Abstract
Cyclic di-GMP (c-di-GMP) has emerged as an important intracellular signaling molecule, controlling the transitions between planktonic (free-living) and sessile lifestyles, biofilm formation, and virulence in a wide variety of microorganisms. The following protocol describes the extraction and quantification of c-di-GMP from Pseudomonas aeruginosa samples. We have made every effort to keep the protocol as general as possible to enable the procedure to be applicable for the analysis of c-di-GMP levels in various bacterial species. However, some modifications may be required for the analysis of c-di-GMP levels in other bacterial species.
Materials and Reagents
I. For procedure I
1. Bacterial culture 2. Phosphate-buffered saline (PBS) (see Recipes) 3. Ethanol (95-100%) 4. Syringes (1 ml) 5. Syringe filters (2 μm; Upchurch Scientific, # B-100) 6. Tris base 7. 0.5 M Ethylenediaminetetraacetic acid (EDTA) disodium salt solution, pH 8.0 8. Microfuge tubes (unpolished, to reduce static)
II. For procedure II
1. Commercially-available bis-(3'-5')-cyclic diguanylic monophosphate (c-di-GMP, Biolog) 2. Ammonium acetate (MS grade) 3. Methanol (HPLC grade) 4. Nanopure water (18 Ohm) 5. Protein determination reagents 6. HPLC Solvent A (see Recipes) 7. HPLC Solvent B (see Recipes) 8. TE buffer (see Recipes)
Equipment
1. Spectrophotometer 2. Refrigerated microcentrifuge 3. Vacuum concentrator/Centrifugal evaporator (e.g.: SpeedVac) 4. Reverse-phase C18 Targa column (2.1 x 40 mm, 5 μm) (The Nest Group, # TR-0421-C185) 5. Heat block or water bath 6. High performance liquid chromatography (HPLC) system and software for HPLC peak analysis (e.g. Agilent 1100 HPLC) 7. Sonicator 8. Homogenizer
Procedure
I. Extraction of c-di-GMP
1. Grow bacterial cells to desired growth stage under required experimental conditions. Proceed directly with the extraction, with no waiting periods or incubation of cells on ice, as this may drastically alter the c-di-GMP levels.
Note: Extraction at mid-exponential phase is recommended, as using early exponential phase cells will yield c-di-GMP levels too low for accurate detection. To extract c-di-GMP from planktonic cells, allow P. aeruginosa to grow in either Lennox broth or Vogel-Bonner minimal medium (VBMM) for 6 hr to mid-exponential phase in flasks at 37°C and 220 rpm. Inoculate using a 5% inoculum size of an overnight culture. Biofilm c-di-GMP levels can be determined from biofilms grown for 3-5 days under flowing conditions; see references 2-4 for more detail.
2. Determine the optical density of the bacterial culture at 600 nm (OD600). Note: If working with biofilm samples, include a step to homogenize (10s on high) the cultures to disrupt cell aggregates prior to OD600 determination.
3. Obtain a bacterial culture volume equivalent to 1 ml of OD600 = 1.8 (e.g.: If the OD600 = 0.9, spin down 2 ml of culture).
Note: This biomass has been optimized for the analysis of c-di-GMP in P. aeruginosa strains PAO1 and PA14 planktonic and biofilm samples. Analysis of c-di-GMP levels in other strains or species may require the initial biomass harvested for extraction to be adjusted.
4. Centrifuge (16,000 x g, 2 min, 4 °C) the respective culture volume. Discard the supernatant.
5. Wash the cell pellet with 1 ml ice-cold PBS (16,000 x g, 2 min, 4 °C). Discard the supernatant.
6. Repeat Step I-5.
7. Resuspend the cell pellet in 100 μl ice-cold PBS and incubate at 100 °C for 5 min.
8. Add ice-cold ethanol (stored at -20 °C until use) to a final concentration of 65% (186 μl of 100% ethanol or 217 μl of 95% ethanol) and vortex for 15 s.
9. Centrifuge sample (16,000 x g, 2 min, 4 °C), and remove and retain the supernatant containing extracted c-di-GMP in a new microfuge tube. Store the supernatant on ice or at -80 °C until step I-10. Retain the cell pellet.
10. Using the cell pellet, repeat twice the extraction procedure in Steps I-7~I-9. Pool the supernatants obtained from the three extractions into one microfuge tube. Retain the cell pellet after the final extraction step. The cell pellet can be stored at -20 °C until step II-3-a.
11. Dry the combined supernatants using a vacuum concentrator/centrifugal evaporator. Following evaporation, a white pellet should be visible. This sample, containing the extracted c-di-GMP, can be stored at -80 °C until step II-2-a.
II. Quantification of c-di-GMP
This procedure has been optimized for the detection of c-di-GMP using an Agilent 1100 HPLC equipped with an autosampler, degasser, pressure regulator, prefilter, and UV/Vis detector set to 253 nm. Separation was carried out using a reverse-phase C18 Targa column (2.1 x 40 mm; 5 μm) and a flow rate of 0.2 ml/min. Solvents containing methanol and ammonium acetate (see recipes for solvents A and B) were used. The following gradient was used to elute c-di-GMP: 0 to 9 min, 1% B (=1% solvent B and 99% solvent A); 9 to 14 min, 15% B; 14 to 19 min, 25% B; 19 to 26 min, 90% B; 26 to 40 min, 1% B. This gradient resulted in the elution of c-di-GMP at approximately 14-15 min.
Note: Analysis of c-di-GMP levels using a different reverse-phase column, flow rate, or HPLC system may require the optimization of HPLC separation gradients.
1. Generation of a standard curve
a. Using commercially available c-di-GMP, prepare the following standards in nanopure water: 1, 2, 5, 10 and 20 pmol/μl
b. To generate the standard curve, inject 20 μl per standard per HPLC run. Use 20 μl of nanopure water as a negative control (0 pmol/μl c-di-GMP). For an example of c-di-GMP detection ( Figure 1).
Figure 1. Example of c-di-GMP standard elution profile.
20 μl of a 10 pmol/μl c-di-GMP standard (200 pmol total) were separated using a reverse-phase C18 Targa column (2.1 x 40 mm; 5 μm) at a flow rate of 0.2 ml/min with the following gradient: 0 to 9 min, 1% B; 9 to 14 min, 15% B; 14 to 19 min, 25% B; 19 to 26 min, 90% B; 26 to 40 min, 1% B. Peak at 15 min corresponds to the elution of c-di-GMP. The c-di-GMP peak was found to have an area of 935.7.
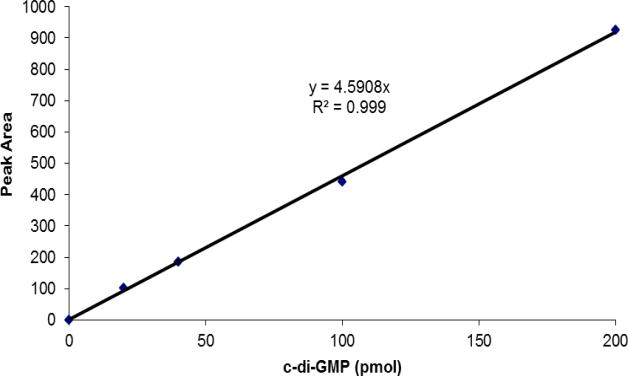
c. Prepare a standard curve by plotting the c-di-GMP amount in pmol (e.g.: 200 pmol for the 20 μl of the 10 pmol/μl standard) vs the peak areas. An example of a standard curve is given in Figure 2. Peak areas can be determined using various programs, including ChemStation for LC (Agilent Technologies), which was used here.
Figure 2. Example of a c-di-GMP HPLC standard curve.
The standard curve was generated by plotting the peak areas obtained following the separation of 20 μl aliquots of c-di-GMP standards (here: 0, 1, 2, 5, and 10 pmol/μl) versus the total c-di-GMP amounts in pmol. Peak areas were obtained using the ChemStation for LC software.
2. Analysis of samples
a. Resuspend the dried extracts from step I-11 in 200 μl nanopure water and vortex for 1min.
b. Centrifuge the solution at max speed (≥16,000 x g) to remove insoluble material.
c. Using a 2 μm HPLC syringe filter attached to a 1 ml syringe, filter the sample supernatants into a new microfuge tube.
Note: Small sample volume loss may occur, but will not interfere with downstream application, as only a limited sample volume (20 μl) is subjected to HPLC analysis.
d. Analyze 20 μl per sample using the HPLC program used for the standards above. Repeat using biological replicates.
Note: This procedure has been optimized for P. aeruginosa PAO1 and PA14. Analysis of cdi-GMP levels in other strains or species may require the adjustment of sample volumes.
e. Determine the peak area for each sample and determine the c-di-GMP amounts using the standard curve established with the commercially available c-di-GMP in Step II-1-c.
3. Calculations
To normalize the c-di-GMP levels, total cellular protein levels from the extraction procedure must be determined.
a. Resuspend the cell pellet from step I-10 in 500 µl TE buffer.
b. Sonicate the suspension for a total of 1 min using 10 s bursts at 5 W followed by 15 s off. Sonication should be carried out on ice.
c. Determine the protein concentrations using a protein determination assay.
d. Normalize the c-di-GMP levels to total cellular protein levels (i.e.: pmol/mg) using the following calculations:
• Total c-di-GMP in pmol
○ = (pmol c-di-GMP) × 10
○ A factor of 10 is used here, as only 1/10th of the c-di-GMP extract (20 μl out of 200 μl) was used for HPLC analysis
• Total protein in mg
○ = (mg/ml protein) * 0.5 ml
→Normalized c-di-GMP (pmol/mg)
○ = Total c-di-GMP/Total protein
Recipes
1. Phosphate Buffered Saline (PBS) 137 mM NaCl 2.7 mM KCl 4.3 mM Na2HPO4·7H2O 1.4 mM KH2PO4, pH 7.2
2. HPLC Solvent A 10 mM ammonium acetate in water Do not adjust pH
3. HPLC Solvent B 10 mM ammonium acetate in methanol (Dissolve ammonium acetate salt in methanol Do not adjust pH)
4. TE Buffer 10 mM Tris-HCl pH 8.0 1 mM EDTA
References
- 1.Amikam D, Steinberger O, Shkolnik T, Ben-Ishai Z. The novel cyclic dinucleotide 3'-5' cyclic diguanylic acid binds to p21ras and enhances DNA synthesis but not cell replication in the Molt 4 cell line. Biochem J. 1995;311(Pt 3):921–927. doi: 10.1042/bj3110921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morgan R, Kohn S, Hwang SH, Hassett DJ, Sauer K. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J Bacteriol. 2006;188(21):7335–7343. doi: 10.1128/JB.00599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petrova OE, Schurr JR, Schurr MJ, Sauer K. Microcolony formation by the opportunistic pathogen Pseudomonas aeruginosa requires pyruvate and pyruvate fermentation. Mol Microbiol. 2012 doi: 10.1111/mmi.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4*.Roy AB, Petrova OE, Sauer K. The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J Bacteriol. 2012;194(11):2904–2915. doi: 10.1128/JB.05346-11. [How to cite this protocol: please cite Reference 4.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simm R, Morr M, Kader A, Nimtz M, Romling U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol. 2004;53(4):1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]