Abstract
The phenomenon that adverse environmental exposures in early life are associated with increased susceptibilities for many adult, particularly metabolic diseases, is now referred to as ‘developmental origins of health and disease (DOHAD)’ or ‘Barker’ hypothesis. Fetal overnutrition and undernutrition have similar long-lasting effects on the setting of the neuroendocrine control systems, energy homeostasis, and metabolism, leading to life-long increased morbidity. There are sensitive time windows during early development, where environmental cues can program persistent epigenetic modifications which are generally assumed to mediate these gene–environment interactions. Most of our current knowledge on fetal programing comes from animal models and epidemiological studies in humans, in particular the Dutch famine birth cohort. In industrialized countries, there is more concern about adverse long-term consequences of fetal overnutrition, i.e. by exposure to gestational diabetes mellitus and/or maternal obesity which affect 10–20% of pregnancies. Epigenetic changes due to maternal diabetes/obesity may predispose the offspring to develop metabolic disease later in life and, thus, transmit the adverse environmental exposure to the next generation. This vicious cycle could contribute significantly to the worldwide metabolic disease epidemics. In this review article, we focus on the epigenetics of an adverse intrauterine environment, in particular gestational diabetes, and its implications for the prevention of complex disease.
Introduction
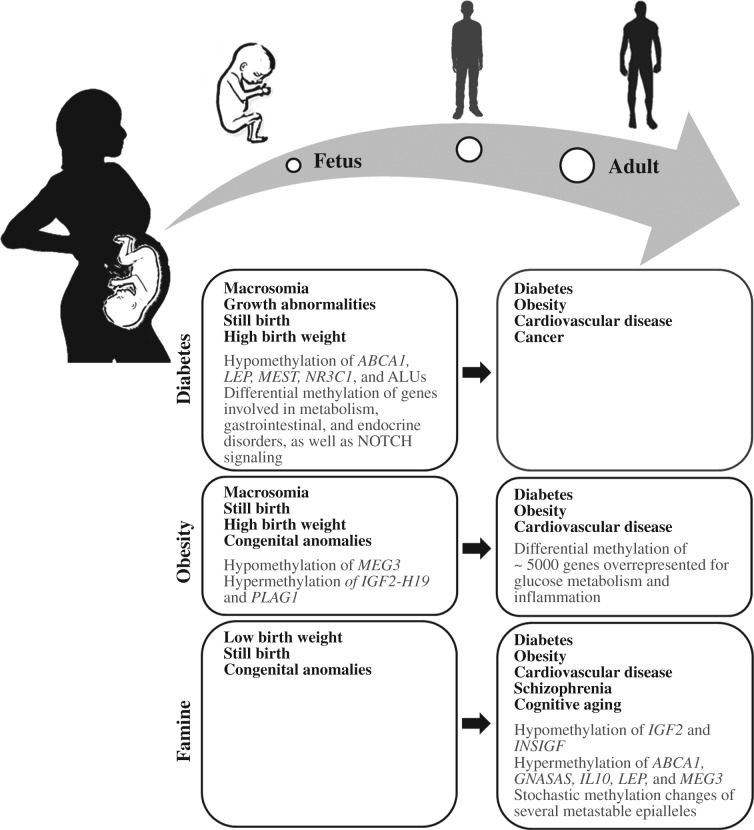
As early as 1880, puerperal diabetes has been associated with severe fetal and neonatal complications (Duncan 1882). In the 1950s, the term ‘gestational diabetes mellitus (GDM)’ was coined for a carbohydrate intolerance that develops during pregnancy and usually resolves after birth (Carrington et al. 1957). Over the course of pregnancy, increasing levels of hormones, such as lactogen, estrogen, and prolactin, are produced by the placenta to shift nutrients from the mother to the fetus and to prevent low blood glucose levels by resisting the action of insulin. Typically, insulin resistance arises during mid-gestation and progresses toward late-gestation. To counteract the diabetogenic effects of placental hormones, the mother's pancreatic islets try to increase insulin production (up to three times) during pregnancy. When the maternal β-cells cannot adapt to the increased food supply to the fetus and increased insulin demands during late pregnancy, the blood glucose levels will rise, stimulating insulin production of the fetal pancreas (Perkins et al. 2007). In GDM pregnancies, the balance between increased insulin resistance and maternal insulin production is disturbed, mostly due to insulin secretion defects of pancreatic β islets (Buchanan & Xiang 2005, Perkins et al. 2007). This results in maternal hyperglycemia, fetal hyperinsulinism, and consequently fetal overnutrition. Insulin gets glucose into cells for energy production and functions as a fetal growth factor, which may explain the increased risk for growth abnormalities, macrosomia, and stillbirth in GDM pregnancies (Fig. 1). The Hyperglycemia and Adverse Pregnancy Outcome (HAPO) Study on 25 000 pregnancies showed a strong correlation between maternal glucose levels and various perinatal complications (HAPO Study Cooperative Research Group et al. 2008, Lowe et al. 2012). Effective treatment of maternal diabetes by diet or insulin therapy can prevent adverse pregnancy outcome. The recommended diet (without fasting) consists of 45% carbohydrate, 30–35% fat, and 20% protein (American Diabetes Association 2003).
Figure 1.
Medical problems and epigenetic changes (in placenta and blood) at birth and later in life, respectively, that have been associated with intrauterine exposure to diabetes mellitus, maternal obesity, and famine.
The GDM shares a common etiology with type 2 diabetes (T2D). The mothers with GDM often develop T2D later in life (Kim et al. 2002). The prevalence of GDM varies among populations and can range from 1 to >10%, depending on ethnicity and diagnosis criteria (American Diabetes Association 2003, Ben-Haroush et al. 2004). The well-known risk factors for GDM include pre-pregnancy obesity, advanced maternal age, and a family history of diabetes. Low physical activity and poor diet increase GDM susceptibility (Buchanan & Xiang 2005). Several sequence variants in genes that modulate pancreatic β cell function and insulin resistance were identified in GDM patients (Robitaille & Grant 2008).
It is now widely accepted that the intrauterine environment influences key developmental processes and has long-lasting effects on health and disease. Already 35 years ago, Freinkel (1980) proposed the concept of ‘fuel-mediated teratogenesis’, postulating that increased nutrients, in particular glucose levels and the resulting fetal hyperinsulinism in GDM pregnancies, have both immediate and long-lasting adverse consequences for the offspring. Since Barker et al. (1993) first reported an association between low birth-weight, which can be considered as a surrogate marker for an adverse intrauterine environment, and increased susceptibilities for metabolic and other complex diseases later in life, this phenomenon is also referred to as ‘Barker hypothesis’ or alternatively ‘developmental origins of health and disease (DOHAD)’ (Gillman 2005). According to the ‘thrifty phenotype’ concept (Hales & Barker 1992), intrauterine undernutrition and overnutrition, respectively, program adaptations of the fetal metabolism to cope with an adverse postnatal environment which lacks or is enriched with suitable nutrients.
Discordant siblings, who were born before and after the mother developed T2D, provide compelling evidence that it is the intrauterine diabetic environment which conveys an increased risk for metabolic disease in the offspring (Dabelea et al. 2000). Additional epidemiological and experimental studies (Wright et al. 2009, Moore 2010, Wu et al. 2012) support the concept that intrauterine hyperglycemia predisposes the offspring to obesity, metabolic and cardiovascular diseases, and a variety of cancers (Fig. 1). This vicious cycle may help to explain the epidemic increase of metabolic diseases in most developed countries.
Non-genetic intergenerational transmission of the metabolic phenotype has been extensively studied in GDM rodent models of intrauterine hyperglycemia. The offspring of rats with mild diabetes, induced by streptozotocin injection, are prone to develop diabetes and show fetal endocrine changes down to the third generation (Aerts & van Assche 1979). Embryo transfer experiments revealed that an euglycemic intrauterine environment cannot alleviate the strong genetic predisposition of Goto-Kakizaki (GK) embryos to develop diabetes, whereas a hyperglycemic environment in GK foster mothers augments the diabetes risk in Wistar embryos that have a low genetic susceptibility (Gill-Randall et al. 2004). Pancreatic islet transplantation in pregnant diabetic rats could improve glucose homeostasis in the offspring (Aerts & van Assche 1992). In a streptozotocin-induced GDM mouse model, intrauterine hyperglycemia was associated with impaired glucose tolerance and abnormal insulin levels in the F1 and F2 offspring, which may be related with abnormal methylation and expression of the IGF2-H19 locus in pancreatic islets and impaired islet structure and function (Ding et al. 2012). It was speculated that dysregulation of IGF2-H19 in sperm contributes to sex-specific transgenerational epigenetic inheritance.
Metabolic phenotype in later life depends on intrauterine nutrition
Phenotypic effects of an adverse nutritional environment in humans
A large number of studies have associated exposures to materno-fetal undernutrition (due to wartimes and wrong political decisions during the last century) with health outcome in later life. The famous ‘Dutch famine birth cohort’ consists of more than 2000 singletons who were born between November 1943 and February 1947 in Amsterdam and systematically followed up since 1996. Under the Nazi embargo of the Western Netherlands in 1944, pregnant women had only 400–800 kcal/day, roughly a quarter of the recommended intake. Individuals exposed to undernutrition during early gestation exhibited an increased risk for metabolic, cardiovascular and other complex diseases, schizophrenia, and accelerated cognitive aging (Roseboom et al. 2006, Schulz 2010; Fig. 1). Similarly, individuals who were born in China from 1958 to 1961 during the ‘Great Leap Forward’ famine displayed higher susceptibility for schizophrenia than comparable pre- and post-famine cohorts (Song et al. 2009). Children born from 1968 to 1970 during the Biafra famine in Nigeria showed a significantly increased risk for obesity and its confounding maladies (Hult et al. 2010).
Fetal overnutrition due to an obesogenic maternal environment also influences the metabolic phenotype later in life. Maternal obesity increases the risk of developing insulin resistance during pregnancy and intrauterine exposure to both maternal obesity and GDM has been linked to increased gestational weight, adiposity, and T2D in youth (Boney et al. 2005, Poston 2012; Fig. 1).
Epigenetic effects
If early-life exposures confer a life-long increased risk for obesity, diabetes, and other complex diseases, they must cause persistent changes in the metabolism, neuroendocrine functions, etc. underlying these disease susceptibilities. One possible explanation is that intrauterine malnutrition interferes with organogenesis and, thus, has a long-lasting negative impact on organ morphology and physiology (Svensson et al. 1992). Another plausible mechanism is environmentally induced epigenetic modifications, leading to persistent changes in gene regulation and pathways (Gluckman et al. 2009, Lehnen et al. 2013). Epigenetic modifications, i.e. in form of DNA methylation and histone modifications, can cause changes in gene expression, which are transmitted to daughter cells during somatic cell division and, perhaps also from one generation to the next. It is well known that the epigenome(s) is highly plastic, particularly during early development (Reik et al. 2001) and susceptible to internal (i.e. during differentiation) and external environmental cues (Faulk & Dolinoy 2011, Feil & Fraga 2012).
A large number of studies on gene–environment interactions focused on 5-methylcytosine, the most prominent epigenetic modification of DNA itself. In non-coding regions of the genome, most CpG sites are methylated to prevent retrotransposition activity and genome instability (Yoder et al. 1997). CpG islands in promoter regions are usually demethylated, and promoter methylation during development or disease processes is associated with gene silencing (Jaenisch & Bird 2003, Weber et al. 2007). In contrast, CpGs in gene bodies are methylated, which is thought to play a role in exon definition and alternative splicing (Gelfman & Ast 2013). Approximately 100–200 of our more than 20 000 genes are expressed in a parent-specific manner. Most of these imprinted genes contain or lie near CpG islands that display differential methylation patterns between the maternal and paternal alleles (Kelsey 2007). Imprinted genes are essential for the regulation of fetal and placental growth, somatic differentiation, as well as neurological and behavioral functions after birth (Reik et al. 2003) and frequently used as a model to study the epigenetic effects of environmental factors during early development (Denomme & Mann 2012, El Hajj & Haaf 2013).
More than 60 years after early gestational exposure, the Dutch famine individuals showed subtle hypomethylation of the imprinted IGF2-H19 locus, compared with their unexposed sibling (Heijmans et al. 2008). A follow-up study on the same cohort reported sex- and exposure timing-dependent changes in blood methylation of several imprinted and non-imprinted genes (Tobi et al. 2009; Fig. 1). A genome-wide screening of Gambian children conceived during the nutritionally poor rainy season revealed stochastic methylation changes in several putative metastable epialleles (Waterland et al. 2010), compared with children conceived in the dry season. The methylation status of metastable epialleles is established during early development, depending on stochastic and environmental factors, and influences the phenotype in later life (Rakyan et al. 2002). Genome-wide blood methylation and expression analyses revealed that maternal obesity/fetal overnutrition also has broad effects on the offsprings’ epigenome (Guénard et al. 2013).
Animal studies
The agouti viable yellow (Avy) mouse is a particularly impressive model for the phenotypic consequences of epigenetic changes during early development. Insertion of an intracisternal A particle upstream of the Avy allele renders the locus susceptible to stochastic and environmentally induced methylation after fertilization. Methylation-dependent silencing of the metastable Avy epiallele suppresses the Avy phenotype, which is characterized by yellow coat color as well as predisposition to obesity, insulin resistance, and cancer (Duhl et al. 1994, Morgan et al. 1999). When fed a western-style diet, the offspring of obese and diabetic Avy mice are more susceptible to develop glucose intolerance, insulin resistance, and hepatic steatosis (Li et al. 2013a ), suggesting epigenetic transmission of the metabolic phenotype to the next generation.
In streptozotocin-induced diabetic mouse models, maternal hyperglycemia interfered with formation of endoplasmic reticulum in maternal oocytes and the resulting embryos, and was associated with time-dependent methylation changes in several imprinted genes (Ge et al. 2013a , Zhang et al. 2013). The effects of an adverse intrauterine environment on oocytes from female offspring appeared to depend on the type of malnutrition. A high-fat maternal diet induced aberrant methylation changes across several metabolism-related genes in both maternal and fetal oocytes, whereas a hyperglycemic environment did not affect fetal germ-cell methylation patterns (Ge et al. 2013b , Ge et al. 2014). In addition, intrauterine exposure to either high-fat diet or maternal diabetes was shown to influence histone H3 and H4 acetylation of embryonal genes implicated in neural tube defects (Salbaum & Kappen 2012).
Similar to an intrauterine environment enriched with nutrients, early postnatal overfeeding can also influence the epigenetic and metabolic status of the offspring. In the small litter Wistar rat model, postnatal overnutrition caused rapid weight gain, early dyslipidemia, hyperinsulinemia, and dysregulated glucose homeostasis (Plagemann et al. 1992). The metabolic syndrome phenotype was associated with hypermethylation of the pro-opiomelanoctin (Pomc) and the insulin receptor (Insr) promoters in the hypothalamus (Plagemann et al. 2009, 2010). Small litter/overfed mice displayed similar pathophysiological changes as displayed by rats, along with a female-specific reduction in physical activity and energy expenditure. Genome-wide methylation analysis revealed persistent and sex-specific epigenetic changes across 900 loci in the hypothalamus associated with neural development (Li et al. 2013b ).
Last but not least, the developmental origin of metabolic disease has also been studied in primates. Japanese macaques exposed to a high-fat maternal diet during pregnancy exhibited increased triglyceride levels and histological changes reminiscent of non-alcoholic fatty liver disease. These metabolic alterations were associated with hyperacetylation of several histone H3 variants which may be due to reduced fetal histone deacetylase 1 activity (Aagaard-Tillery et al. 2008).
Fetal programing by GDM
In GDM pregnancies, the fetus is exposed to increased levels of glucose, free fatty acids, and amino acids, leading to increased fetal insulin production. Hyperglycemia and fetal hyperinsulinism are thought to program metabolic disease predispositions, which manifest later in life (Fig. 1). One plausible mechanism underlying fetal programing by GDM is that nutrient and hormonal levels in utero affect metabolic and neuroendocrine feedback loops, which by themselves are metastabile, but may be stabilized by epigenetic modifications that once established during development are further propagated throughout cell divisions.
Candidate gene studies
Although genome-wide epigenetic studies have become feasible, the candidate gene approach is still valuable, because it avoids the multiple testing problem and, therefore, allows one to detect significant changes of small effect size. Bouchard et al. (2010) measured DNA methylation of leptin (LEP), a hormone that controls energy intake and expenditure, as well as body weight, in pregnant women with impaired glucose tolerance (Fig. 1). Maternal blood glucose levels correlated positively with LEP methylation in maternal placental tissue and negatively in fetal tissue. A follow-up study examined placental methylation of the adiponectin (ADIPOQ) gene promoter, an adipocyte-derived hormone with insulin-sensitizing effects (Bouchard et al. 2012). Lower ADIPOQ methylation in fetal placenta was associated with higher maternal glucose levels and lower methylation on the maternal side of the placenta with higher insulin resistance index. Houde et al. (2013) studied promoter methylation of ATP-binding cassette transporter A1 (ABCA1), a regulator of cholesterol efflux pathways and atherosclerosis. ABCA1 methylation levels in maternal placenta correlated negatively with maternal HDL-cholesterol levels and positively with maternal glucose levels. On the fetal side, placental ABCA1 methylation correlated negatively with cord blood triglyceride levels, whereas cord blood methylation correlated negatively with maternal glucose levels.
In our own study (El Hajj et al. 2013), we compared fetal cord blood as well as placenta methylation levels of seven imprinted genes involved in pre- and postnatal growth, four genes involved in energy metabolism, one anti-inflammatory gene, one tumor suppressor gene, one pluripotency gene, and two repetitive DNA families between GDM and control pregnancies. DNA methylation was quantified by bisulfite pyrosequencing that permits highly accurate measurements at single-base resolution. Of the 14 analyzed genes/repeats, the maternally imprinted MEST gene, the glucocorticoid receptor NR3C1, and ALU interspersed elements (Fig. 1) showed significant hypomethylation in both tissues of newborns of mothers with either dietetically treated or insulin-dependent GDM, compared with controls without exposure to GDM. The largest effect was observed for MEST, which showed a 4% lower methylation in cord blood and a 6% lower methylation in fetal placenta of GDM pregnancies. Recently, it has been reported that MEST hypomethylation in fetal cord blood is also significantly associated with paternal obesity, suggesting transmission of sperm epigenetic signatures for obesity into the next generation (Soubry et al. 2013). Consistent with its suspected role in the programing of an increased metabolic disease risk, MEST methylation was also significantly lower in the blood of obese adults than in normal-weight (sex- and age-matched) controls (El Hajj et al. 2013). In addition, MEST methylation was negatively correlated with BMI and waist circumference, supporting a role for this gene in the development of diet-induced obesity (Carless et al. 2013). In the mouse model, loss of Mest imprinting has been linked to increased body weight and organ size (Shi et al. 2004) and its overexpression promotes fat tissue expansion and adipocyte enlargement (Nikonova et al. 2008).
Genome-wide analyses
One study used a luminometric methylation assay based on methylation-sensitive restriction enzymes to quantify global DNA methylation in pregnancies with different maternal medical risks (Nomura et al. 2014). The placenta but not the cord blood methylation levels were slightly lower in pregnancies with GDM and higher in pregnancies with maternal obesity. Although it is possible that GDM and obesity have a greater impact on placenta than on cord blood and oppositely program the epigenome, these results are preliminary.
To systematically compare cord blood methylation patterns between GDM and non-GDM pregnancies on a genome-wide scale, we used Infinium Human Methylation 450K bead chips, which quantify methylation at >485 000 CpGs distributed across 99% of all RefSeq genes including promoter, 5’UTR, gene body, and 3’UTR. Although we observed numerous between-group methylation differences in the order of one percentage point, following adjustment for multiple testing by the false discovery rate not a single CpG site remained significant. Since the sample size in current genome-wide epigenetic screens does not meet genetic (GWAS) study standards, it is not unexpected that small effects in individual genes do not reach genome-wide significance. Nevertheless, functional annotation enrichment analysis can help to elucidate the biological meaning of such data sets polluted with false positives and negatives.
Ingenuity Pathway Analysis identified ‘Cancer’, ‘Respiratory Disease’, ‘Metabolic Disease’, ‘Cardiovascular Disease’, and ‘Endocrine System Disorder’ as the top five diseases and disorders and ‘Carbohydrate Metabolism, Cardiac Dilation, Cardiovascular System Development and Function’ as top associated network functions. Using Panther Pathway Analysis, the top five pathways ‘Notch Signaling’, ‘Gonadotropin Releasing Hormone Receptor’, ‘PDGF Signaling’, ‘Heterotrimeric G-protein Signaling’, and ‘Integrin Signaling’ as well as the top five biological functions ‘Multicellular Organismal Process’, ‘Metabolic Process’, ‘Developmental Process’, ‘Cell Communication’, and ‘Cellular Process’ were significantly enriched (after Bonferroni's correction for multiple testing) with differentially methylated genes.
In a conceptually related study using the same methylation array, Ruchat et al. (2013) compared the cord blood and fetal placenta methylomes in GDM vs non-GDM pregnancies. The top loci in cord blood were enriched for the pathways ‘Gastrointestinal Disease’, ‘Metabolic Disease’, and ‘Endocrine System Disorder’. In placenta, the enriched pathways included ‘Cardiovascular Disease’, ‘Metabolic Disease’, and ‘Psychological Disorder’. When only considering genes with significant changes in both tissues, the enriched pathways ‘Immunological Disease’, ‘Metabolic Disease’, and ‘Endocrine System Disorder’ were identified. Several differentially methylated genes were correlated with birth weight, linking GDM exposure to macrosomia.
In a genome-wide methylated DNA immunoprecipitation-chip study, comparing blood methylation in offspring from diabetic (T2D) and non-diabetic Pima Indian mothers, pathway analysis of differentially methylated gene promoters identified ‘maturity onset diabetes of the young’, ‘T2D’, and ‘Notch signaling’ (Del Rosario et al. 2014).
Collectively, existing genome-wide analyses support the view that intrauterine diabetic environment epigenetically influences pathways involved in complex diseases. Consistent with a multifactorial disease model, epigenetic changes were reported in a large number of genes, however no changes with large effect size. Owing to small sample sizes and limited statistical power, so far no novel gene(s) has been identified with genome-wide significance in such studies. Concerted efforts (multi-center epigenome studies) are needed to recruit and analyze materials from larger study populations.
Limitations
DNA methylation patterns are copied (by DNA methyltransferase 1) during DNA replication, implying that environmentally induced perturbations that occur during early development can be transmitted through cell divisions to subsequent cell generations. In this light, epigenetic modulations of gene regulation provide a plausible mechanistic link between intrauterine environment and life-long disease risk (Faulk & Dolinoy 2011, Feil & Fraga 2012, Lehnen et al. 2013). However, so far there is only circumstantial evidence that methylation changes that are programed in utero persist into adulthood and are causing metabolic changes. Most studies have only looked at narrow time windows either early or late in life. At present we cannot exclude the formal possibility that the associated epigenetic changes are only secondary effects of the pathophysiological processes predisposing to complex diseases.
Epigenetic variation is regulated in a tissue- and developmental stage-specific manner. Each cell type/tissue in our body is characterized by a specific combination of active and silenced genes, resulting from a complex interplay of stochastic, genetic, and environmental factors. In humans, it is hardly possible to study the target tissues of intrauterine programing, which in the case of GDM include fetal pancreatic islets, adipose tissue, skeletal muscle, liver, and the hypothalamic–pituitary–adrenal axis. Instead, easily accessible tissues such as cord blood and placenta are used to extrapolate the epigenetic effects of an adverse intrauterine environment underlying metabolic programing in the target tissues. Because of the enormous between-tissue differences in DNA methylation patterns, it is a major challenge to identify informative epigenetic signatures in blood or placenta epigenomes and then confirm that they reflect at least to some extent methylation variation in the target tissues. To account for the fact that neither fetal cord blood nor placenta is a crucial target for GDM, in our study (El Hajj et al. 2013) we only considered genes with significant between-group differences in the same direction in both tissues as likely candidates for metabolic programing.
Moreover, the studied tissues are usually composed of many different cell types. Sometimes, it is difficult to exclude that minor methylation changes which have been linked to a specific intrauterine condition are merely due to changes in cell composition of the analysed tissue. For example, whole blood of newborns of mothers with GDM may vary in the differential blood cell counts from normal controls (Yeruchimovich et al. 2000). The development of bioinformatic algorithms using DNA methylation signatures to infer changes in the cellular composition of whole blood samples between individuals (i.e. cases and controls) is a promising strategy to overcome this problem (Houseman et al. 2012).
Outlook
The developmental origins or Barker hypothesis has been supported by a large number of epidemiological and animal studies during the past two decades. It has been proposed that intrauterine programing of the fetal metabolism has evolved to enhance maternal fitness (Wells 2007). On the other hand, the ‘parental conflict hypothesis’ (Moore & Haig 1991) postulates a battle over the allocation of maternal resources during pregnancy and also after birth, using imprinted genes as arms. In general, paternally expressed (maternally methylated) imprinted genes promote growth by extracting maximal maternal resources for a given pregnancy, whereas maternally expressed (paternally methylated) genes restrict growth and allocate resources equally among all offspring, which in polyandrous species may come from different fathers. In this context, it is interesting to speculate that paternally expressed genes, such as MEST (El Hajj et al. 2013), are susceptible to an intrauterine environment enriched with nutrients (overgrowth situation), whereas maternally expressed genes are more important for adaptation to a nutritionally poor environment (growth retardation). In this light, imprinted genes are an obvious target of metabolic programing and can serve as models for studying the long-term effects of an adverse intrauterine environment.
Because embryonal and fetal development exhibit considerable species differences, in particular between humans and rodents, animal models can be useful for proof-of-principle studies but the specific results cannot be directly extrapolated to the human situation. In the mouse model, the expression of several imprinted genes in the liver of offspring was influenced by maternal nutrition during gestation or lactation, but the methylation of the respective imprinting control regions remained largely unchanged (Ivanova et al. 2012).
Is it beneficial to prevent epigenetic changes in utero?
Mothers who develop GDM are usually treated by dietetic measures and/or insulin therapy to prevent fetal hyperglycemia and hyperinsulinism. The methylation changes that are observed in the offspring at birth may result from both GDM and its treatment. In humans, it is difficult to dissect different components of epigenetic variation. Changes in maternal nutrition and pharmacological interventions during pregnancy may have significant effects on the fetal epigenome. During the nutritionally depressed rainy season in Gambia, supplementation of the maternal diet with vitamins and minerals around the time of conception had wide-spread and persisting (at least into early infancy) effects on the epigenome of the offspring (Cooper et al. 2012, Khulan et al. 2012).
Bariatric surgery can be an effective treatment for morbid obesity and diabetes (Edholm et al. 2013). Studies comparing siblings conceived before and after surgical weight loss of the mother showed that children who were not exposed to maternal obesity had a lower risk of developing obesity, improved metabolic status, lipid profiles, and insulin sensitivity (Kral et al. 2006, Smith et al. 2009). Genome-wide analyses of siblings born before and after maternal weight loss found methylation and expression differences in ∼5000 genes enriched in functional categories for glucose metabolism, diabetes signaling, inflammation, and autoimmune disease (Guénard et al. 2013).
In a mouse model of diabetic pregnancy, a high-protein diet was shown to modulate the expression of diet-responsive genes and reduce placental abnormalities (Kappen et al. 2012). Such diet-induced adaptations of the placenta have been implicated in fetal programing (Godfrey 2002) and may help to normalize aberrant methylation patterns and life-long disease risk. However, the materno-fetal interface of the placenta and its role for fetal programing remain to a large extent a black box.
Is it possible to reverse epigenetic modifications after birth?
Eventually, early pharmacological, nutritional, and/or behavioral interventions can be applied to reverse epigenetic changes which have been programed by an adverse intrauterine environment. However, systematic experimental and clinical testing of such treatments in human newborns is difficult to justify. Although breast feeding is usually recommended, early neonatal ingestion of milk from diabetic mothers appears to increase the risk for obesity and impaired glucose tolerance in the offspring (Plagemann et al. 2002), whereas breast feeding after the first week of life and its duration have no influence on the childhood risk (Rodekamp et al. 2005). This argues in favor of the notion that the early neonatal period is a particularly critical time window for therapeutical interventions to break the vicious cycle of metabolic malprograming in utero. In the rat model, it was demonstrated that different postnatal inventions, i.e. neonatal leptin (Vickers et al. 2005, Gluckman et al. 2007), pre-weaning growth hormone (Gray et al. 2013), and juvenile folic acid treatment (Burdge et al. 2009) can prevent the long-term metabolic consequences of fetal undernutrition.
Epigenetic biomarkers for metabolic disease risk?
There are only a few studies demonstrating that epigenetic signatures measured after birth or in childhood may serve as biomarkers for metabolic disease later in life. The methylation status of the retinoid X receptor, alpha (RXRA) and nitric oxide synthase 3 (NOS3) promoters in umbilical cord tissue of healthy newborns were associated with childhood adiposity at 9 years of age (Godfrey et al. 2011). Peroxisomal proliferator-activated receptor-gamma-co-activator-1alpha (PPARGC1A) promoter methylation in the blood of 5–7-year-old children was temporally stable and predicted adiposity up to 14 years (Clarke-Harris et al. 2014). These findings provide a proof-of-principle for the utility of methylation markers in the management of metabolic disease risk and emphasize the need for genome-wide longitudinal studies on well-defined birth cohorts.
If modulations of the fetal epigenome can permanently increase an individual's risk of chronic disorders in later stages of life, we should be much more concerned about ensuring an optimum environment from conception to birth. Following the developmental origins hypothesis, the most important time for the action to prevent metabolic disease epidemics is prenatally and early postnatally (Lehnen et al. 2013), when the epigenome is still highly plastic (Gluckman et al. 2009). Unfortunately, so far there is little knowledge on how to estimate the quality of the intrauterine environment in a given pregnancy and how to improve it. Research in this direction is urgently needed.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the review.
Funding
This study was supported by the German Research Foundation (grant number HA 1374/15-1).
References
- Aagaard-Tillery KM, Grove K, Bishop J, Ke X, Fu Q, McKnight R, Lane RH. Developmental origins of disease and determinants of chromatin structure: maternal diet modifies the primate fetal epigenome. Journal of Molecular Endocrinology. 2008;41:91–102. doi: 10.1677/JME-08-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aerts L, van Assche FA. Is gestational diabetes an acquired condition? Journal of Developmental Physiology. 1979;1:219–225. [PubMed] [Google Scholar]
- Aerts L, van Assche FA. Islet transplantation in diabetic pregnant rats normalizes glucose homeostasis in their offspring. Journal of Developmental Physiology. 1992;17:283–287. [PubMed] [Google Scholar]
- American Diabetes Association. Gestational diabetes mellitus. Diabetes Care. 2003;26(Suppl 1):S103–S105. doi: 10.2337/diacare.26.2007.S103. [DOI] [PubMed] [Google Scholar]
- Barker DJ, Hales CN, Fall CH, Osmond C, Phipps K, Clark PM. Type 2 (non-insulin-dependent) diabetes mellitus, hypertension and hyperlipidaemia (syndrome X): relation to reduced fetal growth. Diabetologia. 1993;36:62–67. doi: 10.1007/BF00399095. [DOI] [PubMed] [Google Scholar]
- Ben-Haroush A, Yogev Y, Hod M. Epidemiology of gestational diabetes mellitus and its association with type 2 diabetes. Diabetic Medicine. 2004;21:103–113. doi: 10.1046/j.1464-5491.2003.00985.x. [DOI] [PubMed] [Google Scholar]
- Boney CM, Verma A, Tucker R, Vohr BR. Metabolic syndrome in childhood: association with birth weight, maternal obesity, and gestational diabetes mellitus. Pediatrics. 2005;115:e290–e296. doi: 10.1542/peds.2004-1808. [DOI] [PubMed] [Google Scholar]
- Bouchard L, Thibault S, Guay SP, Santure M, Monpetit A, St-Pierre J, Perron P, Brisson D. Leptin gene epigenetic adaptation to impaired glucose metabolism during pregnancy. Diabetics Care. 2010;33:2436–2441. doi: 10.2337/dc10-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard L, Hivert MF, Guay SP, St-Pierre J, Perron P, Brisson D. Placental adiponectin gene DNA methylation levels are associated with mothers' blood glucose concentration. Diabetes. 2012;61:1272–1280. doi: 10.2337/db11-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan TA, Xiang AH. Gestational diabetes mellitus. Journal of Clinical Investigation. 2005;115:485–491. doi: 10.1172/JCI200524531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdge GC, Lillycrop KA, Phillips ES, Slater-Jefferies JL, Jackson AA, Hanson MA. Folic acid supplementation during the juvenile–pubertal period in rats modifies the phenotype and epigenotype induced by prenatal nutrition. Journal of Nutrition. 2009;139:1054–1060. doi: 10.3945/jn.109.104653. [DOI] [PubMed] [Google Scholar]
- Carless MA, Kulkarni H, Kos MZ, Charlesworth J, Peralta JM, Göring HH, Curran JE, Almasy L, Dyer TD, Comuzzie AG, et al. Genetic effects on DNA methylation and its potential relevance for obesity in Mexican Americans. PLoS ONE. 2013;8:e73950. doi: 10.1371/journal.pone.0073950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington ER, Shuman CR, Reardon HS. Evaluation of the prediabetic state during pregnancy. Obstetrics and Gynecology. 1957;9:664–669. doi: 10.1097/00006250-195706000-00008. [DOI] [PubMed] [Google Scholar]
- Clarke-Harris R, Wilkin TJ, Hosking J, Pinkney J, Jeffery AN, Metcalf BS, Godfrey KM, Voss LD, Lillycrop KA, Burdge GC. PGC1α promoter methylation in blood at 5–7 years predicts adiposity from 9 to 14 years (EarlyBird 50) Diabetes. 2014;63:2528–2537. doi: 10.2337/db13-0671. [DOI] [PubMed] [Google Scholar]
- Cooper WN, Khulan B, Owens S, Elks CE, Seidel V, Prentice AM, Belteki G, Ong KK, Affara NA, Constância M, et al. DNA methylation profiling at imprinted loci after periconceptional micronutrient supplementation in humans: results of a pilot randomized controlled trial. FASEB Journal. 2012;26:1782–1790. doi: 10.1096/fj.11-192708. [DOI] [PubMed] [Google Scholar]
- Dabelea D, Hanson RL, Lindsay RS, Pettitt DJ, Imperatore G, Gabir MM, Roumain J, Bennett PH, Knowler WC. Intrauterine exposure to diabetes conveys risks for type 2 diabetes and obesity: a study of discordant sibships. Diabetes. 2000;49:2208–2211. doi: 10.2337/diabetes.49.12.2208. [DOI] [PubMed] [Google Scholar]
- Del Rosario MC, Ossowski V, Knowler WC, Bogardus C, Baier LJ, Hanson RL. Potential epigenetic dysregulation of genes associated with MODY and type 2 diabetes in humans exposed to a diabetic intrauterine environment: an analysis of genome-wide DNA methylation. Metabolism. 2014;63:654–660. doi: 10.1016/j.metabol.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denomme MM, Mann MR. Genomic imprints as a model for the analysis of epigenetic stability during ARTs. Reproduction. 2012;144:393–409. doi: 10.1530/REP-12-0237. [DOI] [PubMed] [Google Scholar]
- Ding GL, Wang FF, Shu J, Tian S, Jiang Y, Zhang D, Wang N, Luo Q, Zhang Y, Jin F, et al. Transgenerational glucose intolerance with Igf2/H19 epigenetic alterations in mouse islet induced by intrauterine hyperglycemia. Diabetes. 2012;61:1133–1142. doi: 10.2337/db11-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhl D, Vrieling MH, Miller KA, Wolff GL, Barsh GS. Neomorphic agouti mutations in obese yellow mice. Nature Genetics. 1994;8:59–65. doi: 10.1038/ng0994-59. [DOI] [PubMed] [Google Scholar]
- Duncan JM. On puerperal diabetes. Transactions of the Obstetrical Society of London. 1882;24:256–285. [Google Scholar]
- Edholm D, Svensson F, Näslund I, Karlsson FA, Rask E, Sundbom M. Long-term results 11 years after primary gastric bypass in 384 patients. Surgery for Obesity and Related Diseases. 2013;9:708–713. doi: 10.1016/j.soard.2012.02.011. [DOI] [PubMed] [Google Scholar]
- El Hajj N, Haaf T. Epigenetic disturbances in in vitro cultured gametes and embryos: implications for human assisted reproduction. Fertility and Sterility. 2013;99:632–641. doi: 10.1016/j.fertnstert.2012.12.044. [DOI] [PubMed] [Google Scholar]
- El Hajj N, Pliushch G, Schneider E, Dittrich M, Müller T, Korenkov M, Aretz M, Zechner U, Lehnen H, Haaf T. Metabolic programming of MEST DNA methylation by intrauterine exposure to gestational diabetes mellitus. Diabetes. 2013;62:1320–1328. doi: 10.2337/db12-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulk C, Dolinoy DC. Timing is everything: the when and how of environmentally induced changes in the epigenome of animals. Epigenetics. 2011;6:791–797. doi: 10.4161/epi.6.7.16209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nature Reviews. Genetics. 2012;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- Freinkel N. Banting Lecture 1980. Of pregnancy and progeny. Diabetes. 1980;29:1023–1035. doi: 10.2337/diab.29.12.1023. [DOI] [PubMed] [Google Scholar]
- Ge ZJ, Liang QX, Luo SM, Wei YC, Han ZM, Schatten H, Sun QY, Zhang CL. Diabetic uterus environment may play a key role in alterations of DNA methylation of several imprinted genes at mid-gestation in mice. Reproductive Biology and Endocrinology. 2013a;11:119. doi: 10.1186/1477-7827-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge ZJ, Liang XW, Guo L, Liang QX, Luo SM, Wang YP, Wei YC, Han ZM, Schatten H, Sun QY. Maternal diabetes causes alterations of DNA methylation statuses of some imprinted genes in murine oocytes. Biology of Reproduction. 2013b;88:117. doi: 10.1095/biolreprod.112.105981. [DOI] [PubMed] [Google Scholar]
- Ge ZJ, Luo SM, Lin F, Liang QX, Huang L, Wei YC, Hou Y, Han ZM, Schatten H, Sun QY. DNA methylation in oocytes and liver of female mice and their offspring: effects of high-fat-diet-induced obesity. Environmental Health Perspectives. 2014;122:159–164. doi: 10.1289/ehp.1307047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfman S, Ast G. When epigenetics meets alternative splicing: the roles of DNA methylation and GC architecture. Epigenomics. 2013;5:351–353. doi: 10.2217/epi.13.32. [DOI] [PubMed] [Google Scholar]
- Gillman MW. Developmental origins of health and disease. New England Journal of Medicine. 2005;353:1848–1850. doi: 10.1056/NEJMe058187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill-Randall R, Adams D, Ollerton RL, Lewis M, Alcolado JC. Type 2 diabetes mellitus – genes or intrauterine environment? An embryo transfer paradigm in rats. Diabetologia. 2004;47:1354–1359. doi: 10.1007/s00125-004-1464-x. [DOI] [PubMed] [Google Scholar]
- Gluckman PD, Lillycrop KA, Vickers MH, Pleasants AB, Phillips ES, Beedle AS, Burdge GC, Hanson MA. Metabolic plasticity during mammalian development is directionally dependent on early nutritional status. PNAS. 2007;104:12796–12800. doi: 10.1073/pnas.0705667104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluckman PD, Hanson MA, Buklijas T, Low FM, Beedle AS. Epigenetic mechanisms that underpin metabolic and cardiovascular diseases. Nature Reviews. Endocrinology. 2009;5:401–408. doi: 10.1038/nrendo.2009.102. [DOI] [PubMed] [Google Scholar]
- Godfrey KM. The role of the placenta in fetal programming – a review. Placenta. 2002;23(Suppl A):S20–S27. doi: 10.1053/plac.2002.0773. [DOI] [PubMed] [Google Scholar]
- Godfrey KM, Sheppard A, Gluckman PD, Lillycrop KA, Burdge GC, McLean C, Rodford J, Slater-Jefferies JL, Garratt E, Crozier SR, et al. Epigenetic gene promoter methylation at birth is associated with child's later adiposity. Diabetes. 2011;60:1528–1534. doi: 10.2337/db10-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray C, Li M, Reynolds CM, Vickers MH. Pre-weaning growth hormone treatment reverses hypertension and endothelial dysfunction in adult male offspring of mothers undernourished during pregnancy. PLoS ONE. 2013;8:e53505. doi: 10.1371/journal.pone.0053505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guénard F, Deshaies Y, Cianflone K, Kral JG, Marceau P, Vohl MC. Differential methylation in glucoregulatory genes of offspring born before vs. after maternal gastrointestinal bypass surgery. PNAS. 2013;110:11439–11444. doi: 10.1073/pnas.1216959110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales CN, Barker DJ. Type 2 (non-insulin-dependent) diabetes mellitus: the thrifty phenotype hypothesis. Diabetologia. 1992;35:595–601. doi: 10.1007/BF00400248. [DOI] [PubMed] [Google Scholar]
- HAPO Study Cooperative Research Group , Metzger BE, Lowe LP, Dyer AR, Trimble ER, Chaovarindr U, Coustan DR, Hadden DR, McCance DR, Hod M, et al. Hyperglycemia and adverse pregnancy outcomes. New England Journal of Medicine. 2008;358:1991–2002. doi: 10.1056/NEJMoa0707943. [DOI] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. PNAS. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houde AA, Guay SP, Desgagné V, Hivert MF, Baillargeon JP, St-Pierre J, Perron P, Gaudet D, Brisson D, Bouchard L. Adaptations of placental and cord blood ABCA1 DNA methylation profile to maternal metabolic status. Epigenetics. 2013;8:1289–1302. doi: 10.4161/epi.26554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hult M, Tornhammar P, Ueda P, Chima C, Bonamy AK, Ozumba B, Norman M. Hypertension, diabetes and overweight: looming legacies of the Biafran famine. PLoS ONE. 2010;5:e13582. doi: 10.1371/journal.pone.0013582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova E, Chen JH, Segonds-Pichon A, Ozanne SE, Kelsey G. DNA methylation at differentially methylated regions of imprinted genes is resistant to developmental programming by maternal nutrition. Epigenetics. 2012;7:1200–1210. doi: 10.4161/epi.22141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature Genetics. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Kappen C, Kruger C, MacGowan J, Salbaum JM. Maternal diet modulates placenta growth and gene expression in a mouse model of diabetic pregnancy. PLoS ONE. 2012;7:e38445. doi: 10.1371/journal.pone.0038445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsey G. Genomic imprinting – roles and regulation in development. Endocrine Development. 2007;12:99–112. doi: 10.1159/000109637. [DOI] [PubMed] [Google Scholar]
- Khulan B, Cooper WN, Skinner BM, Bauer J, Owens S, Prentice AM, Belteki G, Constancia M, Dunger D, Affara NA. Periconceptional maternal micronutrient supplementation is associated with widespread gender related changes in the epigenome: a study of a unique resource in the Gambia. Human Molecular Genetics. 2012;21:2086–2101. doi: 10.1093/hmg/dds026. [DOI] [PubMed] [Google Scholar]
- Kim C, Newton KM, Knopp RH. Gestational diabetes and the incidence of type 2 diabetes. Diabetics Care. 2002;25:1862–1868. doi: 10.2337/diacare.25.10.1862. [DOI] [PubMed] [Google Scholar]
- Kral JG, Biron S, Simard S, Hould FS, Lebel S, Marceau S, Marceau P. Large maternal weight loss from obesity surgery prevents transmission of obesity to children who were followed for 2 to 18 years. Pediatrics. 2006;118:e1644–e1649. doi: 10.1542/peds.2006-1379. [DOI] [PubMed] [Google Scholar]
- Lehnen H, Zechner U, Haaf T. Epigenetics of gestational diabetes mellitus and offspring health: the time for action is in early stages of life. Molecular Human Reproduction. 2013;19:415–422. doi: 10.1093/molehr/gat020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CC, Young PE, Maloney CA, Eaton SA, Cowley MJ, Buckland ME, Preiss T, Henstridge DC, Cooney GJ, Febbraio MA, et al. Maternal obesity and diabetes induces latent metabolic defects and widespread epigenetic changes in isogenic mice. Epigenetics. 2013a;8:602–611. doi: 10.4161/epi.24656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Kohorst JJ, Zhang W, Laritsky E, Kunde-Ramamoorthy G, Baker MS, Fiorotto ML, Waterland RA. Early postnatal nutrition determines adult physical activity and energy expenditure in female mice. Diabetes. 2013b;62:2773–2783. doi: 10.2337/db12-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe LP, Metzger BE, Dyer AR, Lowe J, McCance DR, Lappin TR, Trimble ER, Coustan DR, Hadden DR, Hod M, et al. Hyperglycemia and Adverse Pregnancy Outcome (HAPO) study: associations of maternal A1C and glucose with pregnancy outcomes. Diabetics Care. 2012;35:574–580. doi: 10.2337/dc11-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore TR. Fetal exposure to gestational diabetes contributes to subsequent adult metabolic syndrome. American Journal of Obstetrics and Gynecology. 2010;202:643–649. doi: 10.1016/j.ajog.2010.02.059. [DOI] [PubMed] [Google Scholar]
- Moore T, Haig D. Genomic imprinting in mammalian development: a parental tug-of-war. Trends in Genetics. 1991;7:45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nature Genetics. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- Nikonova L, Koza RA, Mendoza T, Chao PM, Curley JP, Kozak LP. Mesoderm-specific transcript is associated with fat mass expansion in response to a positive energy balance. FASEB Journal. 2008;22:3925–3937. doi: 10.1096/fj.08-108266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura Y, Lambertini L, Rialdi A, Lee M, Mystal EY, Grabie M, Manaster I, Huynh N, Finik J, Davey M, et al. Global methylation in the placenta and umbilical cord blood from pregnancies with maternal gestational diabetes, preeclampsia, and obesity. Reproductive Sciences. 2014;21:131–137. doi: 10.1177/1933719113492206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins JM, Dunn JP, Jagasia SM. Perspectives in gestational diabetes mellitus: a review of screening, diagnosis, and treatment. Clinical Diabetes. 2007;25:57–62. doi: 10.2337/diaclin.25.2.57. [DOI] [Google Scholar]
- Plagemann A, Heidrich I, Götz F, Rohde W, Dörner G. Obesity and enhanced diabetes and cardiovascular risk in adult rats due to early postnatal overfeeding. Experimental and Clinical Endocrinology. 1992;99:154–158. doi: 10.1055/s-0029-1211159. [DOI] [PubMed] [Google Scholar]
- Plagemann A, Harder T, Franke K, Kohlhoff R. Long-term impact of neonatal breast-feeding on body weight and glucose tolerance in children of diabetic mothers. Diabetics Care. 2002;25:16–22. doi: 10.2337/diacare.25.1.16. [DOI] [PubMed] [Google Scholar]
- Plagemann A, Harder T, Brunn M, Harder A, Roepke K, Wittrock-Staar M, Ziska T, Schellong K, Rodekamp E, Melchior K, et al. Hypothalamic proopiomelanocortin promoter methylation becomes altered by early overfeeding: an epigenetic model of obesity and the metabolic syndrome. Journal of Physiology. 2009;587:4963–4976. doi: 10.1113/jphysiol.2009.176156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagemann A, Roepke K, Harder T, Brunn M, Harder A, Wittrock-Staar M, Ziska T, Schellong K, Rodekamp E, Melchior K, et al. Epigenetic malprogramming of the insulin receptor promoter due to developmental overfeeding. Journal of Perinatal Medicine. 2010;38:393–400. doi: 10.1515/JPM.2010.051. [DOI] [PubMed] [Google Scholar]
- Poston L. Maternal obesity, gestational weight gain and diet as determinants of offspring long term health. Best Practice & Research. Clinical Endocrinology & Metabolism. 2012;26:627–639. doi: 10.1016/j.beem.2012.03.010. [DOI] [PubMed] [Google Scholar]
- Rakyan VK, Blewitt ME, Druker R, Preis JI, Whitelaw E. Metastable epialleles in mammals. Trends in Genetics. 2002;18:348–531. doi: 10.1016/S0168-9525(02)02709-9. [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Reik W, Constância M, Fowden A, Anderson N, Dean W, Ferguson-Smith A, Tycko B, Sibley C. Regulation of supply and demand for maternal nutrients in mammals by imprinted genes. Journal of Physiology. 2003;547:35–44. doi: 10.1113/jphysiol.2002.033274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robitaille J, Grant AM. The genetics of gestational diabetes mellitus: evidence for relationship with type 2 diabetes mellitus. Genetics in Medicine. 2008;10:240–250. doi: 10.1097/GIM.0b013e31816b8710. [DOI] [PubMed] [Google Scholar]
- Rodekamp E, Harder T, Kohlhoff R, Franke K, Dudenhausen JW, Plagemann A. Long-term impact of breast-feeding on body weight and glucose tolerance in children of diabetic mothers: role of the late neonatal period and early infancy. Diabetics Care. 2005;28:1457–1462. doi: 10.2337/diacare.28.6.1457. [DOI] [PubMed] [Google Scholar]
- Roseboom T, De Rooij S, Painter R. The Dutch famine and its long-term consequences for adult health. Early Human Development. 2006;82:485–491. doi: 10.1016/j.earlhumdev.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Ruchat SM, Houde AA, Voisin G, St-Pierre J, Perron P, Baillargeon JP, Gaudet D, Hivert MF, Brisson D, Bouchard L. Gestational diabetes mellitus epigenetically affects genes predominantly involved in metabolic diseases. Epigenetics. 2013;8:935–943. doi: 10.4161/epi.25578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salbaum JM, Kappen C. Responses of the embryonic epigenome to maternal diabetes. Birth Defects Research. Part A, Clinical and Molecular Teratology. 2012;94:770–781. doi: 10.1002/bdra.23035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz LC. The Dutch Hunger Winter and the developmental origins of health and disease. PNAS. 2010;107:16757–16758. doi: 10.1073/pnas.1012911107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Lefebvre L, Yu Y, Otto S, Krella A, Orth A, Fundele R. Loss-of-imprinting of Peg1 in mouse interspecies hybrids is correlated with altered growth. Genesis. 2004;39:65–72. doi: 10.1002/gene.20027. [DOI] [PubMed] [Google Scholar]
- Smith J, Cianflone K, Biron S, Hould FS, Lebel S, Marceau S, Lescelleur O, Biertho L, Simard S, Kral JG, et al. Effects of maternal surgical weight loss in mothers on intergenerational transmission of obesity. Journal of Clinical Endocrinology and Metabolism. 2009;94:4275–4283. doi: 10.1210/jc.2009-0709. [DOI] [PubMed] [Google Scholar]
- Song S, Wang W, Hu P. Famine, death, and madness: schizophrenia in early adulthood after prenatal exposure to the Chinese Great Leap Forward Famine. Social Science & Medicine. 2009;68:1315–1321. doi: 10.1016/j.socscimed.2009.01.027. [DOI] [PubMed] [Google Scholar]
- Soubry A, Murphy SK, Wang F, Huang Z, Vidal AC, Fuemmeler BF, Kurtzberg J, Murtha A, Jirtle RL, Schildkraut JM, et al. Newborns of obese parents have altered DNA methylation patterns at imprinted genes. International Journal of Obesity. 2013 doi: 10.1038/ijo.2013.193. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson AM, Borg LA, Eriksson UJ. Glucose metabolism in embryos of normal and diabetic rats during organogenesis. Acta Endocrinologica. 1992;127:252–257. doi: 10.1530/acta.0.1270252. [DOI] [PubMed] [Google Scholar]
- Tobi EW, Lumey LH, Talens RP, Kremer D, Putter H, Stein AD, Slagboom PE, Heijmans BT. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Human Molecular Genetics. 2009;18:4046–4053. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers MH, Gluckman PD, Coveny AH, Hofman PL, Cutfield WS, Gertler A, Breier BH, Harris M. Neonatal leptin treatment reverses developmental programming. Endocrinology. 2005;146:4211–4216. doi: 10.1210/en.2005-0581. [DOI] [PubMed] [Google Scholar]
- Waterland RA, Kellermayer R, Laritsky E, Rayco-Solon P, Harris RA, Travisano M, Zhang W, Torskaya MS, Zhang J, Shen L, et al. Season of conception in rural Gambia affects DNA methylation at putative human metastable epialleles. PLoS Genetics. 2010;6:e1001252. doi: 10.1371/journal.pgen.1001252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, Schübeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nature Genetics. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Wells JC. The thrifty phenotype as an adaptive maternal effect. Biological Reviews of the Cambridge Philosophical Society. 2007;82:143–172. doi: 10.1111/j.1469-185X.2006.00007.x. [DOI] [PubMed] [Google Scholar]
- Wright CS, Rifas-Shiman SL, Rich-Edwards JW, Taveras EM, Gillman MW, Oken E. Intrauterine exposure to gestational diabetes, child adiposity, and blood pressure. American Journal of Hypertension . 2009;22:215–220. doi: 10.1038/ajh.2008.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Nohr EA, Bech BH, Vestergaard M, Olsen J. Long-term health outcomes in children born to mothers with diabetes: a population-based cohort study. PLoS ONE. 2012;7:e36727. doi: 10.1371/journal.pone.0036727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeruchimovich M, Mimouni FB, Green DW, Dollberg S. Nucleated red blood cells in healthy infants of women with gestational diabetes. Obstetrics and Gynecology. 2000;95:84–86. doi: 10.1016/S0029-7844(99)00511-6. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends in Genetics. 1997;13:335–340. doi: 10.1016/S0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- Zhang CH, Qian WP, Qi ST, Ge ZJ, Min LJ, Zhu XL, Huang X, Liu JP, Ouyang YC, Hou Y, et al. Maternal diabetes causes abnormal dynamic changes of endoplasmic reticulum during mouse oocyte maturation and early embryo development. Reproductive Biology and Endocrinology. 2013;11:31. doi: 10.1186/1477-7827-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]



 This work is licensed under a
This work is licensed under a