782–788 (2009). [DOI] [PubMed] [Google Scholar]
- Ungar A. R., Kelly G. M. & Moon R. T. Wnt4 affects morphogenesis when misexpressed in the zebrafish embryo. Mech Dev 52, 153–164 (1995). [DOI] [PubMed] [Google Scholar]
- Meyer A. & Van de Peer Y. From 2R to 3R: Evidence for a fishspecific genome duplication (FSGD). Bioessays 27, 937–945 (2005). [DOI] [PubMed] [Google Scholar]
- Delsuc F., Brinkmann H. & Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet 6, 361–375 (2005). [DOI] [PubMed] [Google Scholar]
- Matsui T. et al. Noncanonical Wnt signaling regulates midline convergence of organ primordia during zebrafish development. Genes Dev 19, 164–175 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. S., Pask A. J., Shaw G. & Renfree M. B. Differential expression of WNT4 in testicular and ovarian development in a marsupial. BMC Dev Biol 6, 44 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan B. K., Shen J. H., Olaso R., Ingraham H. A. & Vilain E. Wnt4 over-expression disrupts normal testicular vasculature and inhibits testosterone synthesis by repressing steroidogenic factor 1/beta-catenin synergy. Proc Natl Acad Sci USA 100, 10866–10871 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan B. K. et al. Up-regulation of WNT-4 signaling and dosage-sensitive sex reversal in humans. Am J Hum Genet 68, 1102–1109 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Li J., Brost B., Cheng W. & Jiang S. W. Decreased expression and DNA methylation levels of GATAD1 in preeclamptic placentas. Cell Signal. 26, 959–967 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Martín L. et al. DNA methylation of the gonadal aromatase (cyp19a) promoter is involved in temperature-dependent sex ratio shifts in the European sea bass. PLoS Genet 7, e1002447 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen A. Y. et al. CpG methylation of dmrt1 and cyp19a promoters in relation to their sexual dimorphic expression in the Japanese flounder Paralichthys olivaceus. J Fish Biol 84, 193–205 (2014). [DOI] [PubMed] [Google Scholar]
- Hughes A. L. & Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335, 167–170 (1988). [DOI] [PubMed] [Google Scholar]
- Yang Z. H. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- Rensch B. Evolution Above The Species Level. New York:Columbia Univ.Press. 124P(1960).
- Yen P. T. & N. Bart A. Salinity effects on reproduction of giant freshwater prawn Macrobrachium rosenbergii (de Man). Aquaculture 280, 124–128 (2008). [Google Scholar]
- L. Ohsa C L. Rhyneb A. W. & Grabe S. Effects of salinity on reproduction and survival of the calanoid copepod Pseudodiaptomus pelagicus. Aquaculture 307, 219–224 (2010). [Google Scholar]
- Nissling A., Johansson U. & Jacobsson M. Effects of salinity and temperature conditions on the reproductive success of turbot (Scophthalmus maximus) in the Baltic Sea. Fish Res 80, 230–238 (2006). [Google Scholar]
- Bhat N. & Hong Y. H. Cloning and expression of boule and dazl in the Nile tilapia(Oreochromis niloticus). Gene 540, 140–145 (2014). [DOI] [PubMed] [Google Scholar]
- Roy A., Kucukural A. & Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary data