Abstract
N-acetylglucosaminidases (GlcNAcases) play an important role in the remodeling and recycling of bacterial peptidoglycan. Inhibitors of bacterial GlcNAcases can serve as antibacterial agents and provide an opportunity for the development of new antibiotics. We report the synthesis of triazole derivatives of N-acetylglucosamine using a copper promoted azide-alkyne coupling reaction between 1-azido-N-acetylglucosamine and a small library of terminal alkynes prepared via the Ugi reaction. These compounds were evaluated for their ability to inhibit the growth of bacteria. Two compounds that show bacteriostatic activity against Bacillus were identified, with MIC values of approximately 60 μM in both cases. Bacillus subtilis cultured in the presence of sub-MIC amounts of the glycosyl triazole inhibitors exhibit an elongated phenotype characteristic of impaired cell division. This represents the first report of inhibitors of bacterial cell wall GlcNAcases that demonstrate inhibition of cell growth in whole cell assays.
The microbial glycome contains numerous attractive targets for antibiotic discovery.1 Peptidoglycan (PG), the mesh-like heteropolymer that surrounds all bacterial cells (with the exception of mycoplasma), confers strength, support, and shape to bacteria, as well as providing resistance to internal turgor pressure.2 Maintaining the integrity of PG is essential to bacterial viability, which is reflected by the number of different classes of clinically important antibiotics that target its biosynthesis. The PG sacculus is composed of the amino sugars N-acetylglucosamine (GlcNAc) and N-acetyl muramic acid (MurNAc), linked via a β-(1,4)-glycosidic linkage to form an alternating copolymer.3 Attached to the C-3 lactyl moiety of MurNAc is a highly variable pentapeptide composed of alternating L- and D- amino acids (Figure 1). Adjacent strands of the glycan copolymer are inter-connected through cross-links between these peptides. The biosynthesis of PG, particularly the cytoplasmic assembly and periplasmic cross-linking steps, is fairly well understood. Some of the most successful antibiotics to date, including the β-lactams and vancomycin, inhibit enzymes involved in PG biosynthesis. Conversely, remodeling of the invariant glycan backbone of PG by glycosidases and lyases is poorly understood despite the important roles of these enzymes in cell growth and division.1,4 Many current cell wall acting antibiotics, which act on the highly variable stem peptide, are plagued by resistance issues. Because of the invariant glycan backbone, enzymes that act on it are particularly attractive antibiotic targets.
Figure 1.
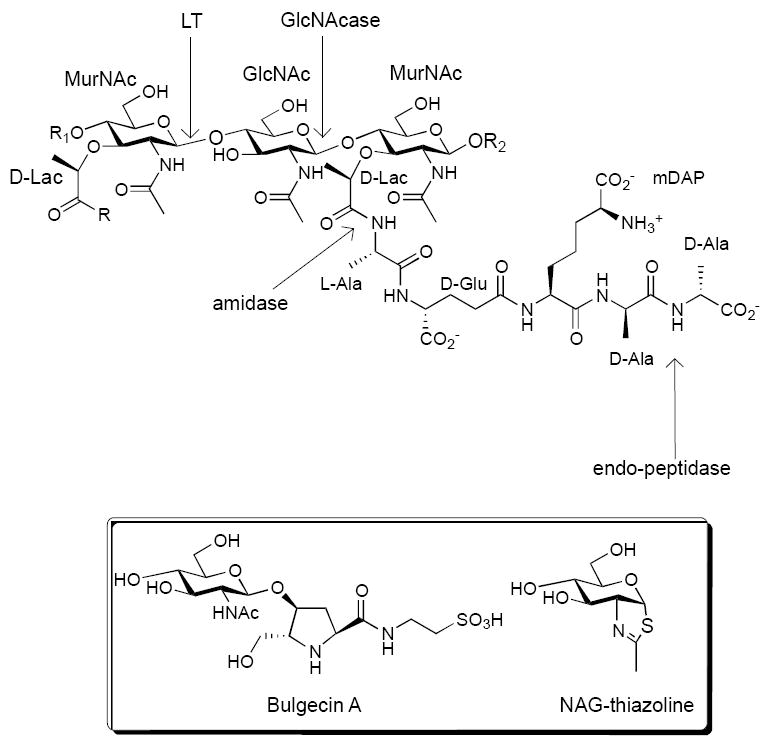
Repeat structure of peptidoglycan (A1-γ chemotype) and cleavage sites of major bacterial autolysins; Box - structures of Bulgecin A and NAG-thiazoline, inhibitors of cell wall glycosyl hydrolases.
Autolysins, also referred to as peptidoglycan hydrolases, are a family of enzymes that include glycosidases and peptidases and are important for cell wall remodeling (Figure 1).2,4,5 These enzymes play particularly important roles in cell division, motility and macromolecular assembly (e.g. pili). There are two classes of autolysins that act on the glycan backbone of PG - lytic transglycosylases (LT) and N-acetylglucosaminidases (GlcNAcase). GlcNAcases can be further categorized as endo-glycosidases, such as LytD from Bacillus subtilis, or exo-glycosidases like LytG.3,6,7 In Gram-positive bacteria, genetic and phenotypic analysis has identified an important role for GlcNAcases in vegetative cell growth and division.8,9 Additionally, bacteria possess a cytosolic exo-acting GlcNAcase (e.g. NagZ) that acts on a disaccharide glycopeptide intermediate generated during PG recycling.10,11 Attempts to inhibit cleavage of the PG glycan backbone have focused on LTs from Gram-negative bacteria, which can be inhibited by the substrate analog NAG thiazoline and the natural product bulgecin A (Figure 1).12-15 While these inhibitors show activity in vitro, they do not demonstrate a measurable minimum inhibitory concentration in whole cell assays. NAG thiazoline treatment of E. coli resulted in the formation of shorter cells without affecting cell viability.15 NAG thiazoline has also been shown to inhibit a number of Gram-positive enzymes that are involved in N-glycan modification.16,17 Treatment of Pseudomonas aeruginosa with a NagZ inhibitor attenuated β-lactam resistance, demonstrating the potential for modulating antibiotic activity by inhibiting PG recycling.18 An iminosugar based inhibitor of NagZ has recently been reported. 19 To date, no inhibitors targeting GlcNAcases that act on the cell wall have demonstrated antimicrobial activity in susceptibility tests. We have previously reported the synthesis of galactosyl and glucosyl triazoles as inhibitors for galactosidases and glucosidases.20 Numerous subsequent studies have demonstrated the generality of the glycosyl triazole pharmacophore for glycosidase inhibition.21-26 Based on this precedent, we sought to examine whether glycosyl triazole derivatives of GlcNAc would inhibit bacterial GlcNAcase activity. GlcNAc triazoles (GNTs) have previously been shown to inhibit O-GlcNAcase and have been examined as inhibitors for human hexosaminidase, but to the best of our knowledge have not been examined for anti-bacterial effects.27,28
Results
We prepared a 21-member GNT library by coupling 1-azido-N-acetylglucosamine (1) to a series of terminal alkynes that were prepared using a multicomponent Ugi reaction (Figure 2). Each Ugi reaction was carried out using either propargyl amine or propiolic acid as the alkyne component to provide either propionamides or N-propargyl amides as the diamide products. While Ugi-reactions can sometimes require prolonged reaction times, we identified conditions that reduced the reaction time to 4-5 hours at elevated temperature and in many cases proceeded to completion, enabling use of the Ugi product directly in the next reaction.29-31 The Ugi-derived terminal alkynes were then subjected to a copper accelerated azide-alkyne coupling reaction using a redox couple of copper powder and copper (II) sulfate. To further facilitate synthesis of the library we simplified the purification of the compounds by using the alkyne-building blocks in excess for the copper accelerated azide-alkyne coupling reaction. This ensured complete consumption of the azido sugar 1, and the unreacted alkyne residue could be easily removed using a silica plug after the reaction, eliminating the need for column chromatography. Removal of trace copper salts from the reaction was carried out by incubating the reaction solution with a commercially available copper chelating resin prior to purification using the silica plug.
Figure 2.
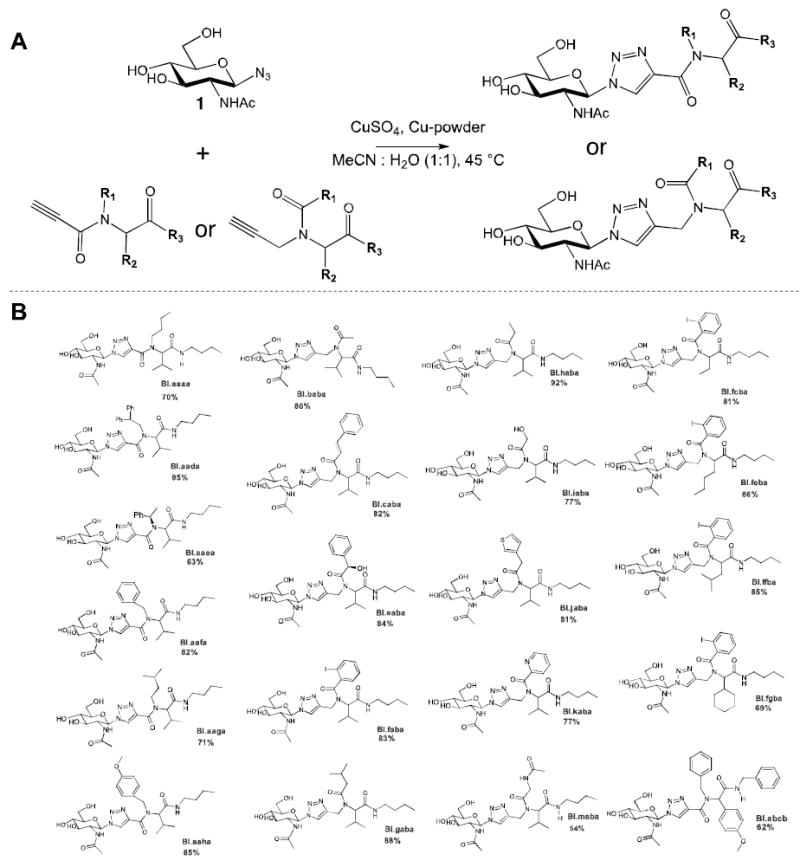
A) Synthetic route for preparation of of GNTs using copper accelerated azide-alkyne coupling; B) Structures of GNTs synthesized and evaluated for antibacterial activity. Yields provided correspond those for the azide-alkyne coupling after purification using a silica plug.
The library of GNTs was screened for its ability to inhibit the growth of a panel of different Gram-positive bacteria. Glycosyl triazole compounds were screened in a whole cell assay against a number of Gram positive organisms using a resazurin microtiter assay.32 The library was initially screened against all test organisms at a single concentration (250 μM) (Supplementary data). This high initial concentration was chosen as many soluble derivatives of PG bind to bacterial autolysins with KD values in the high micromolar range.33 Compounds showing at least 40% inhibition in growth were selected for further investigation. Follow up studies with 6 compounds that inhibited growth by more than 40% indicated that not all of these compounds reduced bacterial growth in a concentration dependent manner. We identified two compounds that inhibited the growth of Bacillus cereus and Bacillus subtilis in the micromolar range (Figure 3). GNT B1.abcb inhibited B. cereus with an MIC value of 39 μg/mL (60μM), and B1.fgba inhibited B. subtilis with an MIC value of 45 μg/mL (63 μM). Both B1.abcb and B1.fgba were screened in a standard bactericidal assay.34 Briefly, B. subtilis and B. cereus were grown in either the absence or presence of inhibitor (at the MIC) for 4 hours. The cells were harvested, washed and subject to serial dilution prior to plating on nutrient broth agar. Colony counts for growth in the presence of BI.fgba or BI.abcb (8.50 ± 3.00 × 107 cfu/mL and 7.00 ± 1.70 × 108 cfu/mL respectively) were similar to controls in the absence of the compounds (9.40 ± 2.88 × 107 cfu/mL for B. subtilis, and 2.67 ± 1.53 × 108 cfu/mL for B. cereus) indicating that these compounds are bacteriostatic in nature. In order to assess the role of the glycone and aglycone components in inhibition, the galactose derivatives of BI.fgba and BI.abcb were synthesized and tested, and exhibited no antimicrobial activity.
Figure 3.
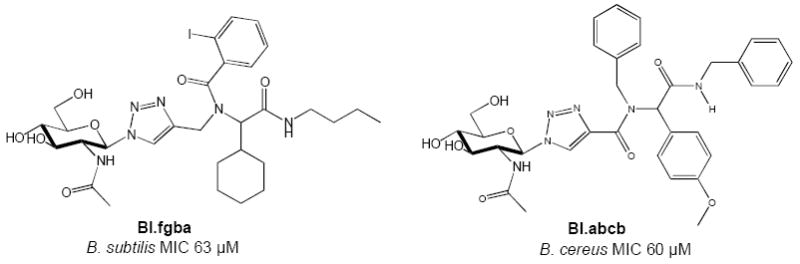
GNTs with antibacterial activity against Bacillus
Incubation of B. subtilis cells with the synthetic substrate β-p-nitrophenyl GlcNAc (pNP-GlcNAc) in the presence of B1.fgba resulted in a concentration dependent inhibition of nitrophenol release (Figure 4A), with complete inhibition observed at 250 μM inhibitor. This result confirms that the likely bacterial target is indeed a GlcNAcase. The differences in potency between the in vitro assay and the anti-bacterial assay may reflect poor access to the target in the whole cell assay. Additionally, it should be noted that pNP-GlcNAc is not the natural substrate for a bacterial GlcNAcase, and lacks many protein-substrate contacts such as the stem peptide as well as an extended glycan chain. Thus, the ability of BI.fgba to inhibit pNP-GlcNAc hydrolysis does not necessarily reflect its ability to inhibit peptidoglycan hydrolysis. Treatment of B. subtilis with B1.fgba at a concentration below its MIC (0.8 × MIC), resulted in a phenotype with highly elongated cells and the appearance of chains of cells (Figure 4B). This is suggestive of a disruption in the cell division/septation machinery, processes that are known to require GlcNAcase activity. This phenotype is reminiscent of that observed in Lactococcus lactis that lacks AcmA or AcmD, which are orthologs of LytG in B. subtilis and involved in cleavage of the septum during cell division.35,36
Figure 4.
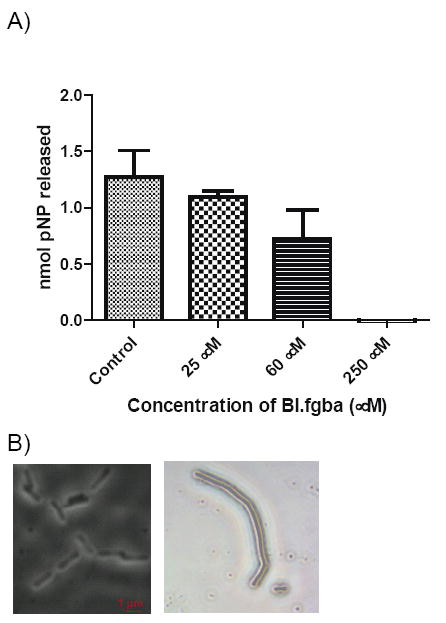
(A) Dose dependent inhibition of pNP-GlcNAc hydrolysis by intact B. subtilis cells in the presence of BI.fgba confirming that the target is a GlcNAcase. (B) Morphological changes to B. subtilis upon exposure to BI.fgba. Control cells (left) were grown in the presence of 1 % DMSO and show the typical rod shaped cells. Cells on the right were treated with 0.8 × MIC (48 μM) BI.fgba and exhibit an elongated phenotype.
In summary we have identified two inhibitors of bacterial GlcNAcases based on a glycosyl triazole scaffold. The bacteriostatic activity of both of these GNTs, in conjunction with biochemical evidence for inhibition for pNP-GlcNAc hydrolysis and the impaired cell division phenotype of cells treated with BI.fgba, is strongly suggestive of disrupted autolysin activity. There have been prior reports of compounds that inhibit purified bacterial GlcNAcases in vitro, as well as compounds that are known GlcNAcase inhibitors that exhibit antibacterial activity indirectly by sensitizing the bacteria to β-lactam antibiotics. However, to the best of our knowledge, this is the first report of compounds that exhibit inhibition of GlcNAcase activity that also directly reduce bacterial growth. While the MIC values exhibited by BI.fgba and BI.abcb are modest, our results demonstrate an important proof of concept and validate the glycosyl triazole scaffold as a viable one for further optimization and development. Our current efforts are directed at identifying the molecular target(s) of these compounds and improving the potency of the GNTs.
Supplementary Material
Acknowledgments
This research was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number 5 P20 GM103430-13.
Contributor Information
Amit Basu, Email: abasu@brown.edu.
Christopher Reid, Email: creid@bryant.edu.
References Cited
- 1.Reid CW, Fulton KM, Twine SM. Future Microbiol. 2010;5:267–288. doi: 10.2217/fmb.09.103. [DOI] [PubMed] [Google Scholar]
- 2.Scheurwater E, Reid CW, Clarke AJ. Int J Biochem Cell Biol. 2008;40:586–591. doi: 10.1016/j.biocel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 3.Ostash B, Walker S. Curr Opin Chem Biol. 2005;9:459–466. doi: 10.1016/j.cbpa.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 4.Uehara T, Bernhardt TG. Curr Opin Microbiol. 2011;14:698–703. doi: 10.1016/j.mib.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Heijenoort J. Microbiology and Molecular Biology Reviews. 2011;75:636–663. doi: 10.1128/MMBR.00022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rashid MH, Mori M, Sekiguchi J. Microbiology. 1995;141:2391–2404. doi: 10.1099/13500872-141-10-2391. [DOI] [PubMed] [Google Scholar]
- 7.Horsburgh GJ, Atrih A, Williamson MP, Foster SJ. Biochemistry. 2003;42:257–264. doi: 10.1021/bi020498c. [DOI] [PubMed] [Google Scholar]
- 8.Camiade E, Peltier J, Bourgeois I, Couture-Tosi E, Courtin P, Antunes A, Chapot-Chartier MP, Dupuy B, Pons JL. J Bacteriol. 2010;192:2373–2384. doi: 10.1128/JB.01546-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Layec S, Decaris B, Leblond-Bourget N. Res Microbiol. 2008;159:507–515. doi: 10.1016/j.resmic.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 10.Votsch W, Templin MF. J Biol Chem. 2000;275:39032–39038. doi: 10.1074/jbc.M004797200. [DOI] [PubMed] [Google Scholar]
- 11.Litzinger SS, Fischer SS, Polzer PP, Diederichs KK, Welte WW, Mayer CC. J Biol Chem. 2010;285:35675–35684. doi: 10.1074/jbc.M110.131037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonis M, Williams A, Guadagnini S, Werts C, Boneca IG. Microbial Drug Resistance. 2012;18:230–239. doi: 10.1089/mdr.2011.0231. [DOI] [PubMed] [Google Scholar]
- 13.Reid CW, Blackburn NT, Clarke AJ. FEMS Microbiology Letters. 2004;234:343–348. doi: 10.1016/j.femsle.2004.03.047. [DOI] [PubMed] [Google Scholar]
- 14.Shinagawa S, Maki M, Kintaka K, Imada A, Asai M. J Antibiot. 1985;38:17–23. doi: 10.7164/antibiotics.38.17. [DOI] [PubMed] [Google Scholar]
- 15.Reid CW, Blackburn NT, Legaree BA, Auzanneau FI, Clarke AJ. FEBS Lett. 2004;574:73–79. doi: 10.1016/j.febslet.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 16.Pluvinage B, Stubbs KA, Hattie M, Vocadlo DJ, Boraston AB. Org Biomol Chem. 2013;11:7907. doi: 10.1039/c3ob41579a. [DOI] [PubMed] [Google Scholar]
- 17.Abbott DW, Macauley MS, Vocadlo DJ, Boraston AB. J Biol Chem. 2009;284:11676–11689. doi: 10.1074/jbc.M809663200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asgarali A, Stubbs KA, Oliver A, Vocadlo DJ, Mark BL. Antimicrob Agents Chemother. 2009;53:2274–2282. doi: 10.1128/AAC.01617-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamaguchi T, Blázquez B, Hesek D, Lee M, Llarrull LI, Boggess B, Oliver AG, Fisher JF, Mobashery S. ACS Med Chem Lett. 2012;3:238–242. doi: 10.1021/ml2002746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossi L, Basu A. Bioorg Med Chem Lett. 2005;15:3596–3599. doi: 10.1016/j.bmcl.2005.05.081. [DOI] [PubMed] [Google Scholar]
- 21.Bokor É, Docsa T, Gergely P, Somsák L. Bioorg Med Chem. 2010;18:1171–1180. doi: 10.1016/j.bmc.2009.12.043. [DOI] [PubMed] [Google Scholar]
- 22.Carvalho I, Andrade P, Campo VL, Guedes PMM, Sesti-Costa R, Silva JS, Schenkman S, Dedola S, Hill L, Rejzek M, Nepogodiev SA, Field RA. Bioorg Med Chem. 2010;18:2412–2427. doi: 10.1016/j.bmc.2010.02.053. [DOI] [PubMed] [Google Scholar]
- 23.Dedola S, Hughes DL, Nepogodiev SA, Rejzek M, Field RA. Carbohyd Res. 2010;345:1123–1134. doi: 10.1016/j.carres.2010.03.041. [DOI] [PubMed] [Google Scholar]
- 24.Slã Movã K, Marhol P, Bezouška K, Lindkvist L, Hansen SG, Křen V, Jensen HH. Bioorg Med Chem Lett. 2010;20:4263–4265. doi: 10.1016/j.bmcl.2010.04.151. [DOI] [PubMed] [Google Scholar]
- 25.Weïwer M, Chen C-C, Kemp MM, Linhardt RJ. Eur J Org Chem. 2009;2009:2611–2620. doi: 10.1002/ejoc.200900117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilkinson BL, Long H, Sim E, Fairbanks AJ. Bioorg Med Chem Lett. 2008;18:6265–6267. doi: 10.1016/j.bmcl.2008.09.082. [DOI] [PubMed] [Google Scholar]
- 27.Li T, Guo L, Zhang Y, Wang J, Li Z, Lin L, Zhang Z, Li L, Lin J, Zhao W, Li J, Wang PG. Carbohyd Res. 2011;346:1083–1092. doi: 10.1016/j.carres.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Numa MMD, Liu H, Huang S-J, Sears P, Shikhman AR, Wong C-H. J Org Chem. 2004;69:6273–6283. doi: 10.1021/jo049355h. [DOI] [PubMed] [Google Scholar]
- 29.Akritopoulou-Zanze I, Gracias V, Djuric SW. Tetrahedron Lett. 2004;45:8439–8441. [Google Scholar]
- 30.Basso A, Banfi L, Guanti G, Riva R, Riu A. Tetrahedron Lett. 2004;45:6109–6111. [Google Scholar]
- 31.Linderman RJ, Binet S, Petrich SR. J Org Chem. 1999;64:336–337. [Google Scholar]
- 32.Palomino JC, Martin A, Camacho M, Guerra H, Swings J, Portaels F. Antimicrob Agents Chemother. 2002;46:2720–2722. doi: 10.1128/AAC.46.8.2720-2722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reid CW, Brewer D, Clarke AJ. Biochemistry. 2004;43:11275–11282. doi: 10.1021/bi049496d. [DOI] [PubMed] [Google Scholar]
- 34.Arnold RR, Brewer M, Gauthier JJ. Infect Immun. 1980;28:893–898. doi: 10.1128/iai.28.3.893-898.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Visweswaran GRR, Steen A, Leenhouts K, Szeliga M, Ruban B, Hesseling-Meinders A, Dijkstra BW, Kuipers OP, Kok J, Buist G. PLoS ONE. 2013;8:e72167. doi: 10.1371/journal.pone.0072167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steen A, Buist G, Horsburgh GJ, Venema G, Kuipers OP, Foster SJ, Kok J. FEBS Journal. 2005;272:2854–2868. doi: 10.1111/j.1742-4658.2005.04706.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


