Abstract
The Gram-positive bacterium Staphylococcus pseudintermedius is a leading cause of canine bacterial pyoderma, resulting in worldwide morbidity in dogs. S. pseudintermedius also causes life-threatening human infections. Furthermore, methicillin-resistant S. pseudintermedius is emerging, resembling the human health threat of methicillin-resistant Staphylococcus aureus. Therefore it is increasingly important to characterize targets for intervention strategies to counteract S. pseudintermedius infections. Here we used biophysical methods, mutagenesis, and X-ray crystallography, to define the ligand-binding properties and structure of SitA, an S. pseudintermedius surface lipoprotein. SitA was strongly and specifically stabilized by Mn2+ and Zn2+ ions. Crystal structures of SitA complexed with Mn2+ and Zn2+ revealed a canonical class III solute-binding protein with the metal cation bound in a cavity between N- and C-terminal lobes. Unexpectedly, one crystal contained both apo- and holo-forms of SitA, revealing a large side-chain reorientation of His64, and associated structural differences accompanying ligand binding. Such conformational changes may regulate fruitful engagement of the cognate ABC (ATP-binding cassette) transporter system (SitBC) required for metal uptake. These results provide the first detailed characterization and mechanistic insights for a potential therapeutic target of the major canine pathogen S. pseudintermedius, and also shed light on homologous structures in related staphylococcal pathogens afflicting humans.
Keywords: class III solute binding protein, divalent metal cation, manganese, staphylococcal disease, transport, zinc
Abbreviations: ABC, ATP-binding cassette; CV, column volume; DSC, differential scanning calorimetry; FhuD2, ferric hydroxamate uptake D2; HIC, hydrophobic interaction chromatography; ITC, isothermal titration calorimetry; MR, molecular replacement; PDB, Protein Data Bank; rmsd, root mean square deviation; RP-HPLC, reverse-phase HPLC; SBP, solute-binding protein; SEC, size-exclusion chromatography; TEV, tobacco etch virus
INTRODUCTION
Staphylococcus pseudintermedius is a Gram-positive bacterium emerging as the most important pathogen causing canine pyoderma [1,2]. Moreover, S. pseudintermedius is associated with other canine diseases such as wound infections, urinary tract infections and otitis externa, thus cumulatively resulting in significant morbidity in dogs on a global scale. S. pseudintermedius was found to be one of the most common pathogens causing disease in thousands of small animal veterinary consultations [3]. Treatment of S. pseudintermedius in dogs is often ineffective, unless strong systemic antibacterials are used, thus risking an increase of antimicrobial-resistant bacteria in pets that can act as reservoirs for such pathogens, with a risk for transmission to humans [4,5]. Methicillin-resistant strains of S. pseudintermedius are emerging [1,2,6], frequently via acquisition of the mecA gene, thus resembling the MRSA (methicillin-resistant Staphylococcus aureus) infections currently increasing in humans and which caused over 18000 patient deaths prior to 2007 in the U.S.A. alone [7]. Although relatively rare, there are several reports of life-threatening S. pseudintermedius infections occurring in humans following dog bite wounds [8]. Consequently, the development of safe and effective therapeutics against S. pseudintermedius would have benefit in alleviating pathogen-induced disease and suffering, primarily in animals, and potentially also in humans [2].
Recently, the whole genomes have been sequenced for S. pseudintermedius strains ED99 [9] and HKU10-03 [10], providing important new resources for investigations of the molecular basis of canine bacterial pyoderma. Both strains exhibit a single circular chromosome with over 2400 predicted protein-coding sequences including numerous virulence factors, such as leucotoxins, exotoxins, adherence factors, superantigens, and other surface-exposed proteins, many of which may be interesting therapeutic targets [11,12]. For example, the cell-wall-associated proteins SpsD and SpsL are expressed by S. pseudintermedius during canine infection, are immunogenic and mediate binding to host extracellular matrix proteins, suggesting a role in disease pathogenesis [13].
Considering the similarities between S. pseudintermedius infections in dogs, and S. aureus infections in humans, increased knowledge of canine host–pathogen interactions involving S. pseudintermedius may potentially inform areas of human health and infectious disease, as well as veterinary medicine. Likewise, the wealth of information derived from extensive research into human infection by S. aureus may also promote an understanding of S. pseudintermedius. In particular, one set of surface-exposed proteins that has captured much attention in studies of staphylococci, and of pathogenic bacteria in general, is that of the SBPs (solute-binding proteins) [14,15]. SBPs act in concert with an integral membrane protein that mediates transmembrane solute transport, powered by an associated cytoplasmic ATP-binding protein, thus comprising the canonical bacterial ABC (ATP-binding cassette) transporter. A number of SBPs from both Gram-negative and Gram-positive bacteria have generated protective immunogenicity in animal models. Moreover, despite the possibility of steric hindrance due to the bacterial cell wall, anti-SBP antibodies can clear some bacterial infections and may inhibit bacterial growth by reducing their nutrient uptake. As such, ABC transporter components are generally considered interesting candidates for antibacterial vaccines or therapies [16].
In Gram-positive bacteria (including the staphylococci) SBPs are processed as extracellular surface lipoproteins, whereas in Gram-negative bacteria SBPs are localized in the periplasm and therefore were initially known as periplasmic binding proteins [14,15]. The SBPs typically share an overall architecture made of a ligand-binding groove located between two interfacing globular domains. Based on structure, the SBP family has been divided into three classes, depending largely on the linker(s) connecting the two globular domains [15]. The class I SBPs have three connecting segments between the two domains. In contrast, the class II SBPs have two interdomain crossovers, whereas the class III proteins only have one segment connecting the two domains [14,17]. Different SBPs function by binding to different solutes, including siderophores, carbohydrates, peptides, amino acids, and ions, and by shuttling these solute ligands to membrane-bound receptors for active transport into the bacterium; thus providing nutrients essential for survival, growth and virulence [14,15].
Some recently described examples of SBPs from S. aureus include FhuD2 (ferric hydroxamate uptake D2), which plays a role in staphylococcal dissemination [18] and indirectly binds Fe3+ via siderophores accommodated in a central pocket [19]; and MntC, an immunogenic surface lipoprotein [20] which directly binds Mn2+ ions [21]. Furthermore, the SBP structures of MtsA from Streptococcus pyogenes, and PsaA from Streptococcus pneumoniae, also reveal direct binding to Fe2+ and Zn2+, respectively, and their metal-binding ability is thought to have an essential role in virulence [22,23]. Interestingly, some SBPs can bind several different divalent metal cations, and competition by extracellular Zn2+ has been reported to inhibit bacterial uptake of Mn2+ [24], potentially reducing growth and virulence, due to the essential requirement for Mn2+ exhibited by many bacteria, including the staphylococci [25,26]. Indeed, both manganese and zinc are fundamental trace elements present in most organisms and are important for many biological processes, wherein these cations may act as cofactors of metallo-enzymes or as protein-stabilizing factors. Estimates of the zinc concentration in human plasma are at the low micromolar level, with reports ranging from 12 μM [27] to 2 μM bound zinc and as little as 30 nM for free zinc ions [28]. In a different study, it was estimated that human whole blood contains manganese at up to approximately 0.2 μM, most of which was found in erythrocytes [29]. Consequently, the potential competition between bacterial and host proteins for binding to metal ions will depend on the complex interplay of several factors, including the availability of free metals in the given circumstances, the abundance of chelating proteins, and their affinities for each metal ion; characteristics which are currently poorly characterized for the precise example occurring during S. pseudintermedius infections of dogs.
Despite this wealth of knowledge, and the sequencing of two S. pseudintermedius genomes, to date there is no detailed information available about the structure or function SBPs potentially present on the surface of S. pseudintermedius. Since such lipoproteins may play roles in virulence by scavenging essential solutes for active uptake from the host environment, we investigated the putative metal-binding protein, SitA, a potential therapeutic target of S. pseudintermedius. On a similar theme, recent studies of the close orthologue MntC from S. aureus showed that it can provide protective immunity in animal models of infection, suggesting that it has properties suitable for a candidate vaccine antigen [20].
Here we performed a detailed structural and functional study of SitA in order to fully characterize this S. pseudintermedius surface-exposed lipoprotein. We observed significant increases in the stability of SitA upon binding to divalent metal cations and took advantage of this to obtain crystals, which ultimately allowed determination of the structure of apo-, Zn2+- and Mn2+-bound forms of SitA. These structures, coupled with biophysical studies and site-directed mutagenesis, enabled precise identification of the key metal-binding determinants and provided insights into ligand specificity and the structural consequences of ligand binding that may govern ABC transporter engagement by SitA. The results discussed enhance our structural and functional understanding of the solute-binding proteins and ligand transport mechanisms in bacterial pathogens and in particular S. pseudintermedius.
EXPERIMENTAL
Molecular cloning and protein preparation
The SitA gene fragment encoding residues G23-Q306 was cloned by PCR, using the PIPE method [30], from S. pseudintermedius strain IV369-1041 (obtained from Quotient Bioresearch Ltd) (GenBank code CP002478). The SitA genes in the fully-sequenced HKU10-03 and ED99 genomes share 100% sequence identity with IV369-1041. The cloned fragment lacked the first 22 residues which encode a signal peptide for protein export and a lipobox cysteine residue. The fragment was inserted into a modified pET-15 vector (Novagen), enabling cytoplasmic expression of the SitA protein with an N-terminal 6-His tag followed by a cleavage site for the TEV (tobacco etch virus) protease. The PIPE method was also used to create two mutants by inserting the following point mutations in the SitA23–306 expression construct: H64A and E203A+D278A. All plasmids were verified by DNA sequencing. The residue numbering employed refers to the full-length SitA protein, Uniprot code F0P9H5.
The SitA expression constructs (wild-type or mutant clones) were transformed into chemically competent Escherichia coli BL21 (DE3) cells and the production of recombinant SitA was performed using the EnPresso Tablet Cultivation Set (BioSilta) growth system supplemented with 100 μg.ml−1 ampicillin. Bacteria were grown at 30°C for a total of 40 h and target protein production was induced by the addition of 1 mM IPTG (isopropyl β-D-thiogalactoside). Cells were harvested by centrifugation (6400 g, 30 min, 4°C), resuspended in 50 mM sodium phosphate pH 8.0, 300 mM NaCl, and lysed by sonication (Qsonica Q700) for 5 min with cycles of 30 s sonication (40% amplitude) interspersed with 30 s on ice. Cell lysates were clarified by centrifugation at 36200 g for 30 min, and the supernatant was filtered using a 0.22 μm membrane (Corning filter system) prior to protein purification.
SitA was purified by affinity chromatography using an AKTA purifier 10 system (GE Healthcare). The filtered supernatant was loaded onto an Ni-NTA resin (10 ml column, GE Healthcare), and SitA was eluted using five steps of imidazole at 0, 25, 50, 75 and 250 mM concentration, at a flow rate of 5 ml.min−1. Fractions containing SitA were identified by a band migrating at ~35 kDa in SDS–PAGE analysis. The N-terminal 6-His tag was removed enzymatically by addition of a 6-His tagged solubility-enhanced TEV protease [31], with cleavage proceeding at room temperature overnight in buffer containing 50 mM sodium phosphate pH 8.0, 300 mM NaCl, 0.5 mM EDTA, 1 mM DTT (dithiothreitol). Subsequently, the sample was reloaded on the Ni-NTA resin to recapture the protease and free His tag, thus allowing elution in the column flow-through of SitA protein in the tagless form, which was used in all studies described herein. The SitA sample was concentrated and loaded onto a HiLoad Superdex 75 (26/60) preparative SEC (size-exclusion chromatography) column equilibrated in buffer containing 20 mM Tris–HCl pH 8.0, 150 mM NaCl, at a flow-rate of 1 ml.min−1. SitA protein was collected and 1 mM EDTA was added to remove divalent cations. The final yield of SitA obtained from 0.45 litre growth medium was over 200 mg (~11 mg protein per g wet biomass). The quality of the final sample was checked using 4–12% SDS–PAGE gradient gels in MES buffer. Sample purity was estimated to be >97%, for wild-type and mutant proteins, using RP-HPLC (reverse-phase HPLC) performed on a C4 column under standard conditions, essentially as described previously [32].
To separate the apo-form from metal-bound forms of SitA, an HIC (hydrophobic interaction chromatography) step was performed using a 5 ml HP butyl sepharose column (GE Healthcare) running at 1 ml·min−1. The column was equilibrated with ten CVs (column volumes) of buffer A (PBS containing 1.55 M sodium citrate, pH 7.2, 10 mM EDTA). The elution of the different forms of the protein was achieved with a linear gradient between buffer A and buffer B (PBS pH 7.2, 10 mM EDTA) in 60 CV. Two distinct peaks were eluted between 22 and 34% and between 51 and 86% of buffer B.
DSC (differential scanning calorimetry)
The thermal stability of SitA proteins was assessed by DSC using a MicroCal VP-Capillary DSC instrument (GE Healthcare). SitA samples were prepared at protein concentration of 0.5 mg·ml−1 (~15 μM) in buffer containing 20 mM Tris–HCl, 150 mM NaCl, pH 7.5, with or without 2 mM MnCl2, ZnCl2, MgCl2 or CaCl2, with or without 10 mM EDTA. The DSC temperature scan ranged from 10 to 110°C, with a thermal ramping rate of 200°C·h−1 and a 4 s filter period. Data were analysed by subtraction of the reference data for a sample containing only buffer, using the Origin 7 software.
CD
Far-UV CD spectra were recorded from 190 to 260 nm at 25°C using a Jasco-810 spectropolarimeter (Jasco). The cuvette chamber temperature was regulated using a PCB-1500 Peltier temperature system controller (Perkin-Elmer). SitA samples with or without 1 mM MnCl2 were prepared at 0.2 mg.ml−1 (~6 μM) in 5 mM KH2NaPO4 buffer, pH 7.2. A quartz cuvette with an optical path length of 1 mm was used. Spectra were acquired at 1 nm bandwidth, 0.5 s response time, 0.2 nm step size and 10 nm.min−1 scan speed. Each spectrum was calculated as the average of five accumulations. The spectra were corrected by subtracting the buffer baseline followed by application of a smoothing function in Spectra Manager v2.0 (Jasco).
Crystallization, data collection and structure determination
For crystallization trials, purified SitA was concentrated to >150 mg.ml−1 using centrifugal concentration devices with 10 kDa molecular weight cut-off membranes (Amicon ultrafree, Millipore). Protein concentration was determined using Bradford assay (Bio-Rad, Protein Assay), and BSA as the reference. SitA in 20 mM Tris–HCl pH 8.0, 150 mM NaCl was prepared in three different ways for crystallographic screenings: (i) SitA apo protein, (ii) SitA+10 mM ZnCl2 and (iii) SitA+10 mM MnCl2. Drops of 200 nl SitA protein (with or without metal ions)+200 nl crystallization reservoir solution were dispensed using a Crystal Gryphon robot (Art Robbins Instruments). Metal ions were not added to the reservoir solution at any point. Crystals were grown in low-profile crystallization plates (Greiner) in a sitting-drop vapour diffusion format. Over 1500 crystallization conditions were screened at 20°C, with automatic imaging performed using a RockImager-182 (Formulatrix).
SitA crystals in presence of ZnCl2 or MnCl2 ions grew after incubation with a reservoir solution containing 30% (v/v) 1,1,1,3,3,3-hexafluoro-2-propanol (condition G3, MIDAS screen, Molecular Dimensions Ltd). Crystals were cryo-protected by addition of 20% (w/v) ethylene glycol prior to flash cooling in liquid nitrogen.
Diffraction data of SitA+Zn2+ were collected on beam line PXIII of the Swiss Light Source (SLS); whereas data of SitA+Mn2+ were collected on ID14-4 of the European Synchrotron Radiation Facility (ESRF). Diffraction data were processed using iMosflm [33] and scaled using AIMLESS in the CCP4 software suite [34]. Structure determination was performed using the Phaser Molecular Replacement software [35] with the structure of PsaA [PDB (Protein Data Bank) entry 1PSZ] as the search model. The 3D structures were refined using Phenix [36] and Buster [37], whereas model building was performed using Coot [38].
Crystals of SitA bound to Zn2+ or bound to Mn2+ belonged to space group I 2, with two chains of SitA in the asymmetric unit and with a solvent content of 39% (Matthews's coefficient 2 Å3.Da−1). Structural quality was assessed using Molprobity [39] and figures were prepared using PyMOL (http://www.pymol.org). Atomic coordinates of the SitA structures have been deposited in the PDB with entry codes 4OXR (Mn2+-bound dataset) and 4OXQ (apo/Zn2+-bound dataset). Pairwise structural comparisons of SitA with PsaA (Zn2+-bound form 1PSZ [22], Mn2+-bound form 3ZTT [24], and apo-form 3ZK7 [40]), TroA (Zn2+-bound form 3MFQ [41]), ZnuA (Zn2+-bound forms: 2OGW [42] and 2OSV [43]), MtsA (Fe2+-bound form: 3HH8 [23]) and MntC (Mn2+-bound form: 4K3V [21]), to obtain Cα rmsd (root mean square deviation) scores, were performed using SSM within Coot [38]. The rmsd values reported are the lowest rmsds obtained after making all possible pairwise comparisons of the different components of the respective asymmetric units, most of which contained two or four SBP molecules.
ITC (isothermal titration calorimetry)
ITC measurements were performed using a MicroCal VP-ITC-200 instrument (GE Healthcare). ITC experiments were performed at 25°C. Both SitA and metal ion solutions were prepared in buffer containing 50 mM Hepes pH 7.5 and 50 mM NaCl. SitA protein concentrations were 10–20 μM. The metal ion solutions (MnCl2 and ZnCl2) were prepared at 100 μM concentration. Metal ions were injected at 5 min intervals, with 19, 37 or 73 injections of 2, 1 or 0.5 μl volumes, with injection times of 4, 2 or 1 s, respectively. Data were analysed using the Origin 7.0 software.
RESULTS
Apo-SitA is strongly stabilized by a subset of divalent metal cations
An expression construct for production of the SitA ectodomain (residues G23–Q306) was prepared from the sitA gene of a clinical isolate of S. pseudintermedius, strain IV369-1041 (see the Experimental section). Soluble recombinant SitA protein was produced in the cytoplasm of E. coli and was purified using standard chromatographic techniques, yielding a final protein purity level of >98%, as determined by RP-HPLC. Purity was estimated by measuring the area under the single sharp peak corresponding to SitA present in the chromatogram, with retention time approximately 3.7 min (Supplementary Figure S1). This first protein sample (termed ‘Prot.1’) was analysed by DSC, which revealed two distinct unfolding transitions with peaks at approximately 46 and 66°C (Figure 1A). This result suggested that the initially purified SitA might have been present in both apo- and holo-forms, potentially due to the partial capture of stabilizing ligands from the E. coli cytoplasm. Similar observations were described previously for the SitA homologue, MntC, from S. aureus [21].
Figure 1. Differential scanning calorimetry profiles of different grades of SitA purifications.
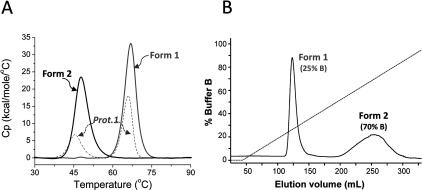
(A) In DSC experiments a Tm value is given by the peak maximum of the scanned melting curve. The DSC profile of the first purified preparation of SitA (‘Prot.1’, dashed grey line), showed that the initial sample generated two peaks (Tm1 45.6°C and Tm2 66.0°C), consistent with two unfolding events, which could potentially be generated either by two different domains within one protein, or by two different forms of the same protein. The latter option was supported by HIC, from which two separate forms of SitA were obtained [Form-1 and Form-2, see panel (B)], each of which generated only one peak in DSC experiments (solid black and grey lines), Tm 47.7°C for Form-2, and Tm 66.9°C for Form-1. (B) The HIC elution profile of SitA, showing clear separation of two forms.
Therefore an additional purification step using HIC was performed, enabling the separation of two distinct forms of SitA (Figure 1B), each of which produced a DSC profile with only a single peak, closely corresponding to one or other of the unfolding transitions originally displayed by Prot.1 (Figure 1A). Since ligand-binding often increases protein thermostability [44], we hypothesized that the peak with higher melting temperature (Tm) was likely to correspond to a metal-bound holo-form of SitA, whereas the form of SitA with the lower Tm corresponded to apo-SitA.
The purified apo-SitA was subsequently examined by DSC for changes in thermostability upon the addition of various potential metal ligands. In the presence of 2 mM Mn2+ or Zn2+ ions, SitA displayed significantly increased Tm values: 66.9°C and 64.1°C, respectively (Figure 2A). In contrast, the stability of SitA was less dramatically increased upon addition of Mg2+ or Ca2+ ions, for which Tm values of only 51.2°C and 53.9°C, respectively, were obtained (Figure 2A). Despite significantly increasing the thermostability of SitA, metal cations did not induce notable changes in CD spectroscopy profiles (Supplementary Figure S2), suggesting that the secondary structure of SitA was not altered by ligand binding.
Figure 2. Different binding and reversibility of divalent metal cations with SitA.
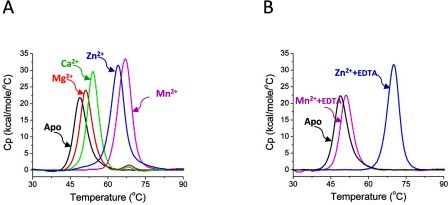
(A) The purified recombinant apo-SitA protein displays a single peak, Tm=48.7°C (black line), the lowest Tm measured in all these experiments. The DSC data for SitA+2 mM MnCl2 (magenta line, Tm 66.9°C) or SitA+2 mM ZnCl2 (blue line, Tm 64.1°C) displayed a single peak, corresponding to an unfolding event with high Tm. The DSC data for SitA+2 mM CaCl2 (green line, Tm 53.9°C) or 2 mM MgCl2 (red line, 51.2°C) showed much smaller binding-induced increases in Tm. (B) A strong chelating agent (EDTA, 10 mM) was added to SitA previously complexed with 2 mM Mn2+ or 2 mM Zn2+ and the DSC profiles were recorded. While the DSC profile of the Mn2+/EDTA-treated SitA sample (magenta line) revealed an unfolding transition which closely resembled that of metal-free apo-SitA (Tm 48.7°C, black line), the Zn2+/EDTA-treated SitA sample (blue line) revealed a Tm of 70.1°C, which approximately matched the profile of metal-bound SitA. (The reason for the minor difference in Tm between SitA+Zn2+ in the presence and absence of EDTA is currently unclear. Nevertheless, EDTA does not appear to stabilize SitA+Mn2+, revealing that SitA has a different propensity to bind the two metals under these conditions.)
SitA binds reversibly to manganese ions
It was demonstrated previously for the S. pneumoniae Mn2+ transporter PsaA that extracellular Zn2+ can competitively inhibit Mn2+ uptake into the bacterium and thereby negatively influence bacterial growth [45]. Moreover, in a recent report describing structural and biochemical studies, it was hypothesized that while the binding of PsaA to Mn2+ displayed an imperfect tetrahedral geometry (coordination number n=4) likely to facilitate Mn2+ ion release to the membrane-bound transporter, the same tetrahedral coordination was better suited for binding of Zn2+, which locked PsaA in a closed state such that Zn2+ binding was essentially irreversible [40]. To understand if SitA might display similarly selective metal-binding properties, apo-SitA was treated with 2 mM MnCl2 or ZnCl2 and, subsequently, was incubated with 10 mM EDTA. Indeed, in accordance with previous results for PsaA [40] and MntC [21], the resulting DSC profiles demonstrated that under these conditions in solution, the binding of SitA to Mn2+ was reversible by EDTA treatment, while binding to Zn2+ was not reversible (Figure 2B).
The crystal structure of SitA with a bound manganese ion reveals a class III SBP fold
Since protein thermostability and, further, ligand-induced stabilization, have been shown to correlate with increased probability of protein crystallization [46,47], we sought to crystallize SitA in the presence of metal ions. A highly concentrated solution of SitA+10 mM MnCl2 was subjected to hundreds of crystallization trials, yielding reproducible crystals only in the presence of hexafluoroisopropanol.
The structure of SitA bound to Mn2+ was determined by MR (molecular replacement). As the MR search model, we used the structure of PsaA (PDB code 1PSZ), which shares 48% sequence identity with SitA. The SitA+Mn2+ structure was refined at 2.0 Å resolution with the R-factor and free R-factor converging at 18.3 and 23.3%, respectively (see Table 1). The final refined model of SitA covered almost the entire protein, spanning residues K29–Q306 (Q306 is the actual C-terminus of SitA), and possessed excellent structure quality statistics (Figure 3A).
Table 1. Data collection and refinement statistics for two SitA structures.
Statistics for the highest-resolution shell are shown in parentheses.
| SitA+Mn2+ | SitA+Zn2+ | |
|---|---|---|
| PDB ID | 4OXR | 4OXQ |
| Data collection | ||
| Wavelength (Å) | 0.9393 | 1.0 |
| Beamline | ESRF, ID14 | SLS, PXIII |
| Resolution range (Å) | 46.45–2.0 (2.05–2.0) | 51.42–2.61 (2.72–2.61) |
| Space group | I 1 2 1 | I 1 2 1 |
| Unit cell | 80.58, 57.49, 112.3, β=101.95 | 81.66, 58.17, 112.3, β=101.8 |
| Total reflections | 114991 (8534) | 43067 (5332) |
| Unique reflections | 33582 (2426) | 15736 (1922) |
| Multiplicity | 3.4 (3.5) | 2.7 (2.8) |
| Completeness (%) | 98.2 (96.9) | 98.9 (95.6) |
| Mean I/sigma(I) | 11.4 (4.7) | 3.7 (1.8) |
| Wilson B-factor | 16.6943.2 | |
| Rsym* | 11.6 (61.4) | 16.8 (44.2) |
| Rmeas** | 16.4 (86.9) | 23.4 (61.1) |
| Refinement | ||
| Resolution range (Å) | 39.4–2.0 | 51.42–2.61 |
| Rwork† | 18.3 | 17.5 |
| Rfree†† | 23.3 | 24.7 |
| Number of atoms | ||
| Macromolecules | 4444 | 4420 |
| Ligands | 2 | 1 |
| Water | 181 | 186 |
| Protein residues | 29–306 (chains A and B) | 29–306 (chain A), 29–125/129–306 (B) |
| RMS(bonds) | 0.010 | 0.010 |
| RMS(angles) | 1.453 | 1.180 |
| Ramachandran (%)§ | ||
| Analysed | 98 | 96 |
| Favoured | 97 | 96 |
| Allowed | 3 | 4 |
| Outliers | 0 | 0 |
| Clashscore | 3.7 | 1.2 |
| Average B-factor | ||
| Macromolecules | 24.9 (chain A and B) | 27.5 (chain A), 30.9 (chain B) |
| Ligands | 7.4, 5.9 (Mn, chain C and D) | 31.0 (Zn, chain C) |
| Solvent | 21.9 (H2O) | 20.8 (H2O) |
| H64 NE2 | 6.20, 5.85 (chain A and B) | 25.16, 33.49 (chain A and B) |
| H137NE2 | 3.74, 9.90 (chain A and B) | 13.97, 18.53 (chain A and B) |
| E203OE1 | 5.43, 11.40 (chain A and B) | 16.16, 19.77 (chain A and B) |
| E2030E1 | 6.32, 6.28 (chain A and B) | 18.51, 21.78 (chain A and B) |
| D278 OD1 | 6.55, 10.66 (chain A and B) | 13.21, 26.17 (chain A and B) |
| D278 OD2 | 8.43, 9.24 (chain A and B) | 30.19, 30.51 (chain A and B) |
*Rsym=Σhkl Σi |Ii(hkl)–<I(hkl)>|/Σhkl Σi Ii(hkl)
**Rmeas=redundancy-independent (multiplicity-weighted) Rmerge as reported from AIMLESS [58].
†Rwork=Σ||F(obs)|−|F(calc)||/Σ|F(obs)|
††Rfree=as for Rwork, calculated for 5.0% of the total reflections, chosen at random and omitted from refinement.
§Figures from Molprobity [39].
Figure 3. X-ray crystallographic structure of SitA bound to a Mn2+ ion.
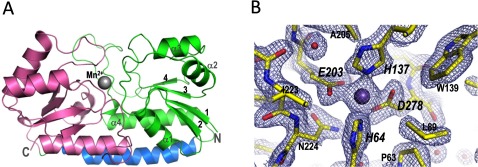
(A) Cartoon representation of SitA showing the typical class III SBP fold: the N-terminal lobe (green) and C-terminal lobe (pink) are connected by a single long α-helix (blue). The Mn2+ ion, shown as a sphere, binds in a cavity between the two lobes. (B) Details in the ligand-binding pocket of Mn2+-bound SitA, with 2Fo–Fc electron density maps contoured at 2σ (shown as cyan mesh). Bond sticks are coloured by element, with carbon, oxygen, and nitrogen atoms coloured in yellow, red and blue, respectively. The Mn2+ ion is shown as a purple sphere; red spheres show water molecules. Key metal-binding residues are labelled in large italics, close neighbours are labelled in smaller non-italic font.
The protein crystals obtained in the presence of Mn2+ contained two chains of SitA per asymmetric unit, arranged in a ‘face-to-face’ manner (Supplementary Figure S3). This dimeric organization is likely to be only a consequence of the very high protein concentration used for crystallization (>4 mM SitA). To allow a complementary non-crystallographic assessment, analytical SEC was performed with SitA at lower concentration (~100 μM), and yet still likely to be above the physiologically relevant concentration, revealing that SitA was monomeric in solution (theoretical monomeric molecular weight: 32 kDa, apparent molecular weight in SEC: 35 kDa). The SEC elution profile of SitA did not change upon addition of 10 mM MnCl2 or ZnCl2 (Supplementary Figure S4). Furthermore, analysis of the crystallographic dimer interface via the PISA (Proteins, Interfaces, Structures & Assemblies) software [48], did not detect any interfaces likely to result in the formation of stable quaternary complexes.
Structurally, the two chains of SitA bound to Mn2+ ions are essentially identical and can be superimposed with rmsd values of only 0.49 Å for all 278 aligned Cα atoms. For each chain, the overall monomeric structure is composed of two different lobes, or domains, connected by a long α-helix (residues A164–K188). The N-terminal domain (K29–T162) is composed of a single β-sheet of four short parallel β-strands partially surrounded by four α-helices. Similarly, the C-terminal domain (residues I192–Q306) is also composed of four short parallel β-strands and four α-helices (Figure 3A). Overall, the protein displays a bilobate bean-like structure, typical of the class III SBP family, with dimensions of ~60×43×43 Å. Electron density was clearly observed for almost the entire protein and for approximately 90 water molecules per chain. Moreover, electron density was clearly observed for a single Mn2+ ion per protein monomer, found in a pocket ~10 Å deep, located between the two lobes. The Mn2+ ion is chelated by nitrogen and oxygen side-chain atoms of His64, His137, Glu203 and Asp278 (Figure 3B).
A search of the PDB revealed several structures similar to SitA, all of which are class III SBPs, most notably including: E. coli ZnuA–rmsd 2.19 Å (for 240 aligned Cα atoms), S. suis TroA–rmsd 1.53 Å (250 Cα atoms), S. pyogenes MtsA–rmsd 1.12 Å (272 Cα atoms), S. pneumoniae PsaA–rmsd 1.03 Å (275 Cα atoms) and S. aureus MntC–rmsd 0.62 Å (276 Cα atoms). The folds of these proteins are well-conserved (for examples, see Supplementary Figure S5) and overall there are relatively few differences when comparing SitA with ligand-free or ligand-bound class III SBPs. Together with our CD spectroscopy data, these observations suggest that the ligands do not induce large changes in the secondary or tertiary structures of these proteins.
SitA structures bound to Mn2+ or Zn2+ are highly similar overall
The DSC experiments described above demonstrated that the thermostability of SitA was most notably increased by Mn2+ and Zn2+ ions, and that upon addition of EDTA the binding in solution to Mn2+ was reversible, while binding to Zn2+ was not appreciably reversible. To understand whether the binding mechanisms were comparable, we sought to crystallize SitA in the presence of Zn2+ ions.
A SitA sample to which Zn2+ had been added produced crystals, again using hexafluoroisopropanol as the reservoir solution (to which metal ions had not been added), and the structure was determined by MR, using our refined structure of SitA bound to Mn2+ as the search model. The SitA+Zn2+ structure was refined at 2.6 Å resolution with the R-factor and free R-factor converging at 17.5 and 24.7%, respectively (see Table 1). As seen for the SitA+Mn2+ crystals, the SitA+Zn2+ crystals contained two monomers per asymmetric unit (despite the existence of SitA as a monomer in solution in the presence of ZnCl2). A pairwise comparison of the SitA monomer structures bound to Mn2+ or Zn2+ revealed that the two structures were almost identical overall (rmsd 0.41 Å). However, unexpectedly, the crystals obtained using the Zn2+-treated SitA revealed two slightly different monomer structures within the same asymmetric unit. Remarkably, while one chain was bound to Zn2+, the other monomer was present in the apo-form, lacking a bound metal ion. A detailed inspection of difference electron density maps around the Zn2+ binding pocket allowed modelling of one Zn2+ ion with 50% occupancy for one chain only (chain A). In contrast, for the other chain present in the asymmetric unit (chain B) the electron density in the binding pocket was compatible only with the presence of a water molecule, not a metal ion, supported also by the B-factor distribution of proximal atoms.
SitA binds Mn2+ and Zn2+ with slightly different coordination geometry
The SitA structures revealed that the Mn2+ or Zn2+ ions were accommodated almost identically in the binding pocket. Electron density was clearly visible for all metal-chelating residues in the pocket, which is well-suited for divalent metal cations (Figure 3B). In both cases, the metal ion is coordinated by side-chain nitrogen and oxygen atoms of four tetrahedrally arranged residues: His64, His137, Glu203 and Asp278 (Figure 4). The metal-chelating residues are located in the lower regions of the binding pocket, and are contributed both by the N-terminal lobe (His64, His137) and the C-terminal lobe (Glu203, Asp278). For the Mn2+-bound structure, the positions of all four chelating residues, coupled with the bidentate nature of the Glu and Asp side chains, enables six coordinate bonds to the metal ion, with nearly perfect octahedral geometry and with protein–metal bond lengths of 2.1–2.4 Å (Figure 4A), as typically seen for Mn2+ [49,50]. In contrast, the binding pocket accommodates the Zn2+ ion in a less canonical manner, in particular because the nitrogen-to-Zn2+ distance involving His64 shows a 3.1 Å bond length, and the carboxylate oxygen-to-Zn2+ distances vary from 2.4 to 2.9 Å (Figure 4B); all of these are longer than the mean distances observed for the coordination of Zn2+ either in high-resolution protein structures [49] or in medium-resolution structures [50]. Although the medium resolution of the Zn2+-bound structure determined here (2.6 Å resolution, compared with 2.0 Å resolution for Mn2+) prompts a cautious interpretation of the precise protein–metal bond distances, the observations above suggest that the binding site is more typical of Mn2+-binding proteins rather than Zn2+-binding proteins, indicating transport of Mn2+ as a more likely biological function of SitA. This function is likely to be shared by other species, since all four metal-chelating residues are fully conserved in the orthologous SitA proteins of S. aureus, S. epidermidis and S. pneumoniae (Figure 5).
Figure 4. Atomic details of the SitA-metal cation interactions.
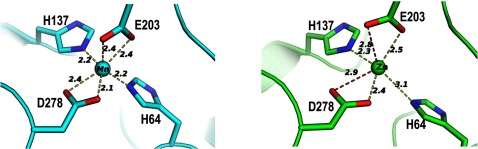
The cation-binding sites of Mn2+-bound SitA (cyan, left panel) and Zn2+-bound SitA (green, right panel) reveal that the binding mechanisms are very similar, with a small but notable difference in metal coordination observed for the His64 side chain. Notably, the side chain NE2 atom of His64 is 2.2 Å from the Mn2+ ion, but is shifted >1 Å further away (3.1 Å) from the Zn2+ ion. For each structure, the B-factors of the metal ions refined to values similar to those of the cognate protein atoms involved in metal chelation.
Figure 5. SitA orthologues show high sequence identity and conservation of metal-binding residues.
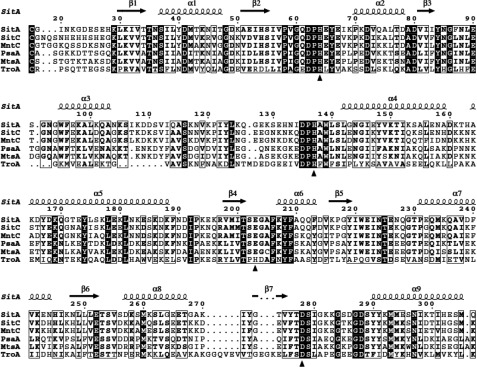
A multiple sequence alignment revealing the high degree of sequence identity (ID) between SitA and its orthologues: SitC in S. epidermidis (68% ID), MntC in S. aureus (65% ID), PsaA in S. pneumoniae (48% ID), MtsA in S. pyogenes (45% ID) and TroA in S. suis (27% ID). The alignments begin at the reactive Cys residue of the lipobox motif conserved in these Gram-positive lipoproteins. Residues are numbered according to full-length SitA, and are shaded black if fully conserved, or are boxed if partially conserved. Metal-chelating residues are indicated with black triangles. Secondary structure elements of SitA (derived from the Mn2+-bound structure) are shown above the sequences. Multiple sequence alignments and figures were prepared using the MAFFT algorithm [59] and ESPript [60].
The apo-SitA structure reveals a gating mechanism for entry to the metal-binding pocket
To gain insights into the putative ligand-induced mechanism resulting in fruitful engagement of the SitBC receptor by the metal-bound form of SitA, we sought to determine the structure of apo-SitA. Generally, it seems plausible to infer that the crystallization of apo-form SBPs is problematic, since relatively few apo-SBP structures are present in the PDB compared with the numerous holo-SBP structures available. Presumably this is because the ligand-enhanced stability promotes crystallization of the holo-forms, as discussed above, while the more dynamic ligand-free apo-forms are recalcitrant to crystallization. Indeed, we were unable to crystallize a purely apo-form of SitA. However, as described briefly above, using a Zn2+-treated sample, SitA crystals were obtained in which the asymmetric unit contained two polypeptide chains in different states, one of which presented SitA in the Zn2+-bound form (chain A), and the other of which presented SitA in the apo-form (chain B).
A first comparison of the apo- and Zn2+-bound SitA structures revealed a very high degree of similarity, rmsd 0.39 Å (for 274 aligned Cα atoms). However, proximal to the metal binding site, in the loop spanning residues N224–N227, the Cα atom positions differed significantly (maximum displacement of 4.5 Å for E226), which appears to be the only notable change in the C-terminal lobe (Figure 6A). In addition, in the N-terminal lobe, also proximal to the metal, significant changes in loop S91-W95 were observed (maximum displacement of 1.9 Å for N93), and these changes are concomitant with small shifts of 0.5–0.7 Å in all Cα atoms in the immediately following α-helix (K98–A103), and in the spatially adjacent α-helix (E67–T77). Furthermore, in the apo-structure, there was increased flexibility (disorder) in the loop connecting K125–K129, for which electron density was not observed. Collectively, these small changes create local differences in the conformation and electrostatic surface profile of SitA in this region that SBPs typically use for binding to their cognate ABC transporters [51]. Most interestingly, the different local loop conformations render the binding pocket considerably more ‘open’ in the apo-structure, and ‘closed’ in the holo-structures. Concomitant with these local structural rearrangements, the side chain of residue His64 undergoes a remarkable reorientation by rotation of >100° around the Cα–Cβ bond, adopting a rotameric state that repositions the His64 NE2 atom >7 Å away from the potential metal-binding site, thus destroying the tetrahedral arrangement of amino acid side chains seen for metal coordination in the ‘closed’ holo-structure (Figure 6A).
Figure 6. Differences proximal to the ligand-binding pocket in apo- and holo-SitA.
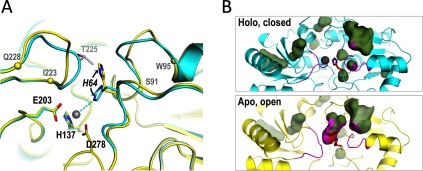
(A) Cartoon representations of the aligned structures of Zn2+-bound SitA (cyan, as in Figure 4) and apo-SitA (yellow) which were observed within the same asymmetric unit. The differences in the metal-binding site of apo- and holo-SitA are most notable in two loops S91–W95 and I223–Q228 (demarcated by Cα spheres on the apo- structure) surrounding the ligand-binding pocket and the outward position of His64 seen in the apo-structure, which is incompatible with the closed holo-structure, since it would clash with Thr225 (pale grey) in the holo-structure. Metal-chelating residues are labelled in black font, with the dynamic His64 in italics. (B) The conformational changes highlighted by the ribbon representations in panel (A) also lead to two rather different surface/cavity landscapes for the apo- and holo-forms of SitA. Here, blob-surfaces show internal cavities and/or surface-exposed tunnels/pockets. Most of the blobs are present in both states, showing that holo-SitA (upper panel) and apo-SitA (lower panel) surfaces/cavities are highly conserved. However, for apo-SitA, the additional large blob in the centre of the lower panel represents the tunnel leading from the apo-SitA surface down into the metal-free binding site. This tunnel disappears upon metal binding and closure, and hence the blob is absent in the upper panel. The flexible loops S91–W95 and I223–Q228 are coloured magenta, while sticks of His64 are coloured red and blue for Cα and nitrogen atoms. The side-chain contributions of the loops and of His64 to lining the walls of the cavities, are shown with magenta and red spots, respectively. All figures were made using Pymol v1.7, using surface ‘cavity’ mode for panel B.
Finally, a comparison of the apo- and holo-SitA structures showed that most of their pockets, surface-exposed tunnels and cavities are conserved. However, unlike the holo-form, the apo-form presents a large tunnel leading from the surface down into the metal binding site. This cavity is eliminated upon metal binding, due to ‘closure’ of the aforementioned loops (especially N224–N227) and repositioning of His64, in what resembles a ‘gating’ mechanism controlling entry of metal ions into the binding pocket (Figure 6A). A similar reorientation of an analogous histidine (His67) was also recently reported for PsaA, where the holo-form shows a ‘closed’ tetrahedral metal coordination geometry [22], whereas the apo-form reveals His67 in an outward-facing rotameric state due to a large rotation of the side chain [40]. These structural insights reveal the major differences between the apo- and holo-forms of SitA, which may underlie the ligand-dependent mechanism governing functional engagement of the SitBC receptor.
Point mutations in the ligand-binding site abolish metal binding
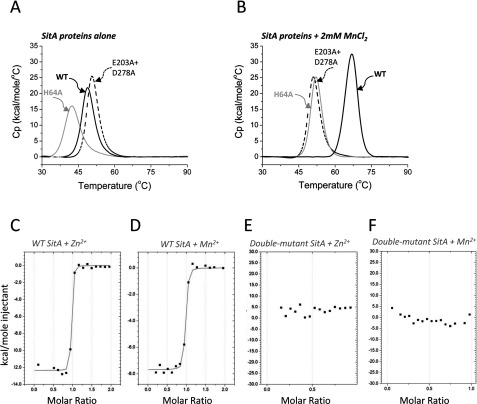
To verify the importance of the metal-chelating residues in the SitA-binding pocket, single and double point mutants of SitA were prepared. The mutant proteins were successfully produced and purified as for wild-type SitA. The metal-binding properties of each mutant were examined by DSC. In the absence of metal ions, the double mutant E203A+D278A displayed a single peak corresponding to an unfolding event with a low Tm of approximately 45°C, equivalent to the low Tm observed for wild type apo-SitA (Figure 7A). In the absence of metal ions, the single mutant H64A showed a slightly reduced Tm (42°C), possibly because the substitution with Ala results in the loss of mildly stabilizing interactions occurring between the imidazole ring and residues in loops S91–W95 and N224–N227 which are proximal to the side chain of His64 in the apo-SitA structure (Figure 6A). Upon addition of 2 mM MnCl2, the wild-type SitA showed the usual large increase in thermostability (Tm=67°C), while the mutants behaved differently. The H64A single mutant was stabilized by Mn2+ (Tm increased from 42 to 53°C)–though to a lesser extent than for wild-type SitA, and the E203A+D278A double mutant displayed essentially no change in Tm upon addition of Mn2+ (Figure 7B). Subsequently, we attempted to use ITC to determine the equilibrium dissociation constant (KD) for the binding of wild type SitA to Mn2+ and Zn2+ ions. The ITC data obtained indicated tight interactions of wild-type SitA with both metal ions. The binding curves relating the heats of injection to the molar ratio metal:SitA displayed sharp transitions, revealing that the interactions were apparently of high affinity, and were close to the upper limits that can be precisely detected by this technique [52]. Under these conditions, we estimated the KD values to be in the low nanomolar range for both metal ions (Figures 7C and 7D). However, owing to the technical limitations discussed, we cannot exclude that the binding interaction of one or both metal ions was even tighter. More importantly, ITC measurements using the double-mutant E203A+D278A did not show any significant interaction with metal ions, confirming that this simultaneous pair of mutations completely abolished the Mn2+ and Zn2+-binding ability of SitA (Figures 7E and 7F). It was also recently shown that the corresponding residues in PsaA, namely E205 and especially D280, were essential for binding both Zn2+ and Mn2+ ions [40].
Figure 7. SitA point mutations abolish Mn2+-induced stabilization.
(A) DSC experiments performed in the absence of metal ions revealed only a single peak, with similarly low Tm values (40–50°C) for WT (solid black line, Tm 48.7°C) and the SitA mutants H64A (grey line, Tm 42.4) and E203A+D278A (dashed black line, Tm 50.6°C). (B) DSC experiments performed in the presence of 2 mM MnCl2 revealed that the H64A mutant (grey line, Tm 52.0°C) shows a small but notable increase in Tm, whereas the double-mutant E203A+D278A (dashed black line, Tm 51.0°C) shows no change in Tm. The WT protein (solid black line, Tm 66.9°C) exhibits the typical large increase in Tm in presence of Mn2+ ions. (C–F) ITC experiments to examine the interaction of wild-type and mutant SitA with Mn2+ and Zn2+ ions. Panels (C) and (D) show the high affinity interaction of Mn2+ and Zn2+ with wild-type SitA; panels (E) and (F) show data for the SitA E203+D278A double-mutant, which presented no detectable interaction with Zn2+ ions or Mn2+ ions.
DISCUSSION
Structural and biochemical studies of SBPs have provided many insights into the molecular mechanisms used by bacteria to obtain a wide range of nutritional solutes [14,15]. In particular, due to the necessity of metal ions in a wide range of important metabolic processes, modes of metal acquisition during staphylococcal infections have captured much attention. In this report, we have presented a series of investigations performed in order to fully characterize SitA, a surface-exposed lipoprotein from the animal and human pathogen, S. pseudintermedius. The structures presented herein are the first reports of protein structures from S. pseudintermedius. As discussed below, the structures, biophysical studies and site-directed mutagenesis, revealed the determinants of ligand binding in SitA, and unveiled subtle ligand-induced conformational changes that may underlie the molecular switch required for fruitful receptor engagement and ion transport.
Our studies were focused on the SitA protein from a clinical isolate of pathogenic S. pseudintermedius. Based on sequence analyses, SitA is annotated in the UniProt Knowledge-Base as a putative metal-binding protein. To explore this hypothesis experimentally, we performed CD and DSC experiments. Although the CD spectra did not reveal differences in the presence or absence of metals, the DSC experiments revealed a large stabilization of SitA in the presence of divalent metal cations. In particular, both Zn2+ and Mn2+ ions increased the Tm of SitA by ~20°C (Figure 2A). Furthermore, Mg2+ and Ca2+ also stabilized SitA, although to a much lesser extent, suggesting that there is distinct specificity for the transition metal ions. Indeed, following an analysis of thousands of metalloprotein structures in the PDB, it was reported previously that the alkali metals Mg2+ and Ca2+ are rarely coordinated by nitrogen (e.g. from histidine imidazole side chains, of which two are present in the SitA-binding pocket), but instead demonstrate a preference for the coordinating element to be oxygen, in particular deriving from an amino acid side-chain carboxylate group [49,50]. The strong intrinsic tendency of SitA to bind metal ions was also evident from the observation that recombinant SitA simply purified from E. coli lysates using EDTA-treated buffers displayed two peaks in DSC experiments–indicating that apo-SitA had partially gathered metal ions during production in the bacterial cytoplasm. The difficulty of precluding metal binding, even in the presence of strong chelating agents, has also been observed previously in studies of the proteins PsaA from S. pneumoniae [40] and MntC from S. aureus [21]. These observations support the hypothesis that SitA is involved in the capture of metal ions during the bacterial lifecycle. The amino acid sequence of the SitA protein studied herein is 100% conserved in the S. pseudintermedius genomes reported to date [9,10], suggesting that transition metal binding by SitA is important at least for these three strains of the pathogen. Interestingly, the binding of SitA to Mn2+ was found to be reversed by EDTA, while this chelating agent was not able to remove Zn2+ from SitA. Since EDTA is only one of many possible metal ion chelating agents, it is conceivable that alternative stronger metal ion binders might have induced different effects. Nevertheless, our observations suggest that Mn2+ may be more readily bound and then released compared to Zn2+, and thus is likely to be the more efficiently transported ligand. However, it cannot be excluded that the release and transport of Zn2+ might be promoted by conformational changes potentially occurring in SitA upon binding to the membrane-bound receptor.
To gain insights into the molecular mechanism of ligand binding, we sought to use X-ray crystallography to obtain the three-dimensional structure of SitA. Since protein crystallization is often facilitated by small-molecule ligands and high thermostability [46,47], we exploited the metal-induced thermostabilization reported above to generate co-crystals and initially determined the structure of Mn2+-bound SitA (Figure 3). The SitA structure revealed all the hallmarks of a class III SBP, presenting a bilobate structure of N- and C-terminal domains connected by an α-helix, with the Mn2+ ion harboured in a central ligand-binding pocket. The large interface between the two lobes, the characteristically long α-helix of 25 residues, and the ‘domain bridging’ effect of the Mn2+ ion coordinated by residues from both the N- and C-terminal lobes, collectively appear to confer rigidity to the overall protein, and presumably account for the high thermostability displayed by holo-SitA. The metal-induced stabilization presumably holds SitA in the optimal conformation required for effective receptor binding and subsequent ligand transport. Here, we used the previously reported coordinates of PsaA in order to determine experimentally the structure of Mn2+-bound SitA. PsaA is one of the class III SBPs produced by S. pneumoniae and is part of the well-characterized manganese ABC transporter system PsBCA [45]. Pairwise comparisons showed that SitA and PsaA share a sequence identity of 48% and exhibited very similar secondary structure compositions with 3D structures that can be superimposed with rmsd values of only 1 Å and revealing only minor structural differences in some loop regions (Supplementary Figure S5). This high degree of structural similarity strongly suggests a shared function in manganese binding and transport. Nevertheless, while Mn2+-bound SitA displays near-perfect octahedral binding geometry, this has not been equally observed in all Mn2+-binding SBPs, including PsaA, and MntC from the cyanobacterium Synechocystis [53]. Indeed, recently it was proposed that the imperfect metal coordination exhibited by PsaA may have a physiological role in aiding ligand release [40]. Further structural and functional studies will be required to better understand the impact of these subtle, intriguing and potentially important differences in metal coordination schemes between the SBPs present in many different bacterial species.
In SitA, the Mn2+ ion is bound in a polar and electrostatic environment, with hexavalent octahedral coordination via the side chains of His64, His137, Glu203 and Asp278 (Figure 4). This direct protein–metal interaction is in contrast with the indirect capture of Fe3+ by siderophore-binding SBPs, as exemplified by the Fe3+–ferrichrome–FhuD2 interaction [19]. Here, in our crystallographic ‘snapshot’, the Mn2+ ion is engulfed and completely buried from the solvent (Figure 6B, upper panel). Presumably, binding of the Mn2+-bound SitA lipoprotein to its membrane-bound receptor, SitBC, can promote conformational changes in SitA sufficient to induce release of its metal cargo, in order to enable nutrient uptake and recycling of the apo-form of the transport protein. We wished to explore this hypothesis by studying interactions of apo- and ligand-bound forms of SitA with recombinant SitB or SitBC. However, despite extensive efforts, we were unable to produce a soluble form of the integral membrane protein SitB alone or in complex with SitC.
Subsequently, we also determined the structure of SitA bound to Zn2+ (Figure 4). The two metal-bound SitA structures were highly similar overall, although in particular the position of the ion-chelating His64 side chain was different. The possibility to undergo these minor local rearrangements, hosted upon an unchanging scaffold presenting four tetrahedrally arranged metal-chelating amino acids, may underlie the observed ability of SitA to bind to several different divalent metal cations. Comparison of the Zn2+-bound SitA structure with another zinc-bound SBP, the E. coli protein ZnuA, revealed considerable fold similarity (rmsd 2.2 Å) but to a lesser extent than when comparing SitA with the Mn2+-binding protein PsaA (see above and Supplementary Figure S5). A characteristic feature that may distinguish the Zn2+-binding SBP is in the cation-binding site residues and their organization. Typically, the Mn2+ binding site is composed by two histidines, one glutamate and one aspartate, whereas the Zn2+-binding site instead typically involves three histidines and one aspartate or glutamate (Supplementary Figure S5) [41–43]. Interestingly, while these SBPs display subtle variations on the same structural motif to determine distinct binding specificities, it has also been hypothesized that the streptococcal Lbp (laminin-binding protein) has undergone convergent evolution to enable Zn2+-coordination via a metal-binding site with a geometry similar to those of several SBPs including ZnuA [54].
Measurements by ITC revealed that SitA binds very tightly to both Mn2+ and Zn2+ ions, although the estimated values (low nanomolar range KD) were close to the limits of the technique [52] and should therefore be considered as apparent affinity estimates. Mutations to alanine in the chelating residues strongly reduced the metal-induced stabilization observed in DSC; and, moreover, the double mutant E203A+D278A was sufficient to completely abolish binding to Zn2+ or Mn2+ in ITC experiments (Figure 7). Overall, these insights coupled with the availability of characterized non-binding mutants of SitA provide a platform for future investigations of the physiological role of SitA during colonization and/or invasion in animal models of disease caused by S. pseudintermedius. The observations discussed suggest that S. pseudintermedius may use the highly conserved SitA metal-binding residues to capture and transport a range of nutrients, likely with a preference for Mn2+ ions for which it displayed the canonical commonly observed geometry and bond lengths [49,50]. Nonetheless, the structure of MtsA bound to Fe2+ [23] showed how the same four conserved and spatially equivalent chelating residues in a different SBP protein can be used to bind yet another different divalent metal cation (iron). Since the overall structures and, more importantly, the spatial arrangement of all four metal-binding residues, are conserved in homologous proteins from other species (Figure 5), it seems likely that SitC in S. epidermidis, MntC in S. aureus, PsaA in S. pneumoniae, and MtsA in S. pyogenes, may also display a degree of promiscuity in ligand binding. Which metals are actually bound and efficiently transported in vivo will likely also depend on metal bioavailability in the niche being colonized. This issue has been studied and discussed to some extent for PsaA, where it was concluded that a greater stability of the Zn2+-bound form, compared with the Mn2+ bound form, of PsaA might represent a mechanism for the antibacterial effect of Zn2+ [24]. Whether this mechanistic hypothesis holds for the SBPs of other species remains to be determined.
Finally, in addition to the Mn2+ and Zn2+ bound structures of SitA, we also obtained a ligand-free, apo-form of SitA. Notably, a search of the PDB using the protein structure comparison service, Fold [55], revealed 12 different (non-redundant) SBPs for which over 60 distinct polypeptide chain structures have been determined and which can all be pairwise-aligned with SitA with rmsds <3.0 Å. All such structures are SBPs with divalent metal-binding properties. The majority of these distinct protein structures (at least eight, to date) have been determined only in the metal-bound state. None of these protein structures have been determined only in the apo-form, and only three of these structures have been determined as wild-type proteins both in the metal-bound and metal-free states, potentially allowing mechanistic insights. However, in the apo-form of TroA from T. pallidum, despite the absence of a metal ion, the protein displays a closed state, with all potential metal-binding residues (His68, His133, His199, Asp279) showing inward-facing positions [56]. Similarly, inspection of apo- and holo-forms of ZnuA from E. coli reveals that both show a closed state with all potential metal-binding residues (Glu59, His60, His143, His207) in inward-facing positions compatible with metal binding [57]. Interestingly, the most recent related study provided the apo-form structure of wild-type PsaA from S. pneumoniae (PDB 3ZK7) [40], revealing an outward-facing histidine analogous to His64 in SitA (Figure 6A). As such, PsaA and the SitA structures determined herein offer a rare possibility to visually assess the detailed structural and conformational effects of ligand binding in an SBP.
The comparison of apo- and holo-forms of SitA shows how the binding of a metal ion coincides with changes in the structures of at least two neighbouring loops. Interestingly, in the SitA apo-form, the side chain of His64 is flipped outwards, adopting a position incompatible with the full canonical metal coordination scheme. It is conceivable that with these two loops and His64 in the outward ‘open’ position, Mn2+ can readily enter the binding pocket of the SitA apo-form via the open tunnel (Figure 6B, lower panel). After metal binding, the large reorientation of His64 into the inward ‘closed’ position, and clamping-down of the two neighbouring loops, effectively close the tunnel (Figure 6B, upper panel), resulting in high-affinity ligand binding. Similarly, ligand-dependent rearrangement of the corresponding residue His60 in the Zn2+-binding site of E. coli ZnuA has been reported [57]. Collectively, these findings suggest that this His residue, located at the outer edge of the binding pocket and very highly conserved in metal-binding SBPs, may mediate a conserved pivotal role in ion binding and release.
These structural rearrangements may be the mechanism triggering the ligand-dependent engagement of the SitBC receptor required for nutrient metal uptake and essential for bacterial virulence. On the basis of sequence and structure conservations, a similar binding and transport mechanism may be conserved in numerous SitA protein homologues. In summary, our results provide the first experimentally determined mechanistic and functional insights into this surface-exposed lipoprotein from S. pseudintermedius, a bacterial pathogen of animals and humans, of growing veterinary and medical importance.
Online data
ACKNOWLEDGEMENTS
We gratefully acknowledge Sandor Brockhauser for beamline support at the ESRF (Grenoble) and Joachim Diez (Expose GmbH) at the Swiss Light Source (Villigen) for assistance in X-ray data collection. We also wish to thank the following for participating in this collaboration and for their kind support; from Novartis Vaccines: Maria Giuliani, Daniele Veggi, Alessia Liguori, Werner Pansegrau, Manuele Martinelli, Silvana Savino, John Telford, Andrea Carfí and Paolo Costantino; from the Novartis Institute of Biomedical Research (Basel, Switzerland): Srinivas Honnappa and Sandra Jacob; from the Novartis PhD Academy: Ilaria Ferlenghi; from the University of Siena (Italy): Professor Cosima T. Baldari; and from Novartis Animal Health: Kristin Bloink and Richard Harland.
AUTHOR CONTRIBUTION
All authors planned and designed studies for the SitA project. Francesca Abate prepared all protein samples and performed all biochemical and crystallization studies, and resolved the structure with support from Enrico Malito and Matthew J. Bottomley. Francesca Abate and Paola Lo Surdo performed CD and DSC analyses. Francesca Abate and Matthew J. Bottomley wrote the manuscript. All authors discussed the results and read, revised and approved the final manuscript.
FUNDING
This work was supported, in part, by a Novartis Academy Ph.D. Fellowship (to F.A.) registered at the University of Siena, Italy.
References
- 1.Bannoehr J., Ben Zakour N. L., Waller A. S., Guardabassi L., Thoday K. L., van den Broek A. H., Fitzgerald J. R. Population genetic structure of the Staphylococcus intermedius group: insights into agr diversification and the emergence of methicillin-resistant strains. J. Bacteriol. 2007;189:8685–8692. doi: 10.1128/JB.01150-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fitzgerald J. R. The Staphylococcus intermedius group of bacterial pathogens: species re-classification, pathogenesis and the emergence of meticillin resistance. Vet. Dermatol. 2009;20:490–495. doi: 10.1111/j.1365-3164.2009.00828.x. [DOI] [PubMed] [Google Scholar]
- 3.Hill P. B., Lo A., Eden C. A., Huntley S., Morey V., Ramsey S., Richardson C., Smith D. J., Sutton C., Taylor M. D., et al. Survey of the prevalence, diagnosis and treatment of dermatological conditions in small animals in general practice. Vet. Rec. 2006;158:533–539. doi: 10.1136/vr.158.16.533. [DOI] [PubMed] [Google Scholar]
- 4.Guardabassi L., Schwarz S., Lloyd D. H. Pet animals as reservoirs of antimicrobial-resistant bacteria. J. Antimicrob. Chemother. 2004;54:321–332. doi: 10.1093/jac/dkh332. [DOI] [PubMed] [Google Scholar]
- 5.Van Hoovels L., Vankeerberghen A., Boel A., Van Vaerenbergh K., De Beenhouwer H. First case of Staphylococcus pseudintermedius infection in a human. J. Clin. Microbiol. 2006;44:4609–4612. doi: 10.1128/JCM.01308-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perreten V., Kadlec K., Schwarz S., Gronlund Andersson U., Finn M., Greko C., Moodley A., Kania S. A., Frank L. A., Bemis D. A., et al. Clonal spread of methicillin-resistant Staphylococcus pseudintermedius in Europe and North America: an international multicentre study. J. Antimicrob. Chemother. 2010;65:1145–1154. doi: 10.1093/jac/dkq078. [DOI] [PubMed] [Google Scholar]
- 7.Klevens R. M., Morrison M. A., Nadle J., Petit S., Gershman K., Ray S., Harrison L. H., Lynfield R., Dumyati G., Townes J. M., et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. J. Am. Med. Assoc. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 8.Guardabassi L., Loeber M. E., Jacobson A. Transmission of multiple antimicrobial-resistant Staphylococcus intermedius between dogs affected by deep pyoderma and their owners. Vet. Microbiol. 2004;98:23–27. doi: 10.1016/j.vetmic.2003.09.021. [DOI] [PubMed] [Google Scholar]
- 9.Ben Zakour N. L., Bannoehr J., van den Broek A. H., Thoday K. L., Fitzgerald J. R. Complete genome sequence of the canine pathogen Staphylococcus pseudintermedius. J. Bacteriol. 2011;193:2363–2364. doi: 10.1128/JB.00137-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tse H., Tsoi H. W., Leung S. P., Urquhart I. J., Lau S. K., Woo P. C., Yuen K. Y. Complete genome sequence of the veterinary pathogen Staphylococcus pseudintermedius strain HKU10-03, isolated in a case of canine pyoderma. J. Bacteriol. 2011;193:1783–1784. doi: 10.1128/JB.00023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganesh V. K., Rivera J. J., Smeds E., Ko Y. P., Bowden M. G., Wann E. R., Gurusiddappa S., Fitzgerald J. R., Hook M. A structural model of the Staphylococcus aureus ClfA-fibrinogen interaction opens new avenues for the design of anti-staphylococcal therapeutics. PLoS Pathog. 2008;4:e1000226. doi: 10.1371/journal.ppat.1000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stranger-Jones Y. K., Bae T., Schneewind O. Vaccine assembly from surface proteins of Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 2006;103:16942–16947. doi: 10.1073/pnas.0606863103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bannoehr J., Ben Zakour N. L., Reglinski M., Inglis N. F., Prabhakaran S., Fossum E., Smith D. G., Wilson G. J., Cartwright R. A., Haas J., et al. Genomic and surface proteomic analysis of the canine pathogen Staphylococcus pseudintermedius reveals proteins that mediate adherence to the extracellular matrix. Infect. Immun. 2011;79:3074–3086. doi: 10.1128/IAI.00137-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu B. C., Vogel H. J. A structural and functional analysis of type III periplasmic and substrate binding proteins: their role in bacterial siderophore and heme transport. Biol. Chem. 2011;392:39–52. doi: 10.1515/bc.2011.012. [DOI] [PubMed] [Google Scholar]
- 15.Quiocho F. A., Ledvina P. S. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: variation of common themes. Mol. Microbiol. 1996;20:17–25. doi: 10.1111/j.1365-2958.1996.tb02484.x. [DOI] [PubMed] [Google Scholar]
- 16.Garmory H. S., Titball R. W. ATP-binding cassette transporters are targets for the development of antibacterial vaccines and therapies. Infect. Immun. 2004;72:6757–6763. doi: 10.1128/IAI.72.12.6757-6763.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krewulak K. D., Vogel H. J. Structural biology of bacterial iron uptake. Biochim. Biophys. Acta. 2008;1778:1781–1804. doi: 10.1016/j.bbamem.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 18.Mishra R. P., Mariotti P., Fiaschi L., Nosari S., Maccari S., Liberatori S., Fontana M. R., Pezzicoli A., De Falco M. G., Falugi F., et al. Staphylococcus aureus FhuD2 is involved in the early phase of staphylococcal dissemination and generates protective immunity in mice. J. Infect. Dis. 2012;206:1041–1049. doi: 10.1093/infdis/jis463. [DOI] [PubMed] [Google Scholar]
- 19.Mariotti P., Malito E., Biancucci M., Lo Surdo P., Mishra R. P., Nardi-Dei V., Savino S., Nissum M., Spraggon G., Grandi G., et al. Structural and functional characterization of the Staphylococcus aureus virulence factor and vaccine candidate FhuD2. Biochem. J. 2013;449:683–693. doi: 10.1042/BJ20121426. [DOI] [PubMed] [Google Scholar]
- 20.Anderson A. S., Scully I. L., Timofeyeva Y., Murphy E., McNeil L. K., Mininni T., Nunez L., Carriere M., Singer C., Dilts D. A., Jansen K. U. Staphylococcus aureus manganese transport protein C is a highly conserved cell surface protein that elicits protective immunity against S. aureus and Staphylococcus epidermidis. J. Infect. Dis. 2012;205:1688–1696. doi: 10.1093/infdis/jis272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gribenko A., Mosyak L., Ghosh S., Parris K., Svenson K., Moran J., Chu L., Li S., Liu T., Woods V. L., Jr., et al. Three-dimensional structure and biophysical characterization of Staphylococcus aureus cell surface antigen-manganese transporter MntC. J. Mol. Biol. 2013;425:3429–3445. doi: 10.1016/j.jmb.2013.06.033. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence M. C., Pilling P. A., Epa V. C., Berry A. M., Ogunniyi A. D., Paton J. C. The crystal structure of pneumococcal surface antigen PsaA reveals a metal-binding site and a novel structure for a putative ABC-type binding protein. Structure. 1998;6:1553–1561. doi: 10.1016/S0969-2126(98)00153-1. [DOI] [PubMed] [Google Scholar]
- 23.Sun X., Baker H. M., Ge R., Sun H., He Q. Y., Baker E. N. Crystal structure and metal binding properties of the lipoprotein MtsA, responsible for iron transport in Streptococcus pyogenes. Biochemistry. 2009;48:6184–6190. doi: 10.1021/bi900552c. [DOI] [PubMed] [Google Scholar]
- 24.McDevitt C. A., Ogunniyi A. D., Valkov E., Lawrence M. C., Kobe B., McEwan A. G., Paton J. C. A molecular mechanism for bacterial susceptibility to zinc. PLoS Pathogens. 2011;7:1–9. doi: 10.1371/journal.ppat.1002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papp-Wallace K. M., Maguire M. E. Manganese transport and the role of manganese in virulence. Annu. Rev. Microbiol. 2006;60:187–209. doi: 10.1146/annurev.micro.60.080805.142149. [DOI] [PubMed] [Google Scholar]
- 26.Horsburgh M. J., Wharton S. J., Karavolos M., Foster S. J. Manganese: elemental defence for a life with oxygen. Trends Microbiol. 2002;10:496–501. doi: 10.1016/S0966-842X(02)02462-9. [DOI] [PubMed] [Google Scholar]
- 27.Walter R. M., Jr, Uriu-Hare J. Y., Olin K. L., Oster M. H., Anawalt B. D., Critchfield J. W., Keen C. L. Copper, zinc, manganese, and magnesium status and complications of diabetes mellitus. Diab. Care. 1991;14:1050–1056. doi: 10.2337/diacare.14.11.1050. [DOI] [PubMed] [Google Scholar]
- 28.Bloxam D. L., Tan J. C., Parkinson C. E. Non-protein bound zinc concentration in human plasma and amniotic fluid measured by ultrafiltration. Clin. Chim. Acta. 1984;144:81–93. doi: 10.1016/0009-8981(84)90041-X. [DOI] [PubMed] [Google Scholar]
- 29.Milne D. B., Sims R. L., Ralston N. V. Manganese content of the cellular components of blood. Clin. Chem. 1990;36:450–452. [PubMed] [Google Scholar]
- 30.Klock H. E., Lesley S. A. The Polymerase Incomplete Primer Extension (PIPE) method applied to high-throughput cloning and site-directed mutagenesis. Methods Mol. Biol. 2009;498:91–103. doi: 10.1007/978-1-59745-196-3. [DOI] [PubMed] [Google Scholar]
- 31.van den Berg S., Lofdahl P. A., Hard T., Berglund H. Improved solubility of TEV protease by directed evolution. J. Biotechnol. 2006;121:291–298. doi: 10.1016/j.jbiotec.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 32.Zingaretti C., Falugi F., Nardi-Dei V., Pietrocola G., Mariani M., Liberatori S., Gallotta M., Tontini M., Tani C., Speziale P., et al. Streptococcus pyogenes SpyCEP: a chemokine-inactivating protease with unique structural and biochemical features. FASEB J. 2010;24:2839–2848. doi: 10.1096/fj.09-145631. [DOI] [PubMed] [Google Scholar]
- 33.Battye T. G., Kontogiannis L., Johnson O., Powell H. R., Leslie A. G. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P., Sharff A., Smart O. S., Vonrhein C., Womack T. O. BUSTER version 2.11.4. Cambridge, U.K.: Global Phasing Ltd; 2011. [Google Scholar]
- 38.Emsley P. C. K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Counago R. M., Ween M. P., Begg S. L., Bajaj M., Zuegg J., O’Mara M. L., Cooper M. A., McEwan A. G., Paton J. C., Kobe B., McDevitt C. A. Imperfect coordination chemistry facilitates metal ion release in the Psa permease. Nat. Chem. Biol. 2014;10:35–41. doi: 10.1038/nchembio.1382. [DOI] [PubMed] [Google Scholar]
- 41.Zheng B., Zhang Q., Gao J., Han H., Li M., Zhang J., Qi J., Yan J., Gao G. F. Insight into the interaction of metal ions with TroA from Streptococcus suis. PLoS One. 2011;6:e19510. doi: 10.1371/journal.pone.0019510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chandra B. R., Yogavel M., Sharma A. Structural analysis of ABC-family periplasmic zinc binding protein provides new insights into mechanism of ligand uptake and release. J. Mol. Biol. 2007;367:970–982. doi: 10.1016/j.jmb.2007.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H., Jogl G. Crystal structure of the zinc-binding transport protein ZnuA from Escherichia coli reveals an unexpected variation in metal coordination. J. Mol. Biol. 2007;368:1358–1366. doi: 10.1016/j.jmb.2007.02.107. [DOI] [PubMed] [Google Scholar]
- 44.Giuliani S. E., Frank A. M., Collart F. R. Functional assignment of solute-binding proteins of ABC transporters using a fluorescence-based thermal shift assay. Biochemistry. 2008;47:13974–13984. doi: 10.1021/bi801648r. [DOI] [PubMed] [Google Scholar]
- 45.McDevitt C. A., Ogunniyi A. D., Valkov E., Lawrence M. C., Kobe B., McEwan A. G., Paton J. C. A molecular mechanism for bacterial susceptibility to zinc. PLoS Pathog. 2011;7:e1002357. doi: 10.1371/journal.ppat.1002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vedadi M., Niesen F. H., Allali-Hassani A., Fedorov O. Y., Finerty P. J., Jr., Wasney G. A., Yeung R., Arrowsmith C., Ball L. J., Berglund H., et al. Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proc. Natl. Acad. Sci. U. S. A. 2006;103:15835–15840. doi: 10.1073/pnas.0605224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dupeux F., Rower M., Seroul G., Blot D., Marquez J. A. A thermal stability assay can help to estimate the crystallization likelihood of biological samples. Acta Crystallogr. D Biol. Crystallogr. 2011;67:915–919. doi: 10.1107/S0907444911036225. [DOI] [PubMed] [Google Scholar]
- 48.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 49.Harding M. M., Nowicki M. W., Walkinshaw M. D. Metals in protein structures: a review of their principal features. Crystallogr. Rev. 2010;16:247–302. doi: 10.1080/0889311X.2010.485616. [DOI] [Google Scholar]
- 50.Zheng H., Chruszcz M., Lasota P., Lebioda L., Minor W. Data mining of metal ion environments present in protein structures. J. Inorg. Biochem. 2008;102:1765–1776. doi: 10.1016/j.jinorgbio.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Korkhov V. M., Mireku S. A., Locher K. P. Structure of AMP–PNP-bound vitamin B12 transporter BtuCD-F. Nature. 2012;490:367–372. doi: 10.1038/nature11442. [DOI] [PubMed] [Google Scholar]
- 52.Velazquez-Campoy A., Ohtaka H., Nezami A., Muzammil S., Freire E. Isothermal titration calorimetry. Current Protocols in Cell Biology/editorial board, Juan S. Bonifacino et al. Chapter 17, Unit 17, 18. 2004. [DOI] [PubMed]
- 53.Rukhman V., Anati R., Melamed-Frank M., Adir N. The MntC crystal structure suggests that import of Mn2+ in cyanobacteria is redox controlled. J. Mol. Biol. 2005;348:961–969. doi: 10.1016/j.jmb.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 54.Linke C., Caradoc-Davies T. T., Young P. G., Proft T., Baker E. N. The laminin-binding protein Lbp from Streptococcus pyogenes is a zinc receptor. J. Bacteriol. 2009;191:5814–5823. doi: 10.1128/JB.00485-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krissinel E., Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 56.Lee Y. H., Dorwart M. R., Hazlett K. R., Deka R. K., Norgard M. V., Radolf J. D., Hasemann C. A. The crystal structure of Zn(II)-free Treponema pallidum TroA, a periplasmic metal-binding protein, reveals a closed conformation. J. Bacteriol. 2002;184:2300–2304. doi: 10.1128/JB.184.8.2300-2304.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yatsunyk L. A., Easton J. A., Kim L. R., Sugarbaker S. A., Bennett B., Breece R. M., Vorontsov II, Tierney D. L., Crowder M. W., Rosenzweig A. C. Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli. J. Biol. Inorg. Chem. 2008;13:271–288. doi: 10.1007/s00775-007-0320-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Evans P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 59.Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gouet P., Courcelle E., Stuart D. I., Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.