Abstract
Animals display diverse colors and patterns that vary within and between species. Similar phenotypes appear in both closely related and widely divergent taxa. Pigment patterns thus provide an opportunity to explore how development is altered to produce differences in form and whether similar phenotypes share a common genetic basis. Understanding the development and evolution of pigment patterns requires knowledge of the cellular interactions and signaling pathways that produce those patterns. These complex traits provide unparalleled opportunities for integrating studies from ecology and behavior to molecular biology and biophysics.
Keywords: chromatophore, neural crest, pigment pattern, animal colour
1. Introduction
In vertebrates, pigmentation and coloration vary widely from black and white stripes of zebra to muted browns in house sparrows to bright colors and bold patterns of tropical fish. Closely related species may show highly divergent patterns, while distantly related species can appear strikingly similar. Understanding the genetic and developmental basis of variation in form is of central importance to developmental biology. Pigment patterns present an ideal system in which to study how developmental changes generate differences in form both within and between species.
Our current understanding of pigment pattern development owes a great deal to amateur “geneticists” of the 17th and 18th century, who cultivated unique or striking coat colors in “fancy” mice [1,2]. Upon the rediscovery of Mendel's work, these mice afforded opportunities to study modes of inheritance and genetic interaction. A century later, over 100 coat color loci have been identified in mouse and nearly half of these have now been cloned [3]. This work identified genes important for the specification, differentiation and morphogenesis of pigment cells as well as those involved in pigment synthesis. Despite this progress, little is known about the molecular or cellular processes that generate complex patterns such as stripes and spots, or how alterations in those processes might produce different phenotypes.
Vertebrate pigment cells are derived from the neural crest, a transient population of cells formed during embryogenesis that gives rise to a wide range of additional cell types including glia, neurons, bone and cartilage [4]. In birds and mammals, unpigmented pigment cell precursors known as melanoblasts migrate from the neural crest to the epidermis and into developing feather or hair follicles [5,6]. Mature pigment cells known as melanocytes synthesize melanin pigment, package it into melanosomes and then transfer those melanosomes to keratinocytes for deposition into developing feathers and hairs. Though mammals and birds have only one pigment cell type, the melanocyte, these cells can produce either eumelanin (black/brown) or pheomelanin (yellow/red) and can switch rapidly between the synthesis of these two pigment types [7]. In contrast to mammals and birds, poikilotherm vertebrates such as fish and amphibians have multiple pigment cell types known as chromatophores: black melanophores, yellow xanthophores, red erythrophores, iridescent iridophores, white leucophores and blue cyanophores [8,9,10]. Rather than transferring their pigments to other cell types, chromatophores retain their pigments intracellularly. Despite these differences, many of the genetic pathways critical for melanocyte differentiation and morphogenesis are conserved in melanophores and other chromatophore types [8,11].
In addition to their utility for studying genetic and cellular mechanisms, pigment patterns also provide ample opportunities to explore relationships between phenotype and environment. While the diverse patterns of domestic animals have mainly aesthetic value, the pigment patterns of wild animals are of great importance to their fitness. These patterns serve a variety of functions including camouflage and warning coloration and influence aspects or behavior such as mate recognition, mate choice, and shoaling preference [12,13,14,15]. Their clear ecological significance makes pigment patterns potential targets of both natural selection and sexual selection. Thus far, the genetic bases of adaptive traits have been identified only in a few cases [16,17,18,19]. Pigment patterns present many exciting opportunities to study the genetics and development of these important traits.
In birds and mammals, pigmentation can vary across the entire body or across individual hairs or feathers. By varying the type of melanin produced in different regions of the body a wide range of patterns may be achieved. Poikilotherms, likewise, can arrange their chromatophores to achieve broad swaths of uniform color or complex patterns. In the following sections we review how various types of colors are achieved in different vertebrates, and then discuss how colors are arranged to produce various patterns.
2. Colors
2.1. Black, Brown and White: Melanism/Albinism
One of the most common pigment polymorphisms found in vertebrates is melanism [20]. In mammals, increased production of eumelanin and a corresponding reduction in pheomelanin synthesis generates a melanic phenotype, which may be dark brown or entirely black. This switch is primarily controlled by the interaction of two genes: the melanocortin receptor 1 (Mc1r) and agouti. Mc1r is a seven transmembrane G protein-coupled receptor expressed by melanocytes and agouti is a small paracrine signaling molecule expressed by dermal papilla cells [21,22]. In the absence of agouti, α-MSH (α melanocyte-stimulating hormone) stimulates Mc1r to synthesize eumelanin. When agouti is present, it inhibits Mc1r activity, causing the melanocyte to synthesize pheomelanin instead of eumelanin. In many mammals, including mice, agouti is normally expressed during the mid-portion of hair growth resulting in hairs with a black (eumelanin) base and tip and a yellow (pheomelanin) band in the middle.
Alterations to these two proteins are the most frequent causes of melanism in mammals (Table 1). In mice, dominant Mc1r or recessive agouti alleles increase eumelanin synthesis generating a black coat. Deletions in agouti are associated with recessive melanic phenotypes in domestic cats [23], horses [24] and Japanese quail [25]. Dominant black phenotypes are associated with Mc1r mutations in many mammalian species including pocket mice [18], pigs [26], sheep [27], jaguars and juguarundis [23]. Dominant Mc1r mutations in birds also cause melanism, but the effects appear more variable than in mammals (bananaquits [28]; artic skuas and lesser snow geese [29]; chickens [30,31]). While an association between black coloration and Mc1r mutations is observed in many species, in only a few cases has functional work determined that these mutations alter Mc1r activity [27,32]. This type of demonstration is critical since mutations in genes other than Mc1r or agouti can also generate melanism. One notable example is the cause of dominant black in domestic dogs. Linkage analysis and molecular cloning revealed that black coat color in domestic dogs results from a mutation in the β-defensin gene, CBD103 [33,34]. CBD103 is highly expressed in dog skin and competitively inhibits agouti binding to Mc1r. Mice over-expressing CBD103 in an agouti background develop black coats, suggesting that CBD103 expression in skin could produce black coats in other mammals as well [34].
Table 1.
Mutations and polymorphisms affecting pigmentation and patterning.
D = Dorsal; V = Ventral; ( ) = allele
| Color or Pattern | Gene | Common name (Species) | Phenotype | Reference |
|---|---|---|---|---|
| Melanism | Mc1r | chicken (Gallus gallus) | black (E) | [30, 31] |
| sheep (Ovis aries) | black | [27] | ||
| pig (Sus sp) | black | [26] | ||
| bananaquit (Coereba flaveola) | black | [28] | ||
| jaguarundi (Herpailurus yaguarondi) | black | [23] | ||
| artic skua (Stercorarius parasiticus) | gray (intermediate), black | [29] | ||
| lesser snow geese (Anser c. caerulescens) | blue variant | [29] | ||
| jaguar (Panthera onca) | black | [23] | ||
| rock pocket mouse (Chaetodipus intermedius) | black | [18] | ||
| mouse (Mus musculus) | black (Eso) | [98] | ||
| Agouti | horse (Equus caballus) | black | [24] | |
| domestic cat (Felis catus) | black | [23] | ||
| mouse (Mus musculus) | black (a or ae) | [98] | ||
| Japanese quail (Coturnix japonica) | blackish-brown (rb) | [25] | ||
| B-defensin | domestic dog (Canis familiarus) | black | [33, 34] | |
|
| ||||
| Albinism | Tyrosinase | human (Homo sapiens) | albino | [38] |
| Oca2 | mexican tetra (Astyanax sp) | albino | [19] | |
| human (Homo sapiens) | albino | [38] | ||
|
| ||||
| Yellow/Red | Mc1r | Kermode Bear (Ursus americanus) | very pale yellow | [44] |
| pig (Sus sp) | red | [26] | ||
| horse (Equus caballus) | chesnut | [43] | ||
| cow (Bos taurus) | red | [42] | ||
| chicken (Gallus gallus) | uniformly red-yellow | [30] | ||
| Golden Retriever, Irish Setter, Yellow Labrador Retriever (Canis familiarus) | yellow or red coat | [40, 41] | ||
| mouse (Mus musculus) | yellow (e) | [98] | ||
| beach mouse (Peromyscus polionotus) | lighter pigmentation | [45] | ||
| Agouti | Japanese quail (Coturnix japonica) | wheat-straw yellow plumage (Y) | [39] | |
| mouse (Mus musculus) | yellow (Ay or Avy) | [39] | ||
|
| ||||
| Light Ventrum | Agouti | mouse (Mus musculus) | agouti hairs D and V (A) | [98] |
| agouti hairs D, yellow hairs V (Aw) | ||||
| black hairs D, yellow hairs V (at) | ||||
| Steel | three-spine stickleback (Gasterosteus aculeatus) | light ventral skin and gills | [81] | |
|
| ||||
| Stripe/Bar | Csf1r | zebrafish (Danio rerio) | no xanthophores, fewer melanophores | [90] |
| Mitfa | zebrafish (Danio rerio) | no melanophores | [91] | |
|
| ||||
| Spotting/Dominant White | Kit | horse (Equus caballus) | Sabino1 spotting, white coat | [112] |
| white spotting, dominant white | [113] | |||
| pig (Sus sp) | spots, dominant white | [109, 110] | ||
| mouse (Mus musculus) | white forehead, belly spots, dominant white (W, Wv) | [100, 101] | ||
| zebrafish (Danio rerio) | reduced melanophores in stripes | [116] | ||
| Ednrb | horse (Equus caballus) | frame overo white markings, lethal white foal syndrome | [114, 115] | |
| mouse (Mus musculus) | white spots, fully white coat (s, sl) | [99] | ||
| zebrafish (Danio rerio) | reduced melanophores, spots | [117] | ||
In poikilotherms, α-MSH and MCH (melanin-concentrating hormone) have different effects depending on the duration of their signals. Short-term expression causes melanosomes to disperse (making the animal appear darker) or to aggregate (making the animal appear lighter) respectively, a set of processes known as physiological color change [35] (see “Green” section below). Longer-term expression of α-MSH and MCH causes alteration to the number of melanophores, known as morphological color change [36]. These alterations to melanization occur through some of the same pathways as mammalian melanism, but the mechanism in poikilotherms is reversible and does not involve genetic alteration. For example, darkening that occurs during adaptation to a black background in Mozambique tilapia is not associated with a change in the sequence or expression level of Mc1r[37].
Albinism is characterized by reduced melanin in the skin, hair and eyes. Hair follicles of albino mice contain melanocytes, but do not produce melanin. Like melanism, albinism has evolved repeatedly in widely divergent taxa, often through mutations in the same set of genes (Table 1). In humans, albinism is associated with mutations in several genes including Tyrosinase and Tyrosinase related protein-1, two enzymes required for melanin synthesis [38]. Mutations in ocular and cutaneous albinism 2 (Oca2) are another frequent cause of albinism in humans. Cave dwelling populations of Mexican tetra evolved albinism independently multiple times. At least two of these independent populations contain genomic alterations in Oca2 that abrogate the protein's function in cell-culture assays [19].
2.2. Yellow-Orange-Red
Like black phenotypes, changes in a variety of genes can cause yellow, orange, or red coloration in vertebrates (Table 1), but unlike black, these colors can be produced by a wide range of pigments. In mammals and birds, loss of function mutations in Mc1r or dominant mutations in agouti cause increased pheomelanin synthesis, generating a yellow or red coat or feathers. Constitutive activation of agouti in mice and Japanese quail produce pheomelanic phenotypes [39]. Mutations in Mc1r are associated with yellow or red coats in several breeds of domestic dog [40,41], pigs [26], cows [42], horses [43], chickens [30] and Kermode bears [44]. A point mutation in Mc1r causing reduced α-MSH binding affinity is partially responsible for the light coloration of some populations of beach mice [45]. The differences in pigmentation between light beach mice and dark inland mice are quantitative differences and are produced by the differential activity of at least two major genes [46]. These results highlight the difficulties in determining the genes responsible for phenotypic variation between species, since in many cases changes in the activity of multiple genes contribute to the difference in phenotype.
While yellowish or rufous feathers in birds are often colored by some combination of pheomelanin and eumelanin [47], many striking yellow, orange and red feather colors are due instead to the presence of carotenoid pigments [48]. Animals cannot synthesize carotenoid pigments the way they synthesize melanin or other pigments, and therefore must obtain carotenoids from a dietary source. Carotenoids can be metabolized to alter the colors they produce (e.g., [49]) and in some cases, animals preferentially utilize certain dietary pigments over others (e.g., [50]).
In poikilotherms, colors in the yellow-to-red range are produced by xanthophores and erythrophores. Generally, xanthophores are yellow and erythrophores are red pigment-containing cells, although, both colors can be produced by a range of pteridine and carotenoid pigments and both xanthophores and erythrophores can contain both pigment types [48,51]. Xanthophore color can even vary between two closely-related species, for example they appear yellow in zebrafish Danio rerio and orange in its close relative, the pearl danio, D. albolineatus [52]. Differences between xanthophores and erythrophores beyond their color have been difficult to identify in part because models of xanthophore study, such as zebrafish and the medaka Oryzias latipes, both lack erythrophores. Within the Danio clade, the phylogenetic distribution of species with erythrophores suggests this cell type may have arisen or been lost independently multiple times [52,53].
Red coloration is less commonly generated in other ways, such as hemoglobin visible through the skin (e.g., [54]) and other rare pigments (see [55] for review). One additional red pigment type has been discovered: pterorhodin, a dark-red pteridine dimer, contained strangely enough in melanophores! This pigment was identified in only two groups of frogs, the phyllomedusids from the New World and the hylids from Australia [56,57]. Its presence in melanophores of these frogs, located within curiously large melanosomes, raises a number of questions about the evolutionary origin of pigment cells, the evolution of pigment within the cells, and the evolution of the frog groups in question [57] – all of which remain to be answered.
2.3. Blue and other Structural Colors
With the exception of the extremely rare cyanophore [10], blues observed in vertebrates are not due to the presence of pigment. Instead, they are structural colors, caused by the reflection of short-wavelength light off nanostructures in the animal's skin or feathers [58]. Blue-greens, ultraviolet-blues, iridescent colors, and some whites also fall in this category.
In birds and mammals, blue-colored skin is caused by thick arrays of collagen fibers in the dermis [59,60]. These fibers are “quasi-ordered”: closely but not perfectly aligned with one another. The precise color produced is dependent on the diameter of fibers in the array, with smaller-diameter fibers reflecting shorter-wavelength light. The colors produced can therefore range from ultraviolet to (rarely) yellow or orange, although blue is most common. The collagen array is often underlain by melanin, which blocks reflective scatter from tissue beneath the array and absorbs wavelengths of light other than those reflected by the collagen. These color-producing collagen arrays have evolved repeatedly in both mammals and birds [59,60], but neither their development nor their evolution have been studied.
In birds' feathers, a matrix of air bubbles in keratin known as the “spongy layer” in the barbs produces non-iridescent colors dependent on the scale of the quasi-ordered matrix, similar to the scale-dependence of structural skin color [61]. Without underlying melanin, the scatter produced by this matrix appears pale [62], a condition that in some species has been exaggerated in the evolution of bright white feathers from structurally pigmented ones [61]. Iridescence, the color of which changes depending on the angle from which it is viewed, is produced by myriad patterns of alternating layers of keratin, air and melanin in barbules (subdivisions off the barbs) ([63] and references therein). This is the most common type of structural color in birds [61], and it may be particularly important as a source of UV reflectance.
Blue coloration in fish, amphibians and reptiles is due primarily to the presence of iridophores underlain by melanophores [58,64]. Iridophores and leucophores are both described as reflective or shiny pigment cells, with iridophores giving an iridescent shine and leucophores appearing white [10,51] or cream [65]. The appearance of both cell types is due to the presence of guanine and hypoxanthine crystals or platelets within the cells: longer, more horizontally arranged platelets in the iridophores; shorter, more vertically arranged platelets in leucophores [51]. These distinctions are certainly not always used: zebrafish have been occasionally reported to possess two types of iridophores, either gold and silver [66] or ‘S’ and ‘L’ for the short and long platelets in they contain [67,68], and these types may correspond to what other authors describe as iridophores and leucophores. White chromatophores have also been reported in the fins of zebrafish [66] and these may be leucophores [65]. Medaka have both iridophores and leucophores, and the broad array of mutants in which only one cell type or the other is affected offer an exciting opportunity to study the biology of these still-mysterious chromatophores [65].
2.4. Green and physiological color change
Combinations of structural blue color and pteridines or carotenoids can produce green skin or feathers [61,64,69] or purple feathers [48]. In a number of amphibians and reptiles, greens produced in this way can change to other colors like brown in anolis lizards or tree frogs [58,70]. Transitions between these colors are made possible by a three-dimensional arrangement of chromatophores known as a dermal chromatophore unit [71]. In amphibians and reptiles that undergo these significant physiological color changes, the dermal chromatophore unit is stereotypically made up of a single representative each (from top to bottom in the skin) of xanthophore, iridophore, and melanophore. Shifts between colors are caused by dispersion and aggregation of the chromatosomes in each layer of cells such that different wavelengths of light are absorbed or reflected. Changes in chameleon color and patterning are similarly caused by shifts in different layers of chromatophores.
While only a few fish undergo such striking physiological color changes (e.g. [72]), many exhibit lightening and darkening associated with changes in environmental conditions and stress. As mentioned previously, physiological color change in teleosts is controlled in part by the hormone pair MSH and MCH [10,35]. The mechanisms of MSH and MCH signaling in melanosome movement appear to be similar to the melanin stimulating roles played by the corresponding molecules in mammals. Physiological color change in amphibians is slightly different than in teleosts. At high levels, MSH causes melanosome dispersion as in teleosts, but low levels of MSH alone (rather than correspondingly high levels of MCH) are sufficient to drive melanosome aggregation [35]. In addition to responding to these hormones and others, chromatophores are innervated, allowing more rapid response to stimuli than would be possible through hormone signaling alone, and they are even able to respond to light stimulus directly (for review, see [10]).
3. Patterns
Changing the type of pigment produced by melanocytes can lead to dramatic differences in coloration over the entire body of an animal, but many of the most striking pigment patterns found in nature are the result of regional differences in pigmentation, such as splotches of orange on a guppy, the stripes of a zebra, or the white head and tail of a bald eagle. Most research on pigment patterning to date has focused on pigmentation rather than on patterning: how various genes affect the number of pigment cells, or their migration, but not specifically on how the complex patterns are established or maintained. In this section, we cover a few well-studied patterns (Fig. 1, Table 1) and highlight examples of patterns that may be useful for future study (Fig. 2).
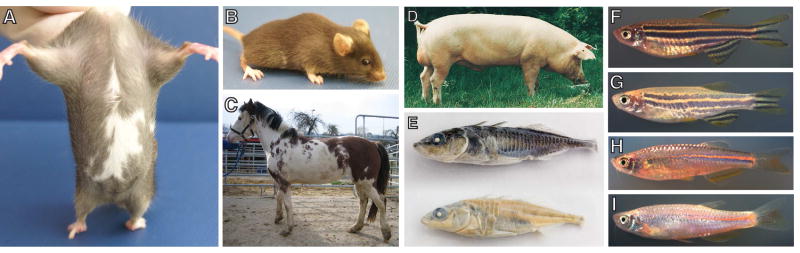
Figure 1.
Pigment patterns caused by KIT pathway mutations.
A, B. Heterozygous kit mice are mostly brown with irregular belly and sometimes forehead spots.
C. Franches-Montagnes horse with p.Y717X mutation in KIT
D. Dominant white Swedish Landrace pig
E. Marine (above) and freshwater (below) threespine sticklebacks (Gasterosteus aculeatus) with higher and lower levels of kit ligand expression
F, G: Wild-type and homozygous kit-mutant Danio rerio
H, I: Wild-type and homozygous kit-mutant Danio albolineatus
Photo credits: A, B: A Incao; C: B Haase; D: L Andersson; E: F Chan, C Miller, D Kingsley; F, G: LBP; H, I: MGM.
Figure 2.
Examples of interesting pigment patterns for which the genetic and developmental bases remain unknown.
A. Tabby stripes on a domestic shorthair cat
B, C. Cryptic dorsum and brilliant ventrum of a male western fence lizard, Sceloporus occidentalis
D. Brilliant coloration and patterning in a poison dart frog, Oophaga pumilio
E. Spots on Danio kyathit, a close relative of the zebrafish, Danio rerio
E. Irregular orange and black spots on a male guppy, Poecilia reticulata
F. Stripe-like patches on a salamander, Ambystoma tigrinum
Photo credits: A: DM Parichy; B, C: A Leache; D: D Gonzalez; E: LBP; F: A Price; F: HB Shaffer.
3.1. Dorsal/Ventral Patterning
A contrasting dark dorsum and light ventrum is one of the most common pigment patterns found among vertebrates. This arrangement is a classic form of crypsis: for land animals, the “countershading” keeps overhead illumination from casting dark shadows on their undersides (e.g. [73]); for fish, a dark dorsum blends with deeper water, while a light ventrum blends with the light surface. In mice, this marked difference in dorsal-ventral pigmentation is caused by variation in the activity of two distinct agouti transcripts [21,22]. A “hair-cycle specific” transcript is expressed throughout the body during the mid-portion of hair growth [22,74]. This transcript is required for the characteristic pheomelanin band of agouti hairs. A second “ventral-specific” transcript is expressed throughout the hair cycle, but only on the ventral side of the body [74]. Mice expressing both “hair-cycle specific” and “ventral-specific” agouti transcripts develop a dorsum with agouti-banded hairs and a yellow or cream belly. Expression of “ventral-specific” agouti begins during mammalian embryogenesis suggesting the genes involved in the establishment and/or maintenance of ventral identity regulate expression of this transcript [22]. Analysis of the mouse droopy ear (deH) mutant provides some insight into the mechanisms that might regulate agouti. droopy ear mutants show a shift in the dorsal-ventral pigmentation boundary, resulting from dorsal expansion of “ventral-specific” agouti expression [75]. Cloning of droopy ear revealed a large deletion in Tbx15, a T-box transcription factor that cues the establishment of dorsal dermis [75]. The effects of Tbx15 on pigment patterning, however, are likely indirect, resulting from alterations in positional identity of the dermis rather than through direct regulation of agouti transcripts.
The combination of a dark dorsum and light ventrum is also common in poikilotherms, and it appears the mechanisms that generate this pattern are similar to those in mammals. In amphibians, a ventral specific factor, melanization inhibiting factor (MIF), represses differentiation and melanization of melanocytes in vitro [76,77]. MIF is highly expressed in unmelanized ventral skin and not only suppresses melanization, but also appears to promote iridophore localization in this region [78,79]. Similar to MIF, an agouti homolog expressed mainly in the golfish ventrum blocks melanization in cell culture [80]. Whether MIF is an amphibian homolog of agouti or a distinct molecule with similar function remains unknown. Regardless of MIF's identity, the expression of a ventral-specific inhibitor of melanization seems like a common mechanism responsible for generating a light ventral side in poikilotherms.
Additionally, other genes or pathways may determine how far pigmentation extends ventrally. Recently, Miller and colleagues showed that differences in gill and ventral skin pigmentation between marine and freshwater three-spine stickleback result from differences in kit-ligand expression [81]. In mice and zebrafish, kit-ligand (also known as Steel Factor, Mast Cell Growth Factor) is required for melanocyte migration and survival (see Spots section below) [82]. kit-ligand expression is reduced in freshwater three-spine stickleback populations with lighter gill and ventral skin pigmentation [81]. It will be interesting to see if reduced kit-ligand expression is always found in lightly pigmented populations of three-spine stickleback and to determine whether changes in kit-ligand expression are responsible for naturally-occurring variation in pigmentation patterns in other vertebrates.
3.2. Stripes and Bars
Stripes may be repeated across the body, like those of a tiger, or found only in discrete regions, like the rings on a raccoon tail. Tabby cats display one of the most familiar mammalian stripe patterns. Tabby stripes are visible only when functional agouti is expressed and result from agouti banded hairs (light stripes) alternating with completely eumelanic hairs (dark stripes) [7]. How the tabby locus interacts with agouti to achieve this pattern is unknown, but its identification will no doubt shed light on the development of striped patterns in other mammalian species. Thus far, identifying the genes responsible for mammalian stripe formation has been difficult because only one mutation has been found to cause stripes in lab mice [83]. Despite the diversity of colors produced by birds, feather patterning resulting in bars, stripes or spots is controlled exclusively by melanin production [47]. Studies in Japanese quail suggest that local cues in the feather papillae act on melanocyte precursors to control their differentiation [84,85]. Differences in the timing or distribution of these differentiation cues could lead to differences in pigment patterns among different species of birds.
Though bird and mammalian models of stripe formation are lacking, good fish and amphibian models exist: adult zebrafish and larval salamanders (Ambystoma tigrinum tigrinum and others) develop horizontal stripes of melanophores separated by interstripes of xanthophores. Microsurgical experiments on salamanders demonstrated that chromatophore migration is influenced by the tissue over which they travel, and that the lateral line in particular is both directly and indirectly responsible for the melanophore-free interstripe region [86,87], although this is not true for all salamander species ([86,88] and references therein). Signals from the underlying tissue affect chromatophore location in embryonic zebrafish [89], but so far a role for such signals has not been demonstrated in adult zebrafish. Studies of stripe formation in adult zebrafish have instead focused on interactions between chromatophores themselves. In mutants lacking either all xanthophores (csf1r [90]) or all melanophores (nacre [91]), the remaining chromatophores fail to organize into stripes. When the missing cell type is added by cell transplantation, stripe formation is restored in the vicinity of the donor cells [92,93].
A recent in silico study modeled pattern formation in zebrafish by varying attractive or repulsive forces of both homotypic (melanophore-melanophore or xanthophore-xanthophore) and heterotypic (melanophore-xanthophore) interactions to examine the conditions that might generate patterns like those observed in wild-type or mutant D. rerio [94]. While previous descriptions of melanophore and xanthophore behaviors during teleost and amphibian pattern formation implied a repulsive action of xanthophores on melanophores [87,89], this model suggests that stripe formation requires both heterotypic attraction and homotypic repulsion. There are difficulties with applying these model conditions directly to stripe formation in D. rerio, but the suggestion that homotypic repulsive interactions may be involved in the formation of stripes is one worth investigating.
Experiments with temperature-sensitive alleles of D. rerio csf1r also suggest that the timing of stripe-formation signals may affect stripe orientation. When fish raised at restrictive temperatures are shifted to permissive temperature later in development, melanophores organize into stripes on the flank and fins, but in the caudal and anal fins, the orientation of these stripes is seemingly random [92]. Several danios and salamanders exhibit vertical stripes or bars, but whether these bars result from differences in the timing of the same stripe forming signals or from different signals remains unclear [95,96]. East African chiclid species present an opportunity to explore differences in stripe forming mechanisms between vertical bars and horizontal stripes. For several decades, two species of cichlids have been distinguished by the orientation of two dark facial stripes: Neolamprologus pulcher has two curved vertical bars on the operculum, while N. brichardi exhibits one vertical bar on the operculum and one horizontal stripe connecting the eye to the operculum bar. A recent phylogenetic study revealed that these species are not monophyletic groups, and that N. brichardi T-shaped markings evolved repeatedly from the ancestral N. pulcher bar pattern [97]. These phenotypes provide an excellent opportunity to investigate the genetic basis of stripe/bar orientation, and further to determine whether the same mechanism has been utilized repeatedly to reorient the anterior stripe in the “brichardi” phenotype.
3.3 Spots
Spots can be regular and repeated, like those found on a cheetah, or broad and irregular, like the black and white pattern of dairy cows. Although regular spotting patterns are not found not in lab mice, several mouse mutants do show irregular white spotting patterns [98]. These mutants develop patches of white hair and skin that are completely devoid of melanocytes. Two white spotting mutants, piebald-lethal and Dominant spotting result from mutations in the seven transmembrane G-protein coupled receptor Ednrb and in the receptor tyrosine kinase Kit, respectively [99,100,101]. piebald-lethal homozygotes are almost completely white and develop megacolon due to defects of the enteric ganglia [99]. These mice lack most melanocyte precursors, suggesting that early Endrb activity is required for the development of these cells [102]. Melanocyte precursors also require Ednrb for dorsolateral migration from the neural crest [103]. Dominant spotting mutants develop completely white coats and are sterile, anemic, and lack mature mast cells. Mice heterozygous at this loci display diluted coats with characteristic white spots on the forehead and belly [98,104]. Kit signaling is required for melanocyte precursor migration and survival and later is required for their entry into developing hair follicles [105,106,107,108].
The pleiotropic effects of Kit and Ednrb make mutations in the coding regions of these genes unlikely candidates for spotting patterns in wild populations, but studies in domestic species reveal several instances in which spotted or completely white patterns are associated with Kit or Ednrb mutations. In domestic pigs, white coat color is dominantly inherited and is caused by the combined effects of two mutations in Kit: a duplication of the coding region of Kit including some, but not all of the regulatory regions; and a splice mutation in one copy of Kit resulting in skipping of exon 17 [109,110,111]. These mutations are not associated with defects in fertility or viability, though a reduction in white blood cells suggests a mild effect on hematompoesis [110]. Pigs with an extra copy of Kit, but no splice mutation develop a patched pattern of white spots interspersed with colored spots [109,110].
Spotting patterns in horses ranging from small regions of the body to nearly complete depigmentation and are often associated with Kit mutations. Exon skipping in Kit is associated with a Sabino spotting pattern in horses, resulting in irregular white patches on the face and feet in heterozygotes and a completely white coat in homozygotes [112]. This mutation does not completely eliminate wildtype Kit transcript, possibly explaining the lack of reported health defects in these horses. Distinct mutations in Kit coding sequence are also associated with depigmentation and dominant white coat color in Franches-Montagnes, Camarillo White Horse and Arabian horse breeds [113]. Additionally, frame overo white markings in American Paint Horses are associated with a dominant mutation in Ednrb [114,115]. Similar to mice, horses homozygous for this allele are completely white, but die shortly after birth due to an absence of enteric ganglia.
The roles of kit and ednrb are conserved in zebrafish melanophore development, but show some intriguing differences compared to mammals. Whereas mouse Kit and Ednrb mutants lack nearly all melanocytes, zebrafish, kit and ednrb1 mutants each develop roughly half the normal number of adult melanophores [116,117]. Double mutants lack all adult body melanophores, demonstrating that kit and ednrb are required by genetically distinct subsets of adult melanophores [66]. Interestingly, several species of Danio develop spotted patterns similar to zebrafish ednrb1 mutants. These species also develop fewer melanophores than D. rerio, and fail to complement zebrafish ednrb1 mutants, making changes in ednrb1 activity a good candidate for species differences in pigment patterning [118] (LBP and DM Parichy, unpublished data).
While Ednrb and Kit mutants demonstrate how differences in melanoblast migration and differentiation can effect pigment pattern formation, interactions among pigment cell types are also likely to play a large role in spotted pattern development. Several zebrafish mutants may provide insight into the types of cell interactions that form spots. Like ednrb or kit mutants, leopard mutants fail to develop a subset of adult melanophores [66]. Pigment patterns of various leopard alleles range from broken stripes to spots to widely dispersed melanophores and are caused by mutations in connexin41.8, a gap junction component [119]. Cell transplants suggest that connexin41.8 promotes both homotypic attractions among melanophores and among xanthophores, as well as repulsive and attractive heterotypic interaction between melanophores and xanthophores [93].
Yellow spots on the anal fins of male haplochromine cichlids act as egg-dummies, and are an important part of the mating system of many species. Salzburger and collaborators report that csf1ra is expressed in sites of xanthophore spot formation on the anal fin of a wide variety of species, but that in one basal riverine haplochromine csf1ra RNA is expressed in pearly spots on the dorsal fin but not in the yellow tissue surrounding the spots [120]. This result indicates that we still have much to learn about the genes that control pigment cell development and pattern formation.
4. Seasonal and Life Cycle Alterations to Colors and Patterns
Changing ecological pressures throughout the ontogeny of an organism can necessitate corresponding changes in the organism's appearance. Animals that experience drastic seasonal ecosystem change, such as snowy winter, change their pigmentation to match: snowshoe hare, artic fox, snowy owl and ptarmigan are well-known examples [20]. Other animals exhibit specific coloration during the mating season: for example, three-spine stickleback males exhibit nuptial coloration that ranges from bright red to black depending on the species or morph [121].
Permanent changes in color or patterning are common as well. In a number of mammals this change is a relatively simple one: puma cubs and white-tailed deer fawns have spots, while older individuals of their species do not. Some fish species, such as wrasse and stoplight parrotfish, undergo a female-to-male transition and exhibit associated color changes [122,123]. Many teleosts and amphibians undergo metamorphosis, and the coloration or patterning changes associated with this can be dramatic [86,96]. Hormones like testosterone or the steroid androgen 11KT are known to be involved in these changes (e.g. [123]) but the mechanisms connecting hormone level and pigment cells have not been studied directly. In addition, while the behaviors of particular populations of cells during metamorphosis or any other coloration change may be known, the signals underlying these behaviors are still largely mysterious. To what extent is the pattern established by the pigment cells themselves, or by a prepattern set in the underlying tissue? Is patterning attributable to direct cell-cell contact or to secreted signaling molecules? Because the changes taking place are so significant, metamorphosis is an excellent time to study these questions of cell migration and behavior, cell-cell interactions, and pattern formation [57,124,125].
5. Evolution of Coloration, and Other Areas for Future Study
The study of animal coloration provides a framework in which to integrate tremendously divergent research interests. Pigment cells can be used to study a wide range of cellular and molecular topics: specification and differentiation of neural crest derivatives; cell migration; pigment synthesis; intracellular organelle transport; cell signaling; and pattern formation – not to mention the many diseases and syndromes associated with the pigmentary system [126].
At the same time, the importance of coloration and color patterning to the ecology of organisms cannot be overstated: predation avoidance and mate attraction in particular have been studied extensively as possible ecological explanations for specific colorations. Guppies provide a classic example of the tradeoffs between mate attraction (sexual selection) and predation avoidance (natural selection). In a classic series of experiments, Endler demonstrated that orange spots on male guppies made them more attractive to potential mates, but also made them easier targets for predators. Therefore, high predation pressure resulted in duller-colored guppies, while lower predation pressure allowed sexual selection to drive the evolution of guppies with larger, brighter orange spots [12,127]. A recent study examined an interesting variation on this tradeoff: sexual selection for increased visibility vs. the incidence of melanoma in male swordtails (Xiphophorus cortezi). In populations with relatively low frequency of the melanoma-causing Xmrk allele, females strongly preferred a male with a large melanophore spot on its caudal fin over a male without this spot. The role of Xmrk in enlarging this spot could explain why this oncogene has been maintained in these populations. In a population with much higher frequency of the Xmrk genotype and higher incidence of malignant melanomas, females still show a strong preference for increased melanization, but actually prefer males without the caudal fin spot, indicating an adaptation away from the pleiotropic effects of Xmrk [128].
The role of pigmentation and pigment patterns in evolution give us the opportunity to tie molecular, cellular, and ecological studies together in the same organisms. Doing so requires adding two more layers of complexity: the visual systems of the organisms involved; and the ecosystem in which the relevant inter- or intraspecific interactions take place. The importance of UV signals in particular (e.g. [129,130]) demonstrate the folly of relying on human perception of an animal's coloration or patterning in determining which components are important [131]. Any analysis of how a particular pigment mutation or adaptation affects a species or group of species must take into account how conspecifics and predators perceive the change, as well as the environment in which the change is viewed [132,133]. So far, these different fields have remained quite separate from one another, but connecting them to each other offers an exciting future direction for research. One very recent paper ties together some of these previously disparate fields [134]. This is by no means the first study to cross boundaries in this way, but it is a particularly elegant example. Seehausen and collaborators focus on the sympatric speciation of haplochromine cichlids by evaluating the evolution of an opsin, correlations of light penetration gradients with depth and turbidity, male nuptial coloration, and female mating preference [134]. The results – that the strength of preference of females with red-biased vision for red males, and of females with blue-biased vision for blue males, is dependent on both the absolute environmental conditions experienced by each population and by the rate of variation in those conditions – show the power of approaching a complicated problem like cichlid speciation from so many angles at once [135]. With increases in the availability of sequenced genomes and an increasing array of pigment pattern mutants to learn from, such integrative studies should become more common and even more informative – a tremendously exciting prospect.
Acknowledgments
This work supported by NSF Graduate Research Fellowships to MGM and LBP. Unpublished results supported by NSF grant IOB 0541733 to DM Parichy. Michael Elizondo, Tiffany Gordon and Mike Nishizaki provided helpful comments on this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jackson IJ. Molecular and developmental genetics of mouse coat color. Annu Rev Genet. 1994;28:189–217. doi: 10.1146/annurev.ge.28.120194.001201. [DOI] [PubMed] [Google Scholar]
- 2.Wolff GL. Regulation of yellow pigment formation in mice: A historical perspective. Pigment Cell Res. 2003;16:2–15. doi: 10.1034/j.1600-0749.2003.00012.x. [DOI] [PubMed] [Google Scholar]
- 3.Bennet DC, Lamoreau ML. The color loci of mice -- a genetic century. Pigment Cell Res. 2003;16:333–4. doi: 10.1034/j.1600-0749.2003.00067.x. [DOI] [PubMed] [Google Scholar]
- 4.Le Douarin NM. The Neural Crest. Cambridge: Cambridge University Press; 1999. [Google Scholar]
- 5.Slominski A, Wortsman J, Plonka PM, Schallreuter KU, Paus R, Tobin DJ. Hair follicle pigmentation. Journal of Investigative Dermatology. 2005;124:13–21. doi: 10.1111/j.0022-202X.2004.23528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamaguchi Y, Brenner M, Hearing VJ. The regulation of skin pigmentation. J Biol Chem. 2007;282:27557–61. doi: 10.1074/jbc.R700026200. [DOI] [PubMed] [Google Scholar]
- 7.Barsh GS. Regulation of pigment type switching by Agoutin, melanocortin signaling, Attractin and Mahoganoid. In: Nordlund J, Boissy R, Hearing V, et al., editors. The Pigmentary System: Physiology and Pathophysiology. 2nd. Malden: Blackwell Publishing Inc; 2006. p. 395. [Google Scholar]
- 8.Parichy DM, Reedy MV, Erickson CA. Regulation of melanoblast migration and differentiation. In: Nordland JJ, Boissy RE, Hearing VJ, et al., editors. The Pigmentary System: Physiology and Pathophysiology. 2nd. Malden: Blackwell Publishing Inc; 2006. [Google Scholar]
- 9.Kelsh RN. Genetics and evolution of pigment patterns in fish. Pigment Cell Res. 2004;17:326–26. doi: 10.1111/j.1600-0749.2004.00174.x. [DOI] [PubMed] [Google Scholar]
- 10.Fujii R. The regulation of motile activity in fish chromatophores. Pigment Cell Res. 2000;13:300–19. doi: 10.1034/j.1600-0749.2000.130502.x. [DOI] [PubMed] [Google Scholar]
- 11.Cooper CD, Raible D. Mechanisms for reaching the differentiated state: insights from neural crest derived melanocytes. Seminars in Cell and Developmental Biology. 2008 doi: 10.1016/j.semcdb.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Endler JA. Natural selection on color patterns in Poecilia reticulata. Evolution. 1980;34:76–91. doi: 10.1111/j.1558-5646.1980.tb04790.x. [DOI] [PubMed] [Google Scholar]
- 13.Endler JA. Natural and sexual selection on color patterns in poeciliid fishes. Environ Biol Fishes. 1983;9:173–90. [Google Scholar]
- 14.Engeszer RE. Learned social preference in zebrafish. Curr Biol. 2004;14:881–4. doi: 10.1016/j.cub.2004.04.042. [DOI] [PubMed] [Google Scholar]
- 15.Reynolds RG, Fitzpatrick BM. Assortative mating in poison-dart frogs based on an ecologically important trait. Evolution. 2007;61:2253–9. doi: 10.1111/j.1558-5646.2007.00174.x. [DOI] [PubMed] [Google Scholar]
- 16.Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin's finches. Science. 2004;305:1462–5. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- 17.Gompel N, Prud'homme B, Wittkopp PJ, Kassner VA, Carroll SB. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433:481–7. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- 18.Nachman MW, Hoekstra HE, D'Agostino SL. The genetic basis of adaptive melanism in pocket mice. Procedings of the National Academy of Sciences, U S A. 2003;100:5268–73. doi: 10.1073/pnas.0431157100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Protas ME. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat Genet. 2006;38:107–11. doi: 10.1038/ng1700. [DOI] [PubMed] [Google Scholar]
- 20.Searle AG. Comparative Genetics of Coat Colour in Mammals. New York: Academic Press; 1968. [Google Scholar]
- 21.Bultman SJ, Michaud EJ, Woychik RP. Molecular characterization of the mouse agouti locus. Cell. 1992;71:1195–204. doi: 10.1016/s0092-8674(05)80067-4. [DOI] [PubMed] [Google Scholar]
- 22.Millar SE, Miller MW, Stevens ME, Barsh GS. Expression and transgenic studies of the mouse agouti gene provide insight into the mechanism by which mammalian coat color patterns are generated. Development. 1995;121:3223–32. doi: 10.1242/dev.121.10.3223. [DOI] [PubMed] [Google Scholar]
- 23.Eizirik E, Yuhki N, Johnson WE, Menotti-Raymond M, Hannah SS, O'Brien SJ. Molecular genetics and evolution of melanism in the cat family. Curr Biol. 2003;13:448–53. doi: 10.1016/s0960-9822(03)00128-3. [DOI] [PubMed] [Google Scholar]
- 24.Rieder S, Taurit S, Mariat D, Langlois B, Guerin G. Mutations in the agouti (ASIP), the extension (MC1R), and the brown (TYRP1) loci and their association to coat color phenotypes in horses (Equus caballus) Mamm Genome. 2001;12:450–5. doi: 10.1007/s003350020017. [DOI] [PubMed] [Google Scholar]
- 25.Hiragaki T, Inoue-Murayama M, Miwa M, Fujiwara A, Mitzutani M, Minvielle F, et al. Recessive black is allelic to the yellow plumage locus in Japanese quail and associated with a frameshift deletion in the ASIP. Genetics. 2008;178:771–5. doi: 10.1534/genetics.107.077040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kijas JMH, Wales R, Tornsten A, Chardon P, Moller M, Andersson L. Melanocortin Receptor 1 (MC1R) mutations and coat color in pigs. Genetics. 1998;150:1177–85. doi: 10.1093/genetics/150.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Våge DI, Klungland H, Lu D, Cone RD. Molecular and pharmacological characterization of dominant black coat color in sheep. Mamm Genome. 1999;10:39–43. doi: 10.1007/s003359900939. [DOI] [PubMed] [Google Scholar]
- 28.Theron E, Hawkins K, Bermingham E, Ricklefs RE, Mundy NI. The molecular basis of an avian plumage polymorphism in the wild: a melanocortin-1-receptor point mutation is perfectly associated with the melanic plumage morph of the bananaquit, Coereba flaveola. Curr Biol. 2001;11:550–7. doi: 10.1016/s0960-9822(01)00158-0. [DOI] [PubMed] [Google Scholar]
- 29.Mundy NI, Badcock NS, Hart T, Scribner K, Janssen K, Nadeau NJ. Conserved genetic basis of quantitative plumage trait involved in mate choice. Science. 303:1870–3. doi: 10.1126/science.1093834. [DOI] [PubMed] [Google Scholar]
- 30.Takeuchi S, Suzuki H, Yabuuchi M, Takahashi S. A possible involvement of melanocortin-1-receptor in regulating feather color pigmentation in the chicken. Biochim Biophys Acta. 1996;1308:164–8. doi: 10.1016/0167-4781(96)00100-5. [DOI] [PubMed] [Google Scholar]
- 31.Kerje S, Lind J, Shutz K, Jensen P, Andersson L. Melanocortin-1-receptor (MC1R) mutations are associated with plumage colour in chicken. Anim Genet. 2003;334:241–8. doi: 10.1046/j.1365-2052.2003.00991.x. [DOI] [PubMed] [Google Scholar]
- 32.Robbins LS, Nadeau JH, Johnson KR, Kelly MA, Roselli-Rehfuss L, Baack E, et al. Pigmentation phenotypes of variant extension locus alleles result from point mutations that alter MSH receptor function. Cell. 1993;72:827–34. doi: 10.1016/0092-8674(93)90572-8. [DOI] [PubMed] [Google Scholar]
- 33.Kerns JA, Cargill EJ, Clark LA, Candille SI, Berryere TG, Olivier M. Linkage and segregation analysis of black and brindle coat color in domestic dogs. Genetics. 2007;176:1679–89. doi: 10.1534/genetics.107.074237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Candille SI, Kaelin CB, Cattanach BM, Yu B, Thomson DA, Nix MA. A β-defensin mutation causes black coat color in domestic dogs. Science. 2007;318:1418–23. doi: 10.1126/science.1147880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Logan DW, Bum SF, Jackson IJ. Regulation of pigmentation in zebrafish melanophores. Pigment Cell Res. 2006;19:206–13. doi: 10.1111/j.1600-0749.2006.00307.x. [DOI] [PubMed] [Google Scholar]
- 36.Sugimoto M. Morphological color changes in fish: regulation of pigment cell density and morphology. Microsc Res Tech. 2002;58:496–503. doi: 10.1002/jemt.10168. [DOI] [PubMed] [Google Scholar]
- 37.van der Salm AL, Metz JR, Wendelaar Bonga SE, Flik G. Alpha-MSH, the melanocortin-1-receptor and background adaptation in the Mozambique tilapia, Oreochromis mossambicus. Gen Comp Endocrinol. 2005;144:140–9. doi: 10.1016/j.ygcen.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 38.Oetting WS, Fryer JP, S S, King RA. Oculocutaneous Albinism Type 1: the last 100 years. Pigment Cell Res. 2003;16:307–11. doi: 10.1034/j.1600-0749.2003.00045.x. [DOI] [PubMed] [Google Scholar]
- 39.Nadeau NJ, Minvielle F, Ito S, Inoue-Murayama M, Gourichon D, Follett SA. Characterization of Japanese quail yellow as a genomic deletion upstream of the avian homolog of the mammalian ASIP (agouti) gene. Genetics. 2008;178:777–86. doi: 10.1534/genetics.107.077073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Everts RE, Rothuizen J, van Oost BA. Identification of a premature stop codon in the melanocyte-stimulating hormone gene (MC1R) in Labrador and Golden retrievers with yellow coat colour. Anim Genet. 2000;31:194–9. doi: 10.1046/j.1365-2052.2000.00639.x. [DOI] [PubMed] [Google Scholar]
- 41.Newton JM, Wilkie AL, He L, Jordan SA, Metallinos DL, Holmes NG. Melanocortin 1 receptor variation in the domestic dog. Mamm Genome. 2000;11:24–30. doi: 10.1007/s003350010005. [DOI] [PubMed] [Google Scholar]
- 42.Klungland H, Våge DI, Gomez-Raya L, Adalsteinsson S, Lien S. The role of melanocyte-stimulating hormone (MSH) receptor in bovine coat color determination. Mamm Genome. 1995;6:636–9. doi: 10.1007/BF00352371. [DOI] [PubMed] [Google Scholar]
- 43.Marklund L, Moller MJ, Sandberg K, Andersson L. A missense mutation in the gene for melanocyte-stimulating hormone receptor (MC1R) is associated with the chestnut coat color in horses. Mamm Genome. 1996;7:895–9. doi: 10.1007/s003359900264. [DOI] [PubMed] [Google Scholar]
- 44.Ritland K, Newton C, Marshall HD. Inheritance and population structure of the white-phased “Kermode” black bear. Curr Biol. 2001;11:1468–72. doi: 10.1016/s0960-9822(01)00448-1. [DOI] [PubMed] [Google Scholar]
- 45.Hoekstra HE, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science. 2006;313:101–4. doi: 10.1126/science.1126121. [DOI] [PubMed] [Google Scholar]
- 46.Steiner CC, Weber JN, Hoekstra HE. Adaptive variation in beach mice is produced by two interacting pigmentation genes. PLoS Biol. 2007;5:1880–9. doi: 10.1371/journal.pbio.0050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McGraw KJ. Mechanics of melanin-based coloration. In: Hill GE, McGraw KJ, editors. Bird Coloration: Mechanisms and Measurements. Cambridge: Harvard University Press; 2006. pp. 243–94. [Google Scholar]
- 48.McGraw KJ. Mechanics of carotenoid-based coloration. In: Hill GE, McGraw KJ, editors. Bird Coloration: Mechanisms and Measurements. Cambridge: Harvard University Press; 2006. pp. 177–242. [Google Scholar]
- 49.McGraw KJ, Hill GE, Navara KJ, Parker RS. Differential accumulation and pigmenting ability of dietary carotenoids in colorful finches. Physiol Biochem Zool. 2004;77:484–91. doi: 10.1086/383506. [DOI] [PubMed] [Google Scholar]
- 50.McGraw KJ, Beebee MD, Hill GE, Parker RS. Lutein-based plumage coloration in songbirds is a consequence of selective pigment incorporation into feathers. Comp Biochem Physiol B Biochem Mol Biol. 2003;135:689–96. doi: 10.1016/s1096-4959(03)00164-7. [DOI] [PubMed] [Google Scholar]
- 51.Ziegler I. The pteridine pathway in zebrafish: regulation and specification during the determination of neural crest cell-fate. Pigment Cell Res. 2003;26:172–82. doi: 10.1034/j.1600-0749.2003.00044.x. [DOI] [PubMed] [Google Scholar]
- 52.Quigley IK, Manuel JL, Roberts RA, Nuckels RJ, Herrington ER, MacDonald EL, et al. Evolutionary diversification of pigment pattern in Danio fishes: differential fms dependence and stripe loss in D. albolineatus. Development. 2005;132:89–104. doi: 10.1242/dev.01547. [DOI] [PubMed] [Google Scholar]
- 53.McClure M. Development and evolution of melanophore patterns in fishes of the genus Danio (Teleostei: Cyprinidae) J Morphol. 1999;241:83–105. doi: 10.1002/(SICI)1097-4687(199907)241:1<83::AID-JMOR5>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 54.Juola FA, McGraw KJ, Dearborn DC. Carotenoids and throat pouch coloration in the great frigatebird (Fregata minor) Comp Biochem Physiol B Biochem Mol Biol. 2007;149:370–7. doi: 10.1016/j.cbpb.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 55.McGraw KJ. Mechanics of uncommon colors: pterins, porphyrins, and psittacofulvins. In: Hill GE, McGraw KJ, editors. Bird Coloration: Mechanisms and Measurements. Cambridge: Harvard University Press; 2006. pp. 354–98. [Google Scholar]
- 56.Bagnara JT, Taylor JD, Prota G. Color changes, unusual melanosomes, and a new pigment from leaf frogs. Science. 1973;182:1034–5. doi: 10.1126/science.182.4116.1034. [DOI] [PubMed] [Google Scholar]
- 57.Bagnara JT. Enigmas of pterhodin, a red melanosomal pigment of tree frogs. Pigment Cell Res. 2003;16:510–6. doi: 10.1034/j.1600-0749.2003.00075.x. [DOI] [PubMed] [Google Scholar]
- 58.Bagnara JT, Fernandez PJ, Fujii R. On the blue coloration of vertebrates. Pigment Cell Res. 2007;20:14–26. doi: 10.1111/j.1600-0749.2006.00360.x. [DOI] [PubMed] [Google Scholar]
- 59.Prum RO, Torres R. Structural colouration of avian skin: convergent evolution of coherently scattering dermal collagen arrays. J Exp Biol. 2003;206:2409–29. doi: 10.1242/jeb.00431. [DOI] [PubMed] [Google Scholar]
- 60.Prum RO, Torres RH. Structural colouration of mammalian skin: convergent evolution of coherently scattering dermal collagen arrays. J Exp Biol. 2004;207:2157–72. doi: 10.1242/jeb.00989. [DOI] [PubMed] [Google Scholar]
- 61.Prum RO. Anatomy, physics and evolution of structural colors. In: Hill GE, McGraw KJ, editors. Bird Coloration: Mechanisms and Measurements. Cambridge: Harvard University Press; 2006. pp. 295–353. [Google Scholar]
- 62.Shawkey MD, Hill GE. Significance of a basal melanin layer to production of non-iridescent structural plumage color: evidence from an amelanotic Stellar's jay (Cyanocitta stelleri) J Exp Biol. 2006;209:1245–50. doi: 10.1242/jeb.02115. [DOI] [PubMed] [Google Scholar]
- 63.Doucet SM, Shawkey MD, Hill GE, Montgomerie R. Iridescent plumage in satin bowerbirds: structure, mechanisms and nanostructural predictors of individual variation in colour. J Exp Biol. 2006;209:380–90. doi: 10.1242/jeb.01988. [DOI] [PubMed] [Google Scholar]
- 64.Kuriyama T, Miyaji K, Sugimoto M, Hasegawa M. Ultrastructure of the dermal chromatophores in a lizard (Scincidae: Plestiodon latiscutatus) with conspicuous body and tail coloration. Zoolog Sci. 2006;23:793–9. doi: 10.2108/zsj.23.793. [DOI] [PubMed] [Google Scholar]
- 65.Lamoreux ML, Kelsh RN, Wakamatsu Y, Ozato K. Pigment pattern formation in the medaka embryo. Pigment Cell Res. 2005;18:64–73. doi: 10.1111/j.1600-0749.2005.00216.x. [DOI] [PubMed] [Google Scholar]
- 66.Johnson SL, Africa D, Walker C, Weston JA. Genetic control of adult pigment stripe development in zebrafish. Dev Biol. 1995;167:27–33. doi: 10.1006/dbio.1995.1004. [DOI] [PubMed] [Google Scholar]
- 67.Hirata M, Nakamura K, Kanemaru T, Shibata Y, Kondo S. Pigment cell organization in the hypodermis of zebrafish. Dev Dyn. 2003;227:497–503. doi: 10.1002/dvdy.10334. [DOI] [PubMed] [Google Scholar]
- 68.Hirata M, Nakamura K, Kondo S. Pigment cell distributions in different tissues of the zebrafish, with special reference to the striped pigment pattern. Dev Dyn. 2005;234:293–300. doi: 10.1002/dvdy.20513. [DOI] [PubMed] [Google Scholar]
- 69.Velando A, Beamonte-Barrientos R, Torres R. Pigment-based skin colour in the blue-footed booby: an honest signal of current conditions used by females to adjust reproductive investment. Oecologia. 2006;149:535–42. doi: 10.1007/s00442-006-0457-5. [DOI] [PubMed] [Google Scholar]
- 70.Bagnara JT, Matsumoto J. Comparative anatomy and physiology of pigment cells in nonmammalian tissues. In: Nordlund JJ, Boissy RE, Hearing VJ, et al., editors. The Pigmentary System: Physiology and Pathophysiology. 2nd. Malden: Blackwell Publishing Inc; 2006. pp. 11–59. [Google Scholar]
- 71.Bagnara JT, Taylor JD, Hadley ME. The dermal chromatophore unit. J Cell Biol. 1968;38:67–79. doi: 10.1083/jcb.38.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sköld HN, Amundsen T, Svensson PA, Mayer I, Bjelvenmark J, Forsgren E. Hormonal regulation of female nuptial coloration in a fish. Horm Behav. 2008;54:549–56. doi: 10.1016/j.yhbeh.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 73.Rowland HM, Cuthill IC, Harvey IF, Speed MP, Ruxton GD. Can't tell the caterpillars from the trees: countershading enhances survival in a woodland. Proc R Soc B. 2008;275:2539–45. doi: 10.1098/rspb.2008.0812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vrieling H, Duhl DMJ, Millar SE, Miller KA, Barsh GS. Differences in dorsal and ventral pigmentation result from regional expression of the mouse agouti gene. Procedings of the National Academy of Sciences, U S A. 1994;91:5667–71. doi: 10.1073/pnas.91.12.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Candille SI, Van Raamsdonk CD, Chen C, Kuijper S, Chen-Tsai Y, Russ A. Dorsoventral patterning of the mouse coat by Tbx15. PLoS Biol. 2004;2:30–42. doi: 10.1371/journal.pbio.0020003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fukuzawa T, Ide H. A ventrally localized inhibitor of melanization in Xenopus laevis skin. Dev Biol. 1988;129:25–36. doi: 10.1016/0012-1606(88)90158-3. [DOI] [PubMed] [Google Scholar]
- 77.Fukuzawa T, Bagnara JT. Control of melanoblast differentiation in amphibia by alpha-melanocyte stimulating hormone, a serum melanization factor, and a melanization inhibiting factor. Pigment Cell Res. 1989;2:171–81. doi: 10.1111/j.1600-0749.1989.tb00183.x. [DOI] [PubMed] [Google Scholar]
- 78.Fukuzawa T, Samaraweera P, Mangano FT, Law JH, Bagnara JT. Evidence that MIF plays a role in the development of pigmentation patterns in the frog. Dev Biol. 1995;167:148–58. doi: 10.1006/dbio.1995.1013. [DOI] [PubMed] [Google Scholar]
- 79.Bagnara JT, Fukuzawa T. Stimulation of cultured iridophores by amphibian ventral conditioned medium. Pigment Cell Res. 1990;3:243–50. doi: 10.1111/j.1600-0749.1990.tb00296.x. [DOI] [PubMed] [Google Scholar]
- 80.Cerdá-Reverter JM, Haitina T, Schiöth HB, Peter RE. Gene structure of the goldfish agouti-signaling protein: a putative role in the dorsal-ventral pigment pattern of fish. Endocrinology. 2005;146:1597–610. doi: 10.1210/en.2004-1346. [DOI] [PubMed] [Google Scholar]
- 81.Miller CT, Beleza S, Pollen AA, Schluter D, Kittles RA, Shriver MD, et al. cis-regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell. 2007;131:1179–89. doi: 10.1016/j.cell.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hultman KA, Bahary N, Zon LI, Johnson SL. Gene duplication of the zebrafish kit ligand and partitioning of melanocyte development functions to kit ligand a. PLoS Genet. 2007;3:e17. doi: 10.1371/journal.pgen.0030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Suzuki N, Hirata M, Kondo S. Traveling stripes on the skin of a mutant mouse. Proc Natl Acad Sci U S A. 2003;100:9680–5. doi: 10.1073/pnas.1731184100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Richardson MK, Hornbruch A, Wolpert L. Mechanisms of pigment pattern formation in the quail embryo. Development. 1990;109:81–9. [Google Scholar]
- 85.Richardson MK, Hornbruch A, Wolpert L. Pigment patterns in neural crest chimeras constructed from quail and guinea fowl embryos. Dev Biol. 1991;143:309–19. doi: 10.1016/0012-1606(91)90082-e. [DOI] [PubMed] [Google Scholar]
- 86.Parichy DM. Pigment patterns of larval salamanders (Ambystomatidae, Salamandridae): the role of the lateral line sensory system and the evolution of pattern-forming mechanisms. Dev Biol. 1996;175:265–82. doi: 10.1006/dbio.1996.0114. [DOI] [PubMed] [Google Scholar]
- 87.Parichy DM. When neural crest and placodes collide: interactions between melanophores and the lateral lines that generate stripes in the salamander Ambystoma tigrinum tigrinum (Ambystomatidae) Dev Biol. 1996;175:283–300. doi: 10.1006/dbio.1996.0115. [DOI] [PubMed] [Google Scholar]
- 88.Parichy DM. Homology and evolutionary novelty in the deployment of extracellular matrix molecules during pigment pattern formation in the salamanders Taricha torosa and T. rivularis (Salamandridae) J Exp Zool. 2001;291:13–24. doi: 10.1002/jez.2. [DOI] [PubMed] [Google Scholar]
- 89.Svetic V, Hollway GE, Elworthy S, Chipperfield TR, Davison C, Adams RJ. Sdf1a patterns zebrafish melanophores and links the somite and melanophore pattern defects in choker mutants. Development. 2007;134:1011–22. doi: 10.1242/dev.02789. [DOI] [PubMed] [Google Scholar]
- 90.Parichy DM, Ransom DG, Paw B, Zon LI, Johnson SL. An orthologue of the kit-related gene fms is required for development of neural crest-derived xanthophores and a subpopulation of adult melanophores in the zebrafish, Danio rerio. Development. 2000;127:3031–44. doi: 10.1242/dev.127.14.3031. [DOI] [PubMed] [Google Scholar]
- 91.Lister JA, Robertson CP, Lepage T, Johnson SL, Ransom DW. nacre encodes a zebrafish micropthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development. 1999;126:3757–67. doi: 10.1242/dev.126.17.3757. [DOI] [PubMed] [Google Scholar]
- 92.Parichy DM, Turner JM. Temporal and cellular requirements for Fms signaling during zebrafish adult pigment pattern development. Development. 2003;130:817–33. doi: 10.1242/dev.00307. [DOI] [PubMed] [Google Scholar]
- 93.Maderspacher F, Nusslein-Volhard C. Formation of the adult pigment pattern in zebrafish requires leopard and obelix dependent cell interactions. Development. 2003;130:3447–57. doi: 10.1242/dev.00519. [DOI] [PubMed] [Google Scholar]
- 94.Caicedo-Carvajal CE, Shinbrot T. In silico zebrafish pattern formation. Dev Biol. 2008;15:397–403. doi: 10.1016/j.ydbio.2007.12.036. [DOI] [PubMed] [Google Scholar]
- 95.Epperlein HH, Löfberg J, Olsson L. Neural crest cell migration and pigment pattern formation in urodele amphibians. Int J Dev Biol. 1996;40:229–38. [PubMed] [Google Scholar]
- 96.Parichy DM. Evolution of danio pigment pattern development. Heredity. 2006;97:200–10. doi: 10.1038/sj.hdy.6800867. [DOI] [PubMed] [Google Scholar]
- 97.Duftner N, Sefc KM, Koblmüler S, Salzburger W, Taborsky M, Sturmbauer C. Parallel evolution of facial stripe patterns in the Neolamprologus brichardi/pulcher species complex endemic to Lake Tanganyika. Mol Phylogenet Evol. 2007;45:706–15. doi: 10.1016/j.ympev.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 98.Silvers WK. The Coat Colors of Mice: A Model for Mammalian Gene Action and Interaction. New York: Springer-Verlag; 1979. [Google Scholar]
- 99.Hosoda K, Hammer RE, Richardson JA, Baynash AG, Cheung JC, Giaid A. Targeted and natural (piebald-lethal) mutations of Endothelin-B receptor gene produce megacolon associated with spotted coat color in mice. Cell. 1994;79:1267–76. doi: 10.1016/0092-8674(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 100.Chabot B, Stephenson DA, Chapman VM, Besmer P, Bernstein A. The proto-oncogene c-kit encoding a transmembrane tyrosine kinase receptor maps to the mouse W locus. Nature. 1988;355:88–9. doi: 10.1038/335088a0. [DOI] [PubMed] [Google Scholar]
- 101.Geissler EN, Ryan MA, Housman DE. The dominant-white spotting (W) of the mouse encodes the c-kit proto-oncogene. Cell. 1998;55:185–92. doi: 10.1016/0092-8674(88)90020-7. [DOI] [PubMed] [Google Scholar]
- 102.Pavan WJ, Tlighman SM. Piebald lethal (sl) acts early to disrupt the development of neural crest-derived melanocytes. Procedings of the National Academy of Sciences, U S A. 1994;91:7159–63. doi: 10.1073/pnas.91.15.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee HO, Levorse JM, Shin MK. The endothelin receptor-B is required for the migration of neural crest-derived melanocyte and enteric neuron precursors. Dev Biol. 2003;259:162–75. doi: 10.1016/s0012-1606(03)00160-x. [DOI] [PubMed] [Google Scholar]
- 104.Wehrle-Haller B. The role of kit-ligand in melanocyte development and epidermal homeostasis. Pigment Cell Res. 2003;16:287–96. doi: 10.1034/j.1600-0749.2003.00055.x. [DOI] [PubMed] [Google Scholar]
- 105.Cable J, Jackson IJ, Steel KP. Mutations at the W locus affect survival of neural crest-derived melanocytes in the mouse. Mech Dev. 1995;50:139–50. doi: 10.1016/0925-4773(94)00331-g. [DOI] [PubMed] [Google Scholar]
- 106.Murphy M, Reid K, Williams DE, Lyman SD, Bartlett PF. Steel factor is required for maintenance, but not differentiation, of melanocyte precursors in the neural crest. Dev Biol. 1992;153:396–401. doi: 10.1016/0012-1606(92)90124-y. [DOI] [PubMed] [Google Scholar]
- 107.Reid K, Nishikawa S, Bartlett PF, Murphy M. Steel factor directs melanocyte development in vitro through selective regulation of the number of c-kit+ progenitors. Dev Biol. 1995;169:568–79. doi: 10.1006/dbio.1995.1170. [DOI] [PubMed] [Google Scholar]
- 108.Wehrle-Haller B, Weston JA. Soluble and cell-bound forms of steel factor activity play distinct roles in melanocyte precursor dispersal and survival on the lateral neural crest migration pathway. Development. 1995;121:731–42. doi: 10.1242/dev.121.3.731. [DOI] [PubMed] [Google Scholar]
- 109.Johansson Moller M, Chaudry R, Hellmén E, Höyheim B, Chowdhary B, Andersson L. Pigs with the dominant white coat color phenotype carry a duplication of the KIT gene encoding the mast/stem cell growth factor receptor. Mamm Genome. 1996;7:822–30. doi: 10.1007/s003359900244. [DOI] [PubMed] [Google Scholar]
- 110.Marklund S, Kijas JMH, Rodriguez-Martinez H, Ronnstrand L, Funa K, Moller M. Molecular basis for the dominant white phenotype in the domestic pig. Genome Res. 1998;8:826–33. doi: 10.1101/gr.8.8.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Giuffra E, Törnsten A, Marklund S, Bongcan-Rudloff E, Chardon P, Kijas JMH. A large duplication associated with dominant white color in pigs originated by homologous recombination between LINE elements flanking KIT. Mamm Genome. 2002;13:569–77. doi: 10.1007/s00335-002-2184-5. [DOI] [PubMed] [Google Scholar]
- 112.Brooks SA, Bailey E. Exon skipping in the KIT gene causes a sabino spotting pattern in horses. Mamm Genome. 2005;16:893–902. doi: 10.1007/s00335-005-2472-y. [DOI] [PubMed] [Google Scholar]
- 113.Haase B, Brooks SA, Schlumbaum A, Azor PJ, Bailey E, Alaeddine F. Allelic heterogeneity at the equine KIT locus in dominant white (W) horses. PLoS Genet. 2007;3 doi: 10.1371/journal.pgen.0030195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Metallinos DL, Bowling AT, Rine J. A missense mutation in the endothelin-B receptor is associated with Lethal White Foal Syndrome: an equine version of Hirschsprung Disease. Mamm Genome. 1998;9:426–31. doi: 10.1007/s003359900790. [DOI] [PubMed] [Google Scholar]
- 115.Santschi EM, Purdy AK, Valberg SJ, Vrotsos PD, Kaese H, Mickelson JR. Endothelin receptor B polymorphism associated with lethal white foal syndrome in horses. Mamm Genome. 1998;9:306–9. doi: 10.1007/s003359900754. [DOI] [PubMed] [Google Scholar]
- 116.Parichy DM, Rawls JF, Pratt SJ, Whitfield TT, Johnson SL. Zebrafish sparse corresponds to an orthologue of c-kit and is required for the morphogenesis of a subpopulation of melanocytes, but is not essential for hematopoiesis or primordial germ cell development. Development. 1999;126:3425–36. doi: 10.1242/dev.126.15.3425. [DOI] [PubMed] [Google Scholar]
- 117.Parichy DM, Mellgren EM, Rawls JF, Lopes SS, Kelsh RN, Johnson SL. Mutational analysis of endothelin receptor b1 (rose) during neural crest and pigment pattern development in the zebrafish Danio rerio. Dev Biol. 2000;227:294–306. doi: 10.1006/dbio.2000.9899. [DOI] [PubMed] [Google Scholar]
- 118.Quigley IK, Turner JM, Nuckels RJ, Manuel JL, Budi EH, MacDonald EL, et al. Pigment pattern evolution by differential deployment of neural crest and post-embryonic melanophore lineages in Danio fishes. Development. 2004;131:6053–69. doi: 10.1242/dev.01526. [DOI] [PubMed] [Google Scholar]
- 119.Watanabe M, Iwashita M, Ishii M, Kurachi Y, Kawakami A, Kondo S. Spot pattern of leopard Danio is caused by mutation in the zebrafish connexin41.8 gene. EMBO Rep. 2006;7:893–7. doi: 10.1038/sj.embor.7400757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Salzburger W, Braasch I, Meyer A. Adaptive sequence evolution in a color gene involved in the formation of the characteristic egg-dummies of male haplochromine cichlid fishes. BMC Biol. 2007;5:51–63. doi: 10.1186/1741-7007-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Boughman JW. Divergent sexual selection enhances reproductive isolation in sticklebacks. Nature. 2001;411:944–8. doi: 10.1038/35082064. [DOI] [PubMed] [Google Scholar]
- 122.Ohta K, Hirano M, Mine T, Mizutani H, Yamaguchi A, Matsuyama M. Body color change and serum steroid hormone levels throughout the process of sex change in the adult wrasse, Pseudolabrus sieboldi. Mar Biol. 2008;153:843–52. [Google Scholar]
- 123.Cardwell JR, Liley NR. Hormonal control of sex and color change in the stoplight parrotfish, Sparisoma yiride. Gen Comp Endocrinol. 1990;81:7–20. doi: 10.1016/0016-6480(91)90120-u. [DOI] [PubMed] [Google Scholar]
- 124.Yasutomi M, Yamada S. Formation of the dermal chromatophore unit (DCU) in the tree frog Hyla arborea. Pigment Cell Res. 1998;11 doi: 10.1111/j.1600-0749.1998.tb00730.x. [DOI] [PubMed] [Google Scholar]
- 125.Parichy DM. Experimental analysis of character coupling across a complex life cycle: pigment pattern metamorphosis in the tiger salamander, Ambystoma tigrinum tigrinum. J Morphol. 1998;237:53–67. doi: 10.1002/(SICI)1097-4687(199807)237:1<53::AID-JMOR5>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 126.Nordland JJ, Boissy RE, Hearing VJ, King DC, Oetting WS, Ortonne JP, editors. The Pigmentary System: Physiology and Pathophysiology. 2nd. Cambridge: Blackwell Publishing Ltd; 2006. [Google Scholar]
- 127.Endler JA. A predator's view of animal color patterns. Evol Biol. 1978;11:319–64. [Google Scholar]
- 128.Fernandez AA, Morris MR. Mate choice for more melanin as a mechanism to maintain a functional oncogene. Proc Natl Acad Sci U S A. 2008;105:13503–7. doi: 10.1073/pnas.0803851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rick IP, Bakker TCM. Color signaling in conspicuous red sticklebacks: do ultraviolet signals surpass others? BMC Evol Biol. 2008;8:189–97. doi: 10.1186/1471-2148-8-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hunt S, Cuthill IC, Bennett ATD, Church SC, Partridge JC. Is the ultraviolet waveband a special communication channel in avian mate choice? J Exp Biol. 2001;204:2499–507. doi: 10.1242/jeb.204.14.2499. [DOI] [PubMed] [Google Scholar]
- 131.Engeszer RE, Wang G, Ryan MJ, Parichy DM. Sex-specific perceptual spaces for a vertebrate basal social aggregative behavior. Proc Natl Acad Sci U S A. 2008;105:929–33. doi: 10.1073/pnas.0708778105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cuthill IC. Color Perception. In: Hill GE, McGraw KJ, editors. Bird Coloration: Mechanisms and Measurements. Cambridge: Harvard University Press; 2006. [Google Scholar]
- 133.Théry M. Effects of light environment on color communication. In: Hill GE, McGraw KJ, editors. Bird Coloration: Mechanisms and Measurements. Cambridge: Harvard University Press; 2006. [Google Scholar]
- 134.Seehausen O, Terai Y, Magalhaes IS, Carleton KL, Mrosso HDJ, Miyagi R. Speciation through sensory drive in cichlid fish. Nature. 2008;455:620–6. doi: 10.1038/nature07285. [DOI] [PubMed] [Google Scholar]
- 135.Kirkpatrick M, Price T. Sensory Ecology: In sight of speciation. Nature. 2008;455:601–2. doi: 10.1038/455601a. [DOI] [PubMed] [Google Scholar]