Abstract
Significance: Protein neddylation is catalyzed by an E1 NEDD8-activating enzyme (NAE), an E2 NEDD8-conjugating enzyme, and an E3 NEDD8 ligase. Known physiological substrates of neddylation are cullin family members. Cullin neddylation leads to activation of cullin-RING ligases (CRLs), the largest family of E3 ubiquitin ligases responsible for ubiquitylation and degradation of many key signaling/regulatory proteins. Thus, through modulating CRLs, neddylation regulates many biological processes, including cell cycle progression, signal transduction, and tumorigenesis. Given that NEDD8 is overexpressed and CRLs are abnormally activated in many human cancers, targeting protein neddylation, in general, and cullin neddylation, in particular, appears to be an attractive anticancer approach. Recent Advances: MLN4924, a small molecule inhibitor of NAE, was discovered that inactivates CRLs and causes accumulation of CRL substrates to suppress tumor cell growth both in vitro and in vivo. Promising preclinical results advanced MLN4924 to several clinical trials for anticancer therapy. Critical Issues: In preclinical settings, MLN4924 effectively suppresses tumor cell growth by inducing apoptosis, senescence, and autophagy, and causes sensitization to chemoradiation therapies in a cellular context-dependent manner. Signal molecules that determine the cell fate upon MLN4924 treatment, however, remain elusive. Cancer cells develop MLN4924 resistance by selecting target mutations. Future Directions: In the clinical side, several Phase 1b trials are under way to determine the safety and efficacy of MLN4924, acting alone or in combination with conventional chemotherapy, against human solid tumors. In the preclinical side, the efforts are being made to develop additional neddylation inhibitors by targeting NEDD8 E2s and E3s. Antioxid. Redox Signal. 21, 2383–2400.
Introduction: Protein Ubiquitylation and Neddylation
Protein ubiquitylation, catalyzed by the ubiquitin–proteasome system (UPS), is a major clearance system for the maintenance of protein homeostasis by degrading unwanted proteins, which include misfolded, damaged, and short-lived proteins (15, 41). A protein destined for degradation via UPS is marked by a polyubiquitin tag, resulting from a biochemical process known as ubiquitylation, which is carried out via a three-step enzymatic cascade (15). First, ubiquitin is activated in an ATP-dependent reaction by a ubiquitin-activating enzyme (UAE) (E1) and is then transferred to a ubiquitin-conjugating enzyme (E2). Finally, a ubiquitin ligase (E3), which recognizes and recruits a target protein, designated as the substrate, transfers and conjugates ubiquitin from the E2 onto a lysine (K) residue on the substrate. Ubiquitin itself contains seven lysine residues, which serve as the acceptors for the second ubiquitin molecule, leading to polyubiquitylation of the substrate after multiple rounds of this reaction. In this process, E3 ubiquitin ligases perform a critical role through the selective binding of protein substrates. The human genome encodes 2 E1s, 38 E2s, and more than 600 E3 ubiquitin ligases, which can be subdivided into four major classes based on their structural and biochemical features (163). The UPS is abnormally regulated in many human diseases, particularly in neurodegenerative diseases and cancers (16, 17). The successful development of bortezomib (VELCADE), a first-in-class proteasome inhibitor for the treatment of multiple myeloma and relapsed mantle cell lymphoma (105), demonstrates that the UPS is an attractive anticancer target.
Protein neddylation is a process of tagging NEDD8 onto a substrate protein, not for degradation, but for modulation of protein activity/function. NEDD8 is one of the most studied ubiquitin-like (UBL) proteins and is 60% identical and 80% homologous to ubiquitin (56). Like ubiquitin, NEDD8 is attached to its substrates by an isopeptide linkage between its C-terminal Gly76 and a lysine residue of the target protein. However, NEDD8 is first synthesized as a precursor that contains five additional residues downstream from Gly76 that need to be cleaved by C-terminal hydrolases, which include UCH-L3 (52, 71) and NEDP1/DEN1/SENP8 (33, 89, 149). After this processing, NEDD8 is activated in an ATP-dependent reaction by an E1 NEDD8-activating enzyme (NAE). Activated NEDD8 is then transferred to an E2 NEDD8-conjugating enzyme, which shuttles it to an E3 ligase and ultimately conjugates NEDD8 to its specific substrates (Fig. 1).
FIG. 1.
The enzymatic cascades for protein neddylation and deneddylation. Schematic representation of each step of the NEDD8 conjugation pathway, including NEDD8 precursor processing, NEDD8 activation by NAE, E2 loading, conjugation to a substrate by an E3 and recycling of NEDD8 by a NEDD8 isopeptidase. The involving enzymes in each step are listed. NAE, NEDD8-activating enzyme. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
The NEDD8 cascade is known to contain a single E1 (NAE), two E2s (UBE2M, also known as UBC12, and UBE2F), and a few E3s (see Fig. 1). NAE is a heterodimer, consisting of NAE1/APPBP1 and UBA3/NAEβ (8) and is structurally and biochemically similar to UAE. NAE1 and UBA3 are homologous to the amino and carboxyl regions of UAE, respectively. UBE2M preferentially neddylates RING box protein-1 (RBX1)-associated cullins (CUL1–3, 4A and 4B), whereas UBE2F promotes neddylation of RBX2-associated CUL5 (43). Except defective in cullin neddylation 1 (DCN1) (66, 67) and DCN1-like proteins (90), the majority of NEDD8 E3 ligases contain really interesting new gene (RING) finger domains, which include RBX1 and RBX2 (also known as regulators of cullins 1 [ROC1] and ROC2/SAG, respectively) (26, 43, 57), murine double minute-2 (MDM2) (150), casitas B-lineage lymphoma (c-CBL) (108, 170), SCFFBXO11 (3), ring finger protein 1111 (RNF111) (84), inhibitors of apoptosis (IAPs) (9, 97), Tfb3 (112), and TRIM40 (100). DCN1 serves as an NEDD8 E3 ligase for cullin neddylation in Caenorhabditis elegans and Saccharomyces cerevisiae (67). Human cells harbor five DCN1-like proteins termed DCNL1–DCNL5, which have distinct N-terminal domains, but share a conserved C-terminal potentiating neddylation (PONY) domain. In yeast, this PONY domain of DCN1 is necessary and sufficient for cullin neddylation in vivo and in vitro (66). DCNL1–DCNL3 have been shown to interact with cullins and modulate cullin neddylation (90). Interestingly, DCN1 does not contain a RING domain for its catalytic activity, rather it directly interacts with the NEDD8 E2 UBE2M on a surface that overlaps with the E1-binding site (66). A recent structural study showed that UBE2M is N-terminal acetylated and this N-acetyl-methionine is completely buried in a hydrophobic pocket of DCN1 E3. This interaction promotes cullin neddylation (121).
A reverse process for protein neddylation is protein deneddylation. Conjugated NEDD8 is removed from a neddylated substrate by the action of an NEDD8 isopeptidase. The best characterized NEDD8 isopeptidase is the COP9 signalosome complex (CSN), a zinc metalloprotease consisting of 8 subunits. CSN5 is the catalytic subunit of CSN, which deneddylates cullins (18, 146). NEDP1, a cysteine protease, is another NEDD8-specific isopeptidase, which also processes NEDD8 precursor (40, 116, 122). Additional proteases with dual specificity for NEDD8 and ubiquitin include USP21 (37), Ataxin-3 (31), PfUCH54 (4), UCH-L1, and UCH-L3 (40) (Fig. 1).
Conservation of the Neddylation Pathway During Evolution
Although NEDD8 was initially identified as a gene that is downregulated in mouse brain during development (64), it was soon demonstrated that NEDD8 is detectable in various mouse tissues and highly conserved in vertebrate species as well as in yeast (65), suggesting that the neddylation pathway is essential during species evolution. Indeed, genetic depletion of components of the NEDD8 pathway in Schizosaccharomyces pombe (106), C. elegans (53), Drosophila (107), or in mouse (137) is lethal, demonstrating that the NEDD8 pathway is essential for the viability of most model organisms. Interestingly, although an intact NEDD8 pathway is not essential for cell growth in S. cerevisiae (74), the combination of pathway mutants with temperature-sensitive mutants of cdc34 caused lethality (69). Mutations in the AXR1 and ECR1 genes in Arabidopsis thaliana, the orthologs of human NAE1 and UBA3, respectively, result in defects in auxin signaling (111). Moreover, plants deficient in both AXR1 (E1) and RCE1, the ortholog of NEDD8 E2, show a seedling lethal phenotype, a characteristic of defective auxin signaling (23). Inactivation of the NEDD8 pathway in TS-41 hamster cells expressing a temperature-sensitive mutant of Smc1, the ortholog of human NAE1, leads to repetitive cycles of G1/S-phases without entering the G2/M-phase at the nonpermissive temperature, suggesting an important role for the NEDD8 pathway in regulating DNA replication, cell cycle progression, and cell division (38). In addition, C. elegans embryos expressing a temperature-sensitive mutant of rfl-1, the ortholog of human UBA3, have a myriad of defects in cytokinesis at a restrictive temperature (68).
In humans, accumulating evidence shows that NEDD8 is overexpressed in some human diseases such as neurodegenerative disorders (24, 96) and cancers (12, 120). Thus, targeting protein neddylation appears to be an attractive approach for targeted anticancer therapy (143). Indeed, MLN4924, a newly discovered small molecule inhibitor of NAE, suppresses tumor cell growth both in vitro and in vivo (127) and has been advanced to several Phase I clinical trials against a number of human malignancies (126, 128). Therefore, it may be very helpful for future drug development to characterize as to which cellular proteins are neddylated, how this modification affects their functions, and how this neddylation is regulated.
Substrates of the Neddylation Pathway and Associated Biological Processes
Cullin-RING ligases
In contrast to a wide range of ubiquitylated proteins, very few proteins are known to be neddylated. Table 1 lists most, if not all, known substrates of the NEDD8 pathway. The best characterized substrate of the NEDD8 pathway is the cullin family of proteins (109). In the human genome, there are eight cullin family members, including Cul-1–3, 4A, 4B, 5, 7, and Cul-9, also known as PARC, with an evolutionarily conserved cullin homology domain (CH domain with ∼150 amino acids) at the C-terminus. Each cullin protein acts as a molecular scaffold that binds to an adaptor protein and a substrate receptor protein at the N-terminus, and a RING protein, RBX1 or RBX2 at the C-terminus to assemble cullin-RING ligases (CRLs), the largest family of E3 ubiquitin ligases. By promoting ubiquitylation and degradation of a variety of key substrates, CRLs control many important biological processes, including cell cycle progression, DNA repair, signal transduction, gene transcription, embryonic development, genomic integrity, and tumorigenesis [for recent review, see Ref. (163)]. Each individual cullin contains a key lysine residue at its C-terminus for targeted NEDD8 modification, which is required for the CRL activity. RBX1 and RBX2 in conjunction with UBE2M and UBE2F, respectively (43, 57), promote cullin neddylation. Recently, DCN1 and DCNL1–DCNL3 proteins were also found to act as NEDD8 E3 ligases for cullins (66, 67, 90).
Table 1.
NEDD8 Pathway Substrates
| Substrates | E3 ligase | Function of neddylation | References |
|---|---|---|---|
| Cullins | RBX1/2 and/or DCN1 | Increases CRL activity | 43, 57, 66, 67, 90, 109 |
| p53 | MDM2 and SCFFBXO11 | Inhibits p53 transcriptional activity | 3, 150 |
| p73 | MDM2 | Inhibits p73 transcriptional activity | 142 |
| MDM2 | MDM2 | Increases Mdm2 stability | 150 |
| Ribosomal proteins (L11, S14) | MDM2 | Increases ribosomal protein stability | 133, 151, 159 |
| HuR | MDM2 | Controls the nuclear localization of HuR and protects it from degradation | 30 |
| EGFR | c-CBL | Enhances the efficiency of EGFR ubiquitination and facilitates its degradation | 108 |
| TGF-βRII | c-CBL | Stabilizes TGF-βRII by antagonizing ubiquitination and degradation | 170 |
| HIF1α/HIF2α | Increases protein stability | 118 | |
| BCA3 | Recruits histone deacetylase SIRT1 that represses NFκB-dependent transcription | 35 | |
| AICD | Inhibits AICD-mediated transcriptional activation via the inhibition of its interaction with the transcription coactivators Fe65 and Tip60 | 72 | |
| E2F-1 | Reduces E2F-1 stability, transcriptional activity, and cell growth | 80 | |
| IKKγ | TRIM40 | Inhibits NF-κB activity | 100 |
| SHC | Facilitates the formation of a ZAP70-Shc-Grb2 signaling complex and affects downstream Erk activation | 51 | |
| Caspases/IAPs/RIP1 | IAPs | Suppresses caspase activity | 9, 97 |
| RCAN1 | Increases RCAN1 stability by inhibiting proteasomal degradation of RCAN1 and increases RCAN1 binding to calcineurin | 101 | |
| pVHL | Prevents pVHL interaction with Cul2-containing complexes, promotes pVHL association with fibronectin, and the assembly of extracellular matrix | 130 | |
| Parkin/PINK1 | Increases parkin E3 ligase activity and stabilizes PINK1 55 kDa fragment | 13, 140 | |
| Histone H4 | RNF111 | Activates DNA damage-induced ubiquitination | 84 |
AICD, APP intracellular domain; BCA3, breast cancer-associated protein 3; c-CBL, casitas B-lineage lymphoma; CRL, cullin-RING ligase; DCN1, defective in cullin neddylation 1; E2F-1, E2F transcription factor 1; EGFR, epidermal growth factor receptor; HIF1, hypoxia-inducible factor-1; HIF2, hypoxia-inducible factor-2; HuR, Hu antigen R; IAPs, inhibitors of apoptosis; MDM2, murine double minute-2; PINK1, PTEN induced putative kinase 1; pVHL, von Hippel-Lindau protein; RCAN1, regulator of calcineurin 1; RING, really interesting new gene; RNF111, ring finger protein 111; TGFβ-RII, transforming growth factor β type II receptor.
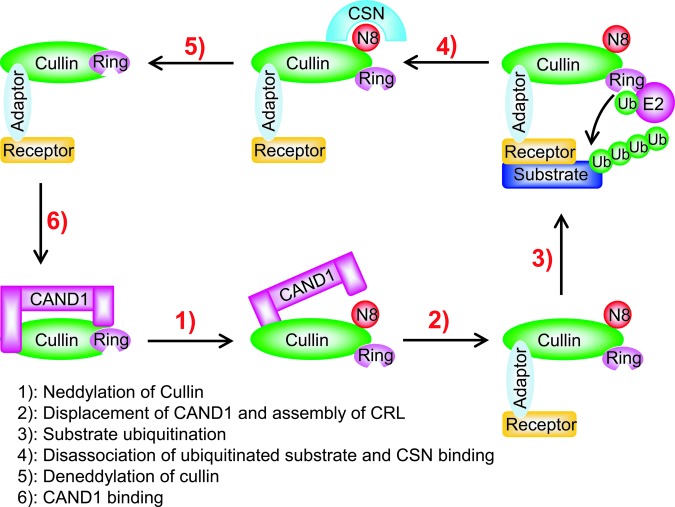
Cullin neddylation is a process known to activate CRLs, thus promoting the ubiquitylation of their protein substrates. In an inactive mode, cullin-associated and neddylation-dissociated-1 (CAND1) binds to non-neddylated cullins and blocks the binding of cullins to the substrate receptor–adaptor module via its N-terminus. Neddylation of cullins disrupts this inhibitory binding by CAND1 and retains the CRLs in an active conformation (28, 36, 57, 78, 152, 166), which (i) increases and stabilizes the recruitment of ubiquitin-charged E2 to CRLs (119), (ii) bridges an ∼50 Å gap between the substrate-docking site and the E2-active site (167), which greatly facilitates the initiation of ubiquitin transfer (119), and (iii) enhances the rate of ubiquitin chain elongation, suggesting that this conformation change also improves the activated E2 access to the end of the nascent polyubiquitin chain (119, 124) (Fig. 2).
FIG. 2.
Dynamic regulation of CRL activity by neddylation and deneddylation. The binding of unmodified cullin to CAND1 inhibits the cullin binding to the substrate receptor–adaptor module at its N-terminus. Neddylation of cullin disrupts the inhibitory binding by CAND1 and retains the CRL in an active conformation to promote substrate ubiquitylation. After dissociation of polyubiquitylated substrate from the CRL complex, CSN binds to the neddylation site of cullin and removes NEDD8 from cullin for recycling. CAND1 then binds to cullin and inactivates CRL. CAND1, cullin-associated and neddylation-dissociated-1; CRL, cullin-RING ligase; CSN, COP9 signalosome complex; RING, really interesting new gene. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
The mechanism, by which cullin neddylation enhances the transfer of ubiquitin (Ub) from ubiquitin charged E2 to the substrate, is fully illuminated by crystal structures of the C-terminal domain (CTD) of CUL5 bound to RBX1 in the unmodified and NEDD8-conjugated states (28). Neddylation induces striking conformational rearrangements in CUL5CTD-RBX1, which eliminates the CAND1 binding site and releases the RBX1 RING domain from its tight cullin binding into an open conformation. Although the RING domain still is tethered to CUL5CTD, the linker is more flexible and readily bridges the ∼50 Å gap between the E2-active site and naked substrate present in the original CRL complex structure, which explains why neddylation promotes the initial ubiquitin transfer. This flexibility also facilitates access of the activated E2 to the end of the nascent ubiquitin chain, allowing efficient polyubiquitylation (21, 22, 113) (Fig. 2).
Upon polyubiquitylation, the substrate is separated from the CRL complex, and the COP9 signalosome complex (CSN) then binds to neddylated cullins and removes NEDD8 from cullins in a reaction known as deneddylation (83). Deneddylated cullins bind to CAND1, which keeps the CRLs in an inactive conformation (Fig. 2). Thus, neddylation promotes the assembly of active CRL E3 complexes and stimulates the ubiquitination of substrates, whereas deneddylation promotes the dissociation of CRL E3 complexes and potentiates cullin binding with inhibitory CAND1. Dynamic neddylation and deneddylation of cullins facilitate the recycling of the cullin-RING core, making it available for assembly with other members of CRLs permitting the ubiquitination of many different substrates in a short time as required by the cell to maintain cellular homeostasis (79). Accumulating data suggest that mutations in the components of the COP9 signalosome (CSN) lead to defects in cell cycle progression, signal transduction, and development (5, 6, 147). Thus, it is very important to understand mechanistically how neddylation and deneddylation are precisely regulated under physiological conditions or in response to both internal and external stimuli.
Other noncullin NEDD8 substrates
In addition to cullin family members, which are the known physiological substrates of neddylation (113), several cellular proteins are also reported to be neddylated, although a more rigorous validation by, for example, the NAE inhibitor MLN4924 is still necessary.
p53
The stability and function of the p53 tumor suppressor is tightly regulated by post-translational modifications, including ubiquitylation and neddylation, in which the MDM2 oncoprotein plays a critical role. Acting as a major ubiquitin E3 ligase, MDM2 promotes p53 polyubiquitylation and proteasomal degradation with its physiological relevance demonstrated by the full rescue of embryonic lethality in Mdm2-null mice by the loss of p53 (20, 55). Significantly, MDM2 also promotes p53 neddylation, which inhibits the p53 transactivation activity (150). A mutational analysis of the RING finger domain of MDM2 showed that mutations, which affect the ubiquitin ligase activity of MDM2, also affect its Nedd8 ligase activity (123). These findings therefore provide a strong piece of evidence that MDM2 is a common component of the ubiquitin and NEDD8 conjugation pathway. However, there is also evidence to suggest that MDM2-mediated ubiquitylation and neddylation of p53 are differentially regulated and elicit different effects on p53. First of all, MDM2-mediated p53 ubiquitylation required six lysine residues in the C-terminus of p53 (Lys370, Lys372, Lys373, Lys381, Lys382, and Lys386), whereas neddylation only required three lysine residues (Lys370, Lys372, and Lys373) (150). Second, endogenous p53 in response to UV-induced DNA damage showed a differential pattern of modification by ubiquitin and NEDD8 (150). Third, the Tip60 acetyl-transferase, a known regulator of the MDM2-p53 axis, was shown to preferentially inhibit MDM2-mediated p53 neddylation, but not ubiquitylation (25). Finally, the p53-Ub fusion protein promotes the cytoplasmic localization of p53, whereas the p53-NEDD8 fusion protein has little effect on nuclear export. Despite the difference in subcellular localization, both the p53-Ub and p53-NEDD8 fusion proteins retain similar transcriptional activity and both induce apoptosis at a similar level to nonfused p53 (11). In addition to MDM2, SCFFBXO11, a CRL E3 ligase, was found to directly bind to p53 that unexpectedly promoted p53 neddylation rather than ubiquitylation. Consistent with an inhibitory role of p53 neddylation, SCFFBXO11 suppresses the p53 transcriptional activity (3).
p73 and MDM2
p73, a member of the p53 family, is also subject to neddylation by MDM2 E3 (142). MDM2 promotes the neddylation of TAp73 (full-length), but not ΔNp73 (N-terminally truncated with N-terminal MDM2-binding domain deleted), leading to accumulation of TAp73 in the cytoplasm, thus suppressing its transactivation activity (142). Interestingly, MDM2 can also catalyze self-neddylation in a manner similar to its autoubiquitylation (150). However, unlike MDM2 autoubiquitylation, which destabilizes it, MDM2 autoneddylation was shown to increase its protein stability (144). Furthermore, NEDP1, identified as a chemotherapy-induced isopeptidase, deneddylates MDM2, leading to MDM2 destabilization with concomitant p53 activation. Likewise, knockdown of endogenous NEDP1 stabilized MDM2, decreased p53, and increased chemoresistance of cancer cells (144).
Ribosomal proteins
Several ribosomal proteins have been identified as potential NEDD8 substrates (151). L11 was found to be neddylated by Mdm2 and deneddylated by NEDP1 (133). Under nucleolar stressed conditions, L11 neddylation was reduced, which triggered relocalization of L11 from the nucleolus to the nucleoplasm. MDM2-mediated L11 neddylation protects L11 from degradation, a process required for p53 stabilization during nucleolar stress (133). Thus, neddylation regulates both the subcellular localization and stability of L11. A recent study showed that the MDM2/NEDP1 pair also regulates the neddylation and deneddylation cycle of the ribosomal protein S14, where neddylation causes protein stabilization and modulates the subcellular localization (159). Given that both L11 (81, 160) and S14 (169) bind to MDM2 and regulate p53 stability, the neddylation–deneddylation cycle of L11 and S14 by the MDM2/NEDP1 pair adds another layer of control of the MDM2/p53 axis to ensure precise regulation of p53 protein levels.
Other cellular proteins
With the advancement of detection technologies, many additional neddylated proteins have been identified, along with some of their corresponding E3s. Table 1 provides a relatively comprehensive listing of these neddylated proteins, known E3s for their neddylation, and the functional consequences of neddylation. Examples include (i) Hu antigen R (HuR), a central RNA-binding protein, highly abundant in many cancers (2), neddylated by Mdm2 (30), (ii) receptor proteins, such as EGFR (108), and transforming growth factor (TGF)-β type II receptor (170), neddylated by the c-Cbl E3, (iii) transcriptional regulators such as HIF1α/HIF2α (118), breast cancer-associated protein 3 (BCA3) (35), APP intracellular domain (AICD) (72), and E2F-1 (80), (iv) signaling molecules such as inhibitor of κB kinase gamma (IKKγ) neddylated by TRIM40 (100), caspases neddylated by IAP (9), Shc (51), and regulator of calcineurin 1 (RCAN1) (101), (v) E3 ubiquitin ligases, such as von Hippel-Lindau (VHL) tumor suppressor (130) and Parkin/PINK1 (13, 140), and (vi) finally, histone H4 neddylated by RNF111 (84). Taken together, these studies suggest that neddylation plays a role beyond cullin modification and associated protein degradation. However, given that these studies were mainly conducted using in vitro biochemical and cell culture systems, the physiological relevance and biological significance of these neddylation modifications await thorough validation by in vivo animal models or in human cancer specimens.
The Regulation of Redox Homeostasis by Neddylation
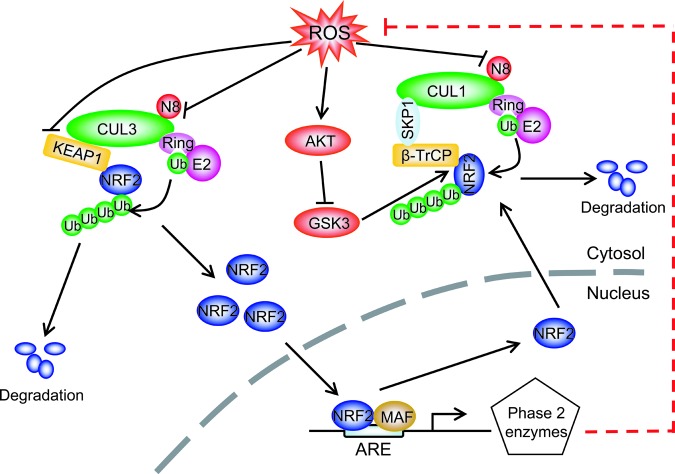
The accumulation of damaging reactive oxygen species (ROS) contributes to a number of pathologies in various human diseases, including cancer (32, 131). ROS levels, therefore, need to be tightly controlled to prevent oxidative stress-induced damages (131). The transcription factor nuclear factor erythroid 2-related factor 2 (NRF2) functions as a master regulator of redox homeostasis by inducing the transcription of a wide array of genes involved in defense against ROS to restore intracellular redox homeostasis (59). Indeed, nearly all NRF2 target genes contain a consensus DNA binding motif (RGTGACNNNGC, where R represents purine and N represents any base) in their promoter region designated as the antioxidant response element (ARE) (44, 117). The ARE is required for NRF2 binding and subsequent transcriptional activation of several antioxidant enzymes (59). By modulation of the cellular levels of NRF2, a well-established substrate of CRL1 and CRL3 E3 ligases, the neddylation process, which activates the CRL activity via cullin neddylation, effectively regulates redox homeostasis (14, 114, 115, 141, 158) (Fig. 3).
FIG. 3.
Redox regulation of NRF2 via CRL E3s: Under normal physiological conditions, the NRF2 level is kept low as a result of targeted degradation by (i) CRL3 upon Keap1 binding and (ii) CRL1 upon β-TrCP binding, following GSK3-mediated NRF2 phosphorylation. Under oxidative stressed conditions, ROS on one hand inhibits cullin neddylation to inactivate CRLs and on the other hand activates AKT to block GSK3-mediated NRF2 phosphorylation, leading to suppression of NRF2 degradation. Accumulated NRF2 then translocates into the nucleus, where it complexes with MAF to bind to the ARE and transactivates the expression of antioxidant enzymes to scavenge ROS. ARE, antioxidant response element; GSK3, glycongen synthase kinase 3; Keap1, Kelch-like ECH-associated protein 1; NRF2, nuclear factor erythroid 2-related factor 2; ROS, reactive oxygen species. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Under a normal unstressed redox environment, the NRF2 level is very low due to targeted ubiquitylation and degradation by two CRL E3s. First, NRF2 is trapped in the cytoplasm by binding to its inhibitor, Kelch-like ECH-associated protein 1 (Keap1), which recruits Cul3 E3 ligase (CRL3), and targets NRF2 for proteasomal degradation. Second, NRF2 is phosphorylated by glycogen syntase kinase 3 (GSK3) on the serine residues with the beta-transducin repeat containing protein (βTrCP) binding motif (DSGIS), which facilitates βTrCP-NRF2 binding and subsequent NRF2 degradation by SCFβTrCP E3 ligase (also known as CRL1) (14, 114, 115). Under the initial phase of oxidative stresses, there are at least three mechanisms by which ROS triggers NRF2 accumulation: (i) ROS oxidizes the cysteine residues on Keap1 to change its conformation, which disrupts the Keap1-NRF2 binding and subsequent NRF2 degradation (44), (ii) ROS inhibits several phosphatases to activate AKT, which sequentially phosphorylates and inactivates GSK3 (138) leading to abrogation of NRF2 phosphorylation and subsequent βTrCP binding and CRL1 degradation (14, 114, 115), and (iii) ROS causes oxidative inactivation of the catalytic cysteine residue on Ubc12, the NEDD8-conjugating enzyme (62, 63), resulting in cullin deneddylation and CRL inactivation. Accumulated NRF2 then translocates to the nucleus, where it becomes transcriptionally active by binding with one of the Maf proteins and induces the transcription of phase II antioxidant enzymes, which deactivate ROS (44). In the late phase of oxidative stresses when ROS levels decline, AKT is inactivated by ceramide-activated phosphatases or by other mechanisms (87) with subsequent activation of GSK3, resulting in SCFβTrCP E3-mediated NRF2 degradation (14, 114, 115). Consequently, NRF2 returns to its basal levels and the intracellular redox balance is restored (Fig. 3). It is noteworthy that a wide variety of somatic mutations of Keap1 and NRF2 are found in human cancers and these mutations disrupt Keap1-mediated negative regulation of NRF2, resulting in constitutive activation of NRF2 (39, 129). Activated NRF2 is associated with resistance to standard chemotherapy and poor survival of cancer patients (39, 129), indicating that NRF2 also has oncogenic functions.
Interestingly, the neddylation inhibitor MLN4924 can either decrease or increase the generation of intracellular levels of ROS. On one hand, through inactivation of CRLs, MLN4924 causes NRF2 accumulation (99, 127, 145) to scavenge ROS. On the other hand, through inactivation of NFκB, MLN4924 generates ROS, which is required for DNA damage-induced apoptosis (99, 134). Thus, the net outcome of neddylation effects on ROS generation is likely to be cell-type dependent and context dependent. To date, our understanding of how neddylation regulates redox homeostasis is mainly through the modulation of CRL activity (141). Future studies should be directed toward exploring other potential mechanisms, including investigation of whether the proteins involved in redox homeostasis are direct neddylation targets and if so, under what physiological and/or pathological conditions they are neddylation, and whether and how neddylation affects their functions.
Targeting Neddylation Pathway for Anticancer Therapy
MLN4924 is a newly discovered investigational inhibitor of the NAE (127) (Fig. 4A) currently under clinical development. As an adenosine sulfamate derivative, MLN4924 forms an MLN4924-NEDD8 adduct catalyzed by NAE. With tight binding to the NAE-active site, this MLN4924-NEDD8 adduct resembles adenylated NEDD8, the first intermediate in the NAE reaction cycle, and thus prevents subsequent intraenzyme reactions and blocks the NAE enzymatic activity (10). Given that there is only one NAE known to catalyze this first step of the neddylation reaction, its inhibitor MLN4924 should block the entire neddylation pathway. Indeed, we found that MLN4924 effectively inhibits neddylation of multiple cullins, the only known physiological substrates (21, 113), as evidenced by complete deneddylation of all cullins tested, including Cul1–Cul3, Cul4A, Cul4B, and Cul5 after 6 h of treatment in SK-BR3 breast cancer cells (Fig. 4B) (10). Given that cullin neddylation is required for the activity of CRLs, whereas CRLs are abnormally activated in human cancers (49, 163), MLN4924, by blocking cullin neddylation, inactivates the entire family of CRL E3 ligases and serves as a first-in-class agent, which suppresses tumor cell growth in preclinical models via multiple mechanisms described below.
FIG. 4.
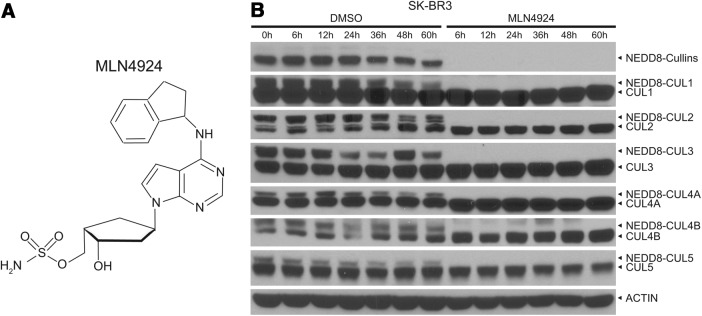
MLN4924 as an inhibitor of NAE. (A) Chemical structure of MLN4924. (B) MLN4924 blocks neddylation of all cullins tested. SK-BR3 cells were treated with 1 μM MLN4924 or DMSO vehicle control for the indicated time periods, followed by western blotting with the indicated antibodies.
MLN4924 induces apoptosis
MLN4924 was first reported in 2009 as a potent growth suppressing agent against a variety of cancer cell lines derived from solid tumors (colon and lung) and hematological malignancies (myeloma and lymphoma) both in in vitro cell culture and in in vivo xenograft models (127). Subsequent studies showed that MLN4924 effectively induces apoptosis in leukemia (91, 93, 125, 127, 134), hepatocellular (82), and Ewing sarcoma cells (85). Consistent with these findings, MLN4924 also induces apoptosis in breast cancer cells (MCF7), as evidenced by increases in the cleaved forms of PARP and caspase-7 (Fig. 5A). MLN4924-induced apoptosis is mediated by several mechanisms, all involving accumulation of CRL substrates. First, by inactivating CRL1SKP2 and CRL4CDT2, which promote chromatin licensing and DNA replication factor 1 (CDT1) degradation (42, 73), MLN4924 caused accumulation of CDT1 to trigger DNA re-replication and S phase arrest, ultimately leading to induction of apoptosis (76, 91, 127). A recent genomewide siRNA screen revealed that induction of apoptosis by MLN4924 involves multiple DNA damage response (DDR) pathways beyond those involving CDT1 stabilization (7). Second, by inhibiting CRL1β-TrCP, MLN4924 causes accumulation of IκBα to block nuclear factor-κB (NF-κB) activation (148), resulting in apoptosis induction (93, 134). Third, by inactivating SAG/RBX2-associated CRLs (50), MLN4924 causes accumulation of proapoptotic NOXA, a known p53 target gene, resulting in apoptosis in some settings (135, 145). Indeed, we found that CDT1, pIκBα, and NOXA were all accumulated significantly after MLN4924 treatment to induce apoptosis in MCF7 cells (Fig. 5A).
FIG. 5.
MLN4924 suppresses tumor cell growth via inducing apoptosis, autophagy, and senescence. (A) Induction of apoptosis: MCF7 cells were treated with 1 μM MLN4924 for the indicated time periods, followed by western blotting with the indicated antibodies. (B–D) Induction of autophagy: MDA-MB231 cells stably expressing EGFP-LC3 were treated with 1 μM MLN4924 or DMSO vehicle control for 24 h and 48 h before photography under a fluorescent microscope (B). Detection of autophagosomes by electron microscopy (EM). SK-BR3 cells were treated for 24 h with MLN4924 (1 μM), along with DMSO vehicle control, followed by the EM analysis. Autophagosomes (arrows) are indicated in MLN4924-treated cells (C). SK-BR3 cells were treated with MLN4924 (1 μM), along with DMSO vehicle control for the indicated time periods, followed by western blotting using the indicated antibodies (D). (E, F) Induction of senescence: SK-BR3 cells were treated with MLN4924 (1 μM) for 8 h and stained with β-Gal after the drug washout for 72 h (E). SK-BR3 cells were treated with 1 μM MLN4924 for the indicated time periods, followed by western blotting with the indicated antibodies (F). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
MLN4924 induces autophagy
Recently, we found that in addition to apoptosis, MLN4924 also effectively induced autophagy in a concentration- and time-dependent manner in multiple human cancer cell lines derived from carcinomas of the breast, colon, liver, brain, and cervix, indicating that it is likely a universal phenomenon in cancer cells (82, 164). As an example, shown in Figure 5, MLN4924 significantly induced punctate structures, a well-defined surrogate for autophagy (94), in EGFP-LC3-expressing MDA-MB231 cells (Fig. 5B). Furthermore, autophagosomes were readily detectable by electron microscopy in SK-BR3 cells upon MLN4924 exposure (Fig. 5C). Finally, MLN4924 induction of autophagy was biochemically demonstrated by a time-dependent conversion of LC3-I to LC3-II and degradation of p62, two well-established biomarkers of autophagy (94) in SK-BR3 cells (Fig. 5D, top panels). Our detailed mechanistic studies revealed that MLN4924-induced autophagy is mainly caused by inactivation of mammalian target of rapamycin complex 1 (mTORC1), as evidenced by reduced phosphorylation of mTOR itself and two mTORC1 substrates, S6K1 and 4E-BP1 (Fig. 5D, middle panels). mTORC1 inactivation is likely mediated by accumulation of (i) DEP domain containing mTOR interacting protein (DEPTOR), a naturally occurring inhibitor of mTORCs (110) and a recently characterized substrate of CRL1β-TrCP (27, 34, 165) and (ii) HIF1α, a well-known substrate of CRL2VHL (45, 46), followed by activation of the HIF1-REDD1-TSC1 axis (164) (Fig. 5D, bottom panels). We further demonstrated that mTORC1 inactivation and subsequent autophagy induction act as an overall survival signal, since abrogation of autophagy via genetic and pharmacological means led to an increased suppression of tumor growth by enhancing apoptosis induction (153, 155, 164). Thus, our findings provide proof-of-concept evidence supporting further investigation of a combination of MLN4924 with an autophagy inhibitor (155, 162, 164).
MLN4924 induces senescence
In addition to induction of apoptosis and autophagy, we and others showed that MLN4924 can also induce irreversible senescence in multiple cancer cell lines (48, 76, 77). We further show here that MLN4924 treatment induces characteristics associated with senescence in SK-BR3 breast cancer cells, as evidenced by enlarged and flattened cellular morphology and positive staining of senescence-associated β-Gal (Fig. 5E). Mechanistic studies revealed that senescence occurs in p21-dependent manner (48), which is logical given that p21 is a known substrate of CRL1SKP2 (157) and CRL4CDT2 (1, 61). Although it was shown that senescence induced by MLN4924 is independent of the pRB/p16 axis in HCT116 cells (48), p16, along with p21 and p27, was remarkably induced in response to MLN4924 in SK-BR3 cells (Fig. 5F), indicating that the role of p16 in senescence could be cell-type dependent.
MLN4924 as a potential sensitizer to chemotherapy and radiation
In addition to suppressing tumor cell growth as a single agent via the mechanism described above, a number of recent studies have shown that MLN4924 could sensitize a variety of otherwise resistant cancer cells to chemotherapeutic and biological agents. For example, we found that MLN4924 sensitized leukemia cells to retinoic acid-induced apoptosis via accumulation of c-Jun and NOXA (135). MLN4924 also sensitized head and neck cancer cells to TRAIL-induced apoptosis through promotion of c-FLIP degradation (161). Moreover, MLN4924 significantly increased the efficacy of cisplatin against cisplatin-resistant ovarian cancer cells by enhancing DNA damage and oxidative stress, and by increasing the expression of the proapoptotic protein, Bcl-2-interating killer (BIK) (99), as well as by inactivating CRL3 (47). MLN4924 was also shown to increase cellular sensitivity to DNA interstrand crosslink inducing agents, such as mitomycin C and hydroxyurea in several cancer cell lines derived from carcinomas of the cervix (HeLa) and colon (HCT116) by a mechanism involving the suppression of DNA damage-induced FANCD2 monoubiquitylation and CHK1 phosphorylation (58). Finally, we tested the combination of MLN4924 with gemcitabine, a standard chemotherapy in pancreatic cancer (95) in two pancreatic cancer cell lines. Our unpublished data showed that MLN4924 significantly sensitized pancreatic cancer cells to gemcitabine.
We also determined the radiosensitizing activity of MLN4924 in pancreatic cancer cells and found that MLN4924 effectively inhibited cullin neddylation and sensitized pancreatic cancer cells to ionizing radiation both in in vitro cell culture and in in vivo xenograft models (145). Mechanistic studies revealed that MLN4924 treatment induced an accumulation of several CRL substrates, including CDT1, WEE1, and NOXA, in parallel with an enhancement of radiation-induced DNA damage, aneuploidy, G2/M phase cell cycle arrest, and apoptosis. Importantly, knockdown of accumulated CDT1 or WEE1 partially rescued MLN4924-mediated radiosensitization, indicating their causal roles (145). We further found that MLN4924 radiosensitization can also be observed in few breast cancer cell lines with mechanisms involving p21 accumulation (154). Thus, MLN4924 is a potent sensitizer to gemcitabine and radiation, two clinically used standard therapies for the treatment of pancreatic cancer patients.
Taken together, accumulated data, including our own studies, clearly demonstrate that MLN4924 effectively inhibits tumor cell growth by inducing all three common types of death, namely apoptosis, autophagy, and senescence, as outlined in Figure 6. Induction of apoptosis, as often seen in both hematologic cancer and solid tumor cells (82, 88, 91, 93, 127, 134, 135, 164), is mainly caused by (i) NF-κB inactivation, resulting from IκBα accumulation, (ii) accumulation of proapoptotic NOXA, and (iii) DDR, resulting from CDT1 accumulation. On the other hand, induction of autophagy, mainly seen in solid tumor lines (82, 164), is largely mediated by inactivation of mTORC1, resulting from accumulation of DEPTOR and HIF1α. Induction of senescence, also generally observed in solid tumor cells, is mainly mediated by p21 accumulation (48). Accumulation of p27 and p16 may also contribute to MLN4924-induced senescence at least in SK-BR3 cells (Fig. 5F). It is noteworthy that in response to MLN4924 treatment, cancer cells are subjected to one or multiple types of death dependent on their own molecular characteristics. For example, in SK-BR3 breast cancer cells, MLN4924 preferentially induces autophagy and senescence, but not apoptosis (Fig. 5F). Thus, these various effects of MLN4924 can be seen in different cancer cell lines, but not all of them would co-occur in every setting. Finally, MLN4924 renders multiple cancer cell lines sensitive to chemotherapy and radiation therapy with mechanisms mainly involving enhanced DNA damage and subsequent induction of apoptosis (Fig. 6).
FIG. 6.
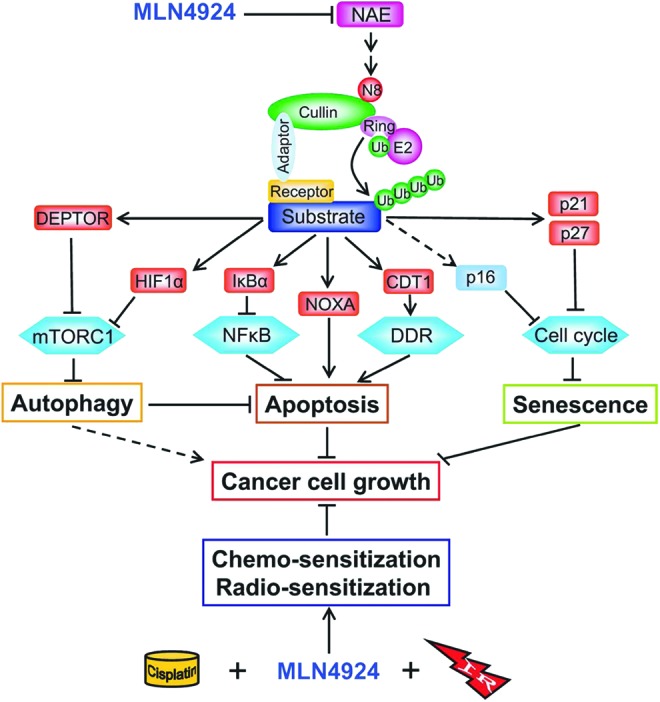
A model for suppression of cancer cell growth and sensitization of chemo- and radiation therapies by MLN4924 (see text for description). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
MLN4924 as a Tool in Identification and Validation of Novel CRL Substrates
Improvements in mass spectrometry methodology and instrumentation are likely to facilitate the discovery of additional CRL substrates. Stable isotope labeling with amino acids in cell culture (SILAC) presents one such advancement, which has been validated as a promising approach to quantitate protein abundance (86, 103, 104) and has been used to study a wide range of cellular protein responses in entire organisms and even in human tissues, including identification of protein/DNA interaction partners, protein post-translational modifications, subcellular protein localization, and changes in protein levels resulting from drug treatments, stress responses, and tumorigenesis (19, 102). Thus, SILAC may serve as a very attractive approach to identify CRL substrates affected by MLN4924. Indeed, a modified SILAC technique in combination with a diGly monoclonal antibody (which recognizes the cleaved C-terminal of the Arg-Gly-Gly sequence in ubiquitin) has been used to systematically and quantitatively assess the ubiquitin modified proteome, including neddyomics (60). The SILAC technique was also used to identify many regulatory proteins as potential novel CRL substrates in A375 melanoma cells treated with MLN4924 (75), followed by validation of MRFAP1 as a true CUL4B substrate (70). Most significantly, both genetic (dominant negative cullins) and pharmacologic (MLN4924) approaches and techniques, including cullin inactivation, coupled with genetic assays (or GPS for global protein stability profiling) (156) and SILAC-MS-based proteomics (or QUAINT for quantitative ubiquitylation interrogation) were used to identify hundreds of proteins whose stabilities or ubiquitylation status are controlled by CRLs (29). Through these approaches, the authors identified and validated NUSAP1 as a CRL1CyclinF substrate, which is degraded in response to UV radiation and is responsible for resistance to antitubulin therapeutics (29).
Clinical Development of MLN4924 as a Novel Class of Anticancer Drug
With promising anticancer activity in preclinical models, MLN4924 has been in clinical investigation since May, 2008. Up to now, there are a total of seven Phase I/II clinical trials for MLN4924 in patients with leukemia, lymphoma, melanoma, and several solid tumors (http://clinicaltrials.gov/ct2/show?term=MLN4924&rank=1) designed with three main trial goals: (i) drug safety and tolerability, (ii) drug pharmacokinetic parameters, and (iii) disease response rate (Table 2). The dosing schedules, major toxicities, and disease response rate have been recently reviewed for the first four earlier trials (98). In general, MLN4924 was administrated via 60-min intravenous infusion on various daily schedules of 21-day cycles (98). The pharmacokinetics of MLN4924 (www.eventureonline.com/eventure/publicAbstractView.do?id=193668&congressId=5650; www.eventureonline.com/eventure/publicAbstractView.do?id=193419&congressId=5650; http://mct.aacrjournals.org/cgi/content/meeting_abstract/10/11_MeetingAbstracts/A38?sid=f99baff2-718b-4552-8e41-c9b9570eb9ff) in plasma or tissue was conducted in serial blood samples, bone marrow aspirates (BMAs), skin punch biopsies, or fine-needle tumor biopsies collected at 3–6 h following a weekly drug dosing. The samples were analyzed by immunohistochemical staining to measure the MLN4924-NEDD8 covalent adduct and expression of CRL substrates, such as CDT1, NRF2, and pIκBα as the biomarkers. The presence of drug was demonstrated by detecting (i) MLN4924-NEDD8 adducts in the BMAs of all 21 patients receiving the drug (www.eventureonline.com/eventure/publicAbstractView.do?id=193668&congressId=5650) and in 100% of the postdose tumor biopsies (http://mct.aacrjournals.org/cgi/content/meeting_abstract/10/11_MeetingAbstracts/A38?sid=f99baff2-718b-4552-8e41-c9b9570eb9ff), and (ii) increased levels of pIκBα- and Nrf-2-regulated gene transcripts in peripheral blood mononuclear cells (www.eventureonline.com/eventure/publicAbstractView.do?id=193419&congressId=5650) and CDT1/NRF2 in skin and tumor biopsies with persistence of up to 24 h (www.eventureonline.com/eventure/publicAbstractView.do?id=193419&congressId=5650; http://mct.aacrjournals.org/cgi/content/meeting_abstract/10/11_MeetingAbstracts/A38?sid=f99baff2-718b-4552-8e41-c9b9570eb9ff). The major side effects reported include fatigue, nausea, vomiting, diarrhea, anemia, neutropenia, and elevated liver enzymes (98). In term of disease response rate, a complete response was seen in four acute myelogenous leukemia (AML) patients, a partial response in one Hodgkin's lymphoma, and one melanoma patient. Nine melanoma patients had prolonged stable disease (98). Since 2011, three MLN4924 combinational phase 1b/2 trials have been launched (http://clinicaltrials.gov/ct2/show?term=MLN4924&rank=1). The first trial is for the treatment of large B-cell lymphoma patients in combination with EPOCH-R chemotherapy; the second is for patients with end stage solid tumors in combination with docetaxel, gemcitabine, and paclitaxel plus carboplatin; and the third one is for adult AML patients in combination with azacitidine (Table 2). Upon completion, these trials will demonstrate the tolerability of MLN4824 in combination with standard chemotherapies and should begin to suggest the potential therapeutic effectiveness of MLN4924 in combination treatment strategies against deadly human cancers.
Table 2.
Clinical Trials of MLN4924
| Clinicaltrials.gov identifier | Phase | Tumor type | Time initiated | With combination | Primary outcome measures | Secondary outcome measures |
|---|---|---|---|---|---|---|
| NCT00677170 | 1 | Advanced nonhematologic | May 2008 | Alone | Determine the safety profile, MTD, and PK/pharmacodynamics of MLN4924 | Disease response |
| NCT00722488 | 1 | Malignancies HM, MM, HL, lymphoma | July 2008 | Alone | Evaluation of safety and tolerability | Disease response |
| NCT00911066 | 1 | AML, ALL, MS | May 2009 | Alone With azacitidine | Adverse events, serious adverse events, assessments of clinical laboratory values, and vital sign measurements | Pharmacokinetic parameters |
| Pharmacodynamic effects | ||||||
| Assess disease response | ||||||
| Heart corrected QT intervals | ||||||
| NCT01011530 | 1 | Metastatic melanoma | November 2009 | Alone | MTD and inform recommended phase 2 dose of MLN4924 | Antitumor activities of MLN4924 |
| Pharmacodynamic effects of MLN4924 on blood and tumor cells | ||||||
| NCT01415765 | 1/2 | Large B-cell lymphoma | August 2011 | Alone With standard EPOCH-R chemotherapy | Assess response of MLN4924 in relapsed/refractory DLBCL | Analyze molecular subtype (ABC and GCB) |
| Assess toxicity and safe tolerated dose of MLN4924 and DA-EPOCH-R | Assess difference in response between ABC and GCB subtypes of relapsed/refractory DLBCL/MLN 4924 alone and w/MLN4924 and DA-EPOCH-R | |||||
| Assess ORR (CR/PR) and PFS of MLN4924 and DA-EPOCH-R in relapsed/refractory DLBCL | Analyze mutations of the ITAM motifs, CARD11, and A20 in DLBC | |||||
| NCT01862328 | 1b | Solid tumors | May 2013 | With docetaxel | Number of adverse events | MLN4924 plasma concentration–time data for population PK analysis |
| With gemcitabine | Disease response rate | |||||
| With carboplatin+paclitaxel | ||||||
| NCT01814826 | 1b | AML | March 2013 | With azacitidine | Number of adverse events | Pharmacokinetic parameters, including but not limited to AUC, Cmax, systemic clearance, volume of distribution, elimination of half-life |
| Assess the safety and tolerability of MLN4924 in combination with azacitidine | Disease response rate | |||||
| Thirty-day mortality rate | ||||||
| Sixty-day mortality rate |
HM, hematologic malignancies; MM, multiple myeloma; HL, Hodgkin lymphoma; AML, acute myelogenous leukemia; ALL, acute lymphoblastic leukemia; MS, myelodysplastic syndrome; EPOCH-R chemotherapy, etoposide, prednisone, vincristine, cyclophosphamide, doxorubicin, and rituximab chemotherapy; MTD, maximum tolerated dose.
Conclusions and Future Perspectives
In summary, the NEDD8 pathway is being validated as a potential anticancer target, mainly because the pathway is overactivated in a number of human cancers (143) and its inactivation by the first-in-class NAE inhibitor, MLN4924, suppresses tumor growth in many preclinical models. Whether MLN4924 will mature to an FDA approved first-in-class anticancer drug will depend on the results of ongoing clinical trials. MLN4924 is anticipated to be less toxic than the FDA approved proteosome inhibitors such as bortezomib and carfilzomib, since MLN4924 selectively blocks degradation of a specific set of cellular proteins regulated by CRLs, whereas bortezomib inhibits degradation of all proteins through inhibition of the 26S proteasome (105). However, tumor cell selectivity issues still exist for MLN4924. First of all, given that CRL activity is required for proliferation, differentiation, and survival of normal cells (132, 136, 168), their inhibition might also be detrimental to normal cells, particularly those with a high proliferation potential, such as bone marrow cells. Second, MLN4924 is an NAE inhibitor and would likely inhibit, in addition to cullin neddylation, other cellular neddylation reactions associated with unknown biological consequences potentially leading to normal tissue toxicity. Of note, despite these potential tumor cell selectivity issues, MLN4924 was well tolerated in mice (127, 145) and demonstrated a manageable toxicity in humans (98).
It has been reported recently that cancer cells can develop resistance to MLN4924 via selection of rare clones with heterozygous mutations in the targeting enzyme NAEβ (92, 139). Nevertheless, MLN4924, which targets multiple CRL-associated signaling pathways, is anticipated to be more effective than targeted therapy using a single kinase inhibitor, since human cancers often harbor multiple mutations with alterations in multiple signaling pathways (54). To overcome the limitations of MLN4924, future basic scientific studies are critical to better understand how the neddylation pathway and its key components regulate the initiation and progression of human tumorigenesis. In the context of drug discovery, specific inhibitors against NEDD8 E2s (UBE2M or UBE2F) and/or a particular E3 with known involvement in human cancer might provide improved tumor cell selectivity and toxicity profiles. It is highly hoped and anticipated that with validation of NEDD8-CRLs as attractive anticancer targets, small molecule inhibitors targeting a unique component of the NEDD8 pathway or a specific CRL E3, known to be activated in human cancer, will be discovered. The development of a novel class of anticancer agents targeting the NEDD8/CRL E3 pathway, acting either alone or in combination with current anticancer therapies is a promising strategy for the treatment of selected sets of cancer patients with tumors bearing overactivation of this pathway.
Abbreviations Used
- βTrCP
beta-transducin repeat containing protein
- AICD
APP intracellular domain
- AML
acute myelogenous leukemia
- APPBP1
amyloid beta precursor protein binding protein 1
- ARE
antioxidant response element
- BCA3
breast cancer-associated protein 3
- BIK
Bcl-2-interating killer
- CAND1
cullin-associated and neddylation-dissociated-1
- c-CBL
casitas B-lineage lymphoma
- CDT1
chromatin licensing and DNA replication factor 1
- CRLs
cullin-RING ligases
- CSN
COP9 signalosome complex
- CTD
C-terminal domain
- CUL1
cullin-1
- CUL2
cullin-2
- CUL3
cullin-3
- CUL4A
cullin-4A
- CUL4B
cullin-4B
- CUL5
cullin-5
- CUL7
cullin-7
- CUL9
cullin-9
- DCN1
defective in cullin neddylation 1
- DCNL
DCN1-like protein
- DDR
DNA damage response
- DEN1
deneddylase 1
- DEPTOR
DEP domain containing mTOR-interacting protein
- E2F-1
E2F transcription factor 1
- EGFP
enhanced green fluorescent protein
- EGFR
epidermal growth factor receptor
- EPOCH-R chemotherapy
etoposide, prednisone, vincristine, cyclophosphamide, doxorubicin, and rituximab chemotherapy
- FANCD2
Fanconi anemia complementation group D2
- FBXO11
F-box protein 11
- GSK3
glycogen synthase kinase 3
- HIF1
hypoxia-inducible factor-1
- HIF2
hypoxia-inducible factor-2
- HuR
Hu antigen R
- IAPs
inhibitors of apoptosis
- IKKγ
inhibitor of κB kinase gamma
- Keap1
Kelch-like ECH-associated protein 1
- LC3
microtubule-associated protein light chain 3
- MDM2
murine double minute-2
- MRFAP1
mortality factor on chromosome 4 associated protein 1
- mTOR
mammalian target of rapamycin
- mTORC1
mammalian target of rapamycin complex 1
- NAE
NEDD8-activating enzyme
- NAE1
NEDD8-activating enzyme E1 Subunit 1
- NEDD8
neural precursor cell expressed, developmentally downregulated 8
- NEDP1
NEDD8-specific protease 1
- NF-κB
nuclear factor-κB
- NRF2
nuclear factor erythroid 2-related factor 2
- PfUCH54
54-kDa plasmodium falciparum ubiquitin C-terminal hydrolase
- PINK1
PTEN induced putative kinase 1
- RBX1
RING box protein-1
- RBX2
RING box protein-2
- RCAN1
regulator of calcineurin 1
- RING
really interesting new gene
- RNF111
ring finger protein 111
- ROC1
regulator of cullins 1
- ROC2
regulator of cullins 2
- ROS
reactive oxygen species
- SAG
sensitive to apoptosis gene
- SCF
Skp1, cullin, and F-box protein
- SENP8
SUMO/sentrin-specific peptidase family member 8
- SILAC
stable isotope labeling with amino acids in cell culture
- SKP1
S-phase kinase-associated protein 1
- SKP2
S-phase kinase-associated protein 2
- TGFβ-RII
transforming growth factor β type II receptor
- TRIM40
tripartite motif containing 40
- UAE
ubiquitin-activating enzyme
- Ub
ubiquitin
- UBA3
ubiquitin-like modifier activating enzyme 3
- UBC12
ubiquitin-conjugating enzyme 12
- UBE2F
ubiquitin-conjugating enzyme E2F
- UBE2M
ubiquitin-conjugating enzyme E2M
- UBLs
ubiquitin-like proteins
- UCH-L1
ubiquitin carboxyl-terminal esterase L1
- UCH-L3
ubiquitin carboxyl-terminal esterase L3
- UPS
ubiquitin–proteasome system
- USP21
ubiquitin-specific peptidase 21
- VHL
von Hippel-Lindau
Acknowledgments
We would like to thank Millennium Pharmaceuticals, Inc. for providing us with MLN4924, and Dr. Allison Berger for stimulating discussions. This work was supported by the NCI grants (CA118762, CA156744, CA170995, and CA171277) to Yi Sun and Susan G. Komen and the Cure PDF Grant (PDF12230424) to Yongchao Zhao.
References
- 1.Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, and Dutta A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev 22: 2496–2506, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdelmohsen K. and Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip Rev RNA 1: 214–229, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abida WM, Nikolaev A, Zhao W, Zhang W, and Gu W. FBXO11 promotes the neddylation of p53 and inhibits its transcriptional activity. J Biol Chem 282: 1797–1804, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Artavanis-Tsakonas K, Misaghi S, Comeaux CA, Catic A, Spooner E, Duraisingh MT, and Ploegh HL. Identification by functional proteomics of a deubiquitinating/deNeddylating enzyme in Plasmodium falciparum. Mol Microbiol 61: 1187–1195, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bech-Otschir D, Seeger M, and Dubiel W. The COP9 signalosome: at the interface between signal transduction and ubiquitin-dependent proteolysis. J Cell Sci 115: 467–473, 2002 [DOI] [PubMed] [Google Scholar]
- 6.Bjorklund M, Taipale M, Varjosalo M, Saharinen J, Lahdenpera J, and Taipale J. Identification of pathways regulating cell size and cell-cycle progression by RNAi. Nature 439: 1009–1013, 2006 [DOI] [PubMed] [Google Scholar]
- 7.Blank JL, Liu XJ, Cosmopoulos K, Bouck DC, Garcia K, Bernard H, Tayber O, Hather G, Liu R, Narayanan U, Milhollen MA, and Lightcap ES. Novel DNA damage checkpoints mediating cell death induced by the NEDD8-activating enzyme inhibitor MLN4924. Cancer Res 73: 225–234, 2013 [DOI] [PubMed] [Google Scholar]
- 8.Bohnsack RN. and Haas AL. Conservation in the mechanism of Nedd8 activation by the human AppBp1-Uba3 heterodimer. J Biol Chem 278: 26823–26830, 2003 [DOI] [PubMed] [Google Scholar]
- 9.Broemer M, Tenev T, Rigbolt KTG, Hempel S, Blagoev B, Silke J, Ditzel M, and Meier P. Systematic in vivo rnai analysis identifies IAPs as NEDD8-E3 ligases. Mol Cell 40: 810–822, 2010 [DOI] [PubMed] [Google Scholar]
- 10.Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt AL, Ma J, Loke HK, Lingaraj T, Wu D, Hamman KB, Spelman JJ, Cullis CA, Langston SP, Vyskocil S, Sells TB, Mallender WD, Visiers I, Li P, Claiborne CF, Rolfe M, Bolen JB, and Dick LR. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol Cell 37: 102–111, 2010 [DOI] [PubMed] [Google Scholar]
- 11.Carter SA. and Vousden KH. p53-Ubl fusions as models of ubiquitination, sumoylation and neddylation of p53. Cell Cycle 7: 2519–2528, 2008 [DOI] [PubMed] [Google Scholar]
- 12.Chairatvit K. and Ngamkitidechakul C. Control of cell proliferation via elevated NEDD8 conjugation in oral squamous cell carcinoma. Mol Cell Biochem 306: 163–169, 2007 [DOI] [PubMed] [Google Scholar]
- 13.Choo YS, Vogler G, Wang D, Kalvakuri S, Iliuk A, Tao WA, Bodmer R, and Zhang Z. Regulation of parkin and PINK1 by neddylation. Hum Mol Genet 21: 2514–2523, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chowdhry S, Zhang Y, McMahon M, Sutherland C, Cuadrado A, and Hayes JD. Nrf2 is controlled by two distinct beta-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene 32: 3765–3781, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciechanover A. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J 17: 7151–7160, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciechanover A. The ubiquitin proteolytic system and pathogenesis of human diseases: a novel platform for mechanism-based drug targeting. Biochem Soc Trans 31: 474–481, 2003 [DOI] [PubMed] [Google Scholar]
- 17.Ciechanover A. and Iwai K. The ubiquitin system: from basic mechanisms to the patient bed. IUBMB Life 56: 193–201, 2004 [DOI] [PubMed] [Google Scholar]
- 18.Cope GA. and Deshaies RJ. COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 114: 663–671, 2003 [DOI] [PubMed] [Google Scholar]
- 19.Cox J. and Mann M. Quantitative, high-resolution proteomics for data-driven systems biology. Annu Rev Biochem 80: 273–299, 2011 [DOI] [PubMed] [Google Scholar]
- 20.de Oca Luna RM, Wagner DS, and Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378: 203–206, 1995 [DOI] [PubMed] [Google Scholar]
- 21.Deshaies RJ, Emberley ED, and Saha A. Control of cullin-RING ubiquitin ligase activity by Nedd8. In: Conjugation and Deconjugation of Ubiquitin Family Modifiers: Subcellular Biochemistry, edited by Groettrup M. New York: Springer, 2010, pp. 41–56 [DOI] [PubMed] [Google Scholar]
- 22.Deshaies RJ. and Joazeiro CAP. RING Domain E3 Ubiquitin Ligases. Annu Rev Biochem 78: 399–434, 2009 [DOI] [PubMed] [Google Scholar]
- 23.Dharmasiri S, Dharmasiri N, Hellmann H, and Estelle M. The RUB/Nedd8 conjugation pathway is required for early development in Arabidopsis. EMBO J 22: 1762–1770, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dil Kuazi A, Kito K, Abe Y, Shin R-W, Kamitani T, and Ueda N. NEDD8 protein is involved in ubiquitinated inclusion bodies. J Pathol 199: 259–266, 2003 [DOI] [PubMed] [Google Scholar]
- 25.Dohmesen C, Koeppel M, and Dobbelstein M. Specific inhibition of Mdm2-mediated neddylation by Tip60. Cell Cycle 7: 222–231, 2008 [DOI] [PubMed] [Google Scholar]
- 26.Duan H, Wang Y, Aviram M, Swaroop M, Loo JA, Bian J, Tian Y, Mueller T, Bisgaier CL, and Sun Y. SAG, a novel zinc RING finger protein that protects cells from apoptosis induced by redox agents. Mol Cell Biol 19: 3145–3155, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duan S, Skaar Jeffrey R, Kuchay S, Toschi A, Kanarek N, Ben-Neriah Y, and Pagano M. mTOR generates an auto-amplification loop by triggering the βTrCP- and CK1α-dependent degradation of DEPTOR. Mol Cell 44: 317–324, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, and Schulman BA. Structural insights into NEDD8 activation of Cullin-RING ligases: conformational control of conjugation. Cell 134: 995–1006, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emanuele Michael J, Elia Andrew EH, Xu Q, Thoma Claudio R, Izhar L, Leng Y, Guo A, Chen Y-N, Rush J, Hsu Paul W-C, Yen H-Chi S, and Elledge Stephen J. Global identification of modular cullin-RING ligase substrates. Cell 147: 459–474, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Embade N, Fernández-Ramos D, Varela-Rey M, Beraza N, Sini M, de Juan VG, Woodhoo A, Martínez-López N, Rodríguez-Iruretagoyena B, Bustamante FJ, de la Hoz AB, Carracedo A, Xirodimas DP, Rodríguez MS, Lu SC, Mato JM, and Martínez-Chantar ML. Murine double minute 2 regulates Hu antigen R stability in human liver and colon cancer through NEDDylation. Hepatology 55: 1237–1248, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferro A, Carvalho AL, Teixeira-Castro A, Almeida C, Tomé RJ, Cortes L, Rodrigues A-J, Logarinho E, Sequeiros J, Macedo-Ribeiro S, and Maciel P. NEDD8: a new ataxin-3 interactor. Biochim Biophys Acta 1773: 1619–1627, 2007 [DOI] [PubMed] [Google Scholar]
- 32.Finkel T. Signal transduction by reactive oxygen species. J Cell Biol 194: 7–15, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gan-Erdene T, Nagamalleswari K, Yin L, Wu K, Pan ZQ, and Wilkinson KD. Identification and characterization of DEN1, a deneddylase of the ULP family. J Biol Chem 278: 28892–28900, 2003 [DOI] [PubMed] [Google Scholar]
- 34.Gao D, Inuzuka H, Tan M-Kwang M, Fukushima H, Locasale Jason W, Liu P, Wan L, Zhai B, Chin YR, Shaik S, Lyssiotis Costas A, Gygi Steven P, Toker A, Cantley Lewis C, Asara John M, Harper JW, and Wei W. mTOR drives its own activation via SCFβTrCP-dependent degradation of the mTOR inhibitor DEPTOR. Mol Cell 44: 290–303, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao F, Cheng J, Shi T, and Yeh ETH. Neddylation of a breast cancer-associated protein recruits a class III histone deacetylase that represses NF[kappa]B-dependent transcription. Nat Cell Biol 8: 1171–1177, 2006 [DOI] [PubMed] [Google Scholar]
- 36.Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu JD, Xiong Y, and Zheng N. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell 119: 517–528, 2004 [DOI] [PubMed] [Google Scholar]
- 37.Gong L, Kamitani T, Millas S, and Yeh ETH. Identification of a novel isopeptidase with dual specificity for ubiquitin- and NEDD8-conjugated proteins. J Biol Chem 275: 14212–14216, 2000 [DOI] [PubMed] [Google Scholar]
- 38.Handeli S. and Weintraub H. The ts41 mutation in Chinese hamster cells leads to successive S phases in the absence of intervening G2, M, and G1. Cell 71: 599–611, 1992 [DOI] [PubMed] [Google Scholar]
- 39.Hayes JD. and McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci 34: 176–188, 2009 [DOI] [PubMed] [Google Scholar]
- 40.Hemelaar J, Borodovsky A, Kessler BM, Reverter D, Cook J, Kolli N, Gan-Erdene T, Wilkinson KD, Gill G, Lima CD, Ploegh HL, and Ovaa H. Specific and covalent targeting of conjugating and deconjugating enzymes of ubiquitin-like proteins. Mol Cell Biol 24: 84–95, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hershko A. and Ciechanover A. The ubiquitin system. Annu Rev Biochem 67: 425–479, 1998 [DOI] [PubMed] [Google Scholar]
- 42.Hu J, McCall CM, Ohta T, and Xiong Y. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat Cell Biol 6: 1003–1009, 2004 [DOI] [PubMed] [Google Scholar]
- 43.Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, Scott DC, Borg LA, Neale G, Murray PJ, Roussel MF, and Schulman BA. E2-RING Expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell 33: 483–495, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Itoh K, Mimura J, and Yamamoto M. Discovery of the negative regulator of Nrf2, Keap1: a historical overview. Antioxid Redox Signal 13: 1665–1678, 2010 [DOI] [PubMed] [Google Scholar]
- 45.Ivan M, Kondo K, Yang HF, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, and Kaelin WG. HIF alpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O-2 sensing. Science 292: 464–468, 2001 [DOI] [PubMed] [Google Scholar]
- 46.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, and Ratcliffe PJ. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O-2-regulated prolyl hydroxylation. Science 292: 468–472, 2001 [DOI] [PubMed] [Google Scholar]
- 47.Jazaeri AA, Shibata E, Park J, Bryant JL, Conaway MR, Modesitt SC, Smith PG, Milhollen MA, Berger AJ, and Dutta A. Overcoming platinum resistance in preclinical models of ovarian cancer using the neddylation inhibitor MLN4924. Mol Cancer Ther 12: 1958–1967, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jia L, Li H, and Sun Y. Induction of p21-dependent senescence by an NAE inhibitor, MLN4924, as a mechanism of growth suppression. Neoplasia 13: 561–569, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jia L. and Sun Y. SCF E3 ubiquitin ligases as anticancer targets. Curr Cancer Drug Targets 11: 347–356, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jia L, Yang J, Hao X, Zheng M, He H, Xiong X, Xu L, and Sun Y. Validation of SAG/RBX2/ROC2 E3 ubiquitin ligase as an anticancer and radiosensitizing target. Clin Cancer Res 16: 814–824, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jin H-s, Liao L, Park Y, and Liu Y-C. Neddylation pathway regulates T-cell function by targeting an adaptor protein Shc and a protein kinase Erk signaling. Proc Natl Acad Sci U S A 110: 624–629, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnston SC, Larsen CN, Cook WJ, Wilkinson KD, and Hill CP. Crystal structure of a deubiquitinating enzyme (human UCH-L3) at 1.8 A resolution. EMBO J 16: 3787–3796, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones D. and Candido EPM. The NED-8 conjugating system in Caenorhabditis elegans is required for embryogenesis and terminal differentiation of the hypodermis. Dev Biol 226: 152–165, 2000 [DOI] [PubMed] [Google Scholar]
- 54.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP, Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR, Kern SE, Hruban RH, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, and Kinzler KW. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321: 1801–1806, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones SN, Roe AE, Donehower LA, and Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378: 206–208, 1995 [DOI] [PubMed] [Google Scholar]
- 56.Kamitani T, Kito K, Nguyen HP, and Yeh ETH. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem 272: 28557–28562, 1997 [DOI] [PubMed] [Google Scholar]
- 57.Kamura T, Conrad MN, Yan Q, Conaway RC, and Conaway JW. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev 13: 2928–2933, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kee Y, Huang M, Chang S, Moreau LA, Park E, Smith PG, and D'Andrea AD. Inhibition of the Nedd8 system sensitizes cells to DNA interstrand cross-linking agents. Mol Cancer Res 10: 369–377, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kensler TW, Wakabayashi N, and Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol 47: 89–116, 2007 [DOI] [PubMed] [Google Scholar]
- 60.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW, and Gygi SP. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell 44: 325–340, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim Y, Starostina NG, and Kipreos ET. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev 22: 2507–2519, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumar A, Wu H, Collier-Hyams LS, Hansen JM, Li T, Yamoah K, Pan ZQ, Jones DP, and Neish AS. Commensal bacteria modulate cullin-dependent signaling via generation of reactive oxygen species. EMBO J 26: 4457–4466, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kumar A, Wu H, Collier-Hyams LS, Kwon YM, Hanson JM, and Neish AS. The bacterial fermentation product butyrate influences epithelial signaling via reactive oxygen species-mediated changes in cullin-1 neddylation. J Immunol 182: 538–546, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumar S, Tomooka Y, and Noda M. Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem Biophys Res Commun 185: 1155–1161, 1992 [DOI] [PubMed] [Google Scholar]
- 65.Kumar S, Yoshida Y, and Noda M. Cloning of a cDNA which encodes a novel ubiquitin-like protein. Biochem Biophys Res Commun 195: 393–399, 1993 [DOI] [PubMed] [Google Scholar]
- 66.Kurz T, Chou Y-C, Willems AR, Meyer-Schaller N, Hecht M-L, Tyers M, Peter M, and Sicheri F. Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol Cell 29: 23–35, 2008 [DOI] [PubMed] [Google Scholar]
- 67.Kurz T, Ozlu N, Rudolf F, O'Rourke SM, Luke B, Hofmann K, Hyman AA, Bowerman B, and Peter M. The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature 435: 1257–1261, 2005 [DOI] [PubMed] [Google Scholar]
- 68.Kurz T, Pintard L, Willis JH, Hamill DR, Gönczy P, Peter M, and Bowerman B. Cytoskeletal regulation by the Nedd8 ubiquitin-like protein modification pathway. Science 295: 1294–1298, 2002 [DOI] [PubMed] [Google Scholar]
- 69.Lammer D, Mathias N, Laplaza JM, Jiang W, Liu Y, Callis J, Goebl M, and Estelle M. Modification of yeast Cdc53p by the ubiquitin-related protein Rub1p affects function of the SCFCdc4 complex. Genes Dev 12: 914–926, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Larance M, Kirkwood KJ, Xirodimas DP, Lundberg E, Uhlen M, and Lamond AI. Characterization of MRFAP1 turnover and interactions downstream of the NEDD8 pathway. Mol Cell Proteomics 11: M111.014407, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Larsen CN, Price JS, and Wilkinson KD. Substrate binding and catalysis by ubiquitin C-terminal hydrolases: identification of two active site residues. Biochemistry 35: 6735–6744, 1996 [DOI] [PubMed] [Google Scholar]
- 72.Lee M-R, Lee D, Shin SK, Kim YH, and Choi CY. Inhibition of APP intracellular domain (AICD) transcriptional activity via covalent conjugation with Nedd8. Biochem Biophys Res Commun 366: 976–981, 2008 [DOI] [PubMed] [Google Scholar]
- 73.Li X, Zhao Q, Liao R, Sun P, and Wu X. The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J Biol Chem 278: 30854–30858, 2003 [DOI] [PubMed] [Google Scholar]
- 74.Liakopoulos D, Doenges G, Matuschewski K, and Jentsch S. A novel protein modification pathway related to the ubiquitin system. EMBO J 17: 2208–2214, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liao H, Liu XJ, Blank JL, Bouck DC, Bernard H, Garcia K, and Lightcap ES. Quantitative proteomic analysis of cellular protein modulation upon inhibition of the NEDD8-activating enzyme by MLN4924. Mol Cell Proteomics 10: M111.009183, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin HK, Chen Z, Wang G, Nardella C, Lee SW, Chan CH, Yang WL, Wang J, Egia A, Nakayama KI, Cordon-Cardo C, Teruya-Feldstein J, and Pandolfi PP. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature 464: 374–379, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lin JJ, Milhollen MA, Smith PG, Narayanan U, and Dutta A. NEDD8-targeting drug MLN4924 elicits DNA rereplication by stabilizing Cdt1 in S phase, triggering checkpoint activation, apoptosis, and senescence in cancer cells. Cancer Res 70: 10310–10320, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu J, Furukawa M, Matsumoto T, and Xiong Y. NEDD8 modification of CUL1 dissociates p120CAND1, an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol Cell 10: 1511–1518, 2002 [DOI] [PubMed] [Google Scholar]
- 79.Lo S-C. and Hannink M. CAND1-Mediated substrate adaptor recycling is required for efficient repression of Nrf2 by Keap1. Mol Cell Biol 26: 1235–1244, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Loftus SJ, Liu G, Carr SM, Munro S, and La Thangue NB. NEDDylation regulates E2F-1-dependent transcription. EMBO Rep 13: 811–818, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, and Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3: 577–587, 2003 [DOI] [PubMed] [Google Scholar]
- 82.Luo Z, Yu G, Lee HW, Li L, Wang L, Yang D, Pan Y, Ding C, Qian J, Wu L, Chu Y, Yi J, Wang X, Sun Y, Jeong LS, Liu J, and Jia L. The Nedd8-activating enzyme inhibitor MLN4924 induces autophagy and apoptosis to suppress liver cancer cell growth. Cancer Res 72: 3360–3371, 2012 [DOI] [PubMed] [Google Scholar]
- 83.Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Shevchenko A, and Deshaies RJ. Promotion of NEDD8-CUL1 conjugate cleavage by COP9 signalosome. Science 292: 1382–1385, 2001 [DOI] [PubMed] [Google Scholar]
- 84.Ma T, Chen Y, Zhang F, Yang C-Y, Wang S, and Yu X. RNF111-Dependent neddylation activates DNA damage-induced ubiquitination. Mol Cell 49: 897–907, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mackintosh C, Garcia-Dominguez DJ, Ordonez JL, Ginel-Picardo A, Smith PG, Sacristan MP, and de Alava E. WEE1 accumulation and deregulation of S-phase proteins mediate MLN4924 potent inhibitory effect on Ewing sarcoma cells. Oncogene 32: 1441–1451, 2013 [DOI] [PubMed] [Google Scholar]
- 86.Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol 7: 952–958, 2006 [DOI] [PubMed] [Google Scholar]
- 87.Martin D, Salinas M, Fujita N, Tsuruo T, and Cuadrado A. Ceramide and reactive oxygen species generated by H2O2 induce caspase-3-independent degradation of Akt/protein kinase B. J Biol Chem 277: 42943–42952, 2002 [DOI] [PubMed] [Google Scholar]
- 88.McMillin DW, Jacobs HM, Delmore JE, Buon L, Hunter ZR, Monrose V, Yu J, Smith PG, Richardson PG, Anderson KC, Treon SP, Kung AL, and Mitsiades CS. Molecular and cellular effects of NEDD8-activating enzyme inhibition in myeloma. Mol Cancer Ther 11: 942–951, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mendoza HM, Shen LN, Botting C, Lewis A, Chen J, Ink B, and Hay RT. NEDP1, a highly conserved cysteine protease that deNEDDylates Cullins. J Biol Chem 278: 25637–25643, 2003 [DOI] [PubMed] [Google Scholar]
- 90.Meyer-Schaller N, Chou Y-C, Sumara I, Martin DDO, Kurz T, Katheder N, Hofmann K, Berthiaume LG, Sicheri F, and Peter M. The human Dcn1-like protein DCNL3 promotes Cul3 neddylation at membranes. Proc Natl Acad Sci U S A 106: 12365–12370, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Milhollen MA, Narayanan U, Soucy TA, Veiby PO, Smith PG, and Amidon B. Inhibition of NEDD8-activating enzyme induces rereplication and apoptosis in human tumor cells consistent with deregulating CDT1 turnover. Cancer Res 71: 3042–3051, 2011 [DOI] [PubMed] [Google Scholar]
- 92.Milhollen MA, Thomas MP, Narayanan U, Traove T, Riceberg J, Amidon BS, Bence NF, Bolen JB, Brownell J, Dick LR, Loke HK, McDonald AA, Ma J, Manfredi MG, Sells TB, Sintchak MD, Yang X, Xu Q, Koenig EM, Gavin JM, and Smith PG. Treatment-emergent mutations in NAEbeta confer resistance to the NEDD8-activating enzyme inhibitor MLN4924. Cancer Cell 21: 388–401, 2012 [DOI] [PubMed] [Google Scholar]
- 93.Mihollen MA, Traore T, Adams-Duffy J, Thomas MP, Berger AJ, Dang L, Dick LR, Garnsey JJ, Koenig E, Langston SP, Manfredi M, Narayanan U, Rolfe M, Staudt LM, Soucy TA, Yu J, Zhang J, Bolen JB, and Smith PG. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-{kappa}B-dependent lymphoma. Blood 116: 1515–1523, 2010 [DOI] [PubMed] [Google Scholar]
- 94.Mizushima N, Yoshimori T, and Levine B. Methods in mammalian autophagy research. Cell 140: 313–326, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Morgan MA, Parsels LA, Maybaum J, and Lawrence TS. Improving gemcitabine-mediated radiosensitization using molecularly targeted therapy: a review. Clin Cancer Res 14: 6744–6750, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mori F, Nishie M, Piao YS, Kito K, Kamitani T, Takahashi H, and Wakabayashi K. Accumulation of NEDD8 in neuronal and glial inclusions of neurodegenerative disorders. Neuropathol Appl Neurobiol 31: 53–61, 2005 [DOI] [PubMed] [Google Scholar]
- 97.Nagano T, Hashimoto T, Nakashima A, Kikkawa U, and Kamada S. X-linked inhibitor of apoptosis protein mediates neddylation by itself but does not function as a NEDD8–E3 ligase for caspase-7. FEBS Lett 586: 1612–1616, 2012 [DOI] [PubMed] [Google Scholar]
- 98.Nawrocki ST, Griffin P, Kelly KR, and Carew JS. MLN4924: a novel first-in-class inhibitor of NEDD8-activating enzyme for cancer therapy. Expert Opin Investig Drugs 21: 1563–1573, 2012 [DOI] [PubMed] [Google Scholar]
- 99.Nawrocki ST, Kelly KR, Smith PG, Espitia CM, Possemato A, Beausoleil SA, Milhollen M, Blakemore S, Thomas M, Berger A, and Carew JS. Disrupting protein NEDDylation with MLN4924 is a novel strategy to target cisplatin resistance in ovarian cancer. Clin Cancer Res 19: 3577–3590, 2013 [DOI] [PubMed] [Google Scholar]
- 100.Noguchi K, Okumura F, Takahashi N, Kataoka A, Kamiyama T, Todo S, and Hatakeyama S. TRIM40 promotes neddylation of IKKγ and is downregulated in gastrointestinal cancers. Carcinogenesis 32: 995–1004, 2011 [DOI] [PubMed] [Google Scholar]
- 101.Noh EH, Hwang HS, Hwang HS, Min B, Im E, and Chung KC. Covalent NEDD8 conjugation increases RCAN1 protein stability and potentiates its inhibitory action on calcineurin. PLos One 7: e48315, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ong S-E. The expanding field of SILAC. Anal Bioanal Chem 404: 967–976, 2012 [DOI] [PubMed] [Google Scholar]
- 103.Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, and Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 1: 376–386, 2002 [DOI] [PubMed] [Google Scholar]
- 104.Ong SE. and Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat Protoc 1: 2650–2660, 2006 [DOI] [PubMed] [Google Scholar]
- 105.Orlowski RZ. and Kuhn DJ. Proteasome inhibitors in cancer therapy: lessons from the first decade. Clin Cancer Res 14: 1649–1657, 2008 [DOI] [PubMed] [Google Scholar]
- 106.Osaka F, Saeki M, Katayama S, Aida N, Toh-e A, Kominami K-i, Toda T, Suzuki T, Chiba T, Tanaka K, and Kato S. Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J 19: 3475–3484, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ou C-Y, Lin Y-F, Chen Y-J, and Chien C-T. Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes Dev 16: 2403–2414, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Oved S, Mosesson Y, Zwang Y, Santonico E, Shtiegman K, Marmor MD, Kochupurakkal BS, Katz M, Lavi S, Cesareni G, and Yarden Y. Conjugation to Nedd8 instigates ubiquitylation and down-regulation of activated receptor tyrosine kinases. J Biol Chem 281: 21640–21651, 2006 [DOI] [PubMed] [Google Scholar]
- 109.Pan Z-Q, Kentsis A, Dias DC, Yamoah K, and Wu K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene 23: 1985–1997, 2004 [DOI] [PubMed] [Google Scholar]
- 110.Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, and Sabatini DM. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 137: 873–886, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pozo JCd, Timpte C, Tan S, Callis J, and Estelle M. The ubiquitin-related protein RUB1 and auxin response in arabidopsis. Science 280: 1760–1763, 1998 [DOI] [PubMed] [Google Scholar]
- 112.Rabut G, Le Dez G, Verma R, Makhnevych T, Knebel A, Kurz T, Boone C, Deshaies Raymond J, and Peter M. The TFIIH subunit Tfb3 regulates cullin neddylation. Mol Cell 43: 488–495, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rabut G. and Peter M. Function and regulation of protein neddylation. ‘Protein modifications: beyond the usual suspects' review series. EMBO Rep 9: 969–976, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rada P, Rojo AI, Chowdhry S, McMahon M, Hayes JD, and Cuadrado A. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol 31: 1121–1133, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rada P, Rojo AI, Evrard-Todeschi N, Innamorato NG, Cotte A, Jaworski T, Tobon-Velasco JC, Devijver H, Garcia-Mayoral MF, Van Leuven F, Hayes JD, Bertho G, and Cuadrado A. Structural and functional characterization of Nrf2 degradation by the glycogen synthase kinase 3/beta-TrCP axis. Mol Cell Biol 32: 3486–3499, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Reverter D, Wu K, Erdene TG, Pan Z-Q, Wilkinson KD, and Lima CD. Structure of a complex between Nedd8 and the Ulp/Senp protease family member Den1. J Mol Biol 345: 141–151, 2005 [DOI] [PubMed] [Google Scholar]
- 117.Rushmore TH, Morton MR, and Pickett CB. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J Biol Chem 266: 11632–11639, 1991 [PubMed] [Google Scholar]
- 118.Ryu J-H, Li S-H, Park H-S, Park J-W, Lee B, and Chun Y-S. Hypoxia-inducible factor α subunit stabilization by NEDD8 conjugation is reactive oxygen species-dependent. J Biol Chem 286: 6963–6970, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Saha A. and Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell 32: 21–31, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Salon C, Brambilla E, Brambilla C, Lantuejoul S, Gazzeri S, and Eymin B. Altered pattern of Cul-1 protein expression and neddylation in human lung tumours: relationships with CAND1 and cyclin E protein levels. J Pathol 213: 303–310, 2007 [DOI] [PubMed] [Google Scholar]
- 121.Scott DC, Monda JK, Bennett EJ, Harper JW, and Schulman BA. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science 334: 674–678, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shen L-n, Liu H, Dong C, Xirodimas D, Naismith JH, and Hay RT. Structural basis of NEDD8 ubiquitin discrimination by the deNEDDylating enzyme NEDP1. EMBO J 24: 1341–1351, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Singh RK, Iyappan S, and Scheffner M. Hetero-oligomerization with MdmX rescues the ubiquitin/Nedd8 ligase activity of RING finger mutants of Mdm2. J Biol Chem 282: 10901–10907, 2007 [DOI] [PubMed] [Google Scholar]
- 124.Skaar JR. and Pagano M. Control of cell growth by the SCF and APC/C ubiquitin ligases. Curr Opin Cell Biol 21: 816–824, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Smith MA, Maris JM, Gorlick R, Kolb EA, Lock R, Carol H, Keir ST, Reynolds CP, Kang MH, Morton CL, Wu J, Smith PG, Yu J, and Houghton PJ. Initial testing of the investigational NEDD8-activating enzyme inhibitor MLN4924 by the pediatric preclinical testing program. Pediatr Blood Cancer 59: 246–253, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Soucy TA, Dick LR, Smith PG, Milhollen MA, and Brownell JE. The NEDD8 conjugation pathway and its relevance in cancer biology and therapy. Genes Cancer 1: 708–716, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, Cullis CA, Doucette A, Garnsey JJ, Gaulin JL, Gershman RE, Lublinsky AR, McDonald A, Mizutani H, Narayanan U, Olhava EJ, Peluso S, Rezaei M, Sintchak MD, Talreja T, Thomas MP, Traore T, Vyskocil S, Weatherhead GS, Yu J, Zhang J, Dick LR, Claiborne CF, Rolfe M, Bolen JB, and Langston SP. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458: 732–736, 2009 [DOI] [PubMed] [Google Scholar]
- 128.Soucy TA, Smith PG, and Rolfe M. Targeting NEDD8-activated cullin-RING ligases for the treatment of cancer. Clin Cancer Res 15: 3912–3916, 2009 [DOI] [PubMed] [Google Scholar]
- 129.Sporn MB. and Liby KT. NRF2 and cancer: the good, the bad and the importance of context. Nat Rev Cancer 12: 564–571, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Stickle NH, Chung J, Klco JM, Hill RP, Kaelin WG, and Ohh M. pVHL Modification by NEDD8 is required for fibronectin matrix assembly and suppression of tumor development. Mol Cell Biol 24: 3251–3261, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sun Y. Free radicals, antioxidant enzymes, and carcinogenesis. Free Radic Biol Med 8: 583–599, 1990 [DOI] [PubMed] [Google Scholar]
- 132.Sun Y. and Li H. Functional characterization of SAG/RBX2/ROC2/RNF7, an antioxidant protein and an E3 ubiquitin ligase. Protein Cell 4: 103–116, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sundqvist A, Liu G, Mirsaliotis A, and Xirodimas DP. Regulation of nucleolar signalling to p53 through NEDDylation of L11. EMBO Rep 10: 1132–1139, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Swords RT, Kelly KR, Smith PG, Garnsey JJ, Mahalingam D, Medina E, Oberheu K, Padmanabhan S, O'Dwyer M, Nawrocki ST, Giles FJ, and Carew JS. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood 115: 3796–3800, 2010 [DOI] [PubMed] [Google Scholar]
- 135.Tan M, Li Y, Yang R, Xi N, and Sun Y. Inactivation of SAG E3 ubiquitin ligase blocks embryonic stem cell differentiation and sensitizes leukemia cells to retinoid acid. PLoS One 6: e27726, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tan M, Zhao Y, Kim S-J, Liu M, Jia L, Saunders Thomas L, Zhu Y, and Sun Y. SAG/RBX2/ROC2 E3 ubiquitin ligase is essential for vascular and neural development by targeting NF1 for degradation. Dev Cell 21: 1062–1076, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tateishi K, Omata M, Tanaka K, and Chiba T. The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J Cell Biol 155: 571–580, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tonks NK. Redox redux: revisiting PTPs and the control of cell signaling. Cell 121: 667–670, 2005 [DOI] [PubMed] [Google Scholar]
- 139.Toth Julia I, Yang L, Dahl R, and Petroski Matthew D. A gatekeeper residue for NEDD8-activating enzyme inhibition by MLN4924. Cell Rep 1: 309–316, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Um JW, Han KA, Im E, Oh Y, Lee K, and Chung KC. Neddylation positively regulates the ubiquitin E3 ligase activity of parkin. J Neurosci Res 90: 1030–1042, 2012 [DOI] [PubMed] [Google Scholar]
- 141.Villeneuve NF, Lau A, and Zhang DD. Regulation of the Nrf2-Keap1 antioxidant response by the ubiquitin proteasome system: an insight into cullin-ring ubiquitin ligases. Antioxid Redox Signal 13: 1699–1712, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Watson IR, Blanch A, Lin DCC, Ohh M, and Irwin MS. Mdm2-mediated NEDD8 modification of TAp73 regulates its transactivation function. J Biol Chem 281: 34096–34103, 2006 [DOI] [PubMed] [Google Scholar]
- 143.Watson IR, Irwin MS, and Ohh M. NEDD8 pathways in cancer, Sine Quibus Non. Cancer Cell 19: 168–176, 2011 [DOI] [PubMed] [Google Scholar]
- 144.Watson IR, Li BK, Roche O, Blanch A, Ohh M, and Irwin MS. Chemotherapy induces NEDP1-mediated destabilization of MDM2. Oncogene 29: 297–304, 2009 [DOI] [PubMed] [Google Scholar]
- 145.Wei D, Li H, Yu J, Sebolt JT, Zhao L, Lawrence TS, Smith PG, Morgan MA, and Sun Y. Radiosensitization of human pancreatic cancer cells by MLN4924, an investigational NEDD8-activating enzyme inhibitor. Cancer Res 72: 282–293, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wei N. and Deng XW. The COP9 signalosome. Annu Rev Cell Dev Biol 19: 261–286, 2003 [DOI] [PubMed] [Google Scholar]
- 147.Wei N, Serino G, and Deng XW. The COP9 signalosome: more than a protease. Trends Biochem Sci 33: 592–600, 2008 [DOI] [PubMed] [Google Scholar]
- 148.Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, and Harper JW. The SCFβ-TRCP–ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev 13: 270–283, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wu K, Yamoah K, Dolios G, Gan-Erdene T, Tan P, Chen A, Lee CG, Wei N, Wilkinson KD, Wang R, and Pan ZQ. DEN1 is a dual function protease capable of processing the C terminus of Nedd8 and deconjugating hyper-neddylated CUL1. J Biol Chem 278: 28882–28891, 2003 [DOI] [PubMed] [Google Scholar]
- 150.Xirodimas DP, Saville MK, Bourdon J-C, Hay RT, and Lane DP. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell 118: 83–97, 2004 [DOI] [PubMed] [Google Scholar]
- 151.Xirodimas DP, Sundqvist A, Nakamura A, Shen L, Botting C, and Hay RT. Ribosomal proteins are targets for the NEDD8 pathway. EMBO Rep 9: 280–286, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yamoah K, Oashi T, Sarikas A, Gazdoiu S, Osman R, and Pan Z-Q. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1's C-terminal tail. Proc Natl Acad Sci U S A 105: 12230–12235, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yang D, Li L, Liu H, Wu L, Luo Z, Li H, Zheng S, Gao H, Chu Y, Sun Y, Liu J, and Jia L. Induction of autophagy and senescence by knockdown of ROC1 E3 ubiquitin ligase to suppress the growth of liver cancer cells. Cell Death Differ 20: 235–247, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yang D, Tan M, Wang G, and Sun Y. The p21-dependent radiosensitization of human breast cancer cells by MLN4924, an investigational inhibitor of NEDD8 activating enzyme. PLos One 7: e34079, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yang D, Zhao Y, Liu J, Sun Y, and Jia L. Protective autophagy induced by RBX1/ROC1 knockdown or CRL inactivation via modulating the DEPTOR-MTOR axis. Autophagy 8: 1856–1858, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yen H-CS, Xu Q, Chou DM, Zhao Z, and Elledge SJ. Global protein stability profiling in mammalian cells. Science 322: 918–923, 2008 [DOI] [PubMed] [Google Scholar]
- 157.Yu Z-K, Gervais JLM, and Zhang H. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21CIP1/WAF1 and cyclin D proteins. Proc Natl Acad Sci U S A 95: 11324–11329, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhang DD, Lo SC, Cross JV, Templeton DJ, and Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol 24: 10941–10953, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhang J, Bai D, Ma X, Guan J, and Zheng X. hCINAP is a novel regulator of ribosomal protein-HDM2-p53 pathway by controlling NEDDylation of ribosomal protein S14. Oncogene 33: 246–254, 2014 [DOI] [PubMed] [Google Scholar]
- 160.Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, and Xiong Y. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol 23: 8902–8912, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhao L, Yue P, Lonial S, Khuri FR, and Sun SY. The NEDD8-activating enzyme inhibitor, MLN4924, cooperates with TRAIL to augment apoptosis through facilitating c-FLIP degradation in head and neck cancer cells. Mol Cancer Ther 10: 2415–2425, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhao Y. and Sun Y. Targeting the mTOR-DEPTOR pathway by CRL E3 ubiquitin ligases: therapeutic application. Neoplasia (New York, NY) 14: 360–367, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhao Y. and Sun Y. Cullin-RING ligases as attractive anti-cancer targets. Curr Pharm Des 19: 3215–3225, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhao Y, Xiong X, Jia L, and Sun Y. Targeting Cullin-RING ligases by MLN4924 induces autophagy via modulating the HIF1-REDD1-TSC1-mTORC1-DEPTOR axis. Cell Death Dis 3: e386, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhao Y, Xiong X, and Sun Y. DEPTOR, an mTOR inhibitor, is a physiological substrate of SCFβTrCP E3 ubiquitin ligase and regulates survival and autophagy. Mol Cell 44: 304–316, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zheng J, Yang X, Harrell JM, Ryzhikov S, Shim E-H, Lykke-Andersen K, Wei N, Sun H, Kobayashi R, and Zhang H. CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol Cell 10: 1519–1526, 2002 [DOI] [PubMed] [Google Scholar]
- 167.Zheng N, Schulman BA, Song LZ, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, and Pavletich NP. Structure of the Cul1-Rbx1-Skp1-F box(Skp2) SCF ubiquitin ligase complex. Nature 416: 703–709, 2002 [DOI] [PubMed] [Google Scholar]
- 168.Zhou W, Wei W, and Sun Y. Genetically engineered mouse models for functional studies of SKP1-CUL1-F-box-protein (SCF) E3 ubiquitin ligases. Cell Res 23: 599–619, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhou X, Hao Q, Liao J, Zhang Q, and Lu H. Ribosomal protein S14 unties the MDM2-p53 loop upon ribosomal stress. Oncogene 32: 388–396, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zuo W, Huang F, Chiang YJ, Li M, Du J, Ding Y, Zhang T, Lee Hyuk W, Jeong Lak S, Chen Y, Deng H, Feng X-H, Luo S, Gao C, and Chen Y-G. c-Cbl-mediated neddylation antagonizes ubiquitination and degradation of the TGF-β Type II Receptor. Mol Cell 49: 499–510, 2013 [DOI] [PubMed] [Google Scholar]