Abstract
Poly-(3-hydroxybutyrate) [P(3HB)] is a polyester synthesized as a carbon and energy reserve material by a wide number of bacteria. This polymer is characterized by its thermo-plastic properties similar to plastics derived from petrochemical industry, such as polyethylene and polypropylene. Furthermore, P(3HB) is an inert, biocompatible and biodegradable material which has been proposed for several uses in medical and biomedical areas. Currently, only few bacterial species such as Cupriavidus necator, Azohydromonas lata and recombinant Escherichia coli have been successfully used for P(3HB) production at industrial level. Nevertheless, in recent years, several fermentation strategies using other microbial models such as Azotobacter vinelandii, A. chroococcum, as well as some methane-utilizing species, have been developed in order to improve the P(3HB) production and also its mean molecular weight.
Introduction
Poly-(3-hydroxybutyrate) [P(3HB)] is produced and intracellularly accumulated as a carbon and energy reserve material. It can be produced by various bacteria, such as Cupriavidus necator, several species of Pseudomonas, Bacillus, Azotobacter and also recombinant Escherichia coli, expressing the P(3HB) biosynthetic genes from C. necator and A. vinelandii (Centeno-Leija et al., 2014). Since its discovery, P(3HB) has been used as substitute for bulk plastics, such as polyethylene and polypropylene, in the chemical industry. More recently, and based on its properties of biocompatibility and biodegradability, new attractive applications for P(3HB) have been proposed in the medical and pharmaceutical fields, where chemical composition and product purity are critical (Williams and Martin, 2005). In the medical field, P(3HB) has been used in artificial organ construction, drug delivery, tissue repair and nutritional/ therapeutic uses (Chen and Wang, 2013).
In all these applications, the molecular mass of P(3HB) is a very important feature to consider, because this determines the mechanical properties of the polymer, and in turn, the final applications. From a biotechnological point of view, the manipulation of the molecular mass of P(3HB) by means of the use of new strains and manipulating the culture conditions, seems to be a convenient method that could considerably improve the properties of P(3HB), expanding the potential application of this polymer, especially in the medical field.
Poly-(3-hydroxybutyrate) is produced by fermentation, either in batch, fed batch or continuous cultures using improved bacterial strains, cultured on inexpensive carbon sources such as beet and cane molasses, corn starch, alcohols and vegetable oils, combined with multi-stage fermentation systems (Lee, 1996; Chen and Page, 1997; Chen, 2009; 2010; Chanprateep, 2010; Peña et al., 2011). All these strategies have been attempted to improve both the yields and process productivity in order to have a more competitive process.
There are several reviews regarding the properties and applications of P(3HB); as well as about the different microorganisms producing P(3HB) (Byrom, 1987; Sudesh et al., 2000; Chen, 2009; 2010; Grage et al., 2009; Chanprateep, 2010; Peña et al., 2011); however, there are not recent reviews about the fermentation strategies for improving the P(3HB) production.
This review aims to summarize the recent trends in the bacterial production of P(3HB) using novel fermentation strategies combined with the use of genetic engineering to improve productivity and quality (in terms of its molecular weight) of P(3HB) that could be applied for its commercial production.
P(3HB): structure and properties
Polyhydroxyalkanoates (PHAs) are linear polyesters conformed by hydroxyacyl units. They can be found as homopolymers or as copolymers containing combined 2-, 3-, 4-, 5- or 6-hydroxyacids (Sudesh et al., 2000; Kessler and Witholt, 2001; Chen, 2010). Polyhydroxyalkanoates classification depends on the number of carbon atoms present in their monomers as short-chain-length PHAs (scl-PHA; three to five C-atoms) and medium-chain-length PHAs (mcl-PHA; with six or more C-atoms) (Pan and Inoue, 2009).
Interest in these polymers has increased in the last decades due to their thermoplastic properties, which make them a biodegradable and environmentally friendly alternative to petroleum based plastics, such as polyethylene and polypropylene. Although PHAs include a broad number of polymers of diverse monomeric composition, only few of them have been incorporated into the large-scale production: P(3HB); poly-(3-hydroxybutyrate-co-3-hydroxyvalerate) [P(3HB-co-3HV)] and poly-(3-hydroxybutyrate-co-3-hydroxyhexanoate) [P(3HB-co-3HHx)] (Chen, 2009; Chanprateep, 2010; Fig. 1).
Fig. 1.
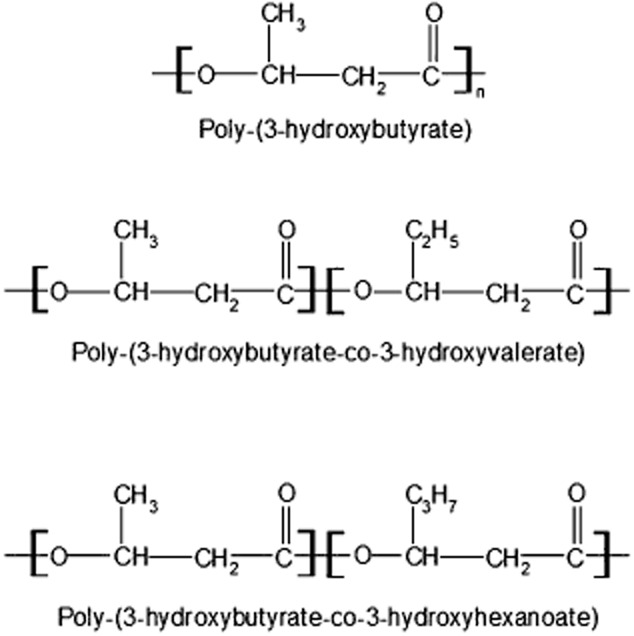
Chemical structure of poly-(3-hydroxybutyrate), poly-(3-hydroxybutyrate-co-3-hydroxyvalerate) and poly-(3-hydroxybutyrate-co-3-hydroxyhexanoate).
Poly-(3-hydroxybutyrate) is the homopolymer of (R)-3-hydroxybutyrate units. It can be obtained within a wide range of molecular masses fluctuating from 200 to up to 20 000 KDa (Kusaka et al., 1998; Sudesh et al., 2000). The thermoplastic properties of P(3HB) and its biodegradability, without generation of toxic by-products, make it a sustainable alternative to petroleum-based plastics. In addition, this polymer is produced by biotechnological strategies allowing the control of its chemical composition, and therefore its physicochemical properties. Besides, this polymer shows interesting properties such as a high biocompatibility with mammalian cells, making them suitable for medical applications (Chen, 2009; 2010; Grage et al., 2009; Pan and Inoue, 2009; Shishatskaya et al., 2011; Bornatsev et al., 2013).
P(3HB) is a semi-crystalline polymer, characterized by a polymorphic crystallization, that is able to crystallize into two forms, α and β (Pan and Inoue, 2009). The α-form which consists in lamellar crystals, being the most common conformation for P(3HB) crystals (Pan and Inoue, 2009; Kabe et al., 2012) and the β-form characterized as a planar zigzag conformation which has been reported in films and fibres with high tensile strength (Iwata, 2005; Pan and Inoue, 2009; Kabe et al., 2012). It must be emphasized that the crystallization process affects the thermal and mechanical properties, as well as biodegradability of biopolymers (Pan and Inoue, 2009).
The thermoplastic and crystallization properties of P(3HB) are highly dependent of its molecular mass. Poly-(3-hydroxybutyrates) of low molecular masses (< 1 × 103 kDa) are characterized by their brittleness and an early thermal degradation, near their melting temperature (above 180°C) (Hong et al., 2013). This behaviour has been explained as a result of its α-form crystallization (Kabe et al., 2012); however, increasing P(3HB) molecular mass improves the mechanical properties of films and fibres by promoting the β-form crystallization (Kabe et al., 2012). In this line, using P(3HB) of ultra-high molecular weight (UHMW; Mw = 5.3 × 103 kDa), Iwata (2005) reported that the tensile strength of the polymer could be manipulated from 38 to 1320 MPa, only by modifying the drawing method. This last value (1320 MPa) is higher than the tensile strength reported for polyethylene, polypropylene, polyvinyl alcohol and polyglycolic acid used at industrial level (Iwata, 2005).
However, up to now the UHMW-P(3HB) production has been restricted only for cultivations of low cell density, such as the cultures of recombinant E. coli XL-1 Blue (pSLY105), harbouring the Cupriavidus necator P(3HB) biosynthetic genes phbCAB (Kusaka et al., 1998; Iwata, 2005; Murakami et al., 2014; Kabe et al., 2012), mixed cultures of methane-utilizing bacteria (Helm et al., 2008) and Azotobacter cultivations (Peña et al., 2014). Therefore, several strategies have been designed to improve the thermo-mechanical properties of P(3HB) including: P(3HB) composites with other PHAs [P(3HV) or P(3HHx)] or other biopolymers (i.e.: cellulose, chitosan; Rajan et al., 2012), the addition of chemical plasticizers (i.e.: polyethylene glycol, glycerol, glycerol triacetate, 4-nonylphenol; Hong et al., 2013), as well as the blending of P(3HB) of different molecular masses (Kabe et al., 2012; Hong et al., 2013).
As shown in Table 1, it is possible to modify and improve the thermo-mechanical properties of P(3HB) for specific applications by combining P(3HB) of medium molecular weight with UHMW-P(3HB) (Sharma et al., 2004; Kabe et al., 2012) or P(3HB) of very low molecular weight [LMW-P(3HB); Mw = 1.76 kDa] (Hong et al., 2013). In this line, blending P(3HB) of medium molecular weight with only 5% of UHMW-P(3HB) increased the tensile strength and elongation at break up to 33% and 48%, reaching values similar to those of conventional plastic films (Kabe et al., 2012). In contrast, addition of LMW-P(3HB) reduces polymer crystallinity, as well as the melting and crystallization temperature of P(3HB), but positively affects elongation at break and degradation rate (Hong et al., 2013), being this last characteristic of great interest for biomedical applications.
Table 1.
Thermo-mechanical properties of P(3HB) and its composites with UHMW-P(3HB) or LMW-P(3HB)
| Compound | Drawn ratio | Tg (°C) | Tc (°C) | Tm (°C) | Tensile strength (MPa) | Elongation at break (%) | Young's modulus (GPa) | Crystallinity (%) | Reference |
|---|---|---|---|---|---|---|---|---|---|
| P3HB | 12a | 1.8 | 53 | 170 | 161 | 45 | 2.8 | 78 | Kabe et al., 2012 |
| UHMW | 10a | 2.4 | 57 | 172 | 191 | 56 | 1.6 | 73 | Kabe et al., 2012 |
| UHMW/P3HB (5/95) | 12a | 2.2 | 53 | 170 | 242 | 88 | 1.5 | 75 | Kabe et al., 2012 |
| UHMW | 60b | n.d. | n.d. | n.d. | 1320 | 35 | 18.1 | n.d. | Iwata, 2005 |
| P3HB/LMW (87.5/12.5) | None | −2.6 | 93 | 162.3 | 23.4 | 4.2 | n.d. | 44.8 | Hong et al., 2013 |
| P3HB/LMW (83.3/16.6) | None | −4.8 | 82 | 160.5 | 24.3 | 9.8 | n.d. | 40.4 | Hong et al., 2013 |
| P3HB/LMW (75/25) | None | −7.3 | 76 | 155.8 | 11.6 | 3.8 | n.d. | 37.8 | Hong et al., 2013 |
Processed by cold drawing.
Processed by cold drawing/two step drawing.
Tg, temperature to glass transition; Tc, crystallization temperature; Tm, melting temperature; n.d., not described.
Biomedical applications of P(3HB)
Previous reviews have focused on novel applications of P(3HB) and other PHAs in several biomedical areas (Chen and Wu, 2005; Chen, 2009; Grage et al., 2009; Peña et al., 2011; Chen and Wang, 2013) which can be described as follows: material for sutures and tissue engineering, including heart valves, bone scaffolding, scaffolds for skeletal myotubes and nerve tissue (Grage et al., 2009; Ricotti et al., 2012; Masaeli et al., 2013); nano or micro beads for drug delivery and target-specific therapy for treatment of illness such as cancer and tuberculosis (Grage et al., 2009; Parlane et al., 2012; Althuri et al., 2013); and finally, its possible application as biomarker or biosensor (Grage et al., 2009). Table 2 summarizes some of the more recent attempts to apply P(3HB) in these fields, mainly as tissue engineering scaffolds and micro or nanoparticles for drugs delivery. It must be emphasized that, for these applications, P(3HB)s of a wide range of molecular weights (MW) have been used. For applications such as nano- or microparticles, the MW did not affect the production yield of particles (Shishatskaya et al., 2011). On the other hand, P(3HB) used for tissue engineering, in some cases requires to be mixed with materials such as chitosan (Cao et al., 2005; Medvecky et al., 2014; Mendonca et al., 2013), other PHAs (Masaeli et al., 2013), polyethylene glycol (PEG) (Chan et al., 2014), hydroxyapatite (Shishatskaya et al., 2006; Ramier et al., 2014) or even cell growth inductors (Filho et al., 2013). Addition of those materials allows to improve not only the mechanical properties of P(3HB) but also its degradability, hydrophilicity and its cell attachment capabilities.
Table 2.
Biomedical applications of P(3HB) with different molecular weights
| Applications | P(3HB) MW (kDa) | Preparation procedure | Reference | |
|---|---|---|---|---|
| P(3HB) LMW | Osteoblast scaffolds | 220 | P3HB and hydroxyapatite were mixed using mechanical and physical methods | Shishatskaya et al., 2006 |
| Scaffolds | 89–110 | Blends of P3HB and chitosan at different ratios were evaluated | Medvecky et al., 2014 | |
| Nanofibrous scaffolds for bone tissue engineering | 144 | Electrospinning/electrospraying, P3HB and hydroxyapatite | Ramier et al., 2014 | |
| P(3HB) | Nanoparticles for retinoic acid (RA) delivery | 350 | 50 nm particles of P3HB/RA were prepared by dialysis | Errico et al., 2009 |
| Microcapsules for drugs delivery | 300 | Microcapsules of 0.5–1.5 μm with P3HB and smectite clays were formed | da Silva-Valenzuela et al., 2010 | |
| Scaffolds of PHB and otholits (osteoinductor) for bone tissue regeneration | 300 | Solutions of P3HB and otholits (1% w/w) were electrospinning | Filho et al., 2013 | |
| Scaffolds 3D for osteoblasts engineering | 524 | P3HB and chitosan blends were evaluated | Mendonca et al., 2013 | |
| Scaffolds for tissue engineering | 300 | P3HB scaffolds were prepared by salt leaching and electrospinning | Masaeli et al., 2012 | |
| Nanofibrous scaffolds nerve tissue engineering | 437 | Blends of P3HB (50)/PHBV (50) were treated by electrospinning | Masaeli et al., 2013 | |
| P(3HB) UHMW | Scaffolds for tissue engineering | 890 | Chitosan and P3HB films were prepared by emulsion blending | Cao et al., 2005 |
| Scaffolds for nerve cells | 1143 | P3HB was treated with PEG reducing 10 fold-times its MW but promote cell growth | Chan et al., 2014 |
Producers of P(3HB)
The ability to synthesize and accumulate P(3HB) and other PHAs as a carbon and energy reserve material is widespread among the prokaryotes. More than 300 species, mainly of bacteria, have been reported to produce these polymers (Olivera et al., 2001; Chanprateep, 2010). However, not all of these microorganisms have been shown to accumulate sufficient P(3HB) for large-scale production. Among the bacteria that are able to accumulate large amounts of PHA are C. necator (formerly known as Ralstonia eutropha or Alcaligenes eutrophus), Azohydromonas lata (also known as Alcaligenes latus), Pseudomonas oleovorans, Pseudomonas putida, Aeromonas hydrophila, Paracoccus denitrificans, Methylobacterium extorquens, Bacillus spp., Azotobacter vinelandii and recombinant E. coli, expressing the P(3HB) biosynthetic genes from C. necator, A. lata or A. vinelandii (Lee, 1996; Olivera et al., 2001; Chen, 2009; Centeno-Leija et al., 2014). Figure 2 shows A. vinelandii cells with granules of P(3HB). From the microorganisms mentioned, the more successful species for production at pilot or large scale are C. necator, A. lata and recombinant E. coli, being able to accumulate up to 80% of the polymer from a final dry cell weight of up to 200, 60 and 150 g l−1 respectively (Chen, 2009).
Fig. 2.
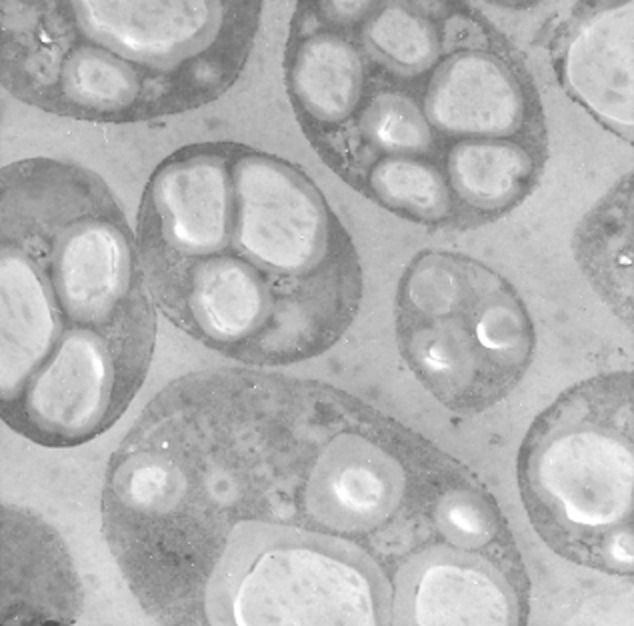
Transmission electron micrograph of a thin section of A. vinelandii containing P(3HB) granules (white inclusions).
Many species of Archaea have also been shown to be PHA producers, particularly members of Haloarchaea (Legat et al., 2010; Poli et al., 2011). These organisms could present important advantages as PHA producers because they can utilize cheap carbon sources (Huang et al., 2006), they do not need strict sterilization (they are able to grow in hypersaline conditions, in which very few organisms can survive), and because they can release the polymer produced easily because they lyse in distilled water, facilitating its isolation and lowering the production costs (Hezayen et al., 2000; Poli et al., 2011). The carbohydrate-utilizing species Haloferax mediterranei is particularly interesting because it accumulates large amounts of P(3HB) on glucose or starch, it grows optimally with 25% (w/v) salts and accumulates 60–65% of polymer (w/w) (Rodriguez-Valera and Lillo, 1992). H. mediterranei, shows the highest potential for industrial application because it can reach cell concentrations of 140 g l−1, with a PHA content of 55.6% reaching a PHA concentration of 77.8 g l−1 in a repeated fed-batch fermentation (Huang et al., 2006), and it is also able to produce a P(3HB-co-P3HV) copolymer (10.4 mol% 3HV) from enzymatic extruded starch (Chen et al., 2006).
Metabolic pathways and genetics involved in production of P(3HB)
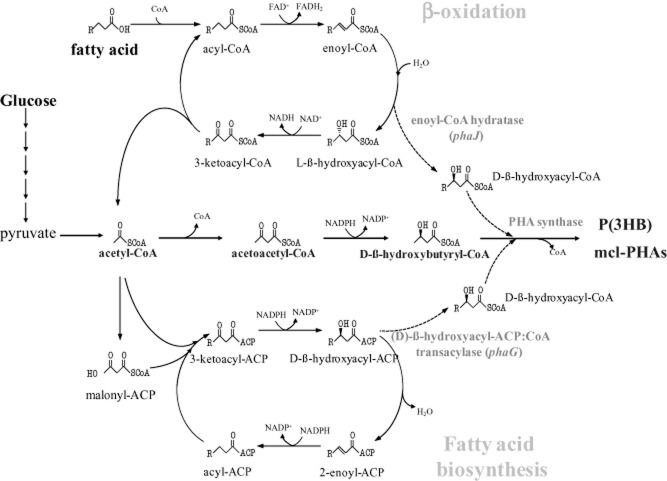
The biosynthetic pathway for P(3HB) (Fig. 3) starts with the condensation of two molecules of acetyl-CoA to form acetoacetyl-CoA. The enzyme catalyzing this reaction is 3-ketothiolase, encoded by the phbA gene. An acetoacetyl-CoA reductase (gene phbB) coverts the acetoacetyl-CoA to 3-hydroxybutyryl-CoA using NADPH. Finally, the enzyme PHA synthase (encoded by phbC) polymerizes the 3-hydroxybutyryl-CoA monomers to P(3HB), liberating CoA (Rehm, 2003; Stubbe et al., 2005) (Fig. 3). In some species, the P(3HB) biosynthetic genes phbA, phbB and phbC are clustered and are presumably organized in one operon phbCAB (Reddy et al., 2003); although this gene order varies from species to species, and the genes can also be unlinked. More than 60 PHA synthase genes (phbC or phaC) from eubacteria have been cloned and sequenced, and many more have been revealed in the bacterial genomes sequenced (Steinbüchel and Lütke-Eversloh, 2003). Other genes whose products are also involved in PHA metabolism and their specific metabolic roles have been reviewed by Chen (2010).
Fig. 3.
Metabolic pathways and genetics involved in the production of P(3HB).
Besides P(3HB), other PHAs containing 150 different monomers have been reported. This PHA diversity is due to the broad substrate range exhibited by the PHA synthases, the PHA polymerizing enzymes (Steinbüchel and Lütke-Eversloh, 2003; Stubbe et al., 2005; Volova et al., 2013). The different PHAs are synthesized depending also on the carbon source provided; the metabolic routes present to convert that carbon source into the hydroxyacyl-CoA monomers, and the specificity of the PHA synthase of that particular organism. The biosynthetic pathways reported up to date have been reviewed recently (Lu et al., 2009; Chen, 2010; Panchal et al., 2013), so we only present a brief description of the routes involved. For the synthesis of PHAs composed of 3-hydroxyalkanoic acids of C6–C16 (referred to as mcl-PHAs) the hydroxyacyl-CoA precursors are derived from fatty acid metabolism (Fig. 3). These precursors can be obtained either from ß-oxidation of alkanes, alkanols or alkanoic acids (De Smet et al., 1983; Brandl et al., 1988; Lagaveen et al., 1988), mainly by an enantioselective enoyl-CoA hydratase (encoded by phaJ) that produces the (R)-hydroxyacyl-CoA (Tsuge et al., 2003), or from fatty acid de novo biosynthesis using an (R)-3-hydroxyacyl-ACP:CoA transacylase (encoded by phaG) to produce the substrates for the PHA synthase from a non-related carbon source, such as carbohydrates (Rehm et al., 1998; Hoffmann et al., 2000a,b; Matsumoto et al., 2001).
Molecular strategies to improve P(3HB) production
Although many of the P(3HB) production systems use non-genetically modified bacterial strains, some efforts have been undertaken to increase the production of these polymers by genetic manipulation. These efforts include mainly the modification of the metabolism to favour P(3HB) synthesis, the modification of regulatory systems controlling P(3HB) synthesis and recombinant phb gene expression.
The P(3HB) biosynthetic routes compete for precursors with central metabolic pathways, such as the tricarboxylic acid (TCA) cycle, fatty acid degradation (ß-oxidation) and fatty acid biosynthesis. They also compete with other biosynthetic pathways that use common precursors. Three examples of genetic modifications that favour P(3HB) synthesis by metabolism modification of the producer strain were reported in A. vinelandii. Page and Knosp (1993) reported a strain (UWD), which has a mutation in the respiratory NADH oxidase that resulted in the ability to accumulate P(3HB) during the exponential phase without the need of nutrient limitation. The second example is found in the inactivation of pyruvate carboxylase, the anaplerotic enzyme catalyzing the ATP-dependent carboxylation of pyruvate, to generate oxaloacetate that replenishes the TCA cycle (Segura and Espín, 2004). This mutation increased three times the specific production of P(3HB) (gP(3HB) gprotein-1), in contrast with the wild type strain A. vinelandii UW136, probably as a result of a diminished flux of acetyl-CoA into TCA cycle, leaving it available for P(3HB) synthesis. In the same bacterium, a mutation blocking the synthesis of alginate, an exopolysaccharide produced by this organism, increased the P(3HB)-specific production up to five times, depending on the growth conditions evaluated, with a higher yield based on glucose as compared with the wild type strain ATCC9046. The mutation not only increased the capacity of the bacterium to produce P(3HB) per biomass unit, but also allowed an increased growth, raising the volumetric production of the polymer up to 10-fold (Segura et al., 2003).
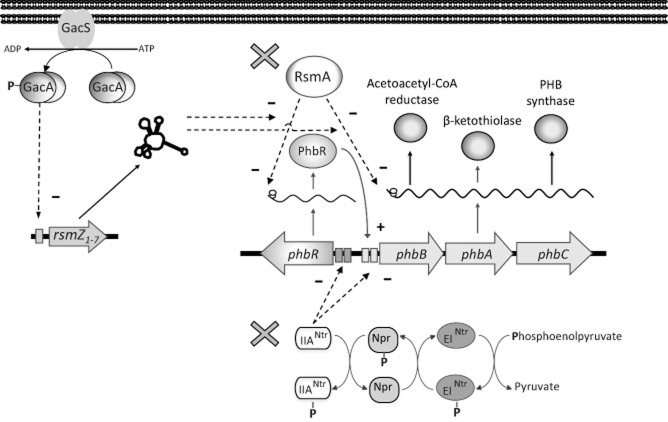
Regarding the modification of regulatory systems controlling PHA synthesis to increase their production, some interesting examples are also found in A. vinelandii. Poly-(3-hydroxybutyrate) synthesis in this bacterium is regulated by the nitrogen-related phosphotransferase system (PTSNtr), where the non-phosphorylated form of the IIANtr protein negatively regulates the expression of the P(3HB) biosynthetic operon (Segura and Espín, 1998; Noguez et al., 2008). Another system regulating P(3HB) synthesis in A. vinelandii is the post-transcriptional regulatory system RsmZ/Y-A, where the RsmA protein represses translation of the mRNAs of the phbBAC biosynthetic operon and of phbR that codes for its transcriptional activator (Hernández-Eligio et al., 2012). In each case, negative regulators IIANtr and RsmA were identified (Fig. 4). In order to have P(3HB) overproducing strains of A. vinelandii OP, the gene coding for the IIANtr (ptsN) was inactivated. This mutation increased 77% the specific production of P(3HB), equivalent to 4.1 g l−1 of PHB (3.5 g l−1 in the case of the wild type), with a 36% higher yield of product based on the consumed substrate (Peña et al., 2014). Later, a mutant where both negative regulators (IIANtr and RsmA) were inactivated was constructed (Fig. 4), further increasing the P(3HB) accumulation capacity of A. vinelandii. This strategy, together with the implementation of a fermentation strategy allowed to produce 27 g l−1 of P(3HB) (García et al., 2013).
Fig. 4.
Model of the regulatory systems controlling the expression of the phb genes in A. vinelandii. (+) indicate positive regulation; (−) indicate negative regulation. Promoters are indicated as rectangles. The regulators inactivated in the A. vinelandii improved strains OPN and OPNA are indicated by a grey cross.
Another case illustrating production improvement by manipulation of regulatory systems is found in the cianobacterium Synechocystis sp. PCC 6803. In this bacterium, the overexpression of the sigma factor SigE, previously shown to activate the expression of many sugar catabolic genes and to enhance the levels of acetyl-CoA, increased the production of P(3HB) two or three times (Osanai et al., 2013).
Fast growth on simple media and the possibility to reach a high cell density in the culture with a high-content P(3HB) are important factors to consider for a successful P(3HB) production process. Because E. coli is an extensively studied bacterium with well-established technologies for genome manipulation, cultivation and downstream processing, many studies have focused on the use of E. coli to efficiently produce these polymers. This bacterium is a non-PHA producer; however, the genes of the P(3HB) producer C. necator H16 were cloned in E. coli for the first time by Slater et al1988. in 1988, enabling the production of P(3HB) in this organism. Since then, many different genetic modifications have been attempted, both to improve the accumulation of P(3HB) at low-cost with high productivity and to produce diverse copolymers using metabolic engineering and synthetic biology strategies. These strategies have been reviewed recently (Li et al., 2007; Wang et al., 2013).
Fermentation strategies to improve the production of P(3HB)
Effect of carbon source on P(3HB) production
The mayor expenses in the production of P(3HB) are determined by the cost of the fermentation substrate, the polymer extraction from the cells and the treatment of fermentation and extraction wastes (Chen, 2010). Of all these factors, the cost of the carbon source has the greatest influence on the price of P(3HB). Because of the above, new alternatives have been proposed to reduce the costs of raw materials. It is important to note that the selection of carbon sources should not focus only on the market prices but also on the availability and on global price (Chanprateep, 2010).
Table 3 summarizes different carbon sources used for the P(3HB) production. Fortunately, most P(3HB) producers can metabolize a wide range of raw materials. For example, it is known that several species of Azotobacter can use corn syrup, cane molasses, beet molasses or malt extract as carbon sources (Kim, 2000; Myshkina et al., 2008; Peña et al., 2011). For example, Kim (2000) reported the use of two inexpensive substrates, starch and whey, to produce P(3HB) in fed-batch cultures of A. chroococcum H23 and recombinant E. coli. These authors found that in fed-batch culture of A. chroococcum H23 a cell concentration of 54 g l−1 with 46% (w/w) P(3HB) was obtained with oxygen limitation, whereas 71 g l−1 of cells with 20% (w/w) P(3HB) was achieved without oxygen limitation. In the case of whey as carbon source, using recombinant E. coli 6576, Kim (2000) reported a P(3HB) content of 80%, with a cell concentration of 31 g l−1.
Table 3.
Comparison of P(3HB) volumetric production, content and yields using different carbon sources
| Organism | Carbon source | Quantity of carbon source (g) employed | DCW (g l−1) | P(3HB) concentration (g l−1) | P(3HB) content (%) | P(3HB) yield based on carbon source (g g−1) | Reference |
|---|---|---|---|---|---|---|---|
| A. lata | Sucrose | 72.9 | 10.78 | 5.25 | 48 | n.d. | Zafar et al., 2012a |
| C. necator DSM545 | Glucose | 523 | 164 | 125 | 76.2 | 0.22 | Mozumder et al., 2014 |
| Waste glycerol | n.d | 104.7 | 65.6 | 62.7 | 0.52 | Mozumder et al., 2014 | |
| Waste glycerol | 170.8 | 76.2 | 38.1 | 50 | 0.34 | Cavalheiro et al., 2009 | |
| Pure glycerol | 249 | 82.5 | 51.2 | 62 | 0.36 | Cavalheiro et al., 2009 | |
| A. chrococcum H23 | Alpechin/acetate | 30/0.06 | 7.36 | 6.10 | 82.9 | n.d | Pozo et al., 2002 |
| Starch | 200 | 54 | 25 | 46 | n.d | Kim, 2000 | |
| E. coli recombinant GCSC 6576 | Whey | 340 | 31 | 25 | 80 | n.d. | Kim, 2000 |
n.d., data not described.
On the other hand, P(3HB) and P[3HB-co-3HV]) copolymers were produced by A. chroococcum strain H23, when growing in culture media supplemented with wastewater from olive oil mills (alphechin), as the sole carbon source (Pozo et al., 2002). A maximal concentration of P(3HB) of 6.2 g l−1 was reached when the cells were cultured in shaken flasks at 250 r.p.m. for 48 h at 30°C in liquid medium supplemented with 60% (v/v) alpechin and 0.12% (v/v) ammonium acetate (Table 3). Production of PHAs by A. chroococcum strain H23 using alpechin looks promising, as the use of a cheap substrate for the production of these materials is essential if bioplastics are to become competitive products.
In this context, crude glycerol (a by-product of the large-scale production of diesel oil from rape) has been evaluated for its potential use as a cheap feedstock for P(3HB) production (Cavalheiro et al., 2009; Mozumder et al., 2014). Bacteria used has been C. necator DSM 545, which accumulated P(3HB) from pure glycerol up to a content of 62.5% (w/w) of cell dry mass, reaching a volumetric production of 51.2 g l−1, with a yield on glycerol of 0.36 g P(3HB) g gly-1 (Cavalheiro et al., 2009). On the other hand, when this by-product was used by Mozumder and colleagues (2014), a maximal biomass concentration of 104.7 g l−1 was reached, with a P(3HB) concentration in the broth culture of 65.6 g l−1. In addition, the molecular weight of P(3HB) produced with C. necator from glycerol varies between 7.86 × 102 kDa (with waste glycerol) and 9.57 × 102 kDa (with pure glycerol), which allows the processing by common techniques of the polymer industry (Cavalheiro et al., 2009).
A recent report on P(3HB) production using A. lata has been published (Zafar et al., 2012a). In this study, the optimization of P(3HB) production process using A. lata MTCC 2311 was carried out. By using a genetic algorithm on an artificial neural network, the predicted maximum P(3HB) production of 5.95 g l−1 was found, using 35.2 g l−1 of sucrose and 1.58 g l−1 of urea (Zafar et al., 2012a); however, the highest experimental P(3HB) concentration (5.25 g l−1) was achieved using 36.48 g l−1 of sucrose. The same authors reported that the use of propionic acid together with cane molasses allowed the synthesis of the copolymer P(3HB-co-3HV) in maximal concentrations of 7.2 g l−1 in shaken flasks and of 6.7 g l−1 in 3-L bioreactor (Zafar et al., 2012b).
Fermentation strategies
Only a few species of bacteria producing P(3HB) have been used at industrial scale to produce the polymer. These include C. necator, A. lata and recombinant E. coli (Khanna and Srivastava, 2005). On the other hand, there are some bacteria, such as A. vinelandii and A. chroococcum which can accumulate a high P(3HB) content and therefore could be used for the synthesis of this polymer at large scale.
Several studies have been carried out which described the P(3HB) production by several microbial strains, either in batch, fed batch or continuous cultures. Batch fermentation for P(3HB) production is a popular process due to its flexibility and low operation costs. However, batch cultures have the disadvantage that, usually, the yields and productivities of P(3HB) are low. In this sense, the P(3HB) production in batch cultures of C. necator ATCC 17699 has been studied using acetic acid as a carbon source (Wang and Yu, 2001). In this study, the P(3HB) productivity was of only 0.046 g l−1 h−1 employing a carbon/nitrogen (C/N) weight ratio of 76, with a maximal accumulation of P(3HB) close to 50% (w/w).
The systems more often employed for P(3HB) production are those involving two or three stages. These fermentations have been widely used for the production of P(3HB) and other PHAs (Ruan et al., 2003; Rocha et al., 2008). The fed-batch cultures have been employed to achieve high cell densities and a high concentration of P(3HB) (Kulpreecha et al., 2009). Fed-batch cultivations are systems where one or more nutrients are supplied to the bioreactor and the products and other components are kept within the system until the end of fermentation. This means that there is an inflow but no outflow, and the volume changes with respect to time (Mejía et al., 2010). There are several ways to feed the cultures, and it is possible to add one or more components.
Currently there are reports in the literature about the use of exponentially fed-batch cultures for P(3HB) production with microorganisms as A. lata (Grothe and Chisti, 2000). These authors obtained a maximal biomass concentration of 36 g l−1 with a P(3HB) volumetric production of 20 g l−1 by varying the components in the culture. More recent studies have shown that a total concentration of 4.5 g l−1 of P(3HB) was obtained using limiting conditions of dissolved oxygen with processed cheese whey supplemented with ammonium sulfate in fed-batch culture of Methylobacterium sp. ZP24 (Nath et al., 2008). This investigation reflects the possibility of developing a cheap biological route for production of green thermoplastics.
Recently, an integrated model was used for the optimization of the production of P(3HB) with tailor-made molecular properties in A. lata. A single-shot feeding strategy with fresh medium free of nitrogen was designed and experimentally tested. Using this strategy, a maximal concentration of P(3HB) of 11.84 g l−1 was obtained, equivalent to polymer content equal to 95% (w/w) of dry cell weight (Penloglou et al., 2012a).
Table 4 shows the more recent results reported about of maximal concentration and productivity of P(3HB) reached using different microorganism and fed-batch systems. From these studies, the cases for P(3HB) production using Bacillus megaterium, C. necator, recombinant E. coli and Azotobacter are highlighted. For example, Kulpreecha and colleagues (2009) reported a high P(3HB) production (30.5 g l−1) and P(3HB) productivity (1.27 g l−1 h−1) in a fed-batch culture of B. megaterium BA-019 using sugarcane molasses as a carbon source. More recently, Kanjanachumpol and colleagues (2013) found that in cultures of B. megaterium BA-019 with intermittent feeding of the sugarcane molasses and an increase of the C/N ratio at 12.5 improved the biomass and volumetric productivity of P(3HB), reaching a maximal biomass concentration of 90.7 g l−1 with 45.84% (w/w) of P(3HB) content and a productivity of 1.73 g l−1 h−1 P(3HB).
Table 4.
Comparison of P(3HB) production using different microorganism and fed-batch strategies
| Organism | Feeding strategy | DWC (g l−1) | P(3HB) (g l−1) | P(3HB) productivity (g l1 h−1) | P(3HB) content (% wt) | Reference |
|---|---|---|---|---|---|---|
| B. megabacterium BA-019 | pH stat | 72.6 | 30.5 | 1.27 | 42 | Kulpreecha et al., 2009 |
| Intermittent | 90.7 | 41.6 | 1.73 | 46 | Kanjanachumpol et al., 2013 | |
| C. necator | Pulses | 75 | 53 | 0.92 | 71 | Tanadchangsaeng and Yu, 2012 |
| Pulses | 83 | 67.2 | 2.5 | 81 | Pradella et al., 2012 | |
| Pulses | 82.5 | 51.2 | 1.52 | 62 | Cavalheiro et al., 2009 | |
| Exponential + coupled to alkali addition monitoring + constant with N2 limitation | 164 | 125 | 2.03 | 76.2 | Mozumder et al., 2014 | |
| E. coli | pH stat | 119.5 | 96.2 | 2.57 | 80 | Ahn et al., 2000 |
| A. vinelandii | Exponential + pulses | 37.2 | 27.3 | 0.5 | 73.3 | García et al., 2013 |
In the case of C. necator, Tanadchangsaeng and Yu (2012) reported a significant increase in P(3HB) volumetric production and productivity (53 g l−1 and 0.92 g l−1 h−1 respectively) in a fed batch using glycerol as a carbon source. Considering this, they suggested that glycerol is an ideal feedstock for producing bioplastics via bacterial fermentation due to its ubiquity, low price and high degree of reduction. However, the productivities reported using glycerol as carbon source (Cavalheiro et al., 2009) are still relatively low compared to other reports. An example is the high P(3HB) productivity reached by C. necator, using soybean oil in fed-batch culture (Pradella et al., 2012). In this study, the authors reported a maximal P(3HB) concentration of 67.2 g l−1 with a volumetric productivity of 2.5 g l−1 h−1. On the other hand, Mozumder and colleagues (2014) using C. necator, developed a three-stage feeding strategy using glucose as the sole carbon source that resulted in a P(3HB) concentration of 125 g l−1, with a P(3HB) content of 76% achieving a productivity of 2.03 g l−1 h−1.
Another successful case is that reported by Ahn and colleagues (2000), who developed fermentation strategies for P(3HB) production from whey by recombinant E. coli strain CGSC 4401. Using a pH stat fed-batch cultures, adding a concentrated whey solution containing 280 g l−1 was possible to reach final cell and P(3HB) concentrations of 119 and 96 g l−1 respectively, at 37.5 h, with a maximal productivity of 2.57 g l−1 h−1 (Table 4). The strategies developed in this study provide an attractive solution to whey disposal and utilization of this raw material for the P(3HB) production at large scale.
For several decades the synthesis of P(3HB) by Azotobacter strains has been the subject of studies, either in batch (Page and Knosp, 1993; Page et al., 2001; Myshkina et al., 2008), continuous (Senior et al., 1972; Senior and Dawes, 1973) or fed-batch cultures (Page and Cornish, 1993; Chen and Page, 1997; Kim and Chang, 1998; García et al., 2013). However, the information related with the fermentation systems has been scarce in recent years. On the other hand, to our knowledge, none of these processes has yet been adopted for the industrial production of P(3HB).
Recently, our group reported (García et al., 2013) a mixed fermentation strategy based on exponentially fed-batch cultures (EFBC) and nutrient pulses with sucrose and yeast extract to achieve a high concentration of P(3HB) by A. vinelandii OPNA, which carries a mutation on the genes encoding IIANtr (ptsN) and RsmA (rsmA) that negatively regulate the synthesis of P(3HB). Using a strategy of exponential feeding coupled with nutrient pulses (with sucrose and yeast extract), the production of P(3HB) increased sevenfold (with respect to the values obtained in batch cultures) to reach a maximal P(3HB) concentration of 27.5 ± 3.2 g l−1 at 60 h of fermentation (Table 4). Overall, the use of the OPNA mutant of A. vinelandii, impaired in the P(3HB) regulatory systems, in combination with a mixed fermentation strategy, could be a feasible strategy to optimize the P(3HB) production at industrial level (García et al., 2013).
Influence of the culture conditions on the P(3HB) molecular mass
The molecular mass (MM) of P(3HB) determines the elastic behaviour of the material and its mechanical resistance (Iwata, 2005). Fibres of P(3HB) with a MM of about 3.0 × 102 kDa have a tensile strength of 190 MPa and an elongation at break of 5%. In contrast, the tensile strength of fibres of P(3HB)-UHMW with a MM of 5.3 × 103 kDa could be manipulated to increase up to sevenfold (1320 MPa) with an elongation at break of 57% (Iwata, 2005). Therefore, for P(3HB) commercial production, it is desirable to obtain polymers with a suitable molecular mass for their final application, especially in the medical field.
It has been described by several authors how the P(3HB) molecular mass depends on the culture conditions such as: medium composition, pH and oxygen availability. In the next section, the influence of these parameters on the molecular weight of the P(3HB) will be discussed.
Medium composition
The effect of the medium composition on the P(3HB) MM has been reported for Azotobacter species, C. necator, A. lata and for methane-utilizing mixed cultures (Chen and Page, 1994; Wang and Yu, 2001; Helm et al., 2008; Myshkina et al., 2008; Penloglou et al., 2012b).
Wang and Yu (2001) reported that the mean molecular mass (MMM) of P(3HB) produced by C. necator could be altered by the medium composition, under chemically defined conditions and using acetic acid as carbon source. These authors evaluated the effect of C/N ratio on the MMM. The MMM of the polymer was higher (8.2 × 102 kDa) in cultures developed under low C/N ratio, with respect to those obtained under high C/N ratio (MM = 5.2 × 102 kDa) (Table 5). However, the amount of P(3HB) per residual biomass increased from 0.5 to 1.2 g P(3HB) g biomass-1 increasing the C/N ratio.
Table 5.
Influence of culture conditions on the molecular mass of PHB
| Organism | Carbon Source | Condition | MMW (kDa) | PHB content (%) | Reference |
|---|---|---|---|---|---|
| C. necator | Acetic Acid | Low C/N ratio = 4 | 820 | 50 | Wang and Yu. 2001 |
| High C/N ratio = 72 | 520 | ||||
| A. lata | Sucrose | C/N ratio = 20 | 2576 | 15 | Penloglou et al., 2012b |
| C/N ratio = 8 | 596 | 35 | |||
| C/P ratio = 8 | 2076 | 27 | |||
| A. vinelandii UWD | Beet molasses | 5% (w/v) | 4100 | N.S. | Chen and Page, 1994 |
| Beet molasses | 10% (w/v) | 3500 | |||
| Sucrose | 5% (w/v) | 1600 | |||
| A. chroccoccum 7B | Sucrose | 2% (w/v) | 1200–1600 | 74–79 | Myshkina et al., 2008 |
| Sucrose+Molasses | 590 | 60 | |||
| E. coli XL-1 | Glucose | pH = 6.0–7.0 | 2000–2500 | 32–35 | Bocanegra et al., 2013 |
| Xylose | |||||
| A. chroccoccum 6B | Glucose | 0.5 VVM | 1100 | 63.5 | Quagliano and Miyazaki, 1997 |
| 2.5 VVM | 100 | 7.6 | |||
| A. vinelandii OPN | Sucrose | Low aeration | 2020 | 67 | Peña et al., 2014 |
| High aeration | 1010 | 62 |
N.S., not specified.
On the other hand, in A. lata, Penloglou and colleagues (2012b) evaluated in 2-L shaken flasks cultures the effect of the initial C/N ratio and carbon/phosphates (C/P) weight ratio on the MM of P(3HB). These authors reported that the polymer reached highest MMM values (2.5 × 103 kDa) for a C/N ratio of 20 and an MM of 2.0 × 103 kDa when C/P ratio was 8; however, under such growth conditions, the P(3HB) accumulation was lower than 30%. Also, these authors observed that the MM diminished up to 20 and 3 times-fold as the C/N or C/P ratios decreased to 6 and 0.8, respectively.
The role of the potassium, iron and sulfur deficiency on the MM of the P(3HB) has been studied in methane-utilizing mixed cultures by Helm and colleagues (2008). In two-stages cultures (with a continuous-growth phase and a discontinuous P(3HB)-accumulation phase), P(3HB) accumulation was higher in those cultures under potassium deficiency (33.6% w/w) than the accumulation obtained under iron and sulfur-deficiency conditions. With respect to the MM of the P(3HB), the highest value (3.1 × 103 kDa) was obtained in the cultures developed under potassium deficiency, and the lowest value (1.7 × 103 kDa) was achieved in those cultures lacking iron. It must be emphasized that the MM of 3.1 × 103 kDa is up to now the highest value reported for methanotrophic bacteria.
In the case of A. vinelandii, Chen and Page (1994) reported that strain UWD produced a polymer with a high-molecular weight (4.1 × 103 kDa), when this bacterium was grown in 5% w/v beet molasses medium. The polymer MM decreased when the beet molasses concentration was increased. Similar results were obtained in equivalent concentrations of sucrose (as raw sugar), but the polymer MM was not greater than 1.6 × 103 kDa (Table 5).
For the producer strain A. chroococcum 7B, it has been shown that the MM of P(3HB) depends on changes in the medium composition, specially carbon source (Myshkina et al., 2008). These authors described that the MM of P(3HB) obtained using glucose, sucrose or starch as carbon sources, oscillated around 1.2 × 103 and 1.6 × 103 kDa (Table 5). However, the P(3HB) MM decreased to 5.9 × 102 kDa when A. chroococcum 7B was cultured using sucrose complemented with molasses at 2% (w/v). The negative effect of the introduction of molasses suggested that presence of organic acids in this kind of raw material affected P(3HB) biosynthesis. To confirm this behaviour, the MM of P(3HB) was evaluated in cultures of A. chroococcum 7B using sucrose (2% w/w) supplemented with sodium acetate at different concentrations (from 2 to 5 g l−1). Under such conditions, the MM of P(3HB) decreased as the acetate concentration increased. These results, provided an original method for production of P(3HB) with predetermined MM within a wide range, from 2.7 × 102 kDa (using 2% sucrose w/v and acetate 5 g l−1) to 1.5 × 103 kDa (with sucrose as a sole carbon source).
Influence of pH
The pH of the broth culture is a critical parameter for the optimal production of P(3HB). Reports have been published about the influence of this parameter on the concentration and molecular weight of this polymer (Kusaka et al., 1998; Myshkina et al., 2008; Bocanegra et al., 2013).
In this line, Myshkina and colleagues (2008) reported in shake flask cultures using A. chroococcum strain 7B that the mean molecular weight (MMW) of P(3HB) was influenced by the pH of the broth culture, finding that the MMW was maximum (1485 kDa) when the bacterium was grown at neutral pH (7.0). A variation of pH in the interval of 6.0 to 8.0 allowed the synthesis of PHB of predetermined MMW in a wide range from 354 to 1485 kDa, determined by capillary viscometry.
On the other hand, Kusaka and colleagues (1998) reported that in cultures of recombinant E. coli XL-1 Blue (pSYL105), harbouring C. necator P(3HB) biosynthesis phbCAB genes, the MM of P(3HB) could be manipulated by changes in pH, reaching one of the highest values of MM reported for P(3HB) (11 × 103 kDa) when E. coli cultures were grown at pH 6.5, and this value dropped up to 10-fold times (1.1 × 103 kDa) when pH increased to 7.0.
More recently, Bocanegra and colleagues (2013) evaluated P(3HB) production by recombinant E. coli XL-1 Blue harbouring plasmid pSK::phbCAB at three different pHs (6.0, 6.5 and 7.0). Cultures in bioreactor using glucose as the sole carbon source at variable pH values (6.0, 6.5, or 7.0) allowed the production of P(3HB) with MMW varying between 2.0 and 2.5 × 103 kDa. These values were significantly higher than those obtained by natural bacterial strains (0.5–1.0 MDa). However, in contrast to that reported by Kusaka et al., 1998, no influence of pH was observed on the MMW of the polymer produced (Table 5).
Influence of aeration conditions
There are reports in the literature where the influence of the aeration conditions on the MMW of P(3HB) has been evaluated. Quagliano and Miyazaki (1997) evaluated different levels of aeration in a stirred bioreactor for A. chroococcum 6B. These authors reported that at lower aeration (0.5 vvm), the MM of P(3HB) (determined by the intrinsic viscosity) was of 1.1 × 103 kDa. In contrast, at higher aeration (2.5 vvm), the molecular weight significantly decreased at values of 1.0 × 102 kDa. In addition, Myshkina and colleagues (2008) found that by culturing A. chroococcum 7B in shake flasks, the molecular mass of P(3HB) increased from 1.48 × 103 to 1.67 × 103 kDa when the agitation rate decreased from 250 to 190 r.p.m. (Table 5). However, the yield of P(3HB) on biomass was very similar in both conditions evaluated.
Previous studies in our group revealed that the MM of P(3HB) is strongly influenced by both the aeration condition and the strain tested (Peña et al., 2014). In that study, a maximal MM of 2.02 × 103 kDa was observed for the P(3HB) isolated from the cultures of OPN mutant under low aeration conditions at 60 h of cultivation. A similar behaviour was observed in the polymer produced by the OP strain, obtaining a P(3HB) with a MW of 1.65 × 103 kDa at the same time. In contrast, in the cultures at high aeration, the molecular weight of P(3HB) decreased to 1.01 × 103 kDa and 5.51 × 102 kDa for the OPN and parental strain (OP) respectively (Table 5).
Finally, it is important to point out that the MM can be controlled to some extent by genetic manipulation. An interesting example was reported by Hiroe and colleagues (2012). They showed that the concentration of active PHA synthase, relative to that of the enzymes supplying the monomer has a negative correlation with the P(3HB) molecular weight. They were able to construct strains producing a high molecular weight polymer by changing the order of the phaA, phaB and phaC genes within the operon, which in turn determines their relative expression level. Another example illustrating the effect of genetic changes on P(3HB) MM control was reported by Zheng and colleagues (2006). A deletion of 78 amino acid residues from the highly variable N-terminal fragment of the P(3HB) synthase of C. necator, resulted in a 60-fold increase in the average molecular weight, reaching a size of 2.84 × 103 kDa. An α-helix structure was predicted in this region, and mutations disrupting this structure at amino acids 75 and 81 were shown to also increase 50-fold the size of the polymer, allowing simultaneously a higher production of the P(3HB).
Conclusions and future prospects
In this article, several aspects about P(3HB) polymer production using different microorganisms and fermentation strategies have been reviewed. It is clear that the commercial applications of P(3HB) depend on the characteristics of the polymer. In this sense, it has been shown that the strain and culture conditions employed determine the molecular mass of the P(3HB) produced, and that this characteristic can also be further modified by genetic alteration of the producer strain. The understanding of the regulatory mechanisms controlling the synthesis of P(3HB) has also helped in some cases to construct mutants improved for P(3HB) production. In addition, some recombinant strains have shown to produce sufficient P(3HB) for large-scale production. The development of fermentation strategies has also shown promising results in terms of improving the productivity. Undoubtedly, the fed-batch fermentation and the multistage systems seem to be the more suitable strategies for improving the P(3HB) production. By using this kind of systems, it has been possible to reach a very high yields and productivities of P(3HB). Overall, the use of recombinant strains, in combination with a multistage fermentation process and raw materials for low cost could be a feasible strategy to optimize the P(3HB) production at the industrial level. However, the cost of the substrates for P(3HB) production and extraction of these materials is still the bottleneck, which limits the possibility to market them at larger scale. For this reason, the implementation of systems of production by mixed microbial cultures and wastes as substrates seems to have many advantages in the close future. In addition, the use of Archaeabacteria could be a feasible strategy to the PHA production, because they can utilize cheap carbon sources and are able to grow under extreme conditions, in which other microorganisms do not survive.
Acknowledgments
We thank Prof. Guadalupe Espín for her critical review of the manuscript, we also thank to Mario Trejo and Martin Patiño for their technical support.
Conflict of interest
None declared.
References
- Ahn W, Park J, Lee S. Production of poly(3-hydroxybutyrate) by fed-batch culture of recombinant Escherichia coli with a highly concentrated whey solution. Appl Environ Microbiol. 2000;66:3624–3627. doi: 10.1128/aem.66.8.3624-3627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Althuri A, Mathew J, Sindhu R, Banerjee R, Pandey A, Binod P. Microbial synthesis of poly-3-hydroxybutyrate and its application as targeted drug delivery vehicle. Bioresour Technol. 2013;145:290–296. doi: 10.1016/j.biortech.2013.01.106. [DOI] [PubMed] [Google Scholar]
- Bocanegra JK, Pradella JGC, da Silva LF, Taciro MK, Gomez JGC. Influence of pH on the molecular weight of poly-3-hydroxybutiric acid (P3HB) produced by recombinant Escherichia coli. Appl Biochem Biotechnol. 2013;170:1336–1347. doi: 10.1007/s12010-013-0257-4. [DOI] [PubMed] [Google Scholar]
- Bornatsev AP, Yakovlev SG, Zharkova II, Boskhomdzhiev AP, Bagrov DV, Myshkina VL, et al. Cell attachment on poly(3-hydroxybutyrate)-poly(ethylene glycol) copolymer produced by Azotobacter chroococcum 7B. BMC Biochem. 2013;14:12. doi: 10.1186/1471-2091-14-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl H, Gross RA, Lenz RW, Fuller RC. Pseudomonas oleovorans as a source of poly(β-hydroxyalkanoates) for potential applications as biodegradable polyesters. Appl Environ Microbiol. 1988;54:1977–1982. doi: 10.1128/aem.54.8.1977-1982.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrom D. Polymer synthesis by microorganisms: technology and economics. Trends Biotechnol. 1987;5:246–250. [Google Scholar]
- Cao W, Wang A, Jing D, Gong Y, Zhao N, Zhang X. Novel biodegradable films blended with poly(3-hydroxybutyrate) J Biomater Sci Polym Ed. 2005;16:1379–1394. doi: 10.1163/156856205774472308. [DOI] [PubMed] [Google Scholar]
- Cavalheiro JMBT, De Almeida M, Grandfils C, Da Fonseca M. Poly (3-hydroxybutyrate) production by Cupriavidus necato using waste glycerol. Process Biochem. 2009;44:509–515. [Google Scholar]
- Centeno-Leija S, Huerta-Beristain G, Giles-Gomez M, Bolivar F, Gosset G, Martinez A. Improving poly-3-hydroxybutyrate production in Escherichia coli by combining the increase in the NADPH pool and acetyl-CoA availability. Antonie Van Leeuwenhoek. 2014;105:687–696. doi: 10.1007/s10482-014-0124-5. [DOI] [PubMed] [Google Scholar]
- Chan RTH, Rusell RA, Marcal H, Lee TH, Holden PJ, Foster JR. BioPEGylation of polyhydroxybutyrate promotes nerve cell health and migration. Biomacromolecules. 2014;15:339–349. doi: 10.1021/bm401572a. [DOI] [PubMed] [Google Scholar]
- Chanprateep S. Current trends in biodegradable polyhydroxyalkanoates. J Biosci Bioeng. 2010;110:621–632. doi: 10.1016/j.jbiosc.2010.07.014. [DOI] [PubMed] [Google Scholar]
- Chen CW, Don TR, Yen HF. Enzymatic extruded starch as a carbon source for the production of poly (3-hydroxybutyrate-co-hydroxyvalerate) by Haloferax mediterranei. Process Biochem. 2006;41:2289–2296. [Google Scholar]
- Chen G. A microbial polyhydroxyalkanoates (PHA) based bio-and materials industry. Chem Soc Rev. 2009;38:2434–2446. doi: 10.1039/b812677c. [DOI] [PubMed] [Google Scholar]
- Chen G, Wang Y. Medical applications of biopolyesters polyhydroxyalkanoates. Chin J Polym Sci. 2013;31:719–736. [Google Scholar]
- Chen G, Wu Q. The application of polyhydroxyalkanoates as tissue engineering materials. Biomaterials. 2005;26:6565–6578. doi: 10.1016/j.biomaterials.2005.04.036. [DOI] [PubMed] [Google Scholar]
- Chen GQ. Plastics completely synthesized by bacteria: Polyhydroxyalkanoates. In: Chen G-Q, editor. Plastics from Bacteria. Berlin, Germany: Microbiology Monographs, Springer-Verlag; 2010. pp. 17–37. Alexander Steinbüchel, Series (ed.) [Google Scholar]
- Chen GQ, Page W. The effect of substrate on the molecular weight of poly-β-hydroxybutyrate produced by Azotobacter vinelandii UWD. Biotechnol Lett. 1994;16:155–160. [Google Scholar]
- Chen GQ, Page W. Production of poly-β-hydroxybutyrate by Azotobacter vinelandii in a two-stage fermentation process. Biotechnol Tech. 1997;11:347–350. [Google Scholar]
- De Smet MJ, Eggink G, Witholt B, Kingma J, Wynberg H. Characterization of intracellular inclusions formed by Pseudomonas oleovorans during growth on octane. J Bacteriol. 1983;154:870–878. doi: 10.1128/jb.154.2.870-878.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errico C, Bartoli C, Chiellini F, Chiellini E. Poly(hydroxyalkanoates)-based polymeric nanoparticles for drug delivery. J Biomed Biotechnol. 2009 doi: 10.1155/2009/571702. doi: 10.1155/2009/571702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filho LX, Olyveira GM, Basmaji P, Manzine-Costa LM. Novel electrospun nanotholits/PHB scaffolds for bone tissue regeneration. J Nanosci Nanotechnol. 2013;13:4715–4719. doi: 10.1166/jnn.2013.7191. [DOI] [PubMed] [Google Scholar]
- García A, Segura D, Espín G, Galindo E, Castillo T, Pena C. High production of poly-β-hydroxybutyrate (PHB) by an Azotobacter vinelandii mutant altered in PHB regulation using a fed-batch fermentation process. Biochem Eng J. 2014;82:117–123. [Google Scholar]
- Grage K, Jahns AC, Parlane N, Palanisamy R, Rasiah IA, Atwood JA, Rehm BAH. Bacterial polyhydroxyalkanoate granules: biogenesis, structure, and potential use as nano-/micro-beads in biotechnological and biomedical applications. Biomacromolecules. 2009;10:660–669. doi: 10.1021/bm801394s. [DOI] [PubMed] [Google Scholar]
- Grothe E, Chisti Y. Poly (ß-hydroxybutiric acid) thermoplastic production by Alcaligenes latus: behavior of fed-batch cultures. Bioprocess Eng. 2000;22:441–449. [Google Scholar]
- Helm J, Wendlandt KD, Jechorek M, Stottmeister U. Potassium deficiency results in accumulation of ultra-high-molecular weight poly-β-hydroxybutyrate in a methane-utilizing mixed culture. J Appl Microbiol. 2008;105:1054–1061. doi: 10.1111/j.1365-2672.2008.03831.x. [DOI] [PubMed] [Google Scholar]
- Hernández-Eligio A, Moreno S, Castellanos M, Castañeda M, Nuñez C, Muriel-Millan L, Espín G. RsmA post-transcriptionally controls PhbR expression and polyhydroxybutyrate biosynthesis in Azotobacter vinelandii. Microbiology. 2012;158:1953–1963. doi: 10.1099/mic.0.059329-0. [DOI] [PubMed] [Google Scholar]
- Hezayen FF, Rehm BHA, Eberhardt R, Steinbüchel A. Polymer production by two newly isolated extremely halophilic archaea: application of a novel corrosion-resistant bioreactor. Appl Microbiol Biotechnol. 2000;54:319–325. doi: 10.1007/s002530000394. [DOI] [PubMed] [Google Scholar]
- Hiroe A, Tsuge K, Nomura CT, Itaya M, Tsuge T. Rearrangement of gene order in the phaCAB operon leads to effective production of ultra-high-molecular-weight poly[(R)-3-hydroxybutyrate] in genetically engineered Escherichia coli. Appl Environ Microbiol. 2012;78:3177–3184. doi: 10.1128/AEM.07715-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann N, Steinbüchel A, Rehm BHA. The Pseudomonas aeruginosa phaG gene product is involved in the synthesis of polyhydroxyalkanoic acids consisting of medium-chain-length constituents from non-related carbon sources. FEMS Microbiol Lett. 2000a;184:253–260. doi: 10.1111/j.1574-6968.2000.tb09023.x. [DOI] [PubMed] [Google Scholar]
- Hoffmann N, Steinbüchel A, Rehm BHA. Homologous functional expression of cryptic phaG from Pseudomonas oleovorans establishes the transacylase-mediated polyhydroxy- alkanoate biosynthetic pathway. Appl Microbiol Biotechnol. 2000b;54:665–670. doi: 10.1007/s002530000441. [DOI] [PubMed] [Google Scholar]
- Hong SW, Hsu HW, Ye MT. Thermal properties and applications of low molecular weight polyhyxybutyrate. J Therm Anal Calorim. 2013;111:1243–1250. [Google Scholar]
- Huang TY, Duan KJ, Huang SY, Chen CW. Production of polyhydroxyalkanoates from inexpensive extruded rice bran and starch by Haloferax mediterranei. J Ind Microbiol Biotechnol. 2006;33:701–706. doi: 10.1007/s10295-006-0098-z. [DOI] [PubMed] [Google Scholar]
- Iwata T. Strong fibers and films of microbial polyesters. Macromol Biosci. 2005;5:689–701. doi: 10.1002/mabi.200500066. [DOI] [PubMed] [Google Scholar]
- Kabe T, Tsuge T, Kasuya K, Takemura A, Hikima T, Takata M, Iwata T. Physical and structural effects of adding ultra-high-molecular-weight poly[(R)-3-hydroxybutyrate] to wild type poly[(R)-3-hydroxybutyrate] Macromolecules. 2012;45:1858–1865. [Google Scholar]
- Kanjanachumpol P, Kulpreecha S, Tolieng V, Thongchul N. Enhancing polyhydroxybutyrate production from high cell density fed-batch fermentation of Bacillus megaterium BA-019. Bioproc Biosyst Eng. 2013;36:1463–1474. doi: 10.1007/s00449-013-0885-7. [DOI] [PubMed] [Google Scholar]
- Kessler B, Witholt B. Factors involved in the regulatory network of polyhydroxyalkanoate metabolism. J Biotechnol. 2001;86:97–104. doi: 10.1016/s0168-1656(00)00404-1. [DOI] [PubMed] [Google Scholar]
- Khanna S, Srivastava AK. Recent advances in microbial polyhydroxyalkanoates. Process Biochem. 2005;40:607–619. [Google Scholar]
- Kim BS. Production of poly(3-hydroxybutyrate) from inexpensive substrates. Enzyme Microb Technol. 2000;27:774–777. doi: 10.1016/s0141-0229(00)00299-4. [DOI] [PubMed] [Google Scholar]
- Kim BS, Chang HN. Production of poly(3-hydroxybutyrate) from starch by Azotobacter chroococcum. Biotechnol Lett. 1998;20:109–112. [Google Scholar]
- Kulpreecha S, Boonruangthavorn A, Meksiriporn B, Thongchul N. Inexpensive fed-batch cultivation for high poly(3-hydroxybutyrate) production by a new isolate of Bacillus megaterium. J Biosci Bioeng. 2009;107:240–249. doi: 10.1016/j.jbiosc.2008.10.006. [DOI] [PubMed] [Google Scholar]
- Kusaka S, Iwata T, Doi Y. Microbial synthesis and physical properties of ultra-high-molecular-weight poly[(R)-3hydroxybutyrate] Pure Appl Chem A. 1998;35:319–335. [Google Scholar]
- Lagaveen RG, Huisman GW, Preusting H, Ketelaar P, Eggink G, Witholt B. Formation of polyesters by Pseudomonas oleovorans; effect of substrates on formation and composition of poly-(R)-3- hydroxyalkanoates and poly-(R)-3-hydroxyalkenoates. Appl Environ Microbiol. 1988;54:2924–2932. doi: 10.1128/aem.54.12.2924-2932.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY. Bacterial polyhydroxyalkanoates. Biotechnol Bioeng. 1996;49:1–14. doi: 10.1002/(SICI)1097-0290(19960105)49:1<1::AID-BIT1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Legat A, Gruber C, Zangger K, Wanner G, Stan-Lotter H. Identification of polyhydroxyalkanoates in Halococcus and other haloarchaeal species. Appl Microbiol Biotechnol. 2010;87:1119–1127. doi: 10.1007/s00253-010-2611-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Zhang H, Qi Q. The production of polyhydroxyalkanoates in recombinant Escherichia coli. Bioresour Technol. 2007;98:2313–2320. doi: 10.1016/j.biortech.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Lu J, Tappel RC, Nomura CT. Mini-review: biosynthesis of poly(hydroxyalkanoates) Polym Rev. 2009;49:226–248. doi:10.1080/ 15583720903048243. [Google Scholar]
- Masaeli E, Morshed M, Rasekhian P, Karbasi S, Karbalaie K, Karamali F, et al. Does the tissue engineering architecture of poly(3-hydroxybutyrate) scaffold affects cell-material interactions? J Biomed Mater Res A. 2012;100A:1907–1918. doi: 10.1002/jbm.a.34131. [DOI] [PubMed] [Google Scholar]
- Masaeli E, Morshed M, Nasr-Esfahani MH, Sadri S, Hilderink J, van Apeldoorn A, et al. Fabrication, characterization and cellular compatibility of poly (hydroxy alkanoate) composite nonofibrous scaffolds for nerve tissue engineering. PLoS ONE. 2013;8:e57157. doi: 10.1371/journal.pone.0057157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Matsusaki H, Taguchi S, Seki M, Doi Y. Cloning and characterization of the Pseudomonas sp. 61-3 phaG gene involved in polyhydroxyalkanoate biosynthesis. Biomacromolecules. 2001;2:142–147. doi: 10.1021/bm005604+. [DOI] [PubMed] [Google Scholar]
- Medvecky L, Giretova M, Stulajterova R. Properties and in vitro characterization of polyhydroxybutyrate-chitosan scaffolds prepared by modified precipitation method. J Mater Sci Mater Med. 2014;25:777–789. doi: 10.1007/s10856-013-5105-0. [DOI] [PubMed] [Google Scholar]
- Mejía MA, Segura D, Espín G, Galindo E, Peña C. Two stage fermentation process for alginate production by Azotobacter vinelandii mutant altered in poly-B- hydroxybutirate PHB synthesis. J Appl Microbiol. 2010;108:55–61. doi: 10.1111/j.1365-2672.2009.04403.x. [DOI] [PubMed] [Google Scholar]
- Mendonca RH, de Oliveira T, Ferreira de Costa M, da Silva RM. Production of 3D scaffolds applied to tissue engineering using chitosan swelling as a porogenic agent. J Appl Polym Sci. 2013;129:614–625. [Google Scholar]
- Mozumder MSI, De Wever H, Volcke EIP, Garcia-Gonzalez L. A robust fed-batch feeding strategy independent of the carbon source for optimal polyhydroxybutyrate production. Process Biochem. 2014;155:272–280. [Google Scholar]
- Murakami R, Sato H, Dybal J, Iwata T, Ozaki Y. Formation and stability of β-structure in biodegradable ultra-high-molecular weight poly(3-hydroxybutyrate) by infrared, Raman, and quantum chemical calculation studies. Polymer. 2007;48:2672–2680. [Google Scholar]
- Myshkina VL, Nikolaeva DA, Makhina TK, Bonartsev AP, Filatova EV, Ruzhitsky AO, Bonartseva GA. Effect of growth conditions on the molecular weight of Poly-3-hydroxybutyrate produced by Azotobacter chroococcum 7B. Appl Biochem Microbiol. 2008;44:482–486. [PubMed] [Google Scholar]
- Nath A, Dixit M, Bandiya A, Chavda S, Desai AJ. Enhanced PHB production and scale up studies using cheese whey in fed batch cultures of Methylobacterium sp. ZP24. Bioresour Technol. 2008;99:5749–5755. doi: 10.1016/j.biortech.2007.10.017. [DOI] [PubMed] [Google Scholar]
- Noguez R, Segura D, Moreno S, Hernández A, Juárez K, Espín G. Enzyme INtr, NPr and IIANtr are involved in regulation of the poly-β-hydroxybutyrate biosynthetic genes in Azotobacter vinelandii. J Mol Microbiol Biotechnol. 2008;15:244–254. doi: 10.1159/000108658. [DOI] [PubMed] [Google Scholar]
- Olivera E, Carnicero D, Jodra R, Miñambres B, García B, Abraham G, et al. Genetically engineered Pseudomonas: a factory of new bioplastics with broad applications. Environ Microbiol. 2001;3:612–618. doi: 10.1046/j.1462-2920.2001.00224.x. [DOI] [PubMed] [Google Scholar]
- Osanai T, Numata K, Oikawa A, Kuwahara A, Iijima H, Doi Y, et al. Increased bioplastic production with an RNA polymerase sigma factor SigE during nitrogen starvation in Synechocystis sp. PCC 6803. DNA Res. 2013;20:525–535. doi: 10.1093/dnares/dst028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page W, Cornish A. Growth of Azotobacter vinelandii UWD in fish peptone medium and simplified extraction of poly-β-hydroxybutyrate. Appl Microbiol. 1993;59:4236–4244. doi: 10.1128/aem.59.12.4236-4244.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page WJ, Knosp O. Hyperproduction of poly-β-hydroxybutyrate during exponential growth of Azotobacter vinelandii UWD. Appl Environ Microbiol. 1989;55:1334–1339. doi: 10.1128/aem.55.6.1334-1339.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page WJ, Tindale A, Chandra M, Kwon E. Alginate formation in I UWD during stationary phase and the turnover of poly-3-hydroxybutyrate. Microbiology. 2001;147:483–490. doi: 10.1099/00221287-147-2-483. [DOI] [PubMed] [Google Scholar]
- Pan P, Inoue Y. Polymorphism and isomorphism in biodegradable polyesters. Prog Polym Sci. 2009;34:605–640. [Google Scholar]
- Panchal B, Bagdadi A, Roy I. Polyhydroxyalkanoates: the natural polymers produced by bacterial fermentation. In: Thomas S, Visakh PM, Mathew AP, editors. Advances in Natural Polymers. Berlin Heidelberg, Germany: Springer; 2013. pp. 397–421. [Google Scholar]
- Parlane NA, Grage K, Mifune J, Randall J, Basaraba D, Wedlock DN, et al. Vaccines displaying mycobacterial proteins on biopolyester beads stimulate cellular immunity and induce protection against tuberculosis. Clin Vaccine Immunol. 2012;19:37–44. doi: 10.1128/CVI.05505-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penloglou G, Chatzidoukas C, Kiparissides C. Microbial production of polyhydroxybutyrate with tailor-made properties: an integrated modelling approach and experimental validation. Biotechnol Adv. 2012a;30:329–337. doi: 10.1016/j.biotechadv.2011.06.021. [DOI] [PubMed] [Google Scholar]
- Penloglou G, Kretza E, Chatzidoukas C, Parouti S, Kiparissides C. On the control of molecular weight distribution of polyhydroxybutyrate in Azohydromonas lata. Biochem Eng J. 2012b;62:39–47. [Google Scholar]
- Peña C, Castillo T, Nuñez C, Segura D. Bioprocess Design: Fermentation Strategies for Improving the Production of Alginate and Poly-β-Hydroxyalkanoates (PHAs) by Azotobacter vinelandii. Rijeka, Croatia: INTECH- Open Access Publisher; 2011. pp. 217–242. [Google Scholar]
- Peña C, López S, García A, Espín G, Romo-Uribe A, Segura D. Biosynthesis of poly-β-hydroxybutyrate (PHB) with a high molecular mass by a mutant strain of Azotobacter vinelandii (OPN) Ann Microbiol. 2014;64:39–47. [Google Scholar]
- Poli A, Di Donato P, Abbamondi GR, Nicolaus B. Synthesis, production, and biotechnological applications of exopolysaccharides and polyhydroxyalkanoates by archaea. Archaea. 2011 doi: 10.1155/2011/693253. doi: 10.1155/2011/693253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo C, Martínez-Toledo MV, Rodelas B, González-Lopez J. Effects of culture conditions on the production of polyhydroxyalkanoates by Azotobacter chroococcum H23 in media containing a high concentration of alpechín (wastewater from olive oil mills) as primary carbon source. J Biotechnol. 2002;97:125–131. doi: 10.1016/s0168-1656(02)00056-1. [DOI] [PubMed] [Google Scholar]
- Pradella JGC, Ienczak J, Romero C, Taciro M. Carbon source pulsed feeding to attain high yield and high productivity in poly(3-hydroxybutyrate) (PHB) production from soybean oil using Cupriavidus necator. Biotechnol Lett. 2012;34:1003–1007. doi: 10.1007/s10529-012-0863-1. [DOI] [PubMed] [Google Scholar]
- Quagliano J, Miyazaki S. Effect of aeration and carbon/nitrogen ratio on the molecular mass of the biodegradable polymer poly-β-hydroxybutyrate obtained from Azotobacter chroococcum 6B. Appl Microbiol Biotechnol. 1997;48:662–664. [Google Scholar]
- Rajan R, Sreekumar PA, Joseph K, Skrifvars M. Thermal and mechanical properties of chitosan reinforced polyhydroxybutyrate composites. J Appl Polym Sci. 2012;124:3357–3362. [Google Scholar]
- Ramier J, Bouderlique T, Stoilova O, Manolova N, Rashkov I, Langlois V, et al. Biocomposite scaffolds based on electrospun poly(3-hydroxybutyrate) nanofibers and electrosprayed hydroxyapatite nonoparticles for bone tissue engineering applications. Mater Sci Eng C. 2014;38:161–169. doi: 10.1016/j.msec.2014.01.046. [DOI] [PubMed] [Google Scholar]
- Reddy CSK, Ghai R, Rashmi, Kalia VC. Polyhydroxyalkanoates: an overview. Bioresour Technol. 2003;87:137–146. doi: 10.1016/s0960-8524(02)00212-2. [DOI] [PubMed] [Google Scholar]
- Rehm BHA. Polyester synthases: natural catalysts for plastics. Biochem J. 2003;376:15–33. doi: 10.1042/BJ20031254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm BHA, Krüger N, Steinbüchel A. A new metabolic link between fatty acid de novo synthesis and Polyhydroxyalkanoic acid synthesis: the phaG gene from Pseudomonas putida KT2440 encodes a 3-hydroxyacyl-acyl carrier protein-Coenzyme A transferase. J Biol Chem. 1998;273:24044–24051. doi: 10.1074/jbc.273.37.24044. [DOI] [PubMed] [Google Scholar]
- Ricotti L, Polini A, Genchi GG, Ciofani G, Iandolo D, Vazao H, et al. Proliferation and skeletal myotube formation capability of C2C12 and H9c2 cells on isotropic and anisotropic electrospun nanofibrous PHB scaffolds. Biomed Mater. 2012;7:035010. doi: 10.1088/1748-6041/7/3/035010. doi: 10.1088/1748-6041/7/3/035010. [DOI] [PubMed] [Google Scholar]
- Rocha RCS, Silva LF, Taciro MK, Pradella JGC. Production of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) P(3HB-co-3HV) with a broad range of 3HV content at high yields by Burkholderia sacchari IPT 189. World J Microbiol Biotechnol. 2008;24:427–431. [Google Scholar]
- Rodriguez-Valera F, Lillo J. Halobacteria as producers of polyhydroxyalkanoates. FEMS Microbiol Lett. 1992;103:181–186. [Google Scholar]
- Ruan W, Chen J, Lun S. Production of biodegradable polymer by A. eutrophus using volatile fatty acids from acidified waste water. Process Biochem. 2003;39:295–299. [Google Scholar]
- Segura D, Espín G. Mutational inactivation of a gene homologous to Escherichia coli ptsP affects poly-β-hydroxybutyrate accumulation and nitrogen fixation in Azotobacter vinelandii. J Bacteriol. 1998;180:4790–4798. doi: 10.1128/jb.180.18.4790-4798.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura D, Espín G. Inactivation of pycA, encoding pyruvate carboxylase activity, increases poly-beta-hydroxybutyrate accumulation in Azotobacter vinelandii on solid medium. Appl Microbiol Biotechnol. 2004;65:414–418. doi: 10.1007/s00253-004-1611-9. [DOI] [PubMed] [Google Scholar]
- Segura D, Guzmán J, Espín G. Azotobacter vinelandii mutants that overproduce poly-β-hydroxybutyrate or alginate. Appl Microbiol Biotechnol. 2003;63:159–163. doi: 10.1007/s00253-003-1397-1. [DOI] [PubMed] [Google Scholar]
- Senior P, Dawes E. The regulation of poly-β-hydroxybutyrate metabolism in Azotobacter beijerinkii. Biochem J. 1973;134:225–238. doi: 10.1042/bj1340225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior P, Beech G, Ritchie G, Dawes E. The role of oxygen limitation in the formation of poly-β-hydroxybutyrate during batch and continuous culture of Azotobacter beijerinckii. Biochem J. 1972;128:1193–1201. doi: 10.1042/bj1281193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma L, Yoshino O, Toshiji K, Tadahisa I, Yoshiharu D. Fiber formation in medium and ultra high molecular weight Polyhydroxybutyrate blends under shear flow. Macromol Mater Eng. 2004;289:1068–1073. [Google Scholar]
- Shishatskaya E, Khilusov IA, Volova T. A hybrid PHB-hydroxyapatite composite for biomedical application: production, in vitro and in vivo investigation. J Biomater Sci Polym Ed. 2006;17:481–498. doi: 10.1163/156856206776986242. [DOI] [PubMed] [Google Scholar]
- Shishatskaya E, Goreva A, Kalacheva G, Volova T. Biocompatibility and resorption of intravenously administered polymer microparticles in tissues of internal organs of laboratory animals. J Biomater Sci. 2011;22:2185–2203. doi: 10.1163/092050610X537138. [DOI] [PubMed] [Google Scholar]
- da Silva-Valenzuela MDG, Wang SH, Wiebeck H, Valenzuela-Díaz FR. Nanocomposite microcapsules from powders of polyhydroxybutyrate (PHB) and smectite clays. Mat Sci Forum. 2010;660–661:794–798. [Google Scholar]
- Slater SC, Voige WH, Dennis DE. Cloning and expression in Escherichia coli of the Alcaligenes eutrophus H16 poly-beta-hydroxybutyrate biosynthetic pathway. J Bacteriol. 1988;170:4431–4436. doi: 10.1128/jb.170.10.4431-4436.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbüchel A, Lütke-Eversloh T. Metabolic engineering and pathway construction for biotechnological production of relevant polyhydroxyalkanoates in microorganisms. Biochem Eng J. 2003;16:81–96. [Google Scholar]
- Stubbe J, Tian J, He A, Sinskey A, Lawrence A, Liu P. Nontemplate-dependent polymerization processes: polyhydroxyalkanoate synthases as a paradigm. Annu Rev Biochem. 2005;74:433–480. doi: 10.1146/annurev.biochem.74.082803.133013. [DOI] [PubMed] [Google Scholar]
- Sudesh K, Abe H, Doi Y. Synthesis, structure and properties of polyhydroxyalkanoates: biological polyesters. Prog Polym Sci. 2000;25:1503–1555. [Google Scholar]
- Tanadchangsaeng N, Yu J. Microbial synthesis of polyhydroxybutyrate from glycerol: gluconeogenesis, molecular weight and material properties of biopolyester. Biotechnol Bioeng. 2012;109:2808–2818. doi: 10.1002/bit.24546. [DOI] [PubMed] [Google Scholar]
- Tsuge T, Taguchi K, Taguchi S, Doi Y. Molecular characterization and properties of (R)-specific enoyl-CoA hydratases from Pseudomonas aeruginosa: metabolic tools for synthesis of polyhydroxyalkanoates via fatty acid β-oxidation. Int J Biol Macromol. 2003;31:195–205. doi: 10.1016/s0141-8130(02)00082-x. [DOI] [PubMed] [Google Scholar]
- Volova TG, Zhila NO, Shishatskaya EI, Mironov PV, Vasil'ev AD, Sukovatyi AG, Sinskey AJ. The physicochemical properties of polyhydroxyalkanoates with different chemical structures. Polym Science Ser A. 2013;55:427–437. [Google Scholar]
- Wang J, Yu J. Kinetic analysis on formation of Poly(3-hydroxybntrate) by Ralstonia eutropha under chemically defined conditions. J Ind Microbiol Biotechnol. 2001;26:121–126. doi: 10.1038/sj.jim.7000097. [DOI] [PubMed] [Google Scholar]
- Wang Q, Zhuang Q, Liang Q, Qi Q. Polyhydroxyalkanoic acids from structurally-unrelated carbon sources in Escherichia coli. Appl Microbiol Biotechnol. 2013;97:3301–3307. doi: 10.1007/s00253-013-4809-x. [DOI] [PubMed] [Google Scholar]
- Williams S, Martin D. Applications of polyhydroxyalkanoates (PHA) in medicine and pharmacy. Biolymers Online. 2005 doi: 10.1002/3527600035.bpol4004. [Google Scholar]
- Zafar M, Kumar S, Dhiman A. Optimization of poly(3-hydroxybutyrateco- 3-hydroxyvalerate) production process by using Azohydromonas lata MTCC 2311 by using genetic algorithm on artificial neural network on response surface methodology. Biocatal Agric Biotechnol. 2012a;1:70–79. [Google Scholar]
- Zafar M, Kumar S, Dhiman A. Artificial intelligence based modeling and optimization of poly(3-hydroxybutyrateco- 3-hydroxyvalerate) production process by using Azohydromonas lata MTCC 2311 from cane molasses supplemented with volatile fatty acids: a genetic algorithm paradigm. Bioresour Technol. 2012b;104:631–641. doi: 10.1016/j.biortech.2011.10.024. [DOI] [PubMed] [Google Scholar]
- Zheng Z, Li M, Xue X-J, Tian H-L, Li Z, Chen G-Q. Mutation on N-terminus of polyhydroxybutyrate synthase of Ralstonia eutropha enhanced PHB accumulation. Appl Microbiol Biotechnol. 2006;72:896–905. doi: 10.1007/s00253-006-0371-0. [DOI] [PubMed] [Google Scholar]