Summary
The viral accessory protein Vpx, expressed by certain simian and human immunodeficiency viruses (SIVs and HIVs), is thought to improve viral infectivity of myeloid cells. We infected 35 Asian macaques and African green monkeys with viruses that do or do not express Vpx, and examined viral targeting of cells in vivo. While lack of Vpx expression affected viral dynamics in vivo, with decreased viral loads and infection of CD4+ T cells, Vpx expression had no detectable effect on infectivity of myeloid cells. Moreover, viral DNA was only observed within myeloid cells in tissues not massively depleted of CD4+ T-cells. Myeloid cells containing viral DNA also showed evidence of T cell phagocytosis in vivo, suggesting their viral DNA may be attributed to phagocytosis of SIV-infected T cells. These data suggest that myeloid cells are not a major source of SIV in vivo, irrespective of Vpx expression.
INTRODUCTION
Human immunodeficiency viruses (HIV) and simian immunodeficiency viruses (SIV) infect humans and nonhuman primates and these viruses persist for the life of the host. The ability of HIV and SIV to maintain life-long infections is thought to depend on the ability of these viruses to latently infect long-lived cells in vivo (Katlama et al., 2013). Thus understanding which cells are targeted by the virus is of critical importance, as the life span of individual cells infected by the virus will dictate the longevity of latent infection within the host. The cells reported to be targeted by HIV and SIV in vivo are CD4+ T cells and myeloid cells (Alexaki et al., 2008). Neither CD4+ T cells nor myeloid cells represent a homogeneous pool of target cells. Instead, individual subsets of CD4+ T cells and myeloid cells are thought to be differentially infected by the virus in vivo.
Among CD4+ T cells, three large populations exist: naïve CD4+ T cells; memory CD4+ T cells; and terminally differentiated memory CD4+ T cells (Brenchley et al., 2004a). While all three subsets are infected by the virus in vivo, they do not populate all anatomical sites equally (Brenchley et al., 2004b) and are not equally infected (Brenchley et al., 2004a), with memory CD4+ T cells residing within the gastrointestinal (GI) tract infected most frequently (Brenchley et al., 2004b; Mattapallil et al., 2005a). Memory CD4+ T cells are thought to live for decades (Combadiere et al., 2004) and thus represent a very long-lived population of target cells for the virus. Myeloid cells have also been suggested to be an important target cell population for HIV/SIV (Hirsch et al., 1998; Igarashi et al., 2001; Koppensteiner et al., 2012; Orenstein et al., 1997) and while there has been some debate related to their longevity, it is generally thought their half life is approximately one-two months (Gonzalez-Mejia and Doseff, 2009; van Furth, 1989), with the possible exception of brain microglia cells (Greter and Merad, 2013). However, recent findings that macrophages are capable of proliferation in vivo suggest that tissue macrophages may, indeed, have a more limited life span (Jenkins et al., 2011; Robbins et al., 2013).
In fact, one factor that influences the ability of the virus to infect individual cells is the proliferative capacity of the target cells. Indeed, cells that are actively dividing are more prone to infection in vivo than resting cells (Alexaki et al., 2008). One explanation for limited infectivity of resting cells, compared to activated and dividing cells, is low intracellular concentrations of nucleotides within resting cells (Goldstone et al., 2012). In resting cells nucleotides are hydrolyzed by the host protein SAM domain and HD domain-containing protein 1 (SAMHD1) (Goldstone et al., 2012). The activity of SAMHD1 is thought to involve its phosphorylation and is active in resting CD4+ T cells and myeloid cells, and its expression and activity are thought to limit infection of these cells by HIV/SIV (Baldauf et al., 2012; Laguette et al., 2011).
Recent studies have implicated viral protein x (Vpx), a viral accessory protein expressed by some strains of SIV and by HIV-2, in binding to SAMHD1 leading to its proteasomal degradation (Laguette et al., 2011). SIVs used to experimentally infect Asian macaques and HIV-2 originate from SIVsmm, which is a virus that naturally infects sooty mangabeys in western Africa and expresses the viral accessory protein Vpx. HIV-1 and other immunodeficiency lentiviruses, like SIVagm, do not express Vpx (Fregoso et al., 2013). Given the differential expression of Vpx by HIVs and SIVs one prediction might be that these viruses differ in their proclivity to infect resting CD4+ T cells and myeloid cells in vivo. Here we have examined tissue cellular sources for SIV using Asian macaques and African green monkeys infected with different strains of the virus expressing or lacking Vpx. We found viral DNA within myeloid cells only in anatomical sites that were not severely CD4+ T cell depleted, that Vpx expression had little (if any) effect on our ability to detect viral DNA within myeloid cells, and that myeloid cells that contained viral DNA also had evidence of T cell phagocytosis in vivo.
RESULTS
Animals and anatomical sites studied
To investigate cellular sources for primate immunodeficiency viruses, we isolated lymphocytes from liver, spleen, small intestine, large intestine, mesenteric lymph nodes, and broncheoalveolar lavage (BAL) using tissues taken at necropsy from SIV-infected Asian macaques (both rhesus and pigtail) and SIV-infected African green monkeys (35 animals total). These animals were sacrificed at different time points relative to SIV infection, ranging from the acute phase of infection to end stage simian AIDS. As such, the animals had diverse CD4+ T cell counts and plasma viral loads (Table 1). This diverse cohort of SIV-infected animals provided an ideal opportunity to examine cellular targets for viral replication at different stages of disease. Moreover, the different viral inoculum allowed for examination of Vpx-mediated effects of cellular targeting.
Table 1.
Study animal characteristics
| Animal | virus | Species | CD4+ T cellsa | viral loadb | Disease state |
|---|---|---|---|---|---|
| MGE | mac239 | RM | 283 | 7.3×104 | Acute |
| K7M | mac239 | RM | 188 | 2.4×106 | Acute |
| K8Y | mac239 | RM | 1855 | 8.4×107 | Acute |
| A8E084 | mac239 | RM | 513 | 1.1×106 | Acute |
| DB4E | mac239 | RM | 565 | 8.1×105 | Chronic |
| DB07 | mac239 | RM | 317 | 5.5×105 | Chronic |
| DCXX | mac239 | RM | 439 | 1.5×106 | Chronic |
| PSP | mac239 | RM | 194 | 7.8×106 | sAIDS |
| CF5T | mac239 | RM | 216 | 8.0×105 | sAIDS |
| CE5D | mac239 | RM | 532 | 1.2×105 | sAIDS |
| DB17 | mac239 | RM | 122 | 9.2×104 | sAIDS |
| CF4J | mac239 | RM | 241 | 2.0×105 | sAIDS |
| KMO | mac239ΔVpx | RM | 803 | 1.7×102 | Acute |
| EV3 | mac239ΔVpx | RM | 1312 | 3.2×102 | Acute |
| EMJ | mac239ΔVpx | RM | 280 | 7.8×102 | Acute |
| 4364 | mac239ΔVpx | RM | 426 | 9.2×102 | Acute |
| 591 | e543 | RM | 186 | 2.5×105 | Chronic |
| 760 | e543 | RM | 296 | 5.0×103 | Chronic |
| 759 | e543 | RM | 526 | 6.0×102 | Chronic |
| 827 | e543 | RM | 69 | 3.7×105 | sAIDS |
| 833 | e543 | RM | 133 | 1.5×105 | sAIDS |
| DBPX | e660 | RM | 304 | Undetected | Chronic |
| 98P030 | mac239 | PTM | 178 | 7.2×103 | Chronic |
| 99P029 | mac239 | PTM | 8 | 5.4×106 | Chronic |
| 99P030 | mac239 | PTM | 29 | 3.7×104 | sAIDS |
| A1P012 | mac239 | PTM | 5 | 4.2×105 | sAIDS |
| A0P007 | mac239 | PTM | 30 | 4.9×104 | sAIDS |
| 98P005 | mac239 | PTM | 33 | 7.6×103 | sAIDS |
| 99P052 | mac239 | PTM | 30 | 6.4×105 | sAIDS |
| A0P039 | mac239 | PTM | 45 | 1.9×104 | sAIDS |
| AG17 | agm | AGM | 235 | 9.7×104 | Chronic |
| AG24 | agm | AGM | 128 | 1.1×104 | Chronic |
| AG5 | agm | AGM | 251 | Undetected | Chronic |
| AG10 | agm | AGM | 212 | 5.4×104 | Chronic |
| AG12 | agm | AGM | 252 | 4.0×102 | Chronic |
Number of CD4+ T cells / μl of blood
copies of viral RNA / ml of plasma
Memory CD4+ T cells and myeloid cells express SAMHD1
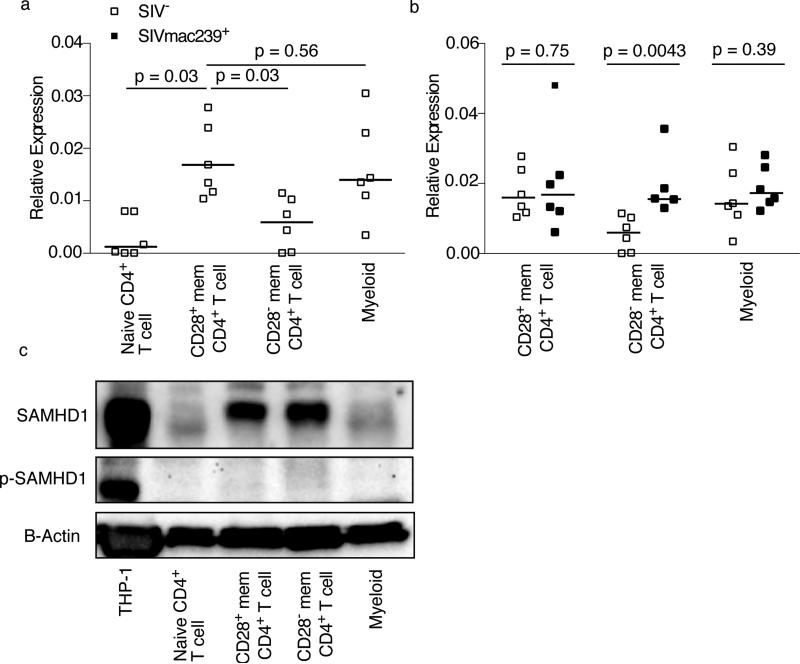
Given that the viral accessory protein Vpx was recently shown to degrade the host restriction factor SAMHD1 (Laguette et al., 2011), we initially studied expression of SAMHD1 in subsets of lymphocytes thought to be targeted by HIV and SIV in vivo. Using flow cytometric sorting, we isolated naïve CD4+ T cells, CD28+ and CD28− memory CD4+ T cells and monocytes from peripheral blood of 6 SIV-uninfected rhesus macaques and examined relative expression of SAMHD1 mRNA by quantitative RTPCR (Figure 1A). We found that naïve CD4+ T cells expressed very low amounts of SAMHD1 mRNA. Meanwhile, CD28+ memory CD4+ T cells expressed higher amounts of SAMHD1 mRNA than CD28− memory CD4+ T cells (p=0.031), and peripheral blood monocytes expressed SAMHD1 at similar amounts to CD28+ memory CD4+ T cells. Thus, we anticipated any Vpx-mediated effects on SAMHD1 to be observed predominantly in myeloid cells and CD28+ memory CD4+ T cells. To confirm that the transcription of SAMHD1 was not affected by the proinflammatory state of the immune system observed in progressive SIV infection, we sorted the same subsets of lymphocytes from peripheral blood of 6 chronically SIVmac239-infected rhesus macaques. Expression of SAMHD1 message was found to be statistically equivalent across lymphocyte subsets irrespective of SIV infection status (Figure 1B) as confirmed by RT-PCR and western blotting (Figure 1C). Moreover, the protein did not appear to be phosphorylated by any of the cells we studied in vivo (Figure 1C). It was therefore possible to examine the proclivity of viruses with and without Vpx to infect different cellular targets. We hypothesized that viruses encoding Vpx would infect CD28+ memory CD4+ T cells and myeloid cells more efficiently than viruses without Vpx.
Figure 1. Memory CD4+ T cells and myeloid cells express SAMHD1.
SAMHD1 mRNA in naïve CD4+ T cells, CD28+ memory CD4+ T cells, CD28− memory CD4+ T cells, and myeloid cells in peripheral blood of SIV-uninfected (A) and SIV-infected (B) rhesus macaques. Expression relative to β-actin mRNA. (C) Total and phosphorylated SAMHD1 protein in naïve CD4+ T cells, CD28+ memory CD4+ T cells, CD28− memory CD4+ T cells, and myeloid cells in peripheral blood of SIV-uninfected animals. Forty μg of primary cell extract or 20 μg of THP-1 cell extract were separated by SDS-PAGE and Western blotted using antibodies against SAMHD1, phosphorylated SAMHD1 or β-actin. Horizontal lines indicate the median. Western blots are representative of three experiments.
Myeloid cells contain little, if any, viral DNA in mucosal sites
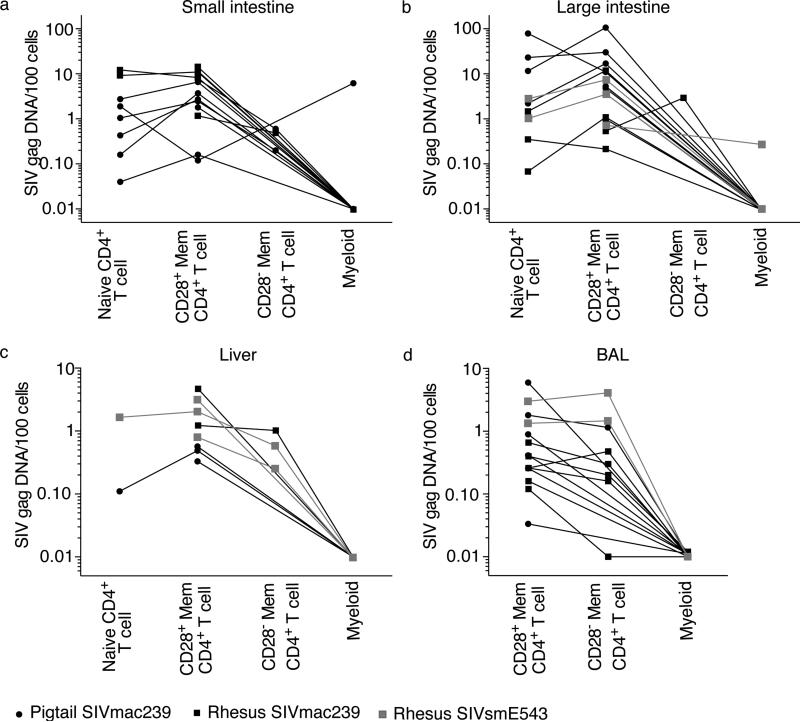
Given that mucosal sites have been shown to be massively depleted of CD4+ T cells during the acute phase of infection and throughout the chronic phase of infection (Brenchley et al., 2004b; Mattapallil et al., 2005a; Picker et al., 2004; Veazey et al., 1998), we hypothesized that without preferred CD4+ T cell targets, viruses expressing Vpx would more efficiently infect myeloid cells at mucosal sites. Therefore, we flow cytometrically sorted the few memory CD28+, CD28− memory CD4+ T cells when possible, and myeloid cells from small intestine, large intestine, liver, and BAL of SIV-infected Asian macaques (Figure 2). The myeloid cells were sorted as to include all myeloid cell types, including macrophages, monocytes, and the various subsets of dendritic cells (gating strategy in Figure S1). Each subset of CD4+ T cells was not equally abundant at each anatomical site. For example, naïve CD4+ T cells and differentiated CD28− memory CD4+ T cells were not abundant in the liver or within the GI tract (Figure 2A-C). Thus we were unable to sort sufficient numbers of cells corresponding to each CD4+ T cell subset. However, it was possible to amplify viral DNA from CD28+ memory CD4+ T cells from all four mucosal sites of every animal we examined. Moreover, we successfully amplified viral DNA from naïve CD4+ T cells from the small and large intestines of approximately 50% of the animals. There were very low frequencies of naïve CD4+ T cells in the liver of all animals, but we were able to obtain sufficient numbers of liver naïve CD4+ T cells from two animals in our cohorts to amplify viral DNA. Although we successfully amplified viral DNA from even small numbers of CD28+ memory CD4+ T cells (on average only 2,000 cells) sorted from GI tract, liver, and BAL samples, we found viral DNA in myeloid cells from the GI tracts of only two animals. The frequencies of CD4+ T cells in the intestines of these animals (99P029 for small intestine and 759 for large intestine) were 10.3% and 36.6%, respectively. Therefore the GI tracts of these animals contained ample CD4+ T cell targets. There were only 5 copies of viral DNA in GI tract myeloid cells of 759, and 15 copies of viral DNA in GI tract myeloid cells of 99P029. We found no viral DNA in myeloid cells from the BAL or liver, despite having been able to flow cytometrically sort abundant numbers of myeloid cells (more than 2 million cumulatively) from both liver and BAL.
Figure 2. Myeloid cells contain little, if any, SIV DNA in mucosal sites.
Copies of viral DNA / 100 naïve CD4+ T cells, CD28+ memory CD4+ T cells, CD28− memory CD4+ T cells, and myeloid cells in SIV-infected Asian macaques (A) small intestine, (B) large intestine, (C) liver, and (D) BAL. See also Figure S1.
Myeloid cells can contain viral DNA in lymphoid tissues
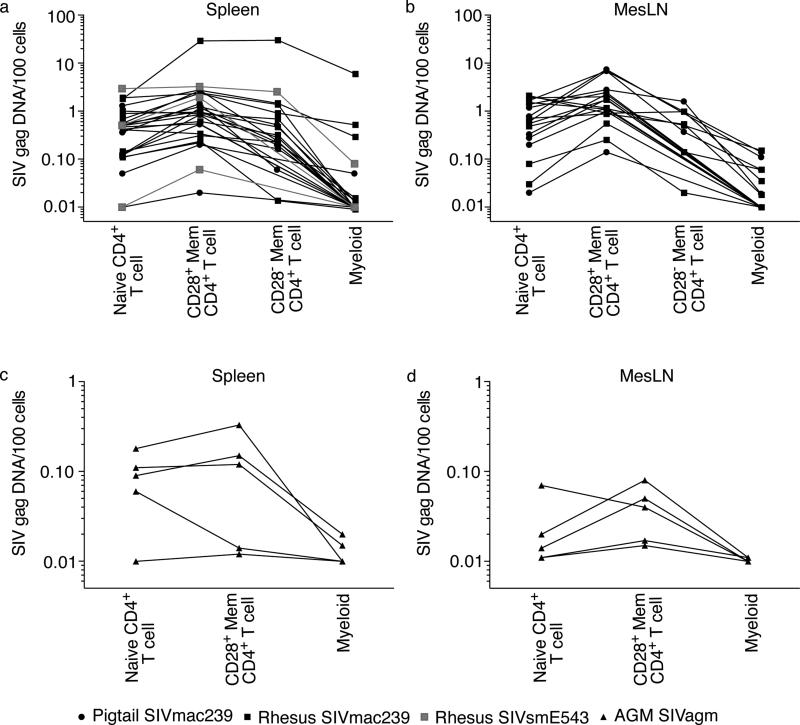
After finding remarkably low amounts of viral DNA within myeloid cells from mucosal tissues, we next examined infection frequencies of the same cellular subsets within spleen and mesenteric lymph nodes (mLNs) where viral replication is likely more rampant in chronically SIV-infected animals. Consistent with our findings from mucosal tissues, the CD28+ memory CD4+ T cells were more frequently infected than any other subset in both rhesus and pigtail macaques, irrespective of which virus lineage was chosen to infect (Figure 3A-B). However, unlike our findings within the GI tract, myeloid cells within the mLNs and spleen of SIV-infected Asian macaques frequently contained viral DNA. In particular, K8Y demonstrated the highest amounts of viral DNA within splenic myeloid cells, after sacrifice during peak viral replication at day 10 post-SIV infection.
Figure 3. Myeloid cells can contain SIV DNA in lymphoid tissues.
Copies of viral DNA / 100 naïve CD4+ T cells, CD28+ memory CD4+ T cells, CD28− memory CD4+ T cells, and myeloid cells in SIV-infected Asian macaques (A) spleen, (B) mLN, or SIVagm-infected African green monkey (C) spleen, or (D) mLN.
Given that Asian macaques infected with viruses encoding Vpx had myeloid cells containing viral DNA in the spleen and mLNs, we next sought to examine the infection frequencies of cell subsets of animals infected with viruses which do not express Vpx. We flow cytometrically sorted naïve CD4+ T cells, CD28+ memory CD4+ T cells and myeloid cells from mLNs and spleens of 5 chronically SIVagm-infected AGM. Similar to our findings in SIV-infected Asian macaques, we found that CD28+ memory CD4+ T cells of the spleen and mLNs were more frequently infected than any other cellular subset (Figure 3C-D). Furthermore, viral DNA was detected in myeloid cells from the spleen and mLNs from two of five AGM (40%) (Figure 3C-D). These data suggest that expression of Vpx by SIV did not allow the virus to infect myeloid cells more efficiently in vivo. Indeed, exactly the same percentage of Asian macaques (40%) had viral DNA within splenic and mLN myeloid cells when the infecting viruses expressed Vpx (Figure 3A-B).
Myeloid cells can contain viral DNA without viral expression of Vpx
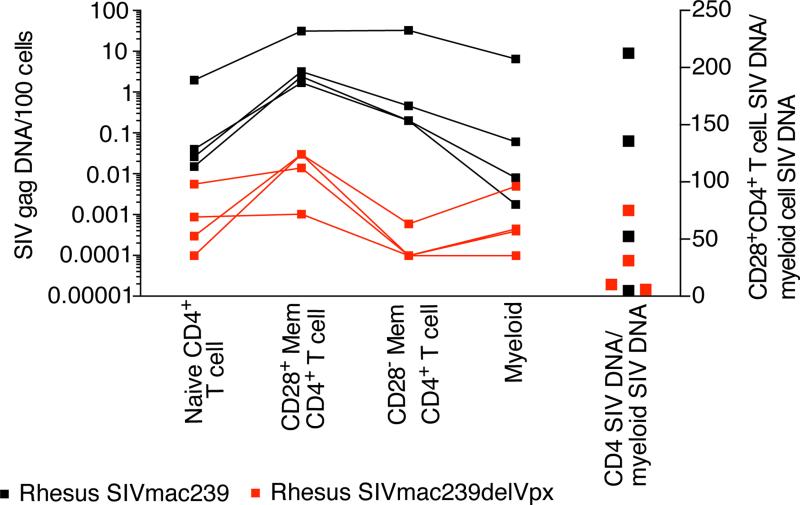
Recent studies have proposed an evolutionary relationship shared by host restriction factors and viral proteins Vpx and Vpr (Fregoso et al., 2013), suggesting that our finding of viral DNA in myeloid cells of SIVagm-infected AGM (Figure 3C-D) might be attributed to Vpr-mediated degradation of SAMHD1 since SIVagm expresses Vpr. Therefore we examined the infection frequencies of cellular targets in animals acutely infected with a virus genetically manipulated to delete Vpx (Berger et al., 2012). Animals infected with SIVmac239ΔVpx clearly manifested lower cellular infection (Figure 4) and correspondingly low plasma viral loads (Table 1).
Figure 4. Myeloid cells can contain viral DNA without viral expression of Vpx by SIV.
Copies of viral DNA /100 naïve CD4+ T cells, CD28+ memory CD4+ T cells, CD28− memory CD4+ T cells, and myeloid cells in spleen of acutely SIVmac239-infected and acutely SIVmac239ΔVpx-infected Asian macaques (left 4 columns). Ratio of viral DNA within CD28+ memory CD4+ T cells compared to viral DNA within myeloid cells (right column).
As revealed by our studies of infected cell subsets in lymphoid tissue, acutely SIV-infected animals had the highest amount of viral DNA within myeloid cells in spleen. Therefore, to investigate whether the high frequency of infected myeloid cells was mediated by expression of Vpx by SIVmac239, we infected 4 animals with SIVmac239ΔVpx and sacrificed during acute infection. We then flow cytometrically sorted the different cell subsets from the spleen and measured viral DNA concentration (Figure 4). While the amount of viral DNA was considerably lower in the animals acutely SIV- mac239ΔVpx infected compared to animals acutely SIV-infected with wild type SIVmac239, we amplified viral DNA from myeloid cells of all acutely-infected animals. To determine if there was a decreased tendency to detect SIVmac239ΔVpx within myeloid cells compared to CD4+ T cells, we measured the ratio of viral DNA in CD28+ memory CD4+ T cells to the level of viral DNA in myeloid cells in animals acutely infected with SIVmac239ΔVpx or SIVmac239 (Figure 4). We found that these ratios completely overlapped among animals infected SIVs with or without Vpx (p=0.46). From these data, it seems unlikely that Vpx was facilitating infection of myeloid cells in vivo. These data suggested to us that the viral DNA found within myeloid cells may either not be attributed to bone fide viral infection in vivo, or other viral adaptations are necessary to render myeloid cells susceptible to productive infection.
Myeloid cells phagocytose T cells in vivo
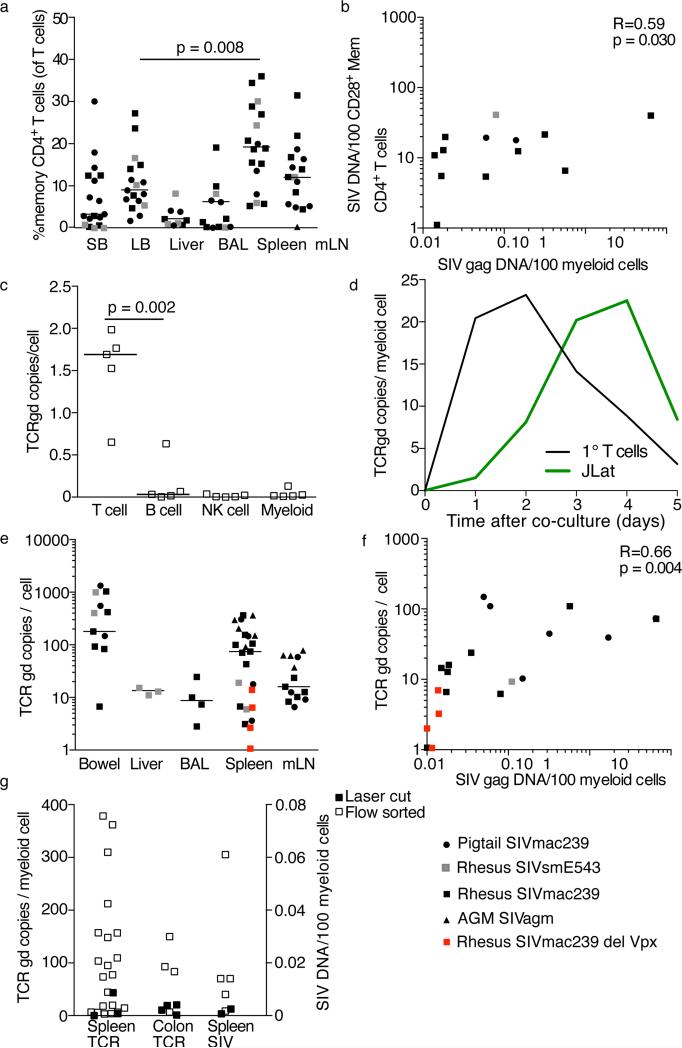
Given the predominant assumption that macrophage tropism develops as a result of CD4+ T cell target loss, we were somewhat surprised to find viral DNA only in myeloid cells from tissues in which CD4+ T cells were more abundant (spleen and mLN, Figure 5A). Moreover, the quantities of viral DNA found in myeloid populations directly correlated with the frequencies of viral DNA+ CD4+ T cells in the same anatomical sites (Figure 5B). This finding suggested that one potential explanation for the viral DNA found in myeloid cells was impure flow cytometric sorting. Indeed, myeloid cells are notoriously difficult to sort purely (Heeregrave et al., 2009). Therefore we performed a post-sort analysis of myeloid cells isolated from the spleens of several animals where the myeloid cells were found to contain viral DNA (Figure S2). While the post-sort purity of myeloid cells was consistently lower than that of CD4+ T cell subsets (data not shown), the number of contaminating memory CD28+ CD4+ T cells was too low to account for all of the viral DNA found within sorted myeloid cells.
Figure 5. Tissue myeloid cells phagocytose T cells.
(A) Frequencies of CD4+ T cells in the small bowel (SB), large bowel (LB), liver, broncheoalveolar lavage (BAL), spleen, and mesenteric lymph nodes (mLN) from our cohort of SIV-infected animals. (B) Correlation between the amount of viral DNA in CD4+ T cells and the amount of viral DNA in myeloid cells. (C) Amount of rearranged TCR DNA in peripheral blood T cells, B cells, NK cells and monocytes from uninfected rhesus macaques. (D) Amount of rearranged TCR DNA in monocytes at indicated time points after co-culture with irradiated autologous primary T cells (black) or J-Lat cells (green). (E) Amount of rearranged TCR DNA in myeloid cells from anatomical sites. (F) Correlation between the amount of viral DNA in myeloid cells and the amount of rearranged TCR DNA in myeloid cells. (G) Amount of rearranged TCR DNA (left columns) or SIV DNA (right column) in myeloid cells isolated by flow cytometry (open symbols) or laser cutting (closed symbols). Horizontal lines indicate the median. See also Figure S2.
A third explanation for viral DNA within myeloid cells is myeloid cell-mediated phagocytosis of SIV-infected CD4+ T cells. Indeed, phagocytosis of apoptotic and necrotic cells is a well-established function of tissue macrophages, especially in the spleen (Wynn et al., 2013). Specifically, we quantified TCR γδ DNA because these loci are rearranged during early T cell development and as a result, many circulating mature T cells express rearranged, but nonfunctional, γδ genes (Spits, 2002). To confirm that our PCR would detect T cell-specific DNA, we also flow cytometrically sorted T cells, B cells, natural killer (NK ) cells, and monocytes from peripheral blood of 5 SIV-uninfected animals. We found between 0.5 and 2 copies of rearranged TCR DNA in T cells, but there was very little, if any, rearranged TCR DNA in B cells, NK cells, or monocytes (Figure 5C). We then confirmed that this PCR assay could be used to measure phagocytosis of T cells by co-culturing peripheral blood monocytes (with very little rearranged TCR DNA, Figure 5C), with irradiated autologous, CFSE labeled, T cells or irradiated, CFSE labeled, J-LAT cells. We then flow cytometrically sorted monocytes from these co-cultures every day after the co-culture until the monocytes began to die (5 days post co-culture). We then measured the amount of rearranged TCR DNA in the monocytes that had been co-cultured. We found that rearranged TCR DNA in monocytes peaked at 2-4 days post co-culture, and then began to decrease (representative data from 4 experiments, Figure 5D).
Having shown that rearranged TCR DNA within myeloid cells could be used as a proxy of T cell phagocytosis, we next sought to investigate phagocytosis of T cells in vivo. We measured the amount of rearranged DNA coding for the T cell receptor within myeloid cells from different anatomical sites (Figure 5E). The number of copies of rearranged TCR DNA in tissue myeloid populations varied widely among animals within the same site and among tissues, particularly at the GI tract and spleen, where variation approached three orders of magnitude (Figure 5E). Thus rearranged TCR DNA was not detected in all tissue myeloid cells equally. To confirm the identity of the rearranged TCR DNA amplified from myeloid cells, we cloned and sequenced the TCR DNA amplified from myeloid cell lysate of two animals, CF4J and AG17. All clone sequences were confirmed to encode rearranged γδ TCR (Table 2). The clones were not homogeneous, with significant sequence diversity evident (Table 2). These data strongly suggest that the TCR DNA amplified from myeloid cells could not be attributed to germ-line TCR DNA and that myeloid cells are phagocytosing T cells in vivo. The number of rearranged TCR copies detected in tissue myeloid cells positively correlated with the quantity of viral DNA measured within the same cells (Figure 5F, r=0.66, p=0.004).
Table 2.
Rearranged TCR DNA sequences in myeloid cells
| TCR VG9 Sequences |
|---|
| -VLFHCQRVSPQ |
| CCSTA-KEFLVI |
| CCSTAKEFPIMH |
| CCSTAKEFPS-H |
| CVVPLPKSF-PH |
| CVVPLPKSFLY- |
| CVVPLPKSFSPH |
| CVVPLPKSFSPH |
Considering that myeloid cells are notoriously difficult to isolate from tissues, and to identify by flow cytometry, we also isolated myeloid cells from paraffin-embedded tissue by immunohistochemical staining for myeloid cells and performing laser capture micro-dissection (LCM) using laser cutting to isolate individual myeloid cells from different tissues. We isolated myeloid cells from the spleen of 3 SIV-infected animals and from the colon of 4 SIV-infected animals and measured the amount of rearranged TCR and viral DNA (Figure 5G). We were able to amplify both viral DNA and rearranged TCR DNA from all LCM samples and the amounts were within the ranges found when we had isolated the cells by flow cytometry (Figure 5G). Taken together, these data suggest that the viral DNA found within myeloid cells could easily be attributed to phagocytosis of SIV-infected CD4+ T cells in vivo.
DISCUSSION
Here we have sampled mucosal and lymphoid tissues from SIV-infected Asian macaques and African green monkeys to examine the cellular sources for SIV based upon differential expression of Vpx. We found that (i) CD28+ memory CD4+ T cells were most frequently viral DNA+ compared to any other cellular target in all anatomical sites; (ii) myeloid cells within mucosal tissues very rarely contained viral DNA; (iii) myeloid cells in the spleen or mLNs of 40% of animals contained viral DNA; (iv) myeloid cells could contain viral DNA irrespective of viral Vpx expression; (v) myeloid cells phagocytosed T cells in vivo; and (vi) viral DNA within myeloid cells was associated with phagotycosis of T cells. Consequently, it was not possible to identify bona fide SIV infection of myeloid cells from any anatomical site of our animals.
Understanding the cellular substrates for HIV/SIV replication in vivo is of importance for developing therapeutic interventions aimed at decreasing the size of the HIV/SIV reservoir (Katlama et al., 2013; Siliciano and Siliciano, 2013). It is fairly well accepted that the half-life of latent reservoirs in HIV/SIV-infected individuals treated with ARVs is longer than the individual's life-span (Finzi et al., 1999). Therefore, ARV regimens that fully suppress viral replication will be insufficient to cure HIV-infected individuals of the infection (Finzi et al., 1999; Katlama et al., 2013; Siliciano and Siliciano, 2013). Given that significant research effort strives to develop therapeutic interventions aimed at stimulating HIV from the latent pool, understanding which cells are infected by the virus is critical (Margolis, 2011).
It is widely accepted that the largest pool of HIV/SIV-infected cells belong to the CD4+ T cell subset, and our data are completely consistent with this supposition. However, another pool of leukocytes posited to be an important source of HIV/SIV- infected cells is myeloid cells, and especially tissue macrophages (Dai et al., 2013; Koenig et al., 1986; Smith et al., 2003; Wang et al., 2001). Consistent with these previous studies, we also found viral DNA within myeloid cells. There are several explanations to account for the finding of viral DNA or RNA within tissue myeloid cells of HIV/SIV-infected individuals. While a popular explanation revolves around bona fide infection of myeloid cells by the virus in vivo, the functional nature of this subset of cells provides for alternative explanations. Myeloid cells, and tissue macrophages in particular, are charged with clearing immunological complexes and phagocytosing dead and dying cells in vivo. During infection, there are high levels of antibodies that bind to the virus and high levels of infected CD4+ T cells, which become necrotic or apoptotic. This suggests that myeloid cells may accumulate viral RNA and viral DNA by the normal clearance functions they have evolved to perform.
Given several recent studies that suggest expression of the accessory protein Vpx by HIV-2 or SIV facilitates viral infectivity of myeloid cells in vitro (Berger et al., 2011; Hrecka et al., 2011; Laguette et al., 2011; Pauls et al., 2013), we used nonhuman primates infected with SIVs either expressing Vpx or lacking Vpx and examined the corresponding amounts of viral DNA within individual subsets of lymphocytes. While viruses that expressed functional Vpx clearly infected CD4+ T cells more efficiently than Vpx deleted viruses, with correspondingly higher plasma viral loads, Vpx expression had no meaningful effect on the presence of viral DNA found within myeloid cells. Our findings are consistent with very recent data suggesting that Vpx expression is insufficient to overcome SAMHD1-mediated HIV-1 restriction among dendritic cells (Bloch et al., 2013). Similarly, Vpx may be insufficient to overcome SAMHD1 in tissue myeloid cells.
Other studies would posit that viruses could evolve towards a macrophage tropism when CD4+ T cell substrates are massively depleted. Indeed, experimental depletion of CD4+ T cells in SIV-infected animals with anti-CD4 antibodies leads to enhanced viral replication within myeloid cells (Ortiz et al., 2011). Moreover, chimeric HIV/SIV viruses which cause rapid and profound depletion of CD4+ T cells might lead to emergence of macrophage tropic viruses (Igarashi et al., 2003). Our animal cohort consisted of 8 animals with less than 50 CD4+ T cells/μl blood (two animals had less than 10 CD4+ T cells/μl). Even in these animals, myeloid cells containing viral DNA also contained rearranged TCR DNA, and myeloid cells contained viral DNA only in tissues that were not dramatically CD4+ T cell depleted. Hence the viral replication attributed to myeloid cells, which almost inevitably immediately precedes death in profoundly CD4+ T cell depleted animals, may in fact be attributed to myeloid cell clearance mechanisms. Moreover, even in these animals with incredibly low numbers of peripheral blood CD4+ T cells, CD4+ T cell numbers in lymphoid tissue were not nearly as low. Notably, despite having only 5 CD4+ T cells/μl of blood, A1P012 had a CD4+ T cell frequency of 43.9% in mLNs, and 39% of these were memory CD4+ T cells.
Our analysis of myeloid cell viral infectivity was, by no means, comprehensive. HIV/SIV-infection of microglia cells in the brain is widely believed to play an important role in virus associated neurological disorders (Burdo et al., 2010; Dang et al., 2012; Gonzalez-Perez et al., 2012; Koenig et al., 1986; Koppensteiner et al., 2012; Thompson et al., 2011). Unfortunately, flow cytometric isolation of lymphoid and myeloid cells from the brain is extremely challenging and we were unable to perform quantitative PCR for viral DNA from brain-resident cell subsets. Hence the immunological clearance mechanisms which seem to underlie viral DNA within GI tract and lymphoid tissue myeloid cells may or may not also occur within myeloid cells of the nervous system and/or other anatomical sites. Moreover, myeloid cells in different anatomical sites are incredibly functionally and phenotypically heterogeneous, and future studies may elaborate whether myeloid cells without phagocytic function contain viral DNA and rearranged TCR DNA.
In addition to myeloid cells, SAMHD1 has also been suggested to restrict HIV/SIV infection of resting CD4+ T cells (Baldauf et al., 2012; Descours et al., 2012). Our data are consistent with viral expression of Vpx leading to enhanced infectivity of resting memory CD4+ T cells given the increased frequencies of viral DNA in CD28+ memory CD4+ T cells sorted from animals infected with Vpx-expressing viruses relative to animals infected with Vpx− viruses. This strongly suggests that Vpx has a functional role in vivo, consistent with recent data demonstrating significantly lower viral loads and delayed progression to simian AIDS in animals infected with Vpx-deleted viruses (Westmoreland et al., 2014). Moreover, that we only routinely found viral DNA in myeloid cells in anatomical sites not massively depleted of CD4+ T cells, could suggest that CD4+ T cell/myeloid cell interactions are required for myeloid cell infectivity or that myeloid cells might become SIV-infected during phagocytosis of CD4+ T cells. Further work is certainly merited to determine if myeloid cells in lymphoid tissues contain replication-competent SIV. However, we believe our data clearly suggest that viral RNA and DNA within myeloid cells can attributed to clearance of virally-infected cells and immune complexes and not, bone fide SIV infection of myeloid cells in vivo.
In summary, our findings suggest that the frequencies of myeloid cells which contain viral DNA within certain anatomical sites of SIV-infected animals are not influenced by viral expression of Vpx. Rather, viral DNA within myeloid cells in the tissues we studied may be attributed to phagocytosis of infected CD4+ T cells. This finding, in turn, strongly suggests that myeloid cells may not represent a significant source of infected cells in vivo and that therapeutic interventions aimed at stimulating latent virus should target CD4+ T cells.
EXPERIMENTAL PROCEDURES
Subjects
To characterize and determine the cellular targets for SIV in vivo, we studied tissues from a diverse cohort of SIVagm-infected African green monkeys and SIV-uninfected and infected RM and PTM. Tissues were obtained at necropsy from 22 rhesus macaques (Macaca mulatta), 8 pigtail macaques (Macaca nemestrina), and 5 African green monkeys (Chlorocebus pygerythrus) at a variety of times after infection with SIVmac239, SIVmac239ΔVpx, SIVsmE543, or SIVagm. Animals with simian AIDS were diagnosed by opportunistic infections, lymphomas, or greater than 15% body mass weight loss. Animals were housed and cared in accordance with American Association for Accreditation of Laboratory Animal Care standards in AAALAC accredited facilities, and all animal procedures were performed according to protocols approved by the Institutional Animal Care and Use Committees of the National Institute of Allergy and Infectious Diseases (Table 1). Monocytes and lymphocytes from 5 healthy humans were obtained by the NIH blood bank. The subjects all gave informed consent in compliance with the NIH institutional review board.
Flow cytometry
The SIV infection frequency of naïve, CD28+ memory and CD28− memory CD4+ T cells and myeloid cells was determined by flow cytometrically sorting each subset. Naïve CD4+ T cells were defined as live, CD45+CD3+CD4+CD28+CD95− lymphocytes; CD28+ memory CD4+ T cells were defined as live, CD45+CD3+CD4+CD28+CD95+ lymphocytes; CD28− memory CD4+ T cells were defined as live, CD45+CD3+CD4+CD28-CD95+/- lymphocytes; myeloid cells were defined as live, CD45+CD3−CD20−NKG2AHLA-DR+. Cells were then stained with the live dead exclusion dye Aqua blue (Invitrogen) then with antibodies to CD3 (clone SP34-2, BDPharmingen), CD4 (clone L200, BDPharmingen), CD95 (clone DX2, BDPharmingen), CD28 (clone CD28.2, Beckman Coulter). CD8 (clone SK8, BDPharmingen), NKG2A (clone Z199 Beckman Coulter), CD20 (clone BDPharmingen), HLADR (clone BDPharmingen), CD14 (clone BDPharmingen), and CD45 (clone BDPharmingen).
Immunohistochemistry and laser cutting microdissection of Myeloid cells
Immunohistochemistry for Myeloid cells was performed on spleen and colon sections (7μm) mounted on PEN membrane frame (MMI) slides using a biotin-free polymer approach (Golden Bridge International, Inc.). Slides were dewaxed and rehydrated with double-distilled H2O and antigen retrieval was performed by heating sections in boiling (100-102°C) 10mM sodium citrate (pH 6.0) for 15 min followed by placing immediately in ddH2O. Nonspecific Ig-binding sites were blocked with blocking buffer (TBS containing 0.05% Tween-20 and 0.25% casein) for 10 min at room temperature. Slides were incubated with a cocktail of myeloid/macrophage specific antibodies including CD163 (1:400; Novocastra), CD68 (1:400; Biocare) and HAM56 (1:400; DAKO) diluted in blocking buffer and incubated for 60 minutes at room temperature. Slides were washed in 1x TBS with 0.05% Tween-20 and were developed with Mouse Polink-2 AP according to manufacturer's recommendations, washed and incubated with Warp Red chromagen (Biocare Medical, Inc.). The slides were washed in ddH2O and air dried prior to laser capture microdisection (LCM) utilizing a laser cutting approach. We determined the optimal laser settings for LCM for colon and spleen tissue in order to prevent potential damage to the myeloid cell DNA was 0.5% transmission, UV cutting speed 91, UV Cut length 400micron. The cutting area was centered on the nucleus of the cell and the size was 16um (14 to 18 um) determined by the size and configuration of each myeloid cell selected. Captured cells (250 from each tissue) on LCM caps with were placed in a GeneAmp tube (Applied Biosystems, Foster City, CA, N8010611) that contained 50 μL of Proteinase K solution (Acturus Picopure DNA Extraction Kit, Applied Biosystems). Each tube was inverted to allow total immersion of the tissue in the proteinase solution and incubated at 65°C for 20 hours. The samples were then stored at -20°C and the total lysate used for DNA PCR analysis.
ΔVpx virus
SIVmac239ΔVpx was created by splice overlap extension PCR as described(Gibbs et al., 1995). Briefly, two PCR products were amplified from SIVmac239 plasmid DNA by using Herculase II Fusion DNA Polymerases (Agilent Technologies) with the following pairs of primers: BstBF (5’-GGAGAGGCCTTCGAATGGCTAAACAG-3’) and DvpxR (5’-CCTGCCCCATGTTACTATTCATCATGCCAGTAT-3’), or DvpxF: (5’- ATACTGGCATGATGAATAGTAACATGGGGCAGG-3’) and AfeR: (5’-CATGAAGAGCGCTCGTTGGAGGATTC-3’). Reaction conditions were as follows: 95°C for 2 minutes, followed by 30 cycles at 95°C for 20 seconds, 60°C for 20 seconds, and 72°C for 30 seconds, followed by 72°C for 2 minutes. A second round PCR was performed using the mixture of these two PCR products as template and the primers BstBF and AfeR with the same reaction conditions. These reactions generated a product containing a truncated SIVmac239 Vpx region in which nt6245-6346 were deleted and replaced by two stop codons TAGTAA (nucleotide numbers are based on the sequence of SIVmac239, GenBank accession M33262). The full length SIVmac239ΔVpx plasmid was constructed by subcloning of this PCR product into wildtype SIVmac239 plasmid with restriction sites Bstb I and Afe I, and sequenced using an Applied Biosystems 3130XL genetic analyzer. Virus stock was obtained by transfection of 293T cells and titrated on TZM-bl cells as previously described(Wu et al., 2012).
Cell-associated SIV DNA quantification
Cell subsets were sorted using a FACS Aria II using FACSDiva software (BDPharmingen). Sorted cells were then lysed using 25 μL of a 1:100 dilution of proteinase K (Roche, Indianapolis, IN) in 10mM Tris buffer. Quantitative PCR was performed using 5 μL of cell lysates per reaction. Reaction conditions were as follows: 95°C holding stage for 5 minutes, and 50 cycles of 95°C for 15 seconds followed by 60°C for 1 minute using the Taq DNA polymerase kit (Invitrogen, Carlsbad, CA). The sequence of the forward primer for SIVmac239 is GTCTGCGTCATYTGGTGCATTC. The reverse primer sequence is CACTAGYTGTCTCTGCACTATRTGTTTTG. The probe sequence is CTTCRTCAGTYTGTTTCACTTTCTCTTCTGCG. The sequence of the forward primer for SIVsmE543 is GGCAGGAAAATCCCTAGCAG, the reverse primer sequence is GCCCTTACTGCCTTCACTCA, and the probe sequence is AGTCCCTGTTCRGGCGCCAA. The sequence of the forward primer for SIVagm is GTCTGCGTCAT(T/C)TGGTGCATTC. The reverse primer sequence is CACTAG(C/T)TGTCTCTGCACTAT(A/G)TGTTTTG. The probe sequence is CTTC(A/G)TCAGT(C/T)TGTTTCACTTTCTCTTCTGCG. For cell number quantitation monkey albumin was measured as previously described (Mattapallil et al., 2005b). The PCR machine used for all real-time PCR was the StepOne Plus (Applied Biosystems, Carlsbad, CA) and the analysis was performed using StepOne software (Applied Biosystems).
Real-time PCR analysis of SAMHD1 expression
Cell subsets were sorted as described above and extracted using TRI reagent (Sigma-Aldrich, St. Louis, MO). Extracted total RNA was retrotranscribed using SuperScript III reverse transcriptase following the manufacturer's instructions (Invitrogen). Oligonucleotide primers used were as follows: for SAMHD1, forward (5’-GCTGCTGAAGAACATCCGAG-3’) and reverse (5’-GAGAGGGTGGAGCTCAATGT-3’); for ACTB, forward (5’-ATTGCCGACAGGATGCAGAA-3’); and reverse (5’-GCTGATCCACATCTGCTGGAA-3’). Quantitative real-time PCR was performed using Power SYBR Green Master Mix (Invitrogen) at the following conditions: 95°C holding stage for 10 minutes, followed by 45 cycles at 94°C for 30 seconds, 58°C for 45 seconds, and 72°C for 30 seconds. Relative expression was calculated using the ΔCt method.
Western blots
Cell extracts were prepared using IP Lysis Buffer (Pierce) according to the manufacturers instructions. Protein concentration was determined using BioRad Protein Assay Reagent (BioRad). Protein extracts were treated with Laemlii buffer with beta-mercaptoethanol and resolved on a 4-12% NuPage Bis-Tris gel (Invitrogen). Proteins were transferred by electro-blotting onto an Invitrolon PVDF membrane (Invitrogen), blocked with PBS with 0.1% Tween-20 5% BSA and incubated overnight with primary antibody (rabbit anti-SAMHD1 from Protein Tech, mouse anti-β-actin clone ACI4 from Sigma, rabbit anti-phospho-SAMHD1 clone T592 was kindly provided by Dr. Benkirane (Cribier et al., 2013)). Membranes were then washed and incubated with appropriate HRP-conjugated detector antibodies, washed and signal detected using Novex ECL HRP reagent (Invitrogen).
In vitro phagocytosis assay
Autologous lymphocytes or J-LAT cells were irradiated with 3,000 rads. 10 million irradiated lymphocytes or irradiated J-LAT cells were labeled with 0.25 μM CFSE and were mixed with 10 million monocytes. Monocytes were flow cytometrically sorted daily after co-culture and levels of rearranged TCR DNA was determined by qPCR.
Rearranged TCR DNA quantification
Cells were sorted and lysed as described above. Quantitative real-time TCR PCR was performed. Reaction conditions were as follows: 95°C holding stage for 7 minutes, followed by 45 cycles at 95°C for 45 seconds, 60°C for 1 minute and 72°C for 90 seconds. Rearranged TCRG DNA detection was accomplished using a proprietary mix of unlabeled primers (Invivoscribe, San Diego, CA), AmpliTaq Gold Polymerase (Applied Biosystems), and SYBR Green dye (Invitrogen) (Yao and Schneider, 2007).
TCRG Cloning and Sequence Analysis
TCR PCR product was purified by gel electrophoresis using a QIAquick gel extraction kit (Qiagen) and cloned into the pCR4-TOPO vector (Invitrogen). Plasmid was purified using a QIAprep Spin Miniprep kit (Qiagen) and sequenced by Eurofins MWG (Operon, Huntsville, AL). Sequence analysis was performed using Gene Codes Sequencher Version 5.1.
Plasma viral loads
Plasma samples were analyzed for SIV RNA using a fluorescent resonance energy transfer probe-based real-time RT-PCR (TaqMan) assay that provides a threshold sensitivity of 10 copy Eq/ml, as previously described (Cline et al., 2005). All PCR reactions were run on ABI Prism 7700 Sequence Detection System and the fluorescent signal-based quantitation of viral RNA copy numbers in test samples was determined by ABI sequence detection software (Applied Biosystems).
Statistical analysis was performed using GraphPad Prism Version 5 using the Mann-Whitney T test.
Supplementary Material
Acknowledgements
We would like to acknowledge Heather Cronise, JoAnne Swerczek, Richard Herbert, and all the veterinary staff at the NIH animal center. We would like to thank CLIC/BBC for advice and helpful discussions. Funding for this study was provided in part by the Division of Intramural Research/NIAID/NIH. The content of this publication does not necessarily reflect the views or policies of DHHS, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Alexaki A, Liu Y, Wigdahl B. Cellular reservoirs of HIV-1 and their role in viral persistence. Curr HIV Res. 2008;6:388–400. doi: 10.2174/157016208785861195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, Schenkova K, Ambiel I, Wabnitz G, Gramberg T, et al. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat Med. 2012;18:1682–1687. doi: 10.1038/nm.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger A, Sommer AF, Zwarg J, Hamdorf M, Welzel K, Esly N, Panitz S, Reuter A, Ramos I, Jatiani A, et al. SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutieres syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 2011;7:e1002425. doi: 10.1371/journal.ppat.1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger G, Turpin J, Cordeil S, Tartour K, Nguyen XN, Mahieux R, Cimarelli A. Functional analysis of the relationship between Vpx and the restriction factor SAMHD1. The Journal of biological chemistry. 2012;287:41210–41217. doi: 10.1074/jbc.M112.403816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch N, O'Brien M, Norton TD, Polsky SB, Bhardwaj N, Landau NR. HIV Type 1 Infection of Plasmacytoid and Myeloid Dendritic Cells Is Restricted by High Levels of SAMHD1 and Cannot be Counteracted by Vpx. AIDS Res Hum Retroviruses. 2013 doi: 10.1089/aid.2013.0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley JM, Hill BJ, Ambrozak DR, Price DA, Guenaga FJ, Casazza JP, Kuruppu J, Yazdani J, Migueles SA, Connors M, et al. T-Cell Subsets That Harbor Human Immunodeficiency Virus (HIV) In Vivo: Implications for HIV Pathogenesis. J Virol. 2004a;78:1160–1168. doi: 10.1128/JVI.78.3.1160-1168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, Nguyen PL, Khoruts A, Larson M, Haase AT, Douek DC. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med. 2004b;200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdo TH, Soulas C, Orzechowski K, Button J, Krishnan A, Sugimoto C, Alvarez X, Kuroda MJ, Williams KC. Increased monocyte turnover from bone marrow correlates with severity of SIV encephalitis and CD163 levels in plasma. PLoS Pathog. 2010;6:e1000842. doi: 10.1371/journal.ppat.1000842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline AN, Bess JW, Piatak M, Jr., Lifson JD. Highly sensitive SIV plasma viral load assay: practical considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. J Med Primatol. 2005;34:303–312. doi: 10.1111/j.1600-0684.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- Combadiere B, Boissonnas A, Carcelain G, Lefranc E, Samri A, Bricaire F, Debre P, Autran B. Distinct time effects of vaccination on long-term proliferative and IFN-gamma-producing T cell memory to smallpox in humans. J Exp Med. 2004;199:1585–1593. doi: 10.1084/jem.20032083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribier A, Descours B, Valadao AL, Laguette N, Benkirane M. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell reports. 2013;3:1036–1043. doi: 10.1016/j.celrep.2013.03.017. [DOI] [PubMed] [Google Scholar]
- Dai L, Lidie KB, Chen Q, Adelsberger JW, Zheng X, Huang D, Yang J, Lempicki RA, Rehman T, Dewar RL, et al. IL-27 inhibits HIV-1 infection in human macrophages by down-regulating host factor SPTBN1 during monocyte to macrophage differentiation. J Exp Med. 2013;210:517–534. doi: 10.1084/jem.20120572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Q, Whitted S, Goeken RM, Brenchley JM, Matsuda K, Brown CR, Lafont BA, Starost MF, Iyengar R, Plishka RJ, et al. Development of neurological disease is associated with increased immune activation in simian immunodeficiency virus-infected macaques. J Virol. 2012;86:13795–13799. doi: 10.1128/JVI.02174-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descours B, Cribier A, Chable-Bessia C, Ayinde D, Rice G, Crow Y, Yatim A, Schwartz O, Laguette N, Benkirane M. SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4(+) T-cells. Retrovirology. 2012;9:87. doi: 10.1186/1742-4690-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- Fregoso OI, Ahn J, Wang C, Mehrens J, Skowronski J, Emerman M. Evolutionary toggling of Vpx/Vpr specificity results in divergent recognition of the restriction factor SAMHD1. PLoS Pathog. 2013;9:e1003496. doi: 10.1371/journal.ppat.1003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JS, Lackner AA, Lang SM, Simon MA, Sehgal PK, Daniel MD, Desrosiers RC. Progression to AIDS in the absence of a gene for vpr or vpx. J Virol. 1995;69:2378–2383. doi: 10.1128/jvi.69.4.2378-2383.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2012;480:379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Mejia ME, Doseff AI. Regulation of monocytes and macrophages cell fate. Frontiers in bioscience. 2009;14:2413–2431. doi: 10.2741/3387. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Perez MP, O'Connell O, Lin R, Sullivan WM, Bell J, Simmonds P, Clapham PR. Independent evolution of macrophage-tropism and increased charge between HIV-1 R5 envelopes present in brain and immune tissue. Retrovirology. 2012;9:20. doi: 10.1186/1742-4690-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greter M, Merad M. Regulation of microglia development and homeostasis. Glia. 2013;61:121–127. doi: 10.1002/glia.22408. [DOI] [PubMed] [Google Scholar]
- Heeregrave EJ, Geels MJ, Brenchley JM, Baan E, Ambrozak DR, van der Sluis RM, Bennemeer R, Douek DC, Goudsmit J, Pollakis G, et al. Lack of in vivo compartmentalization among HIV-1 infected naive and memory CD4(+) T cell subsets. Virology. 2009 doi: 10.1016/j.virol.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch VM, Sharkey ME, Brown CR, Brichacek B, Goldstein S, Wakefield J, Byrum R, Elkins WR, Hahn BH, Lifson JD, Stevenson M. Vpx is required for dissemination and pathogenesis of SIV(SM) PBj: evidence of macrophage-dependent viral amplification. Nat Med. 1998;4:1401–1408. doi: 10.1038/3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi T, Brown CR, Endo Y, Buckler-White A, Plishka R, Bischofberger N, Hirsch V, Martin MA. Macrophage are the principal reservoir and sustain high virus loads in rhesus macaques after the depletion of CD4+ T cells by a highly pathogenic simian immunodeficiency virus/HIV type 1 chimera (SHIV): Implications for HIV-1 infections of humans. Proc Natl Acad Sci U S A. 2001;98:658–663. doi: 10.1073/pnas.021551798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi T, Imamichi H, Brown CR, Hirsch VM, Martin MA. The emergence and characterization of macrophage-tropic SIV/HIV chimeric viruses (SHIVs) present in CD4+ T cell-depleted rhesus monkeys. Journal of leukocyte biology. 2003;74:772–780. doi: 10.1189/jlb.0503196. [DOI] [PubMed] [Google Scholar]
- Jenkins SJ, Ruckerl D, Cook PC, Jones LH, Finkelman FD, van Rooijen N, MacDonald AS, Allen JE. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science. 2011;332:1284–1288. doi: 10.1126/science.1204351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katlama C, Deeks SG, Autran B, Martinez-Picado J, van Lunzen J, Rouzioux C, Miller M, Vella S, Schmitz JE, Ahlers J, et al. Barriers to a cure for HIV: new ways to target and eradicate HIV-1 reservoirs. Lancet. 2013;381:2109–2117. doi: 10.1016/S0140-6736(13)60104-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig S, Gendelman HE, Orenstein JM, Dal Canto MC, Pezeshkpour GH, Yungbluth M, Janotta F, Aksamit A, Martin MA, Fauci AS. Detection of AIDS virus in macrophages in brain tissue from AIDS patients with encephalopathy. Science. 1986;233:1089–1093. doi: 10.1126/science.3016903. [DOI] [PubMed] [Google Scholar]
- Koppensteiner H, Brack-Werner R, Schindler M. Macrophages and their relevance in Human Immunodeficiency Virus Type I infection. Retrovirology. 2012;9:82. doi: 10.1186/1742-4690-9-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis DM. Histone deacetylase inhibitors and HIV latency. Curr Opin HIV AIDS. 2011;6:25–29. doi: 10.1097/COH.0b013e328341242d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattapallil JJ, Douek DC, Hill B, Nishimura Y, Martin M, Roederer M. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature. 2005a;434:1093–1097. doi: 10.1038/nature03501. [DOI] [PubMed] [Google Scholar]
- Mattapallil JJ, Douek DC, Hill B, Nishimura Y, Martin M, Roederer M. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature. 2005b;434:1093–1097. doi: 10.1038/nature03501. [DOI] [PubMed] [Google Scholar]
- Orenstein JM, Fox C, Wahl SM. Macrophages as a source of HIV during opportunistic infections. Science. 1997;276:1857–1861. doi: 10.1126/science.276.5320.1857. [DOI] [PubMed] [Google Scholar]
- Ortiz AM, Klatt NR, Li B, Yi Y, Tabb B, Hao XP, Sternberg L, Lawson B, Carnathan PM, Cramer EM, et al. Depletion of CD4+ T cells abrogates post-peak decline of viremia in SIV-infected rhesus macaques. J Clin Invest. 2011;121:4433–4445. doi: 10.1172/JCI46023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauls E, Jimenez E, Ruiz A, Permanyer M, Ballana E, Costa H, Nascimiento R, Parkhouse RM, Pena R, Riveiro-Munoz E, et al. Restriction of HIV-1 replication in primary macrophages by IL-12 and IL-18 through the upregulation of SAMHD1. J Immunol. 2013;190:4736–4741. doi: 10.4049/jimmunol.1203226. [DOI] [PubMed] [Google Scholar]
- Picker LJ, Hagen SI, Lum R, Reed-Inderbitzin EF, Daly LM, Sylwester AW, Walker JM, Siess DC, Piatak M, Jr., Wang C, et al. Insufficient production and tissue delivery of CD4+ memory T cells in rapidly progressive simian immunodeficiency virus infection. J Exp Med. 2004;200:1299–1314. doi: 10.1084/jem.20041049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins CS, Hilgendorf I, Weber GF, Theurl I, Iwamoto Y, Figueiredo JL, Gorbatov R, Sukhova GK, Gerhardt LM, Smyth D, et al. Local proliferation dominates lesional macrophage accumulation in atherosclerosis. Nat Med. 2013;19:1166–1172. doi: 10.1038/nm.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano JD, Siliciano RF. HIV-1 eradication strategies: design and assessment. Curr Opin HIV AIDS. 2013;8:318–325. doi: 10.1097/COH.0b013e328361eaca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PD, Meng G, Salazar-Gonzalez JF, Shaw GM. Macrophage HIV-1 infection and the gastrointestinal tract reservoir. Journal of leukocyte biology. 2003;74:642–649. doi: 10.1189/jlb.0503219. [DOI] [PubMed] [Google Scholar]
- Spits H. Development of alphabeta T cells in the human thymus. Nature reviews. Immunology. 2002;2:760–772. doi: 10.1038/nri913. [DOI] [PubMed] [Google Scholar]
- Thompson KA, Cherry CL, Bell JE, McLean CA. Brain cell reservoirs of latent virus in presymptomatic HIV-infected individuals. Am J Pathol. 2011;179:1623–1629. doi: 10.1016/j.ajpath.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Furth R. Origin and turnover of monocytes and macrophages. Current topics in pathology. Ergebnisse der Pathologie. 1989;79:125–150. [PubMed] [Google Scholar]
- Veazey RS, DeMaria M, Chalifoux LV, Shvetz DE, Pauley DR, Knight HL, Rosenzweig M, Johnson RP, Desrosiers RC, Lackner AA. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science. 1998;280:427–431. doi: 10.1126/science.280.5362.427. [DOI] [PubMed] [Google Scholar]
- Wang TH, Donaldson YK, Brettle RP, Bell JE, Simmonds P. Identification of shared populations of human immunodeficiency virus type 1 infecting microglia and tissue macrophages outside the central nervous system. J Virol. 2001;75:11686–11699. doi: 10.1128/JVI.75.23.11686-11699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmoreland SV, Converse AP, Hrecka K, Hurley M, Knight H, Piatak M, Lifson J, Mansfield KG, Skowronski J, Desrosiers RC. SIV Vpx Is Essential for Macrophage Infection but Not for Development of AIDS. PLoS One. 2014;9:e84463. doi: 10.1371/journal.pone.0084463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Ourmanov I, Kuwata T, Goeken R, Brown CR, Buckler-White A, Iyengar R, Plishka R, Aoki ST, Hirsch VM. Sequential evolution and escape from neutralization of simian immunodeficiency virus SIVsmE660 clones in rhesus macaques. J Virol. 2012;86:8835–8847. doi: 10.1128/JVI.00923-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R, Schneider E. Detection of B- and T-cell-specific gene rearrangements in 13 cell lines and 50 clinical specimens using the BIOMED-2 and the original InVivoScribe primers. Leukemia & lymphoma. 2007;48:837–840. doi: 10.1080/10428190601187729. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.