Abstract
Background
The efficacy of artemisinin-based combination therapy (ACT) for Plasmodium falciparum malaria may be threatened by parasites with reduced responsiveness to artemisinins. Among 298 ACT-treated children from Mbita, Kenya, submicroscopic persistence of P. falciparum on day 3 posttreatment was associated with subsequent microscopically detected parasitemia at days 28 or 42.
Methods
DNA sequences of resistance-associated parasite loci pfcrt, pfmdr1, pfubp1, and pfap2mu were determined in the Mbita cohort before treatment, on days 2 and 3 after initiation of treatment, and on the day of treatment failure.
Results
Parasites surviving ACT on day 2 or day 3 posttreatment were significantly more likely than the baseline population to carry the wild-type haplotypes of pfcrt (CVMNK at codons 72–76; P < .001) and pfmdr1 (NFD at codons 86, 184, 1246; P < .001). In contrast, variant alleles of the novel candidate resistance genes pfap2mu (S160N/T; P = .006) and pfubp-1 (E1528D; P < .001) were significantly more prevalent posttreatment. No genetic similarities were found to artemisinin-tolerant parasites recently described in Cambodia.
Conclusions
Among treated children in western Kenya, certain P. falciparum genotypes defined at pfcrt, pfmdr1, pfap2mu, and pfubp1 more often survive ACT at the submicroscopic level, and contribute to onward transmission and subsequent patent recrudescence.
Keywords: artemisinin, drug resistance, genetics, malaria
Over the last decade, the widespread deployment of artemisinin-based combination therapy (ACT), with improved vector control and other measures, has contributed to a reduction of malaria-related morbidity and mortality across sub–Saharan Africa [1, 2]. These public health gains may be lost if the efficacy of ACT against Plasmodium falciparum is not sustained. Recent evidence suggests that parasite genotypes have arisen in Cambodia and surrounding countries that are less rapidly cleared by artesunate monotherapy [3–6]. This has been interpreted by some as bona fide emerging artemisinin resistance [4, 6], and by others as “treatment failure,” with full-blown resistance to ACT not yet present [7]. The efficacy of ACT remains high in Africa, with no reports to date of slow clearance as in Cambodia. However, evidence based on classical microscopic parasite detection suggests that a proportion of ACT-treated children in Kenya do not completely clear P. falciparum parasitemia [8]. More recently, quantitative polymerase chain reaction (qPCR)–detectable submicroscopic residual parasitemia on day 2 or 3 post-ACT in Kenyan children was shown to be a significant risk factor for parasite transmission to mosquitoes, and for microscopically detected parasitemia recurring on day 28 or 42 [9]. Thus, residual asexual parasitemia, if genetically determined, may provide an enhanced likelihood of infecting the vector population, and thus further human hosts. Such evolutionary success, where parasites harboring genetic markers of drug resistance exhibit a transmission advantage, has previously been implicated in the development of resistance to chloroquine and antifolate malaria drugs [10–12].
The possibility that P. falciparum populations in Kenya may be less responsive to ACT therapy than previously reported provides a strong rationale for further investigation, for 2 reasons. First, identification of any heritable genetic signature for this phenotype would enable monitoring of its spread by tracking relevant molecular markers. Second, it is important to determine whether the submicroscopic persisting parasite phenotype described in Kenya is related to the more pronounced microscopically detectable slow clearance described from western Cambodia [3, 4], given important differences in transmission intensity, population prevalence of acquired immunity, and treatment policy. Genetic characterization of the persisting Kenyan parasites at candidate loci known or suspected to be under selection by ACT would address both objectives. Candidate loci include, first, markers for resistance to the quinoline drugs chloroquine and amodiaquine, known to be selected by ACT toward quinoline-sensitive alleles [13–16]. Second, new candidates pcubp1 (encoding ubiquitin carboxyl-terminal hydrolase 1) and pcap2mu (encoding clathrin vesicle-associated adaptor 2, µ subunit), both implicated in artemisinin resistance in the rodent parasite P. chabaudi, have been shown to have polymorphic homologues in P. falciparum [17, 18]. These polymorphisms have yet to be fully validated as markers in parasites with reduced sensitivity to artemisinin derivatives in vivo. However, genome-wide association studies of P. falciparum isolates from coastal Kenya in which the pfubp1 variant E1528D was associated with reduced susceptibility to artemisinin in vitro were recently reported [19].
To characterize any genetic signature of slow clearing submicroscopic infections in ACT-treated children from Mbita, Kenya [9], pfcrt, pfmdr1, pfubp1, and pfap2mu genotypes were determined on days 2 and 3, and on day of recrudescence in the case of later treatment failures. These were compared with pretreatment parasite genotypes isolated from children at baseline, and evaluated for evidence of posttreatment selection in vivo.
METHODS
Trial Design and Participants
As previously described in detail [20], 298 children aged 6 months to 10 years were randomly allocated to receive a 6-dose regimen of artemether-lumefantrine (AL) or a 3-dose regimen of dihydro-artemisinin-piperaquine (DP), fully observed, and followed for 42 days, according to a protocol approved by the Kenya Medical Research Institute Scientific Steering Committee (#1556, 2009) and the London School of Hygiene and Tropical Medicine Ethics Committee (#5455, 2009). Detailed parasite clearance dynamics were assessed by qPCR as described elsewhere [9].
Detection of Genetic Polymorphisms in Loci of Interest
Parasite DNA was extracted from approximately 10 µL blood spots collected on Whatman filter paper prior to treatment (day 0) and on days 2, 3, 7, 14, 28, and 42 after treatment as previously described [9]. Polymorphisms in the pfcrt gene (PF3D7_0709000), encoding the P. falciparum chloroquine resistance transporter, were determined in parasite DNA by multiplex qPCR as described [21]. Polymorphisms at codons 86, 184, and 1246 in the pfmdr1 gene (PF3D7_0523000), encoding P-glycoprotein H1, were identified using direct sequencing of PCR products as previously described with minor modifications [16].
Polymorphisms in the pfap2mu gene (PF3D7_1218300), encoding the µ-subunit of clathrin-associated AP2 adaptor protein [18], were determined by PCR amplification and direct sequencing of 3 fragments encompassing codons 1–174, 121–399, and 377–621, as described [22].
Polymorphisms in pfubp-1 (PF3D7_0104300) in the 300 base-pair region encompassing codons 1463–1563 were determined using a PCR strategy designed and optimized for this study. Nested PCR products were generated using the primer sets:
Nest 1 Forward Primer (pfubp1_1452_1F) CGCCCGTACTATGAAGAAGATC
Nest 1 Reverse Primer (pfubp1_1612_1R) GGCTTTTACCTGAACTGTTCAGG
Nest 2 Forward Primer (pfubp1_1463_2F) CGTAAACAGAATATTCAGGATTGC
Nest 2 Reverse Primer (pfubp1_1563_2R) CTAGCCCTTTATTATCATTATCG
Amplification was performed with 2 nested reactions of 40 cycles of 3-step PCR, with 30 seconds annealing at 53°C (nest 1) or 57°C (nest 2) and elongation at 72°C for 45 seconds. Products were characterized by direct sequencing as described [22]. Ambiguous mixed haplotypes of pfmdr1 were excluded from the analysis as previously described [14]. Mixed genotypes were included in the analysis at the pfcrt locus, where only 2 haplotypes were found at codons 72–76, and at both the pfap2mu and pfubp1 loci, for which only a single variable codon was evaluated.
Analysis of Locus Polymorphism in Global P. falciparum Genome Data
Single-nucleotide polymorphic residues in the pfap2mu and pfubp1 loci were examined in Plasmoview (http://pathogenseq.lshtm.ac.uk/plasmoview) [23], in an ordered set of 631 P. falciparum genomes stratified by geographical region.
Statistical Analysis
Binary variables were compared across pairs of categories, and odds ratios (ORs) with 95% confidence intervals (CIs) estimated. Significance was determined using χ2 distribution. Directional selection in paired samples from the same patient at different time points was tested for statistical significance using McNemar's symmetry test. All analyses were performed in STATA 11 (Timberlake Associates, College Station, TX).
RESULTS
Codons 72–76 of pfcrt
The wild-type pfcrt allele, encoding CVMNK at codons 72–76 of the CRT protein, was confirmed as present in 143 of the 278 (51.4%) evaluable parasite isolates prior to treatment (day 0), of which 89 also harbored the CVIET allele. The remaining 135 isolates (48.6%) were CVIET alone. The CVMNK allele was significantly more common among submicroscopic infections at day 3, being detected in 66 of the 73 successfully evaluated (90.4%; OR, 8.90; 95% CI, 3.88–23.7; P < .001). A significant proportion on day 3 (68%) were mixed infections, comprising both the CVMNK and CVIET genotypes. Thirty-five of the 65 children with detectable CVMNK parasites at day 3 did not carry this genotype at day 0, compared to only 3 children presenting with CVMNK but having only CVIET detected at day 3, demonstrating significant asymmetry, interpretable as strong directional within-host selection for the CVMNK allele (χ2 = 26.95, 1 degree of freedom; P < .001). This effect remained significant after stratification for treatment group (Table 1). These findings suggest that in those children with both pfcrt genotypes present prior to treatment, CVMNK significantly increased in relative abundance from day 0 to day 3. There was evidence that carriage of CVMNK prior to treatment was associated with risk of parasite recurrence at day 28 or day 42 for AL (OR, 2.461, 95% CI, 1.004–6.23; P = .030) but not for DP (OR, 1.405, 95% CI, .154–17.3; P = .714).
Table 1.
Prevalence of pfcrt and pfmdr1 Haplotypes Before and After Treatment With Artemether-Lumefantrine or Dihydroartemisinin-Piperaquine
| Gene | Odds Ratio: Occurrence on Day 3 vs Baseline (95% CI) | Directional Selection Within-Hosta |
||
|---|---|---|---|---|
| LOSS (Frequency) | GAIN (Frequency) | P Value | ||
| pfcrt codons 72-76CVMNK | ||||
| AL (N = 43 143) | 10.32 (3.42–41.4) P < .001 | 1 | 15 | P < .001 (N = 43) |
| DP (N = 30 135) | 7.64 (2.17–40.8) P < .001 | 1 | 9 | P = .0114 (N = 30) |
| Combined ACT (N = 73 278) | 8.90 (3.88–23.69) P < .001 | 2 | 24 | P < .001 (N = 73) |
| pfmdr1 codons 86, 184, 1246 NFD | ||||
| AL (N = 26 101) | 3.15 (1.19–8.60) P = .001 | 1 | 9 | P = .0114 (N = 22) |
| DP (N = 15 104) | 8.52 (1.76–80.3) P = .002 | 1 | 6 | P = .0588 (N = 15) |
| Combined ACT (N = 42 206) | 3.59 (1.68–7.95) P < .001 | 2 | 15 | P = .0016 (N = 38) |
Abbreviations: ACT, artemisinin-based combination therapy; AL, artemether-lumefantrine; DP, dihydro-artemisinin-piperaquine.
a Change in genotype in the same patient between day 0 and day 3 is scored as loss or gain; significance tested using McNemar's test of asymmetry.
Codons 86, 184 and 1246 of pfmdr1
Previous studies have proposed that the haplotype NFD at codons 86, 184, and 1246 is associated with parasite recurrence after AL. We collated information at these 3 positions into haplotypes, using strict criteria to eliminate ambiguity due to mixed infections by excluding all isolates with mixed alleles called at 2 or more of the codons of interest [14]. The proportion of children carrying the haplotype NFD prior to treatment was 38.4%, but this rose to 69.1% on day 3 (N = 42; OR, 3.59; 95% CI, 1.68–7.95; P < .001). However, this strict haplotype definition may have led to biased loss of data from pretreatment isolates because of higher parasite density, and thus greater clonal multiplicity. The analysis was repeated using less stringent criteria for assigning pfmdr1 haplotypes (ie, assigning the NFD haplotype to all ambiguous isolates where this may have been present). This also showed significantly higher prevalence of the NFD haplotype (sensu lato) on day 3 than in the baseline population (N = 48 at day 3; OR, 4.31, 95% CI, 2.09–9.30; P < .001). Paired analysis on both days 0 and 3 at all 3 codon positions was available for 38 children, and evidence of significant within-host directional selection was found for the NFD haplotype sensu stricto (P = .0016). Despite loss of statistical power, stratification by treatment group suggested that this effect occurred in both the AL (N = 22) and DP arms (N = 15) (P = .011, .059, respectively; Table 1). Presence of the NFD haplotype at day 0 was not associated with risk of recrudescence in either arm (data not shown).
Polymorphisms in pfap2mu
As this is the first study to examine the role of pfap2mu polymorphisms in a prospective study of ACT sensitivity, sequence data was obtained at all 621 codons for 166 pretreatment isolates, and for codons 121–399 for a further 78 isolates. A variety of mutations, including synonymous and nonsynonymous single nucleotide polymorphisms, deletions, and insertions, were identified within this gene in the study population, and these are summarized in Tables 2 and 3. We were able to assemble 57 distinct haplotypes from the 166 full-length pfap2mu sequences, 38 of which only occurred once. The most common haplotype, found in 32 pretreatment isolates, was identical to the reference sequence from 3D7.
Table 2.
Synonymous and Nonsynonymous Single-Nucleotide Polymorphisms in the pfap2-mu Gene, and Prevalence at Day 0, Day 2/3, and Day of Failure After ACT Treatment
| Codon | Reference Sequence | AA | Nucleotide Substitution | AA Changes | Prevalence Day 0 (N = 244)a | Prevalence Day 2/3 (N = 32) | Prevalence Day Failure (N = 28) |
|---|---|---|---|---|---|---|---|
| 5 | G | Leu | 15A | … | 8.4% | 4.2% | No data |
| 146 | G | Arg | 437A | Lys | 1.2% | 0% | 3.6% |
| 160 | G | Ser | 479A | Asn | 18.0% | 37.5% | 25.0% |
| 479C | Thr | 1.2% | 3.1% | 0% | |||
| 161 | G | Val | 483A | … | 0.4% | 0% | 0% |
| 162 | T | Ile | 486C | … | 9.8% | 3.1% | 0% |
| 163 | A | Glu | 489G | … | 16.8% | 18.8% | 17.9% |
| 188 | A | Arg | 564G | … | 0.8% | 3.1% | 0% |
| 199 | A | Lys | 596C | Thr | 3.3% | 3.1% | 7.1% |
| 200 | A | Asn | 598T | Tyr | 0.4% | 0% | 0% |
| 233 | T | Asn | 699G | Lys | 0.4% | 0% | 0% |
| 337 | C | Ala | 1010G | Gly | 1.2% | 0% | No data |
| 324 | T | Cys | 1026C | … | 0.6% | 0% | No data |
| 437 | C | Phe | 1311A | Leu | 4.2% | 9.4% | No data |
| 476 | T | Ser | 1428G | … | 4.2% | 3.1% | No data |
| 478 | A | Val | 1434T | … | 2.4% | 0% | No data |
Abbreviations: AA, amino acid; ACT, artemisinin-based combination therapy.
a The middle sequencing fragment (codons 121–399) was successfully analyzed in all isolates. Codon 5 was not determined in 2 isolates without fragment 1 data; codons 5, 337, 324, 437, 476, and 478 were not determined in 74 isolates without fragment 1 or 3 data, and codons 337, 324, 437, 476, and 478 were not determined in 1 isolate without fragment 3 data.
Table 3.
Indels Identified in the pfap2-mu Gene, Consequent Amino Acid Replacements, and Prevalence at Day 0, Day 2/3, and Day Fail After Treatment
| Codon | Reference Sequence | Deletions/Insertions | Prevalence Day 0 (N = 244) | Prevalence Day 2/3 (N = 32) | Prevalence Day Failure (N = 28) |
|---|---|---|---|---|---|
| 226 | 7xAsn | 5xAsn/6xAsn | 0.4%/0.4% | 0%/0% | 0%/0% |
| 8xAsn/9xAsn | 17.6%/1.2% | 9.4%/3.1% | 25.0%/0% | ||
| 233 | 1xLys | 2xLys | 2.0% | 3.1% | 7.1% |
| 319 | 5xAsn | 6xAsn | 12.7% | 6.3% | 42.9% |
| 326 | 4xAsn | 5xAsn | 13.1% | 9.4% | 3.6% |
Pilot analysis of the first 20 isolates found to have persisting parasites on day 2 or day 3 identified the nonsynonymous polymorphism at codon 160, Ser to either Asn or Thr (Table 2), as differing in prevalence between pre- and posttreatment infections. We therefore focused on this mutation for detailed analysis. Among 244 evaluable pretreatment samples, 18.3% and 1.2% carried the Asn or Thr alleles, respectively, at codon 160 of pfap2mu. In contrast, combining 32 evaluable day 2 and day 3 isolates, the prevalence of parasites with either the S160N or S160T variant rose immediately following ACT treatment to 40.6% (OR, 2.87, 95% CI, 1.20–6.61; P = .006). Among these 32 individuals, evidence of directional selection for either variant was found between day 0 and day 2/3 (P = .046). This selection was not seen at the day of failure, when the overall prevalence of codon 160 mutations was 25% (Table 2). There was no evidence that carriage of the S160N or S160T variant prior to treatment was associated with risk of recurrent infection (data not shown).
Polymorphisms in pfubp1
An amplification strategy was designed for a region of pfubp1 from codons 1463–1563, encompassing the polymorphic codon 1528 identified by Borrmann et al [19] as being associated with in vitro parasite responses to artemisinin in Kenya. Direct sequencing of this region was successful in 123 pretreatment isolates, and identified 16 distinct haplotypes defined by synonymous and nonsynonymous point mutations, indels of 1 to 15 amino acids and repeat number variations in low-complexity nucleotide tracts (Figure 1). We focused on the codon E1528D substitution, which occurred in 12 of 123 evaluable individuals (9.8%) prior to treatment. Day 3 finger-prick samples had been largely exhausted by investigations of the other 3 loci, and by previous qPCR analysis [9], but sequencing of pfubp1 was possible for 35 samples from day 3, of which 6 (17.1%) encoded Asp at codon 1528. To increase the sample, we amplified this region from 33 parasitological failures identified by microscopy on day 28 or day 42 [20], of which 14 (42.4%) carried 1528D. Thus evidence was found that the E1528D mutation was significantly more common among parasite isolates after treatment (days 3, 28, or 42) than among the pretreatment population (OR, 3.85, 95% CI, 1.63–9.32; P < .001). Paired analysis in the 36 evaluable individuals (i.e. those with pfubp1 sequence data on day 0 and at least 1 of day 3, day 28, or day 42) showed strong evidence of directional selection (P = .002). Carriage of the 1528D variant at day 0 was not associated with risk of recurrent infections (data not shown).
Figure 1.
Haplotype frequencies at codons 1463–1563 of pfubp1 in 123 pretreatment isolates. A 303-nucleotide fragment was amplified from genomic DNA using primers described in the text, and sequenced directly. Sequences were ordered from the most frequent to the least, and aligned in Clustal W and formatted in Boxshade, with additional hand-editing for clarity. Codon 1258, as identified by Borrman et al [19] is arrowed. Haplotype 1 is identical to the sequence found in the 3D7 reference genome. *Haplotypes 7 and 8 carry a synonymous mutation at codon 1518, encoding Asn.
Polymorphism of pfap2mu and pfubp1 in the Global Genome Sequence Database
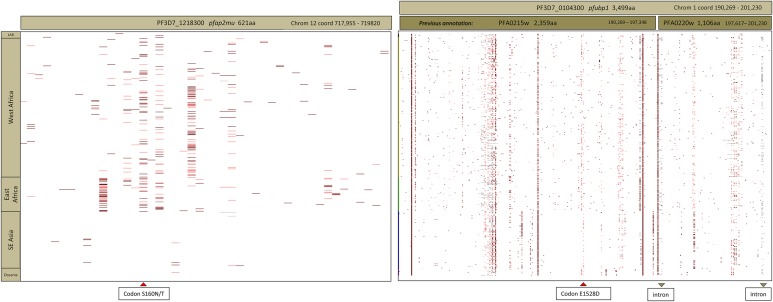
Having identified 2 novel candidate loci contributing to the genetic signature of persisting submicroscopic P. falciparum in our treated Kenyan children, we sought to examine diversity in these 2 loci across 631 genomes using PlasmoView software [23]. For pfap2mu, the S160N/T polymorphism occurred across both West African (Burkina Faso, Gambia, Ghana, Mali, Senegal) and East African (Kenya, Malawi) parasite populations. This polymorphism did not occur in SE Asian populations (Thailand, Cambodia, Vietnam) nor in Papua New Guinea (Figure 2, top). For pfubp1, diversity across the locus was less geographically distinct, and mutations around codons 1520–1540 occurred in all populations (Figure 2, bottom). By contrast, the recently described polymorphisms at codons 493, 539, 543, and 580 of the kelch gene (PF3D7_1343700), associated with artemisinin sensitivity in Cambodia [6], were restricted to SE Asia, and do not occur in available African P. falciparum genomes. In contrast, the kelch polymorphism K189T was very common among African isolates but absent from Asian parasites included in the publicly available sample (data not shown).
Figure 2.
Global sequence diversity in pfap2mu (left) and pfubp1 (right) across genome sequences from 631 P. falciparum isolates mapped to the 3D7 reference genome [23]. Colored bars at any chromosome position (horizontal axis) denote a nonreference substitution at that residue in the relevant isolate (vertical axis). At top is the regional origin of isolates in the analysis, comprising 8 laboratory isolates (L), 367 isolates from West Africa, 88 from East Africa, 151 from SE Asia, and 17 from Oceania (Oc). Codon S160N/T of pfap2mu and codon 1528 of pfupb1 are highlighted by red arrows at the bottom of the figure. For pfubp1, the position of 2 carboxy-terminal introns is also marked. Previous annotation of this region of chromosome 1 as 2 distinct genes (PFA0215w and PFA0220w) is shown above the right-hand figure.
DISCUSSION
We have genetically characterized submicroscopic P. falciparum infections in ACT-treated children from Mbita, Kenya, and described a novel genetic signature at the pfcrt, pfmdr1, pfap2mu, and pfubp1 candidate drug-resistance marker loci. For 3 of these loci, the signature was already present 3 days after treatment with either AL or DP. For pfubp1, selection was seen on day of recrudescence in the case of later treatment failures. By microscopy, all children were parasite-free at day 3 and afebrile, and thus these were persisting, submicroscopic and asymptomatic infections which, in some cases, contributed to mosquito transmission potential on day 7 and patent recrudescent parasitemia on day 28 or day 42 [9, 20]. The evidence that these variant genotypes were overrepresented on day 3, immediately following completion of the full (observed) course of either AL or DP, suggests that selection is exerted partly or primarily by the artemisinin component of combination therapy, and that P. falciparum response to artemisinin in vivo is modulated by variation at these loci. Although not directly shown by our data, this interpretation is supported by a recent study in Uganda [24], which found that lumefantrine and piperaquine did not select for the same haplotypes of pfmdr1 in recurrent infections, implying that early selection for the NFD haplotype is exerted by the artemisinin component in both treatment arms.
Previous studies have documented selection at the pfcrt and pfmdr1 loci in late recurrent infections in ACT-treated African children [13–16], but this is the first demonstration of selection on day 3. The favored CQ-sensitive haplotypes pfcrt CVMNK and pfmdr1 NFD were supplanted in the late 20th century by the spread of CQ resistance. Thus, it is difficult to distinguish between a general disadvantage in vivo caused by the relative lack of fitness of CQ-resistant P. falciparum in the absence of CQ [25, 26], and a specific interaction between these loci and artemisinin. Our study was not powered, nor designed, to explore interlocus interactions, but we observed both pfmdr1-NFD and pfcrt-CVMNK in 43 of 204 evaluable patients on day 0, and in 21 of 34 evaluable submicroscopic infections on day 3 (OR, 6.05, 95% CI, 2.62–14.2; P < .001). This may suggest the benefit of the 2 wild-type genes was enhanced by the presence of artemisinin. Another weakness of our study was that some isolates of interest were not sequenced at all loci, mainly due to exhaustion of small, finite blood samples. Although the limited number of observations in each of the treatment arms precluded comparisons of effect estimates due to lack of power, an exploratory stratification was carried out, generating odds ratios with very wide confidence intervals (data not shown). The directional selection for pfap2mu 160 (AL: P = .015; DP: P = .063) and pfubp1 (AL: P = .0002; DP: P = .029) remained evident in both arms. Future studies with adequate power can now be designed using these estimates of prevalence and effect size. Moreover, age and multiplicity of infection were associated with parasite persistence in the Mbita trial [9], and thus are also potential confounders. These parameters should be investigated by multivariate regression in studies with sufficient power to further dissect the interplay among parasite genotypes, host factors, and different treatment regimens.
This study provides the first preliminary evidence that P. falciparum response to artemisinin in vivo may be modulated by polymorphisms in the loci pfap2mu and pfubp1, both homologues of genes identified in studies of experimentally induced artemisinin resistance in the rodent parasite P. chabaudi [17, 18]. Clathrin-mediated endocytosis is a mechanism common to eukaryotic cells by which membrane-associated proteins and other cargo are transported from the plasma membrane, trans-Golgi network, or endosomes to acceptor compartments; the adaptor complex 2 is associated with cell surface endocytosis, recruiting to the vesicle both cargo and structural components including clathrin, and may have a role in hemoglobin trafficking in malaria parasites. The ubiquitination of proteins modulates endocytosis, and proteosomal recycling, with known substrates including P-glycoproteins such as Pgh1 (encoded in malaria parasites by pfmdr1) [27]. Thus, these 2 loci may both contribute to endosomal digestion of hemoglobin in malaria parasites [18].
The persistence of microscopically undetectable parasites following treatment of clinical malaria does not constitute in vivo antimalarial resistance as normally defined, and may have been equally common in the pre-ACT era. The application of a novel endpoint, qPCR positivity within 72 hours of treatment, has merely permitted this to be observed for the first time. In settings where a significant proportion of malaria patients are semi-immune, newly acquired super-infecting parasites may flourish and cause symptomatic malaria, despite the presence of preexisting “symbiotic” genotypes hitherto stably suppressed by the immune response, but adapted to continuing survival. As we have previously hypothesized [28], it may be these preexisting parasites that remain in some individuals after drug clearance of the symptomatic infection. This hypothesis is only plausible if current ACT regimens are ineffective against low density parasitemia, but recent data do suggest this is the case [28, 29]. Evaluation of current and alternative regimens against asymptomatic parasitemia, using appropriate study protocols specifically designed for this purpose, is required to test this possibility.
Our interpretation is that the phenomenon studied here is not antimalarial resistance per se. Consistent with this, the genetic signature described does not resemble that of the “artemisinin-resistant” parasites associated with the slow-clearance phenotype in Cambodia. First, the latter do not carry the CVMNK haplotype of pfcrt and, second, pfmdr1 polymorphisms are not associated with the phenotype [30]. Finally, we found no evidence of codon 160 polymorphism in pfap2mu occurring in SE Asian isolates in the available genomic sequence database, nor evidence of kelch propeller domain polymorphism in any African isolates [6]. We thus conclude that the persisting submicroscopic parasites observed in our cohort of ACT-treated Kenyan children are not related to slow-clearing parasites as seen in Cambodia and nearby countries. Only polymorphisms in pfubp1 may be shared determinants in Cambodia and Kenya. PF3D7_0104300 encompasses 2 loci, PFA0215w and PFA0220w, from earlier genome annotations (Figure 2B), and variant alleles of the former were identified in subpopulations of P. falciparum in Cambodia associated with slow clearance after artesunate monotherapy [31].
We have described a novel genetic signature at 4 loci in P. falciparum, and implicate these genes in modulating in vivo parasite response to artemisinin. Further evaluation in purpose-designed, adequately powered studies of asymptomatic malaria in vivo, and in genetically modified P. falciparum in vitro, are needed to determine the potential public health implications of these observations, and the utility of these loci as markers of artemisinin sensitivity in natural populations of P. falciparum worldwide.
Notes
Acknowledgments. We thank Dr Henk Schallig for encouragement and support as Coordinator of MALACTRES, and Prof. David C. Warhurst for helpful discussions. We thank the Kenya Medical Research Institute (KEMRI) for assistance in protocol development, and for review of our study documentation. This paper has been approved by the KEMRI Publications Committee.
Financial support. This work was supported by the EU FP7 MALACTRES Consortium; the Fundação para a Ciência e a Tecnologia (FCT) of Portugal (grant number SFRH/BD/63129/2009) to G. H.; the EDCTP WANECAM Project to K. B. B. and C. J. S.; the UK Medical Research Council to T. G. C.; and Public Health England to C. J. S.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Aregawi MW, Ali AS, Al-mafazy AW, et al. Reductions in malaria and anaemia case and death burden at hospitals following scale-up of malaria control in Zanzibar, 1999–2008. Malar J. 2011;10:46. doi: 10.1186/1475-2875-10-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karema C, Aregawi MW, Rukundo A, et al. Trends in malaria cases, hospital admissions and deaths following scale-up of anti-malarial interventions, 2000–2010, Rwanda. Malar J. 2012;11:236. doi: 10.1186/1475-2875-11-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM Artemisinin Resistance in Cambodia 1 (ARC1) Study Consortium. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008;359:2619–20. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 4.Dondorp AM, Nosten F, Yi P, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–67. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheeseman IH, Miller BA, Nair S, et al. A major genome region underlying artemisinin resistance in malaria. Science. 2012;336:79–82. doi: 10.1126/science.1215966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ariey F, Witkowski B, Amaratunga C, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–5. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krishna S, Kremsner PG. Antidogmatic approaches to artemisinin resistance: reappraisal as treatment failure with artemisinin combination therapy. Trends Parasitol. 2013;29:313–7. doi: 10.1016/j.pt.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Borrmann S, Sasi P, Mwai L, et al. Declining responsiveness of Plasmodium falciparum infections to artemisinin-based combination treatments on the Kenyan coast. PLOS One. 2011;6:e26005. doi: 10.1371/journal.pone.0026005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beshir KB, Sutherland CJ, Sawa P, et al. Residual Plasmodium falciparum parasitemia in Kenyan children after artemisinin-combination therapy is associated with increased transmission to mosquitoes and parasite recurrence. J Infect Dis. 2013;208:2017–24. doi: 10.1093/infdis/jit431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sutherland CJ, Alloueche A, Curtis J, et al. Gambian children successfully treated with chloroquine can harbour and transmit Plasmodium falciparum gametocytes carrying resistance genes. Am J Trop Med Hyg. 2002;67:578–85. doi: 10.4269/ajtmh.2002.67.578. [DOI] [PubMed] [Google Scholar]
- 11.Hallett RL, Dunyo S, Ord R, et al. Treatment of malaria in Gambian children with chloroquine plus sulphadoxine-pyrimethamine favours survival and transmission to mosquitoes of multi-drug-resistant Plasmodium falciparum. PLOS Clin Trials. 2006;1:e15. doi: 10.1371/journal.pctr.0010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Méndez F, Herrera S, Murrain B, et al. Selection of antifolate-resistant Plasmodium falciparum by sulfadoxine-pyrimethamine treatment and infectivity to Anopheles mosquitoes. Am J Trop Med Hyg. 2007;77:438–43. [PubMed] [Google Scholar]
- 13.Sisowath C, Stromberg J, Martensson A, et al. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem) J Infect Dis. 2005;191:1014–7. doi: 10.1086/427997. [DOI] [PubMed] [Google Scholar]
- 14.Humphreys GA, Merinopoulos I, Ahmed J, et al. Amodiaquine and artemether-lumefantrine select distinct alleles of the Plasmodium falciparum pfmdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicr Agents Chemother. 2007;51:991–7. doi: 10.1128/AAC.00875-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sisowath C, Petersen I, Veiga MI, et al. In vivo selection of Plasmodium falciparum parasites carrying the chloroquine-susceptible pfcrt K76 allele after treatment with artemether-lumefantrine in Africa. J Infect Dis. 2009;199:750–7. doi: 10.1086/596738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gadalla NB, Adam I, Elzaki SE, et al. Increased pfmdr1 copy number and sequence polymorphisms in Plasmodium falciparum isolates from Sudanese malaria patients treated with artemether-lumefantrine. Antimicr Agents Chemother. 2011;55:5408–11. doi: 10.1128/AAC.05102-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunt P, Afonso A, Creasey A, et al. Gene encoding a deubiquinating enzyme is mutated in artesunate- and chloroquine-resistant rodent malaria parasites. Mol Microbiol. 2007;65:27–40. doi: 10.1111/j.1365-2958.2007.05753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henriques G, Martinelli A, Rodrigues L, et al. Artemisinin resistance in rodent malaria-mutation in the AP2 adaptor μ-chain suggests involvement of endocytosis and membrane protein trafficking. Malar J. 2013;12:118. doi: 10.1186/1475-2875-12-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borrmann S, Straimer J, Mwai L, et al. Genome-wide screen identifies new candidate genes associated with artemisinin susceptibility in Plasmodium falciparum in Kenya. Sci Rep. 2013;3:3318. doi: 10.1038/srep03318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sawa P, Shekalaghe SA, Drakeley CJ, et al. Malaria transmission after artemether-lumefantrine and dihydroartemisinin-piperaquine: a randomized trial. J Infect Dis. 2013;207:1637–45. doi: 10.1093/infdis/jit077. [DOI] [PubMed] [Google Scholar]
- 21.Gadalla NB, Elzaki SE, Mukhtar E, Warhurst DC, El-Sayed B, Sutherland CJ. Dynamics of pfcrt alleles CVMNK and CVIET in chloroquine-treated Sudanese patients infected with Plasmodium falciparum. Malaria J. 2010;9:74. doi: 10.1186/1475-2875-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Schalkwyk DA, Burrow R, Henriques G, et al. Culture-adapted Plasmodium falciparum isolates from UK travellers: in vitro drug sensitivity, clonality and drug resistance markers. Malaria J. 2013;12:320. doi: 10.1186/1475-2875-12-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Preston MD, Assefa SA, Ocholla H, et al. PlasmoView: A web-based resource to visualise global Plasmodium falciparum genomic variation. J Infect Dis. 2014;209:1808–15. doi: 10.1093/infdis/jit812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conrad MD, Leclair N, Arinaitwe E, et al. Comparative impacts over 5 years of artemisinin-based combination therapies on Plasmodium falciparum polymorphisms that modulate drug sensitivity in Ugandan children. J Infect Dis. 2014;210:344–353. doi: 10.1093/infdis/jiu141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laufer MK, Thesing PC, Eddington ND, et al. Return of chloroquine antimalarial efficacy in Malawi. N Engl J Med. 2006;355:1959–66. doi: 10.1056/NEJMoa062032. [DOI] [PubMed] [Google Scholar]
- 26.Ord R, Alexander N, Dunyo S, et al. Seasonal carriage of pfcrt and pfmdr1 alleles in Gambian Plasmodium falciparum implies reduced fitness of chloroquine-resistant parasites. J Infect Dis. 2007;196:1613–9. doi: 10.1086/522154. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Z, Wu J-Y, Hait W, Yang J-M. Regulation of the stability of P-glycoprotein by ubiquitination. Mol Pharmacol. 2004;66:395–403. doi: 10.1124/mol.104.001966. [DOI] [PubMed] [Google Scholar]
- 28.Dinko B, Oguike MC, Larbi JA, Bousema T, Sutherland CJ. Persistent detection of Plasmodium falciparum, P. malariae, P. ovale curtisi and P. ovale wallikeri after ACT treatment of asymptomatic Ghanaian school-children. Int J Parasitol: Drugs and Drug Resistance. 2013;3:45–50. doi: 10.1016/j.ijpddr.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chijioke-Nwauche I, van Wyk A, Nwauche C, Beshir KB, Kaur H, Sutherland CJ. HIV-positive Nigerian adults harbor significantly higher serum lumefantrine levels than HIV-negative individuals seven days after treatment for Plasmodium falciparum infection. Antimicrob Agents Chemother. 2013;57:4146–50. doi: 10.1128/AAC.02508-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imwong M, Dondorp AM, Nosten F, et al. Exploring the contribution of candidate genes to artemisinin resistance in Plasmodium falciparum. Antimicrob Agents Chemother. 2010;54:2886–92. doi: 10.1128/AAC.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miotto O, Almagro-Garcia J, Manske M, et al. Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat Genet. 2013;45:648–55. doi: 10.1038/ng.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]