Abstract
In flowering plants, the arrangement of flowers on a stem becomes an inflorescence, and a huge variety of inflorescence architecture occurs in nature. Inflorescence architecture also affects crop yield. In simple inflorescences, flowers form on a main stem; by contrast, in compound inflorescences, flowers form on branched stems and the branching pattern defines the architecture of the inflorescence. In this review, we highlight recent findings on the regulation of inflorescence architecture by cytokinin plant hormones. Results in rice (Oryza sativa) and Arabidopsis thaliana show that although these two species have distinct inflorescence architectures, cytokinins have a common effect on inflorescence branching. Based on these studies, we discuss how cytokinins regulate distinct types of inflorescence architecture through their effect on meristem activities.
Keywords: branching, cytokinin, floral meristem, inflorescence, shoot apical meristem
INTRODUCTION
Plants have an enormous, striking diversity of forms, with varying numbers and arrangements of organs in different sizes and shapes; this diversity derives from regulation of meristem activity. The aerial organs of a plant come from the shoot apical meristem (SAM) which gives rise to leaves, stem, and axillary meristems during the vegetative stage and transforms into the inflorescence meristem (IM) after the floral transition. The various developmental patterns of the IM in different species produce diverse inflorescence architectures, which not only attract artists and plant scientists, but also draw the attention of plant breeders, because inflorescence traits directly affect crop yields. Branching hierarchy and complexity depend on the species, but are also affected by environmental factors, including nutrition, light, and temperature (Tanaka et al., 2013; Kyozuka et al., 2014; Teo et al., 2014).
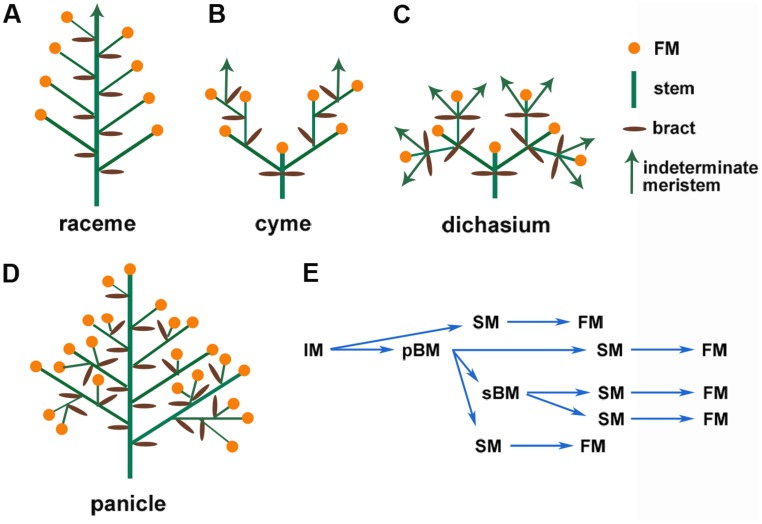
The enormous diversity of inflorescence architecture also leads to difficulties in defining consensus criteria to classify these structures. Following Weberling’s (1989) suggestions, inflorescence architectures can be broadly grouped into inflorescences without branching (simple) and inflorescences with branching (compound). Another key parameter is whether the IM ends in a terminal flower (determinate) or continues to produce structures, including branches and flowers (indeterminate). Following these key distinctions, at least three typical groups of inflorescence architectures are commonly seen, namely the raceme (simple, indeterminate, as in Arabidopsis), the cyme (complex, determinate, as in tomato), and the panicle [complex, determinate, as in wheat (Triticum aestivum); or complex, indeterminate, as in maize (Zea mays), especially tassel; Figure 1; Prusinkiewicz et al., 2007; Kellogg et al., 2013]. These distinct inflorescence architectures result from different developmental programs that are elaborated below.
FIGURE 1.
Schematic representation of common types of inflorescences. (A) Simple raceme, which is indeterminate and unbranched, (B) cyme, (C) dichasium, which are determinate and branched, (D) panicle, which is determinate and branched, and (E) Transition of the reproductive meristems in rice panicle.
Development of the IM conditions the branching of the inflorescence. In Arabidopsis, the IM directly initiates floral meristems (FMs, which are determinate meristems) on its flanks; this forms a simple raceme (Figure 1A; Benlloch et al., 2007; Tanaka et al., 2013; Teo et al., 2014). The grasses have more diverse inflorescence architectures (Kellogg et al., 2013). In a generalized grass inflorescence, the IM gives rise to several branch meristems (BMs, which are usually indeterminate meristems). These BMs may initiate secondary BMs to form lateral branches and spikelet meristems (SMs) that then initiate FMs (Figure 1E). In maize and other Andropogoneae species, determinate spikelet-pair meristems (SPMs) are produced from the IM or BMs, and each SPM makes two SMs. The SM initiates one or more FMs (Kellogg et al., 2013; Kyozuka et al., 2014). These intermediate BMs cause secondary or higher-order branches, which form a compound inflorescence termed the panicle (Benlloch et al., 2007). Therefore, the branch structure determines the final inflorescence pattern, which contributes to the enormous diversity of inflorescence architectures. Specific genetic regulatory networks control every stage and transition of meristem activity, as described in several recent reviews (Tanaka et al., 2013; Kyozuka et al., 2014).
Meristem activity, especially determinacy, fundamentally affects inflorescence architecture. For example, in the raceme-type inflorescence of Arabidopsis, the IM continues to initiate FMs; by contrast, in the cyme-type inflorescence of tomato, the IM forms a terminal flower immediately after developing a new IM below it, which reiterates this pattern (#x1-10011). The panicle-type inflorescence is initially indeterminate and initiates BMs and FMs before it finally terminates in a FM in some species. At least two groups of genes, relatives of Arabidopsis LEAFY (LFY) and TERMINAL FLOWER1 (TFL1), play a central role in meristem determinacy. LFY promotes determinate FM identity and termination of IMs, and TFL1 maintains the indeterminacy of IMs to prevent termination (Prusinkiewicz et al., 2007).
Recent work has identified cytokinins as key regulators of inflorescence architecture in plants with different inflorescence types, through regulation of meristem activity, which is often also associated with meristem identity. Cytokinins have profound effects on plant development and growth, including meristem activity (Kyozuka, 2007; Werner and Schmulling, 2009; Perilli et al., 2010). Accumulating data point to a role for cytokinins in influencing inflorescence complexity by fine-tuning IM and BM determinacy. Also, recent work reveals that cytokinins can regulate the initiation of meristems from floral organ axils (the junction where the floral organ meets the stem), and thus convert a determinate flower into an inflorescence (Han et al., 2014). Here we review these two mechanisms through which cytokinins regulate inflorescence architecture.
CYTOKININS PROMOTE IM ACTIVITY
Increasing cytokinin concentrations and signaling activity increase meristem size and activity. Reduced meristem activity often leads to conversion of an IM or a BM into a terminal flower, which subsequently affects inflorescence architecture.
Work in rice and in Arabidopsis showed that cytokinin levels affects meristem activity and inflorescence complexity. The ATP/ADP isopentenyl transferases (IPTs) catalyze the first step of cytokinin biosynthesis (Miyawaki et al., 2006). Arabidopsis atipt3 5 7 triple mutants and atipt1 3 5 7 quadruple mutants have lower levels of cytokinins, which leads to reduced IM size, formation of a terminal flower, and conversion of an indeterminate inflorescence to a determinate inflorescence (Miyawaki et al., 2006). Rice LONELY GUY (LOG) encodes a cytokinin-activating enzyme catalyzing the final step of cytokinin biosynthesis and LOG is strongly expressed in BMs and FMs of developing panicles. The absence of LOG results in early termination of IM and BMs, which reduces branching complexity (Kurakawa et al., 2007). Arabidopsis has nine LOG homologs and the triple log3 log4 log7 and septuple log1 log2 log3 log4 log5 log7 log8 mutants produce fewer FMs, suggesting reduced IM activity (Kuroha et al., 2009; Tokunaga et al., 2012).
In addition to cytokinin homeostasis, defects in cytokinin signaling also leads to simplified inflorescence architecture. Cytokinins are perceived by transmembrane histidine kinase receptors, such as Arabidopsis HISTIDINE KINASE 2 (AHK2), AHK3, and AHK4. The ahk triple mutants have a smaller IM that terminates early, resulting in a simplified inflorescence with only a few flowers (Nishimura et al., 2004).
Conversely, elevated cytokinin homeostasis results in increased inflorescence complexity. Cytokinin oxidase/dehydrogenase (CKX) plays a major role in the degradation of bioactive cytokinins (Mok and Mok, 2001). Arabidopsis plants overexpressing CKX1 or CKX3 have dramatically reduced cytokinins contents and IMs that produce very few flowers (Werner et al., 2003). CKX overexpression in tobacco plants also leads to fewer flowers and conversion of IMs from indeterminate to determinate (Werner et al., 2001). Similarly, rice varieties with lower OsCKX2 expression have more elaborated and larger panicles with more primary and secondary branches and higher yield, and rice varieties with higher OsCKX2 activity have the opposite phenotype, with fewer branches and lower yield (Ashikari et al., 2005; Li et al., 2013).
Cytokinins promote IM activity and affect inflorescence architecture by promoting expression of the meristematic gene WUSCHEL (WUS) and suppressing the meristem inhibitors CLAVATA1 (CLV1) and CLV3. Plants ectopically treated with cytokinins show a clv-like phenotype with larger IMs and more floral organs (Venglat and Sawhney, 1996; Lindsay et al., 2006). Cytokinins suppress the expression of CLV1; this suppression results in upregulation of WUS expression (Brand et al., 2000; Schoof et al., 2000; Lindsay et al., 2006; Gordon et al., 2009). In addition, cytokinins directly induce WUS expression, independent of CLV1, and WUS enhances cytokinin signaling, forming a positive feedback loop (Leibfried et al., 2005; Gordon et al., 2009). Computational modeling shows that a combination of the negative feedback between WUS and CLV, and the positive feedback of WUS and cytokinin signaling determines the fine-scale positioning of the WUS-expressing stem cell niche domain (Gordon et al., 2009; Chickarmane et al., 2012).
CYTOKININS PROMOTE LATERAL INDETERMINACY IN DETERMINATE FMs
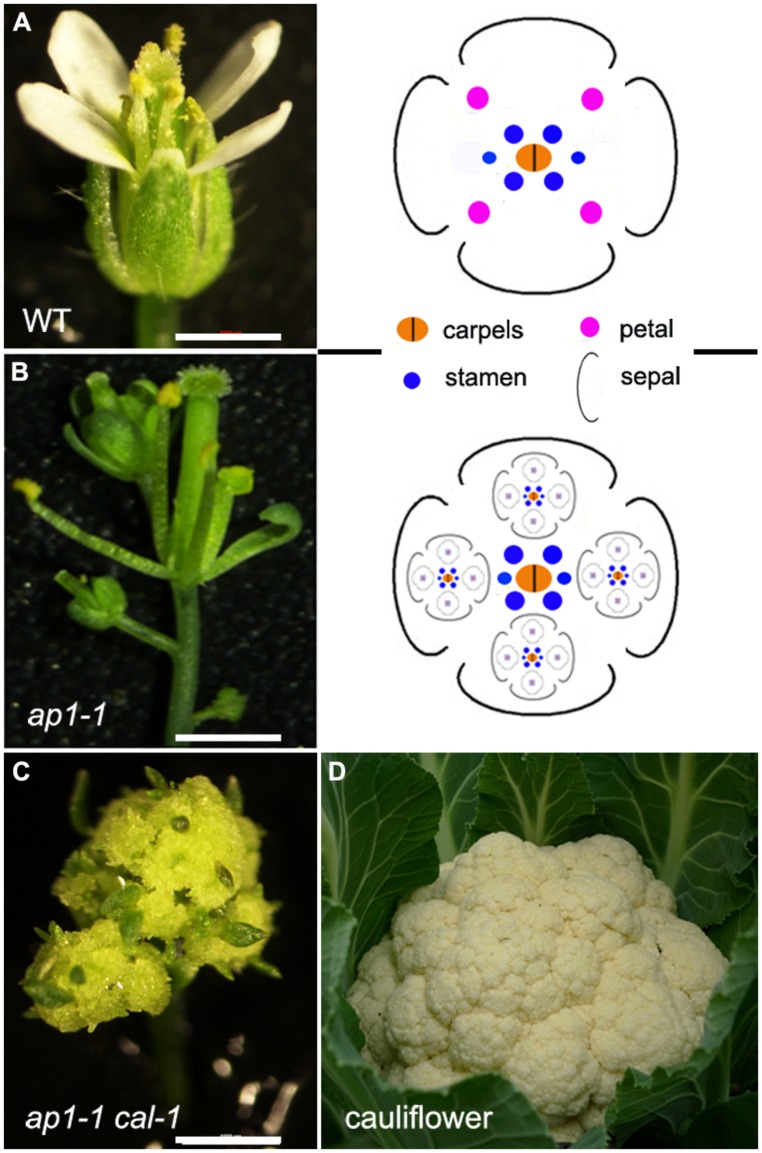
In indeterminate inflorescences, the periphery of the meristem produces BMs (and also SPMs and SMs for grasses) or FMs. In many determinate inflorescences, such as in wheat spikes, BM, SM, and FM can also initiate from the IM before its termination in a FM. In contrast to this initiation pattern, FM and BM can also initiate laterally from a terminal flower, either from the axil of a leaf-like organ (such as petals) or can initiate without subtending lateral organs. These types of inflorescence are termed dichasium and pleiochasium (Figure 1C), depending on the number of lateral branches, and can be considered a specialized cyme. Common examples include cauliflower and broccoli, which have a phenotype similar to that of the Arabidopsis apetala1-1 cauliflower-1 (ap1-1 cal-1) double mutants (Figure 2). In the ap1-1 single mutant (Figure 2B), secondary flowers laterally initiate from sepal axils and from the pedicel. The ap1-1 cal-1 double mutants have the same but more complicated inflorescence branching pattern. This lateral inflorescence branching mechanism has many similarities to vegetative stage lateral shoot branching. In contrast to vegetative shoot branching, inflorescences like the raceme and panicle develop iteratively, similar to frond development in ferns (Sanders et al., 2011). Despite these differences, cytokinins also regulate this type of lateral inflorescence branching.
FIGURE 2.
Conversion of a terminate flower into a pleiochasium-like inflorescence. Phenotype of Arabidopsis (A) Lansberg erecta wild-type, (B) ap1-1 (C) ap1-1 cal-1 flowers, and (D) a cauliflower with a highly elaborated inflorescence, similar to that seen in the Arabidopsis ap1-1 cal-1 mutant. Bars = 1 mm.
Lateral inflorescence branching is controlled by AP1 and related MADS-box transcription factor genes in Arabidopsis and other Brassicaceae species. In the simple indeterminate inflorescence of Arabidopsis, the IM gives rise to FMs and each FM differentiates into four whorls of floral organs that occupy precise positions (Figure 2A). In addition to promoting FM formation and outer floral whorl specification of sepals and petals, AP1 inhibits sepal axil meristem activity (Irish and Sussex, 1990; Mandel et al., 1992). In ap1 mutants, secondary flowers initiate in the axils of sepals, and tertiary flowers can initiate in the sepal axils of secondary flowers, and so on (Figure 2B; Irish and Sussex, 1990; Mandel et al., 1992). This forms a dichasium or pleiochasium-like inflorescence (Figure 1C). The inflorescence phenotype in ap1 is enhanced by cauliflower and fruitful mutants to form a cauliflower-like, highly elaborated pleiochasium inflorescence (Figure 2C; Ferrandiz et al., 2000). Indeed, cauliflower has lost a homolog of AP1 (Figure 2D; Kempin et al., 1995), suggesting that AP1 function is required to inhibit conversion of a simple raceme to a pleiochasium.
A recent study has shown that AP1 inhibits lateral inflorescence branching by reducing cytokinin levels. During vegetative stages, leaf axil axillary meristem formation requires cytokinin signaling (Wang et al., 2014) and during reproductive stages, lateral FM formation similarly requires cytokinin signaling (Han et al., 2014). The ap1 flowers have enhanced cytokinin signaling, as shown by examination of cytokinin-responsive reporter genes, and these flowers also have elevated levels of certain types of cytokinins. In addition, cytokinin treatment or ectopic expression of the cytokinin biosynthesis enzyme IPT8 in the AP1-expressing domain phenocopies the sepal axil secondary flower phenotype (Venglat and Sawhney, 1996; Han et al., 2014). This secondary flower phenotype can be rescued by mutations of cytokinin receptors. Further molecular dissection showed that AP1 suppresses the cytokinin biosynthetic gene LOG1 and activates the cytokinin degradation gene CKX3, through direct binding to the target gene promoters, thus reducing cytokinins levels in the outer whorls of developing flowers. Restoring the expression levels of either LOG1 or CKX3 can partially rescue the ap1 secondary flower phenotype. In addition to affecting cytokinin homeostasis, AP1 also directly downregulates a group of flowering time-related MADS-box genes, including SHORT VEGETATIVE PHASE (SVP), AGAMOUS-LIKE 24 (AGL24), and SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1), to suppress secondary FM formation. Similar to IPT8 overexpression, overexpression of SVP, AGL24, or SOC1 leads to sepal axil secondary FM formation (Liu et al., 2007). There appears to be crosstalk between cytokinin signaling and these flowering time-related MADS-box genes in the regulation of sepal axil secondary FM formation.
Taken together, the results described above show that cytokinins promote inflorescence complexity in different ways, by promoting meristem activity of IMs and BMs in inflorescences that branch iteratively, and by promoting indeterminate lateral meristem formation in inflorescences that branch laterally. Manipulating cytokinin levels directly or indirectly in crops is expected to change inflorescence complexity to increase yields (Kempin et al., 1995; Ashikari et al., 2005; Kurakawa et al., 2007; Zhang et al., 2012; Li et al., 2013).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to those authors whose work could not be cited due to space limitations. The authors’ laboratory is supported by the National Natural Science Foundation of China Grants 31222033 and 31430010, the Strategic Priority Research Program of CAS Grant XDA08020105, and funds from the State Key Laboratory of Plant Genomics (through Grant SKLPG2011B0103).
REFERENCES
- Ashikari M., Sakakibara H., Lin S., Yamamoto T., Takashi T., Nishimura A., et al. (2005). Cytokinin oxidase regulates rice grain production. Science 309 741–745 10.1126/science.1113373 [DOI] [PubMed] [Google Scholar]
- Benlloch R., Berbel A., Serrano-Mislata A., Madueno F. (2007). Floral initiation and inflorescence architecture: a comparative view. Ann. Bot. 100 659–676 10.1093/aob/mcm146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand U., Fletcher J. C., Hobe M., Meyerowitz E. M., Simon R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289 617–619 10.1126/science.289.5479.617 [DOI] [PubMed] [Google Scholar]
- Chickarmane V. S., Gordon S. P., Tarr P. T., Heisler M. G., Meyerowitz E. M. (2012). Cytokinin signaling as a positional cue for patterning the apical-basal axis of the growing Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. U.S.A. 109 4002–4007 10.1073/pnas.1200636109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandiz C., Gu Q., Martienssen R., Yanofsky M. F. (2000). Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127 725–734. [DOI] [PubMed] [Google Scholar]
- Gordon S. P., Chickarmane V. S., Ohno C., Meyerowitz E. M. (2009). Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. U.S.A. 106 16529–16534 10.1073/pnas.0908122106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Zhang C., Yang H., Jiao Y. (2014). Cytokinin pathway mediates APETALA1 function in the establishment of determinate floral meristems in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111 6840–6845 10.1073/pnas.1318532111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish V. F., Sussex I. M. (1990). Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 2 741–753 10.1105/tpc.2.8.741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg E. A., Camara P. E., Rudall P. J., Ladd P., Malcomber S. T., Whipple C. J., et al. (2013). Early inflorescence development in the grasses (Poaceae). Front. Plant Sci. 4:250 10.3389/fpls.2013.00250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempin S. A., Savidge B., Yanofsky M. F. (1995). Molecular basis of the cauliflower phenotype in Arabidopsis. Science 267 522–525 10.1126/science.7824951 [DOI] [PubMed] [Google Scholar]
- Kurakawa T., Ueda N., Maekawa M., Kobayashi K., Kojima M., Nagato Y., et al. (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445 652–655 10.1038/nature05504 [DOI] [PubMed] [Google Scholar]
- Kuroha T., Tokunaga H., Kojima M., Ueda N., Ishida T., Nagawa S., et al. (2009). Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 21 3152–3169 10.1105/tpc.109.068676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyozuka J. (2007). Control of shoot and root meristem function by cytokinin. Curr. Opin. Plant Biol. 10 442–446 10.1016/j.pbi.2007.08.010 [DOI] [PubMed] [Google Scholar]
- Kyozuka J., Tokunaga H., Yoshida A. (2014). Control of grass inflorescence form by the fine-tuning of meristem phase change. Curr. Opin. Plant Biol. 17 110–115 10.1016/j.pbi.2013.11.010 [DOI] [PubMed] [Google Scholar]
- Leibfried A., To J. P., Busch W., Stehling S., Kehle A., Demar M., et al. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438 1172–1175 10.1038/nature04270 [DOI] [PubMed] [Google Scholar]
- Li S., Zhao B., Yuan D., Duan M., Qian Q., Tang L., et al. (2013). Rice zinc finger protein DST enhances grain production through controlling Gn1a/OsCKX2 expression. Proc. Natl. Acad. Sci. U.S.A. 110 3167–3172 10.1073/pnas.1300359110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay D. L., Sawhney V. K., Bonham-Smith P. C. (2006). Cytokinin-induced changes in CLAVATA1 and WUSCHEL expression temporally coincide with altered floral development in Arabidopsis. Plant Sci. 170 1111–1117 10.1016/j.plantsci.2006.01.015 [DOI] [Google Scholar]
- Liu C., Zhou J., Bracha-Drori K., Yalovsky S., Ito T., Yu H. (2007). Specification of Arabidopsis floral meristem identity by repression of flowering time genes. Development 134 1901–1910 10.1242/dev.003103 [DOI] [PubMed] [Google Scholar]
- Mandel M. A., Gustafson-Brown C., Savidge B., Yanofsky M. F. (1992). Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360 273–277 10.1038/360273a0 [DOI] [PubMed] [Google Scholar]
- Miyawaki K., Tarkowski P., Matsumoto-Kitano M., Kato T., Sato S., Tarkowska D., et al. (2006). Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 103 16598–16603 10.1073/pnas.0603522103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok D. W., Mok M. C. (2001). Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 89–118 10.1146/annurev.arplant.52.1.89 [DOI] [PubMed] [Google Scholar]
- Nishimura C., Ohashi Y., Sato S., Kato T., Tabata S., Ueguchi C. (2004). Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16 1365–1377 10.1105/tpc.021477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perilli S., Moubayidin L., Sabatini S. (2010). The molecular basis of cytokinin function. Curr. Opin. Plant Biol. 13 21–26 10.1016/j.pbi.2009.09.018 [DOI] [PubMed] [Google Scholar]
- Prusinkiewicz P., Erasmus Y., Lane B., Harder L. D., Coen E. (2007). Evolution and development of inflorescence architectures. Science 316 1452–1456 10.1126/science.1140429 [DOI] [PubMed] [Google Scholar]
- Sanders H. L., Darrah P. R., Langdale J. A. (2011). Sector analysis and predictive modelling reveal iterative shoot-like development in fern fronds. Development 138 2925–2934 10.1242/dev.065888 [DOI] [PubMed] [Google Scholar]
- Schoof H., Lenhard M., Haecker A., Mayer K. F., Jurgens G., Laux T. (2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100 635–644 10.1016/S0092-8674(00)80700-X [DOI] [PubMed] [Google Scholar]
- Tanaka W., Pautler M., Jackson D., Hirano H. Y. (2013). Grass meristems II: inflorescence architecture, flower development and meristem fate. Plant Cell Physiol. 54 313–324 10.1093/pcp/pct016 [DOI] [PubMed] [Google Scholar]
- Teo Z. W., Song S., Wang Y. Q., Liu J., Yu H. (2014). New insights into the regulation of inflorescence architecture. Trends Plant Sci. 19 158–165 10.1016/j.tplants.2013.11.001 [DOI] [PubMed] [Google Scholar]
- Tokunaga H., Kojima M., Kuroha T., Ishida T., Sugimoto K., Kiba T., et al. (2012). Arabidopsis lonely guy (LOG) multiple mutants reveal a central role of the LOG-dependent pathway in cytokinin activation. Plant J. 69 355–365 10.1111/j.1365-313X.2011.04795.x [DOI] [PubMed] [Google Scholar]
- Venglat S. P., Sawhney V. K. (1996). Benzylaminopurine induces phenocopies of floral meristem and organ identity mutants in wild-type Arabidopsis plants. Planta 198 480–487 10.1007/BF00620066 [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang J., Shi B., Yu T., Qi J., Meyerowitz E. M., et al. (2014). The stem cell niche in leaf axils is established by auxin and cytokinin in Arabidopsis. Plant Cell 26 2055–2067 10.1105/tpc.114.123083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weberling F. (1989). Morphology of Flowers and Inflorescences. Cambridge: Cambridge University Press. [Google Scholar]
- Werner T., Motyka V., Laucou V., Smets R., Van Onckelen H., Schmulling T. (2003). Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15 2532–2550 10.1105/tpc.014928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T., Motyka V., Strnad M., Schmulling T. (2001). Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. U.S.A. 98 10487–10492 10.1073/pnas.171304098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T., Schmulling T. (2009). Cytokinin action in plant development. Curr. Opin. Plant Biol. 12 527–538 10.1016/j.pbi.2009.07.002 [DOI] [PubMed] [Google Scholar]
- Zhang L., Zhao Y. L., Gao L. F., Zhao G. Y., Zhou R. H., Zhang B. S., et al. (2012). TaCKX6-D1, the ortholog of rice OsCKX2, is associated with grain weight in hexaploid wheat. New Phytol. 195 574–584 10.1111/j.1469-8137.2012.04194.x [DOI] [PubMed] [Google Scholar]